Abstract
Manganese superoxide dismutase is an essential component of the mitochondrial antioxidant defense system of most eukaryotes. In the present study, we used a reverse-genetics approach to assess the contribution of the Cryptococcus neoformans manganese superoxide dismutase (Sod2) for antioxidant defense. Strains with mutations in the SOD2 gene exhibited increased susceptibility to oxidative stress as well as poor growth at elevated temperatures compared to isogenic wild-type strains. The sod2Δ mutants were also avirulent in a murine model of inhaled cryptococcosis. Reconstitution of a sod2Δ mutant restored Sod2 activity, eliminated the oxidative stress and temperature-sensitive (ts) phenotypes, and complemented the virulence phenotype. Characterization of the ts phenotype revealed a dependency between Sod2 antioxidant activity and the ability of C. neoformans cells to adapt to growth at elevated temperatures. The ts phenotype could be suppressed by the addition of either ascorbic acid (10 mM) or Mn salen (200 μM) at 30°C, but not at 37°C. Furthermore, sod2Δ mutant cells that were incubated for 24 h at 37°C under anaerobic, but not aerobic, conditions were viable when shifted to the permissive conditions of 25°C in the presence of air. These data suggest that the C. neoformans Sod2 is a major component of the antioxidant defense system in this human fungal pathogen and that adaptation to growth at elevated temperatures is also dependent on Sod2 activity.
Eukaryotic cells possess several specialized enzymes that provide protection against reactive oxygen species produced as a by-product of physiologic metabolic processes (46). The mitochondria are an especially important source of endogenous reactive oxygen species in the cell. Under physiological conditions, a fraction of the oxygen consumed by the mitochondria is incompletely reduced, resulting in the formation of superoxide radical (9). Although superoxide radical can undergo spontaneous degradation, mitochondria depend on a manganese-containing superoxide dismutase (Sod2) to catalyze the rapid conversion of superoxide radical to hydrogen peroxide and oxygen (15). Loss of this enzyme activity has profound consequences for the viability of many organisms. For example, mice with a homozygous SOD2 disruption exhibited heart and liver defects, metabolic acidosis, and early neonatal death (23, 24). Drosophila melanogaster SOD2 mutants exhibited increased sensitivity to oxidative stress and early onset death in young adults (21), and Saccharomyces cerevisiae SOD2 mutants exhibited a rapid loss of viability in the early stationary phase (25). These studies demonstrate a conserved and essential function for Sod2 as a component of the mitochondrial antioxidant defense system of many phylogenetically diverse organisms.
Cryptococcus neoformans is an opportunistic human pathogen. Most cases of C. neoformans disease involve immunocompromised patients, such as those with human immunodeficiency virus infection or receiving corticosteroid therapy. In these patients, C. neoformans can disseminate from the lungs to other organs, with the most common and serious complications arising as a result of meningoencephalitis. In immunocompetent individuals, host defense against infection by C. neoformans is mediated via the mobilization of a cellular immune response, which is largely dependent on the activation of alveolar macrophages (13). Several studies have reported a correlation between resistance to oxidative stress in vitro and the virulence potentials of C. neoformans strains (1, 11, 26, 45), suggesting that C. neoformans must possess a strong antioxidant defense system, which provides some measure of protection against oxygen-dependent host defense mechanisms. However, little is known about the specific role and relative impact of the various constituents of the C. neoformans antioxidant defense system for resistance against oxidative stress and the capacity for virulence.
In the present study, we used a reverse-genetics approach to assess the contribution of the Cryptococcus neoformans var. grubii manganese superoxide dismutase (Sod2) to antioxidant defense. The rationale for studying the C. neoformans var. grubii strain in particular was that these strains account for approximately 95% of all clinical cases of cryptococcosis. We demonstrate in this study that Sod2 is a critical component of the C. neoformans antioxidant defense system and that loss of Sod2 function has deleterious consequences for cell viability. In addition, we provide the first demonstration of a mechanistic relationship between the antioxidant activity of Sod2 and the ability of C. neoformans to adapt to growth at elevated temperatures.
MATERIALS AND METHODS
Strains and media.
C. neoformans var. grubii strains H99 (wild type, serotype A, mating type alpha) and H99R were revived from 15% glycerol stocks stored at −80°C. H99R is a spontaneous ura5 auxotroph isolated by plating strain H99 on 5-fluoroorotic acid agar as described previously (22). The sod2Δ::URA5 transformants were selected on uracil-deficient medium and maintained on yeast extract-peptone-dextrose (YPD; 1% yeast extract, 2% peptone, and 2% dextrose) agar. The sod2Δ SOD2-nat reconstituted strains were selected on YPD medium containing nourseothricin (100 μg/ml) and maintained on YPD agar. Prior to use in mouse studies, yeast strains were incubated in shaken YPD broth for 18 to 20 h at 25°C, harvested, washed three times with sterile phosphate-buffered saline, and counted with a hemocytometer to determine cell concentrations. The inoculum size for all mouse experiments was confirmed by plating dilutions on YPD agar plates and counting CFU. l-Dopa agar (44), YNB agar (37), uridine-deficient agar (22), and Christensen's broth (29) were prepared as described previously. Capsule formation was assessed by India ink staining. Melanin production was assessed by growing cells on l-dopa agar, as described previously (44).
Identification of SOD2.
The amino acid sequence of the S. cerevisiae manganese superoxide dismutase (GenBank accession no. YHR008C) was used to query the C. neoformans H99 genome sequencing project database (Duke Center for Genome Technology, http://cgt.genetics.duke.edu/). TBLASTN analysis revealed the presence of a single C. neoformans SOD2 homolog in the genome. The C. neoformans SOD2 homolog was used to query The Institute for Genomic Research C. neoformans database (http://tigrblast.tigr.org) and the University of Oklahoma C. neoformans cDNA sequencing project database (http://www.genome.ou.edu) to identify SOD2 cDNA sequences. Spidey, which is an mRNA-to-genomic DNA alignment program (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov), was used to identify SOD2 intron-exon boundaries. The SOD2 genomic DNA sequence and SOD2 cDNA sequences were used as input.
Construction of the sod2Δ strain.
PCR overlap was performed to generate the sodΔ::URA5 deletion construct, as described by Davidson et al. (12). The left and right arms of the deletion construct were amplified with the primer pairs sgSOD2OL-1 and sgSOD2OL-2 and sgSOD2OL-3 and sgSOD2OL-4, respectively (Table 1). The URA5 selectable marker was amplified with the primer pair sgSOD2OL-5 and sgSOD2OL-6 (Table 1). The sod2Δ::URA5 deletion construct was amplified with primers sgSOD2OL-1 and sgSOD2OL-4 and gel purified by using the QIAquick gel extraction kit (QIAGEN, Valencia, Calif.), and DNA was concentrated by speed-vac (Savant DNA 110). Biolistic transformation of the ura5 auxotrophic strain H99R was performed by using a Bio-Rad biolistic particle delivery system, as described by Toffaletti et al. (42). Transformants were selected on uridine-dropout medium incubated at 25°C, and colony PCR was performed to identify sod2Δ::URA5 mutant strains with the primer pair sgSOD2 P-5′ and sgSOD2 P-3′. Selection at 25°C was performed because initial attempts to identify sod2Δ mutants under selection at 30°C were only partially successful. Deletion of SOD2 was confirmed by Southern blot analysis, and one sod2Δ::URA5 mutant strain was randomly chosen for further analysis. PCR overlap was performed to generate the reconstitution construct SOD2-nat. The SOD2 fragment of the overlap construct was generated with primers sgSOD2(rc)-OL1 and sgSOD2(rc)-OL2, and the nourseothricin (nat) selectable marker was amplified from plasmid pAI1 (18) using the primer pair sgSOD2(rc)-OL3 and sgSOD2(rc)-OL4. The SOD2-nat reconstitution construct was amplified with primers sgSOD2(rc)-OL1 and sgSOD2(rc)-OL4 and gel purified by using the QIAquick gel extraction kit (QIAGEN), and DNA was concentrated by speed-vac (Savant DNA 110). Biolistic transformation of the sod2Δ::URA5 strain was performed as described above. Following transformation, cells were grown on YPD medium supplemented with 1 M sorbitol for 2 to 4 h and then transferred to YPD medium containing nourseothricin (100 μg/ml). Plates were incubated at 30°C and monitored for 4 to 5 days. Transformants were randomly chosen and screened by colony PCR to identify reconstituted strains. The presence of SOD2 in the genome was confirmed by Southern blot analysis, and one reconstituted strain was randomly chosen for phenotypic analysis.
TABLE 1.
Primers used in this study
| Primer name | Usea | Sequence (5′-3′) |
|---|---|---|
| sgSOD2 OL-1 | A, D | GGTGGGTGATTACAGGTTGG |
| sgSOD2 OL-2 | A, D | GGTCGAGCAACTTCGCTCCATATGACCACCACCGTTGA |
| sgSOD2 OL-3 | A | CCACCTCCTGGAGGCAAGTTGTCTATCTTCGGGTGTGG |
| sgSOD2 OL-4 | A | GAAGGCGCACAAGGTAAGAG |
| sgSOD2 OL-5 | A | TCAACGGTGGTGGTCATATGGAGCGAAGTTGCTCGACC |
| sgSOD2 OL-6 | A | CCACACCCGAAGATAGACAACTTGCCTCCAGGAGGTGG |
| sgSOD2 P-5′ | B | CTCCGCACTTCTCTCGCTAC |
| sgSOD2 P-3′ | B | CGTTCAAATCCACACACCTG |
| sgSOD2(rc)-OL1 | C | GGTGGGTGATTACAGGTTGG |
| sgSOD2(rc)-OL2 | C | GCTCACCTCCCGCAGCCTGAAGGCGCACAAGGTAAGAG |
| sgSOD2(rc)-OL3 | C | CTCTTACCTTGTGCGCCTTCAGGCTGCGGGAGGTGAGC |
| sgSOD2(rc)-OL4 | C | GAAGAGATGTAGAAACGAGTTTC |
The primers were used for the following purposes: A, PCR-OL construction of the sod2Δ::URA5 construct; B, colony PCR detection of sod2Δ mutant strains; C, PCR-OL construction of the SOD2-nat construct; D, probe for Southern blot analysis.
Southern blot analysis.
Genomic DNA was isolated from strains sod2Δ, sod2Δ SOD2, and wild-type H99 as described previously (35). Restriction digestion, gel electrophoresis, DNA transfer, prehybridization, hybridization, and autoradiography were performed as described previously (35). The primer pair sgSOD2OL-1 and sgSOD2OL-2 was used to generate a 754-bp probe that hybridized near the 5′ region of the SOD2 open reading frame. A random primed DNA labeling kit (Boehringer Mannheim) and [32P]dCTP (Amersham) were used to label the probe.
Superoxide dismutase activity assay.
The sod2Δ mutant strain, sod2Δ SOD2 reconstituted strain, and wild-type strain H99 were grown in 30 ml of YPD medium at 25°C with shaking for 48 h. In preliminary experiments, we observed that C. neoformans Sod2 activity was dependent on culture age, with lysates from 48-h cultures exhibiting greater Sod2 activity than cultures incubated for 16 or 24 h. The total protein concentrations of each lysate were standardized prior to analysis; 20 μg of total protein from each lysate was analyzed. Cells were collected by centrifugation and lysed by glass bead disruption as described previously (26). A protease inhibitor (P8340; Sigma) was used to prevent degradation of the Sod2 polypeptide. Cell lysates (30 μg of protein/lane) were separated by native gel electrophoresis (Criterion 10% Tris-HCl gel; Bio-Rad) and subjected to nitroblue tetrazolium staining to assay for Sod2 activity as described previously (6, 14, 26).
Oxidative stress sensitivity.
Disk diffusion assays were performed to assess the sensitivity of sod2Δ, sod2Δ SOD2, and wild-type H99 strains to oxidative stress in vitro. The disk diffusion method has previously been used to assess antifungal drug susceptibility for C. neoformans strains (32). All strains were grown in YPD medium at 25°C overnight with shaking and washed with sterile phosphate-buffered saline, and the concentration of cells was adjusted to an optical density at 600 nm of 0.2. Strains were grown at 25°C because, at higher temperatures, the sod2Δ mutant strain exhibits a temperature-sensitive growth and survival defect. Each strain was diluted 1:10 in YPD agar medium that was cooled but still molten, and plates were poured and allowed to solidify. Sterile paper disks (6-mm diameter) were soaked with 10 μl of 2 M paraquat (Sigma) and placed in the center of each plate. Paraquat is known to generate superoxide anions in the cytosol and mitochondria of yeast cells, which leads to the formation of reactive oxygen species. Plates were incubated at 25°C for 3 to 4 days, at which time the zones of inhibition were photographed and measured. A zone of inhibition was defined as an area of the plate that contained no growth compared to the untreated control plate. Disk diffusion assays were performed for each strain in duplicate and repeated at least three times. Results were reported as the means of the diameters of zones of inhibition ± the standard errors of the means. As a control, a variation of the disk diffusion assay was performed wherein strains were struck on YPD agar plates and a sterile paper disk (6-mm diameter) soaked with 10 μl of 2 M paraquat (Sigma) was placed in the center of the plate. The degree of inhibition was consistent with that of the disk diffusion assays, allowing us to rule out the possibility that the oxidative stress phenotype of sod2Δ mutants was due to heat shock as a result of immersion in molten agar. Student's t test was performed to determine statistical significance (P values of <0.05 were considered significant).
Antioxidants.
The ability of several antioxidants to suppress the ts phenotype of the sod2Δ and wild-type H99 strains was assessed. Mn porphyrins, MnTE-2-PyP5+ (2), MnTnBu-2-PyP5+ (3), MnTnHex-2-PyP5+ (3), MnTDMOE-2-ImP5+ (4), and Mn salen (39) were prepared as previously described. MnTM-4-PyP5+ (MidCentury Chemicals, Chicago, Ill.), MnTBAP3− (Calbiochem), Tempol (Calbiochem), ascorbic acid (Sigma), and MnCl2 · 4H2O (J. T. Baker) were purchased. YPD agar medium was supplemented with various concentrations of each antioxidant. Serial dilutions of the sod2Δ mutant strain and wild-type strain H99 were spotted onto these plates and then incubated at 30 and 37°C.
In vivo testing.
Female A/Jcr mice (20 to 24 g each; NCI/Charles River Laboratories) were used to assess the virulence of the sod2Δ mutant strain and the sod2Δ SOD2 reconstituted strain compared to that of wild-type strain H99. The A/Jcr cryptococcosis infection model is a well-established model that has been used extensively to assess the virulence potential of C. neoformans strains. To assess virulence, groups consisting of 10 A/Jcr mice each were infected via intranasal inhalation with 5 × 105 CFU of each strain (in a volume of 50 μl), as described previously (11). Mice that appeared moribund (i.e., lethargic or exhibiting rapid weight loss) or in pain were humanely euthanized. To assess the ability of the sod2Δ mutant strain to survive in vivo, five mice infected with the sod2Δ mutant strain were sacrificed at day 38, at which time the left lung and brain were collected from each animal and homogenized and CFU determinations were performed (50 μl of total organ homogenate from each animal was inoculated on YPD agar plates and incubated at 25°C). Mice were monitored twice daily. The Duke University Animal Use Committee approved the animal protocol used for these experiments. The Mann-Whitney U test was performed to evaluate survival data for statistical significance.
Nucleotide sequence accession number.
The C. neoformans var. grubii SOD2 sequence has been submitted to GenBank and assigned accession number AY737799.
RESULTS
Identification of SOD2.
To identify the C. neoformans var. grubii SOD2 homolog, TBLASTN analysis of the C. neoformans serotype A strain H99 genome sequence database (Duke University C. neoformans H99 genome database, http://cneo.genetics.duke.edu/) was performed. The S. cerevisiae Sod2 protein sequence was used as the query. We identified a single 1,013-bp region in the C. neoformans genome that contained the SOD2 homolog. The open reading frame is interrupted by 6 introns and encodes a predicted protein of 234 amino acids. The C. neoformans var. grubii Sod2 protein shared significant sequence identities with other fungal Sod2's, including Cryptococcus neoformans var. gattii (95%), Cryptococcus neoformans var. neoformans (98%), Ustilago maydis (65%), Phanerochaete chrysosporium (67%), Schizosaccharomyces pombe (57%), and Aspergillus fumigatus (53%). The C. neoformans var. grubii Sod2 protein contains a characteristic SodA domain (Pfam analysis, http://www.sanger.ac.uk/Software/Pfam/) and a putative mitochondrial localization sequence (PPLPYAYDALEP), which is located at the amino terminus, immediately upstream of the mature peptide. This putative mitochondrial localization sequence was identical to the C. neoformans serotype D Sod2 amino terminus sequence reported by Tesfa-Selase and Hay (41), the predicted serotype B Sod2 mitochondrial localization sequence, and the Sod2 mitochondrial localization sequences of several other fungi, including S. pombe (19).
Construction of sod2Δ and sod2Δ SOD2 mutant strains.
The function of Sod2 was assessed by creating a sod2Δ mutant strain via allele-specific homologous integration of a sod2Δ::URA5 deletion construct at the SOD2 locus. Approximately 80% of the SOD2 open reading frame was deleted. PCR overlap (12) was performed to create a linear sod2Δ::URA5 deletion construct (Fig. 1A), which was introduced into a ura5 auxotrophic strain H99R by biolistic transformation (42). Ten sod2Δ::URA5 mutant strains were identified by colony PCR, and one was randomly chosen for further study. The sod2Δ::URA5 strain was reconstituted following a single random ectopic integration of a linear SOD2-nat construct in the genome (Fig. 1A). Thirty-two sod2Δ SOD2 reconstituted strains were identified by colony PCR and phenotypic analysis. Reconstitution of the sod2Δ strains eliminated the oxidative stress and temperature-sensitive phenotypes. One reconstituted strain (sod2Δ SOD2) was chosen randomly for further study.
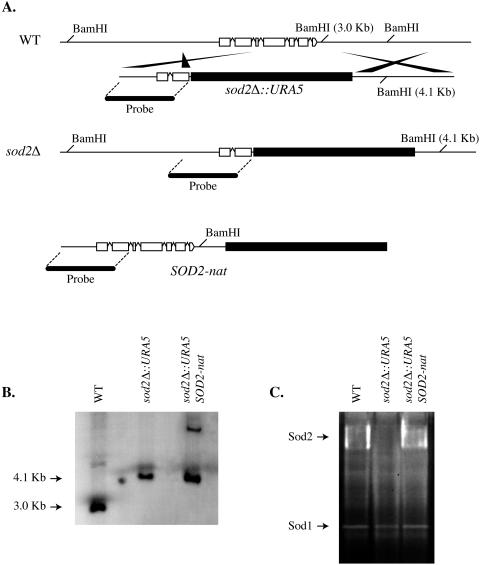
FIG. 1.
Construction of sod2Δ mutant and sod2Δ SOD2 reconstituted strains. (A) PCR overlap was performed to create the sod2Δ::URA5 deletion construct, which consisted of two 1-kb 5′ and 3′ regions of genomic DNA flanking the SOD2 open reading frame and the selectable marker URA5. (B) Southern blot analysis of sod2Δ and the sod2Δ SOD2 reconstituted strains confirmed that a single allele-specific homologous integration of the sod2Δ::URA5 deletion construct occurred at the SOD2 locus and that the SOD2-nat reconstitution construct integrated randomly at a single location in the genome. (C) Cell extracts from strains sod2Δ, sod2Δ SOD2, and wild-type (WT) strain H99 were electrophoresed in a 10% polyacrylamide gel, and superoxide dismutase activity was visualized as described by Beauchamp and Fridovich (6). Strain sod2Δ lacked Sod2 activity, whereas the sod2Δ SOD2 strain and wild-type strain H99 exhibited Sod2 activities. One gel representative of more than five experiments performed is shown.
Southern blot analysis was performed to confirm that a single homologous integration of the deletion construct had occurred at the native SOD2 locus (Fig. 1B, lane 2) and that the SOD2-nat reconstitution construct integrated ectopically at a single location in the genome of the sod2Δ SOD2 reconstituted strain (Fig. 1B, lane 3). Genomic DNA from all strains, including wild-type strain H99, was digested with BamHI. The presence of a 4.1-kb band in lane 2 demonstrated that the majority of the native SOD2 allele was deleted in the sod2Δ mutant strain. The presence of a single high-molecular-weight band in lane 3 indicated that a single integration event of the SOD2-nat reconstitution construct had occurred in the reconstituted strain. There is only one BamHI site in the SOD2-nat reconstitution construct. Thus, the presence of a single high-molecular-weight band demonstrated that a single random ectopic integration event had occurred. In the event that more than one ectopic integration had occurred, we would have expected to see multiple bands, because each integration event would have resulted in placement of the construct in different proximities to other BamHI restriction sites in the genome. It is highly improbable that multiple random integration events at various sites in the genome could occur within equivalent proximities to other BamHI restriction sites.
To confirm the respective loss and restoration of Sod2 function in the sod2Δ deletion strain and the sod2Δ SOD2 reconstituted strain, we assayed cell extracts from these strains and wild-type strain H99 for superoxide dismutase activity (Fig. 1C), as described by Beauchamp and Fridovich (6). The C. neoformans Sod2 is an 80-kDa enzyme that exhibits a high-molecular-weight activity by nitroblue tetrazolium staining (41). As illustrated in Fig. 1C, the sod2Δ mutant strain lacked the high-molecular-weight Sod2 activity, whereas the sod2Δ SOD2 reconstituted strain and wild-type strain H99 exhibited equivalent high-molecular-weight Sod2 activities. There was no relative difference in Sod2 activity between the sod2Δ SOD2 reconstituted strain and wild-type strain H99, as assessed by densitometry analysis. In addition, no relative differences in Sod1 activities were observed between any of the strains, suggesting that Sod1 activity was unaffected as a result of the loss of Sod2 function. These results confirmed the loss and restoration of Sod2 function in the sod2Δ and sod2Δ SOD2 strains, respectively.
sod2Δ is susceptible to oxidative stress in vitro.
Sod2 is an essential component of the antioxidant defense system of many eukaryotes, providing protection against oxidative stress as a result of endogenous and exogenous reactive oxygen species. A disk diffusion assay was performed to assess the susceptibility of the sod2Δ mutant strain to oxidative stress in vitro. In preliminary experiments, we observed that sod2Δ strains exhibited a ts phenotype; therefore, the disk diffusion assays were performed at 25°C. We found that two independent sod2Δ mutant strains were significantly more susceptible to paraquat (disk saturated with 10 μl of 2.0 M paraquat) than wild-type strain H99 and the sod2Δ SOD2 reconstituted strain (P < 0.001) (Fig. 2A). There were no significant differences between the sensitivities of wild-type strain H99 and the sod2Δ SOD2 reconstituted strain to paraquat. As a control, cells were inoculated on solid YPD agar medium and treated with paraquat. The results of these assays were consistent with those of the earlier disk diffusion assay, demonstrating that a brief heat shock due to the immersion of cells in molten agar did not account for the increased sensitivity of the sod2Δ mutant strains to paraquat (Fig. 2B).
FIG. 2.
sod2Δ mutants exhibited increased sensitivity to oxidative stress in vitro. (A) Two independent sod2Δ mutant strains were significantly (P ≤ 0.0001) more susceptible to paraquat (disk saturated with 10 μl of 2.0 M paraquat) than wild-type (WT) strain H99 and the sod2Δ SOD2 reconstituted strain. There was no significant difference between the sensitivities of wild-type strain H99 and the sod2Δ SOD2 reconstituted strain. Disk diffusion assays were performed in duplicate a minimum of three times. Results were reported as the means of the diameters of zones of inhibition ± the standard errors of the means. A zone of inhibition was defined as an area of the plate where no growth was observed (i.e., the agar medium remained clear). One-way analysis of variance was performed to determine statistical significance (P values of <0.05 were considered significant). (B) A modification of the disk diffusion assay was performed as a control to ensure that the oxidative stress phenotype of the sod2Δ strain was not due to a heat shock as a result of immersion in molten agar. Strains were struck on solid YPD agar, and a sterile disk saturated with an equivalent concentration of paraquat, as used in the previous disk diffusion assay, was added to the center of one plate. An untreated plate was included as a control. The results of this assay were consistent with those of the disk diffusion assay. Results from one experiment representative of three performed are shown. *, P < 0.05 versus wild-type strain H99.
Sod2 is essential for growth at 37°C.
C. neoformans possesses several well-characterized phenotypes that are required for virulence, including capsule synthesis, melanin production, urease activity, and the ability to grow at 37°C (40). The sod2Δ mutant strain was assessed to determine whether the loss of Sod2 activity affected any of these phenotypes. We observed no difference between the sod2Δ mutant strain and the wild-type strain H99 in terms of induction of capsule, melanin, or urease activity. However, the sod2Δ mutant strain did exhibit a ts phenotype at 30°C and a more sensitive temperature sensitivity at 37°C (Fig. 3). Reconstitution of the sod2Δ mutant strain restored the ability of cells to grow at 30 and 37°C (Fig. 3). The restoration of Sod2 activity and subsequent rescue of temperature sensitivity in the sod2Δ SOD2 reconstituted strain demonstrated that the ts phenotype was due specifically to a loss of Sod2 activity and not to an unrelated secondary mutation.
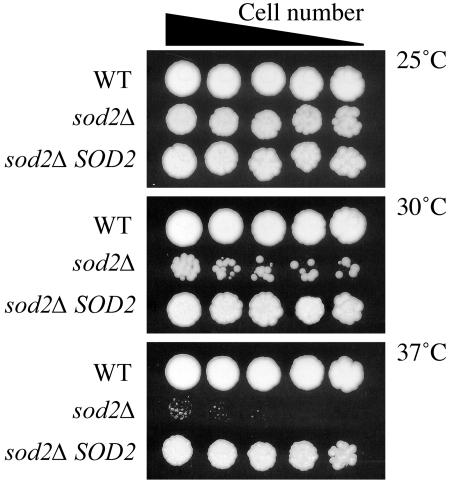
FIG. 3.
Sod2 is required for growth at 30 and 37°C. Strains sod2Δ, sod2Δ SOD2, and wild-type (WT) H99 were serially diluted on YPD agar plates and incubated at 25, 30, and 37°C for up to 3 days. The sod2Δ mutant strain exhibited a progressively severe ts phenotype at 30 and 37°C. The phenotype of the sod2Δ SOD2 reconstituted strain was similar to that of wild-type strain H99.
Antioxidant suppression of ts phenotype.
Given the known function of Sod2, we predicted that the ts phenotype of the sod2Δ mutant strain may be due to an elevation of the endogenous steady-state concentration of reactive oxygen species. To examine this possibility, we assessed whether ascorbic acid and superoxide dismutase mimics Mn salen (20) and Mn porphyrins (MnTE-2-PyP5+, MnTnBu-2-PyP5+, MnTnHex-2-PyP5+, and MnTDMOE-2-ImP5+) could suppress the ts phenotype of the sod2Δ mutant strain at 30 and 37°C (5). The sod2Δ mutant strain and wild-type strain H99 were spotted on agar plates supplemented with various concentrations of each antioxidant and incubated at 30 and 37°C. Ascorbic acid (20 mM) and Mn salen (200 μM) suppressed the ts phenotype at 30°C (Fig. 4) but not at 37°C. Cells were also spotted on plates containing MnCl2 (200 μM) as a control. MnCl2 did not suppress the ts phenotype, suggesting that the antioxidant activity of Mn salen was likely not due to the delivery of Mn to the cells. Surprisingly, the concentrations of ascorbic acid and Mn salen that suppressed the ts phenotype of the sod2Δ mutant strain at 30°C were toxic to wild-type H99 cells at 37°C. Thus, no further testing of these compounds was done at 37°C. None of the Mn porphyrins (MnTE-2-PyP5+, MnTnBu-2-PyP5+, MnTnHex-2-PyP5+, and MnTDMOE-2-ImP5+) suppressed the ts phenotype at 30 or 37°C. These compounds are cationic and highly charged, which may have prohibited them from gaining entry into C. neoformans cells. MnTBAP3− was also unable to suppress the ts phenotype at 30 or 37°C, which may have been due to an inability to gain entry into C. neoformans cells or as a consequence of low catalytic potency. These results are in agreement with our hypothesis that the ts phenotype occurs as a result of oxidative stress, suggesting that an important function of Sod2 is the regulation of the steady-state concentration of endogenous reactive oxygen species in response to environmental changes, such as growth at elevated temperatures.
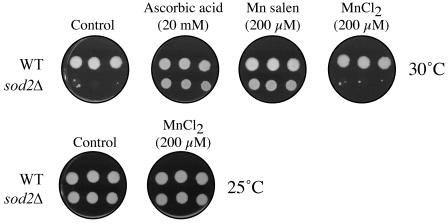
FIG. 4.
The ts phenotype of the sod2Δ mutant strain can be suppressed with antioxidants at 30°C. The ts phenotype of the sod2Δ mutant strain was suppressed at 30°C with the antioxidants ascorbic acid (20 mM) and Mn salen (200 μM). Manganese alone (200 μM, MnCl2) did not suppress the ts phenotype, suggesting that the antioxidant activity of Mn salen, and not that of Mn alone, resulted in the suppression of the ts phenotype. The results from one experiment representative of three performed are shown. WT, wild type.
The ts phenotype is oxygen dependent.
Given the previous results, we next sought a more direct method to assess whether the ts phenotype was oxygen dependent. The sod2Δ mutant strain and wild-type strain H99 were incubated at 37°C under aerobic and anaerobic conditions for 24 h, and then the cells were shifted to 25°C and aerobic conditions. Since C. neoformans is a strict aerobe, under anaerobic conditions oxidative metabolism is arrested and Sod2 activity would be expected to be dispensable (1). If the ts phenotype was oxygen dependent, then the viability of the sod2Δ mutant strain following incubation at 37°C under anaerobic conditions would be expected to be identical to that of wild-type strain H99. If, however, the ts phenotype was oxygen independent, then the sod2Δ mutant should be nonviable under both conditions. We found that the sod2Δ mutant strain exhibited no loss of viability when incubated at 37°C under anaerobic conditions compared to wild-type strain H99. In contrast, the sod2Δ mutant strain was nonviable following incubation at 37°C under aerobic conditions (Fig. 5). These results demonstrated that the ts phenotype of the sod2Δ mutant strain was oxygen dependent, suggesting that Sod2 contributes to the ability of C. neoformans to adapt to growth at elevated temperatures by regulating the steady-state concentration of endogenous reactive oxygen species.
FIG. 5.
The ts phenotype of the sod2Δ mutant strain is oxygen dependent. The sod2Δ mutant strain and wild-type (WT) strain H99 were serially diluted on YPD agar plates and incubated at 37°C under either aerobic or anaerobic conditions for 24 h. Plates were then shifted to aerobic growth conditions at 25°C for 72 h. The sod2Δ mutant cells incubated under aerobic conditions exhibited a severe loss of viability when shifted to 25°C compared to the wild-type strain H99. In contrast, the sod2Δ mutant cells incubated under anaerobic conditions grew as well as wild-type strain H99. The results from one representative experiment are shown.
sod2Δ mutant strain is avirulent.
Given our in vitro results, we hypothesized that the sod2Δ mutant strain would exhibit a severe virulence defect in the murine inhalation model of cryptococcosis. To test this hypothesis 10 A/Jcr mice were infected with 5 × 105 CFU of strain sod2Δ, sod2Δ SOD2, or H99 (wild type) by intranasal inoculation. By days 25 and 38 of infection, all mice infected with wild-type strain H99 or strain sod2Δ SOD2, respectively, had succumbed to infection (Fig. 6). Five mice infected with strain sod2Δ were sacrificed at day 38 postinfection. CFU determination was performed to assess fungal burden in the lungs and brains of these animals. We were unable to recover yeast cells from any of the organs, suggesting that by day 38 postinfection C. neoformans cells had been completely cleared from the mice. Mice infected with the sod2Δ mutant strain survived for up to 80 days, at which point the experiment was ended. These results demonstrate a dependency between antioxidant defense and the ability to adapt to growth at 37°C, which translates into a profound affect on the virulence of C. neoformans.
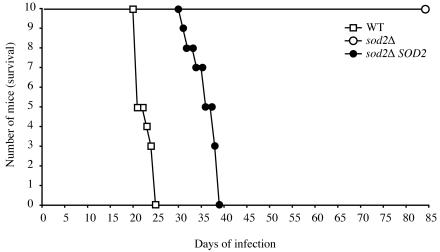
FIG. 6.
Sod2 is required for virulence. (A) Groups of 10 A/Jcr mice were anesthetized by intraperitoneal injection of pentobarbital and infected with 5 × 105 yeast cells of strains sod2Δ, sod2Δ SOD2, and wild-type (WT) H99 via intranasal inhalation. The sod2Δ mutant strain was avirulent: all infected animals survived to 80 days postinfection. Reconstitution of the sod2Δ mutant strain restored virulence. By day 38, all of the mice infected with strain sod2Δ SOD2 had succumbed to infection. All mice infected with wild-type strain H99 had succumbed to infection by day 25.
DISCUSSION
In the present study, we describe the contribution of the C. neoformans manganese superoxide dismutase (Sod2) to antioxidant defense. In yeast cells and other eukaryotes, Sod2 is a nuclear encoded polypeptide that is synthesized in the cytosol and translocated to the matrix of the mitochondria, where it complexes with manganese to form the active holo-Sod2 enzyme (26). The role of Sod2 for antioxidant defense is best illustrated by studies of various phylogenetically diverse eukaryotes in which sod2 null mutants exhibit severe developmental defects and diminished viability (21, 23, 24, 43). Consistent with the results of these studies, in the present study, we demonstrate that C. neoformans Sod2 is an essential component of the antioxidant defense system and that loss of Sod2 function has deleterious consequences for cell viability. In addition, we provide the first demonstration of a mechanistic relationship between the antioxidant activity of Sod2 and the ability of C. neoformans to adapt to growth at elevated temperatures.
Several lines of evidence support a role for Sod2 in mitochondrial antioxidant defense: (i) the C. neoformans Sod2 amino acid sequence contains a highly conserved mitochondrial targeting sequence, (ii) the superoxide dismutase domain of C. neoformans Sod2 exhibits a high degree of conservation with Sod2's from other eukaryotes, and (iii) the C. neoformans sod2Δ mutant strains exhibit phenotypes that are consistent with diminished mitochondrial function. Furthermore, it has been demonstrated that in yeast cells, apo-Sod2 must be transported into the mitochondrial matrix before it can complex with the manganese cofactor, which results in the formation of the active form of the enzyme, holo-Sod2. Given these facts, it is reasonable to assume that Sod2 is an essential component of the C. neoformans mitochondrial antioxidant defense system, which is consistent with what is known about the function of Sod2 in many other eukaryotes.
The observation that ascorbic acid and Mn salen, which is a superoxide dismutase mimic, suppressed the ts phenotype provided the initial evidence that the ts phenotype was oxygen dependent. In the mitochondria, Mn salen may function either as an Mn complex or as the Mn hexaqua ion, Mn(H2O)62+. Although Mn salen may function as a metal transporter, shuttling manganese into the mitochondria, our results suggested that the superoxide dismutase activity of Mn salen accounted for its ability to suppress the ts phenotype because MnCl2 alone did not suppress the ts phenotype of sod2Δ mutants at 30°C. The inability of the cationic Mn porphyrins and MnTBAP3− to suppress the ts phenotype at 30°C may be related to the charge of these compounds, which could prevent them from entering the mitochondria of C. neoformans. The failure of nitroxide Tempol to suppress the ts phenotype may be ascribed to the very low kcat for O2·− dismutation of nitroxides (16). Only very high doses of Tempol have been reported to produce antioxidant affects (17). The inhibitory activities of Mn salen and ascorbic acid for wild-type strain H99 at 37°C could be due to an elevation of superoxide levels in the cell, which can occur when oxygen is reduced by the reduced forms of ascorbic acid (38) and the Mn compounds (7, 38). In addition, Mn(III) salen can reportedly undergo oxidation with H2O2 and ONOO− resulting in O=MnVSalen formation, which is a strong oxidant (36). Direct evidence of the oxygen dependency of the ts phenotype was provided by demonstrating that sod2Δ cells incubated at 37°C under anaerobic conditions did not exhibit reduced viability when shifted to aerobic conditions at 25°C, whereas sod2Δ cells incubated under aerobic conditions exhibited a complete loss of viability. Taken together, these results demonstrated convincingly that the ts phenotype was oxygen dependent and that the antioxidant activity of Sod2 is essential for cells to adapt to growth at elevated temperatures.
Of the known fungi, only a small fraction cause morbidity and mortality in humans and other animals. A requisite characteristic of pathogenic fungi is the ability to grow at 37°C. For some fungal pathogens, such as Blastomyces dermatitidis and Histoplasma capsulatum, a shift from 25 to 37°C triggers a transformation from an infectious form to a pathogenic form (27, 28). The mechanisms involved in this transformation process and the ability to grow at elevated temperatures are poorly understood. Presently, only approximately a dozen C. neoformans genes have been identified that are associated with high temperature growth, with very few understood at the mechanistic level. The results of this study provide the first description of a mechanistic relationship between antioxidant defense and adaptation to growth at 37°C. One interpretation of our results is that Sod2 permits C. neoformans to grow at 37°C by regulating the steady-state concentration of reactive oxygen species in the mitochondria. In the absence of Sod2, we would expect to observe an elevation in the endogenous steady-state concentration of reactive oxygen species, resulting in oxidative damage to macromolecular structures in the mitochondria, such as mDNA, proteins, and lipids. In a previous study, we reported that a C. neoformans var. grubii copper zinc superoxide dismutase null mutant (sod1) exhibited a modest oxidative stress phenotype and a small reduction in its virulence potential (11). In contrast to these results, Sod2 appears to have a much more essential function for C. neoformans. The sod2Δ mutant exhibited oxidative stress and ts phenotypes that were not exhibited by the sod1 mutant, and it was completely avirulent in our murine infection model. We believe that these differences are due to the important antioxidant function of Sod2 in the mitochondrial matrix and highlight the importance of mitochondrial health for C. neoformans viability.
Although the avirulent phenotype of the sod2Δ mutant was not surprising, virulence studies were necessary to rule out the possibility that conditions within the host environment may suppress the oxidative or ts phenotypes, resulting in infection or disease. Previous studies have demonstrated that the C. neoformans calcineurin mutant exhibits a similar ts phenotype in vitro and is avirulent in an animal model of cryptococcal meningitis (31). Despite this severe ts phenotype in vitro, the calcineurin mutant can be recovered from the organs of infected mice up to 2 weeks postinfection, suggesting that although the mutant does not cause disease, it can establish an infection. These results suggest that under some circumstances host conditions can suppress phenotypes in vivo that would otherwise be severe in vitro. Thus, virulence studies are warranted for mutants even in cases in which severe in vitro phenotypes are observed.
The evolution of the C. neoformans antioxidant defense system was likely influenced by a number of factors, including the necessity to regulate the steady-state concentration of endogenous reactive oxygen species and a need to respond to changes in environmental conditions. The three C. neoformans varieties are found in a diverse number of ecological niches ranging from pigeon guano to trees. The association of C. neoformans with certain trees is of particular interest given that trees, like other plants, utilize oxygen-dependent mechanisms to provide protection against invasion by pathogenic microorganisms (8). Several studies have reported the isolation of C. neoformans varieties in association with trees, including Eucalyptus camaldulensis, Butea monosperma, Syzygium cumini, Ficus religiosa, and Tamarindus indica, and other plant material (10, 30, 33, 34). Although C. neoformans var. grubii has historically been less frequently isolated from trees than the other varieties, particularly C. neoformans var. gattii, Nishikawa et al. were able to isolate C. neoformans var. grubii from the hollows of jungle trees in the Amazon forest of Brazil, suggesting that the generally accepted ecological niche of this serotype should be extended to include jungle trees (30). Given the association between some C. neoformans varieties and plants and the ability of plants to utilize oxygen-dependent mechanisms as a component of host defense against pathogenic microorganisms (8), an intriguing possibility is that plant-C. neoformans interactions might have influenced the evolution of the C. neoformans antioxidant defense system.
The results of this study demonstrate a weak point in the antioxidant system of C. neoformans that possibly could be exploited. Most of the present antioxidant targets have a modest impact on the virulence of C. neoformans. However, Sod2 is a possible fungicidal target because of its specific link to antioxidant defense and its added functional implications in high temperature growth. There is a tremendous amount of sequence conservation among the C. neoformans varieties, suggesting that Sod2 possesses an evolutionarily conserved function for C. neoformans. If inhibitors can be identified that are specific for the C. neoformans SOD2 regulatory pathway, then Sod2 could be an excellent target for antifungal drug development.
Acknowledgments
We are grateful to Irwin Fridovich for generously providing the Mn salen and manganese porphyrins used in this work and for critical reading of the manuscript; to Mark Carroll and Valeria Culotta for assistance with the superoxide dismutase activity assay; and to Andrew Alspaugh, Joseph Heitman, and Wiley Schell for comments and critical reading of the manuscript.
This work was supported by Public Health Service grant AI028388. S.S.G. was supported by a National Institutes of Health RSUM grant (AI028388). I.B.-H. is grateful for the financial support of DOD CDMRP (BC024326) and to Aeolus/Incara Pharmaceuticals, Research Triangle Park, N.C.
REFERENCES
- 1.Akhter, S., H. C. McDade, J. M. Gorlach, G. Heinrich, G. M. Cox, and J. R. Perfect. 2003. Role of alternative oxidase gene in pathogenesis of Cryptococcus neoformans. Infect. Immun. 71:5794-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batinic-Haberle, I. 2002. Manganese porphyrins and related compounds as mimics of superoxide dismutase. Methods Enzymol. 349:223-233. [DOI] [PubMed] [Google Scholar]
- 3.Batinic-Haberle, I., I. Spasojevic, R. D. Stevens, P. Hambright, and I. Fridovich. 2002. 2. Manganese(III) meso tetrakis ortho N-alkylpyridylporphyrins. Synthesis, characterization and catalysis of O2.− dismutation. J. Chem. Soc. Dalton Trans. 2002:2689-2696. [Google Scholar]
- 4.Batinic-Haberle, I., I. Spasojevic, R. D. Stevens, P. Hambright, P. Neta, A. Okado-Matsumoto, and I. Fridovich. 2004. New class of potent catalysts of O2.− dismutation. Mn(III) methoxyethylpyridyl- and methoxyethylimidazolylporphyrins. J. Chem. Soc. Dalton Trans 2004:1696-1702. [DOI] [PubMed] [Google Scholar]
- 5.Baudry, M., S. Etienne, A. Bruce, M. Palucki, E. Jacobsen, and B. Malfroy. 1993. Salen-manganese complexes are superoxide dismutase-mimics. Biochem. Biophys. Res. Commun. 192:964-968. [DOI] [PubMed] [Google Scholar]
- 6.Beauchamp, C., and I. Fridovich. 1971. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44:276-287. [DOI] [PubMed] [Google Scholar]
- 7.Bloodsworth, A., V. B. O'Donnell, I. Batinic-Haberle, P. H. Chumley, J. B. Hurt, B. J. Day, J. P. Crow, and B. A. Freeman. 2000. Manganese-porphyrin reactions with lipids and lipoproteins. Free Radic. Biol. Med. 28:1017-1029. [DOI] [PubMed] [Google Scholar]
- 8.Bolwell, G. P. 1999. Role of active oxygen species and NO in plant defense responses. Curr. Opin. Plant Biol. 2:287-294. [DOI] [PubMed] [Google Scholar]
- 9.Boveris, A. 1977. Mitochondrial production of superoxide radical and hydrogen peroxide. Adv. Exp. Med. Biol. 78:67-82. [DOI] [PubMed] [Google Scholar]
- 10.Campisi, E., F. Mancianti, G. Pini, E. Faggi, and G. Gargani. 2003. Investigation in Central Italy of the possible association between Cryptococcus neoformans var. gattii and Eucalyptus camaldulensis. Eur. J. Epidemiol. 18:357-362. [DOI] [PubMed] [Google Scholar]
- 11.Cox, G. M., T. S. Harrison, H. C. McDade, C. P. Taborda, G. Heinrich, A. Casadevall, and J. R. Perfect. 2003. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect. Immun. 71:173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davidson, R. C., J. R. Blankenship, P. R. Kraus, M. de Jesus Berrios, C. M. Hull, C. D'Souza, P. Wang, and J. Heitman. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607-2615. [DOI] [PubMed] [Google Scholar]
- 13.Flesch, I. E., G. Schwamberger, and S. H. Kaufmann. 1989. Fungicidal activity of IFN-gamma-activated macrophages. Extracellular killing of Cryptococcus neoformans. J. Immunol. 142:3219-3224. [PubMed] [Google Scholar]
- 14.Flohe, L., and F. Otting. 1984. Superoxide dismutase assays. Methods Enzymol. 105:93-104. [DOI] [PubMed] [Google Scholar]
- 15.Fridovich, I. 1995. Superoxide radical and superoxide dismutases. Annu. Rev. Biochem. 64:97-112. [DOI] [PubMed] [Google Scholar]
- 16.Goldstein, S., G. Merenyi, A. Russo, and A. Samuni. 2003. The role of oxoammonium cation in the SOD-mimic activity of cyclic nitroxides. J. Am. Chem. Soc. 125:789-795. [DOI] [PubMed] [Google Scholar]
- 17.Hahn, S. M., A. M. DeLuca, D. Coffin, C. M. Krishna, and J. B. Mitchell. 1998. In vivo radioprotection and effects on blood pressure of the stable free radical nitroxides. Int. J. Radiat. Oncol. Biol. Phys. 42:839-842. [DOI] [PubMed] [Google Scholar]
- 18.Idnurm, A., J. L. Reedy, J. C. Nussbaum, and J. Heitman. 2004. Cryptococcus neoformans virulence gene discovery through insertional mutagenesis. Eukaryot. Cell 3:420-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong, J. H., E. S. Kwon, and J. H. Roe. 2001. Characterization of the manganese-containing superoxide dismutase and its gene regulation in stress response of Schizosaccharomyces pombe. Biochem. Biophys. Res. Commun. 283:908-914. [DOI] [PubMed] [Google Scholar]
- 20.Keaney, M., F. Matthijssens, M. Sharpe, J. Vanfleteren, and D. Gems. 2004. Superoxide dismutase mimetics elevate superoxide dismutase activity in vivo but do not retard aging in the nematode Caenorhabditis elegans. Free Radic. Biol. Med. 37:239-250. [DOI] [PubMed] [Google Scholar]
- 21.Kirby, K., J. Hu, A. J. Hilliker, and J. P. Phillips. 2002. RNA interference-mediated silencing of Sod2 in Drosophila leads to early adult-onset mortality and elevated endogenous oxidative stress. Proc. Natl. Acad. Sci. USA 99:16162-16167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwon-Chung, K. J., A. Varma, J. C. Edman, and J. E. Bennett. 1992. Selection of ura5 and ura3 mutants from the two varieties of Cryptococcus neoformans on 5-fluoroorotic acid medium. J. Med. Vet. Mycol. 30:61-69. [PubMed] [Google Scholar]
- 23.Lebovitz, R. M., H. Zhang, H. Vogel, J. Cartwright, Jr., L. Dionne, N. Lu, S. Huang, and M. M. Matzuk. 1996. Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. Proc. Natl. Acad. Sci. USA 93:9782-9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Y., T. T. Huang, E. J. Carlson, S. Melov, P. C. Ursell, J. L. Olson, L. J. Noble, M. P. Yoshimura, C. Berger, P. H. Chan, et al. 1995. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat. Genet. 11:376-381. [DOI] [PubMed] [Google Scholar]
- 25.Longo, V. D., L. L. Liou, J. S. Valentine, and E. B. Gralla. 1999. Mitochondrial superoxide decreases yeast survival in stationary phase. Arch. Biochem. Biophys. 365:131-142. [DOI] [PubMed] [Google Scholar]
- 26.Luk, E., M. Carroll, M. Baker, and V. C. Culotta. 2003. Manganese activation of superoxide dismutase 2 in Saccharomyces cerevisiae requires MTM1, a member of the mitochondrial carrier family. Proc. Natl. Acad. Sci. USA 100:10353-10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maresca, B., and G. S. Kobayashi. 2000. Dimorphism in Histoplasma capsulatum and Blastomyces dermatitidis. Contrib. Microbiol. 5:201-216. [DOI] [PubMed] [Google Scholar]
- 28.Maresca, B., and G. S. Kobayashi. 1989. Dimorphism in Histoplasma capsulatum: a model for the study of cell differentiation in pathogenic fungi. Microbiol. Rev. 53:186-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maslen, L. G. 1952. Routine use of liquid urea medium for identifying Salmonella and Shigella organisms. Br. Med. J. 2:545-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishikawa, M. M., M. S. Lazera, G. G. Barbosa, L. Trilles, B. R. Balassiano, R. C. Macedo, C. C. Bezerra, M. A. Perez, P. Cardarelli, and B. Wanke. 2003. Serotyping of 467 Cryptococcus neoformans isolates from clinical and environmental sources in Brazil: analysis of host and regional patterns. J. Clin. Microbiol. 41:73-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Odom, A., S. Muir, E. Lim, D. L. Toffaletti, J. Perfect, and J. Heitman. 1997. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 16:2576-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfaller, M. A., S. A. Messer, L. Boyken, C. Rice, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2004. Evaluation of the NCCLS M44-P disk diffusion method for determining susceptibilities of 276 clinical isolates of Cryptococcus neoformans to fluconazole. J. Clin. Microbiol. 42:380-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Randhawa, H. S., T. Kowshik, and Z. U. Khan. 2003. Decayed wood of Syzygium cumini and Ficus religiosa living trees in Delhi/New Delhi metropolitan area as natural habitat of Cryptococcus neoformans. Med. Mycol. 41:199-209. [DOI] [PubMed] [Google Scholar]
- 34.Randhawa, H. S., A. Y. Mussa, and Z. U. Khan. 2001. Decaying wood in tree trunk hollows as a natural substrate for Cryptococcus neoformans and other yeast-like fungi of clinical interest. Mycopathologia 151:63-69. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Sharpe, M. A., R. Ollosson, V. C. Stewart, and J. B. Clark. 2002. Oxidation of nitric oxide by oxomanganese-salen complexes: a new mechanism for cellular protection by superoxide dismutase/catalase mimetics. Biochem. J. 366:97-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 38.Spasojevic, I., I. Batinic-Haberle, and I. Fridovich. 2000. Nitrosylation of manganese(II) tetrakis(N-ethylpyridinium-2-yl)porphyrin: a simple and sensitive spectrophotometric assay for nitric oxide. Nitric Oxide 4:526-533. [DOI] [PubMed] [Google Scholar]
- 39.Spasojevic, I., I. Batinic-Haberle, R. D. Stevens, P. Hambright, A. N. Thorpe, J. Grodkowski, P. Neta, and I. Fridovich. 2001. Manganese(III) biliverdin IX dimethyl ester: a powerful catalytic scavenger of superoxide employing the Mn(III)/Mn(IV) redox couple. Inorg. Chem. 40:726-739. [DOI] [PubMed] [Google Scholar]
- 40.Steenbergen, J. N., and A. Casadevall. 2003. The origin and maintenance of virulence for the human pathogenic fungus Cryptococcus neoformans. Microbes Infect. 5:667-675. [DOI] [PubMed] [Google Scholar]
- 41.Tesfa-Selase, F., and R. J. Hay. 1995. Superoxide dismutase of Cryptococcus neoformans: purification and characterization. J. Med. Vet. Mycol. 33:253-259. [PubMed] [Google Scholar]
- 42.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Loon, A. P., B. Pesold-Hurt, and G. Schatz. 1986. A yeast mutant lacking mitochondrial manganese-superoxide dismutase is hypersensitive to oxygen. Proc. Natl. Acad. Sci. USA 83:3820-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, Y., P. Aisen, and A. Casadevall. 1996. Melanin, melanin “ghosts,” and melanin composition in Cryptococcus neoformans. Infect. Immun. 64:2420-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie, Q., K. Kawakami, N. Kudeken, T. Zhang, M. H. Qureshi, and A. Saito. 1997. Different susceptibility of three clinically isolated strains of Cryptococcus neoformans to the fungicidal effects of reactive nitrogen and oxygen intermediates: possible relationships with virulence. Microbiol. Immunol. 41:725-731. [DOI] [PubMed] [Google Scholar]
- 46.Yakes, F. M., and B. Van Houten. 1997. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad. Sci. USA 94:514-519. [DOI] [PMC free article] [PubMed] [Google Scholar]