Abstract
Once deemed heretical, emerging evidence now supports the notion that the inheritance of acquired characteristics can occur through ancestral exposures or experiences and that certain paternally acquired traits can be ‘memorized’ in the sperm as epigenetic information. The search for epigenetic factors in mammalian sperm that transmit acquired phenotypes has recently focused on RNAs and, more recently, RNA modifications. Here, we review insights that have been gained from studying sperm RNAs and RNA modifications, and their roles in influencing offspring phenotypes. We discuss the possible mechanisms by which sperm become acquisitive following environmental–somatic–germline interactions, and how they transmit paternally acquired phenotypes by shaping early embryonic development.
The idea of the inheritance of acquired traits, which suggests that ancestral life experiences acquired in the environment can be inherited by offspring, was once under heated debate1. Recently, great interest in this topic has been rekindled, owing to accumulating evidence from well-designed experiments in different animal species2–8. These studies demonstrated that certain paternal traits that are acquired in response to ancestral exposures, such as toxicant contact, mental stresses and diet changes, can be inherited by the offspring, suggesting that epigenetic inheritance can occur through the sperm. This conclusion was further supported by recent data obtained using in vitro fertilization (IVF) or the injection of sperm heads to fertilize oocytes9–11, which essentially ruled out germline-independent paternal factors (such as seminal fluid and male–female contacts12), thus indicating that sperm could be the carrier of ancestral epigenetic memory. The resurrected idea of germline inheritance of acquired traits has the potential to further our understanding of the aetiology of many modern human diseases that may have originated from environmentally induced transgenerational effects and thus may have widespread medical and social implications.
One of the major reasons for the previous dismissal of the ‘inheritance of acquired traits’ hypothesis was the knowledge that epigenetic reprogramming in the zygote and primordial germ cells resets the epigenome to a naive state by removing most of the epigenetic marks (for example the 5-methylcytosine (5 mC) mark produced by DNA methylation) that result from environmental exposures13. However, changes in DNA methylation in sperm have been detected after exposure to environmental stresses14, presumably having escaped from epigenetic reprogramming, suggesting that these DNA modifications represent a form of molecular carrier for acquired epigenetic memory15. Nevertheless, recent studies based on comprehensive DNA methylation analyses cast doubts on the role of sperm DNA methylation as a molecular mark of epigenetic inheritance that responds to environmental exposure; stochastic variations in sperm DNA methylation were found to make a greater contribution to the sperm methylome than diet16. In other studies, in which sperm DNA methylation patterns showed alterations after environmental exposure, the changed patterns of DNA methylation were not maintained in subsequent generations17–19. These data suggest that other molecular information carriers in sperm are required for the intergenerational transfer and maintenance of acquired traits; such information carriers could include histone marks that can be maintained across generations20–22 and non-coding RNAs (ncRNAs), which can act in trans to escape the reprogramming process23. Recently, proof of a direct causal role of sperm RNAs in transferring acquired traits across generations in mammals has emerged; that is, the injection of total sperm RNAs or a subset of sperm RNAs (for example, microRNAs (miRNAs) or tRNA-derived small RNAs (tsRNAs; also known as tRNA-derived RNA fragments (tRFs)) into normal zygotes can generate offspring that recapitulate paternal phenotypes, including mental stress and metabolic changes9,24–26. Furthermore, sperm RNA modifications and RNA-editing events have been implicated as active participants in the intergenerational transfer of epigenetic information9,27,28, adding layers of complexity to the mechanisms that underlie this process and pointing to a new direction for future studies.
In this Review, we outline the current understanding of acquired epigenetic inheritance through sperm, mainly in mammalian species, focusing on the roles of sperm RNAs and RNA modifications. We discuss the potential mechanisms by which sperm RNAs become acquisitive and the flow of information from the environment to somatic cells and then to sperm. We also discuss known and potential mechanisms by which sperm RNAs reshape embryonic development and offspring phenotypes, and the roles of sperm RNA modifications in this process. We avoid extensive discussion of other candidate epigenetic carriers in sperm (for example, DNA methylation marks and histone modifications, which have been extensively reviewed elsewhere15,29), unless they are highly relevant to the functions and mechanisms of sperm RNAs.
Acquired inheritance through sperm
The first evidence of sperm-mediated epigenetic inheritance of phenotypic traits in mammals was derived from nuclear transplantation experiments in mice30. Such embryo manipulations induced abnormalities in the offspring — including growth defects and aberrant gene expression — which were documented to be inherited by the next generation of offspring through the paternal germ line30, probably owing to altered epigenetic features. A more representative case of environmentally induced epigenetic inheritance through sperm was based on a chemical exposure experiment7: when pregnant female rats were exposed to vinclozolin (a pesticide), the male offspring of the exposed mothers showed a wide range of disease phenotypes, which were inherited by most of the males of the subsequent generations examined that were not directly exposed to vinclozolin. In these early studies, genetic and epigenetic mechanisms cannot be fully distinguished, because nuclear transplantation and chemical exposure can cause gene mutation. Nevertheless, these studies drew great interest in the potential for epigenetic inheritance and triggered further studies to explore whether phenotypes induced by other forms of environmental exposure could also be inherited through the male germ line. Now, data from independent laboratories have demonstrated that paternal characteristics acquired during environmental exposures (including a high-fat diet (HFD)5,9,10,26,31, a low-protein diet5,11, mental stresses8,25,32, odour sensitivity to specific chemicals6 and susceptibility to irradiation33) can indeed be inherited by the offspring through the paternal germ line in a non-Mendelian fashion, thus supporting the idea that certain life experiences and environmental cues can be ‘memorized’ in sperm as epigenetic information.
Is DNA methylation the cause?
At the molecular level, a plausible explanation for this paternal germline inheritance could be environmentally induced aberrant DNA methylation patterns that are maintained in the sperm and escape at least two waves of epigenetic reprogramming13, resulting in the persistence of these patterns in the offspring. Evidence supporting the escape of DNA methylation marks from epigenetic reprogramming comes from well-established mouse studies on intracisternal A particle (IAP) retrotransposon-controlled agouti viable yellow (Avy)34 or axin fused (AxinFu)35 gene activity, in which the methylation state of the IAP was incompletely erased during reprogramming of the male germ line, providing an explanation for the epigenetic inheritance of coat colour or kinky-tail phenotype, respectively. However, such clearly identified inheritance of DNA methylation state in mammals is rare, and consistent changes in gene expression in subsequent offspring resulting from environmentally induced aberrant sperm DNA methylation have not been demonstrated thus far. Furthermore, there is controversy regarding the influence of environmental stresses on DNA methylation14,16, and recent data appear to contradict the role of methylated DNA as a persistent mark that maintains phenotypes in subsequent generations17,36. More comprehensive DNA methylomic analyses with improved coverage and depth37 are required to draw a final conclusion. Uncertainties surrounding the role of DNA methylation have inspired the search for other epigenetic information carriers in sperm that could potentially transfer environmentally acquired traits, which has led to a focus on sperm RNAs.
Sperm RNAs mediate transmission of acquired phenotypes
The existence and functions of sperm RNAs were once a subject of debate (BOX 1). High-throughput technologies, such as microarray analysis and RNA sequencing (RNA-seq), have led to the recent discovery and characterization of various types of sperm RNAs23 (FIG. 1). Sperm RNAs were first implicated in mediating offspring phenotypes in early studies on the transvection38–40 and paramutation41–44 phenomena in mice (BOX 2), and these studies led to the discovery of sperm RNAs as the trans-acting molecules that mediate these phenomena41–43. More recent studies have further demonstrated that sperm RNAs can function as molecular carriers that transmit paternally acquired characteristics to the offspring9,24–26. The first experiment to directly demonstrate the causal relationship between sperm RNAs and inherited characteristics involved the injection of total sperm RNAs from mentally stressed male mice into normal zygotes, which generated offspring that reproduced the behavioural and metabolic alterations observed in the father24. Importantly, it was noted that the stress phenotypes in the offspring were passed onto the subsequent generation, suggesting that transgenerational inheritance is due to the induction of stable, heritable epigenetic marks24. Similarly, two independent groups showed that the injection of total sperm RNAs from males that were fed a chronic HFD9 or a high-fat–high-sugar diet26 into normal zygotes generated offspring that either partially9 or fully26 recapitulated the paternal metabolic disorders. Because mouse sperm contain various types of mRNAs and ncRNAs (for example, miRNAs, PIWI-interacting RNAs (piRNAs), tsRNAs, mitochondrial genome-encoded small RNAs (mitosRNAs) and long ncRNAs (lncRNAs)), identification of the subpopulations of sperm RNAs that are responsible for the transmission of paternal phenotypes is of great interest, and recent emerging evidence has put sperm miRNAs and tsRNAs in the spotlight.
Box 1. Historical view of sperm RNAs.
The mission of a sperm was once believed solely to be the delivery of paternal DNA to the oocyte. Reports of the identification of RNAs in sperm were initially sporadic and controversial. However, the emergence of high-throughput strategies, such as microarray and RNA sequencing (RNA-seq) analyses, has begun to end this controversy by providing solid evidence of the existence of RNA in sperm127. The demonstration of the delivery of sperm RNAs into oocytes unequivocally ended the controversy128 and gave rise to new questions. What are the roles of sperm RNAs? Are sperm RNAs simply negligible remnants of spermatogenesis, or could they represent something more?
Whether sperm RNAs are necessary for early embryonic development remains controversial129–131, particularly given the recent generation of parthenogenetic mice with appreciable survival rates132,133; nonetheless, their potential synergistic action with maternal RNAs have been discussed with interest134. Thus far, the most significant biological functions identified for sperm RNAs are their involvement in non-Mendelian inheritance in mammals, such as the paramutation phenomenon in mice (BOX 2), and their contribution to the intergenerational inheritance of paternally acquired traits, including mental and nutritional stresses9,24–26.
Figure 1. Small non-coding RNAs in mouse male germ cells.
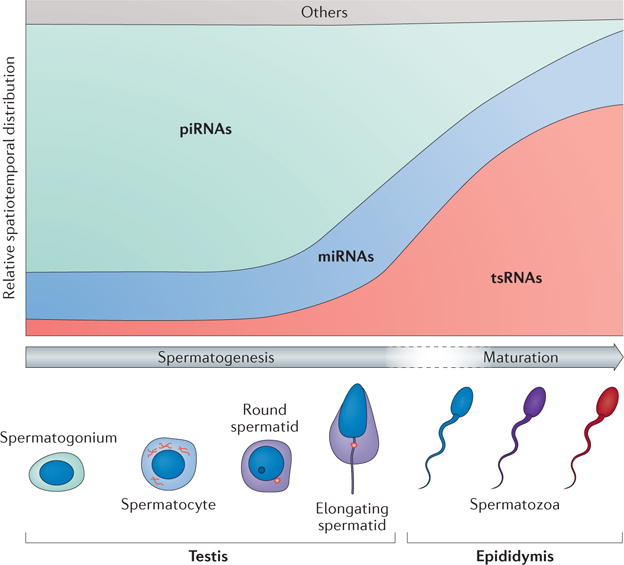
Spatiotemporal distribution of major types of small non-coding RNAs (for example, PIWI-interacting RNAs (piRNAs), microRNAs (miRNAs) and tRNA-derived small RNAs (tsRNAs)) in sperm precursors (for example, spermatogonia, spermatocyte, round spermatid and elongated spermatid) during spermatogenesis in the testis, and in spermatozoa during sperm maturation in the epididymis (which occurs during the transition from the caput epididymis to the cauda epididymis).
Box 2. Transvection and paramutation in mice.
Transvection occurs during chromosome pairing such as during the chromosome synapsis stage of male meiosis in mice, in which an epigenetic modification in one allele is transferred to the other. The transvection phenomenon in mice was first noticed during transgene manipulation38–40. In these cases, induced DNA methylation on one allele (triggered by transgene manipulations at this allele) was transferred to the other allele, the genotype of which remained wild type. The ectopically methylated region of the wild-type allele could then be maintained in subsequent generations in the absence of the initial triggering factors38,39, although no associated phenotype was reported. These phenomena suggest that the manipulation of one allele triggers certain mobile molecules in the cell that could act in trans, thus affecting both the manipulated and the intact alleles, and may also contribute to the maintenance of the effect in the wild-type allele. These trans-acting molecules could be RNAs.
The paramutation phenomenon is conceptually the same as transvection regarding the molecular interactions between two pairing alleles, yet, it is represented by a prominent phenotype. Paramutation was first documented in plants135. In the first paramutation phenomenon reported in mice, a ‘white tail’ phenotype, representing disruption of the Kit gene at one allele, was surprisingly observed in the genotypically wild-type offspring in which the wild-type allele had been exposed to the disrupted Kit allele during meiosis43. It was then demonstrated that abnormally generated RNAs from the Kit loci that were ectopically maintained in wild-type sperm caused the white tail phenotype43, and this finding was repeated by an independent group136. Additional paramutation phenomena in mice have since been reported in other studies28,36,41,42,44. These phenomena implicate diffuse molecules, such as RNAs, that are triggered by one mutated allele acting in trans to induce a heritable change in the other genotypically intact allele, leading to a paramutant phenotype in offspring with a wild-type genotype. The figure depicts the common principle underlying transvection and paramutation phenomena that have been reported in mice. Note that both paramutation and transvection produced changes at the epigenetic level, whereas only paramutation produced a phenotype at the animal level.

Sperm miRNAs
Several studies have reported alterations in sperm miRNA profiles after various types of paternal exposure8,31,33,45, but the functions of sperm miRNAs in transmitting paternally acquired phenotypes were reported only recently25,26. In one mouse study, following the zygotic injection of a combination of nine miRNAs that were altered in sperm after chronic paternal stress, the offspring developed stress-dysregulation phenotypes that recapitulated those of the father25. In another study, zygotic injection of miR-19b (the expression of which is upregulated in the sperm of mice consuming a high-fat–high-sugar diet) induced obesity and glucose intolerance in the offspring, recapitulating paternal phenotypes26. Moreover, the glucose-intolerance phenotype induced in male offspring persisted in subsequent generations despite incomplete penetrance after mating with healthy females26, suggesting that the epigenetic changes induced by miR-19b injection were maintained in the germ line, although concerns were discussed by the authors regarding overdosage of a single miRNA injection, which might not faithfully represent physiological conditions26.
Sperm tsRNAs
The enrichment of tsRNAs in mature sperm was first discovered through sperm RNA-seq analysis in mice46. Sperm tsRNAs are mainly derived from the 5′ end of tRNAs, range in size from 29 nt to 34 nt and are more abundant than miRNAs, constituting the majority of small ncRNAs in sperm9,11,46. Although details of their biogenesis remain unknown, sperm tsRNAs show altered profiles after a HFD9 or a low-protein diet10 in mice, after a HFD45 or an environmental-compound exposure in rats47, and in obese humans48, suggesting that sperm tsRNAs can function as sensitive markers of environmental exposure. Further functional evidence obtained by injecting tsRNA-enriched RNA fragments from the sperm of mice fed a HFD into zygotes indicated that tsRNAs could induce metabolic disorders in offspring, which is similar to the effect of injecting total sperm RNAs9. This effect may be exerted in a synergistic manner along with other sperm or oocyte RNAs or proteins. Moreover, zygotic injection of a combination of tsRNAs9 or a single tsRNA (tsRNAGly(GCC))11 could change early embryonic gene expression, indicating that sperm tsRNAs play an active part in the transmission of paternally acquired traits by reshaping early embryonic development.
A recent study revealed that sperm tsRNAs show a dramatic increase in abundance during late spermatogenesis and epididymal maturation46 (FIG. 1). In addition to the possibility that tsRNA biogenesis occurs through specific cleavage of tRNAs during these stages, recent evidence suggests that sperm may obtain tsRNAs from extracellular vesicles (EVs) that are produced by epididymal epithelial cells11. Moreover, sperm tsRNAs were found to harbour numerous RNA modifications that contribute to the stability of tsRNAs9, and levels of 5-methylcytidine (m5C) and N2-methylguanosine (m2G) in sperm tsRNAs were significantly increased after paternal HFD consumption9. These discoveries raised exciting new possibilities regarding the mechanisms through which sperm gain information from the environment and the potential roles of RNA modifications in mediating epigenetic memory. We discuss these topics in detail below.
Acquisitive sperm — information flow
The production of functional sperm begins with spermato genesis in the testis, which is followed by maturation in the epididymis; each stage involves complex regulatory processes. How can developing sperm sense environmental changes, and how do they become acquisitive and enable the storage of epigenetic memory? Understanding the flow of information (physiological, cellular and genetic) from the environment to somatic cells and then to germ cells is key to answering these questions. Interestingly, heated debate over the flow of hereditary information between the soma and the germ line had already started more than 100 years ago (BOX 3). The once widely accepted ‘Weismann barrier’ theory is now increasingly being challenged49, whereas the once disregarded Darwin’s ‘pangenesis’ theory is receiving increasing attention50, owing, in particular, to advances in research on EVs that can deliver proteins51 and RNAs11 to sperm, and to the discovery of various extracellular mobile RNAs that can be found outside EVs52.
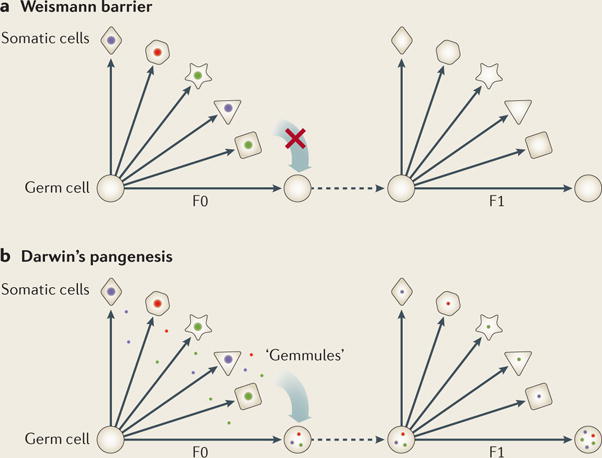
Box 3. Somatic–germline barrier: unbreachable?
The Weismann barrier is a theoretical barrier proposed by August Weismann in 1893; the central idea was that the flow of information can only move from germ cells to somatic cells and not in the reverse direction (see the figure). Based on this theory, traits acquired during environmental exposure cannot be inherited through the germ line.
Darwin’s pangenesis is a theory developed by Charles Darwin in 1868 as a possible mechanism to explain Jean-Baptiste Lamarck’s theory of evolution (proposed in 1801) regarding the inheritance of acquired characteristics. In the pangenesis theory, hypothetical particles called ‘gemmules’ can transfer information between somatic cells and between somatic and germ cells (see the figure), enabling the possible inheritance of acquired traits during ancestral exposure. Interestingly, in a series of debates on pangenesis in 1871 (REF. 137), it was foreseen that “The supposed gemmules must be much more minute than the smallest particles that can be seen by the highest magnifying powers used in these days,” and that “…the gemmules are actual particles suspended and not dissolved in the fluid”. These descriptions were visionary, as the properties of gemmules proposed approximately 150 years ago appear to match well with those of extracellular vesicles (EVs) (for example, exosomes), which are the topic of intense research today. EVs carry a range of active molecules, including various regulatory RNAs (for example, microRNAs (miRNAs) and tRNA-derived small RNAs (tsRNAs)), and indeed show the ability to transfer information between somatic cells53 and from somatic cells to germ cells62.
Transfer of RNAs from soma to sperm
Extracellular vesicles
EVs are membrane-bound microvesicles that are ubiquitously found in biofluids. They display sensitive responses to bodily conditions53 and are frequently found in mammalian reproductive tracts, particularly in the epididymis, semen and uterine fluids54. Cellular communication through EVs represents a new frontier in cell biology, bringing novel approaches for biomarker diagnostics55 and drug delivery56, and raising the possibility of transferring RNA cargoes from somatic cells to germ cells as a way to deliver environmentally obtained information (either harmful or adaptive). Recent data have shown that EVs in the epididymis (called epididymosomes) contain abundant tsRNAs, similar to mature sperm11, and in vitro experiments have demonstrated that epididymosomes can fuse with and transfer tsRNAs into sperm11. This scenario suggests a transfer of tsRNAs between the epididymis and sperm, and is highly intriguing, as a similar transfer might occur in the seminal fluid, where EVs are enriched with mi RNAs and tsRNAs57. Moreover, an enrichment of mi RNAs and tsRNAs is also found in serum58,59 and neuronal synaptic vesicles60, both of which exhibit sensitive physiological changes to bodily conditions, such as inflammation, ageing and calorie restriction58,59,61. These findings implicate potential connections through EVs at the organismal level and suggest the possibility that EVs can be transported from the soma to the germ line, along with their RNA cargoes. Indeed, a recent study reports soma-to-sperm transmission of RNAs in mice with xenografts of human tumour cells that expressed tumour-specific enhanced green fluorescent protein (EGFP) RNAs, which were subsequently found in the bloodstream and sperm of these mice62, although the acquisition of phenotypic features were not examined in their offspring. In another study, soluble serum factors (of unknown identity, but were possibly EVs or RNAs) from rats with liver damage induced chromatin changes at specific loci in the sperm, which then transmitted the liver-specific wound-healing capacity to subsequent generations63. In addition to the evidence from mammals, long-distance transport of RNAs from soma to germ line has been reported in Caenorhabditis elegans and was shown to generate transgenerational gene silencing64–68.
In addition, recent reports have shown that semen samples harbour a range of microbiota69, which can release outer membrane vesicles that are similar to EVs70 and thus may also deliver unexpected RNA cargoes to sperm. It would be of great interest to explore whether sperm could also gain exogenous RNAs from EVs or bacterial outer membrane vesicles during or after ejaculation. Notably, even after ejaculation into the female reproductive tract, sperm remain immersed in an EV-enriched environment that includes EVs from not only the male, but also the female partner or even from another male under polyandrous conditions in which the sperm could experience cryptic female choice, sperm cooperation and competition (BOX 4). It would be interesting to investigate a role under such circumstances for RNA-containing EVs as communication units mediating sperm–sperm and/or female–sperm communication. Studies such as these have the potential to develop new theories in evolution, particularly regarding environmentally driven, rapid or adaptive evolution.
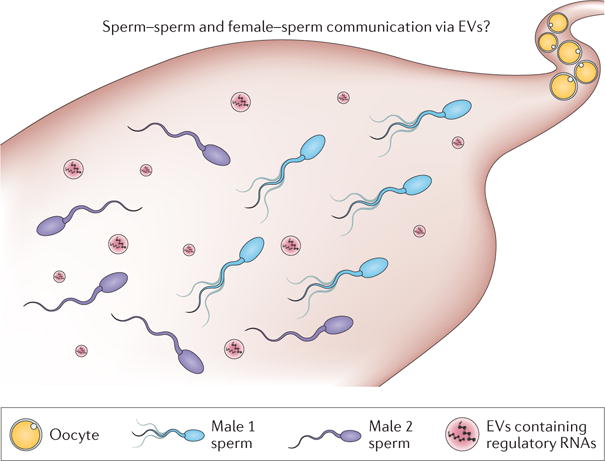
Box 4. Post-copulatory female cryptic choice and sperm behavior.
In polyandrous species, the female can influence the events that occur after copulation leading to biased sperm selection, resulting in the successful fertilization of the oocyte by the sperm of one male over that of another138. This process, termed post-copulatory female cryptic choice, is among the most mysterious biological events ever known; yet, it is important because it biases paternity and influences the trajectory of evolution.
Sperm can also be influenced by each other (see the figure). After ejaculation into the female reproductive tract, individual sperm can group together to form certain networks for mutual advantage in a phenomenon called sperm cooperation that has been well documented in certain species139. By contrast, sperm competition describes the process in polyandrous species in which the sperm of different males compete in the female reproductive tract to fertilize a given set of oocytes, representing an important series of post-copulatory events that drives evolution140.
Despite the evolutionary importance of these well-documented phenomena, the molecular basis of female–sperm and sperm–sperm communication remains enigmatic. Could RNA-containing microvesicles, which are enriched in the semen and uterine fluid, be involved in the mechanism of information exchange during these post-copulatory phenomena?
Specificity and regulation of EV transport
Does the ubiquitous existence of EVs in bodily fluids suggest that cells (including sperm) constantly exchange information via EVs in a non-selective manner? This appears to be counterintuitive. Indeed, recent advances in cancer biology have revealed that EVs are active players in organ-specific metastasis, using distinct membrane integrins as a mechanism for tissue-specific cargo docking71. In addition, pathological conditions have been shown to facilitate the transport of RNA-containing EVs to unanticipated locations. For example, an inflammatory response substantially increased the transport of EVs through the blood–brain barrier, resulting in the direct delivery of functional RNAs from blood cells to neurons, which is rarely observed in healthy mice72. In this regard, it is notable that both obesity73 and mental stress74 are well established to be inflammatory in nature, and both HFD-induced obesity75 and psychological stress76 have been reported to disrupt the blood–testis barrier, which might facilitate the transport of environmentally induced somatic EVs to the developing spermatogenic cells and sperm. The exploration of targeting specificities of RNA-containing EVs under physiological and pathological conditions is still in its infancy, but is of great importance as it should help to unravel the mystery of which types of acquired traits are transmitted to offspring through the germ line and the circumstances under which this is likely to occur. Furthermore, although IVF and sperm head injection protocols have demonstrated that the sperm itself carries sufficient information to transmit paternally acquired phenotypes9–11, it is also possible that non-sperm factors, such as EVs in the semen, contribute to the final phenotype of offspring, especially given the recent experimental evidence for telegony77,78 — a mysterious phenomenon in which offspring can inherit the traits of a male that mated previously with their mother.
Mobile RNAs
Small RNAs are carried as cargo in EVs, but can also be found in bodily fluids outside EVs58,79. Maintenance of their stability outside EVs requires binding proteins, specific secondary structures or RNA modifications58,79, which might also allow their transport into and out of cells. In C. elegans, specific RNA import by cells involves selective transmembrane RNA-gated channels, such as systematic RNA-interference defective protein-1 (SID-1) and SID-2 (REFS 80,81), which allow the transport of RNAs into germ cells, leading to transgenerational gene silencing64. The existence and functions of the mammalian homologues of SID family proteins remain to be determined. A recently defined intriguing aspect of small RNA secondary structure is that certain tsRNAs can adopt an RNA G-quadruplex conformation that contributes to neuroprotective responses82. Most interestingly, tsRNAs with a G-quadruplex structure can be spontaneously taken up by neuronal cells, suggesting that this specific structure could facilitate the transmembrane movement of mobile RNAs. This discovery may be far-reaching because most serum tsRNAs are outside EVs58 and might enter sperm cells by adopting specific RNA structures such as the G-quadruplex. Moreover, evaluating the potential structure of mobile RNAs might also provide fresh insights into the much-debated issue of cross-kingdom transport of small RNAs (for example, the ingestion of plant RNAs by the human body83–87), which is probably affected by RNA modifications because they have been shown to protect mi RNAs88 and tsRNAs9 from degradation.
Alternative ways to alter RNA profiles in sperm
An alternative way of changing sperm RNA profiles in response to environmental stimuli is to alter gene expression during spermatogenesis. Indeed, reports have shown that a HFD can change the testicular transcriptome in mice31, which may lead to altered sperm RNA contents that mirror the environmental exposure. Moreover, injection of total RNAs extracted from the testes of HFD-fed males into normal zygotes induced metabolic disorders in the offspring, similarly to the injection of total RNAs from mature sperm26, suggesting that the HFD led to changes in testis RNAs that were inherited by the sperm and dictated the phenotype of the offspring. The implications of these findings are twofold: the testis may gain somatic RNAs through EVs or mobile RNAs, or testicular transcription may be modulated to generate novel RNA profiles that mirror the HFD conditions, or both. However, it is difficult to imagine how dietary changes can be precisely mirrored by changing the testicular transcriptome (and probably the proteome as well), and how such alterations are transformed into sperm-borne information without loss of the original specificities (that is, metabolic disorders). Nevertheless, such a scenario is possible because recent evidence supports the precise transfer of intergenerational information, ranging from the inheritance of a specific olfactory memory6 to a tissue-specific wound-healing response63. A highly integrated interplay among spermatogenic transcription, sperm RNA biogenesis and storage, DNA modifications and chromatin remodelling might underlie such a possibility. Moreover, recent data also show that mutations in or aberrant expression of certain genes (including RNA-editing enzymes28, chromatin modulators36,44 and RNA methyltransferases27) are responsible for the transgenerational non-Mendelian inheritance of phenotypes and that toxicant-induced epimutations can promote genetic mutations in sperm89. It is likely that some of these genes are modulated during certain types of environmental exposure and contribute to the inheritance of acquired traits. Similar gene-regulation feedback mechanisms have recently been shown to control the duration of transgenerational small RNA inheritance in C. elegans90.
Mechanisms of action
The mechanism by which sperm RNAs reshape early embryonic development to recapitulate a paternally acquired phenotype in offspring remains unclear. In addition, how the initial changes caused by sperm RNAs are converted into a stable form of information to allow transgenerational inheritance remains a major puzzle. Recently, emerging evidence has shed new light on these intriguing questions.
Reshaping embryonic development: the transcriptional cascade
The injection of RNAs into mammalian zygotes has been reported to alter offspring phenotypes41–43 and to mimic paternally acquired traits9,24–26. These studies provide proof-of-principle evidence that perturbations in RNA profiles have a profound impact on early embryonic development, although the amount of RNA injected in these studies was larger than a single sperm could deliver. Under physiological conditions, the amount of RNAs delivered by a single sperm is infinitesimal compared with the RNA reservoir of oocytes. However, data have indeed shown that the incoming sperm can provide information to the host oocyte cytoplasm to functionally influence the order of cell division and spatial patterning in mouse embryos91. A recent single-blastomere RNA-seq analysis of mammalian early embryos suggested that small variations in the transcriptome will inevitably emerge at the two-cell embryo stage, owing to imperfect cleavage division92. These initial biases will subsequently be transformed into a more defined asymmetrical transcriptional pattern upon progression of zygotic transcriptional activation, a dynamic symmetry-breaking process that defines different lineage fates92. Recent evidence showing biased distributions of essential molecules that can bias cell fate in early embryonic stages are supportive of this model93–96. In this regard, even the smallest biases in RNA distribution (for example, sperm miRNAs that can regulate mRNAs) at early embryonic stages (FIG. 2a,b) have the potential to influence the trajectory of development through a butterfly effect. In theory, the altered sperm RNA contents could convey epigenetic information that is specific for paternally acquired traits if the embryonic transcriptional cascade is regulated with analogue and/or digital precision97 (FIG. 2a). Indeed, RNA-seq analyses of early embryos after the injection of sperm tsRNAs from a HFD-fed father revealed a down-regulation of metabolic regulation-related genes in both early embryos and pancreatic islets of offspring, supporting the idea that regulation of the transcriptional cascade continues throughout development9, although the detailed regulatory network remains unknown.
Figure 2. Potential mechanisms involving sperm RNAs in early embryos.
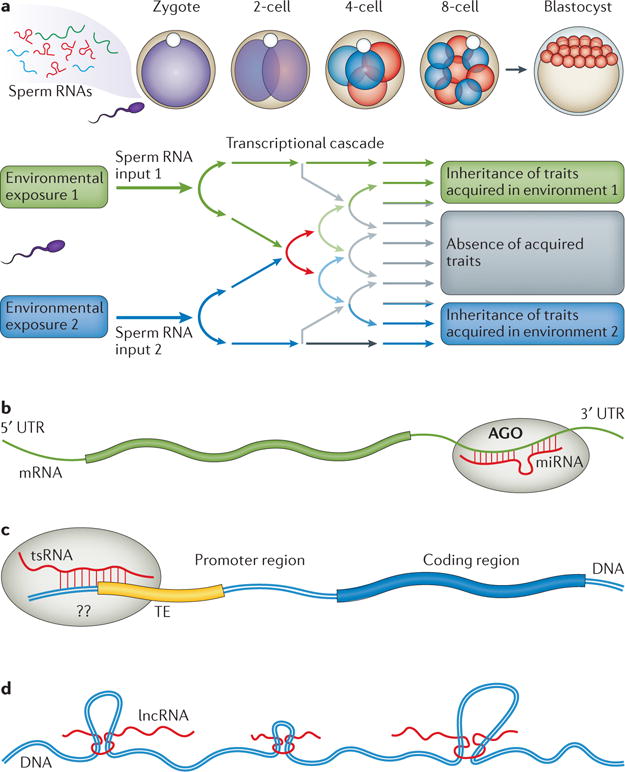
a | Sperm RNA input following certain environmental exposures may trigger a transcriptional cascade in the early embryo that affects the symmetry-breaking process, and may generate phenotypes in the offspring that mirror the paternally acquired phenotypes. b | Sperm microRNAs (miRNAs) could mediate mRNA stability and/or degradation by binding to the 3′ untranslated region (UTR), which is facilitated by Argonaute (AGO), to regulate mRNA metabolism. c | Sperm tRNA-derived small RNAs (tsRNAs) showed sequence matches to gene promoters that are associated with transposable elements (TEs), with unidentified mechanisms or related binding proteins. d | Sperm long non-coding RNAs (lncRNAs) may reshape chromatin 3D structure.
Regulating transposable elements
Transcriptional regulation might explain the phenotypes observed in the immediate offspring following paternal environmental exposure, but how might the phenotypes induced by sperm RNAs be transferred to further generations? RNA-guided DNA methylation has been well established in plants98, and it has been observed in mammals that piRNAs can mediate site-specific DNA methylation99 and retrotransposon silencing15. Therefore, RNA-guided DNA methylation could be one possible mechanism that underlies transgenerational inheritance of acquired phenotypes. Intriguingly, sperm tsRNAs exhibit sequence matches at various gene promoters9, many of which are associated with transposable elements (TEs)100, suggesting that transcriptional regulation of these genes occurs through TEs. Indeed, injection of a single tsRNA (tsRNAGly(GCC)) into a mouse zygote can repress the genes associated with murine endogenous retrovirus with leucine tRNA (MERVL), a type of TE11. Whether the interactions between tsRNAs and TEs in the early embryo can establish a stable mark (for example, altered DNA methylation or histone modifications) and thus change the expression of TE-associated genes in a heritable manner remains an interesting possibility (FIG. 2c). Testing this hypothesis requires comprehensive, high-coverage, genome-wide DNA methylation analysis, such as whole-genome shotgun bisulfite sequencing (WGSBS), in both early embryos and the sperm of offspring. Notably, although recent WGSBS data have suggested that DNA methylation is not the initial molecular carrier for diet-induced epigenetic inheritance in the sperm of exposed males16, whether DNA methylation could act as the secondary effector that relays the initial effects of sperm RNAs remains unresolved. This possibility should be examined carefully with sufficient sequencing coverage and depth37, particularly for TEs in proximity to genes and promoters. In addition to the well-studied 5 mC DNA methylation discussed above, the recent discovery of N6-methyladenine (m6A) DNA methylation in mammalian cells and its association with TEs101 also raise the intriguing possibility of m6A as a potential heritable mark for epigenetic inheritance in mammals, as has been suggested in C. elegans102.
Remodelling chromatin structure
Mature sperm contain numerous mRNAs, small RNAs and lncRNAs23. Both small RNAs and lncRNAs have been shown to regulate chromatin structure98,103. In particular, lncRNAs mediate long-range chromatin interactions, generating unexpected 3D organization of the chromosomes104–107 with extensive topological domains, as revealed by whole-genome chromosome conformation capture (Hi-C) analysis108 (FIG. 2d). Recently, using a transgenic mouse line overexpressing lysine-specific histone demethylase 1 (LSD1; encoded by Kdm1a) it was observed that LSD1 overexpression in one generation impaired offspring development, owing to multiple defects that persisted in subsequent generations in the absence of the initial transgene expression. These transgenerational defects were not transmitted by altered DNA methylation in CpG-rich regions but were associated with abnormal RNA expression in wild-type sperm from the offspring of the transgenic mice36. In this case, the abnormal sperm RNA profiles appeared to be the underlying cause of the defects in offspring, and the sperm of the offspring maintained the aberrant RNA expression. Could this be due to a feedback loop in which abnormal sperm RNAs induce changes in chromatin structure that, in turn, induce the same abnormalities in RNA expression? These are remote but tantalizing possibilities, and a similar RNA-based feedback loop mechanism has been suggested in C. elegans68,109.
Sperm RNA modifications
In addition to RNA sequences, RNA modifications have recently emerged as a new layer in the transcriptional and epigenetic regulation of various biological processes, bringing about a new era of RNA epigenetics110,111. Recently, a study using liquid chromatography–tandem mass spectrometry (LC–MS/MS) to simultaneously identify and quantify multiple RNA modifications in mice112 revealed that sperm small RNAs harbour various RNA modifications. The tsRNA fraction is particularly sensitive in the response to a paternal HFD, showing a significant increase in m5C and m2G (REF. 9) (FIG. 3a). The significance of such alterations remains unclear, yet the RNA modifications present in tsRNAs seem to increase RNA stability, as chemically synthesized tsRNAs without RNA modifications showed more rapid degradation in mouse zygote lysates9 and serum58 (FIG. 3a). This increased RNA stability might prolong the half-life of sperm RNA actions after fertilization and thus preserve their function in transferring paternally acquired traits to offspring9. In addition, RNA modifications could change the structure of RNAs in vivo and alter the specificity of their interactions with other RNAs, DNA or proteins (FIG. 3b), which may represent a new layer of regulation111. By far, the greatest challenge is to reveal the full map of RNA modifications in sperm tsRNAs and other types of RNAs, because the LC–MS/MS approach cannot pinpoint the exact location of each RNA modification. Future combinational use of LC–MS/MS and other methods — for example, antibody-based pull-down and chemical approaches that are already successfully used for genome-wide profiling of RNA modifications (such as, m5C, m6A, m1A and pseudouridine110,111) — as well as the existing databases of RNA modifications113,114, will help to reveal the full map of RNA modifications in sperm RNAs. In particular, RNA-modification maps for tsRNAs and mi RNAs will be of great interest, given their probable roles in the inheritance of acquired traits.
Figure 3. Current knowledge of sperm RNA modifications.
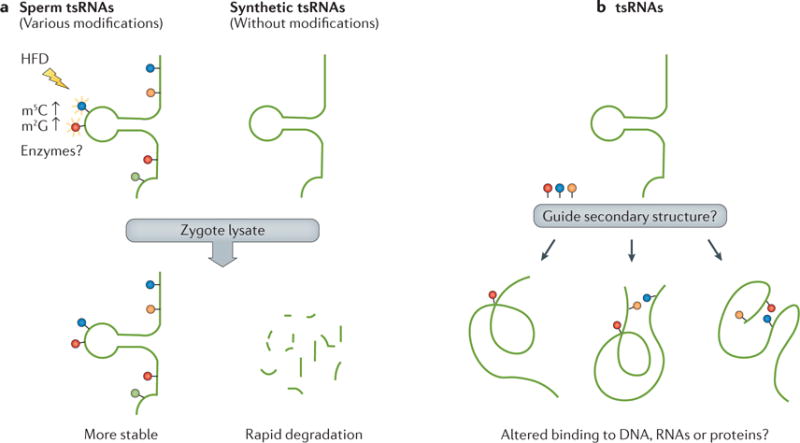
a | Sperm tRNA-derived small RNAs (tsRNAs) contain various RNA modifications (coloured dots) that contribute to their stability in zygote lysate; a high-fat diet (HFD) in male mice increases 5-methylcytidine (m5C) and N2-methylguanosine (m2G) levels in sperm tsRNAs, by unknown enzymes and mechanisms. b | RNA modifications may alter the secondary structure of tsRNAs, which could affect their stability and targeting specificity.
RNA modification enzymes in epigenetic inheritance
The specific enzymes responsible for regulating each sperm RNA modification remain largely unknown, yet several studies have demonstrated the great potential of such enzymes to mediate epigenetic inheritance. One report showed that RNA-mediated epigenetic heredity requires the DNA (cytosine-5)-methyltransferase-like protein 2 (DNMT2)27, which is known to be responsible for adding m5C to several types of tRNAs115,116. Deletion of Dnmt2 also causes increased production of tsRNAs, particularly under stress conditions115,117. Another report showed that the deletion of DEAD box polypeptide 1 (Ddx1), which encodes an RNA helicase that has broad functions in regulating RNA structure and metabolism (including tRNA splicing118 and promoting miRNA maturation119), resulted in transgenerational inheritance of wild-type lethality in wild-type mice derived from heterozygote intercrossing120, which might involve an interplay between abnormal RNAs and a perturbed chromatin structure. Moreover, deletion of apolipoprotein B mRNA-editing enzyme, catalytic polypeptide 1 (Apobec1), an RNA-editing enzyme with cytidine deaminase activity that converts C to U, resulted in the transgenerational inheritance of susceptibility to testicular germ-cell tumours in the first to the third generations of wild-type male offspring of heterozygous female mice28, suggestive of RNA-mediated paramutation effects. All of these transgenerational effects are closely related to abnormal post-transcriptional RNA modifications, including nucleotide modifications, RNA editing, RNA ligation and/or unwinding. This evidence strongly suggests that RNA modifications have a central role in the modulation of epigenetic memory, a hypothesis that warrants future investigations.
Sequencing bias due to RNA modifications
At the technical level, it is important to note that certain RNA modifications can block reverse transcriptase during RNA-seq library preparation, resulting in a failure to detect some modified RNAs and thus generating biased readouts121,122. This is particularly true for tRNAs and tsRNAs, which contain extensive modifications9,113,114,123. Recently, the use of Escherichia coli α-ketoglutarate-dependent dioxygenase (AlkB) in a treatment protocol, which can demethylate reverse-transcriptase-blocking RNA methylations before RNA library constructions, has led to the discovery of abundant, previously undetected tRNAs and tsRNAs121,122. These findings suggest that previously published data sets of small RNAs, particularly those including heavily methylated RNAs (for example, sperm tsRNAs), will require a comprehensive reanalysis using new approaches. Until then, caution should be exercised when interpreting inherited phenotypes based solely on observed changes in one or a few sperm small RNAs, as such observations could be misleading.
Perspectives and conclusion
Evidence for RNA-mediated inheritance of ancestrally acquired traits is now expanding, and solid data have become available from studies on plants, nematodes, fruitflies and mammals. These and other studies that support the inheritance of acquired traits hypothesis have set the stage for a re-evaluation of long-held paradigms that describe the physiological and molecular barriers of hereditary information, as well as the flow of information at both the molecular and physiological levels. The versatility of sperm regulatory RNAs, along with the even more complex forms of RNA modifications and the potential interactions of sperm RNAs with other epigenetic factors, raise important questions regarding the epigenetic inheritance of acquired traits. For example, how many items of information can be stored in the form of sperm RNAs, and how specific can a heritable acquired trait be? Could sensitivity to a specific odour, like or dislike of a specific food, fear of a specific animal or scenario, or adaptation to a lifestyle be inherited? Indeed, recent reports have demonstrated that inheritance of acquired traits could be as precise as the epigenetic memory of a specific odour6, giving rise to wide-ranging possibilities for the inheritance of other life experiences. It would be remarkable if such heritability were to extend beyond passive exposures to environmental factors (for example, chemicals, nutrition or stress) to learned or intentional behaviours or characteristics (such as kindness and charisma).
However, demonstrating the contributions of sperm RNAs to the transfer of acquired traits is only the first step. A fundamental question concerns the nature of the ‘coding mechanism’ through which specific acquired traits become encoded in sperm; could a trait be encoded in the form of a single RNA or a combination of RNAs, or is additional interplay between sperm RNA and other epigenetic factors required? The mechanisms that ‘encode’ this information in the sperm and that ‘decode’ it in the offspring, and the potential transformation of RNA-borne ‘codes’ into other forms of hereditary information (for example, DNA methylation and chromatin structure), are key issues that require investigation. Furthermore, in addition to the flow of information from the environment to sperm RNAs and RNA modifications, and the means by which these factors reshape development, it is important to understand the reversibility of acquired traits over generations, which may have widespread medical and social implications.
Answering these questions will require elegantly designed experiments at both the animal and molecular levels, as well as the improvement of technologies for ‘single-cell omics’ (for example, transcriptomics, DNA methylomics, ChIP–seq (chromatin immunoprecipitation followed by sequencing) and Hi-C)124 for use in sperm and early embryo investigations, which will ideally involve parallel double-125, triple-126 or even panomic approaches. Such advances will probably require at least another decade’s worth of efforts, but may revolutionize our understanding of hereditary information flow, stimulate further interest in old theories (including Lamarckism and Darwin’s pangenesis (BOX 3)) and inspire new waves of research in the fields of genetics and epigenetics.
Acknowledgments
The Chen laboratory is currently supported by start-up funds from the University of Nevada, Reno School of Medicine, USA. This work is in part funded by the Ministry of Science and Technology of the People’s Republic of China (2015CB943000 to Q.C. and 2016YFA0500903, 2015DFG32640 to E.D.), the National Natural Science Foundation of China (81490742, 31671568 to E.D.), the US National Institutes of Health (HD060858, HD071736 and HD085506 to W.Y.) and the John Templeton Foundation, USA (PID: 50183 to W.Y.). The authors thank all Chen laboratory members for critical discussions on the contents of the manuscript and for help in preparing the graphics and illustrations.
Glossary
- Parthenogenetic
A type of asexual reproduction that occurs when a female gamete develops a new individual without being fertilized by a male gamete
- RNA-editing events
Molecular processes by which specific nucleotide sequences in an RNA molecule are changed, such as C-to-U and A-to-I editing
- Kinky-tail phenotype
A mouse phenotype characterized by a kinked tail containing a sharp bend at an angle to the main tail axis
- Transvection
An epigenetic phenomenon that involves the interaction between two homologous chromosomes, resulting in either gene activation or repression at an allele
- Paramutation
Interaction between two alleles at a single locus during meiosis, resulting in epigenetic transfer of information from one allele to another that is heritable for generations
- Epididymal maturation
Spermatozoa from testis undergo a maturation process during transit from the proximal to the distal end of the epididymis, acquiring motility and fertility
- Polyandrous
The mating behaviour of animals in which the females mate with more than one male in a single breeding cycle
- G-Quadruplex conformation
Guanine-rich oligonucleotides that assembled into intra- or inter-molecular guanine tetrad structures, which can be formed by both DNA and RNA
- Lamarckism
The idea that an organism can pass on characteristics that are acquired during its lifetime to its offspring
Footnotes
Competing interests statement
The authors declare no competing interests.
DATABASES
GtRNAdb: http://gtrnadb.ucsc.edu
Modomics: http://modomics.genesilico.pl
RNA Modification Database: http://mods.rna.albany.edu
SpermBase: http://spermbase.org
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Landman OE. The inheritance of acquired characteristics. Annu Rev Genet. 1991;25:1–20. doi: 10.1146/annurev.ge.25.120191.000245. [DOI] [PubMed] [Google Scholar]
- 2.Rechavi O, et al. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell. 2014;158:277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ost A, et al. Paternal diet defines offspring chromatin state and intergenerational obesity. Cell. 2014;159:1352–1364. doi: 10.1016/j.cell.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Ng SF, et al. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature. 2010;467:963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 5.Carone BR, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; References 4 and 5 show that acquired metabolic disorders due to paternal diets could be inherited by the offspring.
- 6.Dias BG, Ressler KJ. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat Neurosci. 2014;17:89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. correction, 328, 690 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodgers AB, Morgan CP, Bronson SL, Revello S, Bale TL. Paternal stress exposure alters sperm microRNA content and reprograms offspring HPA stress axis regulation. J Neurosci. 2013;33:9003–9012. doi: 10.1523/JNEUROSCI.0914-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]; This study demonstrates that zygotic injection of sperm total RNAs or tsRNAs from males fed a HFD can induce metabolic disorders in offspring, and that RNA modifications in sperm tsRNAs are sensitive to a HFD.
- 10.Huypens P, et al. Epigenetic germline inheritance of diet-induced obesity and insulin resistance. Nat Genet. 2016;48:497–499. doi: 10.1038/ng.3527. [DOI] [PubMed] [Google Scholar]
- 11.Sharma U, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper provides evidence suggesting that sperm can gain tsRNAs during epididymal transition, which can influence embryonic gene expression.
- 12.Lane M, Robker RL, Robertson SA. Parenting from before conception. Science. 2014;345:756–760. doi: 10.1126/science.1254400. [DOI] [PubMed] [Google Scholar]
- 13.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei Y, et al. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc Natl Acad Sci USA. 2014;111:1873–1878. doi: 10.1073/pnas.1321195111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet. 2012;13:153–162. doi: 10.1038/nrg3188. [DOI] [PubMed] [Google Scholar]
- 16.Shea JM, et al. Genetic and epigenetic variation, but not diet, shape the sperm methylome. Dev Cell. 2015;35:750–758. doi: 10.1016/j.devcel.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study suggests that DNA methylation is not the primary epigenetic carrier for diet-induced sperm changes that are responsible for reprogramming of offspring metabolism.
- 17.Radford EJ, et al. In utero effects. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science. 2014;345:1255903. doi: 10.1126/science.1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article shows that in utero nutritional exposure during critical windows of germ-cell development alters the sperm methylome of offspring, but the altered methylation does not persist in the next generation.
- 18.Iqbal K, et al. Deleterious effects of endocrine disruptors are corrected in the mammalian germline by epigenome reprogramming. Genome Biol. 2015;16:59. doi: 10.1186/s13059-015-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Waal E, et al. Primary epimutations introduced during intracytoplasmic sperm injection (ICSI) are corrected by germline-specific epigenetic reprogramming. Proc Natl Acad Sci USA. 2012;109:4163–4168. doi: 10.1073/pnas.1201990109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammoud SS, et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brykczynska U, et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- 22.Ragunathan K, Jih G, Moazed D. Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science. 2015;348:1258699. doi: 10.1126/science.1258699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jodar M, et al. The presence, role and clinical use of spermatozoal RNAs. Hum Reprod Update. 2013;19:604–624. doi: 10.1093/humupd/dmt031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gapp K, et al. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci. 2014;17:667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides the first functional evidence for sperm RNAs in transferring the acquired phenotype, showing that the injection of sperm RNAs from a traumatized father induces corresponding phenotypes in offspring.
- 25.Rodgers AB, Morgan CP, Leu NA, Bale TL. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc Natl Acad Sci USA. 2015;112:13699–13704. doi: 10.1073/pnas.1508347112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grandjean V, et al. RNA-mediated paternal heredity of diet-induced obesity and metabolic disorders. Sci Rep. 2015;5:18193. doi: 10.1038/srep18193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiani J, et al. RNA-mediated epigenetic heredity requires the cytosine methyltransferase Dnmt2. PLoS Genet. 2013;9:e1003498. doi: 10.1371/journal.pgen.1003498. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that DNMT2 is required for RNA-mediated non-Mendelian epigenetic inheritance in mice.
- 28.Nelson VR, Heaney JD, Tesar PJ, Davidson NO, Nadeau JH. Transgenerational epigenetic effects of the Apobec1 cytidine deaminase deficiency on testicular germ cell tumor susceptibility and embryonic viability. Proc Natl Acad Sci USA. 2012;109:E2766–E2773. doi: 10.1073/pnas.1207169109. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report shows that APOBEC1 is involved in transgenerational epigenetic inheritance of a phenotype.
- 29.Bohacek J, Mansuy IM. Molecular insights into transgenerational non-genetic inheritance of acquired behaviours. Nat Rev Genet. 2015;16:641–652. doi: 10.1038/nrg3964. [DOI] [PubMed] [Google Scholar]
- 30.Roemer I, Reik W, Dean W, Klose J. Epigenetic inheritance in the mouse. Curr Biol. 1997;7:277–280. doi: 10.1016/s0960-9822(06)00124-2. [DOI] [PubMed] [Google Scholar]
- 31.Fullston T, et al. Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F2 generation and alters the transcriptional profile of testis and sperm microRNA content. FASEB J. 2013;27:4226–4243. doi: 10.1096/fj.12-224048. [DOI] [PubMed] [Google Scholar]
- 32.Wu L, et al. Paternal psychological stress reprograms hepatic gluconeogenesis in offspring. Cell Metab. 2016;23:735–743. doi: 10.1016/j.cmet.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 33.Paris L, et al. Transgenerational inheritance of enhanced susceptibility to radiation-induced medulloblastoma in newborn Ptch1+/− mice after paternal irradiation. Oncotarget. 2015;6:36098–36112. doi: 10.18632/oncotarget.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 35.Rakyan VK, et al. Transgenerational inheritance of epigenetic states at the murine AxinFu allele occurs after maternal and paternal transmission. Proc Natl Acad Sci USA. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siklenka K, et al. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science. 2015;350:aab2006. doi: 10.1126/science.aab2006. [DOI] [PubMed] [Google Scholar]; This study demonstrates that defects induced by genetically disrupting histone methylation cause transgenerational inheritance of phenotypes in mammals.
- 37.Ziller MJ, Hansen KD, Meissner A, Aryee MJ. Coverage recommendations for methylation analysis by whole-genome bisulfite sequencing. Nat Methods. 2015;12:230–232. doi: 10.1038/nmeth.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herman H, et al. Trans allele methylation and paramutation-like effects in mice. Nat Genet. 2003;34:199–202. doi: 10.1038/ng1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rassoulzadegan M, Magliano M, Cuzin F. Transvection effects involving DNA methylation during meiosis in the mouse. EMBO J. 2002;21:440–450. doi: 10.1093/emboj/21.3.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hatada I, et al. Aberrant methylation of an imprinted gene U2af1-rs1(SP2) caused by its own transgene. J Biol Chem. 1997;272:9120–9122. doi: 10.1074/jbc.272.14.9120. [DOI] [PubMed] [Google Scholar]
- 41.Grandjean V, et al. The miR-124-Sox9 paramutation: RNA-mediated epigenetic control of embryonic and adult growth. Development. 2009;136:3647–3655. doi: 10.1242/dev.041061. [DOI] [PubMed] [Google Scholar]
- 42.Wagner KD, et al. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev Cell. 2008;14:962–969. doi: 10.1016/j.devcel.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 43.Rassoulzadegan M, et al. RNA-mediated non-Mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]; This article provides the first evidence that sperm RNAs can mediate epigenetic inheritance in mammals.
- 44.Chong S, et al. Modifiers of epigenetic reprogramming show paternal effects in the mouse. Nat Genet. 2007;39:614–622. doi: 10.1038/ng2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Castro Barbosa T, et al. High-fat diet reprograms the epigenome of rat spermatozoa and transgenerationally affects metabolism of the offspring. Mol Metab. 2016;5:184–197. doi: 10.1016/j.molmet.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng H, et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012;22:1609–1612. doi: 10.1038/cr.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first report that tsRNAs are highly enriched in mature sperm.
- 47.Schuster A, Skinner MK, Yan W. Ancestral vinclozolin exposure alters the epigenetic transgenerational inheritance of sperm small noncoding RNAs. Environ Epigenet. 2016;2:dvw001. doi: 10.1093/eep/dvw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donkin I, et al. Obesity and bariatric surgery drive epigenetic variation of spermatozoa in humans. Cell Metab. 2015;23:369–378. doi: 10.1016/j.cmet.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 49.Surani MA. Breaking the germ line-soma barrier. Nat Rev Mol Cell Biol. 2016;17:136. doi: 10.1038/nrm.2016.12. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y. A new perspective on Darwin’s pangenesis. Biol Rev Camb Philos Soc. 2008;83:141–149. doi: 10.1111/j.1469-185X.2008.00036.x. [DOI] [PubMed] [Google Scholar]
- 51.Koch S, Acebron SP, Herbst J, Hatiboglu G, Niehrs C. Post-transcriptional Wnt signaling governs epididymal sperm maturation. Cell. 2015;163:1225–1236. doi: 10.1016/j.cell.2015.10.029. [DOI] [PubMed] [Google Scholar]
- 52.Sarkies P, Miska EA. Small RNAs break out: the molecular cell biology of mobile small RNAs. Nat Rev Mol Cell Biol. 2014;15:525–535. doi: 10.1038/nrm3840. [DOI] [PubMed] [Google Scholar]
- 53.Lo Cicero A, Stahl PD, Raposo G. Extracellular vesicles shuffling intercellular messages: for good or for bad. Curr Opin Cell Biol. 2015;35:69–77. doi: 10.1016/j.ceb.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 54.Machtinger R, Laurent LC, Baccarelli AA. Extracellular vesicles: roles in gamete maturation, fertilization and embryo implantation. Hum Reprod Update. 2016;22:182–193. doi: 10.1093/humupd/dmv055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mahmoudi K, Ezrin A, Hadjipanayis C. Small extracellular vesicles as tumor biomarkers for glioblastoma. Mol Aspects Med. 2015;45:97–102. doi: 10.1016/j.mam.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 56.Vader P, Mol EA, Pasterkamp G, Schiffelers RM. Extracellular vesicles for drug delivery. Adv Drug Deliv Rev. 2016 doi: 10.1016/j.addr.2016.02.006. http://dx.doi.org/10.1016/j.addr.2016.02.006. [DOI] [PubMed]
- 57.Vojtech L, et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014;42:7290–7304. doi: 10.1093/nar/gku347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, et al. Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J Mol Cell Biol. 2014;6:172–174. doi: 10.1093/jmcb/mjt052. [DOI] [PubMed] [Google Scholar]
- 59.Dhahbi JM, et al. 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics. 2013;14:298. doi: 10.1186/1471-2164-14-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, Wu C, Aramayo R, Sachs MS, Harlow ML. Synaptic vesicles contain small ribonucleic acids (sRNAs) including transfer RNA fragments (trfRNA) and microRNAs (miRNA) Sci Rep. 2015;5:14918. doi: 10.1038/srep14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–4304. doi: 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cossetti C, et al. Soma-to-germline transmission of RNA in mice xenografted with human tumour cells: possible transport by exosomes. PLoS ONE. 2014;9:e101629. doi: 10.1371/journal.pone.0101629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeybel M, et al. Multigenerational epigenetic adaptation of the hepatic wound-healing response. Nat Med. 2012;18:1369–1377. doi: 10.1038/nm.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Devanapally S, Ravikumar S, Jose AM. Double-stranded RNA made in C. elegans neurons can enter the germline and cause transgenerational gene silencing. Proc Natl Acad Sci USA. 2015;112:2133–2138. doi: 10.1073/pnas.1423333112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Curran SP, Wu X, Riedel CG, Ruvkun G. A soma-to-germline transformation in long-lived Caenorhabditis elegans mutants. Nature. 2009;459:1079–1084. doi: 10.1038/nature08106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shirayama M, et al. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buckley BA, et al. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ashe A, et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Javurek AB, et al. Discovery of a novel seminal fluid microbiome and influence of estrogen receptor alpha genetic status. Sci Rep. 2016;6:23027. doi: 10.1038/srep23027. corrigendum 6, 25216 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muraca M, Putignani L, Fierabracci A, Teti A, Perilongo G. Gut microbiota-derived outer membrane vesicles: under-recognized major players in health and disease? Discov Med. 2015;19:343–348. [PubMed] [Google Scholar]
- 71.Hoshino A, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ridder K, et al. Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLoS Biol. 2014;12:e1001874. doi: 10.1371/journal.pbio.1001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Annu Rev Immunol. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 74.Cohen S, et al. Chronic stress, glucocorticoid receptor resistance, inflammation, and disease risk. Proc Natl Acad Sci USA. 2012;109:5995–5999. doi: 10.1073/pnas.1118355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fan Y, et al. Diet-induced obesity in male C57BL/6 mice decreases fertility as a consequence of disrupted blood-testis barrier. PLoS ONE. 2015;10:e0120775. doi: 10.1371/journal.pone.0120775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nargund VH. Effects of psychological stress on male fertility. Nat Rev Urol. 2015;12:373–382. doi: 10.1038/nrurol.2015.112. [DOI] [PubMed] [Google Scholar]
- 77.Crean AJ, Kopps AM, Bonduriansky R. Revisiting telegony: offspring inherit an acquired characteristic of their mother’s previous mate. Ecol Lett. 2014;17:1545–1552. doi: 10.1111/ele.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crean AJ, Adler MI, Bonduriansky R. Seminal fluid and mate choice: new predictions. Trends Ecol Evol. 2016;31:253–255. doi: 10.1016/j.tree.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 79.Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Feinberg EH, Hunter CP. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301:1545–1547. doi: 10.1126/science.1087117. [DOI] [PubMed] [Google Scholar]
- 81.McEwan DL, Weisman AS, Hunter CP. Uptake of extracellular double-stranded RNA by SID-2. Mol Cell. 2012;47:746–754. doi: 10.1016/j.molcel.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ivanov P, et al. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci USA. 2014;111:18201–18206. doi: 10.1073/pnas.1407361111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chin AR, et al. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016;26:217–228. doi: 10.1038/cr.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang J, Farmer LM, Agyekum AA, Hirschi KD. Detection of dietary plant-based small RNAs in animals. Cell Res. 2015;25:517–520. doi: 10.1038/cr.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mlotshwa S, et al. A novel chemopreventive strategy based on therapeutic microRNAs produced in plants. Cell Res. 2015;25:521–524. doi: 10.1038/cr.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang L, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22:107–126. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou Z, et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2015;25:39–49. doi: 10.1038/cr.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ji L, Chen X. Regulation of small RNA stability: methylation and beyond. Cell Res. 2012;22:624–636. doi: 10.1038/cr.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Skinner MK, Guerrero-Bosagna C, Haque MM. Environmentally induced epigenetic transgenerational inheritance of sperm epimutations promote genetic mutations. Epigenetics. 2015;10:762–771. doi: 10.1080/15592294.2015.1062207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Houri-Ze’evi L, et al. A tunable mechanism determines the duration of the transgenerational small RNA inheritance in C. elegans. Cell. 2016;165:88–99. doi: 10.1016/j.cell.2016.02.057. [DOI] [PubMed] [Google Scholar]
- 91.Piotrowska K, Zernicka-Goetz M. Role for sperm in spatial patterning of the early mouse embryo. Nature. 2001;409:517–521. doi: 10.1038/35054069. [DOI] [PubMed] [Google Scholar]
- 92.Shi J, et al. Dynamic transcriptional symmetry-breaking in pre-implantation mammalian embryo development revealed by single-cell RNA-seq. Development. 2015;142:3468–3477. doi: 10.1242/dev.123950. [DOI] [PubMed] [Google Scholar]
- 93.Goolam M, et al. Heterogeneity in Oct4 and Sox2 targets biases cell fate in 4-cell mouse embryos. Cell. 2016;165:61–74. doi: 10.1016/j.cell.2016.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.White MD, et al. Long-lived binding of Sox2 to DNA predicts cell fate in the four-cell mouse embryo. Cell. 2016;165:75–87. doi: 10.1016/j.cell.2016.02.032. [DOI] [PubMed] [Google Scholar]; References 92–94 provide the conceptual framework and experimental evidence that the slightest blastomere–blastomere biases during early embryo development can change the trajectory of developmental potential.
- 95.Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Plachta N, Bollenbach T, Pease S, Fraser SE, Pantazis P. Oct4 kinetics predict cell lineage patterning in the early mammalian embryo. Nat Cell Biol. 2011;13:117–123. doi: 10.1038/ncb2154. [DOI] [PubMed] [Google Scholar]
- 97.Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Watanabe T, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zuo Z, et al. Genome-wide analysis reveals origin of transfer RNA genes from tRNA halves. Mol Biol Evol. 2013;30:2087–2098. doi: 10.1093/molbev/mst107. [DOI] [PubMed] [Google Scholar]
- 101.Wu TP, et al. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature. 2016;532:329–333. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Greer EL, et al. DNA methylation on N6-adenine in C. elegans. Cell. 2015;161:868–878. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Engreitz JM, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hacisuleyman E, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Simon MD, et al. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–469. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xiang JF, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dekker J, Mirny L. The 3D genome as moderator of chromosomal communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Le Thomas A, et al. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev. 2014;28:1667–1680. doi: 10.1101/gad.245514.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 111.Chen K, Zhao BS, He C. Nucleic acid modifications in regulation of gene expression. Cell Chem Biol. 2016;23:74–85. doi: 10.1016/j.chembiol.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yan M, et al. A high-throughput quantitative approach reveals more small RNA modifications in mouse liver and their correlation with diabetes. Anal Chem. 2013;85:12173–12181. doi: 10.1021/ac4036026. [DOI] [PubMed] [Google Scholar]
- 113.Machnicka MA, et al. MODOMICS: a database of RNA modification pathways — 2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cantara WA, et al. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schaefer M, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Goll MG, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 117.Tuorto F, et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19:900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 118.Popow J, Jurkin J, Schleiffer A, Martinez J. Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature. 2014;511:104–107. doi: 10.1038/nature13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Han C, et al. The RNA-binding protein DDX1 promotes primary microRNA maturation and inhibits ovarian tumor progression. Cell Rep. 2014;8:1447–1460. doi: 10.1016/j.celrep.2014.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hildebrandt MR, Germain DR, Monckton EA, Brun M, Godbout R. Ddx1 knockout results in transgenerational wild-type lethality in mice. Sci Rep. 2015;5:9829. doi: 10.1038/srep09829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zheng G, et al. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods. 2015;12:835–837. doi: 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cozen AE, et al. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods. 2015;12:879–884. doi: 10.1038/nmeth.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]; References 121 and 122 report novel methods to discover previously undetected tRNAs and tsRNAs by RNA-seq.
- 123.Kirchner S, Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet. 2015;16:98–112. doi: 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- 124.Schwartzman O, Tanay A. Single-cell epigenomics: techniques and emerging applications. Nat Rev Genet. 2015;16:716–726. doi: 10.1038/nrg3980. [DOI] [PubMed] [Google Scholar]
- 125.Angermueller C, et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat Methods. 2016;13:229–232. doi: 10.1038/nmeth.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hou Y, et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016;26:304–319. doi: 10.1038/cr.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ostermeier GC, Dix DJ, Miller D, Khatri P, Krawetz SA. Spermatozoal RNA profiles of normal fertile men. Lancet. 2002;360:772–777. doi: 10.1016/S0140-6736(02)09899-9. [DOI] [PubMed] [Google Scholar]
- 128.Ostermeier GC, Miller D, Huntriss JD, Diamond MP, Krawetz SA. Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature. 2004;429:154. doi: 10.1038/429154a. [DOI] [PubMed] [Google Scholar]
- 129.Yuan S, et al. mir-34b/c and mir-449a/b/c are required for spermatogenesis, but not for the first cleavage division in mice. Biol Open. 2015;4:212–223. doi: 10.1242/bio.201410959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu WM, et al. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc Natl Acad Sci USA. 2012;109:490–494. doi: 10.1073/pnas.1110368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yuan S, et al. Sperm-borne mi RNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development. 2016;143:635–647. doi: 10.1242/dev.131755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhong C, et al. Parthenogenetic haploid embryonic stem cells efficiently support mouse generation by oocyte injection. Cell Res. 2016;26:131–134. doi: 10.1038/cr.2015.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li Z, et al. Birth of fertile bimaternal offspring following intracytoplasmic injection of parthenogenetic haploid embryonic stem cells. Cell Res. 2016;26:135–138. doi: 10.1038/cr.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Miller D. Confrontation, consolidation, and recognition: the oocyte’s perspective on the incoming sperm. Cold Spring Harb Perspect Med. 2015;5:a023408. doi: 10.1101/cshperspect.a023408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chandler VL. Paramutation: from maize to mice. Cell. 2007;128:641–645. doi: 10.1016/j.cell.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 136.Yuan S, Oliver D, Schuster A, Zheng H, Yan W. Breeding scheme and maternal small RNAs affect the efficiency of transgenerational inheritance of a paramutation in mice. Sci Rep. 2015;5:9266. doi: 10.1038/srep09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Beale LS. Pangenesis. Nature. 1871;4:25–26. [Google Scholar]
- 138.Eberhard WG. Postcopulatory sexual selection: Darwin’s omission and its consequences. Proc Natl Acad Sci USA. 2009;106(Suppl. 1):10025–10032. doi: 10.1073/pnas.0901217106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pizzari T, Foster KR. Sperm sociality: cooperation, altruism, and spite. PLoS Biol. 2008;6:e130. doi: 10.1371/journal.pbio.0060130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Edward DA, Stockley P, Hosken DJ. Sexual conflict and sperm competition. Cold Spring Harb Perspect Biol. 2015;7:a017707. doi: 10.1101/cshperspect.a017707. [DOI] [PMC free article] [PubMed] [Google Scholar]


