Abstract
In vitro studies have shown that the host cytidine deaminase APOBEC3G causes lethal hypermutation in human immunodeficiency virus type 1 reverse transcripts unless its incorporation into virions is blocked by Vif. By examining stably archived sequences in resting CD4+ T cells, we show that hypermutation occurs in most if not all infected individuals. Hypermutated sequences comprised >9% of archived species in resting CD4+ T cells but were not found in plasma virus. Mutations occurred in predicted contexts, with notable hotspots. Thus, defects in Vif function in vivo give rise to hypermutated viral genomes that can be integrated but do not produce progeny viruses.
A component of innate immune defense against viruses is ABOBEC3G, a cytidine deaminase which causes G→A hypermutation in retroviral genomes (9, 11, 16, 18, 26, 35, 36). ABOBEC3G is incorporated into assembling virions and then deaminates cytidines of the single-stranded viral cDNA that is initially synthesized by reverse transcriptase (RT) upon entry of the virus into a new host cell (11, 16, 18, 35, 36). This C→U deamination on the minus strand of the reverse transcript leads to fixation of G→A mutations. Pioneering work by Sheehy et al. identified APOBEC3G as an antiviral factor and showed that human immunodeficiency virus type 1 (HIV-1) Vif overcomes its effects (26). Vif counteracts APOBEC3G by inhibiting its translation and accelerating its degradation, thereby preventing APOBEC3G incorporation into HIV-1 virions (19, 20, 25, 27, 30). Studies of target sequence specificity of ABOBEC3G have revealed a context dependence for the two nucleotides immediately upstream of the targeted dC (1, 11, 35), consistent with reports of G→A hypermutation in HIV-1 sequences from infected individuals (3, 8, 13). In these studies, minus-strand C→U deamination resulted in fixation of G→A mutations within GA and GG dinucleotides with an extreme bias for GGG sequences (3, 13, 31). Within hypermutated sequences, 20 to 94% of guanine nucleotides in these contexts were mutated (13). Recent studies have delineated the preferences of ABOBEC3G and ABOBEC3F, a closely related protein of similar function (33, 34), as GG and GA, respectively (17, 33).
Most studies of the antiviral effects of APOBEC3G have utilized Vif− HIV-1 constructs, and there remains uncertainty about how often G→A hypermutation occurs in HIV-1-infected individuals and about the fate of hypermutated viruses. Janini et al. found hypermutation in 43% of patient samples (13). Hypermutated sequences had in-frame stop codons that would interfere with the production of viral proteins (13). However, the replication defect shown by Vif− viruses may actually operate at an early, postentry stage in viral replication and decrease the formation of proviruses (29, 32). One possibility is that the deoxyuridines produced by deamination undergo uracil excision by uracil-DNA glycosylases, exposing the viral cDNA to nucleases (11).
To understand the nature and distribution of hypermutated sequences in vivo, we analyzed the protease and RT regions of HIV-1 sequences obtained from resting CD4+ T cells of nine patients who had prolonged suppression of viremia to below the limit of detection on highly active antiretroviral therapy (HAART). In patients on successful long-term HAART, labile unintegrated forms of HIV-1 decay (2, 22), and resting CD4+ T cells harbor stably integrated, latent viral genomes, some of which are replication competent (4, 5, 10). This cellular reservoir persists in patients on HAART (6, 21, 28) and continually releases virus into the plasma at low levels (12, 14). We analyzed hypermutation in both the cellular and plasma compartments of these patients.
Resting CD4+ T cells were isolated from peripheral blood mononuclear cells by use of negative selection to remove monocytes, natural killer (NK) cells, B cells, CD8+ T cells, and activated CD4+ T cells as previously described (7). The resulting populations were >90% pure. pol gene sequences from resting cells were obtained by a single genome sequencing method (P. Kwon, M. Wind-Rotolo, and R. F. Siliciano, unpublished data). Plasma sequences were obtained by frequent sampling over a 3-month period and an ultrasensitive RT-PCR capable of separately genotyping RT and protease from patients with viral loads below 50 copies/ml (14). Sequences were analyzed using a program (www.hiv.lanl.gov/HYPERMUT/hypermut.html) that compares each patient sequence to a patient-specific consensus to determine the frequency and context of G→A mutations (23). Hypermutated sequences were defined as having >5% of the total Gs mutated to A but <1% A→G mutations.
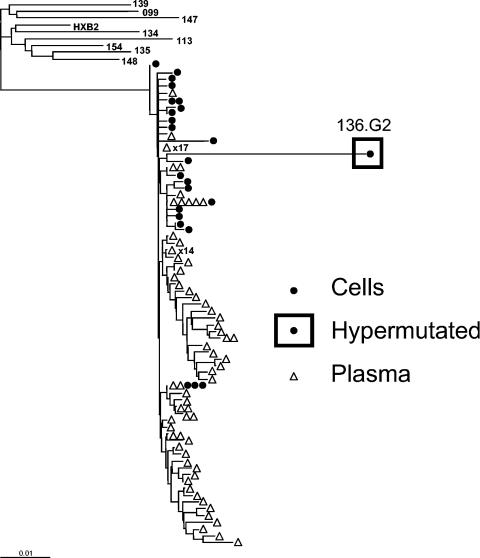
Of 2,024 independent RT and protease sequences from the plasma virus of nine patients, not a single one was hypermutated. In contrast, from a total of 317 independent pol clones from the latent cellular reservoir, hypermutation was detected in 19 of 302 (6.3%) protease sequences and 21 of 309 (6.8%) RT sequences. Both protease and RT were hypermutated in 12 clones. Thus, there were a minimum of 28 (8.8%) hypermutated genomes among the 317 pol clones. This is a minimal estimate, because sequencing repeatedly failed for some pol clones (15 protease and 8 RT sequences), possibly due to hypermutation, and because sequencing additional regions of the viral genome, particularly env and the 5′ half of nef (35), may have revealed additional hypermutation. At least one hypermutated sequence was found with each patient studied. Phylogenetic analysis showed patient-specific clustering and extreme divergence of the hypermutated sequences within patient-specific clusters (Fig. 1). The routine detection of hypermutated sequences in resting CD4+ T cells of patients with prolonged suppression of active viral replication on HAART suggests that hypermutated viral genomes can enter the stable pool of integrated HIV-1 DNA in these cells. The absence of hypermutated plasma sequences indicates that hypermutation blocks virus production from these proviruses.
FIG. 1.
Representative phylogenetic tree of RT sequences from patient 136. Independent sequences obtained from resting CD4+ T cells (•) and from plasma (▵) of patient 136 are displayed along with the consensus sequences from the other patients and the reference sequence HXB2 (15). Independent isolates with identical sequences are represented by one symbol, with the number of isolates indicated next to the symbol. A hypermutated sequence (136.G2) with 21 G→A mutations is enclosed in a square. The reference sequence HXB2 was used as an outgroup. Neighbor-joining phylogenetic analysis of RT positions 2640 to 3233 (relative to HXB2) was carried out as previously described (24).
Within hypermutated sequences, G→A mutations were found at an average of 11% (RT) and 13.5% (protease) of all Gs (range, 5 to 22%), while only 0.18% (RT) and 0.44% (protease) of As were mutated to guanine (Table 1). In 98% of the cases, mutations were found in either GA or GG dinucleotides, with about 20% in GA and 80% in either GG or GGG sequences. In contrast, Janini et al. (13) noted a 3F-like preference for mutation within the GA context in protease sequences from unfractionated peripheral blood mononuclear cells of viremic patients. We found a preference for hypermutation at Gs preceded by T, consistent with a recent study defining TGGG as the tetranucleotide consensus sequence for ABOBEC3G (35). Our results are consistent with the idea of both ABOBEC3G and ABOBEC3F contributing to in vivo hypermutation.
TABLE 1.
Analysis of frequency and sequence context of G → A mutations
| Region | Length (no. of nucleotides) | Total no. of G → A mutations | % Of Gs mutated | Total no. of A → G mutations | % Of As mutated | % Of mutationsa
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| GG | GA | GT | GC | GGG | ||||||
| RT (n = 21) | 710 | 332 | 11 | 11 | 0.18 | 40 | 18 | 1 | 1 | 40 |
| Protease (n = 19) | 297 | 165 | 13.5 | 9 | 0.44 | 59 | 17 | 0 | 2 | 22 |
The percentages of the total G → A mutations occurring within the indicated di- or trinucleotide contexts are indicated. The mutated G is underlined.
Mutational hotspots, defined as G→A mutations in >50% of hypermutated sequences, were identified in the protease and RT genes (Table 2 and Table 3). The substitution at nucleotide (nt) 125 in protease was found in 63% of sequences and resulted in a stop codon. Four other hotspots resulted in a change at a glycine residue. This is as expected, since glycine codons (GGN) contain the targeted GG dinucleotide. In RT, nine hotspots were identified (Table 3). The Gs at nt 151, 263, 333, and 690 occurred at the beginning of GGG trinucleotides and were mutated in 80 to 95% of sequences. RT mutations resulted in substitutions at methionine (n = 4), tryptophan (n = 2), or glycine (n = 2). The mutations in methionine codons (ATG) produced changes to isoleucine (ATA), one of which was at amino acid 184 and corresponded to a known intermediate in the pathway to lamivudine resistance. The mutations within tryptophan codons resulted in stop codons at amino acids 88 and 212. The mutation at nt 333 produced a synonymous change detected in 95% of the sequences.
TABLE 2.
Detailed analysis of G → A mutations in the protease gene of 19 hypermutated sequences from the resting CD4+ T cells of eight patients on HAART
| nt positiona | 42 | 46 | 47 | 48 | 49 | 51 | 60 | 73 | 88 | 100 | 108 | 114 | 118 | 122 | 125 | 126 | 142 | 143 | 144 | 145 | 146 | 151 | 152 | 154 | 178 | 183 | 193 | 202 | 203 | 217 | 232 | 233 | 256 | 257 | 260 | 270 | 280 | 281 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G → A contextb | GAN | GGG | GGG | GGG | GGG | GCN | GGN | GAN | GAN | GAN | GAN | GCN | GGN | GAN | GGN | GAN | GGG | GGG | GGG | GGN | GAN | GGN | GAN | GGN | GAN | GGN | GAN | GGN | GAN | GGN | GGN | GAN | GGN | GAN | GAN | GAN | GGN | GGN | |
| 99.E15 | A | A | A | ||||||||||||||||||||||||||||||||||||
| 99.E19 | A | A | A | A | |||||||||||||||||||||||||||||||||||
| 99.E36 | A | A | A | A | A | A | A | ||||||||||||||||||||||||||||||||
| 113.G2 | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | ||||||||||||||||||||
| 113.I6 | A | A | A | A | A | A | A | A | A | A | |||||||||||||||||||||||||||||
| 113.I16 | A | A | A | A | A | A | |||||||||||||||||||||||||||||||||
| 113.I49 | A | A | A | A | A | A | A | ||||||||||||||||||||||||||||||||
| 113.I105 | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | ||||||||||||||||||||
| 134.D1 | A | A | A | A | A | A | A | ||||||||||||||||||||||||||||||||
| 135.F36 | A | A | A | A | A | ||||||||||||||||||||||||||||||||||
| 135.F39 | A | A | A | A | A | ||||||||||||||||||||||||||||||||||
| 136.G2 | A | A | A | A | A | A | A | A | |||||||||||||||||||||||||||||||
| 139.F1 | A | A | A | A | A | A | |||||||||||||||||||||||||||||||||
| 139.H1 | A | A | A | A | A | A | A | A | |||||||||||||||||||||||||||||||
| 147.E11 | A | A | A | A | A | A | A | ||||||||||||||||||||||||||||||||
| 147.E22 | A | A | A | A | A | A | A | ||||||||||||||||||||||||||||||||
| 154.F7 | A | A | A | A | A | A | A | A | A | A | A | A | |||||||||||||||||||||||||||
| 154.F23 | A | A | A | A | A | A | A | A | A | A | A | A | A | A | |||||||||||||||||||||||||
| 154.G17 | A | A | A | A | A | A | A | A | A | A | A | ||||||||||||||||||||||||||||
| % Of sequences with mutation | 5 | 16 | 16 | 21 | 11 | 5 | 32 | 11 | 11 | 21 | 32 | 5 | 5 | 5 | 63* | 11 | 37 | 42 | 53 | 26 | 11 | 53 | 5 | 47 | 11 | 11 | 16 | 32 | 5 | 42 | 26 | 5 | 74 | 16 | 16 | 5 | 68 | 5 | |
| Amino acid positionc | 42 | 48 | 52 | 86 | 94 | ||||||||||||||||||||||||||||||||||
| Substitutiond | W-* | G-K | G-S | G-R | G-S |
The nucleotide (nt) position of the hypermutated G within the protease gene is given. Nucleotide positions where more than 50% of the hypermutated sequences contained G → A mutations are indicated in boldface type. Designations in first column represent sequences.
The hypermutated G (underlined) and following two 3′ nucleotides are shown.
The corresponding amino acid position within the protease gene where hot spot G → A mutations occur is shown.
The amino acid substitution occurring as a result of a hot spot G → A mutation. A stop codon is labeled with an asterisk (*).
TABLE 3.
Detailed analysis of G → A mutations in the RT gene of 21 hypermutated sequences from the resting CD4+ T cells of nine patients on HAART
| nt positiona
|
48
|
52
|
85
|
123
|
124
|
130
|
133
|
134
|
151
|
157
|
212
|
213
|
215
|
233
|
246
|
263
|
264
|
277
|
295
|
296
|
327
|
333
|
367
|
374
|
375
|
421
|
422
|
453
|
454
|
458
|
463
|
515
|
552
|
568
|
586
|
609
|
615
|
635
|
636
|
637
|
638
|
686
|
690
|
691
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G → A contextb | GGN | GG | GAN | GGN | GAN | GAN | GGG | GGN | GGG | GAN | GGN | GAN | GAN | GAN | GAN | GGG | GGN | GGN | GGG | GGN | GGN | GGG | GAN | GGN | GAN | GGG | GGN | GGG | GGN | GGN | GGN | GAN | GGN | GGN | GGG | GGN | GAN | GGG | GGG | GGG | GGN | GGN | GGG | GGN |
| 99.E15 | A | A | A | A | A | A | A | A | A | A | A | A | ||||||||||||||||||||||||||||||||
| 113.I6 | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | ||||||||||||||||||
| 113.I16 | A | A | A | A | A | A | A | A | A | |||||||||||||||||||||||||||||||||||
| 113.I49 | A | A | A | A | A | A | A | A | A | A | A | A | A | |||||||||||||||||||||||||||||||
| 113.I96 | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | |||||||||||||||||||||||||
| 134.D14 | A | A | A | A | A | A | A | A | A | A | A | A | ||||||||||||||||||||||||||||||||
| 134.D26 | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | ||||||||||||||||||||||||||
| 135.F35 | A | A | A | A | A | A | A | A | A | A | ||||||||||||||||||||||||||||||||||
| 135.F36 | A | A | A | A | A | A | A | A | A | A | A | |||||||||||||||||||||||||||||||||
| 135.F39 | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | ||||||||||||||||||||||||||
| 136.G2 | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | |||||||||||||||||||||||
| 139.F1 | A | A | A | A | A | A | A | A | A | A | A | A | A | A | ||||||||||||||||||||||||||||||
| 139.F21 | A | A | A | A | A | A | A | A | ||||||||||||||||||||||||||||||||||||
| 139.H1 | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | |||||||||||||||||||||||||||
| 147.E8 | A | A | A | A | ||||||||||||||||||||||||||||||||||||||||
| 147.E11 | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | |||||||||||||||||||||||||
| 147.E13 | A | A | A | A | A | A | A | |||||||||||||||||||||||||||||||||||||
| 147.E22 | A | A | A | A | A | A | A | A | A | A | A | A | A | A | ||||||||||||||||||||||||||||||
| 148.G4 | A | A | A | A | A | A | A | A | A | |||||||||||||||||||||||||||||||||||
| 154.F12 | A | A | A | A | A | A | A | A | A | A | A | |||||||||||||||||||||||||||||||||
| 154.F7 | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | |||||||||||||||||||||||
| % Of sequences with mutation | 52 | 24 | 14 | 57 | 10 | 19 | 57 | 10 | 90 | 14 | 24 | 10 | 19 | 14 | 14 | 81 | 24 | 38 | 19 | 14 | 33 | 95 | 14 | 10 | 14 | 19 | 24 | 38 | 19 | 43 | 29 | 19 | 71 | 24 | 19 | 14 | 14 | 43 | 57 | 38 | 10 | 38 | 86 | 19 |
| Amino acid positionc | 16 | 41 | 45 | 51 | 88 | 111 | 184 | 212 | 230 | |||||||||||||||||||||||||||||||||||
| Substitutiond | M-I | M-I | G-R/K | G-R | W-* | None | M-I | W-* | M-I |
The nucleotide (nt) position of the hypermutated G within the RT gene is given. Only positions where at least 10% of the sequences have a mutation are shown. Nucleotide positions where more than 50% of the hypermutated sequences contained G → A mutations are indicated in boldface type. Designation in first column represent sequences.
The hypermutated G (underlined) and following two 3′ nucleotides are shown.
The corresponding amino acid position within the RT gene where hot spot G → A mutations occur is shown.
The amino acid substitution occurring as a result of a hot spot G → A mutation. A stop codon is labeled with an asterisk (*). The amino acid change at position 184 is shown in boldface type to indicate a known drug resistance site.
We have detected hypermutated sequences in resting CD4+ T cells from patients who had prolonged suppression of viremia on HAART. Hypermutated sequences with characteristic APOBEC3G/F-mediated changes constituted >9% of the viral genomes in this compartment and were found in every patient. This level may reflect some degree of accumulation of defective genomes that do not direct the synthesis of a full complement of viral proteins, thereby sparing the infected cell from viral cytopathic effects. Our data suggest that mutated viral genomes are able to integrate into cellular DNA and persist in resting CD4+ T cells even when viral replication is halted with HAART. However, these hypermutated genomes are subsequently incapable of producing virus, as indicated by the fact that the viruses released into the plasma at low level in patients on HAART were completely devoid of hypermutated sequences.
Acknowledgments
This work was supported by the Johns Hopkins University School of Medicine General Clinical Research Center, grant number M01-RR00052, from the National Center for Research Resource/National Institutes of Health and by National Institutes of Health grants AI43222 and AI51178 and a grant from the Doris Duke Charitable Foundation.
REFERENCES
- 1.Beale, R. C., S. K. Petersen-Mahrt, I. N. Watt, R. S. Harris, C. Rada, and M. S. Neuberger. 2004. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 337:585-596. [DOI] [PubMed] [Google Scholar]
- 2.Blankson, J. N., D. Persaud, and R. F. Siliciano. 2002. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 53:557-593. [DOI] [PubMed] [Google Scholar]
- 3.Borman, A. M., C. Quillent, P. Charneau, K. M. Kean, and F. Clavel. 1995. A highly defective HIV-1 group O provirus: evidence for the role of local sequence determinants in G→A hypermutation during negative-strand viral DNA synthesis. Virology 208:601-609. [DOI] [PubMed] [Google Scholar]
- 4.Chun, T. W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y. H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183-188. [DOI] [PubMed] [Google Scholar]
- 5.Chun, T. W., D. Finzi, J. Margolick, K. Chadwick, D. Schwartz, and R. F. Siliciano. 1995. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat. Med. 1:1284-1290. [DOI] [PubMed] [Google Scholar]
- 6.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 7.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 8.Fitzgibbon, J. E., S. Mazar, and D. T. Dubin. 1993. A new type of G→A hypermutation affecting human immunodeficiency virus. AIDS Res. Hum. Retrovir. 9:833-838. [DOI] [PubMed] [Google Scholar]
- 9.Goff, S. P. 2003. Death by deamination: a novel host restriction system for HIV-1. Cell 114:281-283. [DOI] [PubMed] [Google Scholar]
- 10.Han, Y., K. Lassen, D. Monie, A. R. Sedaghat, S. Shimoji, X. Liu, T. C. Pierson, J. B. Margolick, R. F. Siliciano, and J. D. Siliciano. 2004. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 78:6122-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 12.Hermankova, M., S. C. Ray, C. Ruff, M. Powell-Davis, R. Ingersoll, R. T. D'Aquila, T. C. Quinn, J. D. Siliciano, R. F. Siliciano, and D. Persaud. 2001. HIV-1 drug resistance profiles in children and adults with viral load of <50 copies/ml receiving combination therapy. JAMA 286:196-207. [DOI] [PubMed] [Google Scholar]
- 13.Janini, M., M. Rogers, D. R. Birx, and F. E. McCutchan. 2001. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4+ T cells. J. Virol. 75:7973-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieffer, T. L., M. M. Finucane, R. E. Nettles, T. C. Quinn, K. W. Broman, S. C. Ray, D. Persaud, and R. F. Siliciano. 2004. Genotypic analysis of HIV-1 drug resistance at the limit of detection: virus production without evolution in treated adults with undetectable HIV loads. J. Infect. Dis. 189:1452-1465. [DOI] [PubMed] [Google Scholar]
- 15.Kuiken, C., B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, S. Wolinsky, and B. Korber. 2001. HIV sequence compendium 2001. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.M.
- 16.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 17.Liddament, M. T., W. L. Brown, A. J. Schumacher, and R. S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385-1391. [DOI] [PubMed] [Google Scholar]
- 18.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 19.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 20.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 21.Persaud, D., T. Pierson, C. Ruff, D. Finzi, K. R. Chadwick, J. B. Margolick, A. Ruff, N. Hutton, S. Ray, and R. F. Siliciano. 2000. A stable latent reservoir for HIV-1 in resting CD4+ T lymphocytes in infected children. J. Clin. Investig. 105:995-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pierson, T. C., Y. Zhou, T. L. Kieffer, C. T. Ruff, C. Buck, and R. F. Siliciano. 2002. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J. Virol. 76:8518-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rose, P. P., and B. T. Korber. 2000. Detecting hypermutations in viral sequences with an emphasis on G →A hypermutation. Bioinformatics 16:400-401. [DOI] [PubMed] [Google Scholar]
- 24.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 25.Schrofelbauer, B., D. Chen, and N. R. Landau. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 101:3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 27.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 28.Siliciano, J. D., J. Kajdas, D. Finzi, T. C. Quinn, K. Chadwick, J. B. Margolick, C. Kovacs, S. J. Gange, and R. F. Siliciano. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9:727-728. [DOI] [PubMed] [Google Scholar]
- 29.Simon, J. H., and M. H. Malim. 1996. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J. Virol. 70:5297-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 31.Vartanian, J. P., A. Meyerhans, B. Asjo, and S. Wain-Hobson. 1991. Selection, recombination, and G→A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol. 65:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67:4945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu, H., E. S. Svarovskaia, R. Barr, Y. Zhang, M. A. Khan, K. Strebel, and V. K. Pathak. 2004. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. USA 101:5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, Q., R. Konig, S. Pillai, K. Chiles, M. Kearney, S. Palmer, D. Richman, J. M. Coffin, and N. R. Landau. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435-442. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]