Abstract
In Saccharomyces cerevisiae, the G1 cyclin Cln3 initiates the Start of a mitotic cell cycle in response to size and nutrient inputs. Loss of Cln3 delays but does not prevent Start, due to the eventual Cln3-independent transcription of CLN1 and CLN2. When unbudded cells of the human pathogen Candida albicans were depleted of the G1 cyclin Cln3 they increased in size but did not bud. Thus, unlike S. cerevisiae, Cln3 is essential for budding in C. albicans. However, eventually the large unbudded cells spontaneously produced filamentous forms. The morphology was growth medium dependent; on nutritionally poor medium the polarized outgrowths fulfilled the formal criteria for true hyphae. This state is stable, and continued growth leads to a hyphal mycelium, which invades the agar substratum. Interestingly, it is also required for normal hyphal development, as Cln3-depleted cells develop morphological abnormalities if challenged with hyphal inducing signals such as serum or neutral pH. Taken together, these results show that, in C. albicans, Cln3 has assumed a critical role in coordinating mitotic cell division with differentiation.
The human fungal pathogen Candida albicans can grow in a variety of morphological forms (11, 15). These range from unicellular budding yeast cells to true hyphae with parallel-sided walls. In between these two extremes, the fungus can exhibit a variety of growth forms that are collectively referred to as pseudohyphae. During pseudohyphal growth, the daughter bud elongates and, after septum formation, remains attached to the mother cell. As a result, filaments composed of elongated cells with constrictions at the septa are formed. Morphological switching from yeast to hyphae and pseudohyphae can be induced by a variety of environmental conditions, such as a growth temperature above 35°C in the presence of serum or neutral pH (11).
In the related budding yeast Saccharomyces cerevisiae, Start of a mitotic cell cycle is initiated by the G1 cyclin Cln3 in association with the Cdc28 kinase (9). Acting through the MBF (Mcb-binding factor) and SBF (Scb-binding factor) transcription factors, Cdc28-Cln3 then activates the transcription of genes for two further G1 cyclins, CLN1 and CLN2. In association with Cdc28, these cyclins then mediate the critical processes necessary for the formation of a bud, initiation of DNA synthesis, and spindle pole body duplication. Although they have different roles, Cln1, Cln2, and Cln3 are functionally redundant, since viability is not compromised by the deletion of any two of the three; however, deletion of all three is lethal (12). In the absence of Cln3, mother cells increase in size and budding is delayed; however, transcription of CLN1 and CLN2 is activated in the absence of Cln3 and budding eventually occurs (6), probably through the action of Bck2 (7), for which no C. albicans homologue is apparent.
Recently, it was shown that in C. albicans the single Cln1/2 homologue, called Hgc1, is only expressed in the hyphal phase. It is required for the formation of hyphae but not for growth in the budding phase (18). Here we show that cells depleted of the C. albicans Cln3 homologue increase in size but are unable to bud, forming very large unbudded cells and, unlike S. cerevisiae, do not eventually escape the block by forming buds. Instead, Cln3-depleted cells spontaneously form hyphae, even under conditions that would normally favor yeast growth. Thus, not only is Cln3 essential for budding, but it negatively regulates the yeast-hyphal transition. Taken together, these results show that G1 cyclins in C. albicans play different roles in promoting the Start of the cell cycle than in S. cerevisiae. Furthermore, they have assumed a critical role in coordinating mitotic cell division with differentiation.
MATERIALS AND METHODS
Strains.
All strains were derived from BWP17 (17). Gene deletions were constructed as described previously (17). To construct MET3-CLN3, sequences 1 to 374 were amplified by PCR for incorporation into pCaDIS (4). Oligonucleotide primers for all strain constructions are listed in Table 1. The genotypes of these strains are as follows: BWP17, his1Δ/his1Δ arg4Δ/arg4Δ ura3Δ/ura3Δ; MET3-CLN3, cln3Δ::ARG4/URA3-MET3-CLN3 his1Δ/his1Δ.
TABLE 1.
Oligonucleotides used in this study
| Primer | Sequence | Use |
|---|---|---|
| C2-DF | TGA TGC AAT CCC TGA TCA AAG CAA GTA ATT TCA AGT TAC AAT CAA TGG AAT TTA GAT GTC GTT TTC CCA GTC ACG ACG TT | CLN3 disruption primer forward |
| C2-DR | TCT TCG ATC TTT TCA ATG TAT CAT GAT TTT TGA GTG TGT AAC CAG CAA TGG TTG ATG TGC TGT GGA ATT GTG AGC GGA TA | CLN3 disruption primer reverse |
| METp-CLN2-F | GAA GGA TCC ATG TTT CCT AAT TCA CCT GA | Forward primer to amplify 5′ CaCLN3 fragment inserted into pCaDIS |
| METP-CLN2-R | GAA CTG CAG CAA AGA CTA GTC AAT CCC AA | Reverse primer to amplify 5′ CaCLN3 fragment inserted into pCaDIS |
| C2probeF | CAG CAG TAA CTT TGA GGT TG | Forward primer to generate CaCLN3 probe |
| C2probeR | GTC TTG GGT GAT AAT GGT GT | Reverse primer to generate CaCLN3 probe |
| RC8cont | TAT GCG ATT GTG GCT CAT AGT AAC G | Forward primer to confirm that CaCLN3 was under control of MET3 |
| C2-ER | AAG TGT ACC ATA ATT AGC ACT C | Reverse primer to confirm that CaCLN3 was under control of MET3 |
| Hgc1probeF | TAG TCA GCT TCC TGC ACC TC | Forward primer to generate HGC1 probe |
| Hgc1probeR | GTA CCA CTA CCA CCA TTA CC | Reverse primer to generate HGC1 probe |
Media and growth conditions.
SDC medium consists of 1.7 g of Difco yeast nitrogen base without amino acids liter−1, 5 g of ammonium sulfate liter−1, 20 mg of histidine liter−1, and 2% glucose supplemented with 0.7 g of −Met CSM amino acid test mixture (Bio101, Inc., Vista, Calif.) liter−1. SD2C is the same as SDC medium, except it contains 6.7 g of Difco yeast nitrogen base without amino acids liter−1 and does not contain ammonium sulfate. For MET3-repressing conditions, 2.5 mM methionine and 2.5 mM cysteine were added to these media.
Microscopy.
Fluorescence microscopy was carried out as previously described (16).
Northern blotting.
Northern blotting was carried out as described previously (16). PCR amplification was used to prepare probes for HGC1 and CLN3. The primers for CLN3 amplified the sequence between residues 570 and 1099, which were selected to minimize homology to other cyclins. Oligonucleotide primers are listed in Table 1.
Coulter counter analysis.
Cell size profiles were determined with a ZB1 Coulter Counter linked to a C1000 Channelyser (Beckman Coulter, High Wycombe, United Kingdom).
RESULTS AND DISCUSSION
Cln3 is required for budding, but Cln3 depletion results in filamentous growth.
In the C. albicans genome, there are three genes with sequence similarity to those for the G1 cyclins of S. cerevisiae: CCN1 (orf19.3207) (8), HGC1 (orf19.6028) (18), and CLN3 (orf19.1960) (13). The nomenclature used here follows the recommendations of the Candida Genome Annotation working group (www.candidagenome.org/Nomenclature.html). C. albicans CLN3 is the closest homologue of S. cerevisiae CLN3. The two proteins show 40% identity and 57% similarity (P = 1.1e−33) across 182 amino acids in the N-terminal region and 26% identity and 36% similarity (P = 7.6e−33) across 183 amino acids in the C-terminal region. Thus, unlike CCN1, sequence similarity of S. cerevisiae and C. albicans Cln3 proteins extends along their whole length and is not restricted to the N-terminal cyclin box. To investigate the role of Cln3 in C. albicans, we attempted to delete both copies. While we were readily able to delete one copy, we failed in persistent attempts to obtain a homozygous deletion, suggesting that one copy of C. albicans CLN3 is essential. This was confirmed by the identification of C. albicans CLN3 in an independent screen for essential genes based on the UAU cassette (5).
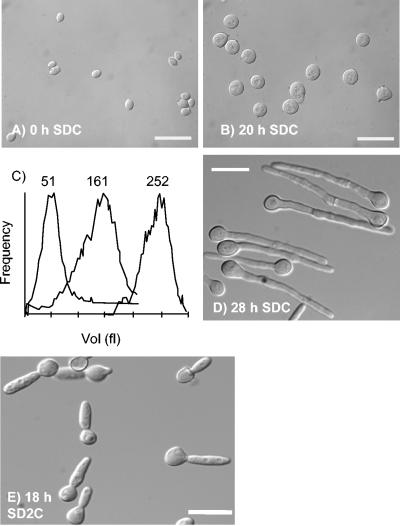
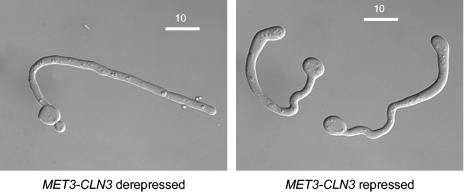
To further investigate the role of Cln3, we constructed a strain (MET3-CLN3) in which a single remaining copy of CLN3 was placed under the control of the MET3 promoter (4). MET3-CLN3 yeast cells were grown to saturation in yeast extract-peptone-dextrose (YEPD) medium. The resulting unbudded cells were then reinoculated into fresh SDC medium at 30°C in the presence or absence of 2.5 mM methionine and 2.5 mM cysteine, which repress and derepress the MET3 promoter, respectively (4). Under these conditions, wild-type cells in derepressing or repressing media grew as yeast cells, as did MET3-CLN3 cells in derepressing medium (data not shown). In repressing medium, the unbudded MET3-CLN3 mother cells grew but did not bud, so they increased in size over the course of 20 h to produce giant cells, approximately fivefold larger than cells at the start of the experiment (Fig. 1A to C). Dramatically, at this time they spontaneously evaginated outgrowths that resembled hyphal germ tubes, although there were some irregularities in the hyphal walls (Fig. 1D). Thus, Cln3 is required for budding, explaining the failure to isolate homozygous cln3Δ/cln3Δ mutants. However, cells can escape the block imposed by the absence of Cln3 by growing as hyphae.
FIG. 1.
Cln3-depleted cells first increase in size without budding and then spontaneously evaginate hyphal germ tubes. (A, B, D, E) Differential interference contrast images of cells at the indicated times after unbudded stationary-phase cells were reinoculated into SDC or SD2C media containing 2.5 mM methionine and 2.5 mM cysteine and incubated at 30°C. Bars, 20 μm. (C) Coulter Counter Channelyser plots of cell volume at (left to right) 0, 4, and 20 h after inoculation.
We found that this phenomenon was very sensitive to formulation of the growth medium. An alternative formulation of SDC medium (SDC2; see Materials and Methods) resulted in a morphology which was uniformly that of pseudohyphae, i.e., constrictions were visible at the bud neck and the polarized extensions were much wider (Fig. 1E). The difference between the two media is that SDC medium has a lower concentration of salts, vitamins, and trace elements than SDC2.
On nutritionally poor medium, Cln3-depleted cells have all the hallmarks of true hyphae.
Hyphae and pseudohyphae are different morphological states, yet polarized growth in pseudohyphae can be so extreme that they superficially resemble hyphae. Recently, formal criteria were proposed to objectively distinguish between hyphae and pseudohyphae (15). We investigated whether the apparent hyphae produced by Cln3 depletion on nutritionally poor SDC medium conformed to these criteria. First, the elongated cells lacked constrictions at the mother cell neck and between adjacent compartments, which is characteristic of hyphae (Fig. 1D). We measured the dimensions of the mother cell and germ tube in Cln3-depleted hyphae and compared them to the dimensions of wild-type cells induced to make hyphae or pseudohyphae. Although the diameter of the Cln3-depleted germ tubes (3.60 ± 0.1 μm, n = 100) was greater than that of normal, serum-induced germ tubes (2.06 ± 0.05 μm, n = 100), the ratio of the diameter of the germ tubes to the diameter of the mother cells was almost identical (0.41 and 0.42, respectively) and much less than that of normal pseudohyphae (0.69). Thus, while the absolute width of Cln3-depleted germ tubes is greater than that of wild-type germ tubes, the proportions relative to the mother cell are very similar.
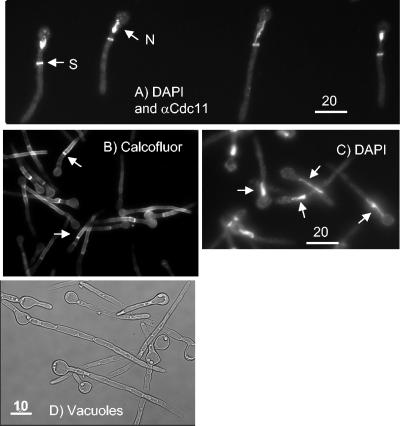
In hyphae, the first septin ring, the first mitosis, and the first septum are located within the germ tube and not at the mother cell neck (14). We visualized the position of the first septin ring by immunocytofluorescence by using a polyclonal antibody to S. cerevisiae Cdc11 (14) (Fig. 2A). We noted that, in repressing SDC medium, 84% of cells displayed a septin ring within the germ tube, 10% of cells had no septin ring, and 6% of cells displayed a septin ring at the mother-bud neck (n = 108). Thus, the first septin ring forms within the germ tube in most cells as it does in true hyphae. We also visualized the septin ring by generating a Cdc10-yellow fluorescent protein fusion (Cdc10-YFP) with similar results, except that in addition to the septin rings within the germ tube we saw patches of delocalized septin within the cell cortex (data not shown). Since these were not visible with the antibody to Cdc11, we presume that the patches are an artifact of the YFP fusion. We visualized the position of the first septum by calcofluor white staining (Fig. 2B). We observed that after 26 h, the septa formed within the germ tube in 76% of cells, no septa were visible in 17% of cells, and the septa formed at the mother-bud neck in 7% of cells (n = 100). Thus, the septum forms within the germ tube as it does in true hyphae.
FIG. 2.
Hyphae induced by Cln3 depletion have the characteristics of true hyphae. Cultures are as described in the legend to Fig. 1. (A) Immunocytofluorescence with an antibody to S. cerevisiae Cdc11 (αCdc11) to visualize septin rings (S) and DAPI to visualize nuclei (N). Note that the septin rings are located within the germ tube and one of the nuclei is starting to migrate into the germ tube. Image taken 23 h after inoculation into repressing SDC medium. (B) Cells stained with calcofluor to visualize septa (examples are noted by arrows). Note that the first septa are formed within the germ tube, not at the mother-bud neck. Image taken 26 h after inoculation into repressing SDC medium. (C) Cells stained with DAPI to visualize nuclei. Arrows indicate examples where nuclei are migrating into the germ tube. Image taken 26 h after inoculation into repressing SDC medium. (D) Vacuoles form in the mother cell and subapical compartments. Image taken 52 h after inoculation into repressing SDC medium.
We visualized the position of nuclei by 4′,6′-diamidino-2-phenylindole (DAPI) staining. We observed that after 26 h, the nucleus had migrated into the germ tube or was dividing within the germ tube in 32% of cells (Fig. 2A and C), there was a single nucleus in the mother cell in 38% of cells, and there were two nuclei, one within the germ tube and one in the mother cell, in 30% of cells (n = 240). Only a single cell was observed where mitosis was taking place across the mother-bud neck. Clearly, it is not possible to classify where mitosis takes place in cells with a single nucleus in the mother cell. Nor is it possible to classify cells with two nuclei, since after mitosis in hyphal germ tubes, one nucleus migrates back into the mother cell (14). However, where the nucleus migrated into the germ tube, mitosis can be classified as taking place within the germ tube. Thus, although it is more difficult to quantify where mitosis takes place, it is clear that, where it is possible to make a judgment, it occurs within the germ tube, as it does in true hyphae.
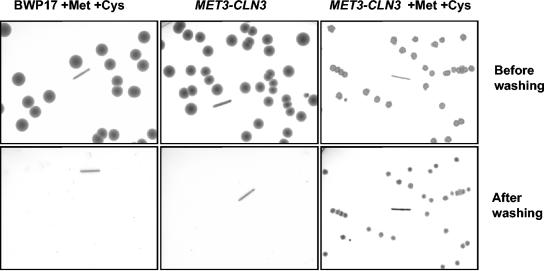
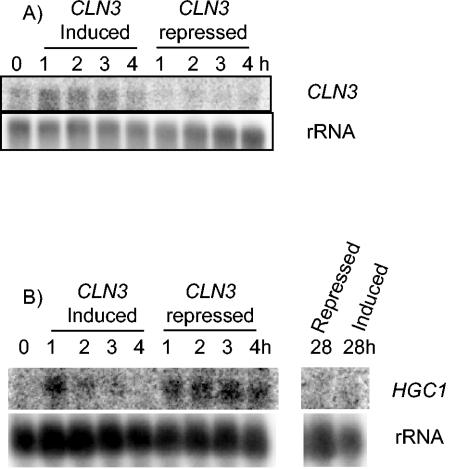
In addition to these formal criteria, we made two further observations, suggesting that hyphae forming upon Cln3 depletion are true hyphae. First, after completion of the first cell cycle, subapical cells in true hyphae become highly vacuolated (15). Differential interference contrast images of the filaments produced by Cln3 depletion also show that the mother cell and subapical cells are vacuolated (Fig. 2D). Secondly, when MET3-CLN3 cells were plated onto SDC plates under MET3-repressing conditions, colonies formed which were composed of filamentous cells and invaded the agar (Fig. 3). Such colonies are characteristic of cells growing in the hyphal state. Thus, the filamentous phenotype caused by Cln3 depletion is stable. Moreover, the formation of colonies under repressing conditions suggests that while Cln3 is essential for budding, it is not required for hyphal growth. However, it is possible that colony formation under repressing conditions is due to leaky expression from the MET3 promoter. We therefore investigated the effectiveness of CLN3 repression when the MET3 promoter was turned off (Fig. 4A). Although the presence of the CLN3 transcript was much lower under repressing conditions, a faint signal persisted, and we cannot completely rule out the possibility that low-level expression of CLN3 from the repressed MET3 promoter is responsible for colony formation.
FIG. 3.
Cln3-depleted cells invade agar. Parental (BWP17) and MET3-CLN3 cells were plated on SDC agar with or without 2.5 mM methionine and cysteine as indicated. After 3 days, the plates were photographed with transmitted light, washed to remove cells above the agar surface, and rephotographed. The line in each image is 5 mm.
FIG. 4.
CLN3 (A) and HGC1 (B) expression in MET3-repressed cells. Unbudded stationary-phase cells were reinoculated into fresh SDC medium with (repressed) or without (induced) methionine and cysteine. Samples were taken after the indicated times and total cellular RNA extracted and used for Northern blot experiments with the indicated probes. Membranes were stripped and rehybridized to an rRNA gene probe to act as a loading control.
Taken together, these results provide strong evidence that Cln3 depletion results in the formation of true hyphae. In an accompanying paper, Bachewich and Whiteway report similar results (1a). They found that the escape from the blocked unbudded state occurred more rapidly (after 4 h) and resulted in a mixture of hyphae and pseudohyphae. The more rapid evagination suggests that cells in their experiments were growing faster than in ours, explaining why they observed a mixture of hyphae and pseudohyphae. One explanation for the difference in growth rates is that we used 2.5 mM cysteine to maximize repression of the MET3 promoter, whereas Bachewich and Whiteway used 0.5 mM. In our experience, the higher concentration of cysteine causes a reduction in growth rate.
Cln3-depleted cells briefly express HGC1.
Expression of HGC1, the homologue of S. cerevisiae CLN1/2 is restricted to the hyphal phase and is required for the formation of hyphae (18). We carried out Northern blot analysis to determine whether the expression of HGC1 is induced during Cln3 depletion. HGC1 was quickly induced in MET-CLN3 cells when stationary-phase cells were reinoculated into fresh medium regardless of whether the MET3 promoter was induced or repressed. However, expression was stronger and lasted longer under repressing conditions (Fig. 4B). Expression of HGC1 in repressed cells had fallen to undetectable levels by 28 h when hyphae formed. Moreover, the expression of HGC1 in serum-induced wild-type cells was much higher than that seen in Cln3-depleted cells (data not shown). Thus, it seems likely that HGC1 expression is not required for hyphal formation induced by Cln3 depletion, suggesting that there is an interaction between Hgc1 and Cln3 during hyphal development.
Cln3 is required for normal hyphal development.
To investigate whether Cln3 is required for normal hyphal growth, we reinoculated stationary-phase MET3-CLN3 cells into repressing medium at 37°C and pH 7.0, conditions that induce hyphal growth in wild-type cells. Initially, hyphal germ tubes developed normally. However, these later developed morphological abnormalities such as kinks and swollen tips (Fig. 5). We observed a similar effect when hyphae were induced by serum rather than neutral pH; however, the effect was less pronounced (data not shown). Thus, Cln3 is also required for normal hyphal development.
FIG. 5.
Cln3 is required for normal hyphal development. MET3-CLN3 cells were grown to saturation in YEPD medium with the MET3 promoter induced. They were reinoculated into fresh SDC medium (pH 7.0) supplemented with or without 2.5 mM methionine and 2.5 mM cysteine, which represses the MET3 promoter, and incubated at 37°C. Under these conditions, control cells grow as hyphae. Images were recorded 6 h after reinoculation.
Concluding remarks.
Loss of a number of different cell cycle components has been shown to result in hyperpolarized bud growth, including the B-type cyclins Clb2 and Clb4 (2), the forkhead transcription factor Fkh2 (3), the Nim1 kinases Hsl1 and Gin4 (16), and the polo kinase CDC5 (1). Moreover, blocking DNA synthesis with hydroxyurea also causes hyperpolarized bud growth (1). The various conditions result in different degrees of polarized growth, but they all result in cells that have a constriction at the mother cell neck, so they must be characterized as pseudohyphae. Cln3 depletion is unique in causing the formation of true hyphae.
Cln3 also plays a role in regulating pseudohyphal formation in S. cerevisiae, as deletion of Cln3 enhances the formation of pseudohyphae during growth on nitrogen-limiting medium (10). However, a major difference is that in S. cerevisiae loss of Cln3 enhances the response to the signal inducing pseudohyphal formation, but it has no effect in the absence of these signals. Thus, the results in this paper show that G1 cyclins play different roles in C. albicans than in S. cerevisiae. Cln3 is essential for growth in the yeast form while Hgc1 is essential for hyphal growth. Moreover, both Hgc1 and Cln3 are intimately linked to the control of differentiation.
Acknowledgments
This work was funded by a project grant from the Wellcome Trust. B.C.y.L. was supported by a fellowship from CONACYT (Mexico) (grant no. 060862/Z/00/Z).
We thank Catherine Bachewich for communicating unpublished data and the Mitchell, Berman, and Gale laboratories for plasmids and strains.
REFERENCES
- 1.Bachewich, C., D. Y. Thomas, and M. Whiteway. 2003. Depletion of a polo-like kinase in Candida albicans activates cyclase-dependent hyphal-like growth. Mol. Biol. Cell 14:2163-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Bachewich, C., and M. Whiteway. 2005. Cyclin Cln3p links G1 progression to hyphal and pseudohyphal development in Candida albicans. Eukaryot. Cell 4:95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bensen, E. S., and J. Berman. 2002. The B-type cyclins Cyb1p and Cyb4p influence morphology in Candida albicans. Mol. Biol. Cell 13:2464. [Google Scholar]
- 3.Bensen, E. S., S. G. Filler, and J. Berman. 2002. A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot. Cell 1:787-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Care, R. A., J. Trevethick, K. M. Binley, and P. E. Sudbery. 1999. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34:792-798. [DOI] [PubMed] [Google Scholar]
- 5.Davis, D. A., V. M. Bruno, L. Loza, S. G. Filler, and A. P. Mitchell. 2002. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162:1573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dirick, L., T. Bohm, and K. Nasmyth. 1995. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 14:4803-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epstein, C. B., and F. Cross. 1994. Genes that can bypass the CLN requirement for Saccharomyces cerevisiae cell cycle START. Mol. Cell. Biol. 14:2041-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ernst, J. F. 2000. Transcription factors in Candida albicans-environmental control of morphogenesis. Microbiology 146:1763-1774. [DOI] [PubMed] [Google Scholar]
- 9.Futcher, B. 1996. Cyclins and the wiring of the yeast cell cycle. Yeast 12:1635-1646. [DOI] [PubMed] [Google Scholar]
- 10.Loeb, J. D. J., T. A. Karentseva, T. Pan, M. Sepulveda-Becerra, and H. P. Liu. 1999. Saccharomyces cerevisiae G1 cyclins are differentially involved in invasive and pseudohyphal growth independent of the filamentation mitogen-activated protein kinase pathway. Genetics 153:1535-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odds, F. C. 1988. Candida and candidosis, p. 42-59. Balliere Tindall, London, United Kingdom.
- 12.Richardson, H. E., C. Wittenberg, F. Cross, and S. I. Reed. 1989. An essential G1 function for cyclin-like proteins in yeast. Cell 59:1127-1134. [DOI] [PubMed] [Google Scholar]
- 13.Sherlock, G., A. M. Bahman, A. Mahal, J. C. Shieh, M. Ferreira, and J. Rosamond. 1994. Molecular cloning and analysis of CDC28 and cyclin homologs from the human fungal pathogen Candida albicans. Mol. Gen. Genet. 245:716-723. [DOI] [PubMed] [Google Scholar]
- 14.Sudbery, P. E. 2001. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localisation. Mol. Microbiol. 41:19-31. [DOI] [PubMed] [Google Scholar]
- 15.Sudbery, P. E., N. A. R. Gow, and J. Berman. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317-324. [DOI] [PubMed] [Google Scholar]
- 16.Wightman, R., S. Bates, P. Amnorrattapan, and P. E. Sudbery. 2004. In Candida albicans, the Nim1 kinases Gin4 and Hsl1 negatively regulate pseudohypha formation and Gin4 also controls septin organization. J. Cell Biol. 164:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson, B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing in Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng, X., Y. Wang, and Y. Wang. 2004. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 23:1845-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]