Abstract
Cryptococcus neoformans is an opportunistic human fungal pathogen that elaborates several virulence attributes, including a polysaccharide capsule and melanin pigments. A conserved Gα protein/cyclic AMP (cAMP) pathway controls melanin and capsule production. To identify targets of this pathway, we used an expression profiling approach to define genes that are transcriptionally regulated by the Gα protein Gpa1. This approach revealed that Gpa1 transcriptionally regulates multiple genes involved in capsule assembly and identified two additional genes with a marked dependence on Gpa1 for transcription. The first is the LAC1 gene, encoding the laccase enzyme that catalyzes a rate-limiting step in diphenol oxidation and melanin production. The second gene identified (LAC2) is adjacent to the LAC1 gene and encodes a second laccase that shares 75% nucleotide identity with LAC1. Similar to the LAC1 gene, LAC2 is induced in response to glucose deprivation. However, LAC2 basal transcript levels are much lower than those for LAC1. Accordingly, a lac2 mutation results in only a modest delay in melanin formation. LAC2 overexpression suppresses the melanin defects of gpa1 and lac1 mutants and partially restores virulence of these strains. These studies provide mechanistic insights into the regulation of capsule and melanin production by the C. neoformans cAMP pathway and demonstrate that multiple laccases contribute to C. neoformans melanin production and pathogenesis.
Cryptococcus neoformans is a human fungal pathogen that primarily infects immunocompromised hosts. In order to establish an infection, pathogenic microorganisms such as C. neoformans must sense host-specific signals and respond with specific adaptive cellular responses that allow their survival in this hostile environment. Accordingly, C. neoformans requires the induction of several factors to be fully virulent. These include an antiphagocytic polysaccharide capsule (19, 27) and production of antioxidant melanin pigments (26).
The importance of capsule and melanin in C. neoformans infections has been studied extensively. The expression of each is induced in response to environmental signals encountered in the host during an infection. For example, capsule formation is induced by severe iron deprivation (55), mammalian physiologic concentrations of CO2/HCO3− (19), or serum (61). Melanin production requires glucose deprivation and the presence of diphenolic substrates, such as catecholamines (39).
Melanins represent a group of dark pigments present in a variety of fungal species. Plant fungal pathogens, such as Magnaporthe grisea, produce melanin in specialized structures known as appresoria to generate the physical pressure required to penetrate plant cells and establish an infection (32). Melanin formation in human fungal pathogens has also been linked to pathogenicity. For example, melanin-deficient mutants of the dematiaceous mold Wangiella dermatitidis are avirulent in animal models compared to wild-type controls (15).
In Cryptococcus neoformans, melanin-deficient mutant strains are also attenuated for virulence (26). Therefore, several investigators have focused on defining the biochemical steps involved in melanin biosynthesis and the potential roles for melanin in the pathogenesis of cryptococcal infections. The synthesis of melanin in C. neoformans involves the oxidation of phenolic substrates into quinones, which then polymerize nonenzymatically into pigmented products (59). The LAC1 gene encoding an enzyme catalyzing the rate-limiting oxidation step in melanin biosynthesis was cloned, and its product was identified as a laccase (59). Laccases are known to have diphenol oxidase activity and utilize a wide array of substrates, oxidizing polyaminobenzenes and mono- or polyphenolic compounds. Diphenol oxidase activity is found in strains producing melanin and is absent in melanin-deficient strains of C. neoformans (59). Strains with a lac1 mutation are attenuated for virulence in animal models but are eventually able to establish a lethal infection (46). Also, lac1 mutant strains can produce melanin after prolonged incubation on melanin-inducing media. Therefore, Lac1 plays a central role in catecholamine oxidation and melanin production, but other proteins or enzymes may function together with this enzyme in melanin biosynthesis.
Studies with C. neoformans reveal that a conserved Gα protein-cyclic AMP (cAMP) pathway regulates the induction of capsule and melanin in response to environmental stimuli detected in the host. The Gpa1-cAMP signaling cascade is conserved in yeast and mammals; however, the functions served by this pathway differ markedly between organisms. In the nonpathogenic fungus Saccharomyces cerevisiae, for example, this cascade plays central roles in filamentation, sporulation, and stress survival (4, 41, 45). In a similar manner, the human fungal pathogen C. neoformans utilizes cAMP signaling to regulate morphological transitions, but it has also coopted this cascade for the regulation of cellular determinants involved in virulence. The conserved components of a cAMP-signaling cascade have been characterized and involve genes encoding the Gα protein (Gpa1), adenylyl cyclase (Cac1), and protein kinase A catalytic subunit (Pka1). Mutant strains lacking these components do not increase capsule or melanin production in response to normal inducing conditions (2, 3, 17). However, little is known about the molecular mechanisms employed by cAMP to induce the expression of these virulence determinants. Here, we used a genomic microarray to identify genes whose expression is dependent upon Gpa1 to define downstream targets of this signal transduction pathway. Using this approach, we demonstrate that the cAMP pathway controls capsule and melanin production by regulating the expression of multiple genes at the transcriptional level. Additionally, we identify and characterize a second gene encoding a laccase homolog involved in melanin production in C. neoformans.
MATERIALS AND METHODS
Strains and media.
The strains used are listed in Table 1. All strains in this study were derivatives of the serotype A wild-type strain H99 (42), with the exception of those in the first microarray experiment, which was performed using the wild-type strain JEC21 and the gpa1 mutant strain BAC21 (54).
TABLE 1.
Strain list
| Strain | Genotype | Reference or source |
|---|---|---|
| H99 | MATα wild-type | 42 |
| AAC51 | MATα ura5 | 3 |
| AAC1 | MATα ade2 gpa1::ADE2 | 2 |
| AAC4 | MATα ade2 gpa1::ADE2 ura5 | This study |
| RPC26 | MATα lac2::neo | This study |
| RPC27 | ||
| RPC28 | ||
| QGC4 | MATα lac2::neo pGPD-LAC2 | This study |
| QGC5 | ||
| QGC6 | ||
| MDJ12 | MATα lac1::nat1 | This study |
| MDJ13 | ||
| MDC16 | MATα ura5 lac1::URA5 | This study |
| MDC17 | ||
| RPC29 | MATα ura5 lac1::URA5 lac2::neo | This study |
| RPC30 | ||
| QGC8 | MATα lac1::nat1 lac2::neo | This study |
| QGC9 | ||
| RPC18 | MATα ade2 gpa1::ADE2 pGPD-LAC2 | This study |
| RPC19 | ||
| RPC20 | ||
| RPC21 | MATα pGPD-LAC2 | This study |
| RPC22 | ||
| RPC23 | ||
| QGC1 | MATα ura5 lac1::URA5 pGPD-LAC2 | This study |
| QGC2 | ||
| QGC3 |
Standard yeast media were used as described previously (49). Niger seed extract and agar were also used (25). As indicated, media were supplemented with 10 mM epinephrine, 0.5 mM dopamine, or 100 μM 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS).
RNA preparation.
The isogenic GPA1 wild-type and gpa1 mutant strains were grown for 24 h at 30°C in synthetic complete (SC) medium with 2% glucose. Strains were pelleted, resuspended in SC medium with 2% glucose, and incubated for 3 h at 30°C. The cells were pelleted and resuspended in either SC medium with 2% glucose, SC with 0% glucose, or SLAD (synthetic low ammonium dextrose) medium. Aliquots were collected by centrifugation at 0, 1, and 2 h and flash frozen on dry ice. Total RNA was extracted from lyophilized cells by using the TRIzol reagent (Invitrogen Life Technologies, Carlsbad, Calif.).
cDNA synthesis and labeling.
Fluorescently labeled cDNA was made by incorporating amino-allyl dUTP during reverse transcription of 10 μg of total RNA. Cy3 or Cy5 fluorescent dyes (Amersham, Piscataway, N.J.) were coupled to the amino-allyl group as previously described (14). We generated a reference sample by pooling an equal amount of RNA from each time point from both strains, converted to cDNA and labeled with Cy3. RNA from each time point was individually labeled with Cy5 and competitively hybridized against the reference sample.
Microarray hybridization and analysis.
The genomic DNA microarray construction was described in detail previously (23). Briefly, 6,144 PCR products were amplified from a 1.6- to 3.2-kb genomic library made with strain H99 genomic DNA and were printed on polylysine-coated glass slides. Slides were prehybridized at 42°C with 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate, and 1% bovine serum albumin, and hybridizations were performed at 42°C with 1× hybridization buffer (50% formamide, 5× SSC, 0.1% sodium dodecyl sulfate). Arrays were scanned with a GenePix 4000B scanner (Axon Instruments, Foster City, Calif.) and analyzed by using GenePix Pro v4.0. Further data analysis was performed with CryptoArray, a Microsoft Excel macro for normalizing and formatting data. The smaller microarray was constructed in a similar manner by printing PCR products from 111 genes from strain JEC21 on polylysine-coated slides and hybridizing, as described above, with fluorescently labeled cDNA from the wild-type strain JEC21 and the gpa1 mutant strain BAC21.
Creation of lac2, lac1, and lac1 lac2 mutants.
To create the lac2::neo disruption allele, the PCR overlap extension method was used as previously described (11). The left fragment was amplified from H99 genomic DNA, using primers AAO304 and AAO306. The right fragment was amplified from H99 genomic DNA, using primers AAO307 and AAO309. The central fragment, containing the neomycin-resistance selectable marker, was amplified from plasmid pJAF1, using primers AAO305 and AAO308. The three PCR amplicons were used collectively as the template for a final PCR with primers AAO304 and AAO309 to generate the 4.0-kb lac2::neo disruption construct. The lac1::URA5 and lac1::nat disruption constructs were similarly created by the PCR overlap method. The constructs were designed such that the neo and the URA5 genes completely replaced the LAC2 and LAC1 genes, respectively, from start to stop codon.
The disruption constructs were used to biolistically transform C. neoformans strains as previously described for gene disruption (52). The lac2::neo and lac1::nat constructs were transformed into strain H99 to generate the lac2 mutant strains RPC26, RPC27, and RPC28 and the lac1 mutant strains MDJ12 and MDJ13. The lac2::neo construct was transformed into the lac1 mutant strain MDJ12 to create the lac1 lac2 double-mutant strain QGC9.
To screen for the lac2 mutation, genomic DNA from each transformant was isolated and used in a PCR with the LAC2-specific primer AAO303 and the neo-specific primer AAO247 (Table 2). Because the recognition sequence of primer AAO303 is outside of the disruption construct, only those transformants that have the disruption construct at the site of the endogenous LAC2 gene will amplify a 1.2-kb product. Three lac2::neo mutant strains and two lac1 lac2 double-mutant strains were identified in this manner. Each strain had phenotypes identical to those of the other genotypically identical mutant strains.
TABLE 2.
Primer list
| Primer | Purposea | Sequence (5′-3′) | Note | Annealing position |
|---|---|---|---|---|
| AAO304 | lac2::neo construct | GGTATCTGACGGCATTAGAAGG | 419 bp upstream of LAC2 start site | LAC2 (−419)-(−398) |
| AAO306 | PCR overlap | CGTGTTAATACAGATAAACCAAGGGT TAGCCTCTATCACAGGTCC | neo marker sequence underlined | LAC2 576-596 |
| AAO307 | PCR overlap | GCTCACATCCTCGCAGCAAGGGAGAA TGCCTGGACATCTCATGC | neo marker sequence underlined | LAC2 2743-2765 |
| AAO309 | lac2::neo construct | CTGCTCCTCTAGATCACTAACGTCAGG | Amplifies out of neo marker | |
| AAO305 | PCR overlap | GACCTGTGATAGAGGCTAACCCTTGG TTTATCTGTATTAACACGG | neo marker sequence underlined | LAC2 576-596 |
| AAO308 | PCR overlap | GCATGAGATGTCCAGGCATTCTCCCT TGCTGCGAGGATGTGAGC | neo marker sequence underlined | LAC2 2743-2765 |
| AAO303 | lac2Δ screen | ATCAGCTATATCACCTGTCAAGGC | Upstream of LAC2 locus | LAC2 (−598)-(−575) |
| AAO247 | neo marker screen | CGTTGAATCCTCAGGATCTTCATGGC | Amplifies out of neo marker | |
| AAO314 | LAC2 probe | CCGGATCCTCTGACACATTCACAACA ATGG | BamHI sequence underlined | LAC2 (−20)-4 |
| AAO315 | LAC2 probe | GCGGTACCGGACGAAGGTAATAGCA GAGAGTCAGG | KpnI sequence underlined | LAC2 3062-3088 |
| AAO463 | LAC2 cDNA | ACATCATATCTCTATCTTCAAGG | Primer sequence specific to LAC2 | LAC2 (−46)-(−24) |
| AAO534 | LAC2 cDNA | CAGGCCATTGAATCTTTTTG | Primer sequence specific to LAC2 | LAC2 2546-2565 |
| AAO570 | LAC2 qRT-PCR | TGTATGGCGCAAGGGGTTACT | Primer sequence specific to LAC2 | LAC2 1657-1677 |
| AAO571 | LAC2 qRT-PCR | AGAACACGACTCTCCAAAGC | Primer sequence specific to LAC2 | LAC2 1948-1967 |
| AAO301 | GPD qRT-PCR | AGTATGACTCCACACATGGTCG | Primer spans intron V of GPD | GPD 405-415; 471-481 |
| AAO302 | GPD qRT-PCR | AGACAAACATCGGAGCATCAGC | Primer spans intron VI of GPD | GPD 693-704; 764-773 |
qRT-PCR, quantitative reverse transcriptase PCR.
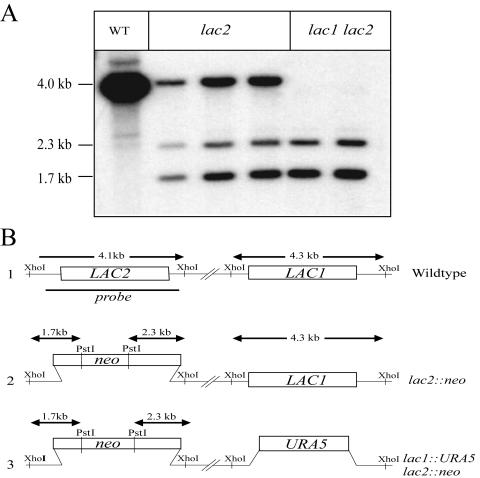
Southern hybridization was performed to confirm gene disruption with genomic DNA digested with PstI and XhoI and using as a probe a 3.1-kb fragment of the LAC2 gene created using primers AAO314 and AAO315. We observed the expected 2.3- and 1.7-kb band in both lac2 mutants as well as a 4.3-kb band corresponding to cross-hybridization at the LAC1 locus. This signal is less intense than the wild-type LAC2 signal at 4.1 kb and is not present in the lac1 lac2 double mutant, in which the coding region of LAC1 has been replaced with URA5.
Overexpression and reconstitution of the LAC2 gene.
Plasmid pRCD83 contains the sequence of the constitutively active promoter of the GPD gene and URA5 selectable marker (36, 58). We cloned the coding sequence of the LAC2 gene into this vector under control of the GPD promoter. The LAC2 gene was amplified from serotype A genomic DNA, using primers AAO314 and AAO315, which contain KpnI and BamHI restriction endonuclease sites. The PCR fragment was ligated into a KpnI- and BamHI-cut pRCD83 vector to create plasmid pRPW1 (pGPD-LAC2). The overexpression construct was biolistically transformed into AAC51 (ura5) and AAC4 (gpa1 ura5). To introduce this plasmid into the lac1 mutant and the lac2 mutant, the dominant selectable marker encoding nourseothricin resistance was subcloned into pRPW1. The 1.8-kb nourseothricin-resistance gene (31) was excised from plasmid pCH233 by using SpeI and XbaI and cloned into an XbaI-cut pRPW1. This new plasmid, pQDG1, was biolistically transformed into RPC32, creating strains QGC4, QGC5, and QGC6. Stable genomic integration of the constructs was documented by repeated culture of the transformants on a nonselective medium, followed by culturing of the strains on either synthetic medium lacking uracil (strains transformed with pRPW1) or yeast-peptone-dextrose (YPD) medium containing nourseothricin (strains transformed with pQDG1). Overexpression of the LAC2 gene in a wild-type strain, the lac1 mutant, the lac2 mutant, the lac1 lac2 mutant, and the gpa1 mutant was confirmed by Northern blot analysis.
Sequencing of LAC2 cDNA.
We amplified LAC2 cDNA from a cDNA library from strain H99 using primers designed to specifically amplify the LAC2 sequence over that of the highly similar LAC1 gene: primer AAO463 (begins at position −46 relative to the predicted start codon) and primer AAO534. The cDNA sequence confirmed predicted intron-exon borders.
Quantification of LAC2 transcript.
C. neoformans strains were incubated in YPD medium with 2% glucose to mid-log phase, pelleted, and resuspended in yeast nitrogen base medium (YNB) without added glucose (or other inducing conditions as listed in Results) for 1 h. The cells were pelleted and flash frozen on dry ice, and total RNA was isolated from these isolates by using TRIzol as previously described (3). The RNA was treated with RNase-free DNase, and cDNA was synthesized using oligo-dT primers from the SuperScript First Strand Synthesis RT kit (Invitrogen). The resulting cDNA was used as a template for quantitative real-time PCR with iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer's specifications. The iCycler iQ Multicolor real-time detection system was used as the fluorescence detector with the following PCR conditions: an initial denaturing cycle of 95°C for 3 min, 40 cycles of denaturation at 95°C for 20 s, and annealing and extension at 53°C for 45 s. These cycles were followed by a standard melt curve from 53 to 93°C with fluorescent monitoring each 0.5°C. These data confirmed the amplification of a single product for each primer pair and the lack of primer dimerization. Reactions were performed in triplicate, and the data were expressed as an average cycle threshold (CT) value, plus or minus standard error. The LAC2 primers used in this reaction (AAO570 and AAO571) amplify a 208-bp amplicon near the 3′ end of the posttranscriptionally modified cDNA. Standard PCRs were run with fivefold dilutions of the cDNA template to determine the optimal amount of template and optimal annealing temperature for the experimental and reference reactions, using a 500 nM concentration of each primer.
A validation curve was also calculated for each cDNA sample to provide an index of the template quality and quantity for each sample. The mean expression level for each gene in each sample was regressed against the overall mean of all samples. The slope provides an estimate of the degree to which the gene is efficiently amplified in the reaction, and applying this r2 value to the statistical evaluation for expression accounts for unpredictable variation between samples.
LAC2 amplification for each strain and condition was normalized against the constitutively expressed GPD gene. Degree of induction was calculated relative to induction for the wild-type strain H99, using the Bio-Rad iCycler software system, which utilizes the comparative CT statistical methods as previously described (54).
Northern analysis.
Total RNA was prepared as described above. Fifteen micrograms of total RNA was analyzed for each sample. Gel electrophoresis, RNA transfer, hybridization, and autoradiography were performed as described previously (47). The probes used for Northern analysis included the entire coding sequences of GPA1 (53), CAS1 (22), CAS2 (34), SMG1 (unpublished data), and ACT1 (10), amplified by PCR from a JEC21 cDNA library. To minimize potential cross-hybridization between the highly related LAC1 and LAC2 genes, we specifically chose as probes the extreme 3′ region of the coding sequences, since this includes the most dissimilar regions of the LAC1 and LAC2 genes. The LAC1 probe corresponded to nucleotides 1392 to 1872 of the LAC1 cDNA sequence, and the LAC2 probe corresponded to nucleotides 1019 to 1715 of the LAC2 cDNA sequence. Although we cannot exclude some degree of cross-hybridization of the LAC1 probe with the LAC2 signal, the absence of detectable LAC2 signal on several Northern blots (see Fig. 3C; also unpublished data) likely minimizes the effect of this possibility. The DNA for probes was labeled using a Random Primed DNA labeling kit (Boehringer Mannheim) and 32P-dCTP (Amersham).
FIG. 3.
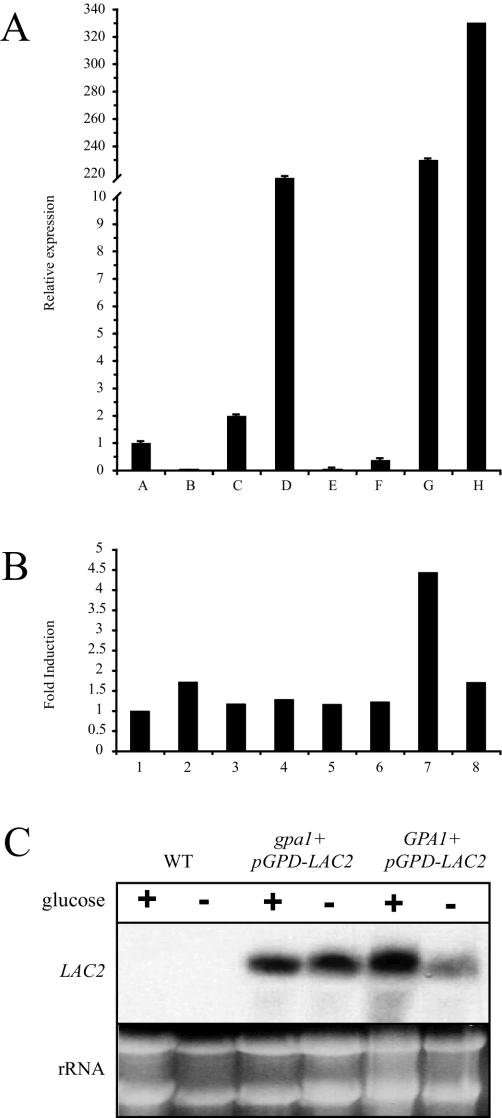
Measuring LAC2 transcript using real-time PCR and Northern analysis. (A) Real-time PCR was used to determine the relative transcript levels of LAC2 in each of the following strains after 1 h of glucose starvation: (A) wild-type (H99), (B) lac2 mutant (RPC27), (C) lac1 mutant (MDC16), (D) lac2+pGPD-LAC2 mutant (QGC1), (E) lac1 lac2 mutant (QGC9), (F) gpa1 mutant (AAC1), (G) gpa1+pGPD-LAC2 mutant (RPC18), and (H) wild type + pGPD-LAC2 (RPC21). The results are demonstrated as expression relative to the wild-type strain H99. Each data point represents the average for triplicate samples with error bars indicated. (B) The wild-type strain H99 was incubated to mid-logarithmic phase in YPD medium and subsequently exposed to several different growth conditions for 1 h: (1) YNB (pH 7), (2) YNB (pH 4), (3) YNB (pH 7) plus 10 mM Na hydro-gen peroxide, (6) YPD plus 10 mM paraquat, (7) YNB plus 0.01% glucose, and (8) YPD plus 100 μM copper sulfate. Total mRNA was isolated from these strains, treated with DNase-free RNase, and converted to cDNA. Relative LAC2 transcript levels, indicated as n-fold induction compared to the baseline condition 1, were determined for each of the samples with real-time PCR. Each data point represents the average for triplicate samples with error bars indicated. (C) Northern analysis documenting LAC2 overexpression. Total RNA was isolated from the wild-type (H99), gpa1+pGPD-LAC2 (RPC18), and wild type + pGPD-LAC2 (RPC21) strains after 1 h of incubation in a glucose-rich (+) or glucose-poor (−) medium and subjected to Northern analysis with the LAC2 gene as a probe. The rRNA signal in the ethidium bromide-stained RNA gel is shown to demonstrate equal RNA loading.
Southern analysis.
Genomic DNA was isolated from strains by using previously described techniques (44). Restriction digestion, gel electrophoresis, DNA transfer, prehybridization, hybridization, and autoradiography were performed as described previously (47).
PCR.
All PCRs were carried out with a Techne Genius thermocycler with 50 ng of template DNA, 100 ng of each oligonucleotide primer, and standard reagents from a TaKaRa kit (Takara Shuzo Co.). The PCR conditions were 95°C for 5 min followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min for each kilobase amplified in the reaction.
Virulence experiments.
In the murine inhalation model of systemic cryptococcosis, A/Jcr mice were intranasally inoculated with 105 cells as previously described (9). Groups of 10 mice were inoculated with each strain in the study and observed twice daily for signs of infection. The statistical significance in the difference between each strain's resulting survival rate was evaluated with the Mann-Whitney statistical model.
In this model, mice develop meningoencephalitis after inhalation of C. neoformans, a course that mimics the natural history of human infection with this organism. Signs consistent with cryptococcosis in this experimental model included lethargy, ruffled fur, and inability to maintain daily care. Moribund mice were sacrificed prior to death, and all studies were performed in compliance with institutional guidelines for animal experimentation. All surviving mice were sacrificed at 80 days after infection.
RESULTS
Transcriptional profiles of the wild-type and gpa1 mutant strains.
Previous studies demonstrated that the Gpa1-adenylyl cyclase signaling cascade in C. neoformans controls the expression of two inducible virulence factors, capsule and melanin. To identify genes whose transcription is dependent on the Gα protein Gpa1, we employed genome microarrays to compare the transcriptional profiles of wild-type and gpa1 mutant strains.
A DNA microarray was created by printing PCR-amplified cDNAs from 111 known C. neoformans genes onto glass slides. Several genes are present multiple times, and all cDNAs were printed in duplicate, resulting in a microarray that contains 260 elements. This slide was simultaneously hybridized with cDNA from a wild-type strain labeled with the Cy3 (green) fluorophore and cDNA from a gpa1 mutant strain labeled with Cy5 (red). cDNA from each strain was synthesized from total RNA extracted from cells after 1 h of glucose starvation. Glucose starvation was predicted to induce some Gpa1-regulated genes because this condition is required for C. neoformans melanin production.
To define genes whose transcription is regulated by Gpa1, we calculated the relative fluorescence intensities for each microarray spot when probed with these different fluorophore-labeled cDNA pools. The expression of 15 genes was increased at least 2.5-fold in the wild-type strain compared to that in the gpa1 mutant. The gene with the greatest difference in expression was the GPA1 gene itself, providing an important internal control for this experiment.
Of the other 14 genes, nine are known or presumed to function in capsule synthesis or assembly (Table 3). The CAS1 gene encodes an O-acetyltransferase, and CAS2 encodes a UDP-xylose synthase, both of which are required for the assembly of glucoronoxylomannan, the primary component of the cryptococcal capsule (22, 34). The CAP10, CAP59, and CAP64 genes encode proteins of unknown function, but each of these genes is required for C. neoformans capsule formation and virulence (5, 7, 8). The other CAS genes were identified in prior genetic screens, and mutations in these genes are associated with capsule defects (G. Janbon, personal communication).
TABLE 3.
Microarray comparison of gene transcript levels in the wild-type strain versus the gpa1 mutant strain
| Gene | Encoded protein | GPA1/gpa1a | Reference |
|---|---|---|---|
| GPA1 | Gα protein | 8.8 | 2, 53 |
| CAS8 | Capsule-associated gene | 6.1 | G. Janbon, unpublished data |
| CAS2 | UDP-xylose synthase | 4.9 | 34 |
| CAP10 | Capsule-associated gene | 4.5 | 7 |
| CAS1 | O-acetyl transferase | 3.7 | 22 |
| CAT2 | Catalase | 3.7 | Unpublished |
| SMG1 | Suppressor of gpa1 mutant phenotypes | 3.6 | Unpublished |
| CAS7 | Capsule-associated gene | 3.4 | G. Janbon, unpublished |
| CAP59 | Capsule-associated gene | 3.2 | 5 |
| CAP64 | Capsule-associated gene | 3.1 | 8 |
| RLM1 | Serum response-like factor protein homolog | 3.1 | Unpublished data |
| THR4 | Threonine synthase | 3.1 | Unpublished data |
| CAS4 | Capsule-associated gene | 2.9 | G. Janbon, unpublished |
| CAS31 | Capsule-associated gene | 2.8 | 33 |
| GPA2 | Gα protein | 2.8 | Unpublished |
GPA1/gpa1 indicates the relative transcript level in the GPA1 wild-type strain compared to the gpa1 mutant strain as assessed by microarray analysis.
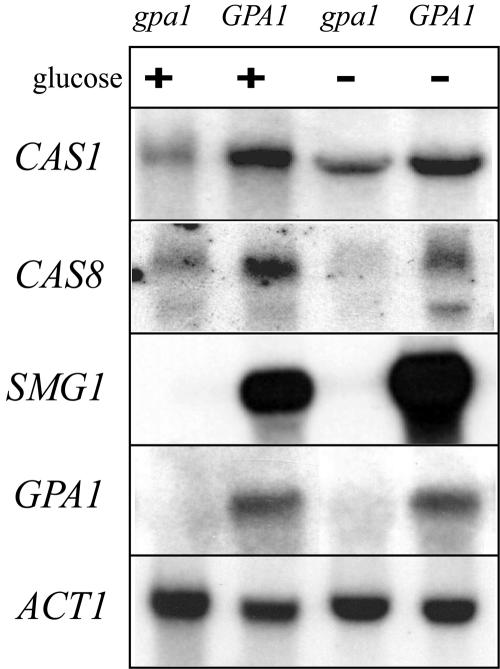
To confirm these findings, we performed Northern analysis of total RNA isolated from the wild-type and gpa1 mutant strains after 1 h of glucose starvation, using several genes from the microarray experiment as probes. In contrast to the case with the wild type, there was no GPA1 signal detected in RNA from the gpa1 mutant strain. As predicted by the microarray analysis data, expression of the CAS1, CAS8, and SMG1 genes was increased in the GPA1 wild-type strain compared to that in the gpa1 mutant (Fig. 1). The SMG1 gene was identified in an independent genetic screen for suppressors of gpa1 mutant phenotypes (unpublished results), and it is therefore reasonable that Gpa1 may regulate the expression of this gene.
FIG. 1.
Northern analysis confirms the gpa1 mutant microarray results. The wild-type (GPA1) and gpa1 mutant strains were incubated to mid-logarithmic phase in YPD and exposed for 1 h to glucose-rich (+) or glucose-poor (−) conditions. Total RNA was extracted from these strains and used for Northern analysis with the CAS1, CAS8, SMG1, and GPA1 genes as probes, with the ACT1 (actin) gene as a loading control.
Identification of a second laccase gene (LAC2) in C. neoformans using a genome microarray.
As a complementary approach, we used a genomic DNA microarray containing more than 6,000 elements to assess Gpa1-regulated expression of potentially unknown genes. This microarray was produced by using PCR products amplified from a 1.6- to 3.2-kb insert genomic library constructed by using strain H99 genomic DNA. The utility of this genomic DNA microarray for assessing gene expression has previously been demonstrated in a study of temperature-regulated gene expression (23). cDNA samples were prepared in the manner described above and hybridized to the genomic microarray.
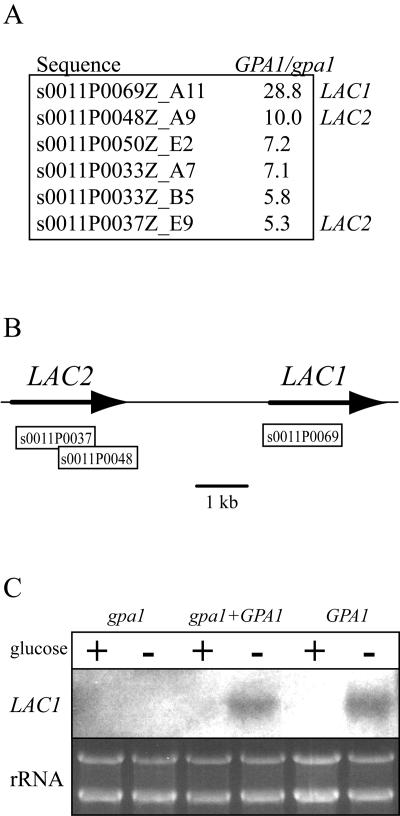
Using the larger microarray, we identified six genomic fragments that demonstrated a fivefold or greater transcription level in the GPA1 wild-type strain compared to the gpa1 mutant strain (Fig. 2A). The genomic fragment s0011P0069Z_A11 demonstrated a 28.8-fold transcriptional dependence on Gpa1. A nucleotide BLAST search of this fragment against the H99 genome database revealed that this genomic fragment is included within the LAC1 locus (Fig. 2B). Northern blot analysis confirmed that the LAC1 gene is transcriptionally regulated by Gpa1. The LAC1 transcript is not detectable in gpa1 mutants after 1 h of glucose starvation, but it can be detected at similar levels in a wild-type strain and in a gpa1+GPA1 reconstituted strain (gpa1 mutant transformed with a wild-type GPA1 gene) (2) incubated under identical conditions (Fig. 2C). This observation is consistent with the melanin-deficient phenotype of the gpa1 mutant strain and provides insight into the molecular mechanism by which the Gpa1 protein controls melanin production.
FIG. 2.
LAC1 and LAC2 transcript levels are regulated by Gpa1. (A) Gene microarray analysis identified six genomic sequences of the serotype A strain H99 that exhibit an increased ratio of cDNA hybridization in the wild-type strain compared to the gpa1 mutant (GPA1/gpa1), suggesting that a gene present in these sequences is differentially expressed in these two strains. (B) One of these sequences (s0011P0069Z_A11) localized to the region of the LAC1 gene, and two sequences (s0011P0048Z_A9 and s0011P0037Z_E9) localized 4 to 5 kb upstream of LAC1, in a region containing a second putative laccase gene, LAC2. (C) To confirm the microarray results, Northern analysis using the LAC1 gene as a probe was performed with total RNA from the wild type (H99), the gpa1 mutant (AAC1), and the gpa1+GPA1 reconstituted strain (AAC3) after 1 h of incubation under glucose-rich (+) or glucose-poor (−) conditions. The rRNA signal in the ethidium bromide-stained RNA gel is shown to demonstrate equal RNA loading.
Two other DNA fragments demonstrated a marked transcriptional dependence on Gpa1 and were transcribed at 10.0- and 5.3-fold-higher levels in the wild-type than in the gpa1 mutant strain, respectively. Both fragments showed perfect nucleotide identity to a region approximately 5 kb upstream of the LAC1 start codon (Fig. 2A and B). Interestingly, these two fragments (s0011P0048Z_A9 and s0011P0037Z_E9) also demonstrated 75 and 76% nucleotide identity, respectively, with the LAC1 locus. Further analysis of this region of the C. neoformans genome revealed the presence of a putative gene encoding a laccase homolog, distinct from LAC1, which we designated LAC2.
The LAC2 gene consists of 2,690 nucleotides from the start to termination codons, contains 13 introns, and encodes a predicted protein of 596 amino acids. The C. neoformans LAC2 cDNA sequence shares 85% nucleotide identity with LAC1 in the first 1,063 nucleotides and 75% identity from nucleotides 1252 to 1788. LAC2 also shares significant homology with other fungal laccases and enzymes involved in pigment formation: 30% amino acid identity with the laccase 2 gene of Botryotinia fuckeliana (48) and 29% identity with a LAC1 precursor gene of Agaricus bisporus (43) and an Neurospora crassa conidial pigment biosynthesis protein (GenBank accession no. CAD 70788). Laccase proteins contain several conserved motifs, including substrate-binding and copper-binding domains. These laccase signature motifs, L1 to L4 (24), are all conserved in the predicted Lac2 protein.
Lac2 is transcriptionally repressed by glucose.
To define the conditions that regulate the expression of the LAC2 gene, we used quantitative real-time PCR. This method was chosen for two reasons. First, we wished to avoid potential cross-hybridization with the highly related LAC1 gene in Northern blots. Also, initial Northern hybridizations that used the most dissimilar regions of LAC1 and LAC2 as probes suggested that the LAC2 gene is transcribed at low levels under several growth conditions (data not shown).
To assess LAC2 transcription by real-time PCR, the wild-type strain H99 was incubated to mid-logarithmic phase in YPD medium and shifted to one of several inducing conditions: YPD (0.01% glucose), YPD plus 100 μM copper sulfate, YPD plus 0.5 mM hydrogen peroxide, YPD plus 10 mM paraquat, YNB (pH 7) (with or without 0.5 mM NaNO2), and YNB (pH 4) (with or without 0.5 mM NaNO2). Acidic aqueous solutions containing NaNO2 (YNB [pH 4] plus NaNO2) provide nitric oxide-inducing conditions (1). Total RNA was isolated from the cell pellets and treated with DNase I, and first-strand cDNA was synthesized with reverse transcriptase. This cDNA was used as the template for quantitative real-time PCRs with LAC2-specific primers. The level of LAC2 transcript in each sample was determined as an n-fold induction relative to the control condition (YNB [pH 7] with 2% glucose), using the comparative CT method and using the GPD gene to normalize for RNA loading. Transcription of the LAC2 gene was not significantly affected by oxidative stress (hydrogen peroxide or paraquat), nitrosative stress (pH 4) (plus NaNO2), low pH, or exogenous copper. The only condition in which LAC2 transcription was induced more than twofold was glucose deprivation (YNB plus 0.1% glucose). In H99 cells incubated in this glucose-poor medium for 2 h, LAC2 was induced four- to fivefold compared to results for H99 cells incubated in a glucose-rich medium (Fig. 3B). These results are similar to those with the LAC1 gene, which is similarly repressed by glucose (59).
The effects of other gene mutations on LAC2 expression were also assessed by real-time PCR. In accordance with the microarray result, LAC2 transcript levels are reduced in a gpa1 mutant strain from wild-type levels (Fig. 3A). This further confirms that this signaling pathway transcriptionally regulates both LAC1 and LAC2. In contrast, LAC2 expression is increased in a lac1 mutant strain compared to wild-type expression (Fig. 3A). However, as evidenced by the melanin defects of a lac1 strain, this increase in LAC2 message is not sufficient to fully restore wild-type levels of laccase activity to this mutant strain.
Disruption of the LAC2 gene results in reduced melanin production.
To determine the relative contribution of the Lac2 protein to melanin production, we used gene disruption and genetic epistasis approaches. The entire coding region of LAC2 was replaced with the dominant selectable marker encoding neomycin resistance (neo) (Fig. 4B). The resulting lac2::neo disruption allele was introduced into the serotype A wild-type strain H99 by biolistic transformation. In three independent transformants (RPC26, RPC27, and RPC28), the LAC2 gene was precisely replaced by integration of the lac2::neo mutant allele, with no ectopic integrations. Two lac1 lac2 double-mutant strains (RPC30 and QGC9) were created in a similar manner, using a lac2 disruption construct to transform a serotype A lac1 mutant strain. The lac1 and lac2 mutations were confirmed by PCR and Southern hybridization (Fig. 4A), and no LAC2 transcript was detected in the lac2 or lac1 lac2 strains by real-time PCR (Fig. 3A). The phenotypes of three independent lac2 mutants were identical, and we chose one of these strains (RPC27) as a representative lac2 mutant for subsequent experiments. Similarly, the lac1 lac2 double-mutant strains demonstrated identical phenotypes, and strain QGC9 was used in subsequent experiments as the lac1 lac2 mutant strain.
FIG. 4.
Southern blot of lac2 and lac1 lac2 mutants. (A) Genomic DNA from the wild type (H99), lac2 mutants (RPC26, RPC27, and RPC28), and lac1 lac2 double mutants (RPC30 and QGC9) was digested with PstI and XhoI and examined by Southern analysis using the indicated region of the LAC2 locus as a probe. (B) Restriction maps of the LAC2 and LAC1 loci.
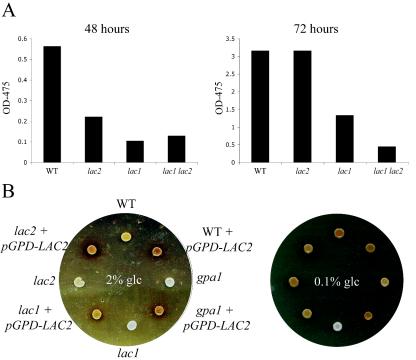
Laccase activity in these strains was quantified by incubating the wild-type, lac1 mutant, lac2 mutant, and lac1 lac2 double-mutant strains in YNB plus 0.1% glucose in the presence of three separate laccase substrates: niger seed extract, dopamine, and epinephrine. Laccase activity was quantified spectrophotometrically by the appearance of pigment in the culture supernatant. The wild-type strain exhibited 2.4-fold-greater laccase activity than the lac2 mutant and fivefold greater laccase activity than the lac1 mutant strain (Fig. 5A). This effect was identical whether Niger seed extract, dopamine, or epinephrine was used as the substrate for melanin production. Therefore, in three separate laccase assays using three different melanin substrates, we observed a consistent, impaired laccase activity due to a lac2 mutation. With longer incubations, melanin formation by the wild-type and lac2 mutant strain reached a saturating level. This observation is consistent with similar melanin levels apparent when these two strains were incubated on solid melanin-inducing media.
FIG. 5.
Laccase activity. (A) The wild-type (H99), lac2 mutant (RPC27), lac1 mutant (MDC16), and lac1 lac2 double mutant (QGC9) were incubated in YNB plus 0.1% glucose with 10 mM epinephrine. Laccase activity was quantified at 48 and 72 h by the appearance of pigment in the supernatant as assessed by measuring the absorbance at 475 nm. (B) The wild-type (H99), wild-type + pGPD-LAC2 (RPC 21), gpa1 (AAC1), gpa1+pGPD-LAC2 (RPC18), lac1 (MDC16), lac1+pGPD-LAC2 (QGC1), lac2 (RPC27), and lac2+pGPD-LAC2 (QGC4) strains were incubated on niger seed medium with either 2 or 0.1% glucose for 72 h at 30°C. Melanin-producing strains make brown pigments on this medium.
The lac1 mutant strain demonstrated a more pronounced decrease in melanin activity than the lac2 mutant strain. However, after prolonged incubation, this strain demonstrated notable pigment formation. This effect was not likely due to autooxidation of the melanin substrates, since cell-free controls demonstrated minimal melanin formation over the course of this experiment. Therefore, enzymes other than Lac1 are involved in C. neoformans melanin formation.
The lac1 lac2 double mutant had the most profound defect in laccase activity, and little detectable pigment was formed in the presence of dopamine, epinephrine, or niger seed extract. Together, these findings support a model in which Lac1 and Lac2 play additive roles in C. neoformans melanin production.
Overexpression of the LAC2 gene restores melanin to the gpa1 and lac1 mutants.
To confirm that the LAC2 sequence represents a functional gene involved in melanin production, we overexpressed this gene in four strain backgrounds. We cloned LAC2 under the control of the constitutively active promoter of the glucose-6-phosphate dehydrogenase gene (GPD) and biolistically transformed this plasmid into the wild-type, gpa1 mutant, lac1 mutant, and lac2 mutant strains. All transformants exhibited wild-type levels of growth on rich (YPD) and minimal (YNB) media, indicating that differences in growth rate or nutrient acquisition do not account for altered melanin production in these strains. Overexpression of LAC2 in these strains was documented by real-time PCR and by Northern hybridization (Fig. 3A and C). The LAC2 transcript was not detectable by this assay in the wild-type strain grown in the presence or absence of glucose. However, the LAC2-overexpressing strains demonstrated LAC2 levels that were clearly detected by Northern analysis (Fig. 3C).
On melanin-inducing niger seed medium, the wild-type strain demonstrates vigorous melanin production that is repressed by higher glucose concentration (Fig. 5B). Overexpression of LAC2 in the wild-type and lac2 strains results in increased melanin production. This activity is especially evident on the medium containing 2% glucose, consistent with the constitutive activity of the GPD promoter. As previously noted (2), little melanin is produced by the gpa1 mutant strain on either medium. LAC2 overexpression completely suppresses the melanin-deficient phenotype of the gpa1 mutant strain.
After 2 days of incubation on niger seed medium, no melanin was apparent in the lac1 mutant strain. However, LAC2 overexpression completely restored wild-type levels of melanin production in this strain. These findings indicate that the LAC2 gene encodes a functional protein involved in melanin biosynthesis and that the Lac2 protein shares a redundant or overlapping function with Lac1. Also, the observation that the melanin defect of the gpa1 mutant can be completely suppressed by LAC2 overexpression further supports our hypothesis that the Gpa1-cAMP pathway controls C. neoformans melanin production at the level of transcription of laccase genes rather than at multiple steps in this pathway.
Substrate specificity of Lac2.
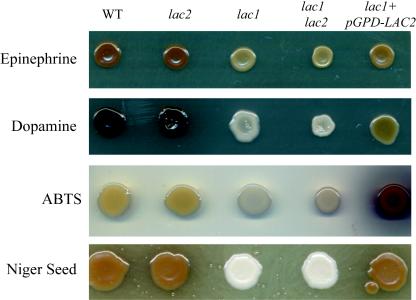
C. neoformans cannot synthesize melanin de novo; rather, it requires the presence of diphenolic substrates, such as epinephrine, dopamine, L-DOPA, ABTS, or poorly characterized compounds present in niger seed extract. By selectively introducing different diphenols into the culture medium, we began to study the substrate specificity of laccases in different C. neoformans strains. We incubated the wild-type, lac1 mutant, lac2 mutant, lac1 lac2 double mutant, and lac1+LAC2 strains on media containing different substrates for melanin production (Fig. 6). Guizotia abysinnica seed (niger seed) extract contains a mixture of substrates used by C. neoformans to produce melanin. When incubated on a medium containing either niger seed extract or epinephrine, the lac2 mutant strain makes melanin almost as efficiently as the wild-type strain, while the lac1 mutant and the lac1 lac2 double mutant do not produce visible melanin after 2 days of incubation. LAC2 overexpression restored wild-type levels of melanin to the melanin-deficient lac1 mutant strain (Fig. 5B and 6).
FIG. 6.
Substrate utilization for melanin. The wild-type, lac2 mutant (RPC27), lac1 mutant (MDC16), lac1 lac2 mutant (QGC9), and lac1+pGPD-LAC2 strains (QGC1) were incubated for 72 h on YNB medium with 0.1% glucose containing one of the following substrates for melanin production: epinephrine, dopamine, ABTS, or niger seed extract.
When these strains are incubated on medium containing ABTS, a different pattern of melanin production was observed. The lac1 and lac1 lac2 mutant strains remained melanin deficient compared with the isogenic wild-type and lac2 mutant strains. However, the LAC2 overexpression strain produced much more melanin than the wild type, resulting in a darker patch of cells than the wild type with a surrounding halo of green pigment (Fig. 6).
In contrast, strains overexpressing the LAC2 gene do not make melanin efficiently when incubated on a medium containing dopamine as the substrate for melanin production (Fig. 6). Although melanin pigment is evident in the lac1+LAC2 strain after 2 days of incubation on dopamine-containing medium, the degree of melanin production is much less than that of the wild-type strain.
A lac1 mutation results in severely melanin-deficient strains on each of these media, and the Lac1 protein is therefore responsible for the majority of melanin production in vitro in the wild-type strain. Therefore, comparing wild-type and LAC2 overexpression strains yields insight into the substrate specificities of the Lac1 and Lac2 proteins. Taken together, these results suggest that Lac1 and Lac2, when expressed at nearly equivalent levels, demonstrate similar substrate utilization for either epinephrine or the diphenols present in niger seed extract. The Lac2 protein is able to utilize ABTS as a melanin substrate more efficiently than the combination of laccases at the levels present in the wild-type strain. However, Lac2 poorly oxidizes dopamine for melanin biosynthesis. Such substrate specificity suggests that multiple laccases may allow C. neoformans to more efficiently produce melanin from different substrates encountered in a variety of environments.
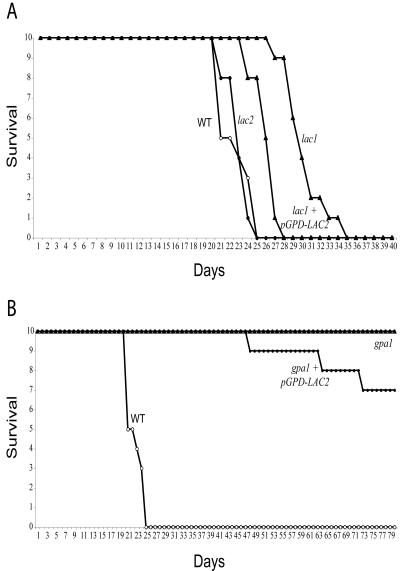
LAC2 and virulence of C. neoformans in an animal model of cryptococcosis.
The murine inhalation model of systemic C. neoformans infection was used to determine whether Lac2 is involved in virulence (Fig. 7) (3, 9). Female A/Jcr mice were intranasally inoculated with 105 C. neoformans cells, and animals were monitored for survival. As previously demonstrated, infection with the wild-type strain H99 results in a consistent lethal effect, with all mice succumbing to the infection between 23 and 27 days postinfection. Mice infected with a lac2 mutant strain (RPC27) demonstrated no statistically significant difference in survival compared to the wild type (P = 0.73), suggesting that Lac2 is dispensable for C. neoformans virulence in this model system. Animals infected with a lac1 mutant (MDC16) demonstrated prolonged survival compared either to the wild type (P = 0.0002) or to the lac2 mutant (P = 0.0002), although all of the mice succumbed to a lethal infection by 35 days. Overexpression of the LAC2 gene in the lac1 mutant background, which restores melanin production, also suppressed the lac1 mutant virulence defect (P = 0.0004).
FIG. 7.
LAC2 and the murine model of inhalational cryptococcosis. Female A/Jcr mice were intranasally inoculated with 105 cells of the following strains: wild type (H99), lac2 mutant (RPC27), lac1 mutant (MDC16), lac1+pGPD-LAC2 strain (QGC1), gpa1 mutant (AAC1), or gpa1+pGPD-LAC2 strain (RPC18). The mice were monitored for clinical signs of cryptococcal infection and sacrificed at predetermined clinical end points that predict imminent mortality.
In contrast, the gpa1 mutant strain did not cause a lethal infection in any animal, even after 80 days of observation. Previously, the gpa1 virulence defect was complemented in the rabbit model of C. neoformans meningitis by reintroduction of the GPA1 gene (2). LAC2 overexpression completely restored melanin production in the gpa1 mutant, but it was unable to restore virulence to this strain. This result is not unexpected, since the gpa1 mutant strain also has a capsule defect, and capsule-deficient C. neoformans strains are attenuated for virulence (27). We observed three lethal infections in mice inoculated with the gpa1+pGPD-LAC2 strain and none in the gpa1 mutant, but the difference in these two virulence curves was not statistically significant. Therefore, constitutive Lac2 activity did not restore virulence to the hypocapsular C. neoformans gpa1 mutant strain in this model system in contrast to its ability to complement the virulence defect of a lac1 mutant.
DISCUSSION
In contrast to other microbial pathogens that are uniquely differentiated to survive within a specific host, many human fungal pathogens must be able to exist in the external environment as well as in an infected animal. In fact, many fungi that are pathogens in humans are unlikely to encounter a mammalian host during their life cycle. This has led to the hypothesis that phenotypes offering a selective advantage for fungi within infected tissue might also play roles for survival in the environment. For example, melanized C. neoformans strains are more resistant to UV killing than identical strains in which melanin is not induced (57). Additionally, melanin-deficient strains are attenuated for survival when confronted with nonmammalian hosts, such as free-living amoebae and nematodes, which may be frequently encountered by fungi in the environment (35, 50). Encapsulated C. neoformans strains are also more resistant to killing by amoebae than nonencapsulated strains (50). Therefore, whether living within an infected host or in the external environment, the ability of microorganisms to coordinately regulate such apparently disparate phenotypes as laccase activity and capsule formation may be advantageous for survival.
We have previously demonstrated that the signal transduction pathway that controls C. neoformans cAMP metabolism regulates numerous cellular traits, including encapsulation, melanin formation, and mating (2, 3, 17, 20). Additionally, our aim is to define how one signaling pathway can link specific input signals to the induction of distinct phenotypic outputs. For example, nitrogen starvation is required for mating in C. neoformans and also for the transcriptional induction of GPA1. However, this environmental signal does not result in the induction of other cAMP-dependent phenotypes, such as melanin and capsule production. Therefore, the cAMP signal appears to be necessary, but not sufficient, for many of the phenotypes it regulates.
This model is perhaps most clearly illustrated in the case of C. neoformans mating. The ability of C. neoformans strains to mate efficiently requires both starvation and the presence of an appropriate mating partner. Recent studies have demonstrated that mutations either in the pheromone response pathway or in the cAMP pathway render the cell sterile (12, 13, 56). However, these two signaling pathways are quite distinct. It is likely that the cAMP pathway transduces nutrient deprivation signals required for mating, and the mitogen-activated protein kinase pheromone response pathway signals the presence of a mating partner. Similarly, melanin and capsule formation may also require multiple environmental signals for induction, and the cAMP pathway likely provides only one of these important inputs.
Numerous proteins are involved in capsule synthesis in C. neoformans. In fact, several capsule genes have been identified whose encoded protein has a yet-unidentified function (5-8, 16, 22). However, a coordinated regulation of proteins involved in capsule synthesis and assembly would likely benefit the cell. Our observation that the Gα protein Gpa1 regulates the transcription of multiple capsule genes begins to define a mechanism by which this signaling pathway allows synchronized capsule gene expression. Initial analysis of the promoter regions of the coregulated capsule genes failed to demonstrate large regions of identity. However, more-detailed promoter evaluations will likely be necessary to elucidate cis- and trans-acting regulatory elements that control capsule gene expression in response to a cAMP signal.
Fungal melanins are also required for survival of several fungal species within mammalian hosts. In human-pathogenic fungi, such as Aspergillus fumigatus, Paracoccidioides brasiliensis, Sporothrix schenckii, W. dermatitidis, and C. neoformans, melanins play a protective role against effectors of cellular immunity and possibly a mechanical role in pathogenesis (28). Therefore, the synthesis of melanin has large implications from the mechanisms of fungal virulence to treatment of infection in the host.
In these studies we identified a second C. neoformans laccase-encoding gene, LAC2, whose product is involved in the melanin biosynthetic pathway. Laccases (benzendiol:oxygen oxidoreductases) are blue, multicopper-containing enzymes that catalyze oxidation of a range of aromatic compounds in different fungal species. Laccases are important catalysts in the oxidative steps of melanin formation. In wood-rotting fungi, laccases also play a prominent role in lignin degradation, allowing parasitism of woody plants (30). Aside from this catalytic role, they are also involved in other physiological processes. For example, fruiting body formation in ascomycetes is dependent on laccases (21, 51). Extracellular laccase-like, multicopper oxidases have also been shown to have ferroxidase activity in the basidiomycete Phanerochaete chrysosporium (29). Laccase activity may also participate in combating oxidative stress under normal quiescent conditions (18).
Analysis of the genomes of the serotype A strain H99, the serotype D strain JEC21, and the serotype B strain WM276 indicates that LAC1 and LAC2 are the result of an ancient duplication event which occurred prior to the differentiation of the C. neoformans varieties. In all of these strains of divergent varieties, the two laccase genes are adjacent to one another and display the same gene orientation. Additionally, phylogenetic analysis of the two laccase genes clearly indicates that all of the LAC1 homologs are more closely related to one another than to other laccase genes within the same strain. Similarly, the LAC2 genes in these three strains are more homologous to each other than to LAC1. Further studies defining the different functions of these laccases will illuminate why C. neoformans maintained two such highly similar genes as the three varieties differentiated and came to occupy different ecological niches.
Several aspects of C. neoformans laccase function have already been described. For example, the Lac1 enzyme is well characterized for its role in the synthesis of melanin (60). However, the presence of other, similar enzymes in this organism was implied by several observations. First, although lac1 mutant strains have a striking melanin defect, these strains eventually produce melanin pigments. This observation may be explained by autooxidation of melanin precursors or the presence of alternative phenoloxidases capable of catalyzing this step in melanin formation. Also, in other basidiomycetes, multiple laccase genes have been identified. For example, eight nonallelic laccases have been defined in Coprinus cinereus (21). Several of these genes are certain to encode oxidases involved in iron metabolism. However, the presence of a gene family with eight apparent members raises interesting questions about the importance of this group of enzymes. Do all of these enzymes perform completely separate and distinct functions, or is there some degree of functional redundancy present among fungal laccases?
In our studies, we demonstrate that the Lac2 protein, though expressed at low levels under the conditions tested thus far in vitro, serves a redundant function in melanin formation with Lac1. When overexpressed, the LAC2 gene suppressed the melanin defect of the lac1 mutant strain. This suggests some degree of functional redundancy, rather than completely distinct functions, among these fungal laccases.
Because fungi are likely to encounter a variety of phenolic compounds in the environment and in the host, the ability to utilize many different compounds as precursors for melanin would offer survival advantages under these very different conditions. Others have suggested that multiple laccases would offer such functional elasticity to pigmented fungi, rather than requiring each laccase to oxidize various substrates (21). We demonstrated that a C. neoformans strain overexpressing the LAC2 gene is able to use several catecholamines with differing efficiencies for melanin production. Additionally, this strain displays different substrate utilization patterns compared with a wild-type strain in which the Lac1 protein is the predominant laccase. This result supports the hypothesis that the presence of multiple laccases provides fungal cells with a broader array of potential substrate utilization patterns.
Four diphenols are present in human brain tissue: L-DOPA, norepinephrine, epinephrine, and dopamine. C. neoformans likely uses these substrates in vivo for melanin biosynthesis, since this yeast has demonstrated production of melanin in animal models of cryptococcosis and in human infections (37, 38). It will be interesting to determine which laccases are functioning in vivo to use these catecholamines for melanin biosynthesis.
More evidence of differential laccase function within this family is suggested by the different expression patterns of the genes. By Northern blot and quantitative PCR analysis, the level of LAC2 transcript is significantly lower than that of LAC1. Likewise, laccases in C. cinereus are shown to have wide variation in expression patterns. Three of the eight isolated laccases in this organism are differentially regulated by nutrients and the presence of metallic and aromatic inducers (21). In other fungi, laccases are induced by such varied signals as temperature, osmotic pressure, and plant secondary metabolites (40, 48). Defining the inducing conditions for cAMP-regulated genes will help to determine the specific upstream signals that activate the cAMP cascade in C. neoformans.
Acknowledgments
This work was supported by PHS grants AI050128 (J.A.A.) and P01 AI44975 (J.H.). Joseph Heitman is a Burroughs Wellcome Fund Scholar in molecular pathogenic mycology and an Associate Investigator of the Howard Hughes Medical Institute. Andrew Alspaugh is a Burroughs Wellcome Fund New Investigator in molecular pathogenic mycology.
The following C. neoformans genome databases were used for gene identification: (i) C. neoformans Genome Project, Stanford Genome Technology Center, funded by the NIAID/NIH under cooperative agreement AI47087; (ii) the genome sequencing project of the serotype B strain WM276, funded by Genome Canada; (iii) the serotype A strain H99 genome project at the Center for Genome Technology at Duke University; and (iv) the Fungal Genome Initiative at the Whitehead Institute at M.I.T. We are indebted to Guilhem Janbon for providing us with the sequences of the CAS genes prior to their publication for inclusion in the microarray. We also acknowledge that Jennifer Lodge's laboratory independently and concurrently identified the C. neoformans LAC2 gene.
REFERENCES
- 1.Alspaugh, J. A., and D. L. Granger. 1991. Inhibition of Cryptococcus neoformans replication by nitrogen oxides supports the role of these molecules as effectors of macrophage-mediated cytostasis. Infect. Immun. 59:2291-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alspaugh, J. A., J. R. Perfect, and J. Heitman. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 11:3206-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alspaugh, J. A., R. Pukkila-Worley, T. Harashima, L. M. Cavallo, D. Funnell, G. M. Cox, J. R. Perfect, J. W. Kronstad, and J. Heitman. 2002. Adenylyl cyclase functions downstream of the G-alpha protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell 1:75-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broach, J. R., and R. J. Deschenes. 1990. The functions of RAS genes in Saccharomyces cerevisiae. Adv. Cancer Res. 54:79-138. [DOI] [PubMed] [Google Scholar]
- 5.Chang, Y. C., and K. J. Kwon-Chung. 1994. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 14:4912-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, Y. C., and K. J. Kwon-Chung. 1998. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans. Infect. Immun. 66:2230-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y. C., and K. J. Kwon-Chung. 1999. Isolation, characterization, and localization of a capsule-associated gene, CAP10, of Cryptococcus neoformans. J. Bacteriol. 181:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y. C., L. A. Penoyer, and K. J. Kwon-Chung. 1996. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect Immun. 64:1977-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, G. M., J. Mukherjee, G. T. Cole, A. Casadevall, and J. R. Perfect. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 68:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, G. M., T. H. Rude, C. C. Dykstra, and J. R. Perfect. 1995. The actin gene from Cryptococcus neoformans: structure and phylogenetic analysis. J. Med. Vet Mycol. 33:261-266. [DOI] [PubMed] [Google Scholar]
- 11.Davidson, R. C., J. R. Blankenship, P. R. Kraus, M. de Jesus Berrios, C. M. Hull, C. D'Souza, P. Wang, and J. Heitman. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607-2615. [DOI] [PubMed] [Google Scholar]
- 12.Davidson, R. C., T. D. Moore, A. R. Odom, and J. Heitman. 2000. Characterization of the MFα pheromone of the human fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 38:1017-1026. [DOI] [PubMed] [Google Scholar]
- 13.Davidson, R. C., C. B. Nichols, G. M. Cox, J. R. Perfect, and J. Heitman. 2003. A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 49:469-485. [DOI] [PubMed] [Google Scholar]
- 14.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 15.Dixon, D. M., J. Migliozzi, C. R. Cooper, O. Solis, B. G. Breslin, and P. J. Szaniszlo. 1992. Melanized and nonmelanized multicellular-form mutants of Wangiella dermatitidis in mice: mortality and histopathology studies. Mycoses 35:17-21. [DOI] [PubMed] [Google Scholar]
- 16.Doering, T. L. 1999. A unique alpha-1,3 mannosyltransferase of the pathogenic fungus Cryptococcus neoformans. J. Bacteriol. 181:5482-5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Souza, C. A., J. A. Alspaugh, C. Yue, T. Harashima, G. M. Cox, J. R. Perfect, and J. Heitman. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21:3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez-Larrea, J., and U. Stahl. 1996. Isolation and characterization of a laccase gene from Podospora anserina. Mol. Gen. Genet. 252:539-551. [DOI] [PubMed] [Google Scholar]
- 19.Granger, D. L., J. R. Perfect, and D. T. Durack. 1985. Virulence of Cryptococcus neoformans: regulation of capsule synthesis by carbon dioxide. J. Clin. Investig. 76:508-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hicks, J. K., C. A. D'Souza, G. M. Cox, and J. Heitman. 2004. Cyclic AMP-dependent protein kinase catalytic subunits have divergent roles in virulence factor production in two varieties of the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell 3:14-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoegger, P. J., M. Navarro-Gonzalez, S. Kilaru, M. Hoffmann, E. D. Westbrook, and U. Kues. 2004. The laccase gene family in Coprinopsis cinerea (Coprinus cinereus). Curr. Genet. 45:9-18. [DOI] [PubMed] [Google Scholar]
- 22.Janbon, G., U. Himmelreich, F. Moyrand, L. Improvisi, and F. Dromer. 2001. Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol. Microbiol. 42:453-467. [DOI] [PubMed] [Google Scholar]
- 23.Kraus, P. R., M.-J. Boily, S. S. Giles, J. E. Staijich, A. Allen, G. M. Cox, F. S. Dietrich, J. R. Perfect, and J. Heitman. 2004. Identification of Cryptococcus neoformans temperature-regulated genes with a genomic DNA microarray. Eukaryot. Cell 3:1249-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar, S. V., P. S. Phale, S. Durani, and P. P. Wangikar. 2003. Combined sequence and structure analysis of the fungal laccase family. Biotechnol. Bioeng. 83:386-394. [DOI] [PubMed] [Google Scholar]
- 25.Kwon-Chung, K. J., and J. E. Bennett. 1992. Cryptococcosis, p. 397-446. In Medical Mycology. Lea & Febiger, Malvern, Pa.
- 26.Kwon-Chung, K. J., I. Polacheck, and T. J. Popkin. 1982. Melanin-lacking mutants of Cryptococcus neoformans and their virulence for mice. J. Bacteriol. 150:1414-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon-Chung, K. J., and J. C. Rhodes. 1986. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect. Immun. 51:218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langfelder, K., M. Streibel, B. Jahn, G. Haase, and A. A. Brakhage. 2003. Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet. Biol. 38:143-158. [DOI] [PubMed] [Google Scholar]
- 29.Larrondo, L. F., L. Salas, F. Melo, R. Vicuna, and D. Cullen. 2003. A novel extracellular multicopper oxidase from Phanerochaete chrysosporium with ferroxidase activity. Appl. Environ. Microbiol. 69:6257-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leonowicz, A., N. S. Cho, J. Luterek, A. Wilkolazka, M. Wojtas-Wasilewska, A. Matuszewska, M. Hofrichter, D. Wesenberg, and J. Rogalski. 2001. Fungal laccase: properties and activity on lignin. J. Basic Microbiol. 41:185-227. [DOI] [PubMed] [Google Scholar]
- 31.McDade, H. C., and G. M. Cox. 2001. A new dominant selectable marker for use in Cryptococcus neoformans. Med. Mycol. 39:151-154. [DOI] [PubMed] [Google Scholar]
- 32.Money, N. P. 1997. Mechanism linking cellular pigmentation and pathogenicity in rice blast disease. Fungal Genet. Biol. 22:151-152. [DOI] [PubMed] [Google Scholar]
- 33.Moyrand, F., Y. C. Chang, U. Himmelreich, K. J. Kwon-Chung, and G. Janbon. 2004. Cas3p belongs to a seven member family of capsule structure designer proteins. Eukaryot. Cell 3:1513-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moyrand, F., B. Klaproth, U. Himmelreich, F. Dromer, and G. Janbon. 2002. Isolation and characterization of capsule structure mutant strains of Cryptococcus neoformans. Mol. Microbiol. 45:837-849. [DOI] [PubMed] [Google Scholar]
- 35.Mylonakis, E., F. M. Ausubel, J. R. Perfect, J. Heitman, and S. B. Calderwood. 2002. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc. Natl. Acad. Sci. USA 99:15675-15680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nichols, C. B., J. A. Fraser, and J. Heitman. 2004. PAK kinases Ste20 and Pak1 govern cell polarity at different stages of mating in Cryptococcus neoformans. Mol. Biol. Cell. 15:4476-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nosanchuk, J. D., A. L. Rosas, S. C. Lee, and A. Casadevall. 2000. Melanisation of Cryptococcus neoformans in human brain tissue. Lancet 355:2049-2050. [DOI] [PubMed] [Google Scholar]
- 38.Nosanchuk, J. D., P. Valadon, M. Feldmesser, and A. Casadevall. 1999. Melanization of Cryptococcus neoformans in murine infection. Mol. Cell Biol. 19:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nurudeen, T. A., and D. G. Ahearn. 1979. Regulation of melanin production by Cryptococcus neoformans. J. Clin. Microbiol. 10:724-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohga, S., and D. J. Royse. 2001. Transcriptional regulation of laccase and cellulase genes during growth and fruiting of Lentinula edodes on supplemented sawdust. FEMS Microbiol. Lett. 201:111-115. [DOI] [PubMed] [Google Scholar]
- 41.Pan, X., and J. Heitman. 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4874-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perfect, J. R., S. D. Lang, and D. T. Durack. 1980. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am. J. Pathol. 101:177-194. [PMC free article] [PubMed] [Google Scholar]
- 43.Perry, C. R., S. E. Matcham, D. A. Wood, and C. F. Thurston. 1993. The structure of laccase protein and its synthesis by the commercial mushroom Agaricus bisporus. J. Gen Microbiol 139:171-178. [DOI] [PubMed] [Google Scholar]
- 44.Pitkin, J. W., D. G. Panaccione, and J. D. Walton. 1996. A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology 142:1557-1565. [DOI] [PubMed] [Google Scholar]
- 45.Robertson, L. S., and G. R. Fink. 1998. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. USA 95:13783-13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salas, S. D., J. E. Bennett, K. J. Kwon-Chung, J. R. Perfect, and P. R. Williamson. 1996. Effect of the laccase gene, CNLAC1, on virulence of Cryptococcus neoformans. J. Exp. Med. 184:377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Schouten, A., L. Wagemakers, F. L. Stefanato, R. M. van der Kaaij, and J. A. van Kan. 2002. Resveratrol acts as a natural profungicide and induces self-intoxication by a specific laccase. Mol. Microbiol. 43:883-894. [DOI] [PubMed] [Google Scholar]
- 49.Sherman, F. 1991. Getting started with yeast, p. 3-21. In C. Guthrie and G. R. Fink (ed.), Methods in enzymology, vol. 194. Academic Press, Inc., San Diego, Calif.. [DOI] [PubMed]
- 50.Steenbergen, J. N., H. A. Shuman, and A. Casadevall. 2001. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc. Natl. Acad. Sci. USA 98:15245-15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suguimoto, H. H., A. M. Barbosa, R. F. Dekker, and R. J. Castro-Gomez. 2001. Veratryl alcohol stimulates fruiting body formation in the oyster mushroom, Pleurotus ostreatus. FEMS Microbiol. Lett. 194:235-238. [DOI] [PubMed] [Google Scholar]
- 52.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tolkacheva, T., P. McNamara, E. Piekarz, and W. Courchesne. 1994. Cloning of a Cryptococcus neoformans gene, GPA1, encoding a G-protein alpha-subunit homolog. Infect. Immun. 62:2849-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vartivarian, S. E., E. J. Anaissie, R. E. Cowart, H. A. Sprigg, M. J. Tingler, and E. S. Jacobson. 1993. Regulation of cryptococcal capsular polysaccharide by iron. J. Infect. Dis. 167:186-190. [DOI] [PubMed] [Google Scholar]
- 56.Wang, P., J. R. Perfect, and J. Heitman. 2000. The G-protein beta subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol. Cell. Biol. 20:352-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, Y., and A. Casadevall. 1994. Decreased susceptibility of melanized Cryptococcus neoformans to UV light. Appl. Environ. Microbiol. 60:3864-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waugh, M. S., C. B. Nichols, C. M. DeCesare, G. M. Cox, J. Heitman, and J. A. Alspaugh. 2002. Ras1 and Ras2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology 148:191-201. [DOI] [PubMed] [Google Scholar]
- 59.Williamson, P. R. 1994. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J. Bacteriol. 176:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williamson, P. R. 1997. Laccase and melanin in the pathogenesis of Cryptococcus neoformans. Front. Biosci. 2:e99—e107. [DOI] [PubMed] [Google Scholar]
- 61.Zaragoza, O., B. C. Fries, and A. Casadevall. 2003. Induction of capsule growth in Cryptococcus neoformans by mammalian serum and CO2. Infect. Immun. 71:6155-6164. [DOI] [PMC free article] [PubMed] [Google Scholar]