Abstract
Cyclophilin A is conserved from yeast to humans and mediates the ability of cyclosporine to perturb signal transduction cascades via inhibition of calcineurin. Cyclophilin A also catalyzes cis-trans peptidyl-prolyl isomerization during protein folding or conformational changes; however, cyclophilin A is not essential in yeast or human cells, and the true biological functions of this highly conserved enzyme have remained enigmatic. In Saccharomyces cerevisiae, cyclophilin A becomes essential in cells compromised for the nuclear prolyl-isomerase Ess1, and cyclophilin A physically interacts with two nuclear histone deacetylase complexes, Sin3-Rpd3 and Set3C, which both control meiosis. Here we show that cyclophilin A is localized to the nucleus in yeast cells and governs the meiotic gene program to promote efficient sporulation. The prolyl-isomerase activity of cyclophilin A is required for this meiotic function. We document that cyclophilin A physically associates with the Set3C histone deacetylase and analyze in detail the structure of this protein-protein complex. Genetic studies support a model in which cyclophilin A controls meiosis via Set3C and an additional target. Our findings reveal a novel nuclear role for cyclophilin A in governing the transcriptional program required for the vegetative to meiotic developmental switch in budding yeast.
Cyclophilin A was originally identified as the intracellular receptor of the immunosuppressive drug cyclosporine (Cs), and the two molecules form a complex that binds to and inhibits the protein phosphatase calcineurin, preventing T-cell activation in mammals (36, 52, 53). Although cyclophilin A is highly conserved from yeast to humans, establishing the normal biological functions of this nonessential protein has proven elusive. Cyclophilin A is the founding member of a class of ubiquitous and highly conserved enzymes collectively known as peptidyl-prolyl cis-trans isomerases, or prolyl-isomerases, which catalyze cis-trans isomerization of the peptide bonds preceding proline residues. The prolyl-isomerase group spans three structurally unrelated protein families: the cyclophilins, FKBPs (for FK506 binding proteins), and parvulins (34, 40, 69, 81). All three families are present in the budding yeast Saccharomyces cerevisiae, which expresses eight different cyclophilins (Cpr1 to Cpr8), four FKBPs (Fpr1 to Fpr4), and a single parvulin (Ess1), which is the only essential prolyl-isomerase in this organism (20, 37, 39).
Cellular roles have been established for several prolyl-isomerases in both mammals and yeast. In mammals, cyclophilin A associates with the human immunodeficiency virus type 1 Gag polyprotein and is incorporated into virion particles, where it is required for human immunodeficiency virus type 1 infectivity (7, 9, 28, 84). Cyclophilin A also binds to and regulates the tyrosine kinase Itk in T lymphocytes (11, 55). FKBP12 regulates the activity of the ryanodine receptor, a calcium release channel in the sarcoplasmic reticulum (61, 85, 86), and both FKBP12 and cyclophilin A interact with and regulate the zinc finger transcription factor YY1 (96).
In yeast, the mitochondrial cyclophilin Cpr3 accelerates protein refolding after mitochondrial import, particularly at elevated temperature (19, 21, 59, 72, 73). Two larger cyclophilins, Cpr6 and Cpr7, were identified in a search for proteins interacting with the histone deacetylase Rpd3 (24), although the functional significance of these interactions is not yet clear. Both Cpr6 and Cpr7, like their human homolog cyclophilin 40, interact with and regulate activity of the molecular chaperone Hsp90 (17, 23, 76). Cpr7 is required for normal cell growth, and mutation or inhibition of Hsp90 causes a severe growth defect in cpr7 mutant cells (22, 23, 58, 83). Ess1, the first eukaryotic parvulin, was originally found to be associated with pre-mRNA processing and termination (38, 39). More recent reports have forged a link between Ess1 and the general transcription machinery and also revealed a connection with chromatin modification (3, 65, 93-95; for a recent review on the cellular functions of yeast prolyl-isomerases, see reference 4).
Cyclophilin A is conserved in yeast and encoded by the CPR1 gene (35). Similar to its mammalian counterpart, yeast cyclophilin A mediates calcineurin inhibition by Cs (12, 27, 66, 88). However, the endogenous functions of the yeast cyclophilin homolog Cpr1 have only recently begun to be elucidated. Cpr1 becomes essential in cells compromised for Ess1 function, and overexpression of Cpr1 suppresses ess1Δ mutations, suggesting a functional overlap between these two structurally unrelated prolyl-isomerases and providing the first evidence that the enzymatic activity of yeast cyclophilin A is important for biological function (3). Cpr1 is required for the glucose-stimulated transport of fructose-1,6-bisphosphatase into Vid (for vacuole import and degradation) vesicles, leading to degradation of this glucolytic enzyme (13). Finally, Cpr1 promotes the proper subcellular localization of an essential zinc finger protein, Zpr1 (2).
A physical association between Cpr1 and the Set3C histone deacetylase complex, which transcriptionally represses meiosis-specific genes, was recently reported (67). Cpr1 has also been previously implicated in physical and functional interactions with the Sin3-Rpd3 histone deacetylase complex (3). Here we show that cyclophilin A is localized to the nucleus in yeast and functions in sporulation by governing the meiotic transcriptional program. We confirm that cyclophilin A physically interacts with the Set3 complex and provide genetic evidence that cyclophilin A promotes meiosis via both Set3C and other targets. Our studies reveal a novel nuclear role for cyclophilin A, shedding insight into the normal cellular functions of this highly conserved heretofore enigmatic enzyme.
MATERIALS AND METHODS
Oligonucleotides.
Oligonucleotides used in the present study are listed in Table S1 in the supplemental material.
Plasmids.
Centromere-based vector pRS316 was described previously (75). Centromere-based plasmid pTB4a and 2μ plasmid pTB3 expressing the wild-type CPR1 gene were also described previously (15, 16). Plasmid pCPR1-GFP, expressing a C-terminal green fluorescent protein (GFP)-tagged version of Cpr1 was obtained from plasmid pTB3 by homologous recombination-mediated insertion of a PCR product amplified with the primers JOHE6791 and JOHE6792 from plasmid pFA6a-GFP(S65T)-kanMX6 (54). Plasmids pCPR1-GFP-wtSV40 (expressing a Cpr1-GFP-APGPPKKKRKVA nuclear localization signal [NLS] fusion), pCPR1-GFP-mutSV40 (expressing a Cpr1-GFP-APGPPKTKRKVA NLS* fusion), pCPR1-GFP-wtPKI (expressing a Cpr1-GFP-APGLALKLAGLDINKT nuclear export signal [NES] fusion), and pCPR1-GFP-mutPKI (expressing a Cpr1-GFP-APGLALKLAGADTNKT NES* fusion) were all obtained from pCPR1-GFP according to a gap-repair method similar to that described previously (64). Residues in boldface correspond to those originally found in SV40 and PKI proteins, respectively; residues not in boldface are added to the fusion protein as a linker sequence. Underscoring denotes residues changed in the mutant NLS and NES versions. To this end, plasmid pCPR1-GFP was linearized by digestion with AscI and cotransformed into an ura3 yeast strain together with PCR fragments obtained with 3′-complementary primer pairs JOHE7564 and JOHE7565 (pCPR1-GFP-wtSV40), JOHE7566 and JOHE7567 (pCPR1-GFP-mutSV40), JOHE7568 and JOHE7569 (pCPR1-GFP-wtPKI), or JOHE7570 and JOHE7571 (pCPR1-GFP-mutPKI). The resulting plasmids, isolated from the corresponding Ura+ transformants, were sequence verified. Plasmids pCPR1-GFP-wtSV40-nat and pCPR1-GFP-wtPKI-nat were obtained by homologous recombination-mediated replacement of the kanamycin resistance module (kanMX4) with the nourseothricin resistance module (nat) and PCR amplified with primers JOHE8033 and JOHE8034 from plasmid pAG25 (33). 2μ plasmids pHS103 and pAM405, expressing IME1 and IME2, respectively, were as described previously (78). Plasmid pCPR1(R53A), expressing the yeast cyclophilin A mutant Cpr1R53A was obtained by PCR overlap-mediated site-directed mutagenesis (42). To this end, PCR products obtained from plasmid pTB3 with the primer pair JOHE9631 and JOHE9634 and the primer pair JOHE9632 and JOHE9633 were mixed together and used as a template in a new PCR with the primers JOHE9631 and JOHE9632. The resulting product was digested with EcoRI and cloned in the EcoRI site of vector YEplac195 (32). Plasmid pRS415-hCyPA, expressing human cyclophilin A (hCyPA) was as described previously (2). Plasmid pMA-FLAG-kanMX6 was constructed by replacing the GFP coding sequence present in plasmid pFA6a-GFP(S65T)-kanMX6 with the 5′-GATTACAAGGATGACGACGATAAG-3′ sequence, encoding the FLAG peptide amino acid sequence DYKDDDDK (25). To this end, pFA6a-GFP(S65T)-kanMX6 was digested with PacI and AscI and ligated to a DNA segment obtained by annealing the complementary primers JOHE8634 and JOHE8635. Plasmid pCPR1-FLAG, expressing a C-terminal FLAG-tagged version of Cpr1 was obtained from pTB4a by homologous recombination-mediated insertion of a PCR product amplified with primers JOHE6791 and JOHE6792 from plasmid pMA-FLAG-kanMX6.
Strains.
Yeast strains used in the present study are listed in Table S2 in the supplemental material. With the exception of the ess1-ts mutant H164RW303 (95), all of the yeast strains used are derivatives of the isogenic, S288C-derived strains BY4741, BY4742, or BY4743 (10). Strains MAY152, MAY153, MAY154, MAY155, MAY156, MAY157, and MAY158 were obtained by replacing the kanMX4 module in strains 17046, 17182, 12271, 13242, 13513, 14561, and 11760, respectively, with the nat module from plasmid pAG25 (33), which was PCR amplified with the primers JOHE8033 and JOHE8034. Strain MAY159 was obtained from BY4743 by replacing the open reading frame (ORF) of one of the copies of CPR1 with the ORF of URA3, which was PCR amplified with the primers JOHE7426 and JOHE7427 from plasmid YEplac195. Strain MAY160 was obtained from MAY159 by disrupting the remaining CPR1 allele with the nat module from plasmid pAG25 PCR amplified with the primers JOHE6732 and JOHE6733. Strains MAY161, MAY163, MAY165, MAY168, and MAY169 were obtained from MAY159 by chromosomal integration of PCR products amplified with primers JOHE6791 and JOHE6792 from plasmids pFA6a-GFP(S65T)-kanMX6, pCPR1-GFP-wtSV40, pCPR1-GFP-mutSV40, pCPR1-GFP-wtPKI, and pCPR1-GFP-mutPKI, respectively. Strain MAY173 was obtained from MAY159 by chromosomal integration of a PCR product amplified with primers JOHE7212 and JOHE6792 from plasmid pFA6a-GFP(S65T)-kanMX6. Strain MAY178 was obtained from BY4741 by replacing the ORF of CPR1 with the ORF of URA3 as in MAY159. Strains MAY225 and MAY199 were obtained from MAY178 by homologous recombination-mediated replacement of the URA3 ORF, in the cpr1Δ::URA3 allele of this strain, with the CPR1(R53A) ORF carried in the EcoRI fragment of plasmid pCPR1(R53A) and with the hCyPA ORF carried in the XbaI-XhoI fragment of plasmid pRS415-hCyPA, respectively. Diploid strains MAY195, MAY196, MAY226, and MAY211 were obtained by crossing strain 13513 with strains BY4741, MAY178, MAY225, and MAY199, respectively. Strains MAY217, MAY243, and MAY289, expressing C-terminal 3HA-tagged versions of Hos2, Sif2, and Yil112w, respectively, were obtained from BY4741 by chromosomal integration of PCR products amplified from plasmid pFA6a-3HA-kanMX6 (54) with the primers JOHE8029 and JOHE8030, JOHE9789 and JOHE9790, and JOHE10310 and JOHE10311, respectively. Strains MAY224, MAY242, and MAY291, expressing C-terminal 13Myc-tagged versions of Set3, Snt1, and Hst1, respectively, were obtained from BY4741 by chromosomal integration of PCR products amplified from plasmid pFA6a-13Myc-kanMX6 (54) with the primers JOHE8027 and JOHE8028, JOHE9791 and JOHE9792, and JOHE10312 and JOHE10313, respectively. set3Δ, snt1Δ, yil112wΔ, sif2Δ, cpr1Δ, hos2Δ, or hst1Δ strains expressing one or more epitope-tagged Set3 complex member were obtained as meiotic products or diploids between these meiotic products derived from crosses between epitope-tagged strains MAY217, MAY224, MAY242, MAY243, MAY289, and MAY291 and strains with deletions in the genes encoding Set3C components, i.e., MAY152, MAY153, MAY154, MAY155, MAY156, MAY157, and MAY158. Strains MAY192, MAY218, MAY219, MAY220, MAY221, MAY222, and MAY223 were obtained from strains BY4741, 17046, 17182, 12271, 13242, 14561, and 11760, respectively, by disruption of CPR1 with the nat module PCR-amplified from plasmid pAG25 with the primers JOHE6732 and JOHE6733. Strains MAY234, MAY235, MAY236, MAY237, MAY238, and MAY239 were all obtained from MAY192 by disruption of SET3, SNT1, YIL112w, SIF2, HOS2, and HST1 with the kanMX2 module PCR amplified from plasmid pFA6-kanMX2 (89) with the primers JOHE7925 and 7926, JOHE9678 and JOHE9679, JOHE9680 and JOHE9681, JOHE9682 and JOHE9683, JOHE7929 and JOHE7930, and JOHE7933 and JOHE7934, respectively. Strains MAY245 to MAY263 were obtained as diploids from crosses between the strains with deletions in the genes encoding Set3C components described above.
Media.
Growth media for S. cerevisiae (synthetic complete medium [SC] and rich complex medium [YPD]) were as described previously (74). The sporulation medium was 1.5% potassium acetate (pH 7.5) supplemented with uracil and the required amino acids.
Localization of Cpr1-GFP fusion proteins.
Subcellular localization of the GFP fusion proteins was carried out in live cells by using a Zeiss Axioskop 2 microscope (Carl Zeiss, Jena, Germany). Strains were cultured in liquid YPD medium to logarithmic growth phase and analyzed directly by fluorescence microscopy. Nuclear DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole). Strains used were MAY161 (Cpr1-GFP), MAY173 (GFP), MAY163 (Cpr1-GFP-NLS), MAY165 (Cpr1-GFP-NLS*), MAY168 (Cpr1-GFP-NES), and MAY169 (Cpr1-GFP-NES*).
Western blot analysis of cyclophilin A and cyclophilin A fusion proteins.
For Western blot analysis of expression of the Cpr1-GFP fusions described above and the active site Cpr1R53A mutant protein (MAY226), yeast strains expressing these proteins were cultured in liquid YPD. Whole-cell protein extracts were prepared by glass bead disruption in lysis buffer A (20 mM HEPES [pH 7.4], 20 mM KCl, 0.5 mM EDTA, and a cocktail of protease inhibitors consisting of 0.5 mM phenylmethylsulfonyl fluoride, 1 μg of pepstatin ml−1, 1 mM benzamidine, and 0.001% aprotinin) by using a FastPrep instrument (FP120; Bio 101/Savant, La Jolla, Calif.). Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a polyvinylidene difluoride membrane (Immun-Blot; Bio-Rad), probed with rabbit polyclonal antiserum against Cpr1 (15), and detected by enhanced chemiluminescence (Amersham Biosciences). As controls, extracts from strains expressing wild-type Cpr1 (MAY195) or with the CPR1 gene deleted (cpr1Δ, MAY196), were analyzed. Expression of hCyPA in yeast was analyzed with rabbit polyclonal antiserum (Biomol).
ess1 suppression assays.
To test ess1 suppression by using the Cpr1-GFP fusion proteins, the ess1-ts strain H164RW303 was transformed with multicopy plasmids expressing the fusion proteins Cpr1-GFP (pCPR1-GFP), Cpr1-GFP-NLS (pCPR1-GFP-wtSV40), Cpr1-GFP-NLS* (pCPR1-GFP-mutSV40), Cpr1-GFP-NES (pCPR1-GFP-wtPKI), Cpr1-GFP-NES* (pCPR1-GFP-mutPKI), or wild-type Cpr1 (pTB3) as a control and tested for growth at the nonpermissive temperature (37°C). To test ess1 suppression by coexpression of two different Cpr1-GFP fusion proteins, H164RW303 was cotransformed with pCPR1-GFP-wtSV40-nat and pCPR1-GFP-wtPKI (Cpr1-GFP-NLS+Cpr1-GFP-NES), with pCPR1-GFP-mutSV40 and pCPR1-GFP-wtPKI-nat (Cpr1-GFP-NLS*+Cpr1-GFP-NES), or with pCPR1-GFP-wtSV40-nat and pCPR1-GFP-mutPKI (Cpr1-GFP-NLS+Cpr1-GFP-NES*). Transformants grown in media for plasmid selection were fivefold serially diluted, spotted on solid YPD medium, and incubated for 2 days at 30 or 37°C.
Sporulation assays.
Sporulation assays were performed essentially as described in Pijnappel et al. (67). Liquid YPD cultures from diploid strains were grown to an optical density at 600 nm of 2.0, and cells were harvested by centrifugation, washed once with sterile water, resuspended in three culture volumes of sporulation medium, and incubated at 25°C. Asci formation was monitored by microscopic observation of at least 200 cells from samples taken at the times indicated. All sporulation experiments included a wild-type strain as a control, and assays were repeated when this control strain showed poor or delayed asci formation. The strains used in experiments with the results shown in Fig. 3A were 33513 (cpr1Δ/cpr1Δ), 37046 (set3Δ/set3Δ), and BY4743 (wild type). The strains used in experiments with results shown in Table 1 were 33513 and BY4743. These strains were transformed with multicopy plasmids expressing IME1 (pHS103), IME2 (pAM405), or CPR1 (pTB4a) or with an empty vector (pRS316) and then assayed for sporulation. Asci formation was determined 100 h after transfer of the transformants from selective medium (SC lacking uracil) to sporulation medium. The strains used in the experiments with the results shown in Fig. 4B were MAY226 (CPR1-R53A), MAY211 (cpr1Δ::hCyPA), MAY195 (wild-type CPR1), and MAY196 (cpr1Δ). The strains used in the experiments with the results shown in Fig. 5 were MAY159 (Cpr1), MAY160 (cpr1Δ), MAY161 (Cpr1-GFP), MAY163 (Cpr1-GFP-NLS), MAY165 (Cpr1-GFP-NLS*), MAY168 (Cpr1-GFP-NES), and MAY169 (Cpr1-GFP-NES*). The strains used in the experiments with results shown in Fig. 7A were MAY245 (set3Δ/set3Δ), MAY246 (set3Δ/set3Δ cpr1Δ/cpr1Δ), MAY247 (SET3/set3Δ cpr1Δ/cpr1Δ), MAY248 (snt1Δ/snt1Δ), MAY249 (snt1Δ/snt1Δ cpr1Δ/cpr1Δ), MAY250 (SNT1/snt1Δ cpr1Δ/cpr1Δ), MAY251 (yil112wΔ/yil112wΔ), MAY252 (yil112wΔ/yil112wΔ cpr1Δ/cpr1Δ), MAY253 (YIL112w/yil112wΔ cpr1Δ/cpr1Δ), MAY254 (sif2Δ/sif2Δ), MAY255 (sif2Δ/sif2Δ cpr1Δ/cpr1Δ), MAY256 (SIF2/sif2Δ cpr1Δ/cpr1Δ), MAY257 (hos2Δ/hos2Δ), MAY258 (hos2Δ/hos2Δ cpr1Δ/cpr1Δ), MAY259 (HOS2/hos2Δ cpr1Δ/cpr1Δ), MAY260 (hst1Δ/hst1Δ), MAY261 (hst1Δ/hst1Δ cpr1Δ/cpr1Δ), MAY262 (HST1/hst1Δ cpr1Δ/cpr1Δ), and the wild-type strain BY4743 as a control.
FIG. 3.
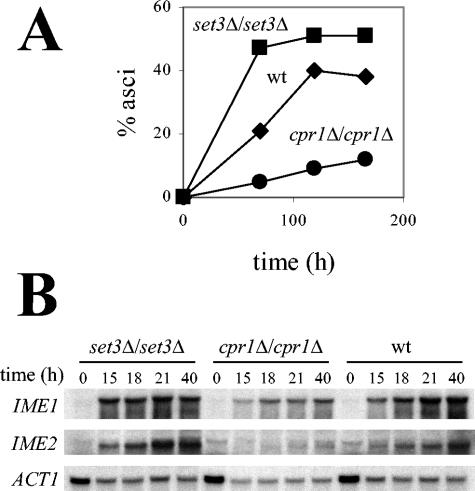
Cyclophilin A promotes sporulation. (A) Sporulation was measured at different time points after transfer of the cells to sporulation medium and plotted as the percentage of asci over the total number of cells in a cpr1Δ/cpr1Δ mutant (•), a set3Δ/set3Δ mutant (▪), and a wild-type strain (♦). (B) Induction of IME1 and IME2 is defective in a cpr1Δ/cpr1Δ mutant. Expression of IME1 and IME2 was analyzed by Northern blot in the strains described above, at the time points indicated, after transfer of cells to sporulation medium. ACT1 gene expression served as a control.
TABLE 1.
IME1 and IME2 overexpression suppresses the sporulation defect of a cpr1Δ/cpr1Δ mutant
| Gene | % Asci (t = 100 h)a
|
|
|---|---|---|
| cpr1Δ/cpr1Δ mutant | Wild type | |
| CPR1 | 19 ± 1.5 | 23 ± 2 |
| IME1 | 25 ± 2 | 37 ± 0.5 |
| IME2 | 27 ± 5.5 | 42 ± 0.5 |
| Vector | 5 ± 1 | 19 ± 2 |
The average ± the standard deviation is shown. wt, Wild type.
FIG. 4.
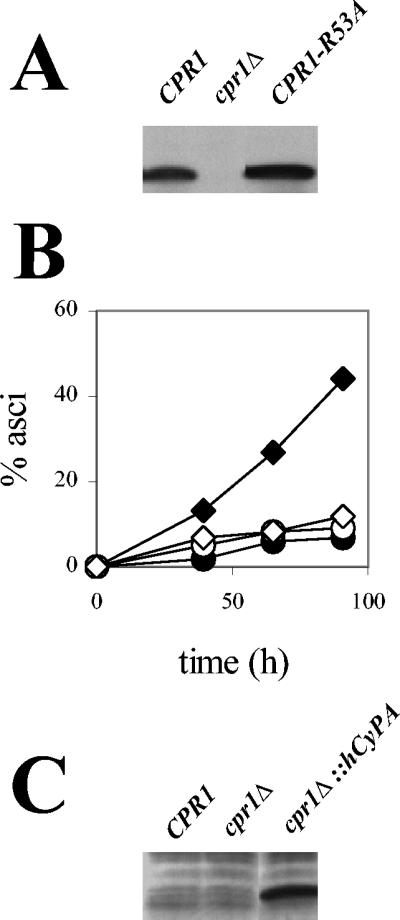
Cyclophilin A prolyl-isomerase activity is required for efficient sporulation. (A) Western blot analysis of Cpr1R53A expression. Proteins extracted from a diploid strain expressing the mutant CPR1-R53A allele were resolved by SDS-PAGE and analyzed with antibodies against Cpr1. Protein extracts from strains expressing wild-type CPR1 or with this gene deleted (cpr1Δ) served as controls. (B) Sporulation of diploid strains expressing Cpr1R53A (⋄) or hCyPA (cpr1Δ::hCyPA) (○). Wild-type CPR1 (♦) and cpr1Δ strain (•) served as controls. (C) Western blot analysis of hCyPA expression. Proteins from the cpr1Δ::hCyPA strain described above were resolved as in panel A and analyzed with antibodies to hCyPA.
FIG. 5.
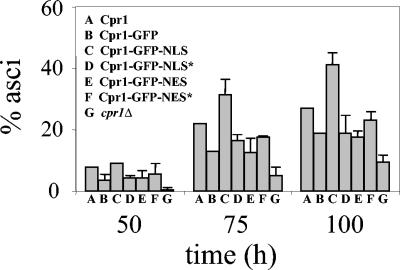
Targeting cylophilin A to the nucleus enhances sporulation. Asci formation was plotted as a percentage over time for strains expressing different Cpr1-GFP fusion proteins. Strains expressing wild-type Cpr1 or lacking this protein (cpr1Δ) were included as controls. Bars represent the standard deviation of values obtained in two independent experiments.
FIG. 7.
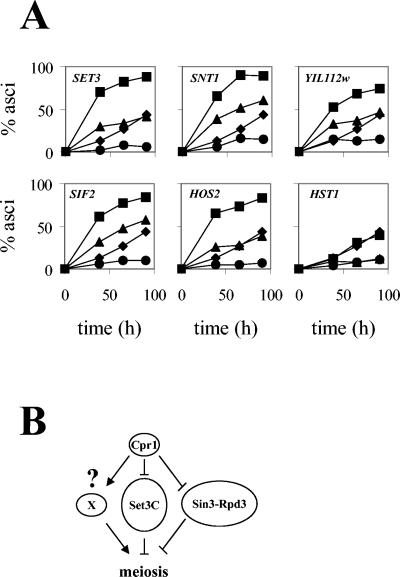
(A) Suppression of the sporulation defect of a cpr1Δ/cpr1Δ mutant by deletion of Set3C genes. Asci formation in diploid strains with both CPR1 and a Set3C-encoding gene deleted (▴) was assayed and compared to that of strains lacking cyclophilin A alone (•). As controls, asci formation in strains with individual Set3C genes deleted (▪) or in the wild-type strain (♦) was assayed. (B) Model for the role of cyclophilin A in meiosis.
Northern blot analysis.
RNA isolation and Northern analysis was as described previously (14). DNA probes hybridizing to IME1, IME2, and ACT1 transcripts were PCR amplified from yeast genomic DNA with the primers JOHE7918 and JOHE7919, JOHE7920 and JOHE7921, and JOHE2287 and JOHE2288, respectively.
Immunoprecipitation of the Set3 complex.
Yeast strains expressing epitope-tagged Set3C components were cultured to logarithmic growth phase in 100 ml of liquid YPD. Whole-cell protein extracts were prepared by glass bead disruption in lysis buffer B (50 mM HEPES [pH 7.4], 120 mM NaCl, 1 mM EDTA, 0.3% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 1 μg of pepstatin ml−1, 1 mM benzamidine, and 0.001% aprotinin), and the protein concentration in the extracts was determined by Bradford (Bio-Rad).
To precipitate the Cpr1-FLAG fusion, 2 mg of total protein was incubated in 0.7 ml of lysis buffer with 30 μl of anti-FLAG M2 affinity gel (EZview; Sigma) for 1 h at 4°C in a nutator mixer. Gel beads were recovered by centrifugation, washed four times with 1 ml of lysis buffer, and once with a buffer containing 50 mM HEPES (pH 7.4), 100 mM KCl, 1 mM EDTA, and 5% glycerol. Beads were resuspended in 50 μl of the same buffer without glycerol, and immunocomplexes were eluted by addition of FLAG peptide (Sigma) to a final concentration of 200 μg/ml.
To precipitate the hemagglutinin (HA) epitope-tagged Set3C proteins, 2 mg of total protein in 0.7 ml of lysis buffer was first incubated with 30 μl of protein A-Sepharose (Amersham Biosciences) for 1 h at 4°C. Samples were then centrifuged, and the supernatant was transferred to new vials and incubated for 1 h at 4°C with 3 μg of anti-HA antibody. To precipitate the immunocomplexes, binding reactions were further incubated with 30 μl of protein A-Sepharose for 1 h at 4°C. Beads were recovered by centrifugation and washed as described above for immunoprecipitation of Cpr1-FLAG, and protein was eluted by heating the beads in SDS-PAGE loading buffer.
Immunocomplexes were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membrane, and epitope-tagged proteins were detected with anti-FLAG monoclonal antibody (M2; Sigma), anti-Myc monoclonal antibody (9E10; Santa Cruz), or anti-HA monoclonal antibody (F-7; Santa Cruz).
To assay coimmunoprecipitation of Set3-Myc, Hos2-HA, Snt1-Myc, or Sif2-HA with Cpr1-FLAG in Set3C mutants, strains expressing a Myc- or HA-tagged Set3C component and with a gene encoding one of the remaining Set3C proteins deleted were transformed with plasmid pCPR1-FLAG, expressing a Cpr1-FLAG fusion protein. The Cpr1-FLAG fusion was immunoprecipitated from extracts obtained from these transformants, and immunocomplexes were resolved and analyzed as described above. Control immunoprecipitation assays were carried out with strains transformed with plasmid pTB4a, expressing untagged, wild-type Cpr1 (no FLAG). Set3-Myc-Cpr1-FLAG interaction was tested in transformants from strains MAYX84-3C (snt1Δ), MAYX85-2C (yil112wΔ), MAYX86-5C (sif2Δ), MAYX87-1B (cpr1Δ), MAYX88-4A (hos2Δ), MAYX89-1C (hst1Δ), and MAYX83-4A (no FLAG). Hos2-HA-Cpr1-FLAG interaction was tested in transformants from strains MAYX76-3A (set3Δ), MAYX77-1A (snt1Δ), MAYX78-1B (yil112wΔ), MAYX79-5A (sif2Δ), MAYX80-3B (cpr1Δ), MAYX82-7C (hst1Δ), and MAYX81-1C (no FLAG). Snt1-Myc-Cpr1-FLAG interaction was tested in transformants from strains MAYX90-1D (set3Δ), MAYX92-9C (yil112wΔ), MAYX93-1A (sif2Δ), MAYX94-2B (cpr1Δ), MAYX95-1B (hos2Δ), MAYX96-4B (hst1Δ), and MAY242 (no FLAG). Sif2-HA-Cpr1-FLAG interaction was tested in transformants from strains MAYX97-1A (set3Δ), MAYX98-2C (snt1Δ), MAYX99-2B (yil112wΔ), MAYX101-4D (cpr1Δ), MAYX102-5B (hos2Δ), MAYX103-2A (hst1Δ), and MAY243 (no FLAG). Yeast strains expressing Gpb2-Myc or Esa1-HA fusion proteins and transformed with plasmid pCPR1-FLAG were included as controls in the assays.
Coimmunoprecipitation of Set3-Myc, Snt1-Myc, or Hst1-Myc with Hos2-HA, Sif2-HA, or Yil112w-HA in Set3C mutants was assayed in strains expressing a pair of Set3C components tagged with the HA and Myc epitopes, respectively, and with a gene encoding one of remaining Set3C members deleted. The HA-tagged Set3C component was immunoprecipitated, and the proteins associated with it were resolved and analyzed as described above. Set3-Myc-Hos2-HA interaction was tested in strains MAY227 (snt1Δ), MAY228 (yil112wΔ), MAY229 (sif2Δ), MAY230 (cpr1Δ), MAY231 (hst1Δ), and MAY232 (wild type). Snt1-Hos2 interaction was tested in strains MAY271 (set3Δ), MAY272 (yil112wΔ), MAY273 (sif2Δ), MAY274 (cpr1Δ), MAY275 (hst1Δ), and MAY276 (wild type). Set3-Myc-Sif2-HA interaction was tested in strains MAY277 (snt1Δ), MAY278 (yil112wΔ), MAY279 (cpr1Δ), MAY280 (hos2Δ), MAY281 (hst1Δ), and MAY282 (wild type). Snt1-Myc-Sif2-HA interaction was tested in strains MAY283 (set3Δ), MAY284 (yil112wΔ), MAY285 (cpr1Δ), MAY286 (hos2Δ), MAY287 (hst1Δ), and MAY288 (wild type). Snt1-Myc-Yil112w-HA interaction was tested in strains MAY294 (set3Δ), MAY295 (sif2Δ), MAY296 (cpr1Δ), MAY297 (hos2Δ), MAY298 (hst1Δ), and MAY299 (wt). Hst1-Myc-Yil112w-HA interaction was tested in strains MAY300 (set3Δ), MAY302 (sif2Δ), MAY303 (cpr1Δ), MAY304 (hos2Δ), and MAY305 (wild type).
RESULTS
Cyclophilin A is nuclear localized.
Cyclophilin A has been traditionally considered a cytoplasmic protein, in part because the target of the cyclophilin A-Cs complex, calcineurin, is cytoplasmic. To investigate the localization of cyclophilin A in S. cerevisiae, we constructed a diploid yeast strain in which the Aequorea victoria GFP (68) was fused in frame to the C terminus of cyclophilin A and expressed from the endogenous CPR1 chromosomal locus. Based on direct fluorescence microscopy, the cyclophilin A-GFP fusion protein is predominantly localized within the nucleus, although a weaker cytoplasmic distribution was also apparent. In a control strain in which GFP alone was expressed from the CPR1 promoter, a uniform intracellular distribution was observed (Fig. 1A). The localization pattern of the cyclophilin A-GFP fusion was the same in a haploid strain and did not change during the cell cycle or in response to Cs addition (data not shown). These results reveal that cyclophilin A accumulates in the nucleus in yeast cells. These observations are consistent with a recent report of global protein localization in yeast that appeared when the present report was in preparation (44).
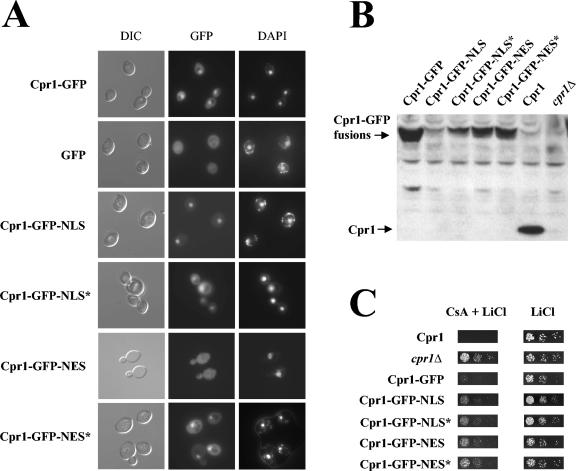
FIG. 1.
Cyclophilin A is localized to the nucleus in yeast. (A) Subcellular localization of cyclophilin A-GFP fusion proteins. Cells expressing different Cpr1-GFP fusion proteins or GFP alone as a control were analyzed by fluorescence microscopy to detect GFP or DAPI signals as described in Materials and Methods. Images with differential interference contrast (DIC) microscopy are included as a reference. (B) Expression analysis of the Cpr1-GFP fusion proteins. Proteins extracted from strains expressing Cpr1-GFP fusions described above were resolved by SDS-PAGE and analyzed by Western blotting with antibodies against Cpr1. As controls, extracts from strains expressing wild-type Cpr1 or with the CPR1 gene deleted (cpr1Δ) were analyzed. (C) Cpr1-GFP fusions mediate Cs toxicity. Cell suspensions from the strains described in panel B were fivefold serially diluted, spotted onto solid YPD medium containing 400 mM LiCl with or without 100 μg of Cs/ml, and incubated for 2 days at 30°C.
Perturbing cyclophilin A localization compromises function.
Cyclophilin A has been implicated in several cellular processes involving genetic or physical interactions with known nuclear proteins. To explore the physiological significance of the observed nuclear accumulation of cyclophilin A, we tested the effects of perturbing cellular localization on cyclophilin A function. We have previously shown that overexpression of cyclophilin A restores viability of yeast mutants defective in the essential parvulin Ess1, revealing a functional overlap between the two enzymes (3). Ess1 interacts with the carboxyl-terminal domain of RNA polymerase II, and both cyclophilin A and Ess1 interact with and regulate the activity of the Sin3-Rpd3 histone deacetylase complex, supporting a nuclear role for these two prolyl-isomerases (3, 95).
To test the importance of localization of cyclophilin A for biological function, the Cpr1-GFP fusion was fused to the nuclear localization signal (NLS) derived from SV40 T antigen (PPKKKRKVA) (47), or to the nuclear export signal (NES) from human protein kinase A inhibitor (PKI; LALKLAGLDINKT) (91). As controls, fusions with nonfunctional, mutant NLS or NES (denoted as NLS* and NES*, respectively) were constructed and assayed. Direct fluorescence microscopy confirmed that the cellular localization of the fusion proteins was appropriately perturbed: cyclophilin A-GFP-NLS was almost exclusively localized to the nucleus, cyclophilin A-GFP-NES was predominantly localized to the cytoplasm, and the cyclophilin A-GFP-NLS* and cyclophilin A-GFP-NES* fusion proteins exhibited a nuclear and cytoplasmic distribution similar to the wild-type Cpr1-GFP protein. These cyclophilin A-GFP fusions were expressed from the chromosome in a single copy (Fig. 1A), and Western blot analysis verified that all were expressed, albeit at levels lower than wild-type Cpr1-GFP (Fig. 1B).
We assayed the ability of these Cpr1-GFP fusion proteins to mediate Cs action and to suppress an ess1-ts mutation. As expected, the wild-type Cpr1-GFP fusion bound Cs and inhibited calcineurin to prevent yeast growth during cation stress (Fig. 1C). The Cpr1-GFP-NLS, Cpr1-GFP-NLS*, Cpr1-GFP-NES, or Cpr1-GFP-NES* fusions also mediated Cs toxicity in this assay (Fig. 1C), although not to the full wild-type level. Thus, perturbing cyclophilin A localization does not prevent Cs binding or inhibition of calcineurin, possibly because calcineurin is known to shuttle between the cytoplasm and nucleus associated with its transcription factor targets and might therefore be subject to cyclophilin A-Cs inhibition in either cellular compartment.
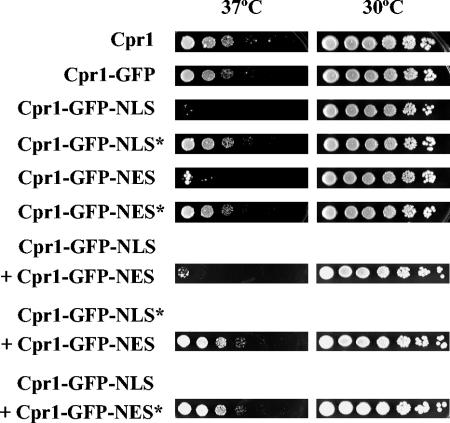
Expression of the Cpr1-GFP fusion restored viability of an ess1-ts mutant at the nonpermissive temperature to the same extent as wild-type Cpr1, indicating that the fusion protein is functional (Fig. 2). In contrast, the addition of a functional NLS or NES (but not of their mutant versions) abolished suppression of the ess1-ts mutation, suggesting that suppression requires cyclophilin A localization to both cellular compartments. To test this hypothesis, we analyzed the suppression of the ess1-ts mutation in cells expressing more than one Cpr1 fusion. As shown in Fig. 2, the coexpression of two different Cpr1-GFP fusions containing functional NLS and NES tags failed to suppress the ess1-ts mutant. Expression of the NLS- or NES-tagged Cpr1-GFP fusions was not deleterious per se to the viability of an ess1-ts mutant and also did not interfere with suppression by another Cpr1-GFP fusion tagged with a nonfunctional NLS or NES (Fig. 2). Taken together, these results suggest suppression may require shuttling of cyclophilin A between the two cellular compartments.
FIG. 2.
Targeting cyclophilin A to the nucleus or cytoplasm blocks ess1 suppression. An ess1-ts mutant strain was transformed with multicopy plasmids expressing different Cpr1-GFP fusion proteins. Transformants were fivefold serially diluted, spotted onto solid YPD medium, and incubated for 2 days at 30 or 37°C (nonpermissive temperature).
Cyclophilin A is required for efficient sporulation.
Cyclophilin A was recently implicated as a physically associated component of a novel histone deacetylase complex, Set3C, which functions to repress early and middle sporulation genes (67). However, whereas Cpr1 was identified by mass spectrometry in the Set3C native complex, no further studies have been reported, nor has it been established whether this interaction is physiologically relevant or merely a spurious protein-protein interaction. Deletion of the SET3 or HOS2 genes, which encode protein subunits of the Set3 complex, causes premature expression of key meiotic regulatory genes early after initiation of sporulation, resulting in accelerated sporulation.
These findings prompted us to test whether cyclophilin A also functions in meiosis and sporulation. The strain background chosen for these experiments, S288c, has the advantage of being a widely used background, mutants are readily available from the collection produced by the Saccharomyces Deletion Project, and strains sporulate at an efficiency (∼40%) that is in the range of many laboratory backgrounds, and this enabled analysis of mutant strains with enhanced and delayed sporulation efficiency. Here we analyzed the kinetics of asci formation in a diploid strain deleted for both alleles of the CPR1 gene compared to the isogenic wild-type and set3Δ/set3Δ mutant strains. As shown in Fig. 3A, deletion of SET3 accelerated sporulation, a finding in accord with previous observations (67). In contrast, deletion of CPR1 conferred a delay in sporulation (Fig. 3A), indicating that Set3 and Cpr1 play opposing roles during sporulation. Similar results were obtained with a cpr1Δ/cpr1Δ mutant in a different genetic background (data not shown). Transformation with a centromere-based plasmid harboring the wild-type CPR1 gene restored sporulation in both cpr1Δ/cpr1Δ mutant strain backgrounds (data not shown). We note that cyclophilin A mutant cells were still capable of undergoing sporulation at a reduced level, indicating that cyclophilin A is important for sporulation but not strictly essential.
We tested the effect of individually deleting each other yeast cyclophilin (CPR2, -3, -4, -5, -6, -7, and -8) or FKBP-encoding gene (FPR1, -2, -3, and -4) on sporulation. Only the cpr7Δ/cpr7Δ mutant exhibited a significant decrease in sporulation rate (data not shown). Cpr7 is required for normal cell growth (22, 23), suggesting that the sporulation defect observed in the cpr7Δ/cpr7Δ mutant might be an indirect effect of impaired growth. Further studies will be required to establish if Cpr7 plays a physiologically relevant role in meiosis.
IME1 and IME2 induction requires cyclophilin A.
Initiation of the yeast sporulation program requires activation of the key meiotic regulatory genes IME1 and IME2 (49, 63, 78, 97; for recent reviews, see references 43 and 48). A recent report (67) described premature induction of IME2 in meiotic cells defective in the Set3C histone deacetylase, linking this complex to regulation of the meiotic transcriptional program. Based on the sporulation-defective phenotype observed in a cpr1Δ/cpr1Δ strain, we next examined transcriptional induction of IME1 and IME2 under sporulation conditions (Fig. 3B). We made the following observations: first, we confirmed that the set3Δ/set3Δ mutant exhibits premature induction of IME1 and IME2 expression, as observed previously, although the wild-type strain used in our studies exhibited faster IME1 and IME2 induction kinetics than the wild-type strain used by Pijnappel et al., which has a different genetic background (67). Second, we found that cpr1Δ/cpr1Δ mutant cells exhibit the opposite phenotype, in which IME1 expression was reduced significantly and IME2 induction was impaired severely.
Since activation of IME1 and IME2 expression is required for entry into meiosis, the decreased expression of these genes caused by the cyclophilin A mutation could be responsible for the sporulation defect of cpr1Δ/cpr1Δ mutant cells. To test this hypothesis, we assayed asci formation in cpr1Δ/cpr1Δ mutant cells expressing IME1 or IME2 from multicopy plasmids. As shown in Table 1, overexpression of either IME1 or IME2 was sufficient to restore sporulation of cpr1Δ/cpr1Δ cells to a wild-type level. Because the function of Ime1 is to activate transcription of IME2 (56, 77), these results suggest that increasing the levels of Ime1 may also overcome the IME2 expression defect of the cyclophilin A mutant. Taken together, these results indicate that cyclophilin A promotes meiosis by governing proper IME1 and IME2 induction and that the meiotic-specific signaling cascade downstream of these genes is functional in the cyclophilin A mutant.
Ime1 induces IME2 by interacting with the C6 zinc cluster protein Ume6. The Ume6 transcriptional repressor binds to the common URS1 cis-element that is located upstream of IME2 and also most early meiotic and several nonmeiotic genes (1, 70, 80, 90, 92). We therefore tested whether cyclophilin A is required for induction of two other genes controlled by Ume6, CAR1 and CAR2, which are involved in arginine catabolism (62). Translational lacZ fusions to these genes (kindly provided by John York) were used to monitor induction in the presence of arginine. Neither the kinetics nor the induction levels of the CAR1-lacZ and CAR2-lacZ reporters were affected in the cpr1Δ/cpr1Δ mutant (data not shown), indicating that cyclophilin A does not control transcription from other Ume6-regulated promoters.
Cyclophilin A prolyl-isomerase activity is required for sporulation.
Suppression of an ess1-ts mutation by cyclophilin A requires prolyl-isomerase activity, supporting an in vivo enzymatic role for Cpr1 (3). Cyclophilin A prolyl-isomerase activity is also required for proper localization of the essential zinc finger protein Zpr1 in yeast (2) and for regulation of the tyrosine kinase Itk in mammalian T cells (11). To test whether the prolyl-isomerase activity of cyclophilin A is also important for its role in sporulation, we assayed asci formation in a diploid strain expressing a Cpr1 active-site mutant as the only source of cyclophilin A. By site-directed mutagenesis an alanine substitution was introduced for arginine 53, a conserved residue in the cyclophilin active site that plays an essential catalytic role in hCyPA (6, 8, 26, 98) and also in yeast Cpr3 (21, 72). Expression of the Cpr1R53A active-site mutant was confirmed by Western blot analysis (Fig. 4A). As shown in Fig. 4B, Cpr1R53A failed to complement the sporulation defect of the cpr1Δ/cpr1Δ mutant, indicating that the prolyl-isomerase activity of cyclophilin A is required for its role in sporulation.
To study further the molecular requirements of cyclophilin A function in meiosis, we tested if human cyclophilin A (hCyPA) could complement the sporulation defect of a cpr1Δ/cpr1Δ mutant. hCyPA was expressed from the chromosomal CPR1 locus under the control of the CPR1 gene promoter and terminator, and expression was verified by Western blot analysis (Fig. 4C). The primary structures of Cpr1 and hCyPA share 65% identity, and previous reports have shown that certain Cpr1 functions can be accomplished by its human homolog (2, 3). However, expression of hCyPA failed to suppress the sporulation defect of the cpr1Δ/cpr1Δ mutant (Fig. 4B). Thus, although the yeast and human proteins are highly conserved, hCyPA was unable to substitute for Cpr1 in yeast sporulation, indicating unique structural constraints for the yeast Cpr1 cyclophilin A homolog.
Targeting cyclophilin A to the nucleus improves sporulation.
The role of Cpr1 in transcriptional regulation of the meiotic program supports a model in which nuclear localization is important for function. To test this hypothesis further, we studied the effect of increasing Cpr1 nuclear localization on sporulation. As shown in Fig. 5, the Cpr1-GFP protein fusion used in the assays described above partially complemented the sporulation defect of a cpr1Δ/cpr1Δ mutant. Targeting the Cpr1-GFP fusion to the nucleus by the addition of an NLS significantly improved its ability to support sporulation and enhanced sporulation beyond the level observed in a control strain expressing wild-type Cpr1-GFP, even though Cpr1-GFP-NLS was expressed at a lower level based on Western blot analysis (Fig. 1B). These observations suggest that cyclophilin A normally functions in the nucleus to control sporulation.
Protein-protein interactions within the Set3 complex.
In previous studies Pijnappel et al. discovered that cyclophilin A is a component of the Set3 complex and analyzed the importance of different Set3C proteins to the integrity of the Set3 complex. Their results indicate that Set3 and Hos2 play pivotal roles in bridging protein interactions within the complex. To elucidate the architecture of the Set3 complex and to address the possible roles of cyclophilin A, we studied the requirement for individual Set3C members in mediating interactions between other proteins in the complex. Our approach was to assay coimmunoprecipitation of pairs of Set3C proteins in the absence of other Set3C components. To this end, we constructed a series of strains expressing pairs of epitope-tagged Set3C proteins and in which the gene encoding one of the remaining Set3C components had been deleted.
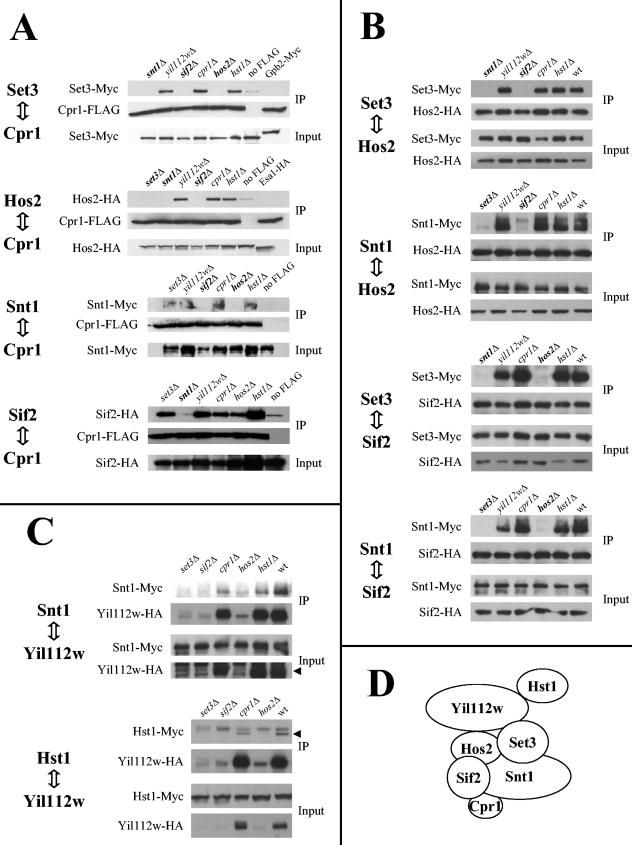
The results of these experiments are shown in Fig. 6. First, we tested interactions between cyclophilin A and four Set3C components: Set3, Hos2, Snt1, and Sif2 (Fig. 6A). Strains expressing epitope-tagged version of these proteins and with genes encoding the other Set3C components deleted were transformed with a centromere-based plasmid expressing a Cpr1-FLAG fusion protein. Immunoprecipitation of the Cpr1-FLAG fusion resulted in precipitation of the Set3-Myc, Hos2-HA, Snt1-Myc, and Sif2-HA fusion proteins. In control experiments, two unrelated proteins fused to the same epitopes, Gpb2-Myc and Esa1-HA, did not associate with Cpr1-FLAG. Deletion of the chromosomal CPR1 gene did not enhance the interactions described above, suggesting that the Cpr1-FLAG fusion interacts with the Set3C proteins as efficiently as endogenous Cpr1.
FIG. 6.
Protein-protein interactions in the Set3 complex. Interactions between pairs of epitope-tagged Set3C components were tested by coimmunoprecipitation. (A) Coimmunoprecipitation of Set3-Myc, Hos2-HA, Snt1-Myc, or Sif2-HA fusion proteins with a Cpr1-FLAG fusion in different Set3C mutants. Control assays with strains lacking a FLAG-tagged protein (no FLAG) were included. (B) Coimmunoprecipitation of Set3-Myc or Snt1-Myc with Hos2-HA or Sif2-HA in Set3C mutants. (C) Coimmunoprecipitation of Snt1-Myc or Hst1-Myc with Yil112w-HA in Set3C mutants. Arrowheads indicate the bands corresponding to Yil112w-HA and Hst1-Myc. (D) Model for the structure the Set3 complex, based on the physical interactions observed between the components of this complex. IP, immunoprecipitation.
Our results support those of Pijnappel et al. (67), confirming that Cpr1 is a component of the Set3 complex. Our findings also extend their studies since they were unable to detect precipitation of this complex with a Cpr1 fusion protein. As shown in Fig. 6A, deletion of YIL112W or HST1 did not prevent the interaction of Cpr1-FLAG with any of the other Set3C protein fusions, indicating that neither Yil112w nor Hst1 mediates any of these interactions. Furthermore, the results obtained from the experiments carried out to test binding between Cpr1 and either Snt1 or Sif2 suggest that Set3 is also dispensable for these interactions, and Hos2 is required only for the Cpr1-Snt1 interaction. In addition, Snt1 and Sif2 were found to be mutually required for their interaction with Cpr1. Taken together, these results support a model in which Snt1 and Sif2 play a major role in mediating physical interactions between Cpr1 and the rest of the Set3 complex.
We extended our analysis of the protein interactions within the Set3 complex to other protein pairs formed between Set3, Hos2, Snt1, and Sif2. As shown in Fig. 6B, binding within any of these protein pairs required the presence of the other two proteins, namely, Set3-Hos2 interaction required Snt1 and Sif2, Snt1-Hos2 interaction required Set3 and Sif2, Set3-Sif2 interaction required Snt1 and Hos2, and Snt1-Sif2 interaction required Set3 and Hos2. None of these interactions required Cpr1, Yil112w, or Hst1. Our results suggest that Set3, Hos2, Snt1, and Sif2 constitute the core of the Set3 complex and that each of these proteins is required for the integrity of the complex (Fig. 6A). Our data also indicate that Cpr1, Yil112w, and Hst1 do not play a major structural role in the Set3 complex. The data reported by Pijnappel et al. (67) also suggest that Yil112w and Hst1 might occupy a more peripheral position in Set3C.
To study a possible role for Cpr1 and other Set3C members in mediating interactions between Yil112w and the rest of Set3C, we tested the coprecipitation of Snt1 and Hst1 with Yil112w in the absence of other Set3C components. As shown in Fig. 6C, expression of Yil112w is dramatically reduced in the absence of Set3, Hos2, or Sif2. In contrast, deletion of HST1 affected neither the expression of Yil112w nor its interaction with Snt1. Similarly, deletion of CPR1 did not have any effect on Yil112w expression or its interactions with Snt1 and Hst1. These results show that Set3, Hos2, and Sif2 are all required for Yil112w expression and suggest that Yil112w is unstable and undergoes proteolytic degradation unless bound to the Set3 complex.
Sporulation defect of cyclophilin A-deficient strain is suppressed by Set3-complex mutations.
Our results shown in Fig. 3 indicate opposing sporulation phenotypes for the set3Δ/set3Δ and cpr1Δ/cpr1Δ mutants, supporting a model in which Cpr1 might function as a negative regulator of Set3. In this model, the repression of IME2 by the Set3 complex would be enhanced in cpr1 mutant cells. One prediction of this model is that disruption of Set3C should suppress the sporulation defect of cpr1Δ/cpr1Δ mutant cells.
We tested this hypothesis by using genetic epistasis analysis. A set of double mutant diploid strains was constructed in which CPR1 and a gene encoding one of the remaining Set3C components were deleted. The sporulation kinetics of these double-mutant strains were then compared to the individual mutants and to the wild type. As shown in Fig. 7A, deletion of SET3 or HOS2 alone conferred an accelerated sporulation phenotype, whereas the sporulation kinetics of an hst1Δ/hst1Δ mutant were similar to the wild type, in accord with a previous report (67). In addition, our results show that deletion of SNT1, SIF2, or YIL112W also accelerates asci formation, extending the meiotic role of SET3 and HOS2 to other Set3C components. Combination of a cpr1 mutation with snt1, sif2, yil112w, set3, or hos2 mutations produced double-mutant strains (i.e., cpr1Δ/cpr1Δ snt1Δ/snt1Δ) whose sporulation rates were higher than that of the cpr1Δ/cpr1Δ mutant and intermediate between those of the wild-type and snt1Δ/snt1Δ, sif2Δ/sif2Δ, yil112wΔ/yil112wΔ, set3Δ/set3Δ, or hos2Δ/hos2Δ mutant strains. Thus, the sporulation defect conferred by a cpr1Δ/cpr1Δ mutation is suppressed by deletion of SNT1, SIF2, YIL112W, SET3, or HOS2. In contrast, the deletion of HST1 did not suppress the sporulation defect of a cpr1Δ/cpr1Δ mutant (Fig. 7A). Similar results were obtained with an independent set of diploid strains constructed by using haploid single mutants from the original Saccharomyces Deletion Project strain collection and also when sporulation rates were examined at earlier time points (t = 24 and 48 h) during the sporulation assays (data not shown). Our data show that mutations in SNT1, SIF2, YIL112W, SET3, or HOS2 counteract the negative effects of the cpr1 mutation on sporulation, providing genetic evidence that the physical interactions observed between Cpr1 and the Set3 complex are functional. However, the finding that the set3 single mutants and set3 cpr1 double mutants do differ in phenotype suggests that the meiotic defect of a cpr1Δ/cpr1Δ mutant is not solely mediated by the Set3 complex.
DISCUSSION
Nuclear role for cyclophilin A.
Cyclophilin A has been traditionally considered a cytoplasmic protein based on biochemical properties and its known ability to bind Cs and inhibit calcineurin, which is known to be cytoplasmic in quiescent cells. However, several previous immunolocalization studies hinted that cyclophilin A might be, at least in part, nuclear in mammalian cells (18, 51, 57, 71). Here we show that Cpr1 is predominantly nuclear localized in yeast. Our finding is consistent with previous studies linking cyclophilin A with known nuclear proteins and nuclear functions. Cpr1 was classified as a cytoplasmic protein in a recent effort to immunolocalize a large number of yeast proteins (50). This discrepancy could be explained by the fact that, in our localization studies, we used yeast strains expressing Cpr1-GFP fusions from the chromosomal, endogenous CPR1 promoter, whereas Kumar et al. analyzed an epitope-tagged version of Cpr1 expressed from the strong galactose-inducible GAL1 promoter from a multicopy plasmid. In addition, our approach confers the advantage of a direct, in vivo visualization of the fluorescence signal in contrast to the more invasive immunolocalization procedures described by Kumar et al. In a more recent study describing the subcellular localization of the yeast proteome by GFP tagging (44), cyclophilin A was reported to exhibit a localization pattern that supported our conclusions.
The Cpr1-GFP fusion protein used here was functional in all assays tested. Cpr1-GFP bound Cs and inhibited calcineurin in vivo, suppressed an ess1-ts mutation, and complemented the sporulation defect of a cpr1Δ/cpr1Δ mutant. These observations indicate that the presence of the GFP domain does not interfere with cyclophilin A in vivo functions and support the conclusion that the observed nuclear localization of the Cpr1-GFP fusion protein is biologically relevant.
The relatively small molecular size of the Cpr1-GFP fusion protein (<50 kDa) could in principle allow it to freely diffuse through the nuclear pore (60, 82). However, protein accumulation in the nucleus requires nuclear import. Analysis of the amino acid sequence of Cpr1 did not reveal any canonical NLS, suggesting that Cpr1 may passively permeate the nuclear pore and then interact with a nuclear protein or protein complex to be retained within the nucleus. Alternatively, Cpr1 could be coimported into the nucleus via interaction with another protein containing an NLS. Our finding that the addition of either an NLS or an NES to Cpr1 abolished suppression of an ess1-ts mutation provides evidence that this function of cyclophilin A may require shuttling between the nucleus and cytoplasm. Although expressed at lower levels (as detected by Western blotting), the Cpr1-GFP-NLS fusion used in these studies accumulates inside the nucleus at levels comparable to those observed for the Cpr1-GFP fusion, indicating that the inability of Cpr1-GFP-NLS to suppress an ess1-ts mutation is not caused by a reduced abundance of this fusion protein in the nuclear compartment. In contrast, targeting Cpr1 to the nucleus enhanced sporulation, and the Cpr1-GFP-NLS fusion protein complemented the sporulation defect of the cpr1/cpr1 mutant, indicating that ess1 suppression and sporulation represent independent, separable cyclophilin A roles with different localization requirements.
Cyclophilin A drives sporulation.
We discovered that cpr1Δ/cpr1Δ mutant strains exhibit a sporulation defect that results from impaired induction of IME1 and IME2 under sporulation conditions. Induction of IME1 precedes and is required for IME2 induction, and both Ime1 and Ime2 are necessary for induction of the meiosis-specific genes downstream in the pathway (43, 48). One possibility is that IME1 expression levels in a cyclophilin A mutant under sporulation conditions are not sufficient to properly induce IME2 transcription. This model provides a parsimonious explanation for the sporulation defects observed in cyclophilin A mutants and is supported by our results showing that overexpression of IME1 suppresses these sporulation defects. Our finding that overexpression of IME1 or IME2 suppresses the cpr1Δ/cpr1Δ mutant sporulation defect indicates that no other essential meiotic signaling element downstream of IME1 and IME2 is defective in mutants lacking cyclophilin A.
An effect of Cs on sporulation proved difficult to assess since sporulation was strongly inhibited by the standard Cs solvent (90% ethanol, 10% Tween 20) (data not shown). Expression of the cyclophilin A active site mutant Cpr1R53A failed to complement the sporulation defect of a cpr1Δ/cpr1Δ mutant, indicating that the prolyl-isomerase activity of cyclophilin A is required for its role in sporulation. It would be important to determine whether the R53A substitution prevents cyclophilin A binding to the Set3 complex, in addition to reduce prolyl-isomerase activity. Human CyPA also failed to complement the sporulation defect of a cpr1Δ/cpr1Δ mutant and, since human CyPA complements other Cpr1 functions (ess1 suppression, viability of zpr1 mutants, etc. [2, 3]), the lack of complementation in this case is significant. hCyPA might not interact properly with the Set3 complex or fail to localize to the yeast nucleus. These and other possibilities should be addressed in future studies.
How does cyclophilin A regulate expression of IME1 and IME2? Cpr1 is a component of the Set3 complex, and yet the meiotic phenotype of a cpr1Δ/cpr1Δ mutant is opposite to that of mutations affecting other components of the Set3C complex, with the exception of hst1. One hypothesis to explain these results is that Cpr1 is a negative regulator of the Set3 complex, and thus, in the absence of Cpr1, the remaining Set3 complex might prevent IME1 and IME2 from being properly induced. In this regard, the role of Cpr1 would be to release meiotic genes from repression by Set3 during sporulation. If this were the only role for Cpr1 during meiosis, then the sporulation defect of a cpr1Δ/cpr1Δ mutant would be imposed by the action of the Set3 complex. In this model, disruption of Set3C genes should suppress the meiotic defect of a cpr1Δ/cpr1Δ mutant and yield a phenotype indistinguishable from that of set3 mutants. In fact, the phenotype of the cpr1 set3 double mutants is more similar to the wild type than to the enhanced sporulation phenotype of the set3 mutant alone. Another way to view this result is that deletion of CPR1 partially counteracts the enhanced sporulation phenotype conferred by mutations in SNT1, SIF2, YIL112W, SET3, or HOS2, indicating that cyclophilin A is required for full expression of this phenotype and therefore plays a positive role in meiosis regulation outside of its physical interactions with the Set3 complex.
We have previously shown that Cpr1 also interacts physically with the Sin3-Rpd3 histone deacetylase complex and regulates its activity in gene silencing (3). The Sin3-Rpd3 complex is a transcriptional corepressor that is recruited by the C6 zinc-cluster protein Ume6 to the promoters of IME2 and other meiotic genes, repressing their expression during mitotic growth (1, 41, 45, 46, 79, 80). Transcriptional activation of IME2 during sporulation requires release from Sin3-mediated repression and concomitant conversion of Ume6 to an activator by interaction with Ime1 (56, 77, 90). One plausible model is that Cpr1 prolyl-isomerase activity mediates conformational changes in the Sin3-Rpd3 complex, leading to release of IME2 from transcriptional repression. We tested epistasis between CPR1 and RPD3, and between CPR1 and SIN3 by assaying sporulation of sin3Δ/sin3Δ cpr1Δ/cprΔ and rpd3Δ/rpd3Δ cpr1Δ/cpr1Δ double mutant diploid strains. These diploids exhibited the same phenotype as individual sin3Δ/sin3Δ and rpd3Δ/rpd3Δ mutants (no asci production), indicating that rpd3 and sin3 mutations are epistatic over cpr1 mutations in sporulation (data not shown). Although sin3Δ/sin3Δ and rpd3Δ/rpd3Δ individual mutants express IME1 and IME2, it has been suggested that the Sin3-Rpd3 complex is required downstream of IME1 and IME2 for adequate expression of other genes involved in asci formation, making it difficult to use epistasis analysis to study the role of Sin3 and Rpd3 in the sporulation defect of cpr1Δ/cpr1Δ or set3CΔ/set3CΔ cpr1Δ/cpr1Δ mutants. Figure 7B depicts our model in which cyclophilin A acts via Set3C and Sin3-Rpd3 to regulate meiosis.
What is the role of cyclophilin A in the Set3C histone deacetylase complex? Cyclophilin A was first shown to be associated with this complex via a proteomics mass spectrometry analysis that revealed a novel 18-kDa protein copurified with epitope-tagged, affinity-purified Set3 (67). However, in these same studies, Set3C components failed to copurify with an epitope-tagged version of cyclophilin A. In our own studies, we have confirmed that cyclophilin A is physically associated with the Set3c complex through extensive coimmunoprecipitation studies. Our studies allow us to exclude several models for how cyclophilin A might control Set3C. First, none of the Set3C components required cyclophilin A for their stable expression. Second, we found no evidence that cyclophilin A is required for assembly of the complex or for the association of specific components with the core of the complex. Our interaction studies implicate Snt1 and Sif2 as primary binding targets of cyclophilin A in the complex and also indicate that the core of the complex is comprised of four proteins—Hos2, Set3, Sif2, and Snt1—whose interactions are mutually interdependent.
There are several possible models by which cyclophilin A might regulate Set3. First, cyclophilin might control activity of the complex by catalyzing a cis-trans peptidyl-prolyl isomerization event in one subunit to effect a native state conformational switch. Alternatively, cyclophilin A might mediate conformational changes that affect Set3C interactions with chromatin or other factors. Because Set3C is a negative regulator of sporulation whereas cyclophilin A plays a positive role, the simplest hypothesis is that cyclophilin A antagonizes the negative role played by Set3C. It is important to note that all of our interaction studies were conducted under vegetative growth conditions. Either the function or the interactions of cyclophilin A with the complex might differ between vegetative and meiotic growth, and conducting mass spectrometric analysis with FLAG-tagged cyclophilin A under both growth states would be one approach to begin to address these and other models.
Cellular functions for cyclophilin A are still being elucidated. Yeast cyclophilin A activity becomes indispensable to cells defective in the essential parvulin Ess1, showing that the biological functions of Cpr1 can be unveiled in certain mutant backgrounds (3). In accord with this idea, Ansari et al. (2) discovered that cyclophilin A also becomes essential in cells harboring temperature-sensitive alleles of ZPR1, which encodes an essential zinc finger protein that redistributes between the cytoplasm and the nucleus in response to proliferation signals and interacts with the essential eukaryotic translation elongation factor 1α (29-31). Taking a similar approach, we have tested deletions of each nonessential ORF in S. cerevisiae for synthetic lethality with a cpr1Δ mutation by using a previously described method (87) and a novel high-throughput method developed by X. Pan and J. D. Boeke (66a). These studies have revealed no further genetic interactions between CPR1 and any other nonessential gene. In summary, extensive analysis reveals cyclophilin A becomes essential in cells compromised for either Zpr1 or Ess1 function and that cyclophilin A physically and functionally interacts with two different histone deacetylase complexes and plays a central role in the precise regulation of the meiotic transcriptional program during sporulation. A recent study revealed a role for the Caenorhabditis elegans nuclear cyclophilin homolog MOG-6 in germ line sex determination, suggesting that this protein is involved in the decision between mitosis and meiosis (5). Because meiosis and sporulation are central steps in the life cycle of many organisms, including budding yeast, cyclophilin A may serve a central nuclear role in the developmental transitions that yeast cells undergo to survive and evolve.
Supplementary Material
Acknowledgments
We thank Aaron Mitchell, John York, Jeremy Luban, John Rohde, and Toshiaki Harashima for plasmids and yeast strains; Bryan Cullen for discussions on NLS and NES sequences; and Emily Wenink and Cristl Arndt for excellent technical assistance.
These studies were supported by National Institutes of Health 1R01 grant AI-50438 from the NIAID to J.H. J.H. is an associate investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Anderson, S. F., C. M. Steber, R. E. Esposito, and J. E. Coleman. 1995. UME6, a negative regulator of meiosis in Saccharomyces cerevisiae, contains a C-terminal Zn2Cys6 binuclear cluster that binds the URS1 DNA sequence in a zinc-dependent manner. Protein Sci. 4:1832-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansari, H., G. Greco, and J. Luban. 2002. Cyclophilin A peptidyl-prolyl isomerase activity promotes Zpr1 nuclear export. Mol. Cell. Biol. 22:6993-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arevalo-Rodriguez, M., M. E. Cardenas, X. Wu, S. D. Hanes, and J. Heitman. 2000. Cyclophilin A and Ess1 interact with and regulate silencing by the Sin3-Rpd3 histone deacetylase. EMBO J. 19:3739-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arevalo-Rodriguez, M., X. Wu, S. D. Hanes, and J. Heitman. 2004. Prolyl isomerases in yeast. Front. Biosci. 9:2420-2446. [DOI] [PubMed] [Google Scholar]
- 5.Belfiore, M., P. Pugnale, Z. Saudan, and A. Puoti. 2004. Roles of the Caenorhabditis elegans cyclophilin-like protein MOG-6 in MEP-1 binding and germline fates. Development 131:2935-2945. [DOI] [PubMed] [Google Scholar]
- 6.Bosco, D. A., E. Z. Eisenmesser, S. Pochapsky, W. I. Sundquist, and D. Kern. 2002. Catalysis of cis/trans isomerization in native HIV-1 capsid by human cyclophilin A. Proc. Natl. Acad. Sci. USA 99:5247-5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braaten, D., C. Aberham, E. K. Franke, L. Yin, W. Phares, and J. Luban. 1996. Cyclosporine A-resistant human immunodeficiency virus type 1 mutants demonstrate that Gag encodes the functional target of cyclophilin A. J. Virol. 70:5170-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braaten, D., H. Ansari, and J. Luban. 1997. The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 71:2107-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J. Virol. 70:3551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 11.Brazin, K. N., R. J. Mallis, D. B. Fulton, and A. H. Andreotti. 2002. Regulation of the tyrosine kinase Itk by the peptidyl-prolyl isomerase cyclophilin A. Proc. Natl. Acad. Sci. USA 99:1899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breuder, T., C. S. Hemenway, N. R. Movva, M. E. Cardenas, and J. Heitman. 1994. Calcineurin is essential in cyclosporin A- and FK506-sensitive yeast strains. Proc. Natl. Acad. Sci. USA 91:5372-5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown, C. R., D.-Y. Cui, G. G.-C. Hung, and H.-L. Chiang. 2001. Cyclophilin A mediates Vid22p function in the import of fructose-1,6-bisphosphatase into Vid vesicles. J. Biol. Chem. 276:48017-48026. [DOI] [PubMed] [Google Scholar]
- 14.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. Di Como, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13:3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cardenas, M. E., E. Lim, and J. Heitman. 1995. Mutations that perturb cyclophilin A ligand binding pocket confer cyclosporin A resistance in Saccharomyces cerevisiae. J. Biol. Chem. 270:20997-21002. [DOI] [PubMed] [Google Scholar]
- 16.Cardenas, M. E., R. S. Muir, T. Breuder, and J. Heitman. 1995. Targets of immunophilin-immunosuppressant complexes are distinct highly conserved regions of calcineurin A. EMBO J. 14:2772-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang, H., and S. Lindquist. 1994. Conservation of Hsp90 macromolecular complexes in Saccharomyces cerevisiae. J. Biol. Chem. 269:24983-24988. [PubMed] [Google Scholar]
- 18.Chiu, R., O. Rey, J. Q. Zheng, J. L. Twiss, J. Song, S. Pang, and K. K. Yokoyama. 2003. Effects of altered expression and localization of cyclophilin A on differentiation of p19 embryonic carcinoma cells. Cell Mol. Neurobiol. 23:929-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis, E. S., A. Becker, J. Heitman, M. N. Hall, and M. B. Brennan. 1992. A yeast cyclophilin gene essential for lactate metabolism at high temperature. Proc. Natl. Acad. Sci. USA 89:11169-11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dolinski, K., S. Muir, M. Cardenas, and J. Heitman. 1997. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:13093-13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dolinski, K., C. Scholz, R. S. Muir, S. Rospert, F. X. Schmid, M. E. Cardenas, and J. Heitman. 1997. Functions of FKBP12 and mitochondrial cyclophilin active site residues in vitro and in vivo in Saccharomyces cerevisiae. Mol. Biol. Cell 8:2267-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dolinski, K. J., M. E. Cardenas, and J. Heitman. 1998. CNS1 encodes an essential p60/Sti1 homolog in Saccharomyces cerevisiae that suppresses cyclophilin 40 mutations and interacts with Hsp90. Mol. Cell. Biol. 18:7344-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duina, A. A., H.-C. J. Chang, J. A. Marsh, S. Lindquist, and R. F. Gaber. 1996. A cyclophilin function in Hsp90-dependent signal transduction. Science 274:1713-1715. [DOI] [PubMed] [Google Scholar]
- 24.Duina, A. A., J. A. Marsh, and R. F. Gaber. 1996. Identification of two CyP-40-like cyclophilins in Saccharomyces cerevisiae, one of which is required for normal growth. Yeast 12:943-952. [DOI] [PubMed] [Google Scholar]
- 25.Einhauer, A., and A. Jungbauer. 2001. The FLAG(TM) peptide, a versatile fusion tag for the purification of recombinant proteins. J. Biochem. Biophys. Methods 49:455-465. [DOI] [PubMed] [Google Scholar]
- 26.Eisenmesser, E. Z., D. A. Bosco, M. Akke, and D. Kern. 2002. Enzyme dynamics during catalysis. Science 295:1520-1523. [DOI] [PubMed] [Google Scholar]
- 27.Foor, F., S. A. Parent, N. Morin, A. M. Dahl, N. Ramadan, G. Chrebet, K. A. Bostian, and J. B. Nielsen. 1992. Calcineurin mediates inhibition by FK506 and cyclosporin of recovery from alpha-factor arrest in yeast. Nature 360:682-684. [DOI] [PubMed] [Google Scholar]
- 28.Franke, E. K., H. E. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 29.Galcheva-Gargova, Z., L. Gangwani, K. N. Konstantinov, M. Mikrut, S. J. Theroux, T. Enoch, and R. J. Davis. 1998. The cytoplasmic zinc finger protein ZPR1 accumulates in the nucleolus of proliferating cells. Mol. Biol. Cell 9:2963-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galcheva-Gargova, Z., K. N. Konstantinov, I.-H. Wu, F. G. Klier, T. Barrett, and R. J. Davis. 1996. Binding of zinc finger protein ZPR1 to the epidermal growth factor receptor. Science 272:1797-1802. [DOI] [PubMed] [Google Scholar]
- 31.Gangwani, L., M. Mikrut, Z. Galcheva-Gargova, and R. J. Davis. 1998. Interaction of ZPR1 with translation elongation factor-1α in proliferating cells. J. Cell Biol. 143:1471-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 33.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 34.Gothel, S. F., and M. A. Marahiel. 1999. Peptidyl-prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. Cell Mol. Life Sci. 55:423-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haendler, B., R. Keller, P. C. Hiestand, H. P. Kocher, G. Wegmann, and N. R. Movva. 1989. Yeast cyclophilin: isolation and characterization of the protein, cDNA and gene. Gene 83:39-46. [DOI] [PubMed] [Google Scholar]
- 36.Handschumacher, R. E., M. W. Harding, J. Rice, R. J. Drugge, and D. W. Speicher. 1984. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science 226:544-547. [DOI] [PubMed] [Google Scholar]
- 37.Hanes, S. D., P. R. Shank, and K. A. Bostian. 1989. Sequence and mutational analysis of ESS1, a gene essential for growth in Saccharomyces cerevisiae. Yeast 5:55-72. [DOI] [PubMed] [Google Scholar]
- 38.Hani, J., B. Schelbert, A. Bernhardt, H. Domdey, G. Fischer, K. Wiebauer, and J.-U. Rahfeld. 1999. Mutations in a peptidylprolyl-cis/trans-isomerase gene lead to a defect in 3′-end formation of a pre-mRNA in Saccharomyces cerevisiae. J. Biol. Chem. 274:108-116. [DOI] [PubMed] [Google Scholar]
- 39.Hani, J., G. Stumpf, and H. Domdey. 1995. PTF1 encodes an essential protein in Saccharomyces cerevisiae, which shows strong homology with a new putative family of PPIases. FEBS Lett. 365:198-202. [DOI] [PubMed] [Google Scholar]
- 40.Harding, M. W., A. Galat, D. E. Uehling, and S. L. Schreiber. 1989. A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature 341:758-760. [DOI] [PubMed] [Google Scholar]
- 41.Hepworth, S. R., H. Friesen, and J. Segall. 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5750-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 43.Honigberg, S. M., and K. Purnapatre. 2003. Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast. J. Cell Sci. 116:2137-2147. [DOI] [PubMed] [Google Scholar]
- 44.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 45.Kadosh, D., and K. Struhl. 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89:365-371. [DOI] [PubMed] [Google Scholar]
- 46.Kadosh, D., and K. Struhl. 1998. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18:5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalderon, D., B. L. Roberts, W. D. Richardson, and A. E. Smith. 1984. A short amino acid sequence able to specify nuclear location. Cell 39:499-509. [DOI] [PubMed] [Google Scholar]
- 48.Kassir, Y., N. Adir, E. Boger-Nadjar, N. G. Raviv, I. Rubin-Bejerano, S. Sagee, and G. Shenhar. 2003. Transcriptional regulation of meiosis in budding yeast. Int. Rev. Cytol. 224:111-171. [DOI] [PubMed] [Google Scholar]
- 49.Kassir, Y., D. Granot, and G. Simchen. 1988. IME1, a positive regulator gene of meiosis in Saccharomyces cerevisiae. Cell 52:853-862. [DOI] [PubMed] [Google Scholar]
- 50.Kumar, A., S. Agarwal, J. A. Heyman, S. Matson, M. Heidtman, S. Piccirillo, L. Umansky, A. Drawid, R. Jansen, Y. Liu, K. H. Cheung, P. Miller, M. Gerstein, G. S. Roeder, and M. Snyder. 2002. Subcellular localization of the yeast proteome. Genes Dev. 16:707-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le Hir, M., Q. Su, L. Weber, G. Woerly, A. Granelli-Piperno, and B. Ryffel. 1995. In situ detection of cyclosporin A: evidence for nuclear localization of cyclosporine and cyclophilins. Lab. Investig. 73:727-733. [PubMed] [Google Scholar]
- 52.Liu, J., M. W. Albers, T. J. Wandless, S. Luan, D. G. Alberg, P. J. Belshaw, P. Cohen, C. MacKintosh, C. B. Klee, and S. L. Schreiber. 1992. Inhibition of T-cell signaling by immunophilin-ligand complexes correlates with loss of calcineurin phosphatase activity. Biochemistry 31:3896-3901. [DOI] [PubMed] [Google Scholar]
- 53.Liu, J., J. D. Farmer, Jr., W. S. Lane, J. Friedman, I. Weissman, and S. L. Schreiber. 1991. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66:807-815. [DOI] [PubMed] [Google Scholar]
- 54.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 55.Mallis, R. J., K. N. Brazin, D. B. Fulton, and A. H. Andreotti. 2002. Structural characterization of a proline-driven conformational switch within the Itk SH2 domain. Nat. Struct. Biol. 9:900-905. [DOI] [PubMed] [Google Scholar]
- 56.Mandel, S., K. Robzyk, and Y. Kassir. 1994. IME1 gene encodes a transcription factor which is required to induce meiosis in Saccharomyces cerevisiae. Dev. Genet. 15:139-147. [DOI] [PubMed] [Google Scholar]
- 57.Marks, W. H., M. W. Harding, R. Handschumacher, C. Marks, and M. I. Lorber. 1991. The immunochemical distribution of cyclophilin in normal mammalian tissues. Transplantation 52:340-345. [DOI] [PubMed] [Google Scholar]
- 58.Marsh, J. A., H. M. Kalton, and R. F. Gaber. 1998. Cns1 is an essential protein associated with the Hsp90 chaperone complex in Saccharomyces cerevisiae that can restore cyclophilin 40-dependent functions in cpr7Δ cells. Mol. Cell. Biol. 18:7353-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matouschek, A., S. Rospert, K. Schmid, B. Glick, and G. Schatz. 1995. Cyclophilin catalyzes protein folding in yeast mitochondria. Proc. Natl. Acad. Sci. USA 92:6319-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-306. [DOI] [PubMed] [Google Scholar]
- 61.Mayrleitner, M., A. P. Timerman, G. Wiederrecht, and S. Fleischer. 1994. The calcium release channel of sarcoplasmic reticulum is modulated by FK-506 binding protein: effect of FKBP-12 on single channel activity of the skeletal muscle ryanodine receptor. Cell Calcium 15:99-108. [DOI] [PubMed] [Google Scholar]
- 62.Messenguy, F., F. Vierendeels, B. Scherens, and E. Dubois. 2000. In Saccharomyces cerevisiae, expression of arginine catabolic genes CAR1 and CAR2 in response to exogenous nitrogen availability is mediated by the Ume6 (CargRI)-Sin3 (CargRII)-Rpd3 (CargRIII) complex. J. Bacteriol. 182:3158-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mitchell, A. P., S. E. Driscoll, and H. E. Smith. 1990. Positive control of sporulation-specific genes by the IME1 and IME2 products in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:2104-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Muhlrad, D., R. Hunter, and R. Parker. 1992. A rapid method for localized mutagenesis of yeast genes. Yeast 8:79-82. [DOI] [PubMed] [Google Scholar]
- 65.Myers, J. K., D. P. Morris, A. L. Greenleaf, and T. G. Oas. 2001. Phosphorylation of RNA polymerase II CTD fragments results in tight binding to the WW domain from the yeast prolyl isomerase Ess1. Biochemistry 40:8479-8486. [DOI] [PubMed] [Google Scholar]
- 66.Nakamura, T., Y. Liu, D. Hirata, H. Namba, S. Harada, T. Hirokawa, and T. Miyakawa. 1993. Protein phosphatase type 2B (calcineurin)-mediated, FK506-sensitive regulation of intracellular ions in yeast is an important determinant for adaptation to high salt stress conditions. EMBO J. 12:4063-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66a.Pan, X., D. S. Yuan, D. Xiong, X. Wang, S. Sokhai-Mahadeo, J. S. Bader, P. Hieter, F. Spencer, and J. D. Boeke. 2004. A robust toolkit for functional profiling of the yeast genome. Mol. Cell 16:487-496. [DOI] [PubMed] [Google Scholar]
- 67.Pijnappel, W. W., D. Schaft, A. Roguev, A. Shevchenko, H. Tekotte, M. Wilm, G. Rigaut, B. Seraphin, R. Aasland, and A. F. Stewart. 2001. The Saccharomyces cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 15:2991-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prasher, D. C. 1995. Using GFP to see the light. Trends Genet. 11:320-323. [DOI] [PubMed] [Google Scholar]
- 69.Rahfeld, J. U., K. P. Rucknagel, B. Schelbert, B. Ludwig, J. Hacker, K. Mann, and G. Fischer. 1994. Confirmation of the existence of a third family among peptidyl-prolyl cis/trans isomerases: amino acid sequence and recombinant production of parvulin. FEBS Lett. 352:180-184. [DOI] [PubMed] [Google Scholar]
- 70.Rubin-Bejerano, I., S. Mandel, K. Robzyk, and Y. Kassir. 1996. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional represssor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol. Cell. Biol. 16:2518-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ryffel, B., G. Woerly, B. Greiner, B. Haendler, M. J. Mihatsch, and B. M. Foxwell. 1991. Distribution of the cyclosporine binding protein cyclophilin in human tissues. Immunology 72:399-404. [PMC free article] [PubMed] [Google Scholar]
- 72.Scholz, C., P. Maier, K. Dolinski, J. Heitman, and F. X. Schmid. 1999. R73A and H144Q mutants of the yeast mitochondrial cyclophilin Cpr3 exhibit a low prolyl isomerase activity in both peptide and protein-folding assays. FEBS Lett. 443:367-369. [DOI] [PubMed] [Google Scholar]
- 73.Scholz, C., T. Schindler, K. Dolinski, J. Heitman, and F. X. Schmid. 1997. Cyclophilin active site mutants have native prolyl isomerase activity with a protein substrate. FEBS Lett. 414:69-73. [DOI] [PubMed] [Google Scholar]
- 74.Sherman, F. 2002. Getting started with yeast. Methods Enzymol. 350:3-41. [DOI] [PubMed] [Google Scholar]
- 75.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Siligardi, G., B. Panaretou, P. Meyer, S. Singh, D. N. Woolfson, P. W. Piper, L. H. Pearl, and C. Prodromou. 2002. Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50cdc37. J. Biol. Chem. 277:20151-20159. [DOI] [PubMed] [Google Scholar]
- 77.Smith, H. E., S. E. Driscoll, R. A. Sia, H. E. Yuan, and A. P. Mitchell. 1993. Genetic evidence for transcriptional activation by the yeast IME1 gene product. Genetics 133:775-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith, H. E., and A. P. Mitchell. 1989. A transcriptional cascade governs entry into meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:2142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Steber, C. M., and R. E. Esposito. 1995. UME6 is a central component of a developmental regulatory switch controlling meiosis-specific gene expression. Proc. Natl. Acad. Sci. USA 92:12490-12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Strich, R., R. Surosky, C. Steber, E. Dubois, F. Messenguy, and R. Esposito. 1994. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 8:796-810. [DOI] [PubMed] [Google Scholar]
- 81.Takahashi, N., T. Hayano, and M. Suzuki. 1989. Peptidyl-prolyl cis-trans isomerase is the cyclosporin A-binding protein cyclophilin. Nature 337:473-475. [DOI] [PubMed] [Google Scholar]
- 82.Talcott, B., and M. S. Moore. 1999. Getting across the nuclear pore complex. Trends Cell Biol. 9:312-318. [DOI] [PubMed] [Google Scholar]
- 83.Tesic, M., J. A. Marsh, S. B. Cullinan, and R. F. Gaber. 2003. Functional interactions between Hsp90 and the co-chaperones Cns1 and Cpr7 in Saccharomyces cerevisiae. J. Biol. Chem. 278:32692-32701. [DOI] [PubMed] [Google Scholar]
- 84.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Gottlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363-365. [DOI] [PubMed] [Google Scholar]
- 85.Timerman, A. P., E. Ogunbumni, E. Freund, G. Wiederrecht, A. R. Marks, and S. Fleischer. 1993. The calcium release channel of sarcoplasmic reticulum is modulated by FK-506-binding protein. Dissociation and reconstitution of FKBP-12 to the calcium release channel of skeletal muscle sarcoplasmic reticulum. J. Biol. Chem. 268:22992-22999. [PubMed] [Google Scholar]
- 86.Timerman, A. P., G. Wiederrecht, A. Marcy, and S. Fleischer. 1995. Characterization of an exchange reaction between soluble FKBP-12 and the FKBP-ryanodine receptor complex. J. Biol. Chem. 270:2451-2459. [DOI] [PubMed] [Google Scholar]
- 87.Tong, A. H. Y., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. V. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 88.Tropschug, M., I. B. Barthelmess, and W. Neupert. 1989. Sensitivity to cyclosporin A is mediated by cyclophilin in Neurospora crassa and Saccharomyces cerevisiae. Nature 342:953-955. [DOI] [PubMed] [Google Scholar]
- 89.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 90.Washburn, B. K., and R. E. Esposito. 2001. Identification of the Sin3-binding site in Ume6 defines a two-step process for conversion of Ume6 from a transcriptional repressor to an activator in yeast. Mol. Cell. Biol. 21:2057-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]
- 92.Williams, R. M., M. Primig, B. K. Washburn, E. A. Winzeler, M. Bellis, C. Sarrauste de Menthiere, R. W. Davis, and R. E. Esposito. 2002. The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc. Natl. Acad. Sci. USA 99:13431-13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu, X., A. Chang, M. Sudol, and S. D. Hanes. 2001. Genetic interactions between the ESS1 prolyl-isomerase and the RSP5 ubiquitin ligase reveal opposing effects on RNA polymerase II function. Curr. Genet. 40:234-242. [DOI] [PubMed] [Google Scholar]
- 94.Wu, X., A. Rossettini, and S. D. Hanes. 2003. The ESS1 prolyl isomerase and its suppressor BYE1 interact with RNA Pol II to inhibit transcription elongation in Saccharomyces cerevisiae. Genetics 165:1687-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu, X., C. B. Wilcox, G. Devasahayam, R. L. Hackett, M. Arevalo-Rodriguez, M. E. Cardenas, J. Heitman, and S. D. Hanes. 2000. The Ess1 prolyl isomerase is linked to chromatin remodeling complexes and the general transcription machinery. EMBO J. 19:3727-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang, W.-M., C. J. Inouye, and E. Seto. 1995. Cyclophilin A and FKBP12 interact with YY1 and alter its transcriptional activity. J. Biol. Chem. 270:15187-15193. [DOI] [PubMed] [Google Scholar]
- 97.Yoshida, M., H. Kawaguchi, Y. Sakata, K. Kominami, M. Hirano, H. Shima, R. Akada, and I. Yamashita. 1990. Initiation of meiosis and sporulation in Saccharomyces cerevisiae requires a novel protein kinase homologue. Mol. Gen. Genet. 221:176-186. [DOI] [PubMed] [Google Scholar]
- 98.Zydowsky, L. D., F. A. Etzkorn, H. Y. Chang, S. B. Ferguson, L. A. Stolz, S. I. Ho, and C. T. Walsh. 1992. Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci. 1:1092-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.