Abstract
The yeast Paf1 complex (Paf1C), composed of Paf1, Ctr9, Cdc73, Rtf1, and Leo1, associates with RNA polymerase II (Pol II) at promoters and in the actively transcribed portions of mRNA genes. Loss of Paf1 results in severe phenotypes and significantly reduced levels of the other Paf1C components. In contrast, loss of Rtf1 causes relatively subtle phenotypic changes and no reduction in the other Paf1C factors but disrupts the association of these factors with Pol II and chromatin. To elucidate the fate of the Paf1C when dissociated from Pol II, we examined the localization of the Paf1C components in paf1 and rtf1 mutant yeast strains. We found that although the Paf1C factors remain nuclear in paf1 and rtf1 strains, loss of Paf1 or Rtf1 results in a change in the subnuclear distribution of the remaining factors. In wild-type cells, Paf1C components are present in the nucleoplasm but not the nucleolus. In contrast, in both paf1 and rtf1 strains, the remaining factors are found in the nucleolus as well as the nucleoplasm. Loss of Paf1 affects nucleolar function; we observed that expression of MAK21 and RRP12, important for rRNA processing, is reduced concomitant with an increase in rRNA precursors in a paf1 strain. However, these changes are not the result of relocalization of the Paf1C because loss of Rtf1 does not cause similar changes in rRNA processing. Instead, we speculate that the change in localization may reflect a link between the Paf1C and newly synthesized mRNAs as they exit the nucleus.
RNA polymerase II (Pol II) is dependent on a multitude of associated protein factors to carry out its functional role in the synthesis of mRNAs. There are specific factors required for formation of an initiation complex at promoters (the mediator and the general initiation factors, or GTFs) (9, 27, 38), factors that associate with the elongating Pol II (1, 47), and factors important for 3′ end formation that associate with Pol II late in the transcription cycle (6, 59). Much of this cyclic interaction is controlled by the phosphorylation state of the repeated amino acid sequence (YSPTSPS) present at the C-terminal domain (CTD) of the largest subunit of Pol II. The unphosphorylated form of the CTD is important for assembly of the initiation complex. Escape from the promoter and the transition to elongation are accompanied by phosphorylation of serine 5 in the CTD repeat, and as Pol II elongates, serine 2 of the repeat is phosphorylated (6, 32, 36). Serine 2 phosphorylation is critical for association of 3′ end formation factors with Pol II (18, 21).
In contrast, some Pol II-associated factors are not sensitive to the phosphorylation state of the CTD, among them elongation factor Spt5 (22) and the factors of the Paf1 complex (Paf1C). The Paf1C was originally identified as a collection of proteins associated with the unphosphorylated form of Pol II (57). Recently, using the technique of chromatin immunoprecipitation, Paf1C components and Spt5 have been shown to be associated with Pol II at promoters and within the coding region of genes (26, 34, 48). Unlike Spt5, which remains associated with Pol II until it terminates transcription, the Paf1C is somehow released from the elongating Pol II as it transits the polyadenylation site (18). Since this transition is not accompanied by known changes in Pol II phosphorylation and because loss of serine 2 phosphorylation caused by loss of the CTD kinase Ctk1 does not affect Pol II association with the Paf1C (18), it appears that the Paf1C associates with Pol II in a CTD modification-independent fashion.
Consistent with its presence at several stages of the Pol II transcription cycle, the Paf1C has been linked to initiation via genetic interactions with TATA-binding protein (51), elongation through genetic interactions with elongation factors Dst1 and Spt5 (50), and polyadenylation by effects on poly(A) tail length (26). It has also been demonstrated that the Paf1C plays an important role in transcription-coupled histone modification and is postulated to serve as a platform for many Pol II-associated factors (14). Because of these connections to many critical aspects of transcription, we were surprised to discover that the connections between Paf1C components and Pol II could be disrupted with little detriment to the yeast cell. Loss of either Rtf1 or Cdc73 results in only subtle phenotypic changes to the yeast cell in contrast to the severe phenotypes observed in strains lacking either Paf1 or Ctr9 (4). However, loss of Rtf1 or Cdc73 disrupts the association of Paf1, Ctr9, and Leo1 with chromatin and the interaction between Paf1 and Pol II (26).
If the Paf1C is still carrying out its major functions when it has lost most of its connections to Pol II and chromatin, where is the complex in the cell and what can its localization tell us about its functional role? To pursue this question, we have used high-resolution immunofluorescence confocal microscopy to determine the location of tagged Paf1C components when the complex has been dissociated from Pol II. We have observed that under normal conditions the Paf1C is present in the nucleoplasm and clearly excluded from the nucleolus. However, in the absence of Rtf1 or Paf1, the remaining Paf1C components are present in both nuclear compartments. This shift in localization is not observed for either Spt5 or Pol II. Although loss of Paf1 does result in changes in rRNA processing, consistent with reduced expression of genes important for rRNA maturation, these changes are not caused by the redistribution of the Paf1C. Instead, we discuss the possibility that the appearance of Paf1C components in the nucleolus may reflect connections between the Paf1C and mRNAs as they are posttranscriptionally processed and move from the nucleus to the cytoplasm.
MATERIALS AND METHODS
Yeast strains and techniques.
The S. cerevisiae strains used in this study were derived from YJJ662 (MATa leu2Δ1 his3Δ200 ura3-52) or an isogenic strain with the α mating type (26, 45). The following strains were described by Mueller et al. (26), and the relevant genotypes are provided here for clarity: YJJ576 (paf1Δ::HIS3), YJJ1303 (rtf1Δ::Kanr), YJJ1330 (LEO1::LEO16HA), YJJ1367 (rtf1Δ::Kanr trp1Δ::hisG), YJJ1368 (paf1Δ::HIS3 trp1Δ::hisG), YJJ1374 (LEO1::LEO16HA rtf1Δ::Kanr), YJJ1378 (LEO1::LEO16HA paf1Δ::HIS3), YJJ1503 (RPB3::RBP36HA), YJJ1505 (RPB3::RPB36HA paf1Δ::HIS3), YJJ1506 (RPB3::RPB36HA rtf1Δ::Kanr), YJJ1578 (SPT5::SPT56HA paf1Δ::HIS3), YJJ1580 (SPT5::SPT56HA rtf1Δ::Kanr), YJJ1583 (CDC73::CDC736HA trp1Δ::hisG), YJJ1585 (CDC73::CDC736HA paf1Δ::HIS3), YJJ1587 (CDC73::CDC736HA rtf1Δ::Kanr), YJJ1589 (RTF1::RTF16HA), YJJ1590 (CTR9::CTR96HA paf1Δ::HIS3), YJJ1592 (CTR9::CTR96HA rtf1Δ::Kanr), YJJ1598 (PAF1::PAF16HA), YJJ1599 (CTR9::CTR96HA). Yeast strains were grown in yeast extract-peptone-dextrose with 4% glucose (15).
Construction of additional HA-tagged yeast strains.
Strain YJJ1697 (his3Δ200 ura3-52 leu2Δ1 trp1Δ::hisG NUP49::NUP496HA/K. lactis TRP1) was constructed from YJJ1655 (MATa his3Δ200 ura3-52 leu2Δ1 trp1Δ::hisG) by using the pYM3 plasmid as described previously (26) but with NUP49-specific sequences. This strain is wild type and hemagglutinin (HA) tagged at the C terminus of NUP49. Strain YJJ1718 (MATa his3Δ200 ura3-52 leu2Δ1 trp1Δ::hisG rtf1Δ::Kanr NUP49::NUP496HA/K. lactis TRP1) is the rtf1Δ version constructed from YJJ1367 with the pYM3 plasmid. Strain YJJ1716 (MATα his3Δ200 ura3-52 leu2Δ1 trp1Δ::hisG paf1Δ::HIS3 NUP49::NUP496HA/K. lactis TRP1) is the paf1Δ version constructed similarly from strain YJJ1368. Strains were verified by both PCR and Western blot analysis (2).
Imaging.
Cells from mid-log-phase cultures (optical density at 600 nm, 0.5 to 1.0) were fixed with formaldehyde, permeabilized with Zymolyase, and attached to coverslips coated with 2-mg/ml poly-l-lysine (37). HA-tagged proteins were visualized by incubating coverslips with an anti-HA antibody directly coupled to the fluorescent dye Alexa 488 (A-21287; Molecular Probes). The nucleolar protein Nop1 was visualized by incubating the coverslips first with an anti-Nop1 antibody (EnCor 28-N2) and subsequently with the secondary antibody comprised of a goat anti-mouse immunoglobulin G3 (IgG3) conjugated to the fluorescent dye Alexa 555 (A21157; Molecular Probes). Note that in double-labeled specimens, the primary antibodies were used in the same incubation step, and the anti-mouse IgG3-Alexa 555 conjugate was incubated with the coverslips in a second step. Although the anti-HA-Alexa 488 antibody was also generated in mice, the antibody subtype is different (IgG1), so there was no cross-reactivity with the Alexa 555 secondary antibody. The coverslips were mounted on slides by using Vectashield H-1200 mounting medium containing 4′,6′-diamidino-2-phenylindole (DAPI; Vector Laboratories). Samples were visualized with an Olympus IX70 inverted microscope by using a 100× oil immersion objective and a Quantix camera. The images were analyzed with DeltaVision software on a Silicon Graphics O2 computer (Applied Precision, Seattle, Wash.) at the University of Colorado Health Sciences Center Light Microscopy Core Facility.
RNA analysis.
RNA was isolated by the hot phenol method (42). Ten micrograms of total RNA was fractionated by formaldehyde gel electrophoresis (2). RNA was transferred to Zeta-Probe GT membranes by standard capillary transfer methods. 32P-labeled probes were generated by PCR amplification of specific yeast genes from genomic yeast DNA (primers: MAK21 forward, 5′-AGTGACAACAACGGCAATCCG; reverse, 5′-GACCATTTAAGAATAAACCTTCAA; RRP12 forward, 5′-ATGGATCAAGACAAAGTTG; reverse, 5′-ATAAGCGGTAACAACCAA), followed by gel purification and random prime labeling with a RadPrime kit (Invitrogen). Oligonucleotides specific for 18S rRNA (5′-GCTTATACTTAGACATGAAT) and 35S and 27S rRNA precursors (5′-CGCCTAGACGCTCTCTTCTTA) (41) were 5′ end labeled (39). Hybridization and washing were performed at 45°C for random prime probes and 38°C for oligonucleotide probes as described previously (35). Blots were exposed to a phosphorimaging screen or to film, and bands were quantitated by using Quantity One imaging software. Signals were normalized to those of 18S rRNA.
RESULTS
Paf1C components are nuclear in wild-type cells.
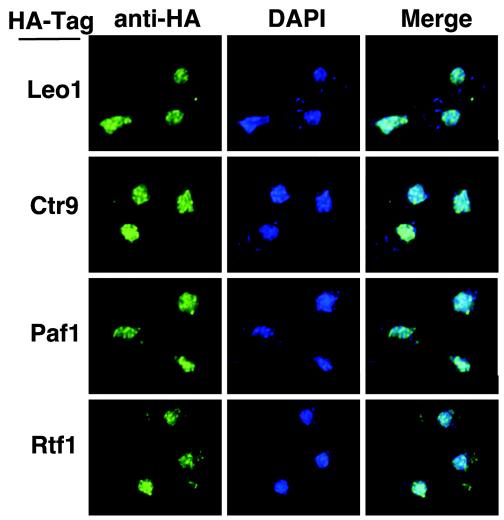
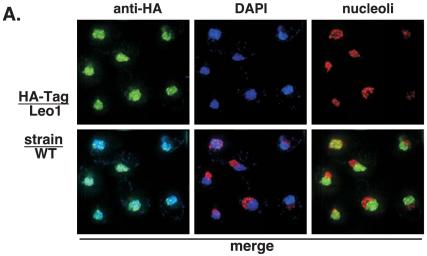
Using biochemical fractionation, we have previously reported that Paf1 is a nuclear protein (46). To determine the localization of the other components of the Paf1C, we utilized a collection of isogenic yeast strains containing HA-tagged forms of Paf1, Ctr9, Cdc73, Rtf1, and Leo1 (Fig. 1A). We also used HA-tagged forms of Pol II subunit Rpb3 and elongation factor Spt5. In each case, the tagged form of the protein functionally replaced the untagged form of the gene (26). The HA-tagged genes were integrated into wild-type, paf1, and rtf1 strains. The HA-tagged proteins were visualized by confocal microscopy (see Materials and Methods) by binding to a highly specific anti-HA antibody directly conjugated to the green-fluorescing dye Alexa 488. There was no signal observed in an untagged yeast strain, consistent with the lack of any HA-cross-reacting material in yeast observed by protein blot analysis (26). We observed that Paf1-HA, Ctr9-HA, Cdc73-HA, Rtf1-HA, and Leo1-HA all colocalized with the DAPI-stained nuclei in wild-type cells (Fig. 2 and data not shown) in a somewhat punctate pattern. These data confirm that all of the Paf1C components have a nuclear localization and are consistent with the results of recent genome-wide localization studies (16).
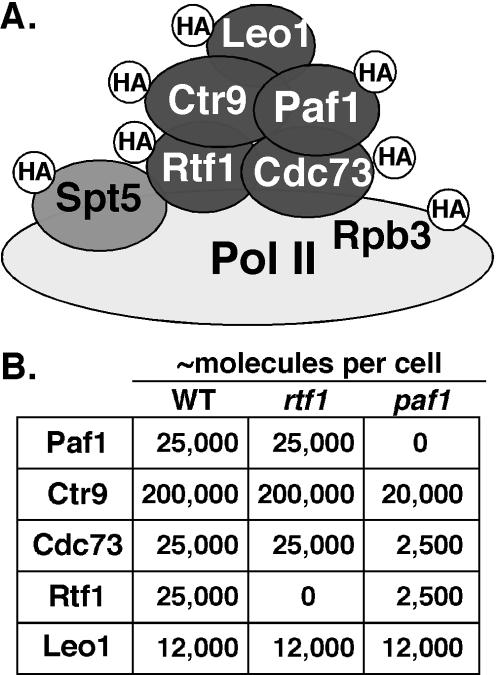
FIG. 1.
The Paf1C and tagged proteins used for microscopy. (A) A schematic of the Paf1C is shown, with designation of HA-tagged proteins constructed as described in Materials and Methods. Rpb3 was tagged as a representative subunit of RNA Pol II (light gray). Spt5 (medium gray) is found associated with the Paf1C but is not considered a core component, as described in the text. Each strain used for fluorescence microscopy contained only one tagged component. (B) Summary of the abundance of Paf1C components and the effects of loss of either Paf1 or Rtf1. The data in the table were derived from those of Mueller et al. (26). WT, wild type.
FIG. 2.
Paf1C components are nuclear in wild-type cells. HA-tagged wild-type strains were labeled with DAPI (blue) and with an anti-HA antibody conjugated to the fluor Alexa 488 (green). Yeast strains shown from top to bottom are the wild type with Leo1-HA (YJJ1330), Ctr9-HA (YJJ1599), Paf1-HA (YJJ1598), and Rtf1-HA (YJJ1589).
Localization of Paf1C components is altered in paf1 and rtf1 mutants.
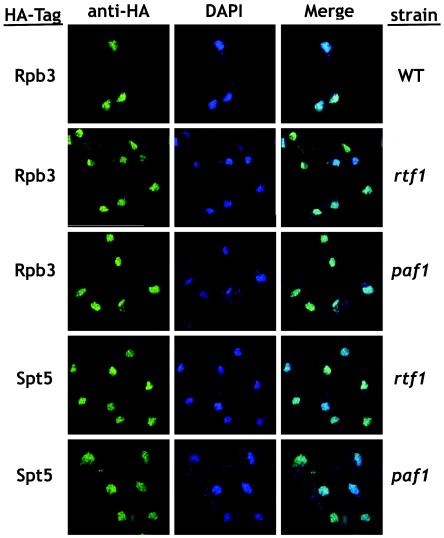
Loss of any of the Paf1C components has no effect on the abundance of Pol II or Spt5, nor does it affect the chromatin distribution of these factors (26). We therefore did not expect to see Paf1C-dependent changes in subcellular localization of Pol II or Spt5. As shown in Fig. 3, we found that Rpb3-HA and Spt5-HA colocalize with the DAPI staining in wild-type, paf1, and rtf1 cells, indicating that neither loss of Paf1C components nor their dissociation from chromatin alters the subcellular localization of Pol II or Spt5.
FIG. 3.
Loss of Paf1 or Rtf1 does not affect localization of Rpb3 or Spt5. HA-tagged Rpb3 or Spt5 were visualized in both paf1 and rtf1 strains by using the anti-HA/Alexa 488 antibody (green) with DAPI to stain the nuclear DNA (blue). A wild-type (WT) strain containing Rpb3-HA (YJJ1503) is shown as a control. The paf1 and rtf1 strains shown from top to bottom are rtf1 with Rpb3-HA (YJJ1506), paf1 with Rpb3-HA (YJJ1505), rtf1 with Spt5HA (YJJ1580), and paf1 with Spt5-HA (YJJ1578).
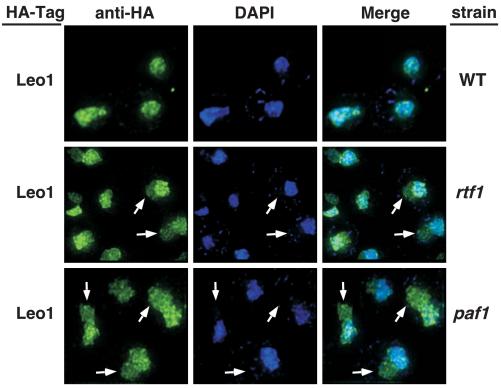
Loss of Paf1 results in a significant reduction in the cellular levels of Ctr9, Cdc73, and Rtf1 but not Leo1 (summarized in Fig. 1B). In contrast, loss of Rtf1 does not change the abundance of the other Paf1C components (Fig. 1B), but it does result in their dissociation from chromatin (26). Because Leo1 levels are unchanged in paf1 and rtf1 cells, we used strains containing HA-tagged Leo1 to assess its distribution when Paf1C is dissociated from chromatin. As shown in Fig. 2, the punctate staining of Leo1-HA colocalized exactly with the nuclear DAPI staining in wild-type cells (Fig. 4, top panel); however, without Paf1 or Rtf1, the Leo1-HA signal was also present in a compartment adjacent to, but distinct from, the DAPI stain (Fig. 4, middle and lower panels). This compartment does not correspond to the cytoplasm based on longer exposures of these images that reveal the outlines of the cells (data not shown).
FIG. 4.
Loss of Paf1 or Rtf1 results in changes in the subnuclear localization of Leo1. Wild-type and mutant strains containing HA-tagged Leo1 were labeled with DAPI (blue) and the anti-HA/Alexa 488 antibody (green). The strains represented are the wild type (WT) with Leo1-HA (YJJ1330), rtf1 with Leo1-HA (YJJ1374), and paf1 with Leo1-HA (YJJ1378). Arrows point to regions of Leo1-HA staining that do not overlap with the DAPI signal.
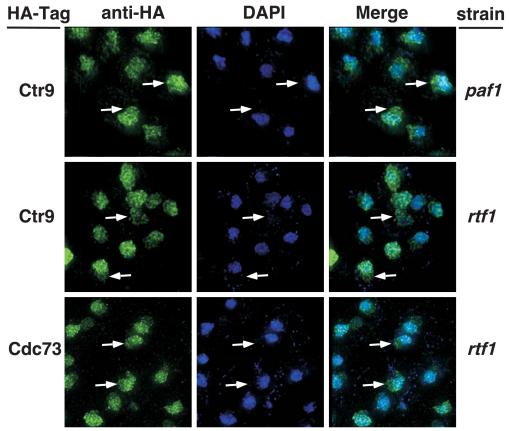
We also examined the localization of Ctr9 and Cdc73 HA-tagged proteins in paf1 and rtf1 mutants. Although Ctr9 levels are reduced in the paf1 strain (Fig. 1B), very high endogenous abundance results in a clear Ctr9-HA signal even when reduced, as shown in the top panel of Fig. 5. Like Leo1-HA, the Ctr9-HA signal also extends outside the DAPI staining region, as demonstrated in the merged signal panel on the right side of the figure. As in paf1, loss of Rtf1 results in the spread of Ctr9-HA staining outside the DAPI-stained area, and Cdc73-HA localization is also sensitive to the loss of Rtf1 (Fig. 5, middle and lower panels). Although Cdc73 levels are reduced in paf1 (Fig. 1B), we observed that the residual signal detected was also present outside the DAPI staining area (data not shown). Thus, although the localization of Pol II and Spt5 was unaffected by the loss of Paf1 or Rtf1 (Fig. 3), the localization of the other Paf1C core components was altered in paf1 and rtf1 strains compared to wild-type cells.
FIG. 5.
Loss of Paf1 or Rtf1 results in changes in the subnuclear localization of Ctr9 and Cdc73. Mutant strains containing HA-tagged Ctr9 or Cdc73 were labeled with DAPI (blue) and the anti-HA/Alexa 488 antibody (green). The strains used were paf1 Ctr9-HA (YJJ1590), rtf1 Ctr9-HA (YJJ1592), and rtf1 Cdc73-HA (YJJ1587). Arrows point to regions of Paf1C component staining that do not overlap with the DAPI signal.
Nuclear pores are unaffected by loss of Paf1 or Rtf1.
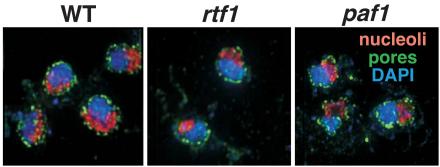
The Paf1C has been shown to associate with the THO/TREX complex component Hpr1, and hpr1 and paf1 strains share many phenotypes (4, 8, 26). Hpr1 and other THO/TREX factors have been shown to play a role in nuclear export of mRNAs (43, 52, 58), and loss of Cdc73 is synthetically lethal with loss of Apq12, a factor required for nucleocytoplasmic mRNA transport (3). In addition, loss of either Hpr1 or Paf1 results in reduced expression of ACC1 (43; data not shown), which is linked to defects in nuclear pores (44). We speculated that the observed mislocalization of Paf1C components upon loss of Paf1 or Rtf1 may correlate with a defect in mRNA export or nuclear pore distribution. To test this idea, we HA tagged the nuclear pore component Nup49, which marks both the location of the pores and delineates the outline of the nuclear membrane (5). In Fig. 6, the green staining corresponds to the location of the HA-tagged Nup49, blue staining is DAPI, and the red signal marks the nucleolus, described further below. The Nup49-HA signal encircles both the DAPI and nucleolar staining in the wild-type, rtf1, and paf1 strains. We did not observe obvious differences in the number or distribution of Nup49-labeled nuclear pores in rtf1 or paf1 cells compared to wild-type. In particular, there is no evidence for a redistribution of pores in the mutant cells that could account for the altered localization of Paf1C components observed above.
FIG. 6.
Loss of Paf1 or Rtf1 does not affect the abundance or distribution of nuclear pores or the appearance of the nucleolus. Wild-type (WT), paf1, and rtf1 strains in which the nuclear pore protein Nup49 was HA tagged were treated with DAPI (blue), the anti-HA/Alexa 488 antibody to stain the nuclear periphery (green), and an anti-Nop1 antibody followed by incubation with a secondary antibody conjugated to Alexa 555 (red) which stains the nucleolus, which is poorly stained with DAPI. The strains used were wild-type (WT) Nup49-HA (YJJ1697), rtf1 Nup49-HA (YJJ1718), and paf1 Nup49-HA (YJJ1716).
Paf1C components are found in the nucleolus in paf1 and rtf1 mutants.
Because the HA-tagged Paf1 components were found in a discrete region adjacent to the DAPI staining region of the nucleus, we investigated whether the complex components could be colocalizing with the nucleolus, which stains poorly with DAPI and is found in a crescent-shaped region next to the DAPI-staining nucleoplasm (reviewed in reference 29). To visualize the nucleolus, we used an antibody to the nucleolar protein Nop1 and a secondary antibody conjugated to the red-fluorescing dye Alexa 555, as described in Materials and Methods. As shown in Fig. 6, we were able to visualize the nucleolus in wild-type and mutant cells as a relatively compact region distinct from the DAPI-staining nucleoplasm. When we compared the localization of Leo1-HA (green) to that of Nop1 (red) in wild-type cells, we observed no overlap between the green and red signals or between the blue DAPI signal and the nucleolar marker (Fig. 7A, lower panels). As described above (Fig. 4), the Leo1-HA signal colocalized with the DAPI nucleoplasmic region in the wild-type cells.
FIG.7.
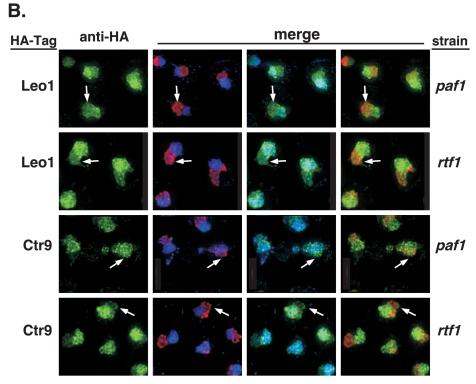
Loss of Paf1 or Rtf1 results in changes in the subnuclear localization of Leo1 and Ctr9. Wild-type (WT), paf1, and rtf1 strains containing HA-tagged Paf1 components were treated with DAPI (blue) and specific antibodies to visualize the Paf1 component plus the nucleolar compartment. Leo1-HA and Ctr9-HA were visualized by treating the samples with an anti-HA/Alexa 488 antibody (green). The nucleolus, represented by the nucleolar marker Nop1, was visualized by treating the samples with an anti-Nop1 antibody and then subsequently treating them with a secondary antibody conjugated to the fluor Alexa 555 (red). (A) A wild-type strain in which Leo1 is HA tagged (YJJ1330) is shown. The top panels show the cells with a single stain in each panel. The bottom panels are the merged pairs of stains (from left to right): DAPI plus Leo1-HA, DAPI plus nucleolus, and Leo1-HA plus nucleolus. (B) paf1 and rtf1 strains in which either Leo1 or Ctr9 is HA tagged are shown. The column on the left represents the single anti-HA stain signal. The next three columns show the merged photos for DAPI plus Nop1 (nucleolus), DAPI plus anti-HA, and Nop1 (nucleolus) plus anti-HA, respectively, from left to right. The strains used were paf1 Leo1-HA (YJJ1378), rtf1 Leo1-HA (YJJ1374), paf1 Ctr9-HA (YJJ1590), and rtf1 Ctr9-HA (YJJ1592). Arrows point to overlaps between nucleolar and Paf1C component staining.
In contrast to the nucleolar exclusion of Paf1C components in wild-type cells (Fig. 7A and data not shown), we found that the green Leo1-HA and Ctr9-HA signals colocalized with both the blue DAPI-stained nucleoplasm and the red Nop1-marked nucleolus in paf1 and rtf1 cells (Fig. 7B) To reduce the complexity of the images, only the green HA-tagged channel is shown as a single image adjacent to all of the pairwise merged images. However, it is clear that the extra staining seen in the paf1 and rtf1 cells is coincident with the nucleolar signal. Therefore, using the technique of immunofluorescence, it is clear that when the Paf1C is disrupted or dissociated from chromatin, it is present in both the nucleoplasm and in the nucleolus.
Quantitative analysis of the altered localization.
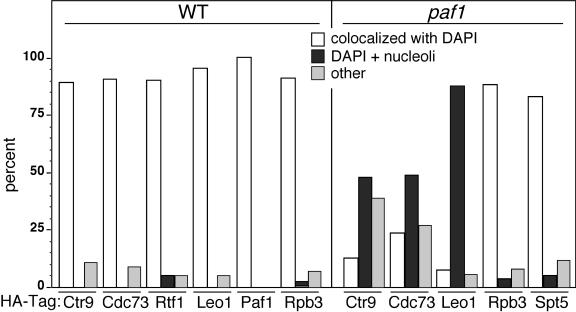
In the absence of biochemical confirmation of the altered subcellular localization and to help establish that the appearance in the nucleolus of the Paf1C components in paf1 and rtf1 strains was not due to sampling a small, nonrepresentative population of cells, we examined larger microscopic fields and categorized the staining pattern of at least 50 cells of each type. Quantitations of the staining patterns from both wild-type and paf1 strains are shown in Fig. 8. In wild-type cells, most images show complete superimposition of the anti-HA staining with that of DAPI (colocalized with DAPI) for all HA-tagged proteins analyzed, Ctr9, Cdc73, Rtf1, Leo1, Paf1, and Rpb3. In contrast, the majority of paf1 cells showed the staining pattern illustrated in Fig. 4 and 5 (DAPI plus nucleoli), in which, although some of the HA-tagged protein colocalizes with DAPI, a distinct portion is also found adjacent to the DAPI staining region, colocalized with the Nop1 nucleolar marker. The paf1 cells also had small numbers of other localization patterns, including poor staining and ring-like staining around the cell periphery (other). The increase in this category for the Ctr9-HA and Cdc73-HA signals is probably due to the significantly reduced abundance of these proteins in the paf1 cells (Fig. 1B). As seen in Fig. 3, the localizations of Rpb3 and Spt5 were unaffected by loss of Paf1. With this quantitative analysis, we have determined that the mislocalization of Paf1C components to the nucleolus correlates with the dissociation of the complex from chromatin and Pol II when either Paf1 or Rtf1 is lost.
FIG. 8.
Ctr9, Cdc73, and Leo1 are relocalized within the nucleus in the absence of Paf1. Large microscopic fields of HA-tagged components in either wild-type (WT) or paf1 strains were examined. Cells exhibiting either complete colocalization with DAPI versus other staining patterns were counted. The following strains were used, with the number in brackets indicating the number of cells counted: wild-type strains, Ctr9-HA (YJJ1599 [72]), Cdc73-HA (YJJ1583 [65]), Rtf1-HA (YJJ1589 [58]), Leo1-HA (YJJ1330 [59]), Paf1-HA (YJJ1598 [58]), Rpb-3HA (YJJ1503 [57]); paf1 strains, Ctr9-HA (YJJ1590 [60]), Cdc73-HA (YJJ1585 [55]), Leo1-HA (YJJ1378 [97]), Rpb3-HA (YJJ1505 [41]), Spt5-HA (YJJ1578 [59]). The “other” category represents a range of cells in which the HA-tagged component showed no staining, poor staining, a ring of staining surrounding damaged or dead cells, and other minor patterns.
Relocalization of the Paf1C to the nucleolus does not correlate with altered nucleolar function.
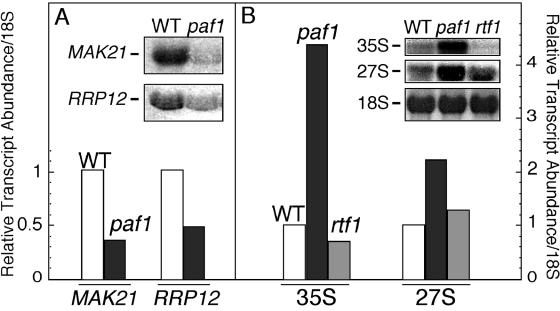
Without Paf1 or Rtf1, the remaining Paf1C components are found in the nucleolus. Does this relocalization disrupt the structure or function of this subnuclear compartment? We first measured the relative volume of the nucleolus in paf1 and rtf1 mutant strains and did not detect any difference in this parameter linked to loss of Paf1 or Rtf1 (data not shown). Loss of Paf1 results in changes in relative mRNA abundance in yeast (8, 35), and we have determined that some of the transcripts affected encode factors important for rRNA processing (K. L. Penheiter, unpublished data). As shown in Fig. 9A, transcripts for the essential genes for two nucleolar factors involved in rRNA processing and ribosome biogenesis, MAK21 (NOC1) (24) and RRP12 (30), are two- to threefold lower in abundance in a paf1 strain than in wild-type cells. Consistent with the reduced expression of these factors, we observed that 35S and 27S rRNA precursors are present in two- to fourfold-elevated amounts in paf1 relative to wild-type cells (Fig. 9B). Although it is interesting that among paf1's, many phenotypes is an effect on rRNA processing, this effect is not caused by the relocalization of the Paf1C to the nucleolus. This conclusion is based on the observation that loss of Rtf1, resulting in relocalization of the Paf1C to the nucleolus (see above), does not affect rRNA processing (Fig. 9B) or the expression of the nucleolar factors (data not shown). Therefore, we do not detect obvious alterations in nucleolar structure or function that correlate with the presence of the Paf1C components in the nucleolus. The relocalization may therefore reflect other aspects of the function of the Paf1C as discussed below.
FIG. 9.
Loss of Paf1, but not Rtf1, results in reduced expression of MAK21 and RRP12 and an increase in rRNA precursors. Total RNA was isolated from isogenic wild-type (WT) (YJJ662), paf1 (YJJ576), and rtf1 (YJJ1303) strains, fractionated, and probed for the indicated transcripts as described in Materials and Methods. The signals were normalized to the 18S rRNA signal and are plotted with the signal from wild-type cells set to 1. (A) Random primed probes were used to detect mRNA transcripts from the MAK21 and RRP12 genes; (B) oligonucleotide probes were used to detect rRNA precursors 35S and 27S as well as mature 18S rRNA.
DISCUSSION
Consistent with the association of the Paf1C with Pol II at promoters and within the coding region of yeast genes, we have found colocalization of Paf1C factors Paf1, Ctr9, Cdc73, Rtf1, and Leo1 with Pol II in the nucleoplasm of wild-type cells. However, when the Paf1C is dissociated from Pol II and chromatin by the loss of Rtf1, the remaining complex components are now also found in the nucleolus.
Although these results are based on immunofluorescence of tagged Paf1C factors, not biochemical fractionation, the fact that the Pol II-associated factor Spt5 does not show a similar relocalization is a strong argument for the specificity of the nucleolar localization. Normally, localization of proteins to the nucleolus requires specific association with components of the rRNA transcription and processing machinery (7). In fact, formation of a recognizable nucleolus is dependent upon active transcription of the rDNA genes (29). The distinct compartmentalization of RNA Pol I and II transcription factors is an active process that requires the RENT/Net1 complex that excludes Pol II (and other factors) from the nucleolar compartment (25, 31, 56). Some factors are known to move between the nucleoplasm and the nucleolus. For example, splicing factors move between nucleoplasmic speckles and the nucleolus in a cell cycle-dependent fashion (reviewed in reference 20), and in mammalian cells, RNA editing enzymes cycle between the nucleolus and the nucleoplasm in response to the appearance of their substrate (10, 40). Overall, the present view of the nucleolus is that localization to this subnuclear region does not appear to be a default mechanism but reflects either a function in this compartment or a sequestration-dependent regulatory mechanism (31, 53, 56).
The appearance of the Paf1C factors in the nucleolus is not accompanied by destabilization of the factors, as evidenced by the unchanged level of all of the other Paf1C factors in the absence of Rtf1 (26). Therefore, the relocalization does not appear to be part of a pathway targeting the proteins for destruction. In addition, the movement of the relatively abundant Paf1C factors into the nucleolus does not obviously disrupt the structure or change the relative size of this compartment. Although loss of Paf1 does result in reduced expression of mRNAs for some essential nucleolar factors (MAK21 and RRP12) and increased accumulation of rRNA precursors, relocalization of the Paf1C caused by loss of Rtf1 does not affect these processes nor result in obvious changes in rRNA processing. In contrast, loss of the Paf1C-associated factor Hpr1 results in changes to nucleolar morphology and a block of mRNA export (43).
The links between mRNA export and the nucleolus are intriguing but only partially characterized. A subset of mRNAs is normally exported through the nucleolus (17, 33), and under certain conditions, yeast heat shock mRNAs accumulate in the nucleolus before moving to the cytoplasm (55). In addition, proteomic studies have demonstrated that the poly(A) binding protein Pab1 associates with the nucleoplasmic Pol II transcription and processing factors Spt5 and Pcf11 as well as the nascent 60S ribosome in the nucleolus (23, 28). These Pab1 associations could be delineating the path of mRNA from synthesis to export and cytoplasmic function. It has in fact been proposed that the nucleolus could be a staging area for the export of mRNAs (31). In contrast, there is also evidence for separation of factors important for nuclear export of mRNAs and the nucleolus (12). Therefore, although the complete path of mRNAs from synthesis to export is not yet clear, it may be that the redistribution of the Paf1C is linked to the movement of some or all mRNAs as they exit the nucleus. It is interesting in this regard that the loss of Cdc73, which, like the loss of Rtf1, results in separation of the Paf1C from chromatin and Pol II (26), and the appearance of the remaining factors in the nucleolus (data not shown) is synthetically lethal in combination with the loss of Apq12, a recently described factor required for proper mRNA export from the nucleus (3).
The Paf1C is normally bound to Pol II and nucleoplasmic chromatin, but perhaps its associations with Hpr1 and the THO/TREX mRNA export complex direct it to follow the mRNA when its direct connections to Pol II have been disrupted by the loss of Rtf1. Alternatively, the Paf1C may normally briefly accompany the nascent mRNA on its export pathway before cycling back to another Pol II molecule. This view could explain the observed disappearance of the Paf1C from chromatin at locations beyond the poly(A) site (18). Reducing the connections to Pol II may then result in a longer association of the Paf1C with mRNA and its appearance in the nucleolus. The presence of the residual Paf1C factors in both the nucleoplasm and the nucleolus may be a reflection of its role in more than one export pathway as proposed by Thomsen and coworkers (55).
A final interesting connection between the Paf1C and the nucleolus is transcriptional silencing. Loss of Paf1 or Rtf1 leads to a reduction in telomeric silencing (19). Silencing can occur at telomeres, the MAT loci, or within the nucleolus (reviewed in reference 13). Part of the control of this process is the distribution of silencing factor and histone deacetylase Sir2 between these compartments (49). Boundaries between active and silenced genes sometimes involve RNA Pol III-transcribed genes (11), which are known to be localized in the nucleolus (54). So the separation of the Pol II nucleoplasmic compartment from the Pol I and III nucleolar compartment is used as a way to regulate the expression of genes. The relocalization of the Paf1C to the nucleolus may therefore be linked to the observed changes in silencing. Although, as described above, we have not observed significant changes to nucleolar morphology linked to the Paf1C relocalization, we have found that the expression level of genes, including MAK21 and RRP12, important for nucleolar function and rRNA maturation, is reduced without Paf1 and that there is an increased accumulation of rRNA precursors. However, these changes do not occur when Rtf1 is lost and the Paf1C is present in the nucleolus. Therefore relocalization of the Paf1C does not result in changes in rRNA processing, but it could impact both mRNA processing and export and the compartmentalization of factors important for gene regulation.
Acknowledgments
We are grateful to M. Winey for insight into our initial observations and suggestions for further experiments, C. Mueller for helpful discussions, J. Betz and E. Amiott for comments on the manuscript, S. Fadul of the Light Microscopy Core Facility at the UCHSC for help with confocal imaging, and T. Deem for construction of strain YJJ1655.
This work was supported by NIH grant RO1-GM38101 to J.A.J.
REFERENCES
- 1.Arndt, K. M., and C. M. Kane. 2003. Running with RNA polymerase: eukaryotic transcript elongation. Trends Genet. 19:543-550. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 3.Baker, K. E., J. Coller, and R. Parker. 2004. The yeast Apq12 protein affects nucleocytoplasmic mRNA transport. RNA 10:1352-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betz, J. L., M. Chang, T. M. Washburn, S. E. Porter, C. L. Mueller, and J. A. Jaehning. 2002. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol. Genet. Genomics 268:272-285. [DOI] [PubMed] [Google Scholar]
- 5.Bucci, M., and S. R. Wente. 1998. A novel fluorescence-based genetic strategy identifies mutants of Saccharomyces cerevisiae defective for nuclear pore complex assembly. Mol. Biol. Cell 9:2439-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buratowski, S. 2003. The CTD code. Nat. Struct. Biol. 10:679-680. [DOI] [PubMed] [Google Scholar]
- 7.Carmo-Fonseca, M., L. Mendes-Soares, and I. Campos. 2000. To be or not to be in the nucleolus. Nat. Cell Biol. 2:E107-112. [DOI] [PubMed] [Google Scholar]
- 8.Chang, M., D. French-Cornay, H. Y. Fan, H. Klein, C. L. Denis, and J. A. Jaehning. 1999. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol. 19:1056-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conaway, R. C., and J. W. Conaway. 1993. General initiation factors for RNA polymerase II. Annu. Rev. Biochem. 62:161-190. [DOI] [PubMed] [Google Scholar]
- 10.Desterro, J. M., L. P. Keegan, M. Lafarga, M. T. Berciano, M. O'Connell, and M. Carmo-Fonseca. 2003. Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci. 116:1805-1818. [DOI] [PubMed] [Google Scholar]
- 11.Donze, D., and R. T. Kamakaka. 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 20:520-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galy, V., O. Gadal, M. Fromont-Racine, A. Romano, A. Jacquier, and U. Nehrbass. 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116:63-73. [DOI] [PubMed] [Google Scholar]
- 13.Gartenberg, M. R. 2000. The Sir proteins of Saccharomyces cerevisiae: mediators of transcriptional silencing and much more. Curr. Opin. Microbiol. 3:132-137. [DOI] [PubMed] [Google Scholar]
- 14.Gerber, M., and A. Shilatifard. 2003. Transcriptional elongation by RNA polymerase II and histone methylation. J. Biol. Chem. 278:26303-26306. [DOI] [PubMed] [Google Scholar]
- 15.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194:1-931. [PubMed] [Google Scholar]
- 16.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 17.Kadowaki, T., R. Schneiter, M. Hitomi, and A. M. Tartakoff. 1995. Mutations in nucleolar proteins lead to nucleolar accumulation of polyA+ RNA in Saccharomyces cerevisiae. Mol. Biol. Cell 6:1103-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, M., S. H. Ahn, N. J. Krogan, J. F. Greenblatt, and S. Buratowski. 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23:354-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, O. W. Ryan, A. Golshani, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11:721-729. [DOI] [PubMed] [Google Scholar]
- 20.Leung, A. K., J. S. Andersen, M. Mann, and A. I. Lamond. 2003. Bioinformatic analysis of the nucleolus. Biochem. J. 376:553-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Licatalosi, D. D., G. Geiger, M. Minet, S. Schroeder, K. Cilli, J. B. McNeil, and D. L. Bentley. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell 9:1101-1111. [DOI] [PubMed] [Google Scholar]
- 22.Lindstrom, D. L., and G. A. Hartzog. 2001. Genetic interactions of Spt4-Spt5 and TFIIS with the RNA polymerase II CTD and CTD modifying enzymes in Saccharomyces cerevisiae. Genetics 159:487-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindstrom, D. L., S. L. Squazzo, N. Muster, T. A. Burckin, K. C. Wachter, C. A. Emigh, J. A. McCleery, J. R. Yates III, and G. A. Hartzog. 2003. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23:1368-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milkereit, P., O. Gadal, A. Podtelejnikov, S. Trumtel, N. Gas, E. Petfalski, D. Tollervey, M. Mann, E. Hurt, and H. Tschochner. 2001. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 105:499-509. [DOI] [PubMed] [Google Scholar]
- 25.Moss, T., and V. Y. Stefanovsky. 2002. At the center of eukaryotic life. Cell 109:545-548. [DOI] [PubMed] [Google Scholar]
- 26.Mueller, C. L., S. E. Porter, M. G. Hoffman, and J. A. Jaehning. 2004. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell 14:447-456. [DOI] [PubMed] [Google Scholar]
- 27.Myers, L. C., and R. D. Kornberg. 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69:729-749. [DOI] [PubMed] [Google Scholar]
- 28.Nissan, T. A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21:5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nomura, M. 2001. Ribosomal RNA genes, RNA polymerases, nucleolar structures, and synthesis of rRNA in the yeast Saccharomyces cerevisiae. Cold Spring Harbor Symp. Quant. Biol. 66:555-565. [DOI] [PubMed] [Google Scholar]
- 30.Oeffinger, M., M. Dlakic, and D. Tollervey. 2004. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev. 18:196-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olson, M. O., M. Dundr, and A. Szebeni. 2000. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 10:189-196. [DOI] [PubMed] [Google Scholar]
- 32.Palancade, B., and O. Bensaude. 2003. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur. J. Biochem. 270:3859-3870. [DOI] [PubMed] [Google Scholar]
- 33.Pederson, T. 1998. The plurifunctional nucleolus. Nucleic Acids Res. 26:3871-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pokholok, D. K., N. M. Hannett, and R. A. Young. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9:799-809. [DOI] [PubMed] [Google Scholar]
- 35.Porter, S. E., T. M. Washburn, M. Chang, and J. A. Jaehning. 2002. The yeast pafl-RNA polymerase II complex is required for full expression of a subset of cell cycle-regulated genes. Eukaryot. Cell 1:830-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prelich, G. 2002. RNA polymerase II carboxy-terminal domain kinases: emerging clues to their function. Eukaryot. Cell. 1:153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pringle, J. R., A. E. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565-602. [DOI] [PubMed] [Google Scholar]
- 38.Reinberg, D., G. Orphanides, R. Ebright, S. Akoulitchev, J. Carcamo, H. Cho, P. Cortes, R. Drapkin, O. Flores, I. Ha, J. A. Inostroza, S. Kim, T. K. Kim, P. Kumar, T. Lagrange, G. LeRoy, H. Lu, D. M. Ma, E. Maldonado, A. Merino, F. Mermelstein, I. Olave, M. Sheldon, R. Shiekhattar, and L. Zawel. 1998. The RNA polymerase II general transcription factors: past, present, and future. Cold Spring Harbor Symp. Quant. Biol. 63:83-103. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Sansam, C. L., K. S. Wells, and R. B. Emeson. 2003. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc. Natl. Acad. Sci. USA 100:14018-14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saveanu, C., D. Bienvenu, A. Namane, P. E. Gleizes, N. Gas, A. Jacquier, and M. Fromont-Racine. 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 20:6475-6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneiter, R., C. E. Guerra, M. Lampl, G. Gogg, S. D. Kohlwein, and H. L. Klein. 1999. The Saccharomyces cerevisiae hyperrecombination mutant hpr1D is synthetically lethal with two conditional alleles of the acetyl coenzyme A carboxylase gene and causes a defect in nuclear export of polyadenylated RNA. Mol. Cell. Biol. 19:3415-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneiter, R., M. Hitomi, A. S. Ivessa, E. V. Fasch, S. D. Kohlwein, and A. M. Tartakoff. 1996. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol. Cell. Biol. 16:7161-7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi, X., M. Chang, A. J. Wolf, C. H. Chang, A. A. Frazer-Abel, P. A. Wade, Z. F. Burton, and J. A. Jaehning. 1997. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol. Cell. Biol. 17:1160-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi, X., A. Finkelstein, A. J. Wolf, P. A. Wade, Z. F. Burton, and J. A. Jaehning. 1996. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol. Cell. Biol. 16:669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shilatifard, A. 2004. Transcriptional elongation control by RNA polymerase II: a new frontier. Biochim. Biophys. Acta 1677:79-86. [DOI] [PubMed] [Google Scholar]
- 48.Simic, R., D. L. Lindstrom, H. G. Tran, K. L. Roinick, P. J. Costa, A. D. Johnson, G. A. Hartzog, and K. M. Arndt. 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22:1846-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, J. S., C. B. Brachmann, L. Pillus, and J. D. Boeke. 1998. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics 149:1205-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Squazzo, S. L., P. J. Costa, D. L. Lindstrom, K. E. Kumer, R. Simic, J. L. Jennings, A. J. Link, K. M. Arndt, and G. A. Hartzog. 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21:1764-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stolinski, L. A., D. M. Eisenmann, and K. M. Arndt. 1997. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4490-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strasser, K., S. Masuda, P. Mason, J. Pfannstiel, M. Oppizzi, S. Rodriguez-Navarro, A. G. Rondon, A. Aguilera, K. Struhl, R. Reed, and E. Hurt. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417:304-308. [DOI] [PubMed] [Google Scholar]
- 53.Tani, T., R. J. Derby, Y. Hiraoka, and D. L. Spector. 1996. Nucleolar accumulation of poly(A)+ RNA in heat-shocked yeast cells: implication of nucleolar involvement in mRNA transport. Mol. Biol. Cell 7:173-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson, M., R. A. Haeusler, P. D. Good, and D. R. Engelke. 2003. Nucleolar clustering of dispersed tRNA genes. Science 302:1399-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomsen, R., D. Libri, J. Boulay, M. Rosbash, and T. H. Jensen. 2003. Localization of nuclear retained mRNAs in Saccharomyces cerevisiae. RNA 9:1049-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Visintin, R., and A. Amon. 2000. The nucleolus: the magician's hat for cell cycle tricks. Curr. Opin. Cell Biol. 12:372-377. [DOI] [PubMed] [Google Scholar]
- 57.Wade, P. A., W. Werel, R. C. Fentzke, N. E. Thompson, J. F. Leykam, R. R. Burgess, J. A. Jaehning, and Z. F. Burton. 1996. A novel collection of accessory factors associated with yeast RNA polymerase II. Protein Expr. Purif. 8:85-90. [DOI] [PubMed] [Google Scholar]
- 58.Zenklusen, D., P. Vinciguerra, J. C. Wyss, and F. Stutz. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 22:8241-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zorio, D. A., and D. L. Bentley. 2004. The link between mRNA processing and transcription: communication works both ways. Exp. Cell Res. 296:91-97. [DOI] [PubMed] [Google Scholar]