SUMMARY
53BP1 is a multi-functional double-strand break (DSB) repair protein that is essential for class switch recombination in B lymphocytes and for sensitizing BRCA1-deficient tumors to PARP inhibitors. Central to all 53BP1 activities is its recruitment to DSBs via the interaction of the tandem Tudor domain with dimethylated lysine 20 of histone H4 (H4K20me2). Here we identify an uncharacterized protein, TIRR (Tudor Interacting Repair Regulator) that directly binds the tandem Tudor domain and masks its H4K20me2 binding motif. Upon DNA damage, ATM phosphorylates 53BP1 and recruits RIF1 to dissociate the 53BP1–TIRR complex. However, over-expression of TIRR impedes 53BP1 function by blocking its localization to DSBs. Depletion of TIRR destabilizes 53BP1 in the nuclear soluble fraction and also alters the DSB-induced protein complex centering 53BP1. These findings identify TIRR as a new factor that influences DSB repair utilizing a unique mechanism of masking the histone methyl-lysine binding function of 53BP1.
INTRODUCTION
P53-binding protein 1 (53BP1) is a multi-faceted double-stranded DNA break (DSB) repair protein which transcends many fields1,2 as it impacts telomere dynamics, antibody genesis and efficacy of cancer therapy. 53BP1 contributes to both the repair3 and the orientation of the broken DNA ends4 during class-switch recombination (CSR), and its loss almost completely abrogates CSR5,6. The function of 53BP1 in the choice of DSB repair pathway is manifested in breast cancer associated gene 1 (Brca1)-deficient cells. Brca1-mutant cells are deficient in homologous recombination (HR)-mediated DSB repair and exquisitely sensitive to treatment with Poly-ADP-Ribose Polymerase-1 (PARP) inhibitors (PARPi)7,8. Loss of 53BP1 restores HR in Brca1-mutant cells 9–11, desensitizing them to PARPi. The minimal focus-forming region of 53BP1 (53BP1-FFR) required for localization of 53BP1 to DSBs includes an oligomerization domain, a tandem Tudor domain12, and the ubiquitin-dependent recruitment (UDR) motif13,14. The Tudor domain binds the dimethylated lysine 20 of histone H4 (H4-K20me2)15 while UDR binds the mono-ubiquitylated lysine 15 of histone H2A14. In response to DSBs, ATM phosphorylates 53BP116,17 and also initiates a signaling cascade which leads to the RNF168-mediated ubiquitylation of chromatin in the vicinity of the DSB18,19. Phosphorylation of 53BP1 and ubiquitylation of histone H2A are both DNA damage-dependent and are necessary for 53BP1 recruitment and retention at DSBs.
Here we describe a functionally uncharacterized protein, TIRR (Tudor Interacting DNA Repair Regulator) that directly interacts with 53BP1 and regulates its recruitment to chromatin. TIRR (alias Syndesmos or Nudt16L1) has 46% sequence homology with Nudt16, a member of the nudix hydrolase family and associates with Syndecan-420,21. However, TIRR lacks enzymatic activity22,23 and its function in cells is unclear. We observe that TIRR directly interacts with the tandem Tudor domain of 53BP1, and the H4K20me2/Tudor interaction interface overlaps with that of TIRR/Tudor. Upon DNA damage, ATM-induced phosphorylation of 53BP1 and RIF1 recruitment are necessary for the dissociation of the TIRR–53BP1 complex. This allows 53BP1 localization to chromatin, and its function in DSB repair and CSR. Over-expression of TIRR blocks 53BP1 recruitment to DSBs and impairs CSR. Conversely, loss of TIRR destabilizes nuclear soluble 53BP1 and the association of 53BP1 with its binding partners is altered in these cells. Thus, TIRR has a dual role in regulating 53BP1 function, first by stabilizing 53BP1 and maintaining its sub-nuclear localization prior to DNA damage and second, by influencing the interaction of 53BP1 with effector proteins in response to DSBs.
RESULTS
TIRR as a new partner of 53BP1
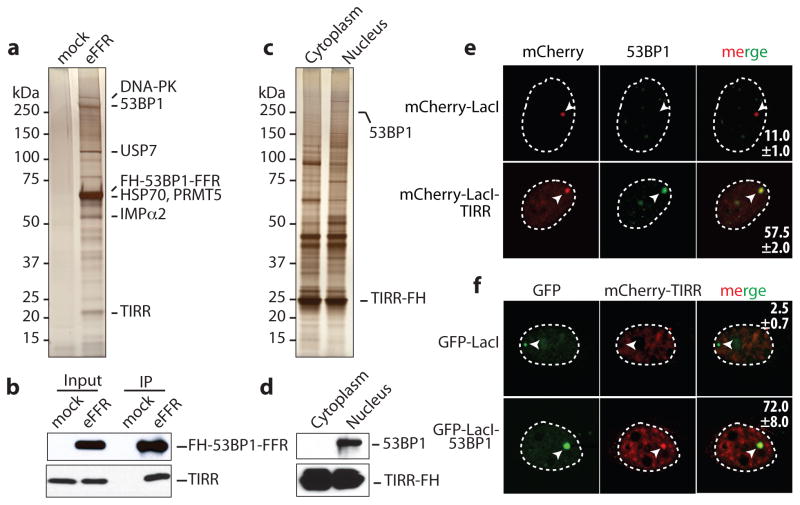
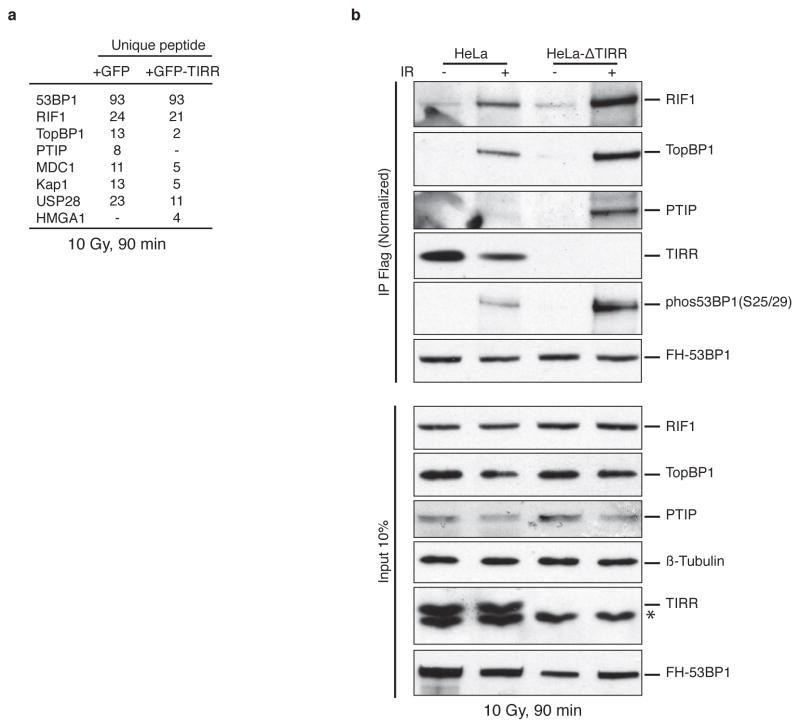
To identify regulatory factors of 53BP1 recruitment to chromatin, proteins associated with 53BP1-FFR were analyzed by mass spectrometry (Fig. 1a, b, Supplementary Table1). Based on percentage of coverage (47.0%) and the number of unique peptides (14), TIRR was one of the most abundant 53BP1-FFR binding partners. Reciprocally 53BP1 was co-purified with a tagged version of TIRR only in the nucleus (Fig. 1c, d). Mass spectrometry analyses further confirmed the presence of 53BP1 within the TIRR nuclear complex together with established 53BP1 interacting proteins such as MDC1 (Supplementary Table 2). Moreover endogenous 53BP1 and TIRR co-immunoprecipitated (Extended Data Fig. 1a).
Figure 1. TIRR is a novel partner of 53BP1.
Silver-staining (a) and immunoblot (b) of FH-53BP1-FFR complex from U2OS soluble nuclear extract. Mock-transfected cells used as control. Silver-staining (c) and immunoblot (d) of TIRR-FH partners purified from U2OS cells. e, Co-localization of mCherry-LacI empty or fused to TIRR with 53BP1 on the LacO array in U2OS19 cells (mean ± s.d., n=2). f, Co-localization of mCherry-TIRR with GFP-LacI either empty or fused to 53BP1 on the LacO array (mean ± s.d., n=2).
To investigate the 53BP1/TIRR interaction in intact cells the LacO/LacI tethering system was utilized24. TIRR was fused to the Escherichia coli lac-repressor (LacI) and tagged with Cherry-red fluorescent protein (mCherry-LacI-TIRR). mCherry-LacI-TIRR co-localized with endogenous 53BP1 in 57.5±2% of the cells analyzed (Fig. 1e). Conversely, GFP-LacI-53BP1 co-localized with TIRR in 72±8% of the cells (Fig. 1f). We concluded that in undamaged cells, TIRR associates with 53BP1 via the FFR region.
53BP1 tandem Tudor domain is required to interact with TIRR
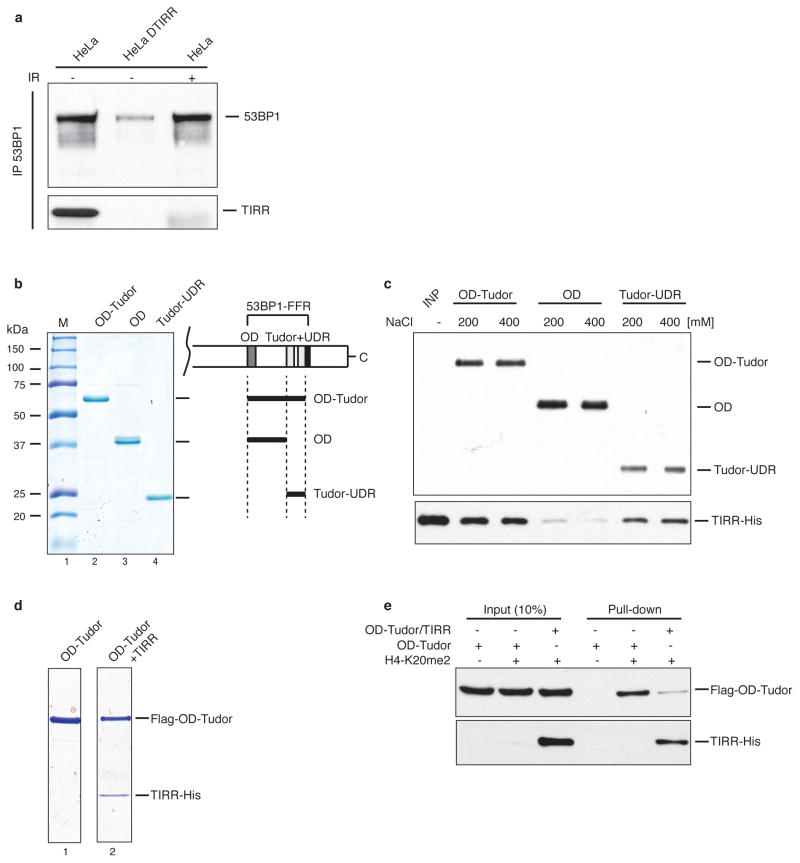
Next, to map the binding site of 53BP1 with TIRR, interaction of recombinant fragments of 53BP1-FFR and recombinant TIRR was assessed at different salt concentrations. TIRR interacted with the Tudor-UDR as well as with the tandem Tudor alone (Extended Data Fig. 1b, c) and it impaired the binding of the Tudor domain with an H4K20me2 peptide (Extended Data Fig. 1e, f). Using isothermal titration calorimetry (ITC), we derived a dissociation constant (Kd) of 0.79 μM for the interaction of Tudor with TIRR (Extended Data Fig. 1d). The ITC results are consistent with a 1:1 stoichiometry for the Tudor–TIRR complex, most likely corresponding to two Tudor molecules binding to one TIRR homodimer.
Binding surface of 53BP1 tandem Tudor for TIRR
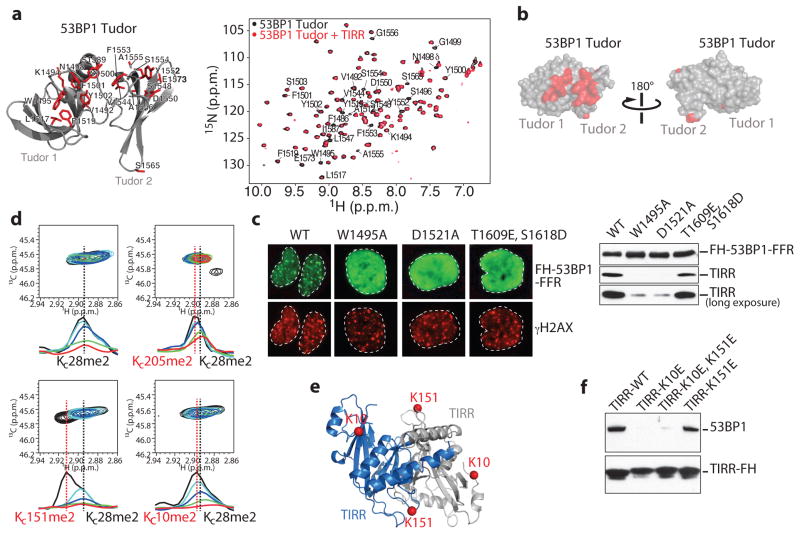
We used nuclear magnetic resonance (NMR) spectroscopy to map the binding interface of TIRR on the tandem Tudor domain surface. Upon serial addition of unlabeled TIRR to 15N-labeled Tudor, we observed progressive decrease of all 1H-15N signal intensities in the 1H-15N heteronuclear single quantum coherence (HSQC) spectrum of Tudor, consistent with the increase in molecular weight resulting from interaction with TIRR. In addition, several signals in the 1H-15N HSQC spectrum were preferentially broadened at sub-stoichiometric amounts of TIRR (Fig. 2a). These preferentially broadened signals belong to residues that map to one side of the tandem Tudor that includes the aromatic cage in the first Tudor domain which accommodates H4 dimethylated lysine 20 and to additional residues in the second Tudor domain (Fig. 2a, b). 53BP1 residues for which signal intensities were preferentially decreased are similar to those affected by interaction with an H4K20me2 peptide, indicating that H4K20me2 and TIRR binding surfaces on Tudor overlap even if there is no H4-like motif in TIRR. The interaction with TIRR is, however, more extensive as it involves residues from the second Tudor domain which do not interact with H4K20me2 (Fig. 2a). To extend this observation in cells, we tested Tudor domain mutants W1495A and D1521A that disrupt binding to H4K20me215 and the phosphomimetic mutants S1609E/T1618D in the UDR motif that inhibits binding to H2AK15ub25,26. As anticipated, these mutants do not form foci in response to IR when transfected into U2OS cells (Fig. 2c). Unlike the S1609E/T1618D mutant, the two Tudor mutants W1495A and D1521A impaired the interaction of 53BP1-FFR with TIRR. This suggested that the Tudor domain, in contrast to the UDR motif, is required for binding TIRR.
Figure 2. TIRR associates with the tandem Tudor domain of 53BP1.
a, (Left) Representation of 53BP1 Tudor (PDB, 2G3R) highlighting in red residues with preferential signal broadening in the 1H-15N HSQC spectrum of Tudor upon titration with unlabeled TIRR. (Right) Overlay of the 1H-15N HSQC Tudor spectra in the absence (black) and presence (red) of TIRR (Tudor:TIRR molar ratio of ~1:0.3). 53BP1 residues with preferential signal broadening are labeled. b, 53BP1 Tudor surface representation showing residues with preferential decrease in signal intensities (see a). c, Immunofluorescence (Left) and Flag immunoprecipitation (Right) using indicated cells. d, Overlay of 1H-13C HMQC spectra of 13C-labeled dimethylated lysine analogs in TIRR (KC10me2, KC28me2 KC151me2 and KC205me2) in the absence (black) and presence of unlabeled 53BP1 Tudor [TIRR:53BP1 molar ratios: 1:0.1 (cyan), 1:0.25 (blue), 1:0.5 (green), 1:1 (red)]. Also shown are 1D slices of the 1H-13C HMQC spectra in the 1H dimension highlighting the preferential broadening of KC10me2 and KC151me2 compared to KC28me2 and KC205me2. e, Residues K10 and K151 highlighted in TIRR crystal structure (PDB, 4ZG0). TIRR gene accession # is NC_000016.10. f, Flag immunoprecipitation from indicated U2OS extracts.
The binding interface of TIRR and 53BP1 Tudor
We also used NMR spectroscopy to probe the Tudor binding surface in TIRR. The standard 1H-15N TROSY HSQC approach could not be used owing to the high molecular weight of TIRR. Therefore, we initially investigated the changes in intensities in the 1H-13C heteronuclear multiple quantum coherence (HMQC) spectrum of TIRR 13C-dimethylated at lysine residues resulting from interaction with 53BP1 Tudor. While all 1H-13C correlation signals in TIRR tended to decrease upon addition of Tudor, of the 9 lysine residues, we noticed the preferential disappearance of signals assigned to K10 and K151, suggesting that these two amino acids are in the vicinity of the binding interface. This approach is difficult owing to extensive signal overlap. Nevertheless, we were able to confirm the preferential disappearance of K10 and K151 signals after selective installation of 13C-labeled dimethyl-lysine analogs to generate TIRR proteins with KC10me2, KC151me2 and KC205me2. For this approach, the two solvent-accessible cysteines in TIRR (C88 and C171) were replaced by serines. A partially buried cysteine (C28) was not mutated. Upon addition of 53BP1, there was preferential disappearance of KC10me2 and KC151me2 signals in comparison to KC205me2 which is not affected by the interaction (Fig. 2d). Unexpectedly, residue C28 was also modified (KC28me2). While this complicates the analysis because the KC28me2, KC6me2 and KC203me2 signals overlap, KC28me2 also serves as an internal control (Fig. 2d). We note that to interact with a single 53BP1 tandem Tudor, K10 and K151, which are far apart within one protomer, necessarily belong to two different protomers of the TIRR homodimer to be sufficiently close in space to both contact one Tudor molecule (Fig. 2e). Having demonstrated that K10 and K151 were in the vicinity of the binding interface, we next asked whether these two residues were involved in the interaction with 53BP1 in cells. As shown in Fig. 2f, the mutation of these two lysine residues into glutamic acid (E) drastically reduced the interaction of TIRR with 53BP1 in undamaged U2OS cells. However, individual mutants revealed that K151E mutant continues to associate with 53BP1 in cells, while K10E mutation is sufficient to disrupt the interaction with 53BP1. We concluded that the residue K10 is crucial for the interaction with 53BP1.
TIRR impairs 53BP1 functions
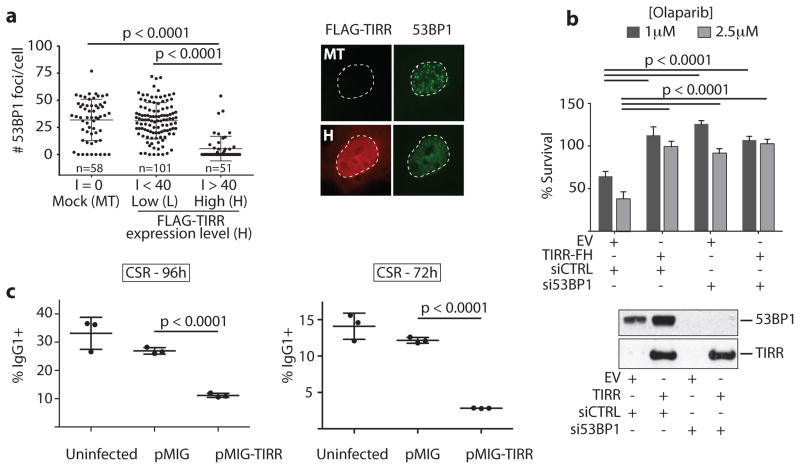
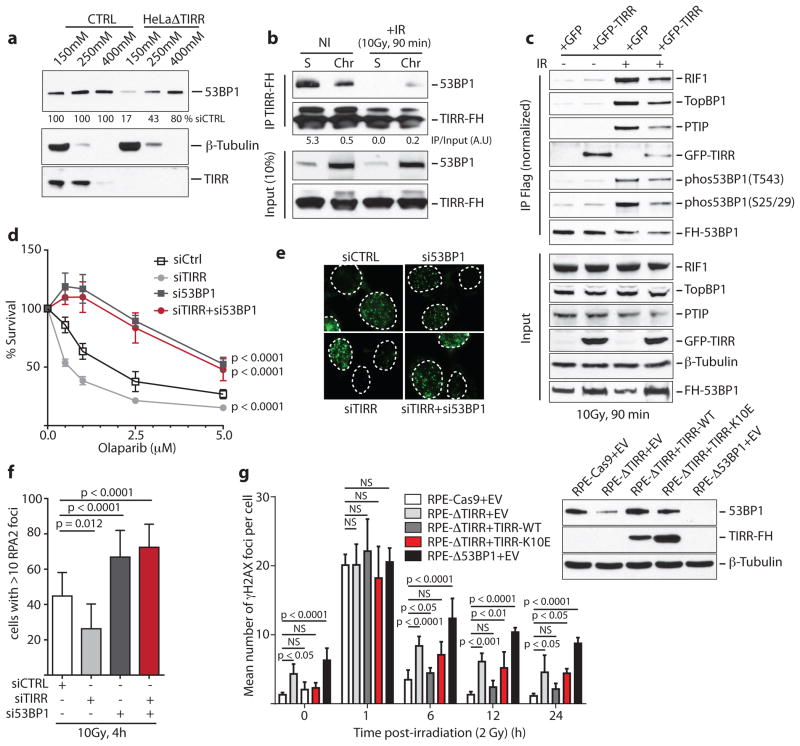
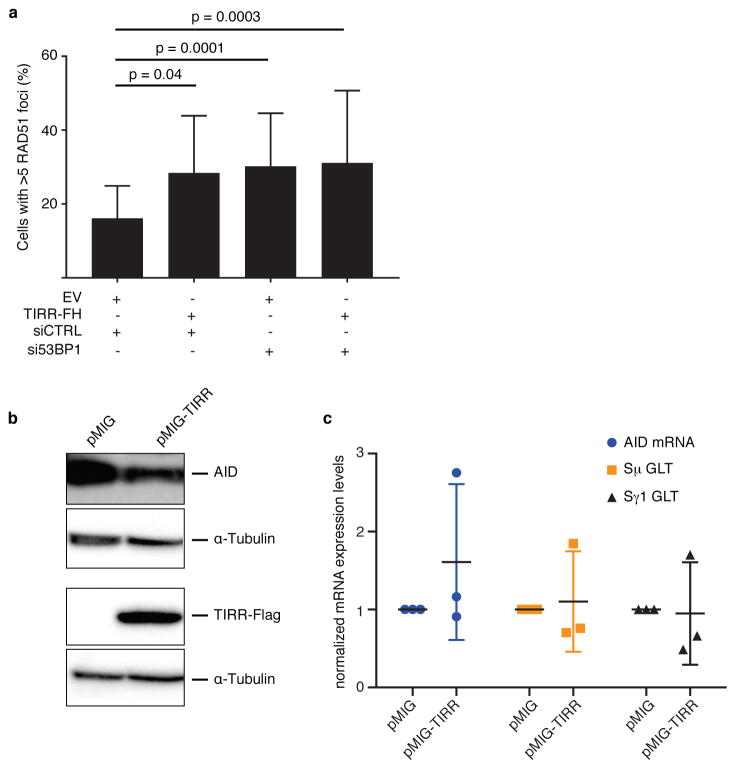
Next we tested whether TIRR interaction with the Tudor domains influences 53BP1 function. First, we observed that high TIRR expression almost completely abolishes 53BP1 foci formation (Fig. 3a). Second, TIRR over-expression conferred resistance to the PARP inhibitor, olaparib (Fig. 3b) and restored HR in MEFs carrying hypomorphic Brca1-Δ11 alleles (Extended Data Fig. 2a). Furthermore, silencing 53BP1 in cells over-expressing TIRR showed the same extent of olaparib resistance as 53BP1-depleted cells, indicating that TIRR-induced olaparib resistance was likely mediated by loss of 53BP1 function. Finally we tested whether TIRR compromised 53BP1 function in CSR of primary B cells. CSR to IgG1 was significantly reduced by TIRR (Fig. 3c and Extended Data Fig. 2b), and was not caused by reduced levels of AID or expression of germline transcripts (Extended Data Fig. 2c, d). Taken together, this data demonstrated that TIRR over-expression can radically impair the function of 53BP1 in DSB repair.
Figure 3. TIRR overexpression compromises 53BP1 functions.
a, Quantification of 53BP1 foci and intensity (I) of TIRR protein expression in FLAG-TIRR- or mock-transfected (MT) U2OS cells (mean ± s.d., n=3). Representative image is shown. b, Survival assay of indicated BRCA1-deficient MEFs (empty vector = EV) treated with Olaparib (mean ± s.d., n=3). Immunoblot shows TIRR-FH and 53BP1expression. c, Quantification of CSR at 96h or 72h post-stimulation (mean ± s.d., n=3 mice).
DNA damage-dependent ATM phosphorylation disrupts the 53BP1–TIRR complex
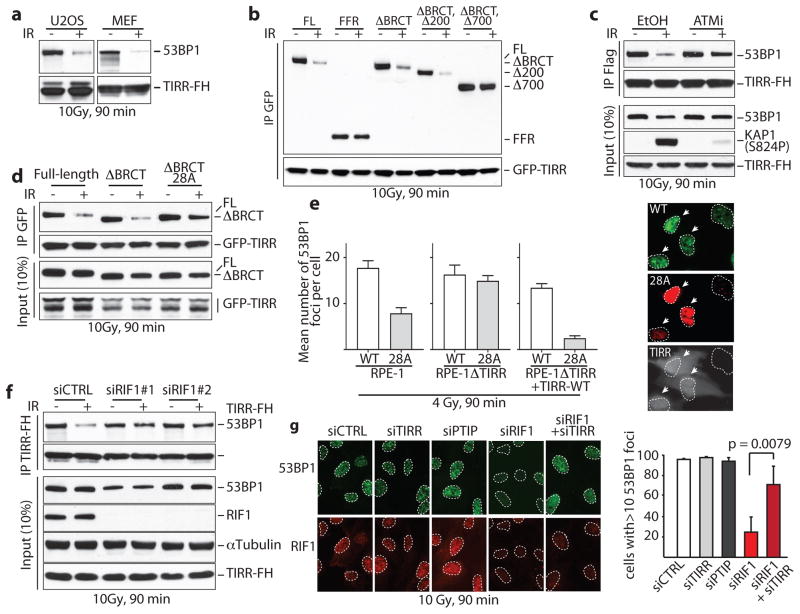
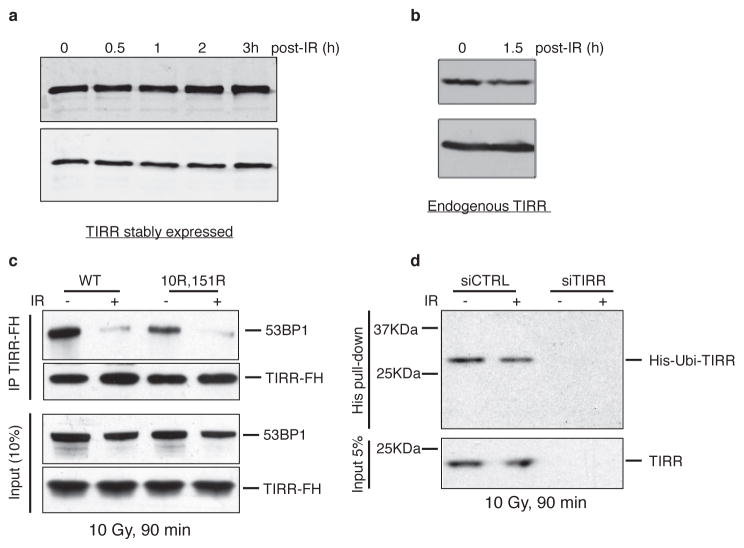
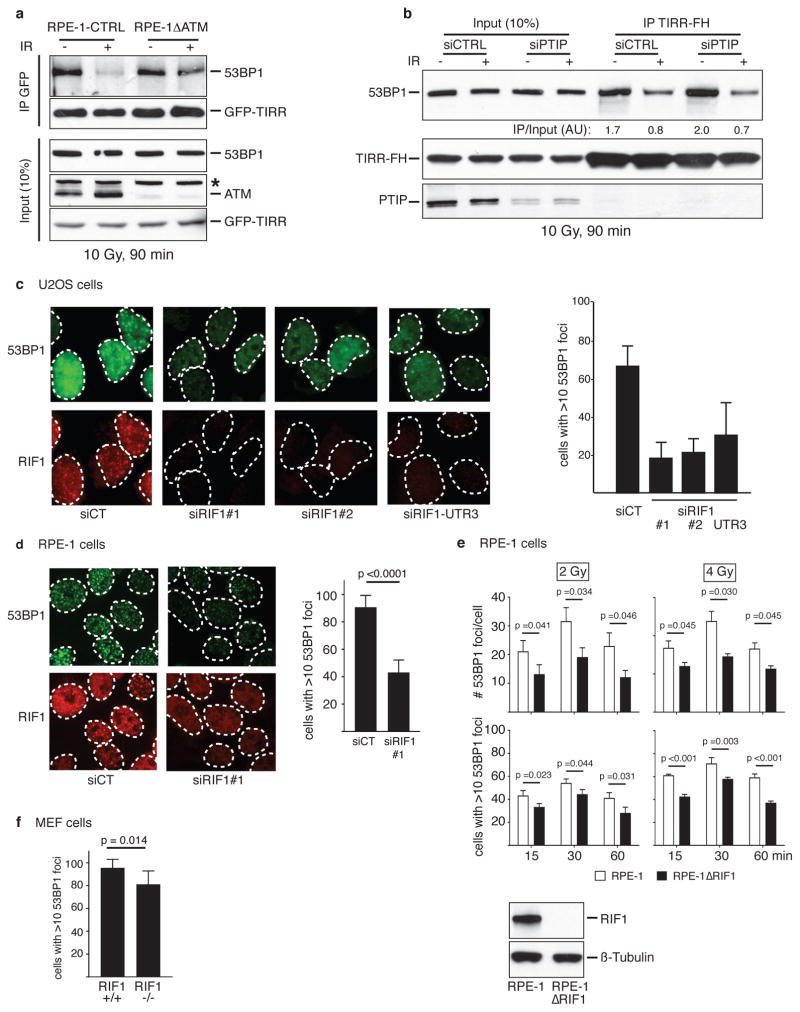
The interaction of 53BP1 and TIRR was reduced following ionizing radiation (IR) (Fig. 4a) and this was not due to degradation of TIRR or 53BP1 (Extended Data Fig. 3a, b). Consistent with this TIRR is not recruited to sites of DNA damage induced by laser micro-irradiation (Extended Data Fig. 3e). The two TIRR residues K10 and K151 that belong to the interaction interface with 53BP1 (Fig. 2) are constitutively ubiquitylated27, however, the double TIRR mutant K151R/K10R dissociated from 53BP1 in response to damage (Extended Data Fig. 3c). More broadly, IR did not alter the TIRR ubiquitylation level (Extended Data Fig. 3d). We then focused on 53BP1 and observed that unlike the full-length (FL) 53BP1 or the 53BP1 lacking the BRCT motif (ΔBRCT), the 53BP1-FFR did not dissociate from TIRR after IR (Fig. 4c). A deletion analysis revealed that a domain encompassing residues 200 to 700 was required for IR-mediated dissociation of 53BP1 from TIRR (Fig. 4c).
Figure 4. 53BP1/TIRR complex disruption by DNA damage-dependent ATM phosphorylation.
a, Immunoblotting of TIRR-FH partners from indicated cells. b, Schematic of FH-53BP1 constructs. c, GFP immunoprecipitation from HeLa cells stably expressing GFP-TIRR and the indicated FH-53BP1 form. d, Flag immunoprecipitation from TIRR-FH-expressing RPE-1 cells pretreated with DMSO or ATMi before irradiation. e, Same as c with the indicated 53BP1 construct. f, Foci formation of GFP-53BP1ΔBRCT-WT and mCherry-53BP1ΔBRCT-28A co-expressed in the indicated cells (mean ± s.d., n=3). Arrows, TIRR-positive cells. g, Flag immununoprecipitation from siRNA-transfected TIRR-FH-expressing RPE-1 cells. h, Immunofluorescence of 53BP1 and RIF1 in siRNA-transfected U2OS cells (mean ± s.d., n=3).
The N-terminus of 53BP1 contains 28 S/TQ consensus sites phosphorylated by ATM following IR mediating the interaction with RIF1 (RAP1-interacting factor 1)28–32 and PTIP33 (PAX transactivation domain–interacting protein). We observed that ATM inhibition (Fig. 4d) or deletion (Extended Data Fig. 4a) impaired the IR-induced dissociation of TIRR from 53BP1. Dissociation was also impeded when the 28 S/TQ sites were mutated to alanine (ΔBRCT-28A) (Fig. 4e). These results suggest that endogenous TIRR would preferentially block recruitment of the 53BP1ΔBRCT-28A mutant to DSB foci relative to its wild-type counterpart. Co-expression of GFP-53BP1ΔBRCT and mCherry-53BP1ΔBRCT-28A in RPE1 cells demonstrated that indeed 53BP1ΔBRCT-28A has limited ability to form foci (Fig. 4f). Deletion of TIRR in these cells restores the ability of 53BP1ΔBRCT-28A to form foci at wild-type levels. Furthermore, introduction of exogenous TIRR selectively impaired the ability of 53BP1ΔBRCT-28A to form foci (Fig. 4f). Collectively, our data suggests that DNA damage-dependent ATM phosphorylation of 53BP1 is required not only for the interaction with RIF1 and PTIP, but also to promote dissociation from TIRR.
RIF1 is involved in dissociation of the 53BP1–TIRR complex following IR
Since the N-terminal phosphorylation of 53BP1 recruits RIF1 and PTIP we investigated whether these factors are involved in the IR-mediated disruption of 53BP1–TIRR complex. Dissociation of the 53BP1–TIRR complex was selectively impaired by the depletion of RIF1 (Fig. 4g) and not by silencing PTIP (Extended Data Fig. 4b). Consistent with this, depletion of RIF1 in RPE-1 and U2OS cells (Fig. 4h and Extended Data Fig. 4c, d) and deletion of RIF1 in RPE1 cells significantly diminished the formation of 53BP1 foci at low doses (2 Gy and 4 Gy) and early time points (15 min and 30 min) after IR (Extended Data Fig. 4e). The effect was relatively mild in MEFs RIF1−/− indicating a potential species-specific difference (Extended Data Fig. 4f). Furthermore, co-depletion of TIRR restored the formation of 53BP1 foci in RIF1-depleted cells (Fig. 4h) suggesting that the impact of RIF1 loss on 53BP1 foci is due to the complex with TIRR. PTIP depletion had no effect on 53BP1 foci formation (Fig. 4h). Together, these results suggest that RIF1 functions not only downstream of 53BP1 as an effector protein in DSB repair but also upstream to promote IR-induced 53BP1/TIRR dissociation and thereby facilitate 53BP1 recruitment to DSB sites.
TIRR regulates 53BP1 activity
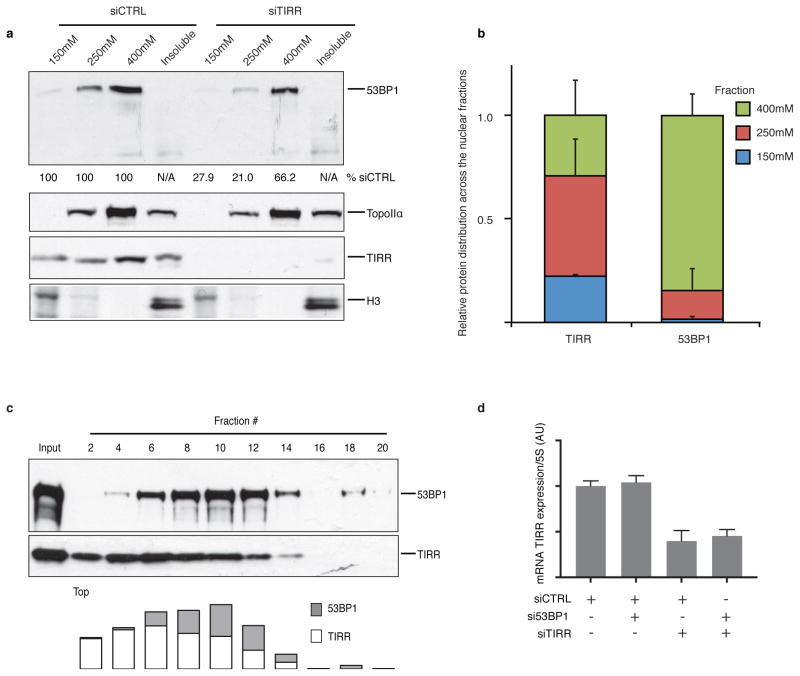
TIRR depletion or deletion significantly reduced 53BP1 protein levels (Fig. 5a, b). In TIRR-depleted cells the relatively soluble form of 53BP1 (150 and 250 mM NaCl fractions) were destabilized (Fig. 5b and Extended Data Fig. 5a). Quantification of relative amounts of TIRR and 53BP1 also revealed that ~75% of TIRR was in the 150 and 250 mM NaCl fractions whereas only ~25% of 53BP1 was in these fractions (Extended Data Fig. 5b). Consequently, in the absence of TIRR the proportion of 53BP1 in the 400 mM fraction (chromatin bound) is significantly increased. Consistent with these results, TIRR interacted almost exclusively with the soluble form of 53BP1 (Fig. 5c). However the total amount of TIRR and 53BP1 varied in different tumor lines (Extended Data Fig. 6a) and this could also impact sub-nuclear distribution of these proteins. Sucrose gradient fractionation of nuclear soluble proteins in undamaged cells indicates that the majority of 53BP1 co-sediments with TIRR (Extended Data Fig. 5c). Collectively, these results suggested that in undamaged cells, the nuclear soluble fraction of 53BP1 was stabilized through an interaction with TIRR. Next we analyzed 53BP1 binding partners by mass spectrometry in TIRR-deleted and GFP-TIRR-expressing cells after irradiation (Supplementary Table 3) and validated the results by immunoblot (Fig. 5d and Extended Data Fig. 7b). In the absence of TIRR, there is a detectable increase in association of 53BP1 with most of its well-characterized binding partners (such as MDC1, PTIP or KAP1) and an increase in IR-induced phosphorylation of 53BP1 on serines 25/29 and threonine 543 (Fig. 5d and Extended Data Fig. 7b). These differences were observed when TIRR-depleted cells were compared with the parental line (Extended Data Fig. 7b) or with the TIRR-depleted cells in which TIRR was re-introduced (Fig. 5d). Taken together our results demonstrate that TIRR depletion had a profound effect on the association of 53BP1 with multiple binding partners.
Figure 5. TIRR regulates 53BP1 functions.
a, Immunoblotting from Hela cell extracts. b, 53BP1 quantification using salt-extracted nuclear proteins prepared from indicated cells. c, Flag immunoprecipitation from nuclear soluble (S) and chromatin (Chr) extracts of TIRR-FH-expressing U2OS cells. d, Immunoblotting of FH-53BP1 partners pull-down from indicated cells. e, Survival assay of siRNA-transfected BRCA1-mutant MEFs treated with Olaparib (mean ± s.d., n=3, 2-Way ANOVA). f, 53BP1 immunofluorescence in BRCA1-mutant MEFs (mean ± s.d., n=3). RPA2 immunofluorescence (g) and quantification (h) in siRNA-transfected BRCA1-mutant MEFs (mean ± s.d., n=3). i, Kinetics of γH2AX foci formation in indicated cell line (mean ± s.d., n=3). NS not significant.
TIRR impacts DNA damage sensitivity
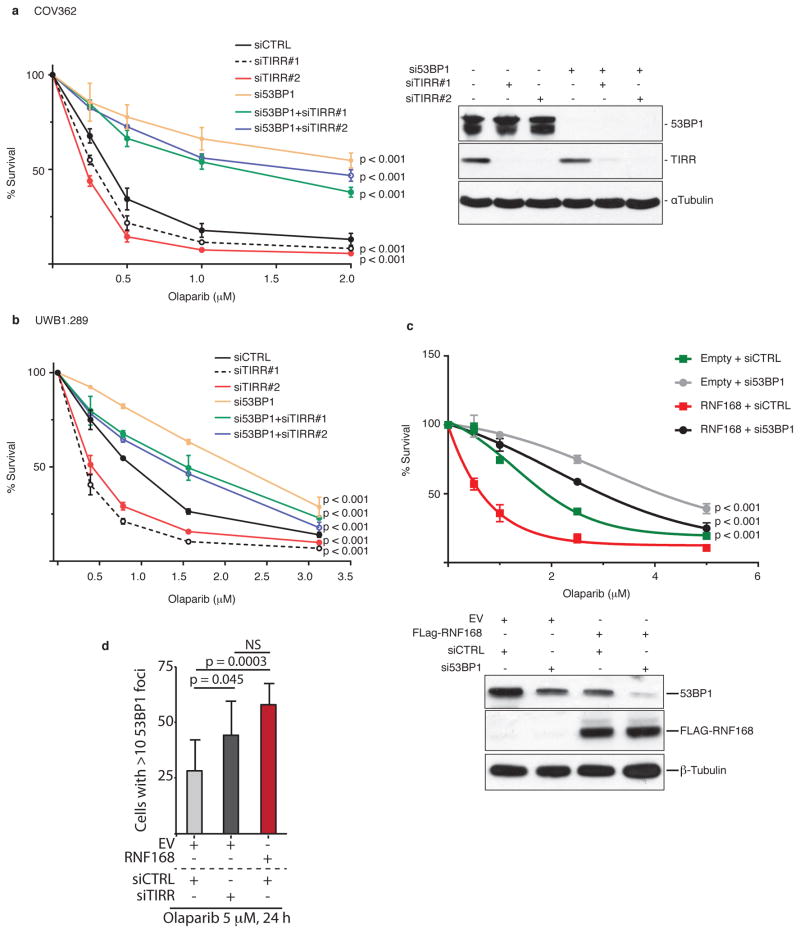
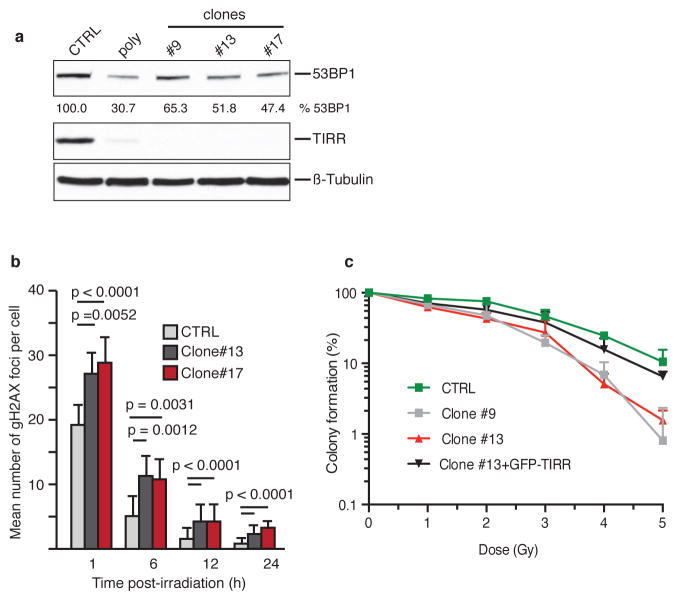
We observed that TIRR-depletion further sensitized Brca1-mutant MEFs and the Brca1-mutant human ovarian carcinoma lines COV362 and UWB289.1 to olaparib (Fig 5e and Extended Data Fig. 8a, b). These results phenocopied the 53BP1-dependent olaparib hypersensitivity induced by over-expressing RNF168 in Brca1-mutant MEFs34 (Extended Data Fig. 8c). TIRR-depletion and RNF168 over-expression increased 53BP1 foci formation (Fig. 5f) and impaired RPA foci formation in Brca1-mutant MEFs (Fig. 5g, h). Co-depletion of 53BP1 rescues all these phenotypes suggesting a 53BP1-mediated impact of TIRR. Furthermore, we observed that deletion of TIRR (Extended Data Fig. 9a) led to persistent DSBs and sensitized cells to radiation (Fig. 5i and Extended Data Fig. 9b, c), and this phenotype is ‘rescued’ by wild-type TIRR but not by the -TIRR mutant (K10E) that does not interact with 53BP1 (Fig. 5i). Together, our results suggested that TIRR influenced 53BP1 activity at multiple levels and broadly impacted DSB repair efficiency in cells.
DISCUSSION
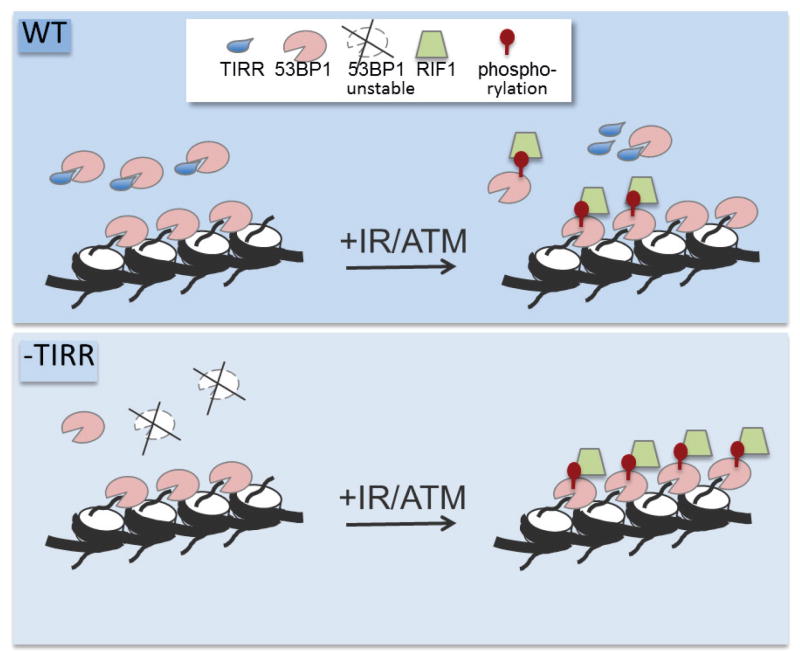
Accumulation of 53BP1 at DSBs requires recognition of the constitutive and highly abundant histone modification, H4K20me235. Selective unmasking of H4K20me2 occurs at DSB sites by removal of L3MBTL136 and by the demethylase Jumonji domain-containing protein 2A (JMJD2A)37 along with concomitant deacetylation of H4K1638. These represents indirect mechanisms restricting 53BP1 access to chromatin. Here we identify a direct regulatory step where TIRR binds the tandem Tudor domain of 53BP1 in the nuclear soluble fraction to restrict its access to chromatin prior to DNA damage. Loss of TIRR leads to a hyper-activation of 53BP1 which pheno-copies the over-expression/ectopic activation of RNF16834,39. We speculate that a subset of chromatin-associated 53BP1 remains inactive in cells (not phosphorylated/no association with effectors) even after DNA damage. Ectopic activation of RNF168 or loss of TIRR activates this subset leading to ‘hyper-active’ 53BP1 (see model in Extended Data Fig. 10).
53BP1 is a representative of Tudor domain-containing histone methyl-lysine readers such as SGF29, Spindlin1, UHRF1, and SHH140. These reader proteins are involved in the pathogenesis of various diseases and identifying small molecules that inhibit interactions between histone methyl-lysine readers and their respective binding partners on chromatin has significant clinical relevance41. Here with TIRR, we have the first example of a bona fide cellular inhibitor of a histone methyl-lysine reader. This is also a unique mechanism by which the activity of this class of proteins maybe broadly regulated. TIRR directly blocks the Tudor/methyl-lysine interface and this observation could be potentially utilized to identify factors that inhibit the methyl-lysine binding function of other Tudor proteins.
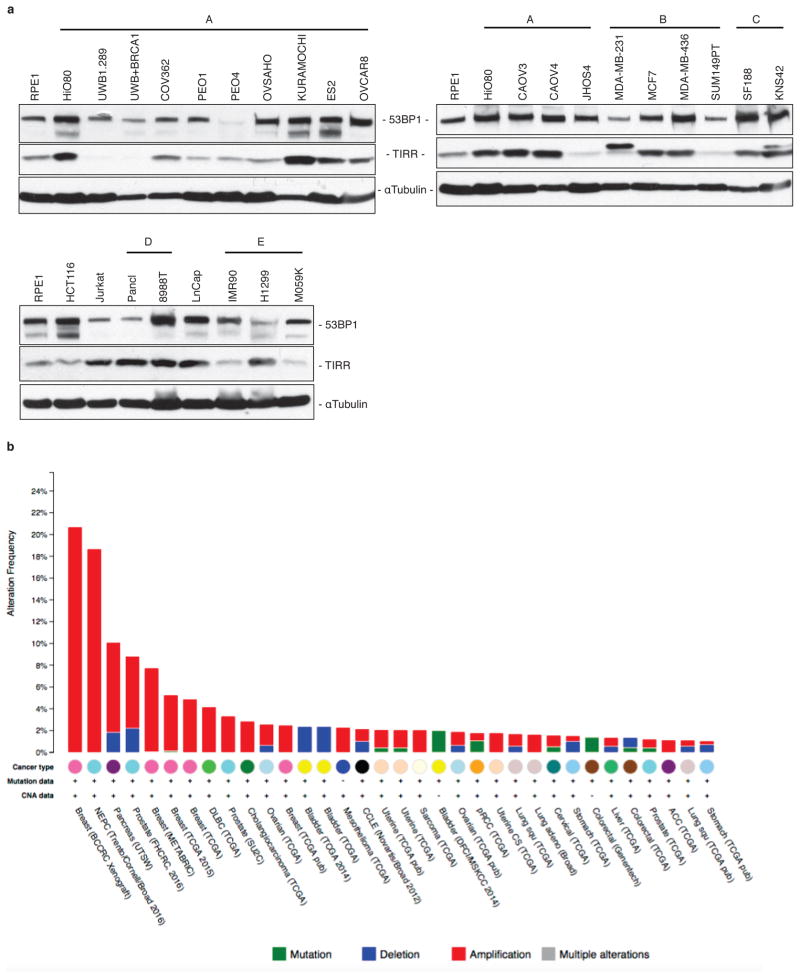
Many clinical trials are underway with PARPi42,43 and therefore resistance to PARPi is an emerging clinical problem44. Over-expression of TIRR causes PARPi resistance in BRCA1-deficient cells. A compilation of 50 studies in the Cancer Genome Atlas (TCGA) shows that the TIRR gene locus (alias Nudt16L1) is amplified in 29 out of the 34 different carcinomas (Extended Data Fig. 6b). BRCA1-mutant tumors may acquire PARPi resistance by amplifying the TIRR gene, and enhancing TIRR expression. Future analysis of BRCA1-mutant tumors from ovarian or breast cancer patients that are resistant to PARPi may reveal the clinical relevance of TIRR in cancer therapy.
METHODS
Cell culture and antibodies
All cells were grown in Dulbecco’s Eagle medium (DMEM) containing 10% Fetal Calf Serum (FCS), except B cells which were grown in RPMI-1640 supplemented with 15% FBS, 1% penicillin/streptomycin, 1% L-glutamine and 50 μM β-mercaptoethanol. Parental cells were tested for mycoplasma contamination.
Mouse antibodies employed were against Flag M2, α- and β-Tubulin (Sigma), γH2AX (Millipore) 6×His (Clontech), GFP (Cell signaling) and ATM (Santa Cruz), rabbit antibodies were against TopBP1, RIF1, PTIP, 53BP1 and phosphoKAP1 (S824) (All Bethyl Laboratories), 53BP1 (Santa Cruz), RNF168 (Millipore), TIRR (Sigma), phospho53BP1 (T543), phospho53BP1 (S25/29), HMGA1, H3 (All Cell Signaling) and AID (generated by the Chaudhuri Lab) and rat antibody against RPA2 (Cell Signaling). The RIF1 antibody used in immunofluorescence is a kind gift of Lifeng Xu (University of California, USA).
Plasmids and transfection
CRISPR guide RNAs were designed using http://crispr.mit.edu/. SgRNA targeting the ATM and TIRR locus were cloned in the pX458 vector carrying the pSpCas9(BB)-2A-GFP (Addgene #48138) or the pLentiGuide-puro vector (Addgene #52963). The following guide sequences were used (PAM). ATM: CTCTATCATGTTCTAGTTGA(CGG); TIRR guide 1: AGATGCAGATGCGTTTCGAC(GGG); TIRR guide 3: CAGTGCCAAGATGTCGACGG(CGG).
Human TIRR and 53BP1 cDNAs were expressed at a moderate level by using the retroviral vector POZ45. Human TIRR cDNA were subcloned into retroviral vector pMIG for class switch experiments and into mCherry-C2 and mCherry-LacI vectors for tethering experiments.
Unless otherwise mentioned, stable and transient transfections were performed using Lipofectamine 2000 (Invitrogen) or Fugene 6 (Promega) following the manufacturer’s instructions.
siRNA-mediated silencing
Cells were transfected with siRNAs using Lipofectamine RNAimax following the manufacturer’s instructions (Invitrogen).
The sequences of the stealth siRNAs (Thermofisher) were as follows:
Human TIRR(#2): UAGCCGUGCUCACGAAGGCGUUGCU; Human TIRR(#3): CACUCUAGAAGCCACACUUAGCAGG; Mouse TIRR: GAGUAGGCGGCUUUCCUAACUUUCU; Human 53BP1: AGAACGAGGAGACGGUAAUAGUGGG; Mouse 53BP1: UGAGCUAUUACUGUCUCCUUGUUCU; Human RIF1(#1): CCUGCUAAGUGUGGCUUCUAGAGUG; Human RIF1(#2): AAUUGAUGAAACUCCCACUUCGAUG; Human RIF1-UTR3: UUAUUCUUAUGACGUAUAGUAUU; Human PTIP(#1) :AGCCAGAATTGAAGACGTA; Human PTIP(#2): GCGACATTCTTCTGGGAAA; Control: AAGCCGGUAUGCCGGUUAAGU
Mice, B cell purification and retroviral infection
C57BL/6J female mice were purchased from The Jackson Laboratory and housed according to the guidelines for animal care of MSKCC Research Animal Resource Center.
Retroviruses were prepared by cotransfecting pMIG or pMIG-TIRR with packaging vector pCL-Eco into HEK293T cells by calcium phosphate method.
Splenic B cell were purified from 8–12 week old mice by negative selection using anti-CD43 magnetic beads (Miltenyi Biotec) according to the manufacturer’s protocol. 2×106 B cells were plated at a density of 1×106 cells/ml and were immediately stimulated with 30 μg/ml LPS (Sigma) plus mouse 25 μg/ml IL-4 (R&D systems). Following stimulation for either 24 h or 48 h, media was aspirated, leaving approximately 1 ml of media per well of a 6-well dish, and retroviruses were added. 6-well dishes were spun at 2000×g for 90 min at 32°C, after which viral supernatants were aspirated and fresh B cell media plus LPS and IL-4 was added.
B cells were harvested at 48 h to purify total RNA for qPCR analysis, or at 72 h and 96h for flow cytometry analysis or to prepare lysates for western blotting.
Flow cytometry class switch recombination analysis
Primary B Cells were transduced with a retrovirus expressing TIRR, stimulated in vitro with lipopolysaccharide (LPS) and interleukin (IL)-4, and switching to IgG1 was measured. Cells were collected at 72 h and 96 h for flow cytometry analysis. Cells were washed with 2 ml FACS buffer (1× PBS, 2.5% FBS), resuspended in 100 μl FACS buffer and stained with the following antibodies for 30 min at 4°C: APC rat anti-mouse IgG1 (BD Biosciences), Alexa Fluor 700 rat anti-mouse B220 (eBiosciences), PE rat anti-mouse IgM (eBiosciences), PerCP/Cy.5 rat anti-mouse CD138 (BioLegend), PE-Cy7 rat anti-mouse CD25 (BD Pharmingen), DAPI (Invitrogen). Cells were washed with 2 ml of FACS buffer, and resuspended in 200 μl FACS buffer. Flow cytometry was performed using an LSR-II flow cytometer (BD Biosciences), and data analysis was performed using FloJo software (version 9.9). P-values for an unpaired t-test were calculated using Graphpad Prism software.
Tethering to the LacO array
The cell line U2OS19 containing 256 lac operator and 96 tetracycline response element copies has been previously described24. TIRR was fused to the Escherichia coli lac-repressor (LacI) and tagged with Cherry-red fluorescent protein (mCherry-LacI-TIRR). 53BP1 was fused to LacI and tagged with GFP-fluorescent protein (GFP-LacI-53BP1). Jet Pei (Polyplus transfection) was used for transient transfections of cells growing on glass coverslips. Twenty-four hours after transfection, soluble proteins were extracted in buffer containing 0.5% Triton-X100, 50 mM Hepes (pH 7.0), 150 mM NaCl, 10 mM EGTA, 2 mM MgCl2 for 30 sec on ice prior to process to immunofluorescence.
Immunofluorescence
Cells were grown on glass coverslips, fixed in 4% formaldehyde in PBS for 15 min at room temperature and blocked/permeabilized for 1 h in PBS containing 0.3% Triton-X100, 1% BSA, 10% fetal bovine (or 3% goat serum). Incubation with primary and secondary antibodies (Alexa Fluor, Molecular Probes) were made in PBS containing 1% BSA and 0.1% Triton-X100 for 1 h at room temperature. Coverslips were mounted using DAPI Fluoromount-G (Southern Biotech). When staining for RPA or RAD51 foci, soluble proteins were preextracted as previously described34 prior to process to immunofluorescence.
Laser microirradiation
Cells were pre-sensitized in phenol red free DMEM medium containing 1 μg/ml of Hoechst 33342 for 15 min. Laser microirradiation was performed with the 405 laser (75% laser power and 30 Hz scan speed) on a Leica SP5X laser scanning confocal microscope equipped with the heated stage. Four random fields were striped along around 15 parallel lines (region of interest) spanning each field of view. Cells were allowed to recover for 15 min, fixed with 4% paraformaldehyde and processed for immunofluorescence.
CTG Assay and clonogenic survival
For assessing cellular cytotoxicity, cells were transfected twice with siRNA in 36-hour intervals. Forty-eight hours after the second transfection, cells were seeded into 96-well plates. Olaparib (Chemietek) was serially diluted in media and added to the wells. Five days later, drug-free media was added to the wells and cells were incubated for 72 h before adding CellTiter-Glo reagent (Promega) to the wells. Plates were scanned using a luminescence microplate reader. Survival at each Olaparib concentration was plotted as a percentage of the survival in drug-free media.
For clonogenic survival, 500 RPE-1 cells were seeded into 10 cm plates. Forty-eight hours later, cells were irradiated with indicated dosage. Colony formation was scored 14 days after irradiation using 2.5% (w/v) crystal violet in methanol. Survival curves were expressed as a percentage and standard error over three independent experiments of colonies formed relative to the non-irradiated control.
RNA purification and qPCR
RNAs were purified from B cells using the TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. cDNAs were generated from 1 μg of purified RNA using Quanta Biosciences qScript cDNA Synthesis Kit according to the manufacturer’s protocol. One microliter out of 20 μl cDNA was used to perform qPCR using the iQ SYBR Green Supermix (Biorad) with the following gene specific primers: AID forward (GCCACCTTCGCAACAAGTCT), AID reverse (CCGGGCACAGTCATAGCAC), Imu forward (CTCTGGCCCTGCTTATTGTTG), Cmu reverse (GAAGACATTTGGGAAGGACTGACT), (GGCCCTTCCAGATCTTTGAG), CG1 reverse (GGATCCAGAGTTCCAGGTCACT). Relative gene expression was calculated according to the delta Ct method.
CRISPR–Cas9 genome editing
For HeLa TIRR knockout line, HeLa cells were transiently transfected with pX458. GFP-positive clones were isolated by fluorescence-activated cell sorting 48 h after transfection, seeded in 96-well plates and further cultured for expansion. For RPE-1 TIRR, 53BP1, RIF1 and ATM knockout lines, RPE-1 cells were first transfected with pLentiCas9-Blast (Addgene #52962) and selected with 40 μg/ml blasticidin for 14 days. The resulting RPE-1/Cas9 stable cell lines were then transduced with pLentiGuide-puro viral particles supplemented with 10 μg/ml polybrene. Cells were selected with 10 μg/ml puromycin 48 h after transduction for 14 days and clonal cell lines were generated by picking single-cell colonies.
Immunoprecipitation
Unless otherwise mentioned, proteins were immunoprecipitated from whole-cell extracts. Briefly, cells were collected, washed and lysed for 30 min in a buffer containing 20 mM Tris-HCl (pH 7.65), 250 mM NaCl, 0.5% NP-40, 5 mM EDTA, 5% glycerol and protease and phosphatase cocktail inhibitors (Roche). Protein concentration from cleared supernatants was estimated by a Bradford assay (Biorad). Five hundred μg of whole-cell extracts were incubated on a roller for 16 h at 4°C with anti-FLAG- (Sigma) or GFP-Trap agarose (Chromotek). Resins were washed five times with TGN buffer [20 mM Tris-HCl (pH 7.65), 150 mM NaCl, 3 mM MgCl2, 10% Glycerol, 0.01% NP-40] before elution in 0.1 M glycine (pH 2.9). Eluted proteins were analyzed by immunoblotting.
Double immunoaffinity purification method
TIRR proteins fused with C-terminal FLAG- and HA-epitope tags (TIRR-FH) and 53BP1 full-length or truncated forms fused with N-terminal FLAG- and HA-epitope tags (FH-53BP1) were stably expressed in cells by retroviral transduction. Transduction, cell fractionation and complexes purification were done as previously described 45. Chromatin extracts were prepared by digestion 10 min at 37°C of pre-extracted nuclear pellets with micrococcal nuclease (Sigma) in 0.34 M sucrose buffer [20 mM Tris-HCl (pH 7.65), 15 mM KCl, 60 mM NaCl, 0.34 M sucrose] containing 2 mM CaCl2. Reaction was stopped by adding EGTA to a final concentration of 5 mM and cleared extracts were used for immunoprecipitation. Mass spectrometry identification of proteins was carried out by Taplin laboratory (Harvard Medical School, Boston, MA).
Preparation of recombinant proteins
Fusion proteins were expressed in Escherichia coli strain BL21(pLysS) and induced overnight with 0.1 mM isopropyl-β-d-thiogalactoside (IPTG) at 16°C. Cells were harvested and resuspended in buffer containing 500 mM NaCl, 20 mM Tris-HCl (pH 8.0), 5 mM DTT and protease inhibitor cocktail (Roche), except for TIRR-His where DTT were omitted. Cells were lysed by sonication and cleared extracts were incubated for 1 h at 4°C with anti-FLAG- (Sigma) or Ni-NTA agarose resin (Life Technologies) to purify FLAG- or His-fusion proteins, respectively. Proteins were eluted in lysis buffer containing 0.4 mg/ml of FLAG peptide (Sigma) (FLAG proteins) or 250 mM imidazole (His proteins) and dialyzed in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl and 10% glycerol. For NMR spectroscopy and ITC measurements, TIRR-His was treated with benzonase (EMD Millipore) after cell lysis and upon binding to Ni-NTA resin, was extensively washed with 50 mM sodium phosphate (pH 7.5), 20 mM imidazole and 1 M NaCl to remove nucleic acids bound to the protein. TIRR was further purified by size-exclusion chromatography using a HiLoad 16/60 Superdex 75 column (GE healthcare) and 50 mM sodium phosphate (pH 7.5), 500 mM NaCl running buffer. 53BP1 was purified as previously reported15,38.
Isothermal titration calorimetry
The ITC data were recorded at 25°C using an iTC200 microcalorimeter (Malvern). TIRR in the calorimeter cell was at concentrations of 8 to 20 μM and 53BP1 tandem Tudor in the injection syringe was at concentrations of 0.1 to 0.4 mM. Protein samples were in 25 mM Tris-HCl (pH 7.5), 500 mM NaCl.
NMR spectroscopy
The NMR data were recorded at 25°C using a Bruker Avance III 700 MHz spectrometer equipped with a cryoprobe. Protein samples were in 50 mM sodium phosphate (pH 7.5), 500 mM NaCl and 10% D2O. 53BP1 tandem Tudor was 15N-labeled using a previously reported procedure15,38. For the titration experiments, TIRR was at concentrations ranging from 20 μM to 50 μM and small aliquots of 3.5 mM 53BP1 Tudor were added. All spectra were recorded using salt-tolerant shaped NMR tubes (Bruker). The reductive alkylation of TIRR to generate 13C-dimethylated lysine residues followed a previously reported protocol making use of 13C-enriched formaldehyde (Sigma)46,47. The lysine residues were assigned by individual mutation to arginine. The synthesis of 13C-methyl alkylation reagent dimethylated 2-chloroethylamine and installation of dimethylated lysine analog followed previously reported protocols47,48. Briefly, two of the three cysteines in TIRR were mutated into serines. Each lysine of interest (K10, K151 and K205) was replaced by a cysteine and converted to the dimethylated lysine analogs KC10me2, KC151me2 and KC205me2. For the reductive alkylation of TIRR and the installation of dimethylated lysine analogs, excess reagent was eliminated by size-exclusion chromatography using a Superdex 75 10/300 GL column (GE healthcare) and 50 mM sodium phosphate (pH 7.5), 500 mM NaCl running buffer. The NMR data were processed with NMRPipe49 and analyzed with NMRView50. PyMol was used to create the molecular representations [WF Delano, The PyMOL Molecular Graphics System, version 1.3r1 (Schrodinger, LLC, New York, 2010)]
TIRR and 53BP1 quantification
Purified TIRR and 53BP1 (aa 1220–1710 fragment) at ~2.5 μg/μl and 125 ng/μl, respectively, were reduced with 10 mM DTT and alkylated with 15 mM iodoacetamide prior to methanol chloroform precipitation followed by digestion with trypsin. Peptides were de-salted on homemade C18 StageTips and analyzed by LC-MS/MS on an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific) to identify a set of target peptides for the analysis of samples. Peptides were separated on a 35 cm microcapillary column with a 100 μm inner diameter and packed with ~0.5 cm Magic C4 resin (5 μm, 100 Å, Michrom Bioresources) and Accucore C18 resin (2.6 μm, 150 Å, Thermo Fisher Scientific). ~4 μl sample were loaded onto the column for each analysis and separated using a 40 min gradient of 7%–26% acetonitrile in 0.125% formic acid with a flow rate of 625 nl/min. MS1 spectra were acquired with the following parameters: resolution 120,000, scan range 400–1500 Th, AGC target 4 × 105, maximum injection time 50 ms, and centroid spectrum data type. Dynamic exclusion was set with exclusion after 1 times with an exclusion duration of 60 s. MS2 spectra were acquired with CID and a resolution of 60,000, AGC 5 × 104, maximum injection time 60 ms isolation window 1.6 Th as centroid spectra. Raw files were converted into mzXML data type and search using Sequest (against all human proteins reported by Uniprot) on an in-house data analysis tool developed by Steve P. Gygi (Harvard Medical School, Boston, USA). Unique peptides were imported into Skyline 3.5.0.9319 and transition lists for heavy and light fragments exported for use on the mass spectrometer. Cells were grown in K8 Lys and lyzed in hypotonic buffer [10mM Tris (pH 7.65), 1.5 mM MgCl2, 10 mM KCl]. Pellet nuclei were resuspended sequentially into 0.34 M sucrose buffer containing 150 mM, 250 mM and 400 mM NaCl. Extraction was performed 10 min on roller and nuclei were pelleted between each extraction by centrifugation at 800 g. TIRR peptides for the different fractions were prepared as for the purified proteins above. For 53BP1, extracts were separated on 4–12% SDS-PAGE gels and the 53BP1 bands excised for in-gel digestion. Bands were reduced with 10 mM DTT and alkylated with 50 mM iodoacetamide. Gel pieces were washed and destained, dried and digested overnight in trypsin. Digests were de-salted on C18 StageTips. Combined samples of light reference peptides and heavy sample peptides were analyzed by LC-MS/MS with the same MS1 settings as before, but without dynamic exclusion and targeting masses as determined and exported from Skyline51. MS2 spectra were collected at a resolution of 15,000, AGC 5 × 104, maximum injection time 120 ms, isolation window 1.6 Th and as centroid spectra. Precursor and fragment ions were assessed in Skyline and ratios of the different fractions determined. TIRR and 53BP1 quantity were determined from the most abundant fraction (400 mM) and approximated for the other fractions.
Pull-down assays
Pull-down of TIRR by 53BP1 truncation mutants was performed by incubating 1 μg of TIRR-His with 2 μg of FLAG-fusion proteins immobilized on anti-FLAG agarose resin in TGN buffer. After 2 h at 4°C, beads were washed four times with TGN buffer containing either 200 or 400 mM NaCl and once with TGN buffer before elution of retained proteins with Laemmli sample buffer and analysis by immunoblotting. Peptide pull downs were performed by incubating the indicated 53BP1 fragment protein with 2 μg of biotinylated histone H4-derived peptide in TNB buffer [50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.05% NP-40, 0.1% BSA]. After 2 h at 4°C, 10 μl of streptavidin-Dynabeads (Dynal) were added and reaction was further incubated 30 min at 4°C. Beads were next treated as described above for pull-down of TIRR-His.
Statistical analysis
Statistics were performed with GraphPad (GraphPad Prism 5.0). Unless mentioned otherwise, all statistics were evaluated by two-tailed Student’s t-test (Mann-Whitney test).
Data Availability Statement
Authors can confirm that all relevant data are included in the paper and or its supplementary information files. The NCBI accession # for TIRR is NC_000016.10.
Extended Data
Extended Data Figure 1. TIRR prevents the interaction of 53BP1 Tudor domain with a peptide derived from H4K20me2.
a, Soluble nuclear proteins were prepared from HeLa and HeLa cells in which TIRR has been knock out using CRISPR/cas9 system (HeLaΔTIRR) undamaged or irradiated at 10 Gy for 90 min. Endogenous 53BP1 was next immunoprecipitated and immunoblotted using the indicated antibody. b, Coomassie staining of FLAG-tagged recombinant proteins purified from bacteria; “OD-Tudor” encompasses the oligomerization, the Tudor and UDR domains, “OD” the oligomerization domain and “Tudor-UDR” the Tudor and UDR domains. c, Recombinant TIRR-His protein was incubated with the indicated FLAG fusion protein in presence of 200 or 400 mM NaCl. Pull-down proteins were subjected to FLAG (top) and His (bottom) immunoblotting. M; molecular size markers. d, Interaction of 53BP1 Tudor with TIRR probed using ITC. e, Coomassie staining of FLAG-OD-Tudor and FLAG-OD-Tudor/TIRR-His complex purified from bacteria coexpressing both proteins. f, Pull-down of the indicated proteins with a biotinylated peptide derived from histone H4-K20me2. Bound proteins were analyzed by immunoblotting.
Extended Data Figure 2. Ectopic TIRR restores RAD51 foci formation in BRCA1-mutant MEFs.
a, Brca1-mutant MEFs stably transduced with an empty vector (EV) or TIRR-FH were transfected with the indicated siRNA. RAD51 was stained six hours after 5 Gy irradiation (mean ± s.d., n=3). b, Quantification of CSR at 96h or 72h post-stimulation from splenic B cells transduced with pMIG or pMIG-TIRR, and stimulated with LPS and IL-4 for 72 h. c, AID, Sμ germline (Sμ GLT) and Sγ1 germline (Sγ1 GLT, black) transcripts in splenic B cells stimulated for 48 h (mean ± s.d., n=3 mice). d, Immunoblotting of AID and TIRR from lysates prepared from splenic B cells stimulated for 72 h.
Extended Data Figure 3. TIRR expression level remains unchanged after irradiation.
a, TIRR-FH-expressing U2OS cells were irradiated at 10 Gy and collected next at the indicated time. Whole-cell extracts were analyzed by immunoblotting using the indicated antibody. b, U2OS whole-cell extracts were prepared 90 min after a 10 Gy irradiation and analyzed by immunoblotting using the indicated antibody. c, RPE-1 cells stably expressing TIRR-FH wild-type (WT) or double mutated (K10R, K151R) were irradiated at 10 Gy for 90 min followed by TIRR-FH immunoprecipitation. Pull-down proteins were subjected to immunoblotting with the indicated antibodies. d, U2OS cells were first transfected with the indicated siRNA. The day after, cells were transfected with a vector encoding Ubiquitin fused to a N-terminal His tag. Forty-eight hours later, cells were irradiated at 10 Gy for 90 min when indicated followed by His pull-down. Pull-down proteins were subjected to immunoblotting with the indicated antibodies. e, Laser stripes examined by immunofluorescence 15min post-irradiation in RPE-1 cells expressing TIRR-FH (left Panel) or RPE-1 cells (Right panel).
Extended Data Figure 4. RIF1 depletion affects 53BP1 foci formation in various cellular models.
a, RPE-1 cells deprived of endogenous ATM using CRISPR/cas9 system (RPE-1 ΔATM) were stably transfected with GFP-TIRR. GFP-TIRR was next pull-down and retained proteins were analyzed by immunoblotting. The star represents a non-specific band. b, Flag immunoprecipitation from extracts prepared from indicated siRNA-transfected TIRR-FH-expressing RPE-1 cells. c, Immunofluorescence of 53BP1 and RIF1 in siRNA-transfected U2OS cells irradiated at 10 Gy for 90 min. The graph represents the percentage of cells harboring more than ten 53BP1 foci. d, Same as c in RPE-1 cells (mean ± s.d., n=2). e, Kinetics of 53BP1 foci formation in RPE-1 ΔRIF1 cells. Results are expressed as number of foci per cell (top) and as percentage of cells harboring more than ten 53BP1 foci (bottom) (mean ± s.d., n=2). Deletion of RIF1 was checked by immunoblotting. f, Same as in e in MEF-RIF1−/− cells.
Extended Data Figure 5. TIRR depletion affects mainly the stability of soluble nuclear 53BP1.
a, Nuclear proteins were sequentially extracted by increasing amount of salt from siRNA-transfected U2OS cells. Following salt extraction, the resulting pellet corresponds to the insoluble material (insoluble). The amount of 53BP1 presents in each fraction was measured by immunoblotting and quantification using ImageJ. b, Relative TIRR and 53BP1 protein distribution in RPE-1 nucleus as measured by mass spectrometry. c, U2OS soluble nuclear extract (1mg) was loaded onto a sucrose gradient 5–30% and ultracentrifuge for 4h at 40,000 rpm. Fractions were collected from the top of the gradient and immunoblotted. The amount of 53BP1 and TIRR was quantified using imageJ. d, Quantification of TIRR mRNA expression in siRNA-transfected Brca1-mutant MEFs. TIRR level was normalized to 5S expression.
Extended Data Figure 6. Expression of TIRR protein in various cancer cell lines and alteration frequency of Nudt16L1 (TIRR) gene in cancer.
a, Immunoblotting of 53BP1 and TIRR from extracts prepared from various human cancer cell lines: A, Ovarian cancer cell lines; B, Breast cancer cell lines; C, pediatric glioma cell lines; D, pancreatic cell lines; E, lung cell lines. HCT116, colon cancer cells; Jurkat, acute T cell leukemia; LnCap, prostate adenocarninoma. Epithelial cells from Retinal pigment RPE-1 and Ovarian surface HiO80 are immortalized cell lines. b, Data extracted from cBioportal (www.cBioportal.org) for alterations in Nudt16L1 (TIRR) gene across different cancer types. 29 out of 34 cancer types exhibit amplifications in the Nudt16L1 gene.
Extended Data Figure 7. TIRR depletion modifies the interaction of 53BP1 with its partners.
a, HeLa cells deprived of endogenous TIRR (HeLaΔTIRR) were stably transduced with FH-53BP1 together with GFP-TIRR or GFP. FH-53BP1 partners were purified from total nuclear extracts (nuclear soluble and chromatin extracts) 90 min after a 10 Gy irradiation and analyzed by mass spectrometry. b, HeLaΔTIRR or parental HeLa cells were stably transduced with FH-53BP1. FH-53BP1 partners were purified from total nuclear extracts from undamaged or irradiated cells and analyzed by immunoblotting after normalization of the amount of 53BP1 pull-down (see results). The star represents a non-specific band.
Extended Data Figure 8. TIRR depletion hypersensitive human BRCA1-mutant cells to PARP inhibitors in a 53BP1-dependent manner.
Human ovarian cancer cell lines Cov362 (a) and UWB1.289 (b) were transfected with the indicated siRNA, followed by Olaparib treatment. Percentage of survival is calculated by normalizing the survival from Olaparib treatment versus untreated cells (mean ± s.d., n=3). Efficiency of siRNAs was checked by immunoblot using the indicated antibodies in Cov362 cell line. c, siRNA-transfected Brca1-mutant MEFs mock-transfected or overexpressing RNF168 were treated with Olaparib. Percentage of survival is calculated as in a and b (mean ± s.d., n=3). Bottom panel. Stable expression of Flag-RNF168 and efficiency of 53BP1 siRNA were checked by immunoblotting.
Extended Data Figure 9. TIRR depletion increases radiosensitivity.
a, Whole cell extracts from RPE-1 cells (CTRL) and from a polyclonal population (poly) and three RPE-1 clones deprived of endogenous TIRR using CRISPR/cas9 system were analyzed by immunoblotting using the indicated antibodies. The amount of 53BP1 was quantified and normalized to β-tubulin using ImageJ. b, Kinetics of γH2AX foci formation in indicated cells after a 2 Gy irradiation. The graph represents the mean number of γH2AX foci per cell (mean ± s.d., n=3). c, Clonogenic survival of indicated cells following irradiations (0 to 5 Gy). TIRR expression has been restored in clone#13 by stable expression of GFP-TIRR. Survival was expressed as a percentage of colonies formed relative to the non-irradiated control (mean ± s.d., n=3).
Extended Data Figure 10. Model depicting the regulation of 53BP1 activity by TIRR.
Supplementary Material
Acknowledgments
We are grateful to Lifeng Xu (University of California, USA) for RIF1 antibody and Simon Boulton (CRUK) for RIF1−/− MEFs. DC is supported by R01 AI101897-01 (NIAID) and R01CA142698-07 (NCI), Leukemia and Lymphoma Society Scholar Grant, Claudia Adams Barr Program for Innovative Cancer Research, DOD Ovarian Cancer Award, Breast SPORE Pilot Award, Robert and Deborah First Fund Award. GM is supported by NIH grants R01 CA132878 and R01 GM116829, and a Mayo Clinic Brain Cancer SPORE Program Pilot Award (P50 CA108961). JWH is supported by add AG011085.
Footnotes
CONTRIBUTIONS
P.D, G.M and D.C designed the study. P.D performed most of the experiments with assistance from MEB, KM, SC, YZH, XFF and NP. AD did the statistical analysis. GC and MVB conducted NMR studies under GM’s supervision. AK conducted microscopy studies with LacO fusion system under ES’s guidance. CM did quantitative MS under JWH’s guidance. WTY did CSR assays under JC’s guidance. P.D, G.M and D.C. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 2.Zimmermann M, de Lange T. 53BP1: pro choice in DNA repair. Trends Cell Biol. 2014;24:108–117. doi: 10.1016/j.tcb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bothmer A, et al. Mechanism of DNA resection during intrachromosomal recombination and immunoglobulin class switching. J Exp Med. 2011;210:115–123. doi: 10.1084/jem.20121975. jem.20121975 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong J, et al. Orientation-specific joining of AID-initiated DNA breaks promotes antibody class switching. Nature. 2015;525:134–139. doi: 10.1038/nature14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward IM, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manis JP, et al. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 7.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 8.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 9.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouwman P, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman JR, Sossick AJ, Boulton SJ, Jackson SP. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. Journal of cell science. 2012;125:3529–3534. doi: 10.1242/jcs.105353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charier G, et al. The Tudor tandem of 53BP1: a new structural motif involved in DNA and RG-rich peptide binding. Structure. 2004;12:1551–1562. doi: 10.1016/j.str.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 13.Iwabuchi K, et al. Potential role for 53BP1 in DNA end-joining repair through direct interaction with DNA. J Biol Chem. 2003;278:36487–36495. doi: 10.1074/jbc.M304066200. [DOI] [PubMed] [Google Scholar]
- 14.Fradet-Turcotte A, et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jowsey P, et al. Characterisation of the sites of DNA damage-induced 53BP1 phosphorylation catalysed by ATM and ATR. DNA Repair (Amst) 2007;6:1536–1544. doi: 10.1016/j.dnarep.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 17.Ward IM, Minn K, Jorda KG, Chen J. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J Biol Chem. 2003;278:19579–19582. doi: 10.1074/jbc.C300117200. [DOI] [PubMed] [Google Scholar]
- 18.Stewart GS, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 19.Doil C, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 20.Baciu PC, et al. Syndesmos, a protein that interacts with the cytoplasmic domain of syndecan-4, mediates cell spreading and actin cytoskeletal organization. Journal of cell science. 2000;113(Pt 2):315–324. doi: 10.1242/jcs.113.2.315. [DOI] [PubMed] [Google Scholar]
- 21.Denhez F, et al. Syndesmos, a syndecan-4 cytoplasmic domain interactor, binds to the focal adhesion adaptor proteins paxillin and Hic-5. The Journal of biological chemistry. 2002;277:12270–12274. doi: 10.1074/jbc.M110291200. [DOI] [PubMed] [Google Scholar]
- 22.Taylor MJ, Peculis BA. Evolutionary conservation supports ancient origin for Nudt16, a nuclear-localized, RNA-binding, RNA-decapping enzyme. Nucleic Acids Res. 2008;36:6021–6034. doi: 10.1093/nar/gkn605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim H, et al. Crystal structure of syndesmos and its interaction with Syndecan-4 proteoglycan. Biochemical and biophysical research communications. 2015;463:762–767. doi: 10.1016/j.bbrc.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Lemaitre C, et al. The nucleoporin 153, a novel factor in double-strand break repair and DNA damage response. Oncogene. 2012;31:4803–4809. doi: 10.1038/onc.2011.638. [DOI] [PubMed] [Google Scholar]
- 25.Orthwein A, et al. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science. 2014;344:189–193. doi: 10.1126/science.1248024. [DOI] [PubMed] [Google Scholar]
- 26.Lee DH, et al. Dephosphorylation enables the recruitment of 53BP1 to double-strand DNA breaks. Mol Cell. 2014;54:512–525. doi: 10.1016/j.molcel.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartlerode AJ, et al. Impact of histone H4 lysine 20 methylation on 53BP1 responses to chromosomal double strand breaks. PloS one. 2012;7:e49211. doi: 10.1371/journal.pone.0049211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mertins P, et al. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods. 2013;10:634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chapman JR, et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell. 2013;49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Virgilio M, et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339:711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Escribano-Diaz C, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Molecular cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. S1097-2765(13)00002-6 [pii] [DOI] [PubMed] [Google Scholar]
- 32.Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 2013;339:700–704. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng L, Fong KW, Wang J, Wang W, Chen J. RIF1 counteracts BRCA1-mediated end resection during DNA repair. The Journal of biological chemistry. 2013;288:11135–11143. doi: 10.1074/jbc.M113.457440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Callen E, et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 2013;153:1266–1280. doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zong D, et al. Ectopic expression of RNF168 and 53BP1 increases mutagenic but not physiological non-homologous end joining. Nucleic Acids Res. 2015;43:4950–4961. doi: 10.1093/nar/gkv336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Acs K, et al. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nature structural & molecular biology. 2011;18:1345–1350. doi: 10.1038/nsmb.2188. [DOI] [PubMed] [Google Scholar]
- 37.Mallette FA, et al. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. The EMBO journal. 2012;31:1865–1878. doi: 10.1038/emboj.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang J, et al. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nature structural & molecular biology. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gudjonsson T, et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012;150:697–709. doi: 10.1016/j.cell.2012.06.039. [DOI] [PubMed] [Google Scholar]
- 40.Lu R, Wang GG. Tudor: a versatile family of histone methylation ‘readers’. Trends Biochem Sci. 2013;38:546–555. doi: 10.1016/j.tibs.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner T, Robaa D, Sippl W, Jung M. Mind the methyl: methyllysine binding proteins in epigenetic regulation. ChemMedChem. 2014;9:466–483. doi: 10.1002/cmdc.201300422. [DOI] [PubMed] [Google Scholar]
- 42.Sandhu SK, Yap TA, de Bono JS. The emerging role of poly(ADP-Ribose) polymerase inhibitors in cancer treatment. Current drug targets. 2011;12:2034–2044. doi: 10.2174/138945011798829438. [DOI] [PubMed] [Google Scholar]
- 43.Yap TA, Sandhu SK, Carden CP, de Bono JS. Poly(ADP-ribose) polymerase (PARP) inhibitors: Exploiting a synthetic lethal strategy in the clinic. CA: a cancer journal for clinicians. 2011;61:31–49. doi: 10.3322/caac.20095. [DOI] [PubMed] [Google Scholar]
- 44.Lord CJ, Ashworth A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nature medicine. 2013;19:1381–1388. doi: 10.1038/nm.3369. [DOI] [PubMed] [Google Scholar]
- 45.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macnaughtan MA, Kane AM, Prestegard JH. Mass spectrometry assisted assignment of NMR resonances in reductively 13C-methylated proteins. J Am Chem Soc. 2005;127:17626–17627. doi: 10.1021/ja056977r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cui G, Botuyan MV, Mer G. Preparation of recombinant peptides with site- and degree-specific lysine (13)C-methylation. Biochemistry. 2009;48:3798–3800. doi: 10.1021/bi900348z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon MD, et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 50.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 51.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Authors can confirm that all relevant data are included in the paper and or its supplementary information files. The NCBI accession # for TIRR is NC_000016.10.