Abstract
Candida albicans is a diploid yeast with a predominantly clonal mode of reproduction, and no complete sexual cycle is known. As a commensal organism, it inhabits a variety of niches in humans. It becomes an opportunistic pathogen in immunocompromised patients and can cause both superficial and disseminated infections. It has been demonstrated that genome rearrangement and genetic variation in isolates of C. albicans are quite common. One possible mechanism for generating genome-level variation among individuals of this primarily clonal fungus is mutation and mitotic recombination leading to loss of heterozygosity (LOH). Taking advantage of a recently published genome-wide single-nucleotide polymorphism (SNP) map (A. Forche, P. T. Magee, B. B. Magee, and G. May, Eukaryot. Cell 3:705-714, 2004), an SNP microarray was developed for 23 SNP loci residing on chromosomes 5, 6, and 7. It was used to examine 21 strains previously shown to have undergone mitotic recombination at the GAL1 locus on chromosome 1 during infection in mice. In addition, karyotypes and morphological properties of these strains were evaluated. Our results show that during in vivo passaging, LOH events occur at observable frequencies, that such mitotic recombination events occur independently in different loci across the genome, and that changes in karyotypes and alterations of phenotypic characteristics can be observed alone, in combination, or together with LOH.
Candida albicans is a commensal diploid yeast and inhabits a variety of niches in human populations. An opportunistic pathogen, it can cause both superficial and disseminated infections. The recently published complete diploid genome sequence of SC5314, a clinical isolate, showed a high degree of heterozygosity, including more than 55,700 single-nucleotide polymorphisms (SNPs) in the 32-Mb diploid genome (21). Clinical isolates of C. albicans show wide variations in karyotype, suggesting that genome rearrangement is relatively common in the host (30). Although C. albicans can be made to mate in the laboratory, there is no evidence for a complete sexual cycle either in vivo or in vitro (4, 20, 29) and the importance of recombination and rearrangement in generating genetic variation is not clear.
One possible mechanism for generating genome-level variation among individuals of this primarily clonal fungus is mutation and mitotic recombination leading to loss of heterozygosity (LOH). For example, Tavanti et al. (44) found evidence for the recombination generating a heterogenous array of haplotypes in clinical isolates of C. albicans. However, the frequency or rate of recombination, and thus its importance, cannot be evaluated by analysis of static haplotypes alone. To that end, we used strains heterozygous at the GAL1 locus to demonstrate that mitotic recombination occurs at a measurable level during the course of experimental infections of mice (15, 16).
Counter-selectable markers such as GAL1 are powerful but of limited utility for studying genome-wide recombination, chromosomal dynamics, or genome rearrangement. For these purposes, we turned to the use of SNP markers. SNPs are the most frequently observed differences at the DNA sequence level across individuals and chromosomes of diploid organisms and thus can be used to study processes involved in the evolution of virulence in C. albicans. To examine LOH across the entire genome, we recently developed an SNP map for strain SC5314 containing 150 marker sequences comprising 561 SNPs and 9 indels (15). The SNPs are spaced about 100 kb apart across the eight chromosomes.
In this paper, we report the development and use of SNP markers, in a microarray format, on the basis of hybridization of allele-specific oligonucleotides to PCR-generated probes from test and control (SC5314) genomes. Our approach is similar to that employed for the high-throughput SNP microarrays used to analyze genome regions of interest in humans (13, 37, 48) and for genetic mapping projects and recombination studies of plants (9, 42). Here, we report the development of microarray approaches for SNP analysis and estimates of recombination frequencies for SNPs located on chromosomes 5, 6, and 7. The array methodology was developed by using strains of known genotype and subsequently used to examine strains previously shown to have undergone mitotic recombination at the GAL1 locus on chromosome 1 during infection in mice (16). The GAL1 recombinant strains were examined for LOH at the SNP loci; in addition, karyotypes and morphological properties of these strains were evaluated. Our results show (i) that during in vivo passaging, LOH events occur at observable frequencies and that such mitotic recombination events occur independently at different loci across the genome and (ii) that changes in karyotypes and alterations of phenotypic characteristics can be observed alone, in combination, or together with LOH events.
MATERIALS AND METHODS
Strains used and analyzed in this study.
To develop and optimize the SNP microarray, we used C. albicans strain SC5314 because its complete genome sequence was available (21). Since all SNP markers were generated using SC5314 sequence (15), we tested the accuracy and discriminatory power of the SNP array by use of control strains SC5314a and SC5314α, which are homozygous for the MTL a or α homologs of chromosome 5, respectively (23, 32). Results for these chromosome 5 homozygous strains were compared to those for SC5314, which is heterozygous for all but two SNPs on chromosome 5. The genotypes were confirmed by sequencing, and these data were used to establish correlations between genotype and array signal.
Previously, we passaged two strains (AF6 and AF7) heterozygous at the GAL1 locus through mice by use of a model of hematogenously disseminated disease (16) and obtained post-mouse-passage isolates that had become homozygous gal−/− from each strain background. A total of 21 of those strains (4 from AF6, 17 from AF7) were characterized for SNP heterozygosity, chromosomal rearrangement, growth rate, colony size, and filamentation on serum. The strain designations for these 21 strains are presented as follows: the last digit of the parental strain name (6 or 7) is followed by the individual strain number (e.g., 6-4205).
Development of an SNP microarray.
For oligonucleotide design and printing of SNP microarrays, 25 SNPs were chosen to represent markers (15) along chromosomes 5 (15 SNPs), 6 (1 SNP), and 7 (9 SNPs). Two 30-mer oligonucleotides per locus were designed to represent the alternate alleles of each SNP (Table 1). Each oligonucleotide consists of a 15-nucleotide T-polylinker and a 15-nucleotide target sequence with the polymorphic base at the middle position (Fig. 1). All oligonucleotides were designed to have similar melting temperatures for optimal microarray hybridization. Oligonucleotides were purchased from Integrated DNA Technologies Inc. (Coralville, Iowa). Printing dilutions (20 μM) were prepared in 1× printing buffer (300 mM sodium phosphate [pH 8.5]). The 50 oligonucleotides (25 SNPs × 2 alleles) were printed in quadruplicate onto Codelink-activated slides (Amersham Biosciences, Piscataway, N.J.) by use of a Genomic Solutions MicroGrid II Total Array system. After printing, oligonucleotides were coupled to the slide surface by overnight incubation in an NaCl humidity chamber by following the instructions of the manufacturer (Amersham Biosciences) and stored at room temperature under desiccating conditions until further use.
TABLE 1.
Allele-specific oligonucleotides and primer pairs for detection of SNPsa
| MP group | Chromosomal location of SNP | Name of oligonucleotide | Allele-specific oligonucleotide (5′-3′)b | Primers for probe generation (5′-3′)c |
|---|---|---|---|---|
| MP-I | 5M | 1855/2172-a1-c | 5AmMC6-ttt ttt ttt ttt ttt TGT TGA CGT TGA TGG | F: CCT GAA GTA ATG CCA CCA |
| 1855/2172-a2-t | 5AmMC6-ttt ttt ttt ttt ttt TGT TGA TGT TGA TGG | R: CCA ATG TCT ATA GAA ACT GCCd | ||
| 5M | SNF1-a1-a | 5AmMC6-ttt ttt ttt ttt ttt AGA GCC AAT AGT CAA | F: CAC CTG AGA GAG AGG TAA C | |
| SNF1-a2-c | 5AmMC6-ttt ttt ttt ttt ttt AGA GCC ACT AGT CAA | R: TTG TAG GCG GTG TGG GAA TTd | ||
| 5M | 1922/2344-a1-c | 5AmMC6-ttt ttt ttt ttt ttt TTA TCG GCG TAG TGA | F: GCC ATG GCC TTA TGC TAT | |
| 1922/2344-a2-t | 5AmMC6-ttt ttt ttt ttt ttt TTA TCG GTG TAG TGA | R: GTT GCA CCA TTC CCA TGG GAd | ||
| 5M | PDE1-a1-c | 5AmMC6-ttt ttt ttt ttt ttt GCA CAA CTA AGG AAC | F: ACG CAC AAC ACC ATA GCA | |
| PDE1-a2-t | 5AmMC6-ttt ttt ttt ttt ttt GCA CAA TTA AGG AAC | R: TAA TAT GCT AGG TGG GGG TTC CTTd | ||
| 5M | 1969/2162-a1-a | 5AmMC6-ttt ttt ttt ttt ttt GAA GTG GAG ATG ATA | F: ATA CGG CCA CTC GGT ATG A | |
| 1969/2162-a2-t | 5AmMC6-ttt ttt ttt ttt ttt GAA GTG GTG ATG ATA | R: CAG CTA ATG CCC CAC GTA ATd | ||
| 5I | DPH5-a1-a | 5AmMC6-ttt ttt ttt ttt ttt TTC TAC AGC AGA GAA | F: CAG CCA ACC AGG AGT CAT TG | |
| DPH5-a2-g | 5AmMC6-ttt ttt ttt ttt ttt TTC TAC GGC AGA GAA | R: ATT GTG GAT GGT CTC GACd | ||
| 5I | 2093/2390-a1-c | 5AmMC6-ttt ttt ttt ttt ttt GAT GAG TAC GAA TCA | F: TCT CTC TTT GGA GTG AGC | |
| 2093/2390-a2-t | 5AmMC6-ttt ttt ttt ttt ttt GAT GAG TAT GAA TCA | R: GAA AGA TGG CTC CTT ATA TTT TGd | ||
| 5I | 1341/2493-a1-a | 5AmMC6-ttt ttt ttt ttt ttt GCA ATT TAG TAG CCA | F: GTC AGC TTC ACC ACA GTT | |
| 1341/2493-a2-g | 5AmMC6-ttt ttt ttt ttt ttt GCA ATT TGG TAG CCA | R: AGT TCC AAC TCC AAA GCCd | ||
| 6O | B15B20-a1-a(rc)e | 5AmMC6-ttt ttt ttt ttt ttt GGT TCA GAA TAA TGA | F: GGA ATT GGA AAG AAG TCAd | |
| B15B20-a2-g(rc) | 5AmMC6-ttt ttt ttt ttt ttt GGT TCA GGA TAA TGA | R: GCA TAT AGT CTA CCC AGT G | ||
| 7C | YDL114-a1-t(rc) | 5AmMC6-ttt ttt ttt ttt ttt ACC TCC GTT GAG TAG | F: CAA CCA CGT TCA GGA CCAd | |
| YDL114-a2-c(rc) | 5AmMC6-ttt ttt ttt ttt ttt ACC TCC GCT GAG TAG | R: CCA CTC TCT CTC AAG TCA | ||
| 7F | YKL084-a1-a | 5AmMC6-ttt ttt ttt ttt ttt TGG CTA TAC ATG TAC | F: TAA CCA TGA ACC AGA CCG | |
| YKL084-a2-t | 5AmMC6-ttt ttt ttt ttt ttt TGG CTA TAC TTG TAC | R: ACC AGG GTT GAA TTG TCCd | ||
| 7F | 2397/2498-a1-c | 5AmMC6-ttt ttt ttt ttt ttt ATT CTG GCT TGA AAG | F: CTC ACC CCT TCG TCA CCA | |
| 2397/2498-a2-t | 5AmMC6-ttt ttt ttt ttt ttt ATT CTG GTT TGA AAG | R: GGT ATT GCT GGT ATC GGTd | ||
| 7F | RBP1-a1-a | 5AmMC6-ttt ttt ttt ttt ttt TGG TTA CAG TAT CAC | F: ATT AGC ACC ACC TTT ACC | |
| RBP1-a2-t | 5AmMC6-ttt ttt ttt ttt ttt TGG TTA CTG TAT CAC | R: TGT CTG AAG AAC TTC CACd | ||
| 7G | YJL54/2-a1-a | 5AmMC6-ttt ttt ttt ttt ttt TGG GAA TAC ACT GAG | F: GGA GAC TGT ACC TTT AGT AG | |
| YJL54/2-a2-c | 5AmMC6-ttt ttt ttt ttt ttt TGG GAA TCC ACT GAG | R: GCT ACT AAA ATA TGC TCA GTGd | ||
| 7G | 6.1345-a1-g(rc) | 5AmMC6-ttt ttt ttt ttt ttt CTC CAT GGC CAT TAG | F: ATC GGC AAT GGA AGT GGGd | |
| 6.1345-a2-a(rc) | 5AmMC6-ttt ttt ttt ttt ttt CTC CAT GAC CAT TAG | R: GTC GTG GGA TTT TCA GCA AG | ||
| 7G | 6.282/2-a1-t(rc) | 5AmMC6-ttt ttt ttt ttt ttt TAT CTA ATG AAG CCA | F: ACC ACC ACA ATT GGC TTC | |
| 6.282/2-a2-c(rc) | 5AmMC6-ttt ttt ttt ttt ttt TAT CTA ACG AAG CCA | R: GAT CAG CCC ATT GTT GATd | ||
| MP-II | 5M | C2F7-a1-t(rc) | 5AmMC6-ttt ttt ttt ttt ttt CAC CAC ATT TGA AAT | F: CGT CGT CTG TTA TAG TCC CCT Td |
| C2F7-a2-c(rc) | 5AmMC6-ttt ttt ttt ttt ttt CAC CAC ACT TGA AAT | R: AGC CGG AAT AGA AAC TTT TGG C | ||
| 5M | HST3-a1-c | 5AmMC6-ttt ttt ttt ttt ttt ACA TCA CCC TCA AAT | F: CCT GGG CAA AAG ACT AAC AG | |
| HST3-a2-t | 5AmMC6-ttt ttt ttt ttt ttt ACA TCA CTC TCA AAT | R: CCA ATG ACT TGA CAC CAG CAd | ||
| 5I | HEX1-a1-g(rc) | 5AmMC6-ttt ttt ttt ttt ttt GGA TTT TTG TAC AAA TC | F: GCT CAA TCC ACA CGC TTG TGd | |
| HEX1-a2-a(rc) | 5AmMC6-ttt ttt ttt ttt ttt GGA TTT TTA TAC AAA TC | R: ACC ACA ACT TCC GTT CCC TT | ||
| 5I | 1817/2082-a1-c | 5AmMC6-ttt ttt ttt ttt ttt AGT TCT CCT CCA TTT | F: GAC CAT TAT TTG AGA GCC AC | |
| 1817/2082-a2-t | 5AmMC6-ttt ttt ttt ttt ttt AGT TCT CTT CCA TTT | R: GGT GCA GTA CTT GTT GAT Gd | ||
| 5M | 1899/2008-a1-c | 5AmMC6-ttt ttt ttt ttt ttt TGT TTT CCG GTT GAT | F: TAC GGC CAA GTT TTC CTC AC | |
| 1899/2008-a2-t | 5AmMC6-ttt ttt ttt ttt ttt TGT TTT CTG GTT GAT | R: GCT CGG GGA TAT AAT TGG CAd | ||
| 5M | 1445/2395-a1-c | 5AmMC6-ttt ttt ttt ttt ttt TGC CTT GCG TTA TAG | F: GGT TTT GGT TTA TCT CCA GGT TTC | |
| 1445/2395-a2-t | 5AmMC6-ttt ttt ttt ttt ttt TGC CTT GTG TTA TAG | R: AGA TAC ATA CCG TGG GAA GCT ATTd | ||
| 5I | 2340/2493-a1-a | 5AmMC6-ttt ttt ttt ttt ttt TTT GGA TAA ACG AGT | F: GCT TGG GGG TTC TGA TAC TT | |
| 2340/2493-a2-g | 5AmMC6-ttt ttt ttt ttt ttt TTT GGA TGA ACG AGT | R: AGC GAC CAT TAC GCA AGG TAd | ||
| 7C | LEU2-a1-c | 5AmMC6-ttt ttt ttt ttt ttt CCC AAT TCA ACG GTT | F: GGT GGT GGT CAA GAA AGA GG | |
| LEU2-a2-t | 5AmMC6-ttt ttt ttt ttt ttt CCC AAT TTA ACG GTT | R: GGT ATT GAG ATT TGG CGT CGd | ||
| 7G | ARG4-a1-t(rc) | 5AmMC6-ttt ttt ttt ttt ttt ACC AAT GTG CCC ATC | F: TCA CGG CAA TTC TTG AAC GGGd | |
| ARG4-a2-a(rc) | 5AmMC6-ttt ttt ttt ttt ttt ACC AAT GAG CCC ATC | R: GCT AAA GCA CCA GAT CCT AAT GGA G |
Allele-specific oligonucleotides (ASOs) are sorted on the basis of their MP-PCR groupings (MP-I and MP-II). The chromosomal location, allele-specific oligonucleotide sequences for both alleles, and primer pairs for amplifcation of SNP markers are given.
Each allele-specific oligonucleotide contains an amino-linker (5AmMC6), a 15-nucleotide poly-T-spacer (ttt ttt ttt ttt ttt), and 15 nucleotides of specific sequence with the polymorphic base at the middle position.
F, forward; R, reverse.
Primer used in PE reaction.
(rc), reverse complement.
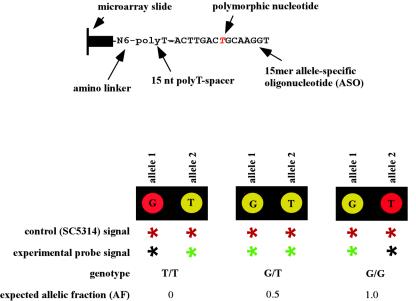
FIG. 1.
Design of allele-specific oligonucleotides and demonstration of the three possible genotypes. The top panel shows the composition of the allele-specific oligonucleotide attached to the microarray slide. The bottom panel depicts the three possible genotypes obtained through competitive hybridization of an experimental and control probe.
Probe generation. (i) Experimental probes.
To amplify each SNP marker region containing the target polymorphism, primer pairs with uniform annealing temperatures were used (15). To generate a probe for the arrays, two multiplex PCRs (MP-PCR) per strain were carried out with 16 and 9 primer pairs (Table 1 and Table 2) and 60 ng of genomic DNA by use of an MP-PCR kit (QIAGEN, Valencia, Calif.) and following the instructions of the manufacturer. PCRs were carried out as follows: initial denaturation step at 95°C for 15 min, denaturation step at 94°C for 30 s, annealing step at 55°C for 1 min 30 s, extension step at 72°C for 1 min 30 s (steps 2 to 4 were cycled 34 times), and final extension step at 72°C for 10 min. A total of 3 μl of each MP-PCR product was run on an agarose gel (3% in 0.5× Tris-acetate-EDTA) to check for proper amplification. MP-PCR products were purified using a QIAGEN PCR purification kit. To generate labeled, single-stranded probes, a previously reported primer extension (PE) reaction procedure (19) was modified. For each MP-PCR a corresponding PE reaction was carried out as follows. A total of 15 μl of purified MP-PCR product was used in a 25-μl PE reaction mixture with 10× PCR buffer (Takara, Madison, Wis.), 12.5 μM concentrations each of dATP, dCTP, dGTP, and dTTP, 0.25 μM concentrations of each primer (Table 1), a 5 μM concentration of Cy3-dUTPs (Amersham), and 1.5 units of Taq Vent exo− polymerase (New England Biolabs, Beverly, Mass.). PE cycling reactions were carried out as follows: initial denaturation step at 95°C for 3 min, denaturation step at 94°C for 20 s, annealing step at 55°C for 20 s, extension step at 72°C for 20 s (the second through fourth steps were cycled 25 times), and final extension step at 72°C for 3 min.
TABLE 2.
Sizes of MP-PCR fragments and SNP positions
| MP group | Chromosomal location of SNP locus | Name of SNP locus | Size of MP product (bp) | SNP position (from 5′ of forward primer) |
|---|---|---|---|---|
| MP-I | 5M | 1855/2172 | 206 | 177 |
| 5M | SNF1 | 117 | 54 | |
| 5M | 1922/2344 | 234 | 207 | |
| 5M | PDE1 | 215 | 190 | |
| 5M | 1969/2162 | 303 | 254 | |
| 5I | DPH5 | 215 | 33 | |
| 5I | 2093/2390 | 175 | 129 | |
| 5I | 1341/2493 | 273 | 174 | |
| 6O | B15B20 | 282 | 52 | |
| 7C | YDL114 | 215 | 38 | |
| 7F | YKL084 | 145 | 112 | |
| 7F | 2397/2498 | 245 | 219 | |
| 7F | RBP1 | 230 | 157 | |
| 7G | YJL54/2 | 188 | 167 | |
| 7G | 6.1345 | 230 | 27 | |
| 7G | 6.282/2 | 282 | 19 | |
| MP-II | 5M | C2F7 | 188 | 77 |
| 5M | HST3 | 182 | 74 | |
| 5I | HEX1 | 146 | 28 | |
| 5I | 1817/2082 | 149 | 54 | |
| 5M | 1899/2008 | 195 | 52 | |
| 5M | 1445/2395 | 207 | 135 | |
| 5I | 2340/2493 | 160 | 134 | |
| 7C | LEU2 | 205 | 174 | |
| 7G | ARG4 | 196 | 92 |
(ii) Control probes.
To account for differences in spotting intensities on the array, variation in hybridization of probes to the oligonucleotides on the array, and possible cross-hybridization, we included an internal control hybridization probe generated from strain SC5314 in all array experiments. The SC5314 probe was generated as described above but using Cy5-dUTPs (Amersham) in the PE reactions. Thus, in all experiments, the experimental probe is Cy3 labeled (green) and the control probe is Cy5 labeled (red).
SNP array probe.
To make the probe used directly on SNP arrays, 8 μl from the experimental (Cy3-labeled) and control (Cy5-labeled) strains were pooled, purified using a Qiaquick PCR purification kit (QIAGEN), and concentrated to a final volume of ∼5 μl by use of Microcon YM30 concentrator columns (Millipore Corporation, Bedford, Calif.) following the manufacturer's instructions. To each concentrated probe, 115 μl of hybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% [SDS], 0.1 mg of salmon sperm DNA/ml) was added and the probes were denatured at 98° for 2 min, cooled by centrifugation at 3,000 rpm in an Eppendorf model 5145C for 4 min, and stored in the dark until use at room temperature.
Slide preparation and hybridization.
Just before hybridization, slides were incubated in blocking solution (50 mM ethanolamine, 1 M Tris, pH 9.0) for 15 min at 50°C to inactivate free amino-linkers. Slides were rinsed twice in distilled water and then washed in 4 × SSC-0.1% SDS for 1 h at 50°C. Finally, they were rinsed in distilled water and then dried by centrifuging for 3 min at 700 rpm (model 5810; Eppendorf, Hamburg, Germany).
A frame-seal chamber (SLF-1201; MJ Research, Waltham, Mass.) was used to enclose the printed surface during the hybridization. A total of 120 μl of hybridization buffer containing the probe was added to the chamber. After the chamber was sealed, the entire slide was transferred to a hybridization cassette (ArrayIt; Telechem International Inc., Sunnyvale, Calif.), and hybridization was carried out at 35°C for 2 h in a water bath (Isotherm [model 202S]; Fisher Scientific, Hanover Park, Ill.) with the hybridization chambers completely immersed. The slide was then removed from the hybridization chamber, the frame-seal chamber was peeled off, and the slide was rinsed briefly with 4× SSC and then washed twice in 2 × SSC-0.1% SDS for 5 min at 35°C, once in 2 × SSC for 1 min at room temperature, and once in 1 × SSC for 1 min at room temperature. Slides were then dried by centrifugation for 3 min at 700 rpm (model 5810; Eppendorf). Slides were scanned using a ScanArray 5000 scanner (Perkin-Elmer) with ScanArray software, version 3.1.
Calculation of relative signal intensity and allele fraction.
Data were analyzed using Genepix Pro software, version 4.0 (Axon Instruments). For each spot and dye label (Cy3 and Cy5), the median pixel intensity was calculated and the median background intensity (calculated using a local background method) was subtracted from each median pixel intensity to yield the total fluorescent intensities (Cy3 and Cy5) for each spot. The ratio of medians was calculated as the ratio of the background-subtracted median pixel intensity of the control probe (SC5314; Cy5) relative to the background-subtracted median pixel intensity of the experimental probe (Cy3). After calculating on the basis of the 1:1 ratio of the reference to the experimental strain in each probe, the ratio of medians was then normalized so that the ratio of medians of all of the features was equal to 1. Allelic fractions (AF) for each SNP locus were calculated from the normalized ratio of medians as follows:
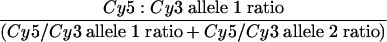 |
In theory, homozygous allele 1 (a1) genotypes will give an AF = 0.0, heterozygous genotypes will give an AF = 0.5, and homozygous a2 genotypes will give AF = 1.0. For example, AF values for a1 homozygotes should be close to 0 because the a2 ratio is a large number while the a1 ratio is close to 1. Because of nonspecific hybridization and variation in labeling, experimentally derived AF values will differ and cutoff values for genotype determination must be determined under the same conditions as those used for the experiments. Consequently, we used data from at least two independent SNP arrays for the SC5314a and SC5314α strains with known homozygous chromosome 5 genotypes to experimentally establish cutoff values as AF < 0.4 for a1 homozygous genotypes, 0.4 < AF < 0.6 for heterozygous genotypes, and AF > 0.6 for a2 homozygous genotypes. As additional sequence-verified genotypes become available, AF data can be incorporated continually to generate robust AF value ranges for genotype determination.
Genotypes determined by SNP microarray were confirmed by sequencing as follows. PCRs were carried out for each primer pair used in the MP-PCR experiments under the following conditions: 10 mM Tris/HCl (pH 8.0), 50 mM KCl, 1.5 mM MgCl2, 100 μM each dATP, dCTP, dGTP and dTTP, 2.5 U of Taq polymerase, 0.5 μl of a 10 μM stock solution of each primer, and 30 ng of genomic DNA in a total volume of 25 μl. PCR cycling conditions were as follows: initial denaturation for 3 min at 94°C, denaturation step for 1 min at 94°C, primer annealing step for 30 s at 55°C, extension step for 1 min at 72°C, and a final extension step for 5 min at 72°C. PCR products were purified using a PCR purification kit (QIAGEN), and 50 to 100 ng of purified DNA was used as a sequencing template. Sequencing reactions were performed with both the forward and reverse primers following the manufacturer's instructions (ABI Big Dye Chemistry; Perkin-Elmer, Boston, Mass.), and run on an ABI 3100 automated sequencer. Sequences were edited and aligned using Sequencer 3.1.1 software (ABI; Perkin-Elmer).
Karyotype analysis.
Pulsed-field gel electrophoresis was used to detect major chromosomal rearrangements in the strains derived in vivo compared to the results seen with their parental strains AF6 and AF7. Samples were prepared as described previously (10). Chromosomes were separated on a CHEF DR-III apparatus (Bio-Rad Laboratories, Hercules, Calif.) at 14°C by use of the following conditions: 1% agarose in 0.5× Tris-borate-EDTA buffer; for block 1, 60- to 120-s switch (120° angle) at 6 V/cm for 36 h; for block 2, 120- to 300-s switch (120° angle) at 4.5 V/cm for 12 h. Gels were stained in ethidium bromide for 15 min, destained in distilled water for 15 min, and photographed.
Determination of colony morphology, growth rate, and filamentation ability.
Growth rates were determined for all strains used in this study by use of AF6 and AF7 as reference controls. Strains were streaked out from freezer stocks onto YEPD medium (Sigma Aldrich, St. Louis, Mo.) (2% glucose, 1% yeast extract, 1% yeast peptone) for 2 days at 30°C. Colony size and morphology were determined. A single colony was then transferred to 5 ml of YEPD liquid medium and grown overnight. A 10−4 dilution was made into 100 ml of fresh medium to give an optical density at 600 nm of 0.001 by use of a Beckman DU-62 spectrophotometer (Beckman Coulter Inc., Fullerton, Calif.). The optical density was determined every hour until the culture reached stationary phase.
Filamentation properties of strains were examined on plates containing 2% agar and 4% bovine calf serum (Sigma). Strains were grown on YEPD medium for 2 days at 30°C. Single colonies of the parental control strain and the recombinant strain were streaked out separately for direct comparison. Plates were incubated for 48 to 72 h at 30°C. Colonies were screened for the presence of filamentous growth at a magnification of ×40 by use of a Eclipse E600 microscope (Nikon, Melville, N.Y.).
RESULTS AND DISCUSSION
Development of the SNP microarray.
For the development of the SNP microarray and to evaluate the correct determination of genotypes, we used probes derived from control strains. Cy5-labeled probes derived from strain SC5314 were hybridized against the Cy3-labeled SC5314 probe, and 23 of the 25 SNPs gave the expected heterozygous genotype and sufficient signal intensities. Two SNP loci which gave ambiguous results were excluded from further analyses. These results were confirmed by sequencing of the SNP markers (data not shown). For each strain analyzed in the study, we performed independent hybridizations using probes derived from different MP-PCRs and at least two slides per hybridization for each strain.
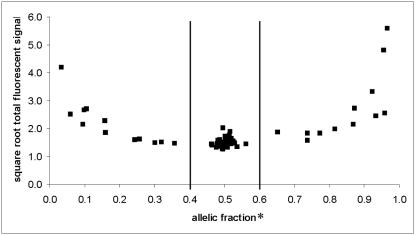
Results for probes derived from strains SC5314a and SC5314α, each carrying a different homologue of chromosome 5, demonstrated that we could distinguish between homozygous and heterozygous genotypes for the 15 SNP chromosome 5 loci (Table 3). We confirmed these results by sequencing the SNP markers as well (data not shown). Haplotypes for SC5314a and SC5314α are listed in Table 4. When plotted, averaged AF values for strains SC5314, SC5314a, and SC5314α fall into three distinct clusters representing homozygous a1 with an average AF = 0.25, the heterozygous a1/a2 with an average AF = 0.50, and homozygous a2 with an average AF = 0.85 (Table 3). The AFs for homozygous a1 and a2 at SNP locus 1445/2395 (0.36 and 0.65, respectively) are close to the homozygous thresholds and were confirmed to be homozygous by sequencing. Several data points with extreme homozygote AFs (e.g., 0.03 and 0.97 for SNP locus 1817/2082) also had high hybridization intensity values. These results are likely due to a homozygous allele that both labeled and hybridized well to the expected array spot combined with low background hybridization or intensity to the array spot representing the absent allele (Fig. 2).
TABLE 3.
Genotypes of parental strains and GAL1 derivatives
| SNP map locationa | SNP locus | Genotype for strain (AF)b:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| SC5314 | SC5314a | SC5314α | AF6 | AF7 | 7-3791d | 6-4191e | 6-4205e | ||
| 1 | C2F7 | a1 | a1 | a1 | a1 | a1 | a1 | a1 | a1 |
| 2 | 1855/2172 | a1/a2 (0.51) | a2 (0.82) | a1 (0.30) | a1/a2 (0.43) | a1/a2 (0.49) | a1/a2 (0.51) | a1/a2 (0.42) | a1/a2 (0.48) |
| 3 | 1899/2008 | a1/a2 (0.50) | a1 (0.26) | a2 (0.87) | a1/a2 (0.41) | a1/a2 (0.47) | a1/a2 (0.46) | a1/a2 (0.59) | a1/a2 (0.50) |
| 4 | SNF1 | a1/a2 (0.52) | a1 (0.11) | a2 (0.74) | a1/a2 (0.52) | a1/a2 (0.50) | a1/a2 (0.44) | a1/a2 (0.54) | a1/a2 (0.49) |
| 5 | HST3 | a1/a2 (0.51) | a1 (0.09) | a2 (0.96) | a1/a2 (0.50) | a1/a2 (0.52) | a1/a2 (0.49) | a2 (0.64) | a1/a2 (0.51) |
| 6 | 1445/2395 | a1/a2 (0.51) | a2 (0.65) | a1 (0.36) | a1/a2 (0.46) | a1/a2 (0.49) | a1/a2 (0.49) | a1/a2 (0.45) | a1/a2 (0.49) |
| 7 | 1922/2344 | a1/a2 (0.49) | a1 (0.32) | a2 (0.77) | a1/a2 (0.50) | a1/a2 (0.50) | a1/a2 (0.51) | a1/a2 (0.59) | a1/a2 (0.50) |
| 8 | PDE1 | a1/a2 (0.50) | a2 (0.93) | a1 (0.16) | a1/a2 (0.48) | a1/a2 (0.50) | a1/a2 (0.51) | a1 (0.37) | a1/a2 (0.51) |
| 9 | 1969/2162 | a1/a2 (0.50) | a1 (0.16) | a2 (0.87) | a1/a2 (0.56) | a1/a2 (0.53) | a1/a2 (0.56) | a2 (0.64) | a1/a2 (0.50) |
| 10 | HEX1c | a1 | a1 | a1 | a1 | a1 | a1 | a1 | a1 |
| 11 | DPH5 | a1/a2 (0.56) | a2 (0.74) | a1 (0.06) | a1/a2 (0.53) | a1/a2 (0.53) | a1/a2 (0.57) | a1 (0.36) | a1/a2 (0.56) |
| 13 | 1817/2082 | a1/a2 (0.50) | a1 (0.03) | a2 (0.97) | a1/a2 (0.46) | a1/a2 (0.50) | a1/a2 (0.46) | a2 (0.61) | a1/a2 (0.50) |
| 14 | 1341/2493 | a1/a2 (0.51) | a1 (0.24) | a2 (0.92) | a1/a2 (0.52) | a1/a2 (0.52) | a1/a2 (0.46) | a2 (0.62) | a1/a2 (0.51) |
| 15 | 2340/2493 | a1/a2 (0.48) | a2 (0.96) | a1 (0.10) | a1/a2 (0.49) | a1/a2 (0.52) | a1/a2 (0.52) | a1 (0.37) | a1/a2 (0.53) |
| 16 | B15B20 | a1/a2 (0.48) | a1/a2 (0.53) | a1/a2 (0.52) | a1/a2 (0.46) | a1/a2 (0.49) | a1/a2 (0.48) | a1/a2 (0.48) | a1 (0.35) |
| 17 | LEU2c | a1 | a1 | a1 | a1 | a1 | a1 | a1 | a1 |
| 18 | YDL114 | a1/a2 (0.49) | a1/a2 (0.49) | a1/a2 (0.48) | a1/a2 (0.51) | a1/a2 (0.51) | a1/a2 (0.51) | a1/a2 (0.50) | a1/a2 (0.48) |
| 19 | YKL084 | a1/a2 (0.53) | a1/a2 (0.49) | a1/a2 (0.49) | a1 (0.02) | a1 (0.02) | a1 (0.02) | a1 (0.05) | a1 (0.03) |
| 20 | 2397/2498 | a1/a2 (0.52) | a1/a2 (0.50) | a1/a2 (0.50) | a1 (0.05) | a1 (0.05) | a1 (0.05) | a1 (0.08) | a1 (0.06) |
| 21 | RBP1 | a1/a2 (0.51) | a1/a2 (0.50) | a1/a2 (0.49) | a1/a2 (0.51) | a1/a2 (0.50) | a1/a2 (0.50) | a1/a2 (0.50) | a1/a2 (0.51) |
| 22 | YJL54/2 | a1/a2 (0.53) | a1/a2 (0.46) | a1/a2 (0.49) | a1/a2 (0.43) | a1/a2 (0.50) | a1/a2 (0.42) | a1/a2 (0.48) | a1/a2 (0.50) |
| 23 | 6.1345 | a1/a2 (0.51) | a1/a2 (0.52) | a1/a2 (0.46) | a1/a2 (0.51) | a1/a2 (0.50) | a1/a2 (0.51) | a1/a2 (0.50) | a1/a2 (0.45) |
| 25 | 6.282 | a1/a2 (0.52) | a1/a2 (0.51) | a1/a2 (0.54) | a1/a2 (0.48) | a1/a2 (0.51) | a1/a2 (0.48) | a1/a2 (0.50) | a1/a2 (0.46) |
Numbering corresponds to the locations of SNPs on the SNP map.
AF values were averaged over a minimum of eight spots (two independent experiments) per strain.
Signal only obtained for homozygous allele; therefore, no calculation of a1/a2 ratios was performed.
Strain after mouse passage in AF7 background.
Strain after mouse passage in AF6 background. Representative SNP genotypes for 19 strains with no LOH.
TABLE 4.
Chromosome 5 haplotypes for strain SC5314 at 15 SNP loci
| SNP map locationa | SNP locus | haplotypeb
|
|
|---|---|---|---|
| a1d | a2 | ||
| 1 | C2F7 | t | t |
| 2 | 1855/2172 | t | c |
| 3 | 1899/2008 | c | t |
| 4 | SNF1 | a | c |
| 5 | HST3 | c | t |
| 6 | 1445/2395 | t | c |
| 7 | 1922/2344 | c | t |
| 8 | PDE1 | t | c |
| 9 | 1969/2162 | a | t |
| 10 | HEX1 | g | g |
| 11 | DPH5 | g | a |
| 12 | 2093/2390c | c | t |
| 13 | 1817/2082 | c | t |
| 14 | 1341/2493 | a | g |
| 15 | 2340/2493 | g | a |
Numbering corresponds to the locations of SNPs on the SNP map (Fig. 3).
Haplotypes were obtained from sorbose-derived strains SC5314a (a1) and SC5314α (a2).
Haplotypes were confirmed by sequencing (data not shown).
Numbering of alleles (a1 versus a2) was random.
FIG. 2.
Cluster diagram showing SNP genotypes (see also Table 3) for strains SC5314, SC5314a, and SC5314α. The AF (see Materials and Methods) (x axis) is plotted against the square root (for scaling purposes only) of the total fluorescent signal (y axis). Each data point in the diagram represents results averaged over 4 (SC5314), 16 (SC5314a), and 14 (Sc5314α) independent experiments. Vertical lines represent the AF cutoff values for genotype determination: homozygous a1 < 0.4; homozygous a2 > 0.6; heterozygous alleles, 0.4 < a1 and a2 < 0.6. Values of 0.4 and 0.6 were scored as ambiguous.
Since all the strains that were analyzed in this study were derived from strain SC5314, there are no homozygous strains to test SNP loci on chromosome 6 and 7 for correct genotype determination for a1 and a2. One way to confirm the homozygous genotypes for these markers would be to clone both alleles for each marker and generate probes from these clones for hybridization to the SNP array. This method is routinely used for haplotype studies of diploid yeast (44, 49). However, with increasing number of SNPs on the SNP array, this procedure would become expensive and time consuming. Another way to test for accurate genotype determination would be to hybridize probes of strains from different genetic backgrounds which are known to carry homozygous alleles. Preliminary results for a limited number of clinical isolates demonstrate correct determination of one of the homozygous alleles at the majority of SNPs on chromosomes 5, 6, and 7 (data not shown).
Cluster or scatter plot analysis has become the method of choice for genotype assignment and determination of cutoff values for the three allelic genotypes (13, 27, 33, 34). Tight clusters indicate good genotype discrimination, and the larger the distance between the clusters the better is the discrimination between homozygous and heterozygous genotypes (27). To determine the cutoff values for our data and to simultaneously test the reproducibility of our SNP array method, strains SC5314, SC5314a, and SC5314α were hybridized in competition with the reference control (SC5314) multiple times. The cutoff values for AF for our data set (see Materials and Methods) differ from the results seen with other studies (1, 19, 33, 37). For example, O'Meara and colleagues developed an SNP microarray method based on apyrase-mediated PE. When this method was used, AFs between 0.29 and 0.71 were scored as heterozygous whereas fractions ≤0.2 and ≥0.8 were scored as homozygous (33). There are two differences between these experiments which may explain the discrepancies. One is that different SNP array technologies were used. The second is that the sizes of the data sets for the determination of the cutoff values differed between the studies. As our data set increases, the cutoff values are constantly recalculated for the highest accuracy in genotype determination. We note that all genotypes determined by SNP microarray analysis were confirmed by sequencing, which further supports the discrimination power of SNP array hybridization.
SNP microarray analysis.
Using the SNP microarray we analyzed both the GAL1/gal1 parental strains AF6 and AF7 for the SNPs found in SC5314. Both parents differed from the standard strain in that they were homozygous for two loci, YKL084 and 2387/2498, on chromosome fragment 7F (Table 3). We found that these same two loci were homozygous in strains CAF-2 (which was derived from SC5314 (14) and CAI4 (derived from CAF-2), the immediate parent of AF6 and AF7 (data not shown). A survey of samples of SC5314 strains obtained from several laboratories showed that these two loci were frequently homozygous. This result indicates that strain SC5314 maintained in different laboratories may become homozygous at loci that are heterozygous in the sequenced strain SC5314.
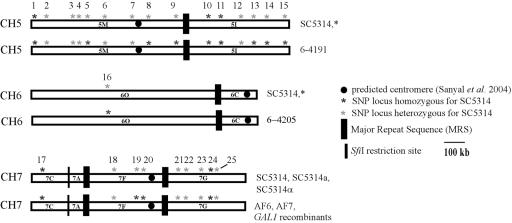
A total of 21 strains derived from AF6 and AF7 after passaging through mice were identified as 2-deoxygalactose resistant (17). LOH at the GAL1 locus which is located on chromosome 1 was confirmed. Of these strains, two (10%) exhibited additional LOH on chromosomes 5 and 6 in the SNP arrays. Strain 6-4205 became homozygous for SNP locus B15B20 on chromosomal fragment 6O, and strain 6-4191 became homozygous for loci HST3, PDE1, 1969/2162, DPH5, 1341/2493, and 2340/2493 on chromosome 5 (Table 3 and Fig. 3). In addition, like their parental strain AF6, both were homozygous for loci YKL084 and 2397/2498 on chromosomal fragment 7F (Table 3 and Fig. 3).
FIG. 3.
Chromosomal location of SNP loci and LOH events of strains 6-4191 and 6-4205 (map modified from Forche et al.) (15). Numbers above chromosomes correspond to SNP order in Table 3. Except for the strains indicated, all strains analyzed exhibited the same SNP genotype as strain SC5314. Two markers on chromosome 7, YKL084 and 2397/2498, are homozygous in CAI-4 and derivative strains.
Among the SNPs exhibiting LOH, the overall LOH frequency was 4.7% since the LOH event for a particular SNP locus was only observed once among the 21 strains analyzed. Since the GAL1 gene is located on chromosomal fragment 1S (http://www-sequence.stanford.edu:8080/haploid19.html), the detected LOH events arose independently in these two strains. LOH events for strain 6-4191 are likely due to gene conversion events, since LOH events along the chromosome are separated by regions which have retained heterozygosity (Fig. 3). Gene conversion has been observed in C. albicans grown both in vivo (16) and in vitro (46). For strain 6-4205, analysis of more SNPs along chromosome 6 is needed to determine the extent of LOH and to make predictions about possible mechanisms. It is important to point out that these strains were preselected by plating on 2-deoxygalactose for one recombination event, homozygosis of the gal1 gene. An important next step will be to determine the frequency of LOH in an unselected population isolated from a mouse.
Cowen and colleagues assayed LOH in several populations that were evolved in vitro in the presence of the antifungal drug fluconazole (11). They observed LOH for two of five markers tested for three populations. One locus that became homozygous in two populations was located in the PDE1 gene on chromosome 5. A second heterozygous locus present in PDE1 is represented on our SNP microarray. Interestingly, LOH for this SNP locus was detected here for strain 6-4191 (Table 3). PDE1 encodes a cyclic nucleotide phosphodiesterase in C. albicans (18).
Karyotype analysis.
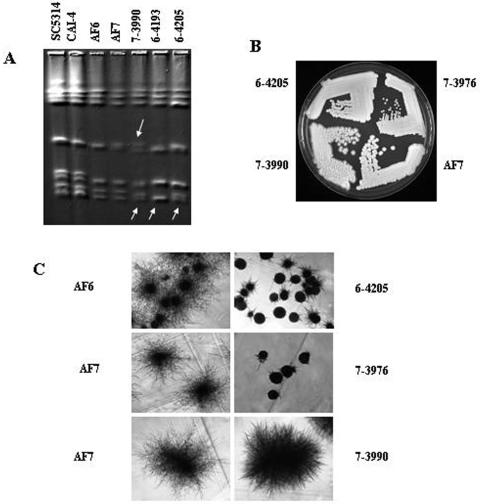
Major chromosomal rearrangements in the AF6 and AF7-derived GAL1 recombinant strains were assessed using pulsed-field gel electrophoresis and were detected for 3 of the 21 strains subjected to karyotypic analysis. Strains 6-4193, 6-4205, and 7-3990 showed size changes for one of the two homologs of chromosome 7 (Fig. 4A). In strains 7-3990 and 6-4205, the larger homolog of chromosome 7 decreased in size compared to the results seen with karyotypes of their parental strains. Strain 6-4193 had chromosome 7 homologs of equal sizes with only one visible band, whereas strains 7-3990 and 6-4205 still had two separable chromosome 7s (Fig. 4A). In addition to chromosome 7 changes, strain 7-3990 also exhibited size differences for the chromosome 5 homologs. In both the parent AF7 and the 7-3990 strains, the two homologs of chromosome 5 are of different sizes but are both visibly larger, although the size difference appears to be larger for strain 7-3990. Strain 6-4205 thus had two genomic changes, a LOH on chromosome 6 and a decrease in the size of chromosome 7. These changes would appear to be unrelated except that both presumably involve recombination. The chromosome 7 changes did not affect heterozygosity at the 7 loci examined in this study.
FIG. 4.
Karyotypes, colony morphology, and filamentation phenotypes of GAL1 recombinant strains. (A) Karyotypes of GAL1 recombinants that exhibit altered chromosomal patterns compared to those seen with the parental strains. Arrows point to chromosomal changes in strains 6-4193, 7-3990, and 6-4205. (B) Colony morphologies of strains 7-3976 and 7-3990 compared to that of strain AF7 on YEPD-uridine plates incubated at 30°C for 2 days. (C) Filamentation phenotypes at a magnification of ×40 for the GAL1 recombinants 6-4205, 7-3976, and 7-3990 compared to the results seen with the parental strains AF6 and AF7. Each set of recombinant and parent strains was streaked onto the same plate for accurate comparison.
Highly variable karyotypes have been observed among clinical isolates of C. albicans (2, 24, 28, 30, 36). Chromosomal rearrangements in vitro and in vivo in C. albicans have been reported previously (11, 24, 35, 38-40). Cowen and colleagues studied the evolution of drug resistance in experimental populations of C. albicans over 330 generations. They observed karyotypic rearrangements for chromosome R in the control populations (evolving without drug treatment) at generation 330. In three of nine populations evolving with drug treatment, chromosomal rearrangements were detected at different time points for several chromosomes, including chromosomes 4 and 5 (11). Rustchenko and coworkers observed chromosomal rearrangements on chromosome R for morphological mutants of C. albicans. They also demonstrated that gain- and loss-of-assimilation function is associated with alterations of chromosomes 2, 5, and 6 (38, 40). Legrand and coworkers detected chromosomal alterations for chromosomes 5, 6, and 7 (24) in a series of clinical isolates from a single patient. These results, together with our findings, indicate that chromosomal changes occur in C. albicans in different in vitro and in vivo environments and can be associated with phenotypic and genotypic alterations.
Phenotypic properties of AF6 and AF7 recombinant strains.
Three of 21 (14.3%) strains were found to have colony morphologies that differed compared to those of their parental strains. Strain 7-3976 had very small colonies, strain 6-4205 showed an intermediate colony size, and strain 7-3990 had a very rough colony morphology, with colonies invading the agar (see Fig. 4B). Strain 7-3976 exhibited a reduced growth rate compared to its parent strain AF7, consistent with the small-colony phenotype. This correlation was also observed for strain 6-4205, which had a growth rate intermediate between those of parental strain AF6 and strain 7-3976. Strain 6-3552 had an increased growth rate compared to parental strain AF6 (Table 5). All other strains had growth rates similar to those of the parental strains (data not shown). Filamentation assays revealed that strains 7-3976 and 6-4205 had a reduced filamentation ability under the conditions used here (Fig. 4C). Strain 3990 exhibited an altered filamentation phenotype, with apparent shorter but denser filaments (Fig. 4C). Filamentation has been shown to be important for invading the host tissue and is therefore an important virulence determinant (7, 32). At least three different pathways leading to the formation of filaments (pseudohyphae and true hypha) have been identified previously (12, 17, 23, 25). The regulation of these pathways overlaps. Therefore, the genetic events leading to the phenotypes observed will be difficult to identify (for an overview of morphogenesis pathways, see reference 25). Differences in colony morphology are frequently observed in C. albicans. They have been associated with chromosomal rearrangements (3, 41) and gene disruption (5, 6, 22, 47).
TABLE 5.
Summary of genotypic and phenotypic results for parental strains and GAL1 derivativesa
| Strain | LOH | Growth rate in YEPD broth (change)b | Colony morphology | Filamentation on serum | Karyotype |
|---|---|---|---|---|---|
| 7-3791 | − | − | − | − | − |
| 7-3972 | − | − | − | − | − |
| 7-3973 | − | − | − | − | − |
| 7-3974 | − | − | − | − | − |
| 7-3975 | − | − | − | − | − |
| 7-3976 | − | + (down) | + | + | − |
| 7-3977 | − | − | − | − | − |
| 7-3978 | − | − | − | − | − |
| 7-3979 | − | − | − | − | − |
| 7-3980 | − | − | − | − | − |
| 7-3981 | − | − | − | − | − |
| 7-3982 | − | − | − | − | − |
| 7-3983 | − | − | − | − | − |
| 7-3990 | − | − | + | + | + |
| 7-4118 | − | − | − | − | − |
| 7-4130 | − | − | − | − | − |
| 7-4164 | − | − | − | − | − |
| 6-3552 | − | + (up) | − | − | − |
| 6-4191 | + | − | − | − | − |
| 6-4193 | − | − | − | − | + |
| 6-4205 | + | + (down) | + | + | + |
“+” and “−” indicate change and no difference, respectively, compared to parental strain results.
up, increase in growth rate compared to parental strain results; down, decrease in growth rate compared to parental strain results.
Recently, SNP microarrays have been adapted for the analysis of gene copy number and to study differential allelic expression in humans (26, 31, 45, 50). These analyses would be very useful in C. albicans studies. Recent reports suggest that alterations in gene copy number (8) as well as differential expression of alleles of certain genes (43) are frequent in the C. albicans genome and that trisomy of chromosome 1 can lead to a decrease in virulence (8). The SNP microarray technology developed here has the potential to be modified to look not only at LOH but also at gene copy number and differential expression of alleles.
The role of mitotic recombination and genome rearrangement in adaptation to a host animal's environment is largely unknown. This study demonstrates that LOH and changes in morphological characteristics can be readily detected in C. albicans at measurable frequencies following infection. The development of a whole-genome SNP array presently under way will provide a high-throughput and accurate method for the detection of LOH across the entire genome and will enable the analysis of large numbers of strains of C. albicans. Using this SNP microarray for genome-wide analysis of strains from different sources (e.g., strains from different host niches, serial isolates of patients, and strains that underwent host passaging) in combination with phenotypic screening should reveal genomic regions or individual genes that are potential players in the adaptation to the host.
Acknowledgments
We thank B. B. Magee and Judy Berman for critical and helpful comments on the manuscript. We thank Julian Naglik for providing SC5314 strains from different laboratories and strain CAF-2.
Research was supported by grant AI46351 awarded to P. T. Magee and G. May.
REFERENCES
- 1.Ahmadian, A., B. Gharizadeh, A. C. Gustafsson, F. Sterky, P. Nyren, M. Uhlen, and J. Lundeberg. 2000. Single-nucleotide polymorphism analysis by pyrosequencing. Anal. Biochem. 280:103-110. [DOI] [PubMed] [Google Scholar]
- 2.Asakura, K., S. Iwaguchi, M. Homma, T. Sukai, K. Higashide, and K. Tanaka. 1991. Electrophoretic karyotypes of clinically isolated yeasts of Candida albicans and C. glabrata. J. Gen. Microbiol. 137:2531-2538. [DOI] [PubMed] [Google Scholar]
- 3.Barton, R. C., and S. Scherer. 1994. Induced chromosome rearrangements and morphologic variation in Candida albicans. J. Bacteriol. 176:756-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, R. J., and A. D. Johnson. 2003. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 22:2505-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105-109. [DOI] [PubMed] [Google Scholar]
- 6.Brega, E., R. Zufferey, and C. B. Mamoun. 2004. Candida albicans Csy1p is a nutrient sensor important for activation of amino acid uptake and hyphal morphogenesis. Eukaryot. Cell 3:135-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calderone, R. A., and W. A. Fonzi. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9:327-335. [DOI] [PubMed] [Google Scholar]
- 8.Chen, X., B. B. Magee, J. Dawson, P. T. Magee, and C. A. Kumamoto. 2004. Chromosome 1 trisomy compromises the virulence of Candida albicans. Mol. Microbiol. 51:551-565. [DOI] [PubMed] [Google Scholar]
- 9.Cho, R. J., M. Mindrinos, D. R. Richards, R. J. Sapolsky, M. Anderson, E. Drenkard, L. Dewdney, T. L. Reuber, M. Stammers, N. Federspiel, A. Theologis, W. H. Yang, E. Hubbell, M. Au, E. Y. Chung, D. Lashkari, B. Lemieux, C. Dean, R. J. Lipshutz, F. M. Ausubel, R. W. Davis, and P. J. Oefner. 1999. Genome-wide mapping with biallelic markers in Arabidopsis thaliana. Nat. Genet. 23:203-207. [DOI] [PubMed] [Google Scholar]
- 10.Chu, W.-S., B. B. Magee, and P. T. Magee. 1993. Construction of an SfiI macrorestriction map of the Candida albicans genome. J. Bacteriol. 175:6637-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cowen, L. E., D. Sanglard, D. Calabrese, C. Sirjusingh, J. B. Anderson, and L. M. Kohn. 2000. Evolution of drug resistance in experimental populations of Candida albicans. J. Bacteriol. 182:1515-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, D., R. B. Wilson, and A. P. Mitchell. 2000. RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan, J. B., X. Q. Chen, M. K. Halushka, A. Berno, X. H. Huang, T. Ryder, R. J. Lipshutz, D. J. Lockhart, and A. Chakravarti. 2000. Parallel genotyping of human SNPs using generic high-density oligonucleotide tag arrays. Genome Res. 10:853-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forche, A., P. T. Magee, B. B. Magee, and G. May. 2004. Genome-wide single-nucleotide polymorphism map for Candida albicans. Eukaryot. Cell 3:705-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forche, A., G. May, J. Beckerman, S. Kauffman, J. Becker, and P. T. Magee. 2003. A system for studying genetic changes in Candida albicans during infection. Fungal Genet. Biol. 39:38-50. [DOI] [PubMed] [Google Scholar]
- 17.Fu, Y., A. Ibrahim, D. Sheppard, Y.-C. Chen, S. French, J. Cutler, S. Filler, and J. Edwards, Jr. 2002. Candida albicans Als1p: an adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol. Microbiol. 61-72. [DOI] [PubMed]
- 18.Hoyer, L. L., L. B. Cieslinski, M. M. McLaughlin, T. J. Torphy, A. R. Shatzman, and G. P. Livi. 1994. A Candida albicans cyclic nucleotide phosphodiesterase: cloning and expression in Saccharomyces cerevisiae and biochemical characterization of the recombinant enzyme. Microbiology 140:1533-1542. [DOI] [PubMed] [Google Scholar]
- 19.Huber, M., A. Muendlein, E. Dornstauder, C. Schneeberger, C. B. Tempfer, M. W. Mueller, and W. M. Schmidt. 2002. Accessing single nucleotide polymorphisms in genomic DNA by multiplex polymerase chain reaction amplification of oligonucleotide microarrays. Anal. Biochem. 303:25-33. [DOI] [PubMed] [Google Scholar]
- 20.Hull, C. M., R. M. Raisner, and A. D. Johnson. 2000. Evidence for mating of the “Asexual” yeast Candida albicans in a mammalian host. Science 289:307-310. [DOI] [PubMed] [Google Scholar]
- 21.Jones, T., N. Federspiel, H. Chibana, J. Dungan, S. Kalman, B. B. Magee, G. Newport, R. Thorstenson, N. Agabian, P. T. Magee, R. W. Davis, and S. Scherer. 2004. The diploid genome of Candida albicans. Proc. Natl. Acad. Sci. USA 101:7329-7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krueger, K. E., A. K. Ghosh, B. P. Krom, and R. L. Cihlar. 2004. Deletion of the NOT4 gene impairs hyphal development and pathogenicity in Candida albicans. Microbiology 150:229-240. [DOI] [PubMed] [Google Scholar]
- 23.Lane, S., C. Birse, S. Zhou, R. Matson, and H. Liu. 2001. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 276:48988-48996. [DOI] [PubMed] [Google Scholar]
- 24.Legrand, M., P. Lepart, A. Forche, F.-M. C. Mueller, T. J. Walsh, P. T. Magee, and B. B. Magee. 2004. Homozygosity at the MTL locus in clinical strains of Candida albicans: karyotypic rearrangements and tetraploid formation. Mol. Microbiol. 52:1451-1462. [DOI] [PubMed] [Google Scholar]
- 25.Liu, H. 2001. Transcriptional control of dimorphism in Candida albicans. Curr. Opin. Microbiol. 4:728-735. [DOI] [PubMed] [Google Scholar]
- 26.Lo, H. S., Z. N. Wang, Y. Hu, H. H. Yang, S. Gere, K. H. Buetow, and M. P. Lee. 2003. Allelic variation in gene expression is common in the human genome. Genome Res. 13:1855-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lovmar, L., M. Fredriksson, U. Liljedahl, S. Sigurdsson, and A.-C. Syvanen. 2003. Quantitative evaluation by minisequencing and microarrays reveals accurate multiplexed SNP genotyping of whole genome amplified DNA. Nucleic Acids Res. 31:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lupetti, A., G. Guzzi, A. Paladini, K. Swart, M. Campa, and S. Senesi. 1995. Molecular typing of Candida albicans in oral candidiasis: karyotype epidemiology with human immunodeficiency virus-seropositive patients in comparison with that with healthy carriers. J. Clin. Microbiol. 33:1238-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magee, B. B., and P. T. Magee. 2000. Induction of mating in Candida albicans by construction of MTL a and MTL alpha strains. Science 289:310-313. [DOI] [PubMed] [Google Scholar]
- 30.Magee, P. T., L. Bowdin, and J. Staudinger. 1992. Comparison of molecular typing methods for Candida albicans. J. Clin. Microbiol. 30:2674-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mei, R., P. C. Galipeau, C. Prass, A. Berno, G. Ghandour, N. Patil, R. K. Wolff, M. S. Chee, B. J. Reid, and D. J. Lockhart. 2000. Genome-wide detection of allelic imbalance using human SNPs and high-density DNA arrays. Genome Res. 10:1126-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell, A. P. 1998. Dimorphism and virulence in Candida albicans. Curr. Opin. Microbiol. 1:687-692. [DOI] [PubMed] [Google Scholar]
- 33.O'Meara, D., A. Ahmadian, J. Odeberg, and J. Lundeberg. 2002. SNP typing by apyrase-mediated allele-specific primer extension on DNA microarrays. Nucleic Acids Res. 30:e75. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pastinen, T., M. Raitio, K. Lindroos, P. Tainola, L. Peltonen, and A.-C. Syvaenen. 2000. A system for specific, high-throughput genotyping by allele-specific primer extension on microarrays. Genome Res. 10:1031-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perepnikhatka, V., F. J. Fischer, M. Niimi, R. A. Baker, R. D. Cannon, Y. K. Wang, F. Sherman, and E. Rustchenko. 1999. Specific chromosome alterations in fluconazole-resistant mutants of Candida albicans. J. Bacteriol. 181:4041-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfaller, M. A., J. Rhine-Chalberg, S. W. Redding, J. Smith, G. Farinacci, A. W. Fothergill, and M. G. Rinaldi. 1994. Variations in fluconazole susceptibility and electrophoretic karyotype among oral isolates of Candida albicans from patients with AIDS and oral candidiasis. J. Clin. Microbiol. 32:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Primdahl, H., F. P. Wikman, H. von der Maase, X.-G. Zhou, H. Wolf, and T. F. Orntorf. 2002. Allelic imbalances in human bladder cancer: genome-wide detection with high-density single-nucleotide polymorphism arrays. J. Natl. Cancer Inst. 94:216-223. [DOI] [PubMed] [Google Scholar]
- 38.Rustchenko, E. P., D. H. Howard, and F. Sherman. 1994. Chromosomal alterations of Candida albicans are associated with the gain and loss of assimilating functions. J. Bacteriol. 176:3231-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rustchenko, E. P., D. H. Howard, and F. Sherman. 1997. Variation in assimilating functions occurs in spontaneous Candida albicans mutants having chromosomal alterations. Microbiology 143:1765-1778. [DOI] [PubMed] [Google Scholar]
- 40.Rustchenko-Bulgac, E. P., and D. H. Howard. 1993. Multiple chromosomal and phenotypic changes in spontaneous mutants of Candida albicans. J. Gen. Microbiol. 139:1195-1207. [DOI] [PubMed] [Google Scholar]
- 41.Rustchenko-Bulgac, E. P., F. Sherman, and J. B. Hicks. 1990. Chromosomal rearrangements associated with morphological mutants provide a means for genetic variation of Candida albicans. J. Bacteriol. 172:1276-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a. Sanyal, K., M. Baum, J. Carbon. 2004. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc. Natl. Acad. Sci. USA 101:11374-11379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmid, K. J., T. R. Sorensen, R. Stracke, O. Torjek, T. Altmann, T. Mitchell-Olds, and B. Weisshaar. 2003. Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res. 13:1250-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staib, P., M. Kretschmar, T. Nichterlein, H. Hof, and J. Morschhauser. 2002. Host versus in vitro signals and intrastrain allelic differences in the expression of a Candida albicans virulence gene. Mol. Microbiol. 44:1351-1366. [DOI] [PubMed] [Google Scholar]
- 44.Tavanti, A., N. A. Gow, M. C. J. Maiden, F. Odds, and D. J. Shaw. 2004. Genetic evidence for recombination in Candida albicans based on haplotype analysis. Fungal Genet. Biol. 41:553-562. [DOI] [PubMed] [Google Scholar]
- 45.Tong, B. C., K. Dhir, P. K. Ha, W. H. Westra, B. P. Alter, D. Sidransky, W. M. Koch, and J. A. Califano. 2004. Use of single nucleotide polymorphism arrays to identify a novel region of loss on chromosome 6Q in squamous cell carcinomas of the oral cavity. Head Neck 26:345-352. [DOI] [PubMed] [Google Scholar]
- 46.Tsang, P. W., B. Cao, P. Y. Siu, and J. Wang. 1999. Loss of heterozygosity, by mitotic gene conversion and crossing over, causes strain-specific adenine mutants in constitutive diploid Candida albicans. Microbiology 145:1623-1629. [DOI] [PubMed] [Google Scholar]
- 47.Uhl, M. A., M. Biery, N. Craig, and A. D. Johnson. 2003. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C. albicans. EMBO J. 22:2668-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, D. G., J. B. Fan, C. J. Siao, A. Berno, P. Young, R. Sapolsky, G. Ghandour, N. Perkins, E. Winchester, J. Spencer, L. Kruglyak, L. Stein, L. Hsie, T. Topaloglou, E. Hubbell, E. Robinson, M. Mittmann, M. S. Morris, N. P. Shen, D. Kilburn, J. Rioux, C. Nusbaum, S. Rozen, T. J. Hudson, R. Lipshutz, M. Chee, and E. S. Lander. 1998. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280:1077-1082. [DOI] [PubMed] [Google Scholar]
- 49.Xu, J., and T. G. Mitchell. 2003. Comparative gene genealogical analyses of strains of serotype AD identify recombination in populations of serotypes A and D in the human pathogenic yeast Cryptococcus neoformans. Microbiology 149:2147-2154. [DOI] [PubMed] [Google Scholar]
- 50.Zhao, X., C. Li, J. G. Paez, K. Chin, P. A. Jaenne, T.-H. Chen, L. Girard, J. Minna, D. Christiani, C. Leo, J. W. Gray, W. R. Sellers, and M. Meyerson. 2004. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 64:3060-3071. [DOI] [PubMed] [Google Scholar]