Abstract
It is widely appreciated that the ends of linear DNA molecules cannot be fully replicated by the conventional replication apparatus. Less well known is that semi-conservative replication of telomeric DNA also presents problems for DNA replication. These problems likely arise from the atypical chromatin structure of telomeres, the GC-richness of telomeric DNA that makes it prone to forming DNA secondary structures, and from RNA-DNA hybrids, formed by transcripts of one or both DNA strands. Given the different aspects of telomeres that complicate their replication, it is not surprising that multiple DNA helicases promote replication of telomeric DNA. This review focuses on one such class of DNA helicases, the Pif1 family of 5′–3′ DNA helicases. In budding and fission yeasts, Pif1 family helicases impact both telomerase-mediated and semi-conservative replication of telomeric DNA as well as recombination-mediated telomere lengthening.
Keywords: Telomere, Helicase, TERRA, Telomerase, DNA replication, Break induced replication, ALT, Pif1
DNA helicases use the energy of NTP hydrolysis to do work on DNA. Their best known function is to separate the two strands of the double helix so they can serve as templates for replication, transcription, or strand exchange, but they also disrupt base pairing of other substrates, such as R-loops or G-quadruplex DNA. In addition, DNA helicases can translocate along a single DNA strand and/or push or even displace proteins or protein complexes from DNA. Owing to these diverse activities, DNA helicases play critical roles in all aspects of DNA metabolism. Here we focus on the role of DNA helicases in replication of telomeric DNA, where their ability to disrupt a variety of structures is put to use.
In eukaryotes and archaea, the replicative DNA helicase is the hexameric MCM (mini-chromosome maintenance) complex, a 3′–5′ DNA helicase that functions in both the initiation and elongation of DNA replication. Although the MCM helicase suffices for the unwinding of most of the genome, accessory DNA helicases are needed to help the fork move past hard-to-replicate sites (reviewed in Refs. [1–3]). Some hard-to replicate sites, such as thymidine dimers, are generated by exogenous DNA damage. However, there are thousands of naturally occurring sites in all genomes that challenge fork progression in every cell cycle. Natural replication impediments include stable protein complexes, DNA secondary structures, highly transcribed genes, and RNA/DNA hybrids. Failure to maneuver past these obstacles results in pausing of the replication fork and increased risk of DNA damage. In this review, we focus on one type of intrinsic replication impediment, telomeres, and the role of Pif1 family 5′–3′ DNA helicases in promoting their replication.
1. Telomeric DNA and telomere binding proteins
Telomeres, the ends of eukaryotic chromosomes, typically consist of many tandem copies of a non-coding short DNA repeat. Although the repeat sequence of telomeric DNA differs among different organisms, certain features are seen in virtually all of them. First, guanines are distributed asymmetrically between the two strands of the double helix, and there is an orientation bias to this distribution, such that the G-rich strand runs 3′–5′ from the end of the chromosome towards its interior (Fig. 1A). Second, in most organisms, the G-rich strand is longer than its complementary C-rich strand so that the telomere bears a single-strand G-rich extension or G-tail. The G-rich strand can be bound by G-strand binding proteins and/or form stable secondary structures, such as G-quadruplex DNA and T-loops (Fig. 1). At least some and perhaps all of these end structures contribute to the capping or end protection function of telomeres.
Fig 1.
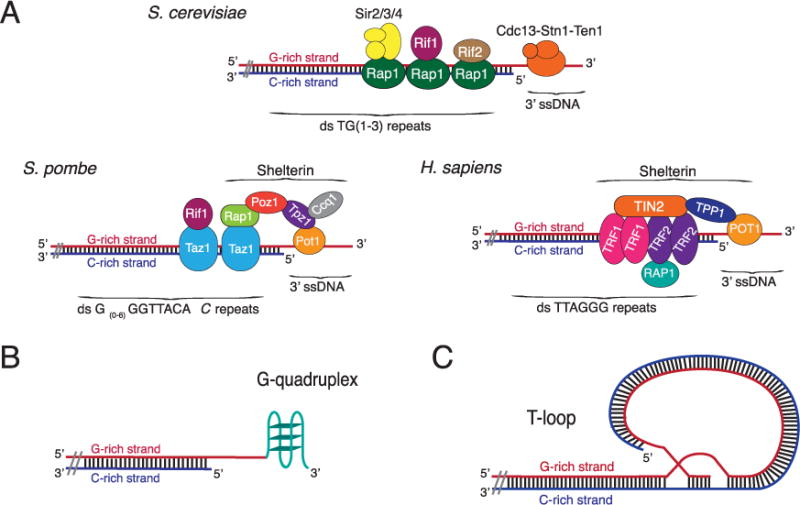
Telomere structures [84].
(A) Structure of telomeric DNA and telomere binding proteins in yeasts and humans. Not all proteins are shown and drawings are not to scale. Budding yeast telomeres consists of ~300 bp of an irregular sequence, 5′ -TG(1–3)-3′ ending in a ~12 nt long G-tail during most of the cell cycle. Rap1 binds duplex telomeric DNA and Cdc13 binds the G-tail. Rif1, Rif2, Sir2, Sir3, Sir4, Stn1, and Ten1 bind via protein–protein interactions. Fission yeast telomeres consist of ~250 bp of an irregular sequence, 5′ -G(0–6)G2T2ACAC-3′ (the terminal C is present in ~13% of repeats). Taz1 binds duplex telomeric DNA and Pot1 binds the G-tail. Rap1, Rif1, Tpz1, Ccq1, and Poz1 bind via protein–protein interactions. At birth, human telomeres have ~15 kb of 5′ -TTAGGG-3′ repeats. TRF1 and TRF2 bind duplex telomeric DNA while the 3′ single-stranded G-tail (~100 nt long) is bound by POT1. Alternatively, the G-tail can be folded into a T-loop (as in panel C). RAP1, TIN1, and TPP1 bind via protein–protein interactions.
(B) G-quadruplex structure [25]. The 3′ G-tail can form a stable four-stranded DNA structure, called a G-quadruplex (G4), held together by G–G Hoogsteen base-pairing. An intra-molecular G4 structure is shown. G4 structures can also form between the G-tails on different telomeres.
(C) T-loop structure [85]. The 3′ overhang forms a lariat-like structure by the G-tail invading the adjacent duplex telomeric DNA. An internal G-strand bubble forms from the displaced strand that can be bound by POT1.
Telomeric DNA is associated with a set of structural proteins that mediate all telomere functions (reviewed in Ref. [4]) (Fig. 1A). These proteins include sequence specific duplex binding proteins such as Rap1 and Taz1 in, respectively, budding and fission yeasts, and TRF1 and 2 in mammals. G-tails are substrates for sequence specific single-strand DNA binding proteins, such as Pot1 in fission yeast and mammals and the CST complex in budding yeast where Cdc13 is the G-strand binding subunit. Other telomere structural proteins are telomere-associated by protein–protein interactions. The G-rich nature of telomeric DNA and its ability to form non-canonical DNA and protein structures contribute to making this part of the chromosome a problem for DNA replication (reviewed in Ref. [5]).
2. End replication problem #1: telomerase and recombination to the rescue
The “end replication problem” refers to difficulties replicating to the very end of a linear DNA molecule (Fig. 2A). This difficulty arises from two features of replicative DNA polymerases: they require a primer to start DNA synthesis and they synthesize DNA only in the 5′ to 3′ direction. The primer is typically an 8–12 nucleotide stretch of RNA, which is removed later in the replication process. Conventional DNA polymerases can fill in the small gaps generated by removal of an internal RNA primer. However, the 8–12 nucleotide gap arising from removing the most terminal RNA primer has no downstream sequence to prime fill-in resynthesis. As a result, small gaps are left at the 5′ ends of newly replicated strands; i.e., linear chromosomes are not completely replicated (Fig. 2A). Hence, a special replication mechanism is needed to prevent progressive loss of DNA from chromosome ends. In almost all eukaryotes, this end replication problem is solved by a specialized reverse transcriptase, telomerase, which uses its integral RNA subunit, telomerase RNA, as a template to lengthen the 3′ ends of chromosomes (Fig. 2B).
Fig. 2.
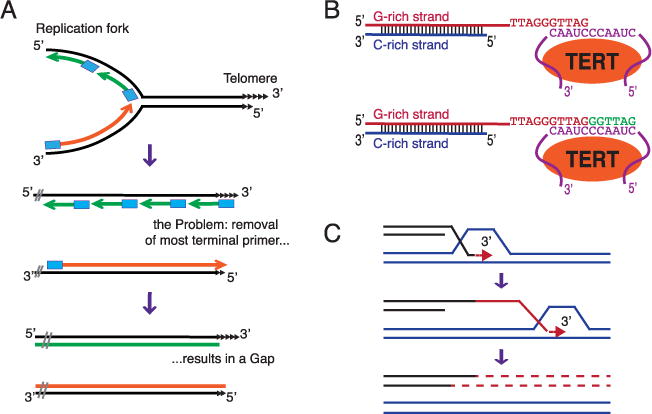
Mechanisms for replicating chromosome ends.
(A) Replicative DNA polymerases cannot replicate the very ends of linear DNA molecules.
Owing to the properties of conventional DNA polymerases, removal of the terminal RNA primer leaves a gap at the 5′ ends of newly replicated strands. Black lines: parental strands of the DNA molecule; Blue rectangles: 8–12 nucleotide RNA primers; Green lines: Okazaki fragments; Orange lines: leading strand synthesis. Only the right end of a chromosome is shown.
(B) In most eukaryotes, telomerase compensates for the loss of terminal sequences.
Telomerase extends the 3′ end of a vertebrate telomere. Orange oval: TERT, the reverse transcriptase subunit of telomerase: Purple: Telomerase RNA and its template region; Green line: newly made telomeric DNA.
(C) Telomeric DNA can be maintained by recombination or ALT (alternative lengthening of telomeres). Shown is break-induced-replication (BIR), which can repair a one-ended DNA break such as an eroded telomere. Red: newly synthesized DNA. The exact mechanism of BIR is still under discussion. Shown is a model where BIR is carried out by a migrating D-loop or bubble.
Although telomerase is by far the most common mechanism to maintain telomeric DNA, alternative (ALT) strategies exist (reviewed in Ref. [6]). Homologous recombination is the most widespread and best studied of the ALT mechanisms (Fig. 2C). Although in some organisms ALT can co-exist with telomerase or even supplant telomerase, ALT is typically studied in cells that normally rely on telomerase but where mutation or developmental regulation eliminates its activity. Budding yeast has two major ALT mechanisms: Type I involves recombination-mediated amplification of sub-telomeric DNA while type II involves lengthening of the telomeric repeats themselves by gene conversion (reviewed in Ref. [7]). Type II-like recombination is the major ALT pathway in humans with ~15% of human tumors using it to maintain telomeres.
3. End replication problem #2
Semi-conservative replication of telomeres is also a challenge (Fig. 3A). As first seen in budding yeast [8], replication forks slow when they approach and move through telomeric chromatin, and this slowing can result in DNA damage. Three different methods have been used to detect fork slowing at telomeres. In budding and fission yeasts, two-dimensional (2D) gel electrophoresis, which separates replicating from non-replicating DNA molecules by their non-linear shape, reveals that replication intermediates accumulate within telomeric DNA [8,9] (Fig. 3B). In both organisms, replication forks slow whether the telomeric DNA is at a chromosome end or at an internal site on the chromosome. Chromatin immuno-precipitation (ChIP) for the leading strand DNA polymerase is a second method that can monitor fork progression [10–12]. With this method, a higher DNA polymerase occupancy within telomeric DNA relative to control sequences detects fork slowing. In mammals, DNA fiber analysis (also called molecular combing) is combined with fluorescent in situ hybridization (FISH) to detect telomeric DNA and anti-dNTP antibodies (e.g., anti-BrdU) to detect newly synthesized DNA [13–16]. These studies reveal that forks slow when they move from sub-telomeric to telomeric DNA.
Fig. 3.
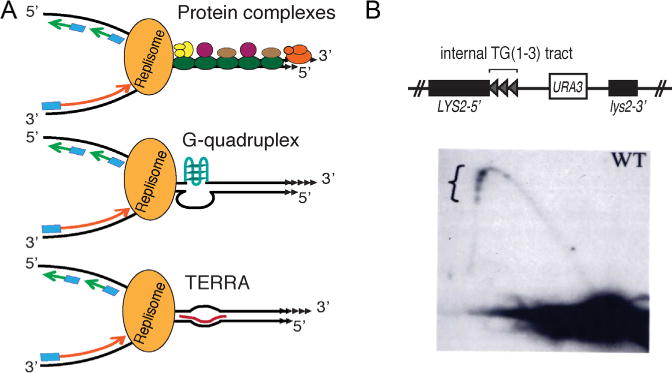
Semi-conservative replication through telomeric DNA.
(A) Replication fork moving towards the right telomere of a chromosome. Fork progression slowed by protein complexes (top), G-quadruplex DNA (middle), TERRA RNA (bottom).
(B) Replication forks slow as they move through an internal tract of telomeric DNA in budding yeast in vivo (adapted from Ref. [8]). Three ~270 bp tracts of TG(1–3) DNA cloned from a telomere and separated from each other by a polylinker were inserted far from a telomere. Top: diagram of the construct; Bottom: two-dimension gel analysis of replication through the tracts. Accumulation of replication intermediates at telomeric tracts (indicated by bracket) is reflected by higher intensity of hybridization signal.
4. Why is telomeric chromatin hard-to-replicate?
As noted, several types of structures can impede fork progression, including stable protein complexes, DNA secondary structures, and RNA/DNA hybrids (Fig. 3A). All three of these structures exist within telomeres. Telomeric DNA is organized into an atypical chromatin structure, which at least in organisms like budding yeast with very short telomeres (~300 bps) is probably devoid of nucleosomes. In budding and fission yeasts, nucleosomes in sub-telomeric DNA have histone marks characteristic of heterochromatin, while in mammals, with their much longer telomeres, the telomeric repeats themselves are assembled into heterochromatin (reviewed in Refs. [17–19]). Their heterochromatic features contribute at least in part to the hard-to-replicate nature of telomeres as does the stable non-nucleosomal structure of the most terminal repeats. For example, fork slowing within budding yeast telomeres is almost eliminated in the absence of Sir proteins that bind telomeric and sub-telomeric DNA and are required for the heterochromatin-like nature of sub-telomeric regions [20].
Telomeric chromatin is by no means the entire explanation for slow fork movement within telomeres. In fission yeast and mice, depleting cells of their respective duplex sequence-specific telomere binding protein, Taz1 and TRF1, paradoxically exacerbates fork slowing within telomeric DNA [9,13]. Depleting budding yeast of Rap1 also has a deleterious impact on fork progression (S. Lim and VAZ, in preparation). The favored explanation for the fork promoting activities of these duplex telomere DNA binding proteins is that the denuded DNA is able to form stable DNA secondary structures, such as G4 DNA, that are known to impede fork progression at non-telomeric sites [12,21–23]. Although experiments with G4 stabilizing drugs provide some support for this hypothesis [24], it is not obvious why loss of a duplex binding protein should expose the G-strand of telomeric DNA to the single-stranded state that is usually needed to form a G4 structure. Nonetheless, there is evidence for G4 formation within telomeric DNA in vivo and its presence is anticipated to cause problems for DNA replication (reviewed in Ref. [25]) (Fig. 3A).
RNA:DNA hybrids are another potential source of fork slowing activity within telomeres (reviewed in Ref. [3]). Even though proximity to a telomere represses transcription of nearby genes, in yeasts and mammals (reviewed in Ref. [19]), the C-rich strand of telomeric DNA is transcribed to generate GT-rich TERRA (telomere repeat-containing RNA) (reviewed in Ref. [26]). (In fission yeast, the G-rich strand of the telomere is also transcribed; Ref. [27]). When TERRA transcripts are base-paired to telomeric DNA, they generate R-loops where the G-rich telomeric DNA strand is displaced to form a single-strand bubble (Fig. 3A). As R-loops impede fork progression at non-telomeric sites, they might also slow forks moving through the telomere. Alternatively, as the G-rich telomeric strand is single-stranded in TERRA-mediated R-loops, it could form G4 structures that stall replication forks.
TERRA results in shortening of the transcribed telomere; i.e., it acts in cis to regulate telomere length. This effect was initially attributed to inhibition of telomerase. Indeed, TERRA can inhibit telomerase in vitro by binding to the template in telomerase RNA and thereby blocking its use [28]. However, TERRA affects telomere length even in telomerase deficient cells [29], and in yeast, TERRA increases recombination-mediated telomere lengthening [30]. In cells lacking both telomerase and homologous recombination, TERRA-mediated telomere shortening still occurs but causes telomere fragility in a replication dependent manner [30]. As with R-loops at non-telomeric sites, mutations in the RNA biogenesis THO/TREX complex increase TERRA and telomere dysfunction.
5. Pif1 family DNA helicases
Pif1 family DNA helicases are found in almost all eukaryotes and are also present in some bacteria and archae (reviewed in Refs. [31–33]). The budding yeast Pif1, for which the family is named, is the prototypical and best-studied member of this helicase family (hereafter called ScPif1; Sc for S. cerevisiae). Budding yeast encodes a second Pif1 family helicase, Rrm3, but most eukaryotes, including fission yeast and humans, encode only one, called Pfh1 (fission yeast) and hPIF1 (humans). However, Trypanosomes express eight distinct Pif1 family helicases, most with non-overlapping functions [34,35]. Neither ScPif1 nor Rrm3 is essential, but a double mutant, pif1Δ rrm3Δ, grows very slowly with many cells arrested at or near the end of S phase, and there is evidence that the two helicases, while having different functions, can serve as back ups for each other in fork progression [36]. The single fission yeast protein, Pfh1, is essential for maintenance of both mitochondrial and nuclear DNA [37] while mice lacking mPIF1 are viable [38]. In contrast, at least six of the eight Trypanosome Pif1 helicases are essential [39].
Pif1 family helicases are multifunctional. Where examined, Pif1 family helicases localize to both mitochondria, where they can be critical for maintaining mitochondrial DNA, as in yeasts [37] and Trypanosomes [34,35,40], and nuclei. So far, all nuclear forms of Pif1 helicases affect telomeres, although not necessarily in the same way. ScPif1, Rrm3, and Pfh1 promote replication and suppress DNA damage at G4 motifs, sequences able to form G4 DNA in vitro, throughout the nuclear genome [12,21], and hPIF1 localizes to sites that are thought to be G4 structures [41]. Rrm3 and Pfh1 also have more general effects on fork progression, being especially critical for promoting replication through RNA Polymerase III transcribed genes [10,12]. Pfh1 also affects forks within the most highly transcribed RNA Polymerase II genes [11]. Other sites of Rrm3 and Pfh1 action are the replication fork barriers in ribosomal DNA, silencers, inactive origins of replication, and centromeres. At some of these sites, dependence on Rrm3 or Pfh1 is imposed at least in part by stable protein complexes [11,20,42] (Fig. 3A). ScPif1 also affects Okazaki fragment maturation [43,44]. As well as roles in DNA replication, ScPif1 and Pfh1 both function in DNA repair [37,45,46].
6. Pif1 family helicases affect telomerase
ScPif1 was first identified because of its critical (but non-essential) role in maintenance of mitochondrial DNA and then rediscovered in a screen for genes that affect telomeres (reviewed in Refs. [31,33]). Loss of Pif1 results in long telomeres, and this lengthening is telomerase dependent. However, the most striking telomere phenotype of pif1Δ cells is a large increase in telomerase-mediated telomere addition (TA) to double strand breaks. The rate of TA is low in wild type cells but increases almost 1000 fold in the absence of ScPif1. Taken together, these data suggest that ScPif1 inhibits telomerase both at telomeres and double strand breaks (DSBs). In vitro and in vivo experiments show that ScPif1 uses its ATPase activity to remove telomerase from telomeres and DSBs [47,48]. This removal results in lower telomerase processivity both in vitro and in vivo. Although ScPif1 acts at both telomeres and DSBs, its action at DSBs, but not at telomeres, requires checkpoint kinase-mediated ScPif1 phosphorylation [49].
In several organisms, including budding yeast, telomerase preferentially elongates short telomeres [50]. ScPif1 contributes to this preference as in its absence telomerase no longer binds preferentially to short telomeres [48]. ScPif1 itself binds better to longer telomeres. Thus, ScPif1 is more likely to remove telomerase from long telomeres, allowing the holoenzyme to act preferentially on the telomeres most in need of lengthening. A preference for removing telomerase from DNA molecules with longer G-tails is also seen in vitro [51].
Rrm3, Pfh1, and hPIF1 were found by searching the database for genes with sequence similarity to ScPif1 [52]. Although Rrm3 affects telomeres, unlike ScPif1, it does not inhibit telomerase [8]. Indeed, deleting RRM3 from pif1Δ cells, partially suppresses the increases in telomere length and telomere addition characteristic of pif1Δ cells. Because Pfh1 is essential, the full impact of its deletion on telomere length is difficult to measure, as telomere length changes due to mutations are often not detected until many generations after the genotypic change. However, pfh1Δ spore clones, which divide only a few times before they arrest in G2 phase, have modestly shorter telomeres than wild type cells [53]. Moreover, over-expressing Pfh1 increases telomere length by a recombination-independent mechanism, and this lengthening does not occur in rad11-D223Y cells [54], in which telomerase action is inefficient [55]. As Pfh1 promotes telomere lengthening in a telomerase-dependent manner, it is unlikely to be an inhibitor of telomerase.
The roles of mammalian Pif1 family helicases are not nearly as well established as that of their fungal counterparts. Deleting mouse PIF1 has no detectable effect on telomere length, telomere addition or chromosome structure, even after multiple mouse generations [38,56]. However, in vivo co-immuno-precipitation experiments suggest that both mPIF1 and hPIF1 are TERT-associated (TERT is the catalytic subunit of mammalian telomerases) [38,57]. One group reported that over-expression of hPIF1 in tissue culture causes telomere shortening (although our lab was unable to reproduce this result) and that hPIF1 reduces telomerase processivity in vitro [58].
7. Rrm3 and Pfh1 helicases promote semi-conservative replication of telomeric DNA
Rrm3 promotes fork progression through terminal and internal tracts of telomeric DNA, as seen by both 2D gels [8] and genome-wide ChIP analyses [10]. The requirement for Rrm3 during telomere replication is not suppressed by deleting Rif1 or Sir proteins (Fig. 1A) so Rrm3 does not promote fork progression through telomeres by displacing these proteins [20,59]. Neither Sir nor Rif proteins bind telomeric DNA directly but rather associate with telomeres via protein–protein interactions with Rap1 (Fig. 1A). Although it is possible that Rrm3 promotes fork progression by acting on Rap1, depleting cells of Rap1 slows fork progression through telomeric DNA (S. Lim and VA Zakian, in preparation). Thus, it seems unlikely that Rap1 is the target of Rrm3.
Pfh1 and Rrm3 have similar effects on semi-conservative replication of telomeres. By both 2D gels and ChIP experiments, depleting cells of Pfh1 slows fork progression within telomeres [11,54].
8. Break induced replication (BIR)
BIR is a recombination-dependent pathway for repairing one-ended DSBs that arise from eroded telomeres or broken replication forks (Fig. 2C). The 3′ single-strand tail on the broken end invades an intact donor molecule by Rad51-dependent strand transfer [60]. This transfer generates a displacement or D-loop in which the invading strand is lengthened by DNA polymerase δ/Pol32 (Pol32 is a non-essential subunit of DNA Pol δ). This lengthening can proceed for very long distances, even to the end of the chromosome (reviewed in Refs. [61–64]). The major model for BIR suggests that as replication proceeds, the bubble retains its original size and migrates down the chromosome, while the newly synthesized DNA is detached from its template and spooled out as single-stranded DNA [46] (Fig. 2C). Ultimately the single-strand is filled in, but synthesis of the complementary strand is delayed relative to first strand synthesis. The long-lived single strand intermediate provides a possible explanation for why BIR is highly mutagenic [65,66]. As both strands from the site of the break are synthesized in the same cell cycle, the repaired chromosome contains a long stretch of conservatively synthesized DNA (Fig. 2C).
ScPif1 is critical for BIR, especially for long distance events. ScPif1 acts on at least two points: it recruits DNA Pol δ to the break and stimulates DNA Pol δ-mediated DNA synthesis and bubble migration. Similar to Pif1 inhibition of telomerase at DSBs [67], DNA damage-induced phosphorylation of Pif1 is required to stimulate its role in BIR [68].
During ALT when telomeres are lengthened by recombination, the substrate, an eroded telomere, is a one-ended break, suggesting that ALT might proceed via BIR. Consistent with this idea, in budding yeast, Pol32 is required for both type I and type II ALT pathways [69], and ScPif1 is essential for the Rad51-dependent type I recombination [70]. In addition, BIR may also lengthen telomeres in telomerase plus cells [68]. Although ALT has been documented as a back-up for telomerase in diverse organisms, so far BIR and Pif1 family helicases are linked to ALT only in budding yeast.
9. Biochemical functions of Pif1 family helicases provide hints for how they affect telomeres
With the exception of ScPif1, most eukaryotic Pif1 family helicases are difficult to purify. Thus, detailed biochemical analyses have largely been limited to ScPif1. Although ScPif1 unwinds tailed duplex DNA, it does so slowly and non-processively (reviewed in Ref. [31]). ScPif1 is more active on forked DNA molecules and even more active on RNA:DNA hybrids, which it unwinds efficiently even under single cycle conditions [71–73]. Likewise, ScPif1 efficiently unwinds a variety of G4 structures (but not all [74]), and the rate of this unwinding is not affected measurably by the presence of a DNA trap [36,72,75]. Indeed, ScPif1 unwinding of G4 structures is so robust that it can counter the effects of G4 stabilizing drugs [72]. In addition, the presence of a G4 structure can increase ScPif1 unwinding of nearby duplex DNA [76]. The relatively modest unwinding by ScPif1of tailed duplexes and the rapid unwinding of RNA/DNA hybrids and G4 structures is seen by both ensemble and single molecule analyses [72]. Single molecule results suggest a patrolling model where ScPif1 sits on a 3′ single-strand tail and reels in the adjacent DNA, removing any G4 or RNA/DNA structures encountered in the process. Consistent with its processivity on G4 DNA in ensemble reactions, ScPif1 can repeatedly unwind G4 structures without dissociating from DNA [72]. Multiple bacterial Pif1 helicases are also much more active on G4 structures than tailed duplex DNA [36].
An in vitro system that mimics early steps in BIR has been used to determine the effects of ScPif1 on this process [45]. A D-loop is assembled on a circular template using Rad51, RPA and Rad54. DNA Pol δ is added with and without ScPif1. Whereas DNA Pol δ alone generates 200–300 nucleotides of DNA, when ScPif1 is present, up to several thousand nucleotides of single-stranded DNA are made. Thus, ScPif1 is not required for initiation of BIR but has a strong stimulatory effect on DNA Pol δ synthesis.
As RNA/DNA hybrids and G4 structures are the preferred substrates for ScPif1, it is tempting to speculate that its diverse effects on telomeres are due to one or both of these activities. If telomeric G-tails fold into an intra-molecular G4 structure, ScPif1 could unwind it, but this unwinding is unlikely to explain ScPif1’s inhibition of telomerase as intra-molecular G4 structures inhibit telomerase [77,78] (although it could explain how Pfh1 promotes fission yeast telomerase [54,79]). A more appealing model is that ScPif1 removes telomerase from DNA ends by unwinding the RNA/DNA hybrid that forms between the template region in telomerase RNA and the single-stranded G-tail (Fig. 2B). Alternatively or in addition, ScPif1 may displace the catalytic subunit, Est2, from DNA, a possibility consistent with genetic experiments that suggest a direct interaction between Est2 and ScPif1 [80].
How do Rrm3 and Pfh1 promote semi-conservative replication of telomeric DNA? At a subset of their many in vivo targets, Rrm3 and Pfh1 promote fork progression past stable non-nucleosomal protein complexes (Fig. 3A). For example, the fork progression activity of both helicases at the ribosomal DNA replication fork barrier (RFB) is no longer needed when protein(s) that bind the RFB are absent [11,42]. However, at budding yeast telomeres, there is no good candidate for a telomere protein whose presence confers Rrm3-sensitivity. Given these considerations, we speculate that Rrm3 and Pfh1 promote semi-conservative replication of telomeric DNA by resolving TERRA-generated R-loops and/or G4 structures. These possibilities can be addressed by studying the effects of G4 stabilizing drugs and RNAseH over-expression on fork progression through telomeres.
During BIR, ScPif1 is thought to recruit DNA Pol δ to the recombination intermediate and to use its ATPase activity to promote opening of the double helix for DNA synthesis and the migration of the D-loop [45]. In vivo, synthesis of the complementary strand is delayed relative to first strand synthesis (Fig. 2C). This single strand intermediate should readily form G4 structures as G4 motifs are found across the lengths of all 16 yeast chromosomes [81]. Thus, we speculate that ScPif1 has an additional role in BIR that involves G4 dissolution. If ALT proceeds via BIR, G4 unwinding might be particularly important for replication-driven recombination of G-rich telomeric DNA.
10. Concluding remarks
Primarily from work in budding and fission yeasts, it is clear that Pif1 helicases act in numerous important ways to promote genome stability. Telomeres are only one of their many substrates, and even their effects on telomeres are multiple and diverse. Pif1 family helicases can promote (fission yeast) or inhibit (budding yeast) telomerase. Rrm3 and Pfh1 promote semi-conservative telomere replication, and ScPif1 is critical for ALT. By detailing these functions, we do not imply that Pif1 family helicases are the only helicases with important roles in telomere maintenance. Indeed there is considerable evidence for telomere specific functions for mammalian helicases such as RTEL1 [15,16,24], BLM [14,82], and WRN [83] in replication of telomeric DNA. Rather studies on the telomeric roles of Pif1 family helicases can define multiple steps in telomere biology that are likely to require specialized activities of specific DNA helicases.
Acknowledgments
Work in the Zakian lab is supported by grants from the NIGMS. CLG is supported by a pre-doctoral fellowship from the NSF. We thank C. Follonier, P.L.T. Tran, and C. Webb for helpful comments on the manuscript.
Abbreviations
- DSB
double strand break
- G4
G-quadruplex
- TERRA
(telomere repeat containing RNA)
- ALT
alternative lengthening of telomeres
- BIR
break induced replication
- Sc
Saccharomyces cerevisiae
- TA
telomere addition
- CST
Cdc13-Stn1-Ten1
- ChIP
chromatin immuno-precipitation
Footnotes
Conflict of interest
The authors have no conflicts of interest.
References
- 1.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. http://dx.doi.org/10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambert S, Carr AM. Checkpoint responses to replication fork barriers. Biochimie. 2005;87:591–602. doi: 10.1016/j.biochi.2004.10.020. http://dx.doi.org/10.1016/j.biochi.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 3.Aguilera A, Garcia-Muse T. R. loops from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. http://dx.doi.org/10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Galati A, Micheli E, Cacchione S. Chromatin structure in telomere dynamics. Front Oncol. 2013;3(46) doi: 10.3389/fonc.2013.00046. http://dx.doi.org/10.3389/fonc.2013.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paeschke K, McDonald KR, Zakian VA. Telomeres: structures in need of unwinding. FEBS Lett. 2010;584:3760–3772. doi: 10.1016/j.febslet.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conomos D, Pickett HA, Reddel RR. Alternative lengthening of telomeres: remodeling the telomere architecture. Front Oncol. 2013;3(27) doi: 10.3389/fonc.2013.00027. http://dx.doi.org/10.3389/fonc.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wellinger RJ, Zakian VA. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics. 2012;191:1073–1105. doi: 10.1534/genetics.111.137851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivessa AS, Zhou JQ, Schulz VP, Monson EM, Zakian VA. Saccharomyces Rrm3p, a 5′–3′ DNA helicase that promotes replication fork progression through telomeric and sub-telomeric DNA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- 10.Azvolinsky A, Giresi P, Lieb J, Zakian V. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell. 2009;34:722–734. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabouri N, McDonald KR, Webb CJ, Cristea IM, Zakian VA. DNA replication through hard-to-replicate sites, including both highly transcribed RNA Pol II and Pol III genes, requires the S pombe Pfh1 helicase. Genes Dev. 2012;26:581–593. doi: 10.1101/gad.184697.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabouri N, Capra JA, Zakian VA. The essential Schizosaccharomyces pombe Pfh1 DNA helicase promotes fork movement past G-quadruplex motifs to prevent DNA damage. BMC Biol. 2014;12(101) doi: 10.1186/s12915-014-0101-5. http://dx.doi.org/10.1186/s12915-014-0101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sfeir A, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. http://dx.doi.org/10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drosopoulos WC, Kosiyatrakul ST, Schildkraut CL. BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J Cell Biol. 2015;210:191–208. doi: 10.1083/jcb.201410061. http://dx.doi.org/10.1083/jcb.201410061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarek G, Vannier JB, Panier S, Petrini JH, Boulton SJ. TRF2 recruits RTEL1 to telomeres in S phase to promote t-loop unwinding. Mol Cell. 2015;57:622–635. doi: 10.1016/j.molcel.2014.12.024. http://dx.doi.org/10.1016/j.molcel.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vannier JB, et al. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science. 2013;342:239–242. doi: 10.1126/science.1241779. http://dx.doi.org/10.1126/science.1241779. [DOI] [PubMed] [Google Scholar]
- 17.Benetti R, Garcia-Cao M, Blasco MA. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat Genet. 2007;39:243–250. doi: 10.1038/ng1952. [DOI] [PubMed] [Google Scholar]
- 18.Kueng S, Oppikofer M, Gasser SM. SIR proteins and the assembly of silent chromatin in budding yeast. Annu Rev Genet. 2013;47:275–306. doi: 10.1146/annurev-genet-021313-173730. http://dx.doi.org/10.1146/annurev-genet-021313-173730. [DOI] [PubMed] [Google Scholar]
- 19.Ottaviani A, Gilson E, Magdinier F. Telomeric position effect: from the yeast paradigm to human pathologies? Biochimie. 2008;90:93–107. doi: 10.1016/j.biochi.2007.07.022. http://dx.doi.org/10.1016/j.biochi.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 20.Ivessa AS. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 21.Paeschke K, Capra JA, Zakian VA. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piazza A. Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res. 2010;38:4337–4348. doi: 10.1093/nar/gkq136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin A Adams, Dionne I, Wellinger RJ, Holm C. The function of DNA polymerase alpha at telomeric G tails is important for telomere homeostasis. Mol Cell Biol. 2000;20:786–796. doi: 10.1128/mcb.20.3.786-796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. http://dx.doi.org/10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 25.Rhodes D, Lipps HJ. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. http://dx.doi.org/10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azzalin CM, Lingner J. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015;25:29–36. doi: 10.1016/j.tcb.2014.08.007. http://dx.doi.org/10.1016/j.tcb.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Bah A, Wischnewski H, Shchepachev V, Azzalin CM. The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res. 2012;40:2995–3005. doi: 10.1093/nar/gkr1153. http://dx.doi.org/10.1093/nar/gkr1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Redon S, Zemp I, Lingner J. A three-state model for the regulation of telomerase by TERRA and hnRNPA1. Nucleic Acids Res. 2013;41:9117–9128. doi: 10.1093/nar/gkt695. http://dx.doi.org/10.1093/nar/gkt695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farnung BO, Brun CM, Arora R, Lorenzi LE, Azzalin CM. Telomerase efficiently elongates highly transcribing telomeres in human cancer cells. PLoS One. 2012;7:e35714. doi: 10.1371/journal.pone.0035714. http://dx.doi.org/10.1371/journal.pone.0035714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balk B, et al. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol. 2013;20:1199–1205. doi: 10.1038/nsmb.2662. http://dx.doi.org/10.1038/nsmb.2662. [DOI] [PubMed] [Google Scholar]
- 31.Bochman ML, Sabouri N, Zakian VA. Unwinding the functions of the Pif1 family helicases. DNA Repair (Amst) 2010;9:237–249. doi: 10.1016/j.dnarep.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bochman ML, Judge CP, Zakian VA. The Pif1 family in prokaryotes: what are our helicases doing in your bacteria? Mol Biol Cell. 2011;22:1955–1959. doi: 10.1091/mbc.E11-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung WH. To peep into Pif1 helicase: multifaceted all the way from genome stability to repair-associated DNA synthesis. J Microbiol. 2014;52:89–98. doi: 10.1007/s12275-014-3524-3. http://dx.doi.org/10.1007/s12275-014-3524-3. [DOI] [PubMed] [Google Scholar]
- 34.Liu B, et al. Trypanosomes have six mitochondrial DNA helicases with one controlling kinetoplast maxicircle replication. Mol Cell. 2009;35:490–501. doi: 10.16/j.molcel.2009.07.004S1097-2765(09)00478-X. [pii][10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu B, Wang J, Yildirir G, Englund PT. TbPIF5 is a Trypanosoma brucei mitochondrial DNA helicase inved in processing of minicircle Okazaki fragments. PLoS Pathog. 2009;5:e1000589. doi: 10.1371/journal.ppat.1000589. http://dx.doi.org/10.1371/journal.ppat.1000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paeschke K. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinter SF, Aubert SD, Zakian VA. The Schizosaccharomyces pombe Pfh1p DNA helicase is essential for the maintenance of nuclear and mitochondrial DNA. Mol Cell Biol. 2008;28:6594–6608. doi: 10.1128/MCB.00191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snow B, et al. Murine pif1 interacts with telomerase and is dispensable for telomere function in vivo. Mol Cell Biol. 2007;27:1017–1026. doi: 10.1128/MCB.01866-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu B, et al. TbPIF1, a Trypanosoma brucei mitochondrial DNA helicase, is essential for kinetoplast minicircle replication. J Biol Chem. 2010;285:7056–7066. doi: 10.1074/jbc.M109.084038. http://dx.doi.org/10.1074/jbc.M109.084038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Futami K, Shimamoto A, Furuichi Y. Mitochondrial and nuclear localization of human Pif1 helicase. Biol Pharm Bull. 2007;30:1685–1692. doi: 10.1248/bpb.30.1685. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez R, et al. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol. 2012;8:301–310. doi: 10.1038/nchembio.780. http://dx.doi.org/10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torres JZ, Bessler JB, Zakian VA. Local chromatin structure at the ribosomal DNA causes replication fork pausing and genome instability in the absence of the S. cerevisiae DNA helicase Rrm3p. Genes Dev. 2004;18:498–503. doi: 10.1101/gad.1154704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossi ML, et al. Pif1 helicase directs eukaryotic Okazaki fragments toward the two-nuclease cleavage pathway for primer removal. J Biol Chem. 2008;283:27483–27493. doi: 10.74/jbc.M804550200. M804550200[pii][10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pike JE, Henry RA, Burgers PM, Campbell JL, Bambara RA. An alternative pathway for Okazaki fragment processing: resolution of fold-back flaps by Pif1 helicase. J Biol Chem. 2010;285:41712–41723. doi: 10.1074/jbc.M110.146894. http://dx.doi.org/10.1074/jbc.M110.146894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson MA, et al. Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013;502:393–396. doi: 10.1038/nature12585. http://dx.doi.org/10.1038/nature12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saini N, et al. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502:389–392. doi: 10.1038/nature12584. http://dx.doi.org/10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boule J, Vega L, Zakian V. The yeast Pif1p helicase removes telomerase from DNA. Nature. 2005;438:57–61. doi: 10.1038/nature04091. [DOI] [PubMed] [Google Scholar]
- 48.Phillips JA, Chan A, Paeschke K, Zakian VA. The pif1 helicase, a negative regulator of telomerase, acts preferentially at long telomeres. PLoS Genet. 2015;11:e1005186. doi: 10.1371/journal.pgen.1005186. http://dx.doi.org/10.1371/journal.pgen.1005186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makovets S, Blackburn EH. DNA damage signalling prevents deleterious telomere addition at DNA breaks. Nat Cell Biol. 2009;11:1383–1386. doi: 10.1038/ncb1985. http://dx.doi.org/10.1038/ncb1985 (ncb1985 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase-extendible and—nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 51.Li JR, Yu YT, Chien IC, Lu CY, Lin JJ, Li HW. Pif1 regulates telomere length by preferentially removing telomerase from long telomere ends. Nucleic Acids Res. 2014;42:8527–8536. doi: 10.1093/nar/gku541. http://dx.doi.org/10.1093/nar/gku541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou JQ, Monson EM, Teng SC, Schulz VP, Zakian VA. The Pif1p helicase, a catalytic inhibitor of telomerase lengthening of yeast telomeres. Science. 2000;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- 53.Zhou JQ. Schizosaccharomyces pombe pfh1+ encodes an essential 5′–3′ DNA helicase that is a member of the PIF1 sub-family of DNA helicases. Mol Biol Cell. 2002;13:2180–2191. doi: 10.1091/mbc.02-02-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McDonald KR, Sabouri N, Webb CJ, Zakian VA. The Pif1 family helicase Pfh1 facilitates telomere replication and has an RPA-dependent role during telomere lengthening. DNA Repair (Amst) 2014;24:80–86. doi: 10.1016/j.dnarep.2014.09.008. http://dx.doi.org/10.1016/j.dnarep.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luciano P, et al. RPA facilitates telomerase activity at chromosome ends in budding and fission yeasts. EMBO J. 2012;31:2034–2046. doi: 10.1038/emboj.2012.40. http://dx.doi.org/10.1038/emboj.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reynolds GE, et al. PIF1 disruption or NBS1 hypomorphism does not affect chromosome healing or fusion resulting from double-strand breaks near telomeres in murine embryonic stem cells. DNA Repair (Amst) 2011;10:1164–1173. doi: 10.1016/j.dnarep.2011.09.002. http://dx.doi.org/10.1016/j.dnarep.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mateyak M, Zakian V. Human PIF helicase is cell cycle regulated and associates with telomerase. Cell Cycle. 2006;23:2796–2804. doi: 10.4161/cc.5.23.3524. [DOI] [PubMed] [Google Scholar]
- 58.Zhang DH, Zhou B, Huang Y, Xu LX, Zhou JQ. The human Pif1 helicase, a potential Escherichia coli RecD homologue, inhibits telomerase activity. Nucleic Acids Res. 2006;34:1393–1404. doi: 10.1093/nar/gkl029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Makovets S, Herskowitz I, Blackburn EH. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol Cell Biol. 2004;24:4019–4031. doi: 10.1128/MCB.24.9.4019-4031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davis AP, Symington LS. RAD51-dependent break-induced replication in yeast. Mol Cell Biol. 2004;24:2344–2351. doi: 10.1128/MCB.24.6.2344-2351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malkova A, Haber JE. Mutations arising during repair of chromosome breaks. Annu Rev Genet. 2012;46:455–473. doi: 10.1146/annurev-genet-110711-155547. http://dx.doi.org/10.1146/annurev-genet-110711-155547. [DOI] [PubMed] [Google Scholar]
- 62.Malkova A, Ira G. Break-induced replication: functions and molecular mechanism. Curr Opin Genet Dev. 2013;23:271–279. doi: 10.1016/j.gde.2013.05.007. http://dx.doi.org/10.1016/j.gde.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anand RP, Lovett ST, Haber JE. Break-induced DNA replication, Cold Spring Harb. Perspect Biol. 2013;5:a010397. doi: 10.1101/cshperspect.a010397. http://dx.doi.org/10.1101/cshperspect.a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sakofsky CJ, Ayyar S, Malkova A. Break-induced replication and genome stability. Biomolecules. 2012;2:483–504. doi: 10.3390/biom2040483. http://dx.doi.org/10.3390/biom2040483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deem A, et al. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. http://dx.doi.org/10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sakofsky CJ, et al. Break-induced replication is a source of mutation clusters underlying kataegis. Cell Rep. 2014;7:1640–1648. doi: 10.1016/j.celrep.2014.04.053. http://dx.doi.org/10.1016/j.celrep.2014.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Makovets S, Blackburn EH. DNA damage signalling prevents deleterious telomere addition at DNA breaks. Nat Cell Biol. 2009;11:1383–1386. doi: 10.1038/ncb1985. http://dx.doi.org/10.1038/ncb1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vasianovich Y, Harrington LA, Makovets S. Break-induced replication requires DNA damage-induced phosphorylation of Pif1 and leads to telomere lengthening. PLoS Genet. 2014;10:e1004679. doi: 10.1371/journal.pgen.1004679. http://dx.doi.org/10.1371/journal.pgen.1004679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. http://dx.doi.org/10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 70.Hu Y, et al. Telomerase-null survivor screening identifies novel telomere recombination regulators. PLoS Genet. 2013;9:e1003208. doi: 10.1371/journal.pgen.1003208. http://dx.doi.org/10.1371/journal.pgen.1003208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boule JB, Zakian VA. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 2007;35:5809–5818. doi: 10.1093/nar/gkm613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou R, Zhang J, Bochman ML, Zakian VA, Ha T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. Elife. 2014;3 doi: 10.7554/eLife.02190. http://dx.doi.org/10.7554/elife.02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chib S, Byrd AK, Raney KD. Yeast helicase Pif1 unwinds RNA:DNA hybrids with higher processivity than DNA:DNA duplexes. J Biol Chem. 2016 doi: 10.1074/jbc.M115.688648. http://dx.doi.org/10.1074/jbc.m115.688648. [DOI] [PMC free article] [PubMed]
- 74.Byrd AK, Raney KD. A parallel quadruplex DNA is bound tightly but unfolded slowly by pif1 helicase. J Biol Chem. 2015;290:6482–6494. doi: 10.1074/jbc.M114.630749. http://dx.doi.org/10.1074/jbc.M114.630749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ribeyre C, et al. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duan XL, et al. G-quadruplexes significantly stimulate Pif1 helicase-catalyzed duplex DNA unwinding. J Biol Chem. 2015;290:7722–7735. doi: 10.1074/jbc.M114.628008. http://dx.doi.org/10.1074/jbc.M114.628008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. http://dx.doi.org/10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 78.Paeschke K, et al. Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nat Struct Mol Biol. 2008;15:598–604. doi: 10.1038/nsmb.1422. http://dx.doi.org/10.1038/nsmb.1422. [DOI] [PubMed] [Google Scholar]
- 79.Audry J, et al. RPA prevents G-rich structure formation at lagging-strand telomeres to allow maintenance of chromosome ends. EMBO J. 2015;34:1942–1958. doi: 10.15252/embj.201490773. http://dx.doi.org/10.15252/embj.201490773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eugster A. The finger subdomain of yeast telomerase cooperates with Pif1p to limit telomere elongation. Nat Struct Mol Biol. 2006;13:734–739. doi: 10.1038/nsmb1126. [DOI] [PubMed] [Google Scholar]
- 81.Capra JA, Paeschke K, Singh M, Zakian VA. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput Biol. 2010;6:e1000861. doi: 10.1371/journal.pcbi.1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zimmermann M, Kibe T, Kabir S, de Lange T. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev. 2014;28:2477–2491. doi: 10.1101/gad.251611.114. http://dx.doi.org/10.1101/gad.251611.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 84.Webb CJ, Wu Y, Zakian VA. DNA repair at telomeres: keeping the ends intact. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012666. http://dx.doi.org/10.1101/cshperspect.a012666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Griffith JD. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]


