Abstract
The tropism of Sindbis virus (SB) for cells of the dendritic cell (DC) lineage and the virulence of SB in vivo are largely determined by the efficacy of alpha/beta interferon (IFN-α/β)-mediated antiviral responses. These responses are essentially intact in the absence of PKR and/or RNase L (K. D. Ryman, L. J. White, R. E. Johnston, and W. B. Klimstra, Viral Immunol. 15:53-76, 2002). In the present studies, we investigated the nature of antiviral effects and identity of antiviral effectors primed by IFN-α/β treatment of bone marrow-derived DCs (BMDCs) generated from mice deficient in PKR and RNase L (TD). IFN-α/β priming exerted significant antiviral activity at very early stages of SB replication and most likely inhibited the initial translation of infecting genomes. The early effect targeted cap-dependent translation as protein synthesis from an SB-like and a simple RNA were inhibited by interferon treatment, but an encephalomyocarditis virus internal ribosome entry site-driven element exhibited no inhibition. Phosphorylation of the α subunit of eukaryotic translation initiation factor 2 was defective after virus infection of TD cells, suggesting other mechanisms of translation inhibition. To identify components of these alternative antiviral pathway(s), we have compared global gene regulation in BMDCs derived from normal 129 Sv/Ev, IFNAR1−/−, and TD mice following infection with SB or treatment with IFN-α/β. Candidate effectors of alternative antiviral pathways were those genes induced by virus infection or IFN-α/β treatment in 129 Sv/Ev and TD-derived BMDC but not in virus-infected or IFN-α/β-treated IFNAR1−/− cells. Statistical analyses of gene array data identified 44 genes that met these criteria which are discussed.
Members of the Alphavirus genus of the family Togaviridae cause significant human and veterinary disease worldwide, with a disturbing potential for natural emergence in epidemic or epizootic outbreaks (19) or for deliberate release during a biological terrorism or warfare attack (3, 35). The alphaviruses are small, enveloped viruses with message-sense, capped, and polyadenylated RNA genomes (47). The model for the alphavirus life cycle predicts that replication begins with cap-dependent translation of the four nonstructural proteins (nsP1 to nsP4) as a polyprotein from the 5′ two-thirds of the genome (for examples, see references 15, 46, and 51). After posttranslational processing, the nsPs transcribe the complementary minus strand, which serves as template for transcription of progeny genome-length RNAs and subgenomic 26S molecules. Nontranslated regions of extensive double-stranded RNA (dsRNA) structure at the 5′ and 3′ ends of the genome are required for translation and replication (10, 11, 18). The structural proteins, capsid (C), and precursor glycoproteins PE2 and 6K/E1 are translated in a second wave from the subgenomic RNA prior to packaging and egress of viral progeny, providing a subtle temporal regulation of protein expression (32).
In nature, the alphaviruses are arthropod borne, vectored between their vertebrate hosts by infected mosquitoes, and deposited in the host's skin as the mosquito takes a blood meal. Subcutaneously inoculated into mice, the virus infects cutaneous dendritic cells (DCs), hijacking the host's immune surveillance program to access regional lymph nodes (13, 33, 36), ultimately facilitating systemic dissemination of the virus by serum viremia. Thus, the ability to establish a productive infection in DCs is of paramount importance in alphavirus virulence and pathogenesis.
As is true for the majority of RNA viruses, it has become apparent that major factors determining alphavirus cell tropism and virulence lie in the sensitivity of the virus to alpha/beta interferon (IFN-α/β)-stimulated antiviral effectors and the relative ability of cells in different tissues to mount an antiviral defense in response to IFN-α/β signaling. IFN-α/β is a primary mediator of protection against infection with Sindbis virus (SB), the prototypic alphavirus (4, 36). The IFN-α/β system rapidly protects DCs from SB infection, as evidenced by the dramatically increased susceptibility of myeloid-lineage cells to infection in the absence of IFN-α/β receptor (IFNAR)-mediated signaling, both in vitro and in vivo (36, 37).
The nonspecific IFN-α/β response for early host defense against virus infection is extremely complex, exhibiting multiple, interconnecting levels of antiviral activity capable of controlling the extent of virus replication, the apparent tissue tropism of the virus and the development of disease (1, 40, 43). Genes whose transcription is upregulated after IFN-α/β treatment of cultured cells and whose products potentially contribute to these effects number in the hundreds (6, 7). Many viruses are susceptible to the two major IFN-α/β-inducible, dsRNA-triggered host translational control programs (17, 40): the coupled 2′-5′ oligoadenylate synthetase (OAS)/RNase L pathway and the dsRNA-dependent protein kinase (PKR) pathway which provide rapid antiviral activity independent of IFN-α/β signaling (40, 43). SB replication activates and is sensitive to both of these antiviral effector mechanisms in murine embryo fibroblasts (38, 39, 41, 42). We have previously demonstrated that early control of SB replication was defective in primary myeloid bone marrow-derived DC (BMDC) cultures generated from mice with targeted disruption of the PKR gene (PKR−/−) or of both the PKR and RNase L genes (triply deficient, or TD), suggesting that the PKR pathway, but not the OAS/RNase L pathway, suppresses virus replication in DCs prior to IFN-α/β induction (37).
Intriguingly, several lines of evidence indicate the existence of IFN-α/β signaling-dependent antiviral pathway(s), able to function independently of PKR and/or OAS/RNase L, with the ability to suppress SB infection (37). First, SB infection of mice TD in RNase L, PKR, and Mx was subclinical, whereas infection of IFNAR1−/− mice was rapidly fatal, indicating that systemic dissemination of virus is restricted by an alternative IFNAR-dependent mechanism. Second, antiviral activity was observed in vitro in immature BMDC from PKR−/− or TD mice at ∼12 h postinfection (h p.i.). The delay in this effect presumably reflects the requirement for synthesis of IFN-α/β and expression of the IFN-α/β-stimulated gene (ISG) effector(s). Third, this effect was sensitive to neutralization of IFN-α/β and was not observed in IFNAR1−/− BMDC. Finally, priming of TD BMDC by pretreatment with IFN-α/β-induced residual antiviral activity against SB that appeared nearly as potent as that observed in cells from wild-type mice. Thus, we hypothesized that IFN-α/β can limit SB replication through PKR-dependent and PKR-independent mechanisms and that the alternative pathway requires IFN-α/β-mediated induction or activation of effector(s). Similar observations have been made for vesicular stomatitis virus (9, 26), encephalomyocarditis virus (EMCV) (26, 56), dengue virus (8), and hepatitis C virus (HCV) (12, 29), and several alternative antiviral effectors have recently been described with activity against these viruses (9, 21, 24).
The present study was undertaken to investigate the mechanisms by which IFN-α/β priming limits SB replication in DCs in the absence of PKR/RNase L-mediated translational control programs. By analyzing global gene regulation under specific experimental conditions in the pathogenesis-appropriate context of the primary immature DC, we have identified candidate effectors of the alternative pathway. In addition, we have gained insight into the nature of this antiviral mechanism by determining the steps at which the alternative pathway interferes with SB replication. We demonstrate that IFN-α/β priming of BMDCs dramatically impacts early virus protein synthesis, either by altering RNA stability or by inhibiting translation, in all probability via a mechanism that targets mRNAs whose translation initiation is 7-methyl guanine cap dependent. This mechanism effectively suppresses the production of the virus replication machinery (nsPs) and blocks initial replication of the genome.
MATERIALS AND METHODS
Cell lines.
Baby hamster kidney cells (BHK-21, ATCC CCL-10) were maintained in alpha minimal essential medium, supplemented with 10% donor calf serum, 2.9 mg of tryptose phosphate/ml, 0.29 mg of l-glutamine/ml, and 100 U of penicillin/ml-0.05 mg of streptomycin/ml. NS47 murine fibroblasts were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 0.29 mg of l-glutamine/ml, 1% nonessential amino acids, 1 mM sodium pyruvate, 20 μg of gentamicin/ml, 5 mM HEPES buffer, and 50 μM β-mercaptoethanol (complete RPMI [cRPMI]). All cells were maintained at 37°C in 5% CO2. Medium and additives were purchased from Mediatech; serum was purchased from Invitrogen.
Mice.
Breeder pairs of IFNAR1−/− and 129 Sv/Ev strain mice were kindly provided by Barbara Sherry (North Carolina State University, Raleigh, N.C.) and Herbert Virgin (Washington University, St. Louis, Mo.). Breeder pairs of TD mice were kindly provided by Bryan Williams and Robert Silverman (Cleveland Clinic Foundation, Cleveland, Ohio). Mice were housed in the Animal Resource Center (Louisiana State University Health Sciences Center, Shreveport [LSUHSC-S]) under specific-pathogen-free conditions. All procedures were carried out in accordance with federal and institutional guidelines for animal care and use.
Primary BMDCs.
Primary immature BMDCs were generated as described previously (37). Briefly, bone marrow cells from femurs and tibia of 8- to 12-week-old mice were aspirated with cRPMI medium. On ice, cells were strained to remove bone fragments, pelleted (60 × g, 5 min), counted, and seeded at 5 × 107 cells per 150-mm-diameter dish. Cells were incubated for 12 to 14 days (37°C, 5% CO2) in cRPMI medium, supplemented with 5% NS47 fibroblast-conditioned culture supernatant and 500 pg of granulocyte-macrophage colony-stimulating factor (Peprotech)/ml (48). Every third day, dishes were gently swirled to dislodge nonadherent cells and two thirds of the supernatant was replaced with fresh, supplemented cRPMI medium. Cells were harvested by using enzyme-free cell dissociation buffer (Invitrogen) with gentle scraping.
Virus cDNA clones, transcripts, and stocks.
The construction of plasmid p39K70, encoding the full-length genomic sequence of a cell culture-adapted SB downstream of an SP6 bacteriophage promoter sequence, has been described previously (28). Plasmid pToto1101/Luc encodes the open reading frame (ORF) for firefly luciferase (fLUC) inserted in frame into the carboxyl-terminal portion of nsP3 in pToto1101 (Fig. 1) (2). 5′-capped infectious viral RNA was generated from XhoI-linearized p39K70 or pToto1101/LUC cDNA templates by in vitro transcription (mMessage mMachine; Ambion). To make virus stocks, RNAs were electroporated into BHK-21 cells (2 pulses at 0.22 kV, 1.0 mF of capacitance, GenePulser II; Bio-Rad). Virus released into the culture supernatant by 24 h was harvested, clarified by centrifugation, and stored at −80°C. Stock titers were determined by standard BHK-21 cell plaque assay and expressed as PFU per milliliter.
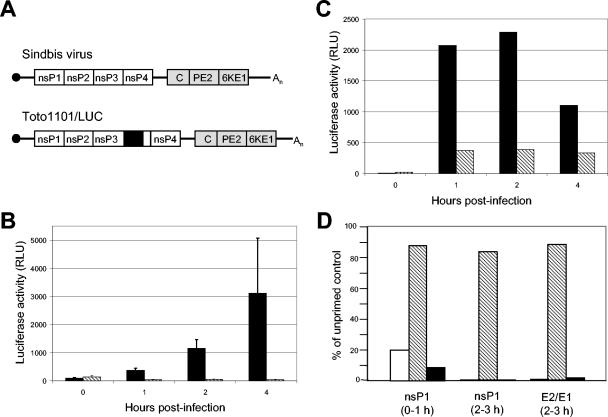
FIG. 1.
(A) Schematic representations of the genomes of unmodified SB and firefly luciferase (fLUC)-expressing Toto1101-LUC virus. NTRs (solid lines), nsP genes (open boxes), structural genes (shaded boxes), 7methyl-GTP cap (filled circle), poly(A) tail (An), and the fLUC ORF (black box within nsP3) are shown. (B) TD BMDC, either untreated (black bars) or primed for 6 h with 1,000 IU of IFN-α/β/ml (hatched bars), were infected with Toto1101/LUC (MOI = 10). At indicated times p.i., cells were lysed and luciferase activity was measured in relative light units (RLU). Data are means (n = 3), with error bars indicating standard deviations. Results are representative of two independent experiments. (C) Untreated (black bars) or IFN-α/β-primed (hatched bars) TD BMDC were harvested by using enzyme-free cell dissociation buffer and electroporated with capped Toto1101-LUC RNA transcripts. Luciferase activity was measured as described for panel B. (D) BMDC, either untreated or IFN-α/β primed, were infected with unmodified SB and pulsed with radiolabeled Cys and Met. Virus proteins were immunoprecipitated, separated by sodium dodecyl sulfate-PAGE, and quantitated on a phosphorimager. The accumulation of nsPs and E2/E1 in IFN-α/β-primed 129 Sv/Ev (white bars), IFNAR1−/− (hatched bars), and TD (black bars) BMDC are presented as a percentage of the untreated control cells. The time of the pulse is indicated.
Translation reporter plasmids.
The SB translation reporter plasmid encoding fLUC, pSBΔnsP/fLUC, was derived from the SB-based replicon p39REP (37). Initially, the fLUC ORF was amplified from pToto1101/LUC with primers to introduce XbaI and NotI sites at the 5′ and 3′ ends, respectively. This fragment was inserted downstream of the 26S subgenomic promoter in p39REP, creating a replicon capable of expressing fLUC, p39REP/fLUC. The fusion of fLUC with truncated nsP1 was completed by overlap PCR mutagenesis. Briefly, primers were designed that amplified the region of p39REP/fLUC from upstream of a SacI site in the vector (including the SP6 promoter and 5′ nontranslated region [5′ NTR]) to nucleotide (nt) 281 (amino acid 74) of the nsP1 gene and, separately, from the 5′ end of the fLUC gene to immediately downstream of the XhoI linearization site of the replicon [including the 3′ NTR and poly(A) tail]. The sense primer annealing to the fLUC 5′ end included 24 nt of nontemplated sequence that was complementary to the nsP1 antisense primer and encoded the fusion of fLUC codon 2 in frame after the methionine (Met) codon at amino acid 74 of nsP1. After first-round overlap PCR primer synthesis and purification, the overlap primers were combined without replicon template in a second round of PCR resulting in deletion of nsP genes downstream of nsP1 nt 281 and the 26S promoter and creating the fused amplicon. This amplicon was inserted with SacI and XhoI into p39REP. The resulting pSBΔnsP/fLUC plasmid was linearized for transcription with XhoI and transcribed from the SP6 promoter (Fig. 2). Plasmid pCITE-Luc-A31 encodes the EMCV internal ribosome entry site (IRES) upstream of the fLUC ORF with a 31-nt poly(A) tail. This plasmid linearizes with SalI and transcribes with T7 polymerase. Construction of plasmid pSP64-Ren-Luc-Poly(A) has been described previously (2). This plasmid encodes the ORF for Renilla luciferase (rLUC) between an SP6 promoter and 3′ poly(A) sequences and is linearized with EcoRI for the production of capped transcripts encoding rLUC. For electroporations, in vitro transcripts were quantitated by spectrophotometry and diluted to equivalent RNA molar ratios. Between 10 and 20 μg of each RNA was used per electroporation reaction.
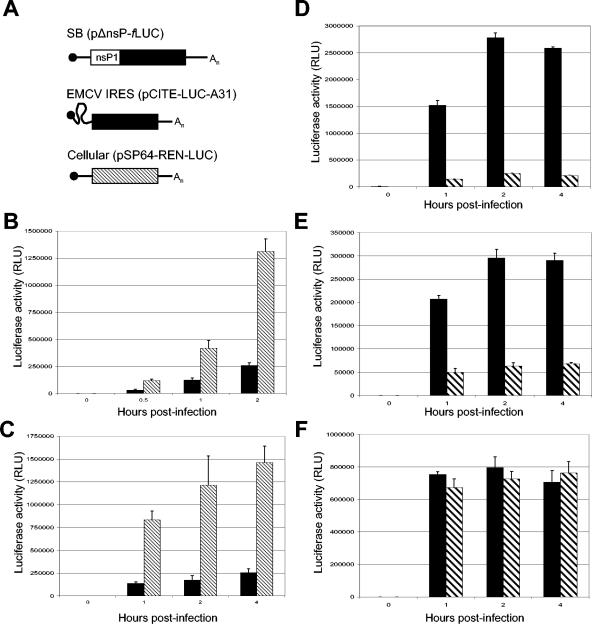
FIG. 2.
Initial translation of virus genomes is suppressed in the absence of PKR. (A) Schematic representations of translational reporter constructs for SB (pSBΔnsP/fLUC), EMCV (pIRES-EMCV-A31), and host cell (pSP64-REN-LUC) mRNAs. (B and C) Capped SB (B) and EMCV IRES (C) translation reporter constructs (black bars) expressing fLUC were coelectroporated into BHK-21 cells with equimolar amounts of capped control RNA encoding rLUC (hatched bars). Cells were lysed at the indicated times, and rLUC and fLUC activities were measured with the dual-luciferase reporter assay system. Data are means (n = 3), with standard deviations indicated by error bars. (D to F) Untreated (black bars) or IFN-α/β-primed (hatched bars) TD BMDC were electroporated with capped SB fLUC-expressing translation reporter RNA transcripts (D), capped control RNA encoding rLUC (E), or EMCV IRES fLUC-expressing translation reporter RNA transcripts (F). Cells were lysed at the indicated times, and rLUC and fLUC luciferase activities are shown as means (n = 3) ± standard deviations.
IFN-α/β treatment and virus infection.
BMDC were primed by the addition of 1,000 IU of IFN-α/β (Access Biomolecular) per ml of cRPMI for 6 h prior to harvest or infection. After removal of the medium, BMDC were infected with 39K70 and Toto1101-LUC virus at a high multiplicity of infection (MOI) (∼10 PFU/cell) for 1 h at 37°C. Cells were washed three times with Dulbecco's phosphate-buffered saline (PBS; Mediatech) supplemented with 1% donor calf serum, and cRPMI was replaced.
Virus growth curves.
Prior to virus infection, BMDC were either left untreated, primed with 1,000 IU of IFN-α/β for 6 h, or treated with 1.0 μg of actinomycin D (ActD, A9415; Sigma)/ml at 37°C for a total of 6 h prior to virus infection, initially for 1 h with ActD alone and then for a further 5 h in the presence of 1,000 IU of IFN-α/β/ml and ActD. At various times p.i., samples of supernatant were collected for plaque assay titration of progeny virions.
Electroporations.
BMDC were harvested as described above, resuspended in OptiMEM (Mediatech) at ∼1 × 107 cells/ml, and electroporated with equimolar amounts of pSBΔnsP/fLUC, pSP64-Ren-Luc-Poly(A), or pCITE-Luc-A31 RNA transcripts (2 pulses at 0.22 kV, 1.0 mF of capacitance). For t = 0, electroporated cells were immediately pelleted via centrifugation and lysed. For later times, cells were either maintained in suspension in OptiMEM medium or diluted into cRPMI and seeded in six-well plates. Cells were harvested for luciferase assay by active lysis (Promega) at different times postelectroporation.
Determination of luciferase activity.
Cell lysates were harvested with 1× luciferase lysis buffer, and luciferase activity was determined by using the dual-luciferase reporter assay system (Promega) according to the manufacturer's recommendations. Luciferase activity was measured on a BMG PolarStar microplate reader (BMG Technologies) and expressed as relative light units.
Pulse-labeling and immunoprecipitation of virus proteins.
BMDC in six-well plates were treated with 1,000 IU of IFN-α/β/ml for 6 h prior to infection with 200 μl of undiluted 39K70 stock for 1 h at 37°C. Cells were resuspended in Met-free, cysteine (Cys)-free minimal essential medium (Gibco). Either immediately after infection (0- to 1-h labeling) or at 2 h p.i. (2- to 3-h labeling), 120 μCi (each) of [35S]Cys and [35S]Met (Amersham)/ml was added. After a further 1 h of incubation, cells were washed three times with PBS-Ca2+/Mg2+ and lysed with scraping in 500 μl of radioimmunoprecipitation assay buffer (22). Supernatants were cleared by brief centrifugation and then preincubated with protein A beads overnight at 4°C with gentle agitation. Protein A beads were removed by centrifugation, aliquots of polyclonal monospecific rabbit anti-nsP1 antiserum (generously provided by Charles Rice, Rockefeller University, New York, N.Y.) (22, 31, 32) or hyperimmune mouse ascites fluid were then added, and the mixture was incubated for 2 h at 4°C. Normal mouse or rabbit sera (Sigma) were used as negative controls. Additional protein A beads were added, and the incubation was continued for another 2 h. After incubation, protein A bead-antibody conjugates were pelleted and washed three times with radioimmunoprecipitation assay buffer. Pellets were resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) loading buffer followed by 10% acrylamide PAGE analysis. Virus protein bands were visualized and quantitated with a Storm phosphorimager (Molecular Dynamics).
Western blotting.
Whole-cell extract lysates were prepared by washing cells twice with PBS and lysing them in whole-cell extract lysis buffer containing protease and phosphatase inhibitor cocktails (Sigma). Protein concentrations were determined by Bradford assay, and equal protein was loaded. After PAGE separation, proteins were transferred to a polyvinylidene difluoride membrane, blocked in Tris-buffered saline-Tween 20-5% nonfat milk, washed two times, and incubated for 1 h with primary antibody: β-actin (Santa Cruz Biotechnology), total α subunit of eukaryotic initiation factor 2 (eIF-2α; Biosource), or phosphospecific eIF-2α (Biosource) at an ∼1:1,000 dilution. Membranes were washed extensively and incubated for 1 h with secondary antibody (∼1:3,000 dilution in Tris-buffered saline-Tween 20-1% milk), biotinylated goat anti-mouse or anti-rabbit as appropriate (Vector Labs). For detection, membranes were incubated for 1 h with streptavidin-horseradish peroxidase conjugate, washed, and developed with ECL-Plus substrate (Amersham) on film and the Storm phosphorimager. Phosphorylated eIF-2α was stained first, and blots were stripped with 0.2 M NaOH and reprobed for total eIF-2α and β-actin sequentially.
RNA isolation, labeling, and microarray hybridization.
129 Sv/Ev-, TD-, and IFNAR1−/−-derived BMDC were treated as described above, and total cellular RNA was isolated by using the RNeasy kit (Qiagen). Five micrograms of total RNA per sample was prepared for hybridization by following the manufacturer's protocols (Affymetrix, Santa Clara, Calif.). Fifteen micrograms of each fragmented cRNA was hybridized to murine U74Av2 GeneChip arrays (Affymetrix), which has ∼6,000 known genes and ∼6,000 expressed sequence tags. Hybridized arrays were scanned with an Agilent GeneArray scanner.
Data interpretation.
Expression data were analyzed with the Micorarray Analysis Suite, version 5.0, and Data Mining Tool, version 3.0 (Affymetrix). Global scaling was performed to compare genes between chips. Each chip was normalized to a target intensity value of 2,500. The absolute call and average difference of 12,488 gene expressions in each sample, as well as the direction of change and relative change of each gene expression between samples, were identified. Each gene is represented by a probe set of 25 probe pairs, consisting of a perfect match oligonucleotide and a mismatch oligonucleotide with a central 1-base mismatch. Absolute call is a qualitative measure in which each gene is assigned a call of present, marginal, or absent, based on the hybridization of the RNA to the probe set. Average difference is a quantitative measure of the level of gene expression, calculated by taking the difference between the mismatch and perfect match of every probe pair and averaging the differences over the entire probe set. Expression analysis files created by Microarray Suite, version 5.0, were exported to Microsoft Excel (Redmond, Wash.) for data formatting. Excel files were imported into Spotfire (Somerville, Mass.) for further filtering. Lists were generated containing genes that were upregulated or downregulated by ≥2- or ≥4-fold in replicate analyses (n = 2). Venn diagrams were generated showing overlaps. Protein BLAST searches (Blastx) were completed on candidate expressed sequence tags by using the National Center for Biotechnology Information Entrez search engine (54).
Confirmatory RT-PCR.
Total cellular RNA was isolated by using Ultraspec (Biotecx) per the manufacturer's instructions. RNA was reverse transcribed (RT) from a dT18 primer in 1× first-strand buffer, 1 mM dithiothreitol, 200 μM deoxynucleoside triphosphates, RNasin (Promega), and SuperScript III (Invitrogen) for 1 h at 42°C. cDNAs were subjected to PCR amplification with gene-specific primers (Integrated DNA Technologies) (Table 1) designed against GenBank sequences. RT-PCR products were resolved by gel electrophoresis and visualized on a VersaDoc 4000 imaging system (Bio-Rad). The specificity of some products was determined by direct sequencing of gel-purified PCR fragments (Sequencing Facility, University of Texas Medical Branch, Galveston, Tex.). The specificity of mouse zinc finger antiviral protein (ZAP) RT-PCR products was confirmed by agarose gel migration analysis compared to a PCR product amplified from a cDNA clone of mouse ZAP (pcDNA4/to/myc-mouseZAP995) provided by Guangxia Gao (Rockefeller University, New York, N.Y.).
TABLE 1.
Primers for RT-PCR microarray confirmation
| Gene | Sense primer | Antisense primer |
|---|---|---|
| B-actin | TAGGCACCAGGGTGTGATGGTGGGAATGGG | GCAGCTCATAGCTCTTCTCCAGGGAGGAAGAGG |
| ISG15 | ATGGCCTGGGACCTAAAGGTGAAG | TTAGGCACACTGGTCCCCTCCCCC |
| ISG20 | ATGGCAGGCATCCCAGAGGTGGTGG | TCAGTCTGACGTCCCAGGGCAAGG |
| IFI54/GARG-39 | ATGAGTACAACGAGTAAGGAGTCACTGGAG | CTAGTATTCAGCACCTGCTTCATCC |
| IFI56/GARG-16 | ATGGGAGAGAATGCTGATGGTGACCAG | TCAGAATGCAGGGTTCATTTCCCC |
| IFI60/GARG-49 | ATGAGTGAGGTCAACCGGGAATCTC | CTATGTTTGCTCTTTAACCTCTTCC |
| IGTP | ATGGATTTAGTCACAAAGTTGCC | TCAGTGAATTTCGGGAGGGAGGAC |
| TGTP | ATGGCTTGGGCCTCCAGCTTTGATGC | TCAAGCTTCCCAGTACTCGGGGGGCTC |
| IIGP | ATGGGTCAGCTGTTCTCTTCACC | CTAGTTTCTTAAACATATCTCTTTAAG |
| mGBP2 | ATGGCCTCAGAGATCCACATGTCGG | TCAGAGTATAGTGCACTTCCCAGACG |
| STAT1 | GTGAAGCCAATGGTGTGGCGAAGAGCG | CAAGACCAGGGGCCTCTGCGGGTGAGTGGG |
| mZAP | ACCAGGCCGGGATCACTCGGTCGGTGGTGG | AGACACATCCTCCAGGGGATCCTTACAGCC |
RESULTS
We have demonstrated previously that elimination of the PKR and OAS/RNase L antiviral pathways does not prevent IFN-α/β-mediated induction of potent antiviral activity against SB (37). Furthermore, the refractory state can be established either by priming of naíve cells or through autocrine activity of IFN-α/β induced by the virus infection. Therefore, activation and/or induction of the major known antiviral proteins, PKR, RNase L, and Mx, does not appear to be required for IFN-α/β-mediated protection against SB infection, revealing the existence of at least one, and perhaps several, IFN-α/β-mediated antiviral pathways that function independently of PKR, RNase L, and Mx protein expression (36, 37). We have used a global approach to identify candidates and characterize the alternative antiviral activity that effectively protects the host from SB.
Of paramount importance to these studies is the isolation of the activities of the PKR-mediated and alternative antiviral pathways by the use of BMDCs derived from genetically modified mice: (i) 129 Sv/Ev-derived BMDCs have functional PKR/RNase L-mediated and IFN-α/β-mediated systems; (ii) TD-derived BMDCs lack PKR and OAS/RNase L pathways but are otherwise IFN-α/β responsive; and (iii) IFNAR1−/−-derived BMDCs possess intact constitutive PKR and OAS/RNase L but lack IFN-α/β-inducible responses including the alternative pathway(s). All of these strains of mice lack the ability to produce functional Mx proteins (56).
Early translation of viral proteins is suppressed by alternative pathway(s).
We have demonstrated previously that synthesis of the virus structural proteins and production of progeny virions are suppressed by IFN-α/β priming of the PKR/RNase L-independent pathway(s) (37). Here, we have evaluated PKR-independent, IFN-α/β-mediated inhibitory effects on SB life cycle events that precede subgenome translation. The first required step in virus infection after entry and uncoating is translation of the incoming genomic RNA to synthesize the proteins necessary to assemble the replication complex. We infected BMDC with Toto1101/Luc virus, a replication-competent virus which expresses fLUC fused into the carboxyl-terminal portion of the nsP3 protein (Fig. 1A) (2) and used changes in luciferase activity in Toto1101/Luc-infected cells as an indicator of translation of the full-length viral RNA. After priming with IFN-α/β, early luciferase synthesis in TD and 129 Sv/Ev-derived BMDC dropped to almost undetectable levels (Fig. 1B). In contrast, luciferase activity was not altered by IFN-α/β treatment of IFNAR1−/−-derived BMDC (data not shown). The IFN-α/β-mediated block to SB RNA translation was still apparent when binding, penetration, and uncoating steps were obviated by directly introducing the RNA into cells. Capped Toto1101/Luc transcripts were transfected into BMDC by electroporation (Fig. 1C). Although the levels of luciferase activity increased more rapidly following electroporation, perhaps because more genomic RNA was introduced per cell, luciferase expression was significantly suppressed by IFN-α/β priming, signifying that the inhibition to virus replication occurred after virus entry and uncoating.
These findings were confirmed for unmodified SB by quantifying the accumulation of radiolabeled virus proteins in the first and third hours of infection (Fig. 1D). Priming of TD BMDC with IFN-α/β reduced the synthesis of nsP1 by ∼10-fold during the first hour of infection and by >100-fold in the third hour compared with untreated controls. Synthesis of the glycoproteins, which requires nsP-dependent replication of the genome, was comparably reduced in the third hour. Taken together, these data suggest that IFN-α/β-mediated virus control, functioning in the absence of PKR, RNase L, and Mx proteins, inhibits the replication of SB during early events in infection but after the genome is released into the cytoplasm.
Initial translation of virus genomes is suppressed in the absence of PKR.
The caveat to these experiments is that measurements of SB protein synthesis, particularly at later times postinfection, represent a combination of protein translated directly from the incoming genomic RNA and from newly synthesized progeny genomes. Accumulation of these proteins would be affected by inhibition of RNA transcription in addition to translation. To address these issues, we generated an SB-based translation reporter construct, pSBΔnsP/fLUC, encoding the N-terminal 74 amino acids of nsP1 as an in-frame fusion with fLUC (Fig. 2A) between the 5′ and 3′ NTR regions required for genome translation and replication (10). Transfection of in vitro-synthesized pSBΔnsP/fLUC or transcripts derived from the pSP64-REN-Luc-Poly(A) plasmid into cells and quantification of the luciferase activity generated allows simple quantitation of translation of incoming viral RNA relative to a host translation control plasmid. The translational competence of these RNAs was demonstrated by cotransfection of BHK-21 cells with control rLUC-expressing RNA and fLUC-expressing SB (Fig. 2B) or EMCV (Fig. 2C) translation reporter RNAs.
To compare translation in the absence of viral RNA replication, RNA transcripts encoding luciferase were electroporated into TD BMDC to allow for the direct quantification of the translation of an RNA that could not be replicated. Data from multiply repeated experiments indicated that translation of incoming SB reporter RNA was reduced ≥10-fold in IFN-α/β-treated cells compared to control cells (Fig. 2D). The translation of the transcript mimicking a host cell RNA, SP64-REN-LUC-poly(A), was also significantly inhibited, however, to a lesser degree (∼5-fold inhibition) than the virus-like RNA (Fig. 2E). Electroporation of 129 Sv/Ev BMDC yielded results similar to TDs, while, as expected, IFN-α/β treatment had no effect on IFNAR1−/− BMDC (data not shown). Results of these experiments were similar regardless of whether electroporated cells were allowed to adhere to tissue culture plates after electroporation or cells were maintained in suspension and pelleted at the time of luciferase assay, and protein assays of collected lysates indicated similar protein contents (data not shown). These results suggested that the reduction in luciferase translation in IFN-treated cells represented inhibition of translation as opposed to wholesale loss or destruction of IFN-treated cells; however, the possibility remained that IFN treatment rendered cells refractory to efficient electroporation.
To address this issue we electroporated an RNA encoding fLUC under the translational control of the EMCV IRES. The rationale for this experiment was based upon recent studies suggesting that IFN-α/β-mediated translational control activities may distinguish between cap-dependent and cap-independent translation initiation mechanisms (29). Surprisingly, IFN-α/β failed to inhibit translation mediated by the EMCV IRES (Fig. 2F). Thus, electroporation efficiency was not diminished for IFN-α/β-treated cells, and notably, the experiment revealed a specificity of early IFN-α/β-mediated translation control for RNAs by cap-dependent initiation. In addition, these experiments suggested that the RNA containing SB 5′ and 3′ replication and translation control sequences may be more sensitive to inhibition than the simple host control RNA.
Phosphorylation of eIF-2α is defective in the absence of PKR.
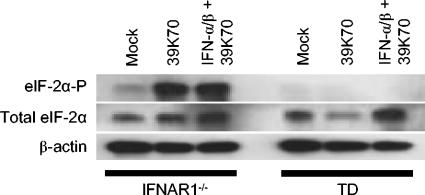
Activation of PKR by dsRNA viral replicative intermediates leads to the PKR-catalyzed phosphorylation of eIF-2α, causing a decline in cap-dependent translation of viral and cellular mRNAs (45). Under conditions of physiological stress, eIF-2α can also be phosphorylated by at least two other kinases: general control nonderepressible-2 kinase and PKR-like endoplasmic reticulum resident kinase (27). Reasoning that the cap-dependent translation of SB transcripts may be suppressed by eIF-2α phosphorylation even in the absence of PKR, we assessed the phosphorylation state of eIF-2α in infected BMDC (Fig. 3). Compared to untreated, mock-infected TD-derived cells, an increase in phosphorylation of eIF-2α was not detectable in response to virus infection regardless of whether or not the cells were primed with IFN-α/β, suggesting that stress-induced eIF-2α phosphorylation does not mediate the alternative antiviral pathway. In contrast, eIF-2α is phosphorylated by virus infection of IFNAR1−/− BMDC, where PKR-mediated inhibition of SB translation is evident (37).
FIG. 3.
eIF-2α phosphorylation in BMDC. IFNAR1−/− (lanes 1 to 3) and TD (lanes 4 to 6) BMDC were left untreated (lanes 1 and 4), infected with SB virus 39K70 (MOI = 10) for 12 h (lanes 2 and 5), or primed with 1,000 IU of IFN-α/β/ml for 6 h prior to infection (lanes 3 and 6). Levels of phosphorylated eIF-2α (eIF2α-P, top panel) and total eIF-2α (middle panel) were compared to levels of β-actin control (bottom panel) in cell extracts by immunoblot analysis. The results shown are representative of three independent experiments.
De novo gene transcription is required for induction of the alternative antiviral response.
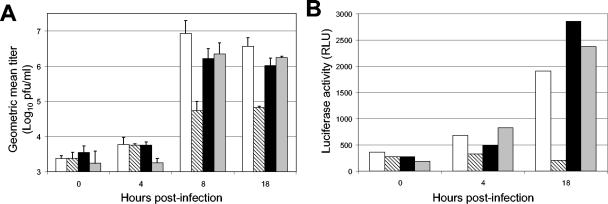
Reasoning that an IFN-α/β-inducible response would most likely require transcriptional upregulation of cellular ISG(s), we determined the effect of an inhibitor of host gene transcription on activity of the alternative pathway. Treatment of TD-derived BMDC with the RNA polymerase inhibitor ActD had no significant effect on the growth of SB compared with untreated controls (Fig. 4A). Confirming our previous results, pretreatment of TD BMDC with 1,000 IU of IFN-α/β/ml suppressed progeny virion production by ∼100-fold. However, if the primed TD cells were pretreated with ActD, SB replication levels were restored to the levels observed in unprimed cells. These data indicate that the predominant IFN-α/β-mediated antiviral effect in the absence of PKR and RNase L pathways requires de novo gene transcription of ISGs. To confirm the association between the requirement for ISG induction and inhibition of genomic cap-dependent translation, TD-derived cells were pretreated with IFN-α/β in the presence or absence of ActD and then infected with Toto1101/Luc virus (Fig. 4B). Results were consistent with the virus titer data in that the inhibitory effects of IFN-α/β were eliminated by ActD treatment.
FIG. 4.
Priming is eliminated by ActD treatment. (A and B) Prior to virus infection, TD BMDC were untreated (white bars), primed with 1,000 IU of IFN-α/β for 6 h (hatched bars), treated with 1.0 μg of ActD/ml for 6 h (black bars), or ActD-treated for 1 h before priming for a further 5 h (shaded bars). (A) Cells were infected (MOI = 10) with unmodified SB virus, and at indicated times after infection, the accumulation of virus released into the cell supernatant was titrated by BHK-21 plaque assay. Data are presented as geometric means (n = 3) of log10 titers (PFU/ml) ± standard deviations. The experiment is representative of two independent experiments. (B) Cells were infected (MOI = 10) with fLUC-expressing Toto1101-LUC virus. Cells were lysed at the indicated times, and fLUC luciferase activities are shown.
Gene profiling for IFN-α/β- and virus-inducible genes in BMDC.
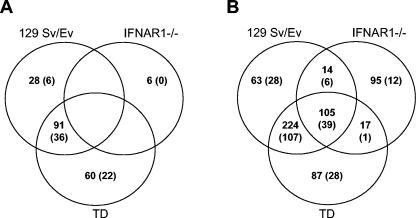
Affymetrix DNA microarray technology was used to analyze global gene induction patterns in BMDC following IFN-α/β priming or virus infection. RNA was harvested at 6 h posttreatment for IFN-α/β priming and at 12 h p.i. for virus infection, as these represent the earliest times at which the alternative antiviral state is fully manifest by the different treatments (37). Data were analyzed to reveal genes upregulated (i) in the presence (TD and 129 Sv/Ev) versus the absence (IFNAR1−/−) of IFN-α/β signaling and (ii) in the presence (129 Sv/Ev and IFNAR1−/−) versus the absence (TD) of PKR/RNase L. Exposure of 129 Sv/Ev- and TD-derived BMDC to IFN-α/β resulted in transcriptional upregulation of 119 and 151 genes, respectively, by ≥2-fold compared to untreated BMDC (Fig. 5A). Of these, 91 genes were upregulated in the presence or absence of PKR/RNase L, and those upregulated in only one cell strain tended to be only marginally induced. The majority of these genes have previously been identified as ISGs (6, 7). Fewer than 6 genes were upregulated by IFN-α/β priming of IFNAR1−/− cells, none of them reproducibly, as would be expected. Absolute call analyses of genes present, marginal, or absent did not reveal a significant overall change in the number of genes being transcribed compared to unprimed cells (data not shown). Thus, the absence of PKR and RNase L does not appear to significantly impact the cell's ability to respond to the presence of IFN-α/β at the time of measurement (6 h posttreatment).
FIG. 5.
Genes similarly and differentially induced by IFN-α/β priming and/or virus infection. Total RNA was isolated from 129 Sv/Ev, TD, and IFNAR1−/− BMDC either untreated, IFN-α/β primed (1,000 IU/ml for 6 h), or SB infected (12 h p.i.) and hybridized to murine U74Av2 GeneChip arrays (Affymetrix) with 12,488 gene expressions in each sample. Expression data were analyzed as described in Materials and Methods. Spotfire software was used to generate lists of genes that were upregulated ≥2- or ≥4-fold over untreated controls in replicate analyses (n = 2). Venn diagrams were generated to show overlaps between genes upregulated ≥2-fold by IFN-α/β priming (A) or by virus infection (B). Similar analyses of ≥4-fold induction are shown in parentheses.
Virus infection of the BMDC cells resulted in the upregulation of significantly more genes than IFN-α/β treatment (Fig. 5B). Virus infection induced the expression of 329 genes in both TD and 129 Sv/Ev cells, 105 of which were also induced in virus-infected IFNAR1−/− cells. This is particularly interesting given the differences in virus replication due to the PKR pathway. Even in 129 Sv/Ev BMDC, where virus replication is extremely restricted, over 400 genes are upregulated by ≥2-fold (180 genes by ≥4-fold). This group includes the IFN-α and IFN-β genes, known ISGs, IFN regulatory factor 1 (IRF-1), IRF-3 and IRF-7, transcription factors, and inflammatory products. The genes upregulated only in TD and 129 Sv/Ev cells are predicted to require signaling of induced IFN-α/β through the IFNAR, whereas the 231 genes upregulated in virus-infected IFNAR1−/− cells are either directly induced by viral products (e.g., through interaction with the Toll-like receptors) or indirectly by a mediator (e.g., cytokine) induced by the virus infection.
Data mining identifies candidate alternative pathway effectors.
As the activity of the alternative pathway is greatly reduced in TD BMDC when de novo gene transcription is blocked by ActD, we reasoned that the effector of the pathway is among the genes shown to be upregulated by IFN-α/β priming of TD-derived BMDC and is likely to be relatively strongly induced. In addition, as an antiviral state can be established by IFN-α/β-mediated priming prior to virus infection or by virus-induced IFN-α/β, alternative pathway effector(s) should also be upregulated in virus-infected TD cells. However, in the absence of IFN-α/β signaling, virus infection alone does not directly elicit an effective alternative response, ultimately permitting high levels of virus replication in vitro and in vivo. Therefore, we inferred that the effector would be among those genes induced in IFN-α/β-primed and virus-infected TD BMDC but not in infected IFNAR1−/− cells. The data were analyzed to reveal ISGs that represented candidate effectors of the IFN-α/β-inducible, PKR-independent antiviral response under the following stringent selection criteria. The final gene set included genes upregulated by ≥4-fold in IFN-α/β-primed and virus-infected TD BMDC that were at least twofold-less-well-induced by virus infection in the absence of the IFNAR in IFNAR1−/− BMDC in two independent experiments (Table 2). These analyses resulted in three sets of genes: (i) 44 genes induced by virus infection and IFN-α/β priming in both experiments, (ii) 11 genes induced by virus infection and IFN-α/β priming in experiment 1 but not by both treatments in experiment 2, and (iii) 9 genes induced by virus infection and IFN-α/β priming in experiment 2 but not by both treatments in experiment 1 (6 of them unknown genes). The genes were grouped according to published activities and relationships (Table 3). Further analyses confirmed that the 44 genes consistently induced in IFN-α/β-primed and virus-infected TD BMDC by ≥4-fold over mock and by ≥2-fold over virus-infected IFNAR1−/− cells were also significantly upregulated (≥2-fold) in IFN-α/β-primed and virus-infected 129 Sv/Ev cells (data not shown), compatible with a role in the PKR/RNase L-independent autocrine response.
TABLE 2.
Functional grouping of candidate effector genes
| Group | Genes |
|---|---|
| GTPases family members | |
| 47-kDa GTPases | IIGP, IGTP, T-cell specific GTPase, GTP1, IFI47 (IRG-47 protein), LRG-47 |
| 67-kDa GTP binding proteins | mGBP-2, IFN-γ induced (Mg11) |
| Mx proteins | Mutant Mx-1 pseudogene, Mx-2 (nonfunctional gene) |
| GARG proteins | GARG-16/IFI56 (p56 protein, interacts with eIE3-3E), GARG-39/IFI54 (p54 protein), GARG-49/IRG2/IFI60 (p60 protein) |
| Transcription factors and accessory proteins | Signal transducing activator of transcription (STAT-1), IFP35 (interacts with STAT-1), IRF-7, IFN-activated 200-series genes (202B, 203, 204, 205), FLN29 (interacts with TRAF, zinc finger protein), Sp100 (interacts with ets family of transcription factors), LySp100 |
| Protein processing and antigen presentation | Ubiquitin-specific protease (UBP43), ISG15 (ubiquitin-like enzyme), ubiquitin-activating enzyme E1 homolog, proteasome subunits, LMP2, LMP7, LMP10, antigen processing transporter (TAP), histocompatibility 2, T region loci 10, 17, 23 (MHC) |
| Chemokines, cytokines, and immune receptors | Monokine induced by IFN-γ (MIG), IFN-inducible protein 10, tumor necrosis factor related apoptosis inducing ligand, monocyte chemoattractant protein 5, interleukin 18 binding protein, Fc-epsilon receptor, Fc-gamma receptor 1 |
| RNA-modifying enzymes | ISG20 (ssRNA-specific RNase), mxRN1p (cytoplasmic 5′-3′ exoribonuclease), mouse vasa homolog (RNA binding protein) |
| Kinases | Deoxycytidine kinase, thymidylate kinase homologue, Cot serine/threonine kinase homologue, Cot serine/threonine kinase homologue |
| Miscellaneous | Inositol polyphosphate-1-phosphatase, inositol 1,4,5-triphosphate receptor (intracellular calcium channel), mvig (viperin homolog), torsin 3A (ER-resident ATP-binding protein), organic anion transporter homolog, T-cell regulatory protein 1, fibrinogen-like protein 2, AIDS-related endonuclease, liver regeneration protein homolog, proprotein convertase subtilisin/kexin type 7, guanine nucleotide binding protein, β4 (G protein), N6-methyl-transferase (70-kDa subunit) |
TABLE 3.
Gene profiling analysis to identify candidate effectors of the alternative pathway
| Probe set identification no. | NCBIf accession no. | Description of gene product | Exp 1 results (induction [fold])
|
Exp 2 results (induction [fold])
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IFNAR1−/− mock versus virus infected (AB-V1) | TD mock versus IFN-α/β treated (TD-IFN1) | TD-IFN1/AB-V1a | TD mock versus virus infected (TD-V1) | TD-V1/AB-V1b | IFNAR1−/− mock versus virus infected (AB-V2) | TD mock versus IFN-α/β treated (TD-IFN2) | TD-IFN2/AB-V2a | TD mock versus virus infected (TD-V2) | TD-V2/AB-V2b | ||||||||
| Fit criteria for candidate effector in two independent microarray experiments
|
|||||||||||||||||
| 103432_atc | AW122677 | ISG20 | 0.4 | 71.0 | 177.5 | 224.4 | 561.0 | 1.3 | 25.6 | 19.7 | 78.8 | 60.6 | |||||
| 104669_at | U73037 | IRF7 | 0.3 | 52.7 | 175.7 | 143.0 | 476.7 | 5.6 | 63.1 | 11.3 | 170.1 | 30.4 | |||||
| 98373_at | A1462516 | FcɛR | 0.2 | 29.2 | 146.0 | 117.8 | 589.0 | 0.2 | 59.7 | 298.5 | 184.8 | 924.0 | |||||
| 102906_at | L38444 | TGTP | 0.1 | 10.1 | 101.0 | 18.3 | 183.0 | 0.1 | 9.5 | 95.0 | 13.5 | 135.0 | |||||
| 96764_at | AJ007971 | IIGP | 0.4 | 37.0 | 92.5 | 68.6 | 171.5 | 0.7 | 25.5 | 36.4 | 37.3 | 53.3 | |||||
| 101436_at | M34815 | MIG | 0.4 | 35.5 | 88.8 | 362.0 | 905.0 | 7.6 | 33.8 | 4.4 | 680.3 | 89.5 | |||||
| 104750_at | M63630 | IRG-47 | 0.2 | 16.3 | 81.5 | 24.4 | 122.0 | 0.4 | 10.3 | 25.8 | 12.9 | 32.3 | |||||
| 95024_at | AW047653 | UBP43 | 2.8 | 162.0 | 57.9 | 467.9 | 167.1 | 1 | 26.7 | 26.7 | 71.0 | 71.0 | |||||
| 103066_at | L32973 | TKH | 3.9 | 107.6 | 27.6 | 243.9 | 62.5 | 22.9 | 173.6 | 7.6 | 354.6 | 15.5 | |||||
| 102879_sat | M31314 | FcγRI | 0.3 | 5.2 | 17.3 | 7.3 | 24.3 | 0.3 | 4.4 | 14.7 | 7.0 | 23.3 | |||||
| 160933_at | U53219 | IGTP | 0.8 | 12.3 | 15.4 | 30.1 | 37.6 | 0.6 | 16.3 | 27.2 | 37.3 | 62.2 | |||||
| 98410_at | AJ007972 | GTPI | 0.6 | 9.2 | 15.3 | 10.1 | 16.8 | 0.7 | 7.2 | 10.3 | 13.5 | 19.3 | |||||
| 93321_at | AF022371 | IFI203 | 1.1 | 16.2 | 14.7 | 31.1 | 28.3 | 1 | 30.9 | 30.9 | 41.1 | 41.1 | |||||
| 98417_at | M21038 | Mx1 | 22.9 | 272.5 | 11.9 | 484.4 | 21.2 | 34.5 | 97 | 2.8 | 148.1 | 4.3 | |||||
| 101876_sat | M35247 | MHC-17 | 0.6 | 6.8 | 11.3 | 12.7 | 21.2 | 0.7 | 6.7 | 9.6 | 11.0 | 15.7 | |||||
| 98472_at | Y00629 | MHC-23 | 0.8 | 9.0 | 11.3 | 13.4 | 16.8 | 0.9 | 8.5 | 9.4 | 12.8 | 14.2 | |||||
| 93949_at | M63658 | GBP-β4 | 0.4 | 4.1 | 10.3 | 9.9 | 24.8 | 0.7 | 3.1 | 4.4 | 7.9 | 11.3 | |||||
| 101465_at | U069240 | STAT-1 | 1.4 | 11.7 | 8.4 | 17.1 | 12.2 | 1.6 | 10.9 | 6.8 | 14.0 | 8.8 | |||||
| 100013_at | AW121732 | IFP35 | 1.0 | 8.2 | 8.2 | 11.1 | 11.1 | 0.8 | 8.4 | 10.5 | 13.6 | 17.0 | |||||
| 104177_at | AA204579 | Mvig | 25.8 | 207.9 | 8.1 | 501.5 | 19.4 | 54.9 | 203.7 | 3.7 | 347.3 | 6.3 | |||||
| 96533_at | AI508931 | Torsin 3A | 1.6 | 12.7 | 7.9 | 17.3 | 10.8 | 1.6 | 9.7 | 6.1 | 10.7 | 6.7 | |||||
| 93717_at | U50712 | MCP-5 | 0.7 | 5.4 | 7.7 | 9.3 | 13.3 | 1.6 | 7.7 | 4.8 | 11.6 | 7.3 | |||||
| 94162_at | D14859 | MVH | 1.6 | 11.2 | 7.0 | 25.3 | 15.8 | 0.8 | 7.4 | 9.3 | 28.2 | 35.3 | |||||
| 97409_at | U19119 | LRG-47 | 0.7 | 4.8 | 6.9 | 8.1 | 11.6 | 0.6 | 5.1 | 8.5 | 6.0 | 10.0 | |||||
| 103080_at | U15635 | Mg11 | 1.3 | 8.6 | 6.6 | 15.7 | 12.1 | 0.9 | 7.4 | 8.2 | 13.1 | 14.6 | |||||
| 94977_at | X15373 | IPR | 1.0 | 6.2 | 6.2 | 5.2 | 5.2 | 1.2 | 5.5 | 4.6 | 5.9 | 4.9 | |||||
| 101848_gat | AF040242 | Sp100 | 1.0 | 6.1 | 6.1 | 4.7 | 4.7 | 0.7 | 5.9 | 8.4 | 5.1 | 7.3 | |||||
| 93865_s_at | M35244 | MHC-10 | 1.1 | 5.8 | 5.3 | 12.7 | 11.5 | 1.1 | 5.4 | 4.9 | 8.6 | 7.8 | |||||
| 98030_at | J03776 | Rpt-1r | 1.5 | 7.9 | 5.3 | 9.6 | 6.4 | 1.1 | 5.6 | 5.1 | 7.1 | 6.5 | |||||
| 93956_at | U43086 | GARG-49 | 14.6 | 76.1 | 5.2 | 137.2 | 9.4 | 22.6 | 68.6 | 3.0 | 115.4 | 5.1 | |||||
| 103035_at | U60020 | TAP | 0.9 | 4.5 | 5.0 | 11.2 | 12.4 | 0.8 | 5.4 | 6.8 | 9.5 | 11.9 | |||||
| 98071_f_at | X77731 | DcK | 1.0 | 4.7 | 4.7 | 5.4 | 5.4 | 0.9 | 4.7 | 5.2 | 5.9 | 6.6 | |||||
| 161511_fat | AV152244 | ISG15 | 76.1 | 340.1 | 4.5 | 694.6 | 9.1 | 36 | 1,389.2 | 38.6 | 2,836.7 | 78.8 | |||||
| 93085_at | D44456 | LMP-2 | 1.1 | 4.8 | 4.4 | 6.3 | 5.7 | 1.1 | 5.7 | 5.2 | 7.4 | 6.7 | |||||
| 104597_at | AJ007970 | mGBP-2 | 6.7 | 26.9 | 4.0 | 37.0 | 5.5 | 9.5 | 26.5 | 2.8 | 37.3 | 3.9 | |||||
| 103254_at | AW049897 | FLN29 | 1.4 | 5.4 | 3.9 | 7.5 | 5.4 | 1.8 | 6 | 3.3 | 9.9 | 5.5 | |||||
| 103517_at | AA822898 | LySp100 | 12.3 | 43.7 | 3.6 | 222.9 | 18.1 | 1.2 | 66.7 | 5.6 | 222.9 | 18.6 | |||||
| 94112_at | U37522 | TRAIL | 2.4 | 7.3 | 3.0 | 16.1 | 6.7 | 1.4 | 26.2 | 18.7 | 79.9 | 57.1 | |||||
| 94774_at | M31418 | IFI202B | 2.8 | 8.1 | 2.9 | 16.3 | 5.8 | 2.9 | 6.6 | 2.3 | 9.6 | 3.3 | |||||
| 103639_at | U43085 | GARG-39 | 26.4 | 75.6 | 2.9 | 118.6 | 4.5 | 28.8 | 106.2 | 3.7 | 154.3 | 5.4 | |||||
| 97949_at | M16238 | FLP2 | 2.0 | 5.1 | 2.6 | 11.6 | 5.8 | 1.6 | 6.1 | 3.8 | 11.0 | 6.9 | |||||
| 98465_f_at | M31419 | IF1204 | 2.8 | 7.1 | 2.5 | 10.3 | 3.7 | 2.5 | 5 | 2.0 | 5.9 | 2.4 | |||||
| 92689_at | AB019505 | IL-18BP | 1.7 | 4.1 | 2.4 | 8.6 | 5.1 | 1 | 6.4 | 6.4 | 11.4 | 11.4 | |||||
| 93858_at | M33266 | IP-10 | 34.3 | 79.9 | 2.3 | 298.2 | 8.7 | 35.8 | 155.4 | 4.3 | 666.3 | 18.6 | |||||
| Fit criteria for candidate effector in microarray experiment 1
|
|||||||||||||||||
| 94224_s_atd | M74123 | IFI205 | 0.8 | 10.9 | 13.6 | 28.1 | 35.1 | 5.5 | 8.2 | 1.5 | 16.8 | 3.1 | |||||
| 98022_at | U48830 | PC-7 | 0.4 | 4.1 | 10.3 | 5.4 | 13.5 | ||||||||||
| 102279_at | AW046479 | UAE-E1 | 0.7 | 4.3 | 6.1 | 6.5 | 9.3 | 1.1 | 3.9 | 3.5 | 7.0 | 6.4 | |||||
| 161448_fat | AV355612 | LMP-7 | 0.7 | 4.2 | 6.0 | 5.2 | 7.4 | ||||||||||
| 161006_at | A1853978 | OAT | 1.6 | 8.2 | 5.1 | 12.7 | 7.9 | 2.1 | 2.9 | ||||||||
| 93942_at | U27295 | InPP | 1.2 | 6.0 | 5.0 | 9.9 | 8.3 | ||||||||||
| 160799_at | AW060549 | ENDO | 1.1 | 4.5 | 4.1 | 39.1 | 35.5 | 12.0 | |||||||||
| 101486_at | Y10875 | LMP-10 | 1.2 | 4.1 | 3.4 | 6.3 | 5.3 | 1.2 | 3.6 | 3.0 | 5.6 | 4.7 | |||||
| 97106_at | D13759 | CotK | 3.3 | 7.5 | 2.3 | 10.7 | 3.2 | ||||||||||
| 100981_at | U43084 | GARG16 | 80.4 | 182.3 | 2.3 | 319.6 | 4.0 | 390.7 | 130.7 | 0.3 | 199.5 | 0.5 | |||||
| 102965_at | AW121646 | LRP | 2.0 | 4.3 | 2.2 | 7.0 | 3.5 | 1.9 | 3.7 | 1.9 | 7.3 | 3.8 | |||||
| Fit criteria for candidate effector in microarray experiment 2
|
|||||||||||||||||
| 160393_ate | AI853226 | EST | 0.6 | 3.1 | 5.5 | 4.8 | 8.6 | 0.7 | 4 | 5.7 | 5.5 | 7.9 | |||||
| 160965_at | AA163960 | EST | 1.5 | 3.6 | 2.4 | 5.5 | 3.6 | 2 | 4.7 | 2.4 | 7.2 | 3.6 | |||||
| 104713_at | AA863717 | N6MT | 1.3 | 2.7 | 2.1 | 6.1 | 4.8 | 1.3 | 4.4 | 3.4 | 7.7 | 5.9 | |||||
| 103202_at | AW047476 | EST | 12.2 | 20.3 | 1.7 | 36.3 | 3.0 | 13.4 | 28.4 | 2.1 | 43.1 | 3.2 | |||||
| 103443_at | AA711704 | EST | 2.4 | 3.4 | 1.4 | 8.1 | 3.3 | 2 | 4 | 2.0 | 12.2 | 6.1 | |||||
| 102699_at | J03368 | EST | 2.4 | 3.1 | 1.3 | 6.7 | 2.8 | 2 | 6.5 | 3.3 | 21.7 | 10.9 | |||||
| 95303_at | AA144469 | 1-8U | 2.1 | 0.4 | 10.6 | 26.5 | 10.5 | 26.3 | |||||||||
| 98283_at | X91617 | mxRN1p | 3.4 | 0.6 | 4.8 | 8.0 | 6.1 | 10.2 | |||||||||
| 104456_at | AAA863742 | EST | 2.3 | 1.8 | 4.1 | 2.3 | 5.2 | 2.9 | |||||||||
To determine whether genes were upregulated at least twofold over virus-infected IFNAR1−/− in IFN-α/β-primed TD BMDC, upregulation in IFN-α/β-primed TD was divided by upregulation in virus-infected IFNAR1−/− BMDC.
To determine whether genes were upregulated at least twofold over virus-infected IFNAR1−/− in virus-infected TD BMDC, upregulation in IFN-α/β-primed TD was divided by upregulation in virus-infected IFNAR1−/− BMDC.
Data in the first group are fit criteria for candidate effectors in two independent microarray experiments.
Data in the second group are fit criteria for candidate effectors in microarray experiment 1.
Data in the third group are fit criteria for candidate effectors in microarray experiment 2.
NCBI, National Center for Biotechnology Information.
Array analyses were verified by RT-PCR.
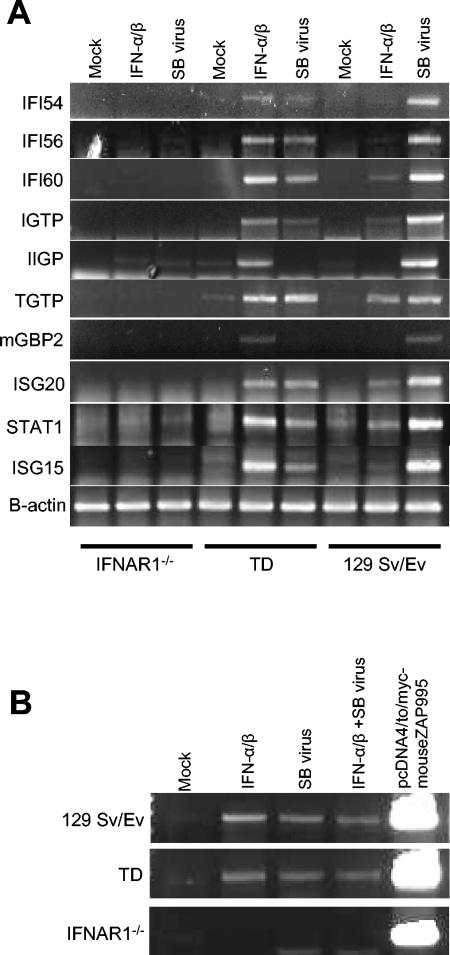
Transcriptional activation of candidate genes, chosen to be representative of some functional groups, was validated by RT-PCR (Fig. 6A). These analyses were repeated three times with independently isolated RNA sample sets. Although considerable variation was observed in mRNA amplification levels, overall the results confirmed the predictions made by the microarray data analyses.
FIG. 6.
RT-PCR confirmation of array. (A) The induction patterns of representative genes identified as candidates for alternative pathway effectors were validated by RT-PCR. Total RNA was isolated from TD and IFNAR1−/− BMDC either untreated, IFN-α/β primed (1,000 IU/ml for 6 h), or SB infected (12 h p.i.) and quantitated. cDNA was synthesized by RT from 50 ng of each RNA with dT(18) primers and amplified by PCR with gene-specific primers (Table 1). (B) cDNA was synthesized from 50 ng of total RNA described above by using a murine ZAP-specific antisense primer, and then amplified by PCR with a ZAP-specific primer pair (Table 1). The same primers were used to amplify a product from plasmid DNA encoding murine ZAP, pcDNA4/to/myc-mouseZAP995, to confirm product size. For both panels, PCR products visualized by gel electrophoresis are indicative of the presence of mRNAs in the original sample. β-Actin mRNA was amplified to confirm the presence of equal RNA, and the results from one experiment representative of three are shown.
Murine ZAP also meets the criteria of candidate effectors.
The rat ZAP was recently demonstrated to potently inhibit the translation of multiple alphaviruses, including SB (2), when overexpressed. Since this gene was not represented on the U74Av2 murine gene chip, a primer set to amplify the mouse homologue of ZAP was also included in these RT-PCR assays (Fig. 6B). In addition to IFN-α/β-primed and virus-infected BMDC, we also measured transcriptional regulation of these genes in BMDC that had been IFN-α/β primed for 6 h, virus infected, and harvested 12 h p.i. The β-actin mRNA was amplified as an RNA loading control. The results established that expression of the murine ZAP gene was IFN-α/β inducible (i.e., an ISG) in myeloid DC, independent of the presence of PKR and/or RNase L. However, transcriptional upregulation was not detected in virus-infected cells in the absence of IFN-α/β signaling, thus identifying ZAP as an additional candidate effector of the alternative antiviral pathway.
DISCUSSION
Although our understanding of IFN-α/β-mediated signaling networks has advanced enormously, many of the mechanisms by which intracellular antiviral activity is exerted are probably yet to be revealed (1, 40, 54). Constitutively expressed PKR and OAS/RNase L provide rapid cellular protection against many viral infections even before IFN-α/β production, so that attempts to delineate activities of other effectors only became feasible with the derivation of PKR- and/or RNase-null mice (54, 55). It has since become apparent that many viruses remain sensitive to the antiviral effects of IFN-α/β even in the absence of PKR/RNase L, including EMCV (26), dengue virus (8), HCV (12, 20, 21, 23, 24, 29, 52), and SB (37).
In the case of SB infection, separation of the IFN-α/β-mediated antiviral response from the PKR/RNase L pathways has exposed a highly effective residual IFN-α/β signaling-dependent, PKR/RNase L-independent antiviral activity (37). In permissive cells, the first step in SB replication after binding, entry, and genome uncoating is translation of the nsPs as a polyprotein from the capped, message-sense genome. In this study, we have demonstrated that IFN-α/β priming of immature myeloid DCs prior to virus infection significantly inhibits accumulation of proteins translated from the genomic RNA. However, this effect was not attributable to the major known host translational control programs mediated through PKR and RNase L because, in their absence, the translation suppressive actions of IFN-α/β priming were not alleviated. Possible effects on virus attachment and entry were ruled out, as translation of in vitro-transcribed, capped viral RNAs transfected directly into the cytoplasm remained significantly impaired in IFN-α/β-treated TD-derived BMDC. In addition to genomic RNA translation inhibition, we have previously demonstrated and confirmed in this study that translation of subgenomic RNA is also impaired. The observed suppression of structural protein expression may result from failure to replicate and accumulate subgenomic RNA or from direct translational inhibition of the subgenomic message.
To eliminate the influence of viral RNA replication or any possible modulating effects of expressed viral proteins, we constructed an SB-based translation reporter construct encoding a truncated nsP1/fLUC fusion protein, flanked by the 5′ and 3′ NTRs. When electroporated into BMDC, translation of 5′-capped SB reporter RNA transcripts and of 5′-capped host-like mRNA transcripts was significantly reduced by IFN-α/β pretreatment of the cells. Translation of the SB reporter was inhibited more efficiently than the host mRNA. In stark contrast, translation initiated by an EMCV IRES was unaffected by IFN-α/β priming of TD BMDC. All of these transcripts were polyadenylated, from which we infer that at least one PKR-independent, IFN-α/β-primed antiviral pathway preferentially targets cap-dependent translation. It remains to be determined whether the block is a result of effects on message stability, RNA targeting, and/or a direct inhibition of the translation process. However, we are able to conclude that the alternative pathway effector(s) does not require activation by virus binding and entry, viral RNA replication, or viral protein synthesis. Indeed, phosphorylation of eIF-2α by an alternative kinase, which has been demonstrated during HCV and bovine viral diarrhea virus infections (25, 34), was not responsible for the inhibition of translation observed in SB-infected BMDCs.
IFN-α/β-mediated signal transduction events increase the abundance of early response genes collectively known as ISGs, the protein products of which mediate the pleiotropic effects of IFN (17). Our data provide compelling evidence that the major alternative pathway mediator(s) is transcriptionally regulated via IFN-α/β signal transduction, as inhibition of host gene transcription by ActD, preventing the induction of ISGs, restores the virus' capacity to replicate and produce progeny virions. Thus, we surmise that an ISG product(s) is the primary effector of the antiviral response against SB in PKR/RNase L-null BMDC. However, a direct linkage has been demonstrated between IFN-α/β receptor signaling, extracellular signal-regulated kinase/phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase pathways, and translation initiation (5, 16, 30, 50). Therefore, it remains possible that direct modulation of cap-dependent translation efficiency could contribute to IFN-α/β-mediated antiviral effects independent of ISG induction.
Importantly, while virus-inducible genes may have antiviral activity against SB and/or other alphaviruses, the presence of dsRNA or other viral products within the cell was not sufficient to induce significant antiviral activity in the absence of IFN-α/β signaling. Consequently, our analyses of global gene induction profiles to identify candidate alternative pathway effectors have focused on ISGs that are induced by IFN-α/β signaling in the presence or absence of PKR/RNase L but not by virus infection in the absence of IFNAR1. Whether or not the alternative pathway(s) has constitutive activity or requires activation by the presence of RNA such as the positive-sense, nonreplicating pSBΔnsP/fLUC virus-like RNA present in our electroporation experiments, remains to be determined. It is worth noting that all of these observations have been made in primary myeloid DC cultures, relevant to the IFN-α/β response in vivo and the pathogenesis of the virus.
The DNA microarray analyses performed in this study have yielded valuable insights on the influence of key components in the IFN-α/β system on the global induction of gene transcription. Many ISGs have antiviral function and are able to limit virus replication at multiple points within the replicative cycle (6, 7). However, many of these genes were also shown to be induced by dsRNA in the absence of IFN-α/β signaling (14). By performing two- and four-way comparisons of our data, we have narrowed the final set of candidate alternative pathway candidates to those genes induced at least fourfold over mock-infected cells in IFN-α/β-treated and virus-infected TD BMDC and at least twofold over virus-infected (or IFN-α/β-treated) IFNAR1−/− cells.
The functional groups of genes induced include viral stress-induced glucocorticoid-attenuated response genes (GARGs), GTPases, RNA-modifying enzymes (RNases and RNA helicases), transcription factors, and protein degradation pathway components. From this set, we have selected genes with appropriate activity (RNA degradation or translation inhibition) and presumptive antiviral activity for further consideration. The recently identified GARG family of genes includes GARG-16 (IFI56), GARG-49/IRG2 (IFI60), and GARG-39 (IFI54). The GARGs are immediate-early response genes containing multiple tetratricopeptide repeat domains, produced in response to viral stress-inducing agents such as IFN-α/β and dsRNA, and involved in protein-protein interactions in a variety of cellular contexts (44). p56 (product of GARG-16) severely inhibits initiation of cap-dependent translation at the level of eIF-3 ternary complex formation (20, 21, 23, 24). Moreover, p56 directs PKR-independent suppression of HCV IRES translation during the IFN-α/β response (12, 21, 23, 24, 52). However, induction of the GARG-16 gene was not consistently observed. The physiological significance of p54 (GARG-39) and p60 (GARG-49/IRG2) induction in these experiments is not clear.
The GTPases are a diverse group of proteins that carry out various cellular functions (49). Although their functions are poorly characterized, two 47-kDa GTPases, TGTP and IGTP, possess some antiviral activity (53). Of particular interest, overexpression of IGTP inhibits coxsackievirus B protein synthesis and functions as a survival factor for the host cell (53). The IFN-α/β-inducible single-stranded-RNA (ssRNA)-specific RNase ISG20 has antiviral activity against some RNA viruses, including vesicular stomatitis virus and EMCV, in the absence of PKR and RNase L (9). Perhaps most intriguingly, the ZAP protein was recently demonstrated to potently inhibit multiple alphaviruses, including SB, by preventing the accumulation of new genomic viral RNA in the cytoplasm (2). RT-PCR analysis of ZAP expression in BMDC also identified this protein as a candidate effector. Like the alternative pathway, the effect of ZAP was limited to 5′-capped, polyadenylated viral RNA genomes (2); however, in contrast, translation of a capped host mRNA was not affected. In ongoing experiments, we are further characterizing the role of these effector candidates in suppression of SB genome translation.
It seems increasingly unlikely that the host employs the same PKR-independent IFN-α/β-inducible pathway for protection against all of the viruses mentioned, especially considering the fundamentally different genome structures and replication strategies they employ. Sensitivity to antiviral effectors is determined by the virus life cycle, requiring a multilayered innate immune response to infection. A growing body of publications present evidence that the nature of the virus genome, whether it is message or complementary sense, capped, and/or polyadenylated, is critical in determining sensitivity to IFN-α/β antiviral effectors (9, 26, 29). A positive-sense RNA virus like SB, in which the genome is capped and polyadenylated and translation is the first required step in replication, is likely to be more sensitive to the effects of an antiviral pathway that suppresses translation than to PKR or RNase L, which must first be activated by dsRNA regions within viral ssRNAs or dsRNA replicative intermediates. Whether or not this antiviral pathway has broad-spectrum activity against the more virulent alphaviruses is currently under investigation, with the ultimate goal of developing rationally designed acute-phase therapeutics.
Acknowledgments
These studies were supported by NIH grants R01 AI22186-18 and R01 GM20818-30.
We thank Robert Silverman, Bryan Williams, Barbara Sherry, and Skip Virgin for generously providing genetically modified mice. We thank Janice Anderson and Sherry Jackson for excellent technical assistance. In addition, we are grateful to Paula Polk in the LSUHSC-S Research Core Facility for assistance with microarray analyses and Robin Bowen in the Molecular Biology Core, Center for Molecular and Tumor Virology.
REFERENCES
- 1.Basler, C. F., and A. Garcia-Sastre. 2002. Viruses and the type I interferon antiviral system: induction and evasion. Int. Rev. Immunol. 21:305-337. [DOI] [PubMed] [Google Scholar]
- 2.Bick, M. J., J. W. Carroll, G. Gao, S. P. Goff, C. M. Rice, and M. R. MacDonald. 2003. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J. Virol. 77:11555-11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgess, T. H., K. E. Steele, B. A. Schoneboom, and F. B. Grieder. 2001. Clinicopathologic features of viral agents of potential use by bioterrorists. Clin. Lab. Med. 21:475-493. [PubMed] [Google Scholar]
- 4.Byrnes, A. P., J. E. Durbin, and D. E. Griffin. 2000. Control of Sindbis virus infection by antibody in interferon-deficient mice. J. Virol. 74:3905-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.David, M., E. Petricoin III, C. Benjamin, R. Pine, M. J. Weber, and A. C. Larner. 1995. Requirement for MAP kinase (ERK2) activity in interferon alpha- and interferon beta-stimulated gene expression through STAT proteins. Science 269:1721-1723. [DOI] [PubMed] [Google Scholar]
- 6.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69:912-920. [PubMed] [Google Scholar]
- 8.Diamond, M. S., and E. Harris. 2001. Interferon inhibits dengue virus infection by preventing translation of viral RNA through a PKR-independent mechanism. Virology 289:297-311. [DOI] [PubMed] [Google Scholar]
- 9.Espert, L., G. Degols, C. Gongora, D. Blondel, B. R. Williams, R. H. Silverman, and N. Mechti. 2003. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. J. Biol. Chem. 278:16151-16158. [DOI] [PubMed] [Google Scholar]
- 10.Frolov, I., and S. Schlesinger. 1996. Translation of Sindbis virus mRNA: analysis of sequences downstream of the initiating AUG codon that enhance translation. J. Virol. 70:1182-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frolov, I., R. Hardy, and C. M. Rice. 2001. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gale, M., Jr. 2003. Effector genes of interferon action against hepatitis C virus. Hepatology 37:975-978. [DOI] [PubMed] [Google Scholar]
- 13.Gardner, J. P., I. Frolov, S. Perri, Y. Ji, M. L. MacKichan, J. zur Megede, M. Chen, B. A. Belli, D. A. Driver, S. Sherrill, C. E. Greer, G. R. Otten, S. W. Barnett, M. A. Liu, T. W. Dubensky, and J. M. Polo. 2000. Infection of human dendritic cells by a Sindbis virus replicon vector is determined by a single amino acid substitution in the E2 glycoprotein. J. Virol. 74:11849-11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geiss, G., G. Jin, J. Guo, R. Bumgarner, M. G. Katze, and G. C. Sen. 2001. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J. Biol. Chem. 276:30178-30182. [DOI] [PubMed] [Google Scholar]
- 15.Gillies, S., and V. Stollar. 1981. Translation of vesicular stomatitis and Sindbis virus mRNAs in cell-free extracts of Aedes albopictus cells. J. Biol. Chem. 256:13188-13192. [PubMed] [Google Scholar]
- 16.Goh, K. C., S. J. Haque, and B. R. Williams. 1999. p38 MAP kinase is required for STAT1 serine phosphorylation and transcriptional activation induced by interferons. EMBO J. 18:5601-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 18.Gorchakov, R., R. Hardy, C. M. Rice, and I. Frolov. 2004. Selection of functional 5′ cis-acting elements promoting efficient Sindbis virus genome replication. J. Virol. 78:61-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffin, D. E. 2001. Alphaviruses, p. 917-962. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 20.Guo, J., K. L. Peters, and G. C. Sen. 2000. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology 267:209-219. [DOI] [PubMed] [Google Scholar]
- 21.Guo, J., D. J. Hui, W. C. Merrick, and G. C. Sen. 2000. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 19:6891-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardy, W. R., and J. H. Strauss. 1988. Processing the nonstructural polyproteins of Sindbis virus: study of the kinetics in vivo by using monospecific antibodies. J. Virol. 62:998-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He, Y., W. Yan, C. Coito, Y. Li, M. Gale, Jr., and M. G. Katze. 2003. The regulation of hepatitis C virus (HCV) internal ribosome-entry site-mediated translation by HCV replicons and nonstructural proteins. J. Gen. Virol. 84:535-543. [DOI] [PubMed] [Google Scholar]
- 24.Hui, D. J., C. R. Bhasker, W. C. Merrick, and G. C. Sen. 2003. Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2.GTP.Met-tRNAi. J. Biol. Chem. 278:39477-39482. [DOI] [PubMed] [Google Scholar]
- 25.Jordan, R., L. Wang, T. M. Graczyk, T. M. Block, and P. R. Romano. 2002. Replication of a cytopathic strain of bovine viral diarrhea virus activates PERK and induces endoplasmic reticulum stress-mediated apoptosis of MDBK cells. J. Virol. 76:9588-9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khabar, K. S., Y. M. Siddiqui, F. al-Zoghaibi, L. al-Haj, M. Dhalla, A. Zhou, B. Dong, M. Whitmore, J. Paranjape, M. N. Al-Ahdal, F. Al-Mohanna, B. R. Williams, and R. H. Silverman. 2003. RNase L mediates transient control of the interferon response through modulation of the double-stranded RNA-dependent protein kinase PKR. J. Biol. Chem. 278:20124-20132. [DOI] [PubMed] [Google Scholar]
- 27.Kimball, S. R., M. J. Clemens, V. J. Tilleray, R. C. Wek, R. L. Horetsky, and L. S. Jefferson. 2001. The double-stranded RNA-activated protein kinase PKR is dispensable for regulation of translation initiation in response to either calcium mobilization from the endoplasmic reticulum or essential amino acid starvation. Biochem. Biophys. Res. Commun. 280:293-300. [DOI] [PubMed] [Google Scholar]
- 28.Klimstra, W. B., K. D. Ryman, and R. E. Johnston. 1998. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J. Virol. 72:7357-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koev, G., R. F. Duncan, and M. M. Lai. 2002. Hepatitis C virus IRES-dependent translation is insensitive to an eIF2alpha-independent mechanism of inhibition by interferon in hepatocyte cell lines. Virology 297:195-202. [DOI] [PubMed] [Google Scholar]
- 30.Lekmine, F., S. Uddin, A. Sassano, S. Parmar, S. M. Brachmann, B. Majchrzak, N. Sonenberg, N. Hay, E. N. Fish, and L. C. Platanias. 2003. Activation of the p70 S6 kinase and phosphorylation of the 4E-BP1 repressor of mRNA translation by type I interferons. J. Biol. Chem. 278:27772-27780. [DOI] [PubMed] [Google Scholar]
- 31.Lemm, J. A., and C. M. Rice. 1993. Assembly of functional Sindbis virus RNA replication complexes: requirement for coexpression of P123 and P34. J. Virol. 67:1905-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemm, J. A., T. Rumenapf, E. G. Strauss, J. H. Strauss, and C. M. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 13:2925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacDonald, G. H., and R. E. Johnston. 2000. Role of dendritic cell targeting in Venezuelan equine encephalitis virus pathogenesis. J. Virol. 74:914-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pavio, N., P. R. Romano, T. M. Graczyk, S. M. Feinstone, and D. R. Taylor. 2003. Protein synthesis and endoplasmic reticulum stress can be modulated by the hepatitis C virus envelope protein E2 through the eukaryotic initiation factor 2alpha kinase PERK. J. Virol. 77:3578-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rotz, L. D., A. S. Khan, S. R. Lillibridge, S. M. Ostroff, and J. M. Hughes. 2002. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 8:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryman, K. D., W. B. Klimstra, K. B. Nguyen, C. A. Biron, and R. E. Johnston. 2000. Alpha/beta interferon protects adult mice from fatal Sindbis virus infection and is an important determinant of cell and tissue tropism. J. Virol. 74:3366-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryman, K. D., L. J. White, R. E. Johnston, and W. B. Klimstra. 2002. Effects of PKR/RNase L-dependent and alternative antiviral pathways on alphavirus replication and pathogenesis. Viral Immunol. 15:53-76. [DOI] [PubMed] [Google Scholar]
- 38.Saito, S. 1989. Possible involvement of virus-induced protein kinase in the antiviral state induced with interferon-gamma against Sindbis virus. J. Interferon Res. 9:23-34. [DOI] [PubMed] [Google Scholar]
- 39.Saito, S. 1990. Enhancement of the interferon-induced double-stranded RNA-dependent protein kinase activity by Sindbis virus infection and heat-shock stress. Microbiol. Immunol. 34:859-870. [DOI] [PubMed] [Google Scholar]
- 40.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawicki, D. L., R. H. Silverman, B. R. Williams, and S. G. Sawicki. 2003. Alphavirus minus-strand synthesis and persistence in mouse embryo fibroblasts derived from mice lacking RNase L and protein kinase R. J. Virol. 77:1801-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schattner, A., G. Merlin, A. Shapira, M. Revel, and W. Wallach. 1982. Comparison of (2′-5′) oligo-adenylate synthetase and interferon blood-levels in mice early after viral infection. J. Interferon Res. 2:285-289. [DOI] [PubMed] [Google Scholar]
- 43.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 44.Smith, J. B., and H. R. Herschman. 1996. The glucocorticoid attenuated response genes GARG-16, GARG-39, and GARG-49/IRG2 encode inducible proteins containing multiple tetratricopeptide repeat domains. Arch. Biochem. Biophys. 330:290-300. [DOI] [PubMed] [Google Scholar]
- 45.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 46.Strauss, E. G., C. M. Rice, and J. H. Strauss. 1984. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology 133:92-110. [DOI] [PubMed] [Google Scholar]
- 47.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takashima, A., S. Xu, K. Ariizumi, and P. R. Bergstresser. 1995. Establishment and characterization of antigen-presenting cell lines (XS series) derived from newborn mouse epidermis. Adv. Exp. Med. Biol. 378:159-162. [DOI] [PubMed] [Google Scholar]
- 49.Taylor, G. A., C. G. Feng, and A. Sher. 2004. p47 GTPases: regulators of immunity to intracellular pathogens. Nat. Rev. Immunol. 4:100-109. [DOI] [PubMed] [Google Scholar]
- 50.Uddin, S., B. Majchrzak, J. Woodson, P. Arunkumar, Y. Alsayed, R. Pine, P. R. Young, E. N. Fish, and L. C. Platanias. 1999. Activation of the p38 mitogen-activated protein kinase by type I interferons. J. Biol. Chem. 274:30127-30131. [DOI] [PubMed] [Google Scholar]
- 51.van Steeg, H., A. Thomas, S. Verbeek, M. Kasperaitis, H. O. Voorma, and R. Benne. 1981. Shutoff of neuroblastoma cell protein synthesis by Semliki Forest virus: loss of ability of crude initiation factors to recognize early Semliki Forest virus and host mRNA's. J. Virol. 38:728-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, C., J. Pflugheber, R. Sumpter, Jr., D. L. Sodora, D. Hui, G. C. Sen, and M. Gale, Jr. 2003. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J. Virol. 77:3898-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, H. M., J. Yuan, P. Cheung, H. Luo, B. Yanagawa, D. Chau, N. Stephan-Tozy, B. W. Wong, J. Zhang, J. E. Wilson, B. M. McManus, and D. Yang. 2003. Overexpression of interferon-gamma-inducible GTPase inhibits coxsackievirus B3-induced apoptosis through the activation of the phosphatidylinositol 3-kinase/Akt pathway and inhibition of viral replication. J. Biol. Chem. 278:33011-33019. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, Z., S. Schwartz, L. Wagner, and W. Miller. 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7:203-214. [DOI] [PubMed] [Google Scholar]
- 55.Zhou, A., J. Paranjape, T. L. Brown, H. Nie, S. Naik, B. Dong, A. Chang, B. Trapp, R. Fairchild, C. Colmenares, and R. H. Silverman. 1997. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 16:6355-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, A., J. M. Paranjape, S. D. Der, B. R. Williams, and R. H. Silverman. 1999. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology 258:435-440. [DOI] [PubMed] [Google Scholar]