Abstract
Cdc28p is the major cyclin-dependent kinase in Saccharomyces cerevisiae. Its activity is required for blocking the reinitiation of DNA replication during mitosis. Here, we show that under conditions where Cdc28p activity is improperly regulated—either through the loss of function of the Schizosaccharomyces pombe wee1 ortholog Swe1p or through the expression of a dominant CDC28 allele, CDC28AF—diploid yeast cells are able to complete several rounds of premeiotic DNA replication within a single meiotic cell cycle. Moreover, a percentage of mutant cells exhibit a “multispore” phenotype, possessing the ability to package more than four spores within a single ascus. These multispored asci contain both even and odd numbers of viable spores. In order for meiotic rereplication and multispore formation to occur, cells must initiate homologous recombination and maintain proper chromosome cohesion during meiosis I. Rad9p- or Rad17p-dependent checkpoint mechanisms are not required for multispore formation and neither are the B-type cyclin Clb6p and the cyclin-dependent kinase inhibitor Sic1p. Finally, we present evidence of a possible role for a Cdc55p-dependent protein phosphatase 2A in initiating meiotic replication.
Meiosis is a complex reorganization of the cell cycle that differs from mitosis in that it consists of a single round of DNA replication followed by two nuclear divisions. The cyclin-dependent kinase (Cdk) Cdc28p associates with various cyclins (Cln or Clb) in order to drive the meiotic process in Saccharomyces cerevisiae. For example, it is believed that Clb5p or Clb6p complexes with Cdc28p to complete premeiotic S phase (46), whereas the Cdc28p-Clb1p protein complex drives the cell out of the recombination checkpoint (40). During mitosis, the Clb-Cdc28p complex plays a role in pre-replication complex (pre-RC) formation and pre-RC disassembly and may be required to regulate the replication machinery (7, 10, 38).
In addition to its role in mitotic DNA replication initiation, Cdc28p activity also seems to be required at several points in S phase to prevent multiple rounds of DNA replication. Nguyen et al. (38) have evidence that Cdc28p-dependent phosphorylation of certain Orc proteins, down-regulation of Cdc6p (a Cln-Cdc28p-dependent function [9]), and nuclear exclusion of Mcm proteins after replication is initiated must all occur together to prevent DNA replication reinitiation. Thus, properly regulating Cdc28p activity during S phase is necessary for both the initiation of DNA replication and the inhibition of a second round of DNA replication. Whether meiotic replication possesses these same mechanisms to regulate initiation firing, and whether Cdc28p regulates these events, is not known.
Proteins other than the cyclin family regulate Cdc28p activity at various times in the cell cycle. Sic1p is an inhibitor of yeast Cdk activity and is required for proper assembly of pre-RCs (26). Sic1p is required to block Cdc28p from prematurely interacting with substrates, such as the ORC (50); thus, it inhibits DNA replication and plays a role in ensuring that origins fire only once during mitotic S phase. The Wee1p ortholog, Swe1p, also inhibits Cdc28p activity through inhibitory phosphorylation at the conserved tyrosine 19 residue of the kinase (29). Swe1p inhibits Cdc28p activity during mitosis prior to spindle pole body separation (29, 33). During meiosis, Swe1p regulates exit from pachytene, the meiotic stage in which recombination is completed, presumably by inhibiting Clb-Cdc28p activity (40).
Currently, the evidence pointing to a role for Cdc28p in initiating premeiotic DNA replication is mixed. ts alleles of cdc28 arrest in meiosis at pachytene, while they arrest in mitosis at G1 (33). Cells with CLB5/6 deleted arrest at G1, implicating Cdc28p in meiotic replication (44, 46). By using the “heat-inducible” Degron system, Guttmann-Raviv et al. (13) demonstrated that Cdc28p was not required for premeiotic replication but that the Cdk-like meiotic kinase Ime2p was required for the correct timing of replication, as well as nuclear divisions and ascus formation. Work from Dirick et al. (8) suggests that the function of Cdk-like Ime2p is to stimulate the degradation of the Cdk inhibitor Sic1p, thus activating a Clb5p/6p-dependent Cdk that can be either Cdc28p or Ime2p itself. A recent report from the laboratory of Ira Herskowitz demonstrated that both Cdc28p and Ime2p have critical roles during meiotic S phase (1).
In an effort to further understand how Cdc28p regulates meiotic DNA replication and uncover regulatory controls governing this event, we have begun to characterize the “multispore” phenotype of cells homozygous for either an activated allele of CDC28 or deletion of SWE1. We find that mutant cells are able to initiate multiple rounds of replication and form multispored asci that in most cases contain between five and eight viable spores. The multispored phenotype requires that homologous recombination be initiated; loss of Rec102p or Spo11p function causes loss of multispore formation in swe1 and CDC28AF cells. There is an additional requirement for Spo13p, suggesting that maintaining proper chromosome cohesion during meiosis I is important for rereplication initiation. Rereplication does not require the RAD9- or RAD17-dependent checkpoints (32, 40, 48, 49) or Sic1p function (34, 52). Finally, we have uncovered a possible role for protein phosphatase 2A (PP2A) in regulating meiotic replication initiation.
MATERIALS AND METHODS
Strain construction and plasmids.
The yeast strains used in this study were derived from the W303 strain background. Yeast transformations were performed using the procedure described by Ito et al. (18). For routine propagation of plasmids, Escherichia coli XL1-Blue cells were used and grown in Luria-Bertani medium supplemented with ampicillin (200 μg/ml). Yeast haploid null mutants were generated by the one-step disruption method of Rothstein (41) with the KanMX cassette (31) for swe1, sic1, spo11, rec102, rad9, rad17, and spo13 strains and pRS406-cdc55::URA3 for cdc55 strains. pRS405-CDC28AF was constructed as previously described (27); alanine and phenylalanine replaced threonine 18 and tyrosine 19, respectively.
Media, growth conditions, and miscellaneous microbial techniques.
Yeast strains were grown in YEPD (1% yeast extract, 2% Bacto peptone, 2% glucose), YPA (1% yeast extract, 2% Bacto peptone, 1% potassium acetate), or SP (0.1% yeast extract, 1% potassium acetate, 0.05% glucose, 20 mg of leucine/liter, 20 mg of tryptophan/liter, 20 mg of histidine/liter, 20 mg of adenine/liter) medium. For sporulation studies, cells were grown to 107 cells/ml at 30°C in YEPD, shifted to YPA for several generations to increase synchrony, and subsequently transferred to SP medium. These growth conditions were required, since cdc55 cells harbor a severe growth defect when grown in the presence of acetate. Data were collected using a Hamamatsu DIG-15 charge-coupled device digital camera and Open Labs software (version 2.1). Final fluorescence images were generated using Adobe Photoshop (version 4.0). Tetrads were dissected using standard methods.
Total RNA isolation.
All solutions were prepared with diethyl pyrocarbonate-treated water. Cells were harvested at various times during sporulation, centrifuged, and pelleted for 30 s. Cell pellets were resuspended in 200 μl of YRL buffer (200 mM Tris [pH 7.5], containing 500 mM NaCl, 10 mM EDTA, and 1% sodium dodecyl sulfate [SDS]) and 200 μl of phenol-chloroform-isoamyl alcohol (PCIAA). Two hundred microliters of nitric acid-washed beads was added, and cells were vortexed for 2.5 min. Three hundred microliters of YRL buffer and 200 μl of PCIAA were then added, and cells were vortexed a second time for an additional 2.5 min. Lysed cells were then centrifuged for 5 min, and the resulting clear lysate was removed and added to 400 μl of PCIAA, vortexed for an additional 2.5 min, and subsequently centrifuged for 5 min. The resulting aqueous layer was added to 500 μl of 100% ethyl alcohol (EtOH), and total RNA was precipitated overnight at −20°C. A total RNA pellet was obtained by centrifugation for 15 min at 4°C, washed twice with 70% EtOH, vacuum dried, and subsequently resuspended in water. Total RNA was stable at −20°C for several weeks.
Northern analysis.
Total RNA was resolved using 6% formaldehyde agarose gels. Twenty micrograms of total RNA in loading buffer (20 mM morpholinepropanesulfonic acid [pH 7.0] containing 10 mM sodium acetate, 2 mM EDTA, 45% formamide, 6% formaldehyde, 1% ethidium bromide, 0.003% bromophenol blue, 0.03% xylene cyanol FF, and 1.5% Ficoll) was used for each sample. Total RNA was blotted onto Hybond-N nitrocellulose (Amersham) overnight at room temperature with 10× SSC (40 mM sodium citrate and 1.8 M NaCl). Church's buffer (500 ml of sodium phosphate buffer, 2 ml of EDTA, 10 g of bovine serum albumin, 70 g of SDS; total volume = 1 liter in distilled water) was used for all hybridization procedures. Hybridization was performed overnight at 65°C. Gel-purified radiolabeled probes were boiled in 200 μl of salmon sperm DNA prior to use. Posthybridization, blots were washed twice in 2× SSC at room temperature, twice in 2× SSC-0.5% SDS at 65°C, and twice in 0.1× SSC at room temperature. Gene expression was determined by autoradiography with X-OMAT film (Kodak). mRNA expression levels were quantified by densitometry with a Bio-Rad Model GS-670 Imaging Densitometer and Molecular Analyst software version 1.4.1.
Fluorescence-activated cell sorting (FACS) analysis.
One milliliter of cells was harvested and pelleted for each time point. Cells were resuspended in 200 μl of 1× phosphate-buffered saline (PBS) and fixed on ice for 1 h with 70% EtOH. After being washed once with 1× PBS, cells were then centrifuged and subsequently treated with RNase solution (1 mg of RNase/ml) for 10 min at 37°C. DNA was stained using propidium iodide solution (5 μg of propidium iodide/ml in 1× PBS) by incubation at 4°C for 20 min.
DAPI staining of DNA.
Cells were fixed in solution A (2.7% formaldehyde and 0.03% Triton X-100) for 30 min at room temperature. Cells were then gently pelleted and subsequently resuspended and incubated in solution B (35 mM sodium phosphate, 5 mM MgCl2, 4% formaldehyde.) for 1 h at room temperature. After being washed three times with 1× PBS, cells were then resuspended in PBS containing DAPI (4′,6′-diamidino-2-phenylindole; 0.10 μg/ml) and incubated on ice for 20 min. Cells were washed three to five times with PBS to remove residual DAPI, and nuclei were visualized by fluorescence microscopy with a Leica DRME fluorescence microscope, UV optics, and a PlanAPO 100× objective.
TetR-GFP analysis.
Various diploid strains heterozygous for the ura3::3xURA3tetO112 locus and expressing a TetR-green fluorescent protein (GFP) fusion protein (35) were made to adhere to poly-l-lysine slides. DAPI solution (0.10 μg/ml) was added and incubated with cells for 15 min at room temperature in the dark. Cells were washed several times with PBS. Chromosomes were visualized through examining the binding of the Tet repressor-GFP fusion to the Tet operator array by fluorescence microscopy and GFP optics. In Table 3, tetO rereplication was determined by calculating the number of TetR-GFP foci within a single nucleus. Nuclei were visualized using DAPI. The values presented are percentages and were determined at 12 h post-induction of sporulation. They represent the average of two independent experiments. An average of 350 cells was examined for each strain. In Table 4, the number of spores containing a GFP-TetR focus was determined at 24 h after inducing sporulation.
TABLE 3.
Intra- and intergenic recombination in wild-type, swe1, and CDC28AF cells
| Strain | Mediuma | Frequency of intragenic recombinants (10−6)b | Frequency (%) of intergenic recombinants
|
|
|---|---|---|---|---|
| CEN3 to MATa/MATαc | CEN2 to HIS3/LYS2d | |||
| SWE1/SWE1 CDC28/CDC28 | Glucose | 2.0 ± 0.1 | 18.5 ± 1.1 | 31.5 ± 2.4 |
| Spore | 650 ± 22 | |||
| swe1/swe1 CDC28/CDC28 | Glucose | 1.8 ± 0.3 | 24.5 ± 1.6 | 27.6 ± 1.4 |
| Spore | 595 ± 17 | |||
| SWE1/SWE1 CDC28AF/CDC28AF | Glucose | 3.8 ± 0.1 | 16.8 ± 1.3 | 29.8 ± 1.8 |
| Spore | 550 ± 30 | |||
Glucose, YEPD; Spore, SP (see Materials and Methods).
Trp+ recombinants per CFU determined at 16 h.
Frequency values were determined using the equation (number of diploid maters × 2)/number of Trp+ diploid recombinants.
Frequency values were determined using the equation (number of His− or Lys− diploid cells × 2)/number of Trp+ diploid recombinants.
TABLE 4.
Genetic requirements for multispore formation
| Mutation | No. of cells forming no. of sporesa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
SWE1/SWE1 CDC28/CDC28
|
swe1/swe1 CDC28/CDC28
|
SWE1/SWE1 CDC28AF/CDC28AF
|
|||||||
| 2 | 4 | >4 | 2 | 4 | >4 | 2 | 4 | >4 | |
| WTc | 7 | 63 | NDb | 8 | 52 | 15 | 3 | 71 | 23 |
| sic1 | <1 | 21 | ND | 2 | 15 | 12 | 5 | 18 | 17 |
| clb5 | <1 | <1 | ND | <1 | <1 | ND | <1 | <1 | ND |
| clb6 | 4 | 62 | ND | 6 | 49 | 17 | 6 | 62 | 21 |
| spo11 | 21 | 47 | ND | 3 | 40 | ND | 2 | 36 | ND |
| spo13 | 45 | <1 | ND | 53 | <1 | ND | 55 | <1 | ND |
| rec102 | 4 | 32 | ND | 2 | 27 | <1 | 3 | 29 | <1 |
| rad9 | 1 | 59 | ND | 4 | 56 | 19 | 2 | 68 | 22 |
| rad17 | 5 | 61 | ND | 6 | 54 | 14 | 7 | 62 | 25 |
| cdc55 | <1 | <1 | ND | <1 | <1 | ND | <1 | <1 | ND |
2, dyads; 4, tetrads; >4, multispores.
ND, none detected.
WT, wild type.
Homologous recombination assays.
All recombination assays were performed on homozygous deletion strains that were constructed using the W303 haploid strains YJN699 [MATa ade2-1 can1::ADE2:CAN1 his3-11,15 lys2(3′del):HIS3:lys2(5′del) leu2-3,112 trp1-1 ura3-1] and YJN700 (MATα ade2-1 can1::ADE2:CAN1 his3-11,15 leu2-3,112 trp1-3′del ura3-1) first described by Lee and Honigberg (16, 25). Intra- and intergenic recombination frequencies were measured using “return to growth” assays (15). The frequency of intragenic recombination between trp1 heteroalleles was measured by determining the number of Trp+ prototrophs. Intergenic recombination frequencies at CEN3 to MATa/MATα and CEN2 to HIS3/LYS2 intervals were determined using prototrophy selection and mating assays as described previously (16, 25).
RESULTS
Diploid yeast cells lacking Cdc28p inhibitory activity form multispores.
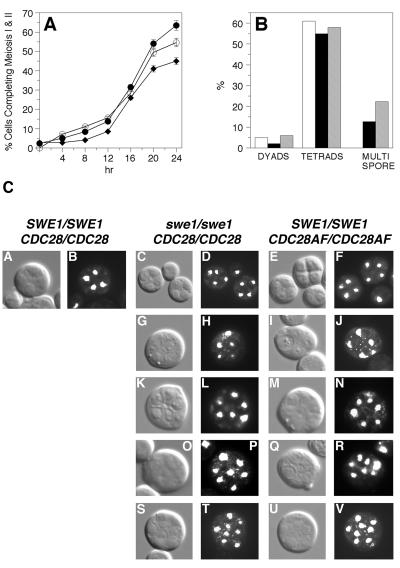
We generated diploid yeast strains lacking Cdc28p inhibitory activity and tested their ability to initiate various meiotic events. Diploid cells lacking SWE1 or expressing the dominant CDC28AF allele were induced to sporulate, and the completion of meiosis I and II was monitored by DAPI staining of nuclei and subsequent fluorescence microscopy. As shown in Fig. 1A, the number of swe1 and CDC28AF cells completing meiosis I and II was similar to that seen in wild-type cells. More importantly, we found that a significant percentage of swe1 (>12%) and CDC28AF (22%) cells were able to form asci that contained greater than four spores (Fig. 1B), whereas this phenotype was not seen in wild-type cells.
FIG. 1.
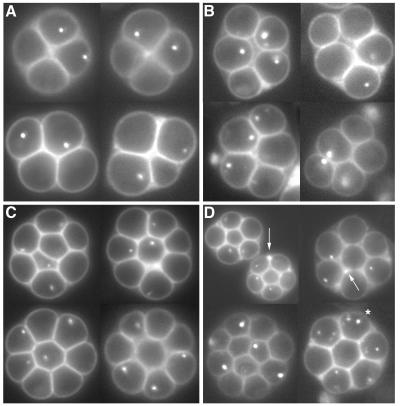
Deregulated Cdc28p activity results in multispore formation. (A) Time-dependent progression through meiosis I and II. Cells were stained with DAPI and visualized by fluorescence microscopy and Nomarski optics. Closed circles, wild type; diamonds, swe1 cells; open circles, CDC28AF cells. (B) At 24 h, cells were fixed in formaldehyde and stained with DAPI, and the percentage of dyads, tetrads, and multispores was determined by fluorescence microscopy with UV optics. Open bars, wild type; black bars, swe1 cells; gray bars, CDC28AF cells. (C) At 24 h, cells were fixed in formaldehyde and then stained with DAPI. Representative images for tetrads (A to F) and asci containing five (G to J), six (K to N), seven (O to R), and eight (S to V) spores were taken using a Leica DRME fluorescence microscope, UV optics, and a PlanAPO 100× objective.
We found that not all multispored asci contained even-numbered spores that were divisible by four (Fig. 1C). The greatest percentage of multispored asci contained either five (pentispored asci) or eight (octispored asci) spores (Fig. 1C, panels G to J and S to V). However, we did routinely observe a small percentage containing six (panels K to N) or seven (panels O to R) spores, and occasionally there were asci with nine spores or higher (data not shown). All spores contained DNA as evidenced by DAPI staining and when examined by light microscopy appeared to be mature.
To determine spore viability, we dissected tetrads, pentispored asci, and octispored asci, with at least 200 spores analyzed for each ascus category. Other multispore ascus categories were very difficult to distinguish by light microscopy and thus were not characterized further. The percentage of viable tetrad spores was 97% for our W303 wild-type strain. The percentage of viable spores in tetrad asci, pentispored asci, and octispored asci from CDC28AF sporulating cells was 95.8, 88.3, and 90.8%, respectively. For the same categories swe1 cells had 81.9, 79.2, and 78.1% viability, respectively. Genotyping of mutant spores demonstrated that the majority were haploid cells that were generated through normal Mendelian segregation of chromosomes. However, we did find that some multispored asci produced nonmating spores; this phenotype was most prevalent in swe1 mutant cells (∼15% of the spores from pentispored asci and ∼12% from octispored asci). Approximately 4% of the spores analyzed from CDC28AF multispored asci exhibited a nonmating phenotype. All nonmaters tested were able to sporulate and form multispored asci, demonstrating that these cells were diploid.
Expression of meiotic genes in swe1 and CDC28AF cells is not drastically altered.
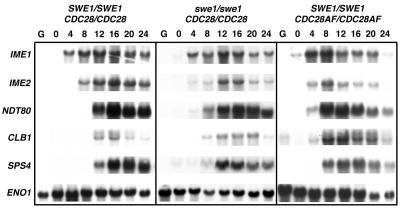
The proper temporal expression of yeast meiosis-specific genes is essential for the correct timing of initiation and completion of sporulation (36). To begin to ascertain why swe1 or CDC28AF cells form multispored asci, we first determined the expression patterns of early, middle, and late meiotic genes. The IME1 and IME2 genes encode a transcriptional activator and Cdk-like kinase, respectively (36). Both are required for early meiotic gene expression and thus are induced very early in meiosis. We found that the timing and levels of IME1 expression in swe1 and CDC28AF cells were similar to those seen for wild-type cells (Fig. 2). IME2 was also induced in all cell types; however, its expression was slightly delayed in swe1 cells (Fig. 2). NDT80 is an early-middle spore gene that encodes a transcription factor required for middle and late spore gene expression (53). We observed that NDT80 expression levels in both mutant cells were similar to those of the wild type. CDC28AF cells did induce the expression of this gene earlier than did either wild-type or swe1 cells. Moreover, Ndt80p was functional, as we observed the Ndt80p-dependent expression of the B-type cyclin gene CLB1. Finally, we found that the expression of the mid-late spore gene SPS4 was induced in all cells examined. The expression of CLB1 and SPS4 was induced earlier in CDC28AF cells. Densitometry analyses of expression levels, compared to ENO1 loading controls, indicated that the levels of all genes analyzed, with the exception of CLB1, were similar in all strains tested. CLB1 levels were sustained longer in swe1 and CDC28AF cells. Thus, the meiotic gene expression patterns in cells harboring defective Cdc28p regulation were similar, but not identical, to that observed in wild-type cells.
FIG. 2.
Cdc28p deregulated cells do not harbor defects in the temporal expression of meiotic genes. Cells were harvested, and total RNA was obtained at the indicated times prior to and post-sporulation induction. Formaldehyde gels were used to resolve RNA, and Northern blot analysis with 32P-labeled probes was used to determine meiotic gene expression levels. The ENO1 gene serves as a loading control.
Multiple rounds of meiotic DNA replication are initiated in swe1 and CDC28AF cells.
Our fluorescence microscopy and tetrad analysis data suggested to us that multiple rounds of DNA replication may have occurred in swe1 and CDC28AF sporulating cells. Each spore in all multispored asci was stained with DAPI (Fig. 1C), and we did find high levels of spore viability. Thus, we determined whether this was the case by examining premeiotic DNA replication in our cells by two different methods. First, we performed FACS analysis on sporulating cells. Second, we constructed strains harboring multiple tandem repeats of the tetO repressor sequence and expressing a TetR-GFP repressor protein and examined replication through the binding of TetR-GFP to chromosomes (35).
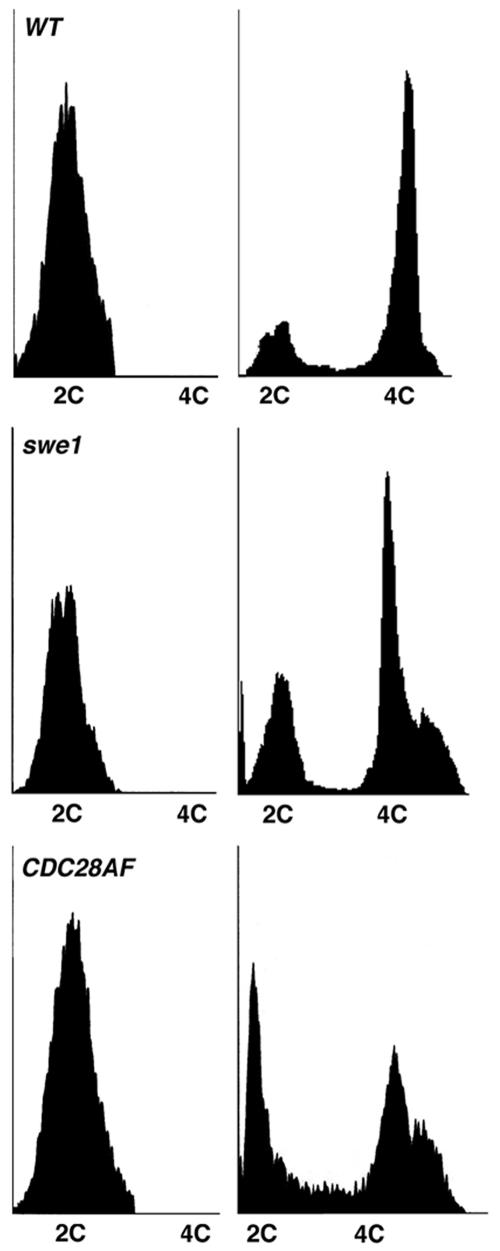
Using FACS analysis, we routinely observed a 4C peak in wild-type cells around 8 h into sporulation, with the majority of cells completing replication by 24 h (Fig. 3). CDC28AF and swe1 cells showed similar replication kinetics but by 24 h had an additional >4C peak, possibly indicative of cells initiating at least one additional round of replication.
FIG. 3.
A subset of Cdc28p deregulated cells have a >4C DNA content post-initiation of meiotic replication. Cells were harvested at 0 h (left panels) and 24 h (right panels) post-sporulation induction, fixed in 70% ethanol, and stained with propidium iodide at 4°C for 1 h. Samples were analyzed by FACS. WT, wild type.
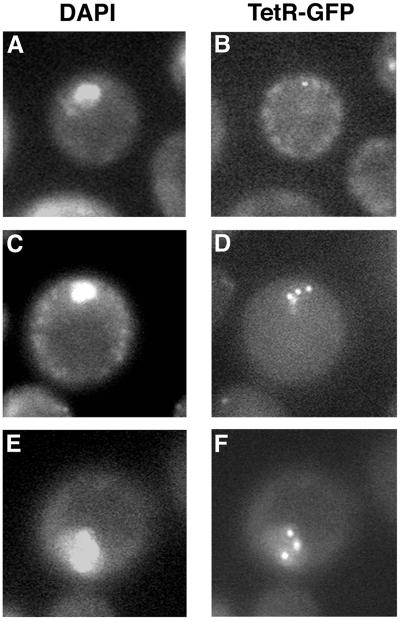
To definitively show that mutant cells had indeed initiated multiple rounds of replication, we directly examined the number of times that the tetO repressor sequence, which had been integrated at a single URA3 locus, was replicated in cells. All strains tested were heterozygous for tetO repressor sequences. In replicating wild-type cells, we routinely observed a single fluorescently marked chromosome (TetR-GFP foci) within a single nucleus at around the time that replication was completed (Fig. 4A and B). We concluded that these cells had gone through a single round of replication, and our inability to see two foci was due to the fact that sister chromatids were still attached. Once meiotic replication was completed, two separate TetR-GFP foci within a single nucleus became visible in wild-type cells just prior to the completion of meiosis I (Table 1). As meiosis progressed, the majority of these foci segregated to two of the four spores within an ascus (Table 2; Fig. 5A). We found that a percentage of swe1 (Fig. 4C and D) and CDC28AF (Fig. 4E and F) cells contained three or more TetR-GFP foci within a single nucleus (Table 1). These segregated into individual spores, as we observed multispored asci containing three or more TetR-GFP foci (Table 2; Fig. 5B to D). Interestingly, in a small percentage of spores, we observed foci that were not packaged into spores (Fig. 5D) or individual spores containing multiple foci (Fig. 5D, asterisk). We also saw that pentispored asci generated from swe1 or CDC28AF cells could contain either two or three individual TetR-GFP foci within five spores (Fig. 5B). Based on these results, we conclude that tetO repressor sequences integrated at URA3 had undergone at least two or more additional replication cycles during a single meiotic S phase.
FIG. 4.
TetO repeats are rereplicated in Cdc28p deregulated cells. Live cells were harvested at 12 h post-sporulation induction and incubated with DAPI. Nuclei (A, C, and E) and TetR-GFP bound to TetO repeats (B, D, and F) were visualized by fluorescence microscopy with UV and fluorescein isothiocyanate optics, respectively. (A and B) Wild type; (C and D) swe1 cells; (E and F) CDC28AF cells.
TABLE 1.
Rereplication of TetO in cells harboring defects in Cdc28p regulation
| Strain | % of cells having no. of GFP foci within a single nucleus
|
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| SWE1 CDC28/SWE1 CDC28 | 34.0 | 66.0 | 0.0 | 0.0 | 0.0 |
| swe1 CDC28/swe1 CDC28 | 30.0 | 50.7 | 9.5 | 6.7 | 3.1 |
| SWE1 CDC28AF/SWE1 CDC28AF | 32.5 | 51.0 | 10.2 | 4.8 | 1.5 |
TABLE 2.
Distribution of TetO within multispores
| Strain | % of cells with:
|
|||||
|---|---|---|---|---|---|---|
| 1/1a | 2/4b | 3/4b | 2/5b | 3/5b | 4/8b | |
| SWE1 CDC28/SWE1 CDC28 | 30.5 | 69.5 | 0.0 | 0.0 | 0.0 | 0.0 |
| swe1 CDC28/swe1 CDC28 | 24.0 | 56.0 | 5.4 | 3.7 | 5.7 | 5.0 |
| SWE1 CDC28AF/SWE1 CDC28AF | 21.3 | 58.7 | 4.7 | 3.0 | 4.7 | 6.8 |
Number of GFP foci/number of nuclei.
Number of spores containing a single GFP focus/number of spores within an ascus.
FIG. 5.
Cdc28p deregulated cells produce multispores that contain rereplicated TetO repeats. Live cells were harvested at 24 h post-sporulation induction. TetR-GFP bound to TetO repeats was visualized by fluorescence microscopy with fluorescein isothiocyanate optics. (A) Wild-type tetrads; (B) pentispored asci; (C) octispored asci; (D) multispored asci. The arrows in panel D indicate TetR-GFP foci not packaged into a spore. The asterisk in panel D denotes a multispore that contains multiple TetR-GFP foci within single spores.
Homologous recombination events are initiated in swe1 and CDC28AF cells.
Evidence suggests that meiotic DNA replication and homologous recombination are linked events (3, 12, 44). Since we observed rereplication block defects in swe1 and CDC28AF cells, we wanted to test whether this defect had any impact on the ability of these cells to initiate homologous recombination. W303 haploid strains first described by Lee and Honigberg (16, 25) were used to construct all diploid strains, as these haploid parents allow for assaying intragenic recombination at TRP1 and intergenic recombination at the CEN3 to MATa/MATα and CEN2 to HIS3/LYS2 intervals. When assayed, the frequencies of intragenic recombination at TRP1 were more or less equivalent in all strains tested (Table 3). Frequencies of recombination varied from 5.5 × 10−2 to 6.5 × 10−2 Trp+ recombinants/CFU. These values are similar to those previously reported (21). The Trp+ diploid recombinants were then assayed for intergenic recombination across two genomic intervals. Once again all cell types had very similar intergenic recombination frequencies (Table 3).
The initiation of homologous recombination is required for multispore formation.
All mutant cells tested initiated and completed various types of homologous recombination. We next wanted to determine whether the initiation of these events was actually required for meiotic rereplication to occur. Deletion of SPO11 or REC102 abolishes double-strand break (DSB) initiation and homologous recombination (2, 6, 11, 21). Spo11p is an endonuclease whose activity catalyzes the DSBs required for homologous recombination. Rec102p is required for meiotic recombination initiation and for the proper timing of the first meiotic division (2, 6). spo11 cells form tetrads; however, individual spores are inviable. rec102 cells produce inviable spores upon being induced to sporulate.
We also wanted to determine whether Spo13p function was important for multispore formation. Loss of SPO13 results in aberrant sister chromatid separation during meiosis I (22, 43). While the catalytic function of Spo13p is not completely understood, its activity is required for stability of the meiotic cohesin Rec8p (24, 43). spo13 cells initiate a single meiotic division, whereby chromosomes can segregate either equationally or reductionally depending on the strain background (17, 21, 22).
We constructed swe1 and CDC28AF strains homozygous for spo11, rec102, or spo13 and determined whether they formed multispores and/or reinitiated meiotic replication upon being induced to sporulate. We found that swe1 and CDC28AF cells lacking SPO11, REC102, or SPO13 were no longer able to form multispores (Table 4). Mutant cells with spo11deleted did form tetrads whose progeny harbored the spo11 inviable phenotype, while those lacking spo13 gave rise to dyad-containing asci. Moreover, FACS analysis showed that the >4C peak previously seen in swe1 and CDC28AF cells was no longer present in double mutant cells (data not shown). We examined meiotic replication for up to 48 h using FACS. Based on our results, we suggest that rereplication requires that homologous recombination be initiated and that proper sister chromatid cohesion be maintained during meiosis I.
CLB6 and SIC1 are not required for multispore formation.
The B-type cyclins Clb5p and Clb6p are required for meiotic S phase (44, 46) and for synaptonemal formation during meiotic recombination (44). Clb5p seems to have a more significant role during meiotic replication than does Clb6p (46). We wanted to determine whether Clb5p and/or Clb6p activities were required for multispore formation and rereplication. swe1 and CDC28AF strains were constructed lacking CLB5 or CLB6 and tested for their ability to form multispores. When induced to sporulate, mutant cells lacking CLB6, but not CLB5, formed multispores (Table 2). FACS analysis demonstrated that mutant and wild-type cells lacking Clb5p function actually failed to initiate meiotic replication (data not shown). Stuart and Wittenberg (46) have shown that clb5 clb6 cells are able to initiate sporulation as demonstrated by the induction of IME1 and IME2 expression. These data indicate that Clb6p function is dispensable for multispore formation, while possibly hinting at a role for a Clb5p-associated Cdk activity. However, we cannot definitively say whether Clb5 is required, since all strains homozygous for CLB5 deletion were unable to initiate meiotic replication.
We next wanted to determine whether transient activation of one or more meiotic checkpoints was required for multispore formation. We tested this by removing the meiotic DNA damage or pachytene checkpoints and examining multispore formation in mutant cells. Deleting RAD9 and RAD17 renders the DNA damage and pachytene checkpoints defective, respectively (32, 48, 49). Mutant cells lacking either one of these checkpoints formed multispores at levels similar to those seen in checkpoint-functional mutant cells (Table 4).
CDC55 encodes one of two B regulatory subunits of PP2A. Yang et al. (54) have demonstrated that Cdc55p-dependent PP2A regulates Swe1p activity during mitosis; thus, it may indirectly regulate Cdc28p. We showed that loss of SWE1 causes diploid cells to reinitiate replication within a single meiotic cell cycle. Thus, we explored the possible role of Cdc55p in regulating multispore formation. We found that swe1 cdc55 and CDC28AF cdc55 diploid cells were incapable of forming any type of spore-containing asci. In fact, cdc55 cells were also defective in normal spore formation, as well. cdc55 cells are able to induce the expression of IME1 and IME2 but are unable to initiate meiotic replication as demonstrated by FACS (L. Rice and J. T. Nickels, unpublished data). These results suggest that Cdc55p may function early in meiosis around the time of meiotic replication initiation. We cannot say whether Cdc55p-dependent PP2A regulates multispore formation.
The Cdc28p inhibitor Sic1p is required for pre-RC formation (19). Thus, we wanted to determine whether a “repriming” of the meiotic replication machinery was required for reinitiation of replication in mutant cells. SIC1 was deleted in swe1 and CDC28AF mutants, and cells were induced to sporulate. CDC28AF sic1 and swe1 sic1 cells were able to sporulate and form multispored asci at levels similar to those seen in single mutants (Table 4). While SIC1 did not seem to be necessary for CDC28AF and swe1 mutants to complete several rounds of DNA replication, it did seem to be required for sporulation efficiency. sic1 cells sporulated with a reduced frequency compared to wild-type cells. Moreover, a large percentage of sic1 spores were inviable, and loss of SWE1 or expression of the CDC28AF allele did not suppress this inviability.
DISCUSSION
Our studies point to Cdc28p activity being tightly regulated during meiotic S phase in order to ensure that only one round of meiotic DNA replication is initiated. One level of regulation requires Swe1p, as diploid cells lacking this kinase activity form multispore asci. Presumably, swe1 cells contain deregulated and possibly hyperactivated Cdc28p activity that lacks Y19 inhibitory phosphorylation. Moreover, our results further support the idea that Cdc28p activity plays a primary role in meiotic replication, most likely through a Cdc28p-Clb5p complex.
We found that swe1 and CDC28AF cells lacking Cdc28p-Clb5p activity did not form multispore asci or initiate meiotic DNA replication, while cells lacking Clb6p were multispore competent. However, cells lacking Clb5p did not even initiate meiotic replication in our strain background. Thus, our results can suggest only that the execution point of Clb5p-Cdk activity and meiotic rereplication initiation are epistatic, as is Cdc55p-dependent PP2A activity. Strich et al. (45) recently demonstrated that constitutive overexpression of CLB1 during meiosis causes multispore formation. swe1 and CDC28AF cells do induce CLB1 earlier and temporally sustain its expression level longer than seen in wild-type cells (Fig. 2). Whether sustained levels of CLB1 play a role in the multispore phenotype of swe1 or CDC28AF cells is currently being pursued.
Yeast strains recalcitrant to Cdc28p-dependent regulation of ORC, Mcm2-7p, and Cdc6p together are defective in blocking mitotic replication reinitiation (38). However, not all origins reinitiate replication in this triply deregulated strain, suggesting that additional regulatory mechanisms exist and help to prevent unscheduled DNA replication. By deregulating Cdc28p activity, we were able to permit a subset of cells to reinitiate meiotic replication. Possibly the additional mechanism alluded to functions through directly regulating Cdc28p activity. Another possibility is that meiotic replication initiation may be under less stringent control, since only a single S phase is initiated prior to the initiation of a terminal ascus phenotype. Therefore, loss of Cdc28p regulation alone can give rise to reinitiation of meiotic, but not mitotic, replication. We need to explore the “activity status” of ORC, Mcm2p-7p, and Cdc6p in Cdc28p deregulated strains in order to better understand how deregulation of Cdc28p leads to replication reinitiation during meiosis.
We would like to examine whether deregulated Cdc28p either inhibits activities necessary to set up a meiotic post-RC or simply keeps the already formed pre-RC in a functional state. Liang and Stillman (28) have shown that cells harboring cdc6-3 alone are able to reinitiate mitotic replication. This allele expresses a Cdc6p that is stable and resistant to Cdc28p-dependent negative regulation. Also it has been shown that high-level expression of the Schizosaccharomyces pombe ortholog Cdc18 induces multiple rounds of DNA replication (37, 39). Cdc6p is degraded through the SCF/Cdc34p ubiquitin-ligase complex (51). Rudner et al. (42) have evidence showing that cells harboring a CDC28 allele nearly identical to CDC28AF have reduced activity of the anaphase-promoting ubiquitin-ligase complex, delaying exit from mitosis. We are currently examining whether Cdc28p has a role in regulating SCF activity during meiosis, or if mutant cells harbor reduced anaphase-promoting complex activity, and whether either contributes to multispore formation.
Why is initiating homologous recombination essential for multispore formation in cells deregulated for Cdc28p activity? The meiotic cell may consider homologous recombination part of meiotic S phase; thus, the post-RC resistant to reinitiation of replication (19) is generated only after recombination has been completed. An active pre-RC therefore remains bound to origins during recombination, and the deregulation of Cdc28p becomes the driving force for multiple rounds of replication. We ruled out the possibility that untimely transient activation of either the DNA damage (32, 48, 49) or pachytene (40, 49) checkpoint during recombination, which could possibly cause the down-regulation of Cdc28p activity and subsequent reformation of a competent pre-RC, was responsible for multispore formation.
Spo11p has a function that is independent of its role in DSB initiation. It, along with Rec8p, is involved in determining the length of meiotic S phase; loss of Spo11p during meiosis shortens meiotic S phase, whereas loss of Rec8p causes an increase (4). It is thought that Spo11p may directly integrate the formation of interhomolog and intersister interactions with ongoing meiotic replication (4), either through a direct interaction with chromatin or through its possibly interacting with specific replication factors (12). Thus, we cannot rule out that the role of Spo11p in initiating meiotic rereplication is to form these stable chromosomal intermediates or interact with replication initiators, rather than initiating DSBs. The fact that loss of REC102 abolishes multispore formation does lend support to the idea that initiating homologous recombination is necessary.
We found that Spo13p was required for rereplication and multispore formation. Interestingly, Spo13p has a role in protecting Rec8p from cleavage by the separase Esp1p (24, 43). spo13 diploid cells have lower levels of Rec8p on centromeres during anaphase of meiosis I (23). Forsburg (12) has suggested that, during meiotic S phase, cells may assemble a “prerecombination complex” that contains Spo11p, with the activity of this complex possibly being regulated by Rec8p through its role in maintaining centromeric cohesion. Loss of Spo13p, and subsequent reduction in Rec8p levels at centromeres, may cause a reduction in the assembly of this complex, which may result in loss of Spo11p on chromosomes and a reduction in the formation of stable interchromosomal interactions necessary for rereplication initiation.
Finally, we have uncovered a potential role for Cdc55p-dependent PP2A during premeiotic DNA replication. Cells lacking Cdc55p express IME1 and IME2 but fail to complete meiotic replication (L. Rice and J. T. Nickels, unpublished data). How might Cdc55p regulate meiosis? CDC55 was originally discovered in a screen aimed at isolating recessive mutations causing cs morphology defects; cdc55 cells have multiple elongated buds that are defective in cytokinesis (14). The cs phenotype can be suppressed by expressing a dominant allele of CDC28, CDC28-Y19F, lacking the conserved inhibitory tyrosine phosphorylation site (Y19) (30). Thus, Cdc55p may regulate the Cdc28p/Clb activity required for meiotic replication through regulating the degree of inhibitory phosphorylation at Y19. It may do so indirectly through regulating the phosphorylation state and/or stability of Swe1p, as this kinase accumulates in mitotically grown cdc55 cells treated with microtubule-destabilizing agents that cause a G2/M arrest (54).
Cdc55p may regulate Cdc45p, which is a component of the pre-RC that is required for replication initiation (19). Xenopus laevis eggs depleted of PP2A have less Cdc45 bound to chromatin (5). Cdc55p may also regulate meiosis through its regulation of protein degradation. The loss of cdc55 can suppress the ts phenotype of cdc20-1 cells (47). Cdc20p is an activator of the anaphase-promoting complex (APC/C), and it interacts with and activates a phosphorylated form of the APC (55). Moreover, cdc55 is synthetic lethal with loss of a specific F-box protein, Grr1p, known to regulate the SCF ubiquitin-ligase complex (20). These results argue that Cdc55p may function to regulate protein proteolysis through its regulation of SCF and/or APC/C complex activities. In any event, our data support the idea that Cdc55p-dependent PP2A activity is essential during meiosis, and we are currently examining how it regulates the meiotic process.
Acknowledgments
This work was supported by March of Dimes Foundation Basil O'Connor Starter Scholarship Grant FY99-277.
We thank Reza Shirzadi for constructing the CDC28AF strain, Jun Yin for generating swe1 and spo13 strains, and Grace Tan for initiating the recombination assays. We thank Monika Jost for performing FACS on our samples. We also thank Kim Nasmyth for providing TetO strains and Saul Honigberg for the recombination strains. We also are grateful to Katrina Cooper, Tom Edlind, Bill Bergman, Randy Strich, and Ed Winter and to their laboratories for many helpful discussions.
REFERENCES
- 1.Benjamin, K. R., C. Zhang, K. M. Shokat, and I. Herskowitz. 2003. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 17:1524-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhargava, J., J. Engebrecht, and G. S. Roeder. 1992. The rec102 mutant of yeast is defective in meiotic recombination and chromosome synapsis. Genetics 130:59-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borde, V., A. H. Goldman, and M. Lichten. 2000. Direct coupling between meiotic DNA replication and recombination initiation. Science 290:806-809. [DOI] [PubMed] [Google Scholar]
- 4.Cha, R. S., B. M. Weiner, S. Keeney, J. Dekker, and N. Kleckner. 2000. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 14:493-503. [PMC free article] [PubMed] [Google Scholar]
- 5.Chou, D. M., P. Petersen, J. C. Walter, and G. Walter. 2002. Protein phosphatase 2A regulates binding of Cdc45 to the prereplication complex. J. Biol. Chem. 277:40520-40527. [DOI] [PubMed] [Google Scholar]
- 6.Cool, M., and R. E. Malone. 1992. Molecular and genetic analysis of the yeast early meiotic recombination genes REC102 and REC107/MER2. Mol. Cell. Biol. 12:1248-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davenport, R. 2001. Cell cycle research: once copied, thrice blocked. Science 292:2415-2417. [DOI] [PubMed] [Google Scholar]
- 8.Dirick, L., L. Goetsch, G. Ammerer, and B. Byers. 1998. Regulation of meiotic S phase by Ime2 and a Clb5,6-associated kinase in Saccharomyces cerevisiae. Science 281:1854-1857. [DOI] [PubMed] [Google Scholar]
- 9.Drury, L., G. Perkins, and J. Diffley. 2000. The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol. 10:231-240. [DOI] [PubMed] [Google Scholar]
- 10.Duncker, B., P. Pasero, D. Braguglia, P. Heun, M. Weinreich, and S. Gasser. 1999. Cyclin B-Cdk1 kinase stimulates ORC- and Cdc6-independent steps of semiconservative plasmid replication in yeast nuclear extracts. Mol. Cell. Biol. 19:1226-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esposito, M. 1984. Molecular mechanisms of recombination in Saccharomyces cerevisiae: testing mitotic and meiotic models by analysis of hypo-rec and hyper-rec mutations. Symp. Soc. Exp. Biol. 38:123-159. [PubMed] [Google Scholar]
- 12.Forsburg, S. L. 2002. Only connect: linking meiotic DNA replication to chromosome dynamics. Mol. Cell 9:703-711. [DOI] [PubMed] [Google Scholar]
- 13.Guttmann-Raviv, N., E. Boger-Nadjar, I. Edri, and Y. Kassir. 2001. Cdc28 and Ime2 possess redundant functions in promoting entry into premeiotic DNA replication in Saccharomyces cerevisiae. Genetics 159:1547-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Healy, A. M., S. Zolnierowicz, A. E. Stapleton, M. Goebl, A. A. DePaoli-Roach, and J. R. Pringle. 1991. CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol. Cell. Biol. 11:5767-5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honigberg, S. M., and R. E. Esposito. 1994. Reversal of cell determination in yeast meiosis: postcommitment arrest allows return to mitotic growth. Proc. Natl. Acad. Sci. USA 91:6559-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honigberg, S. M., and R. H. Lee. 1998. Snf1 kinase connects nutritional pathways controlling meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:4548-4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hugerat, Y., and G. Simchen. 1993. Mixed segregation and recombination of chromosomes and YACs during single-division meiosis in spo13 strains of Saccharomyces cerevisiae. Genetics 135:297-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly, T., and G. Brown. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69:829-880. [DOI] [PubMed] [Google Scholar]
- 20.Kim, Y., L. Francisco, G. C. Chen, E. Marcotte, and C. S. Chan. 1994. Control of cellular morphogenesis by the Ip12/Bem2 GTPase-activating protein: possible role of protein phosphorylation. J. Cell Biol. 127:1381-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klapholz, S., and R. E. Esposito. 1980. Isolation of SPO12-1 and SPO13-1 from a natural variant of yeast that undergoes a single meiotic division. Genetics 96:567-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klapholz, S., and R. E. Esposito. 1980. Recombination and chromosome segregation during the single division meiosis in SPO12-1 and SPO13-1 diploids. Genetics 96:589-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein, F., P. Mahr, M. Galova, S. B. Buonomo, C. Michaelis, K. Nairz, and K. Nasmyth. 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98:91-103. [DOI] [PubMed] [Google Scholar]
- 24.Lee, B., A. Amon, and S. Prinz. 2002. Spo13 regulates cohesin cleavage. Genes Dev. 16:1672-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, R. H., and S. M. Honigberg. 1996. Nutritional regulation of late meiotic events in Saccharomyces cerevisiae through a pathway distinct from initiation. Mol. Cell. Biol. 16:3222-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lengronne, A., and E. Schwob. 2002. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G1. Mol. Cell 9:1067-1078. [DOI] [PubMed] [Google Scholar]
- 27.Leu, J. Y., and G. S. Roeder. 1999. The pachytene checkpoint in S. cerevisiae depends on Swe1-mediated phosphorylation of the cyclin-dependent kinase Cdc28. Mol. Cell 4:805-814. [DOI] [PubMed] [Google Scholar]
- 28.Liang, C., and B. Stillman. 1997. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 11:3375-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim, H., P. Goh, and U. Surana. 1996. Spindle pole body separation in Saccharomyces cerevisiae requires dephosphorylation of the tyrosine 19 residue of Cdc28. Mol. Cell. Biol. 16:6385-6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, F. C., and K. T. Arndt. 1995. The role of Saccharomyces cerevisiae type 2A phosphatase in the actin cytoskeleton and in entry into mitosis. EMBO J. 14:2745-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longtine, M. S., A. R. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 32.Lydall, D., Y. Nikolsky, D. K. Bishop, and T. Weinert. 1996. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383:840-843. [DOI] [PubMed] [Google Scholar]
- 33.Mendenhall, M. D., and A. E. Hodge. 1998. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1191-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendenhall, M. D., C. A. Jones, and S. I. Reed. 1987. Dual regulation of the yeast CDC28-p40 protein kinase complex: cell cycle, pheromone, and nutrient limitation effects. Cell 50:927-935. [DOI] [PubMed] [Google Scholar]
- 35.Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:32-46. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell, A. P. 1994. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol. Rev. 58:56-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muzi-Falconi, M., G. W. Brown, and T. J. Kelly. 1996. Controlling initiation during the cell cycle. DNA replication. Curr. Biol. 6:229-233. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen, V., C. Co, and J. Li. 2001. Cyclin-dependent kinases prevent re-replication through multiple mechanisms. Nature 411:1068-1073. [DOI] [PubMed] [Google Scholar]
- 39.Nishitani, H., and P. Nurse. 1995. p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell 83:397-405. [DOI] [PubMed] [Google Scholar]
- 40.Roeder, G. S., and J. M. Bailis. 2000. The pachytene checkpoint. Trends Genet. 16:395-403. [DOI] [PubMed] [Google Scholar]
- 41.Rothstein, R. J. 1983. One-step gene disruption. Methods Enzymol. 101:20-78. [DOI] [PubMed] [Google Scholar]
- 42.Rudner, A. D., K. G. Hardwick, and A. W. Murray. 2000. Cdc28 activates exit from mitosis in budding yeast. J. Cell Biol. 149:1361-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shonn, M., R. McCarroll, and A. Murray. 2002. Spo13 protects meiotic cohesin at centromeres in meiosis I. Genes Dev. 16:1659-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, K., A. Penkner, K. Ohta, F. Klein, and A. Nicolas. 2001. B-type cyclins CLB5 and CLB6 control the initiation of recombination and synaptonemal complex formation in yeast meiosis. Curr. Biol. 11:88-97. [DOI] [PubMed] [Google Scholar]
- 45.Strich, R., M. J. Mallory, M. Jarnik, and K. F. Cooper. 2004. Cyclin B-cdk activity stimulates meiotic rereplication in budding yeast. Genetics 167:1621-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stuart, D., and C. Wittenberg. 1998. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev. 12:2698-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, Y., and D. J. Burke. 1997. Cdc55p, the B-type regulatory subunit of protein phosphatase 2A, has multiple functions in mitosis and is required for the kinetochore/spindle checkpoint in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber, L., and B. Byers. 1992. A RAD9-dependent checkpoint blocks meiosis of cdc13 yeast cells. Genetics 131:55-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weinert, T. A. 1992. Dual cell cycle checkpoints sensitive to chromosome replication and DNA damage in the budding yeast Saccharomyces cerevisiae. Radiat. Res. 132:141-143. [PubMed] [Google Scholar]
- 50.Weinreich, M., C. Liang, H. Chen, and B. Stillman. 2001. Binding of cyclin-dependent kinases to ORC and Cdc6p regulates the chromosome replication cycle. Proc. Natl. Acad. Sci. USA 98:11211-11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willems, A. R., T. Goh, L. Taylor, I. Chernushevich, A. Shevchenko, and M. Tyers. 1999. SCF ubiquitin protein ligases and phosphorylation-dependent proteolysis. Philos. Trans. R. Soc. Lond. B 354:1533-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wittenberg, C., and S. I. Reed. 1988. Control of the yeast cell cycle is associated with assembly/disassembly of the Cdc28 protein kinase complex. Cell 54:1061-1072. [DOI] [PubMed] [Google Scholar]
- 53.Xu, L., M. Ajimura, R. Padmore, C. Klein, and N. Kleckner. 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:6572-6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, H., W. Jiang, M. Gentry, and R. L. Hallberg. 2000. Loss of a protein phosphatase 2A regulatory subunit (Cdc55p) elicits improper regulation of Swe1p degradation. Mol. Cell. Biol. 20:8143-8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zachariae, W., and K. Nasmyth. 1999. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 13:2039-2058. [DOI] [PubMed] [Google Scholar]