Abstract
Laccases are thought to be important to the virulence of many fungal pathogens by producing melanin, a presumed oxygen radical scavenger. A laccase in Cryptococcus neoformans has been shown to synthesize melanin and contributes to the virulence and the survival in macrophages of this fungal pathogen. One C. neoformans laccase gene, LAC1, previously called CNLAC1, has been extensively studied, and we describe a homologous gene, LAC2, that is found 8 kb away from LAC1 in the genome. In this study we report a role for both laccases, in addition to the thiol peroxidase, Tsa1, in oxidative and nitrosative stress resistance mechanisms of C. neoformans. With use of real-time PCR, similar changes in expression of the two laccase genes occur in response to oxidative and nitrosative stresses, but only the regulation of the LAC2 gene during stress is influenced by Tsa1. Both laccases contribute to melanin production using L-dopa as a substrate and are differentially localized in the cell based on green fluorescent protein fusions. A single deletion of either LAC1 or LAC2 alone had no effect on sensitivity to H2O2 or nitric oxide. However, deletion of either LAC1 or LAC2 in combination with a TSA1 deletion resulted in a slight peroxide sensitivity, and a lac2Δ tsa1Δ deletion strain was sensitive to nitric oxide stress. In addition, the deletion of both laccases reduces survival of C. neoformans in primary macrophages. Based on our expression and functional analysis, we propose a novel model for the interaction of these two systems, which are both important for virulence.
Cryptococcus neoformans is a neurotropic fungal pathogen found worldwide that causes meningoencephalitis, primarily in AIDS patients. In order to cause infection, C. neoformans must evade the host's innate immune system. This opportunistic pathogen has many defense mechanisms to protect itself from the oxidative attack encountered in the host. Professional phagocytes, including macrophages and neutrophils, engulf invading microorganisms and create an oxidant-rich environment in an attempt to protect the host from infection. Some enzymatic defenses shown to be important for stress resistance and virulence in C. neoformans include superoxide dismutase, thiol peroxidase, and flavohemoglobin denitrosylase (2, 4, 16). In addition, melanin has been shown to be important to the virulence of C. neoformans by protecting it and other fungal pathogens from reactive oxygen and nitrogen species produced by macrophages (9, 28; reviewed in reference 15).
The thiol-specific antioxidant Tsa1 is a 2-Cys peroxiredoxin dependent on the thioredoxin system. Recently, we have shown that Tsa1 is important for resistance to oxidative, nitrosative, and temperature stress as well as virulence in mice (16). In addition to its importance to oxidative stress resistance, the Schizosaccharomyces pombe Tsa1 homolog has been shown to be required for peroxide-induced activation of a stress response pathway (26).
One laccase has been described for C. neoformans as a glycosylated 75-kDa copper protein with diphenol oxidase activity (29). In the presence of a diphenolic substrate, melanin is produced by C. neoformans. The LAC1 gene, previously called CNLAC1, is transcriptionally repressed by glucose (29) and is capable of reconstituting melanin-deficient strains (21). Compared to the wild type, strains deficient in LAC1 have been shown to be more susceptible to macrophage killing and less virulent in mice (12, 21).
Since the completion of the genome project for C. neoformans, it has become apparent that another gene highly homologous to LAC1 exists, and we designate this second laccase gene LAC2. We present here the role of the laccases and their interaction with the thioredoxin system-dependent thiol peroxidase Tsa1 in the stress response of C. neoformans. In this study we show that both laccases contribute to melanin production, that there are independent regulatory mechanisms that control expression of the two laccases during nitrosative but not oxidative stress, and that both laccases contribute to survival in macrophages.
MATERIALS AND METHODS
Fungal strains and media.
H99, a well-characterized virulent clinical isolate of C. neoformans serotype A, was used as the wild-type strain. Strain genotypes and sensitivities are listed in Table 1. C. neoformans was grown on rich medium, YPD (1% yeast extract, 1% Bacto-peptone, 2% dextrose) or minimal medium, YNB (pH 4.0) (6.7 g of yeast nitrogen base/liter without amino acids plus 20 g of dextrose/liter and 25 mM sodium succinate at pH 4). Solid media contained 2% Bacto-agar. Selective YPD media contained either 200 U of hygromycin (Calbiochem, San Diego, Calif.)/ml, 100 μg of nourseothricin (Werner BioAgents, Jena-Cospeda, Germany)/ml, 250 μg of phleomycin (InVivoGen)/ml, or 200 μg of Geneticin (Gibco)/ml. For melanin production, glucose-free asparagine medium (1-g/liter l-asparagine, 0.5-g/liter MgSO4, 3-g/liter KH2PO4, 1-mg/liter thiamine) plus 1 mM L-Dopa was used.
TABLE 1.
C. neoformans strains used in these studies and their respective sensitivities to nitric oxide
| Genotypea (reference) | Parental strain or genotype | Construct | NO sensitivity |
|---|---|---|---|
| lac1Δ (17) | H99 | lac1::hyg | wtb |
| lac2Δ | H99 | lac2::nat | wt |
| lac1Δ lac2Δ | lac1Δ | lac2::nat | Slight |
| lac1Δ tsa1Δ | lac1Δ | tsa1::G418 | Slight |
| lac2Δ tsa1Δ | lac2Δ | tsa1::G418 | Moderate |
| lac1Δ lac2Δ tsa1Δ | lac1Δ lac2Δ | tsa1::G418 | Moderate |
| tsa1Δ (16) | H99 | tsa1::hyg | Slight |
| lac1Δ+LAC1 | lac1Δ | LAC1-G418 | wt |
| lac2Δ+LAC2 | lac2Δ | LAC2-G418 | wt |
| lac1Δ lac2Δ+LAC1 | lac1Δ lac2Δ | LAC1-G418 | wt |
| lac1Δ tsa1Δ+TSA1 | lac1Δ tsa1Δ | TSA1-phleo | wt |
| lac2Δ tsa1Δ+TSA1 | lac2Δ tsa1Δ | TSA1-phleo | wt |
| lac1Δ lac2Δ tsa1Δ+TSA1 | lac1Δ lac2Δ tsa1Δ | TSA1-phleo | Slight |
| tsa1Δ+TSA1 | tsa1Δ | TSA1-nat | wt |
Mutant strains that have been complemented by insertion of the wild-type gene into an undetermined location in the genome are indicated by “+” before the wild-type gene.
wt, wild-type sensitivity.
Localization using GFP.
The plasmid containing the coding sequence for green fluorescent protein (GFP) (accession no. U73901) was generously provided by John Perfect. Primers used to amplify this coding region annealed to bases 5 to 24 and 710 to 730. Overlap PCR technology (3) was used to generate the 3′ GFP fusion constructs, which included the entire genomic sequence of the desired gene and ∼1 kb of the upstream sequence, as well as the G418 resistance marker.
RNA extraction and cDNA synthesis.
Following the appropriate treatments, 50 ml of C. neoformans cells were collected by centrifugation at 1,800 × g for 5 min, washed once with distilled water, and lyophilized overnight. The lyophilized pellet was then vortexed with 3 ml of glass beads (1 mm; Biospec, Inc.) and resuspended in 4 ml of TriZol reagent (Invitrogen). After the mixture was left at room temperature for 5 min, 800 μl of chloroform was added and the mixture was shaken for 30 s. This cell lysate was then centrifuged at 4,000 rpm for 10 min, and the supernatant was transferred to a new tube. Two milliliters of isopropanol was added, incubated for 10 min at room temperature, and centrifuged at 2,200 × g for 10 min. After the pellet was washed with 75% ethanol, it was resuspended in water and incubated with DNase I at 37°C for 1 h. The RNA was extracted again with TriZol and chloroform and precipitated with isopropanol as described above. The dried pellet was resuspended in 300 μl of RNase-free water (Gibco) and stored at −80°C.
First-strand cDNA was made by using the First Strand cDNA synthesis kit for reverse transcriptase PCR (RT-PCR) (Roche). The cDNA was used in a PCR to generate the cDNA from LAC2, and the PCR products were sequenced on an ABI377 automated DNA sequencer.
Real-time PCR.
C. neoformans H99 was grown in minimal medium at 30°C with shaking overnight. Exponentially growing cells were treated with H2O2 or NaNO2 and allowed to grow at 30°C with shaking for 2 h. RNA was extracted and first-strand cDNA was made as described above. This cDNA was used as a template in a real-time PCR using SYBR Green PCR reagents (Sigma) according to the manufacturer's recommendations. The LAC1 primers annealed to bases 1316 to 1336 and 1725 to 1744 of the 1,872-bp gene, and the LAC2 primers annealed to bases 1352 to 1371 and 1725 to 1744 of the 1,782-bp gene. Base numbering is of the genomic sequence starting with the start codon ATG. The DNA Engine Opticon (MJ Research, Inc.) was used as the fluorescence detector with the following protocol for the PCR: 35 s at 94°C, 50 s at 53°C, and 50 s at 72°C, and a plate reading was repeated for a total of 40 cycles after a hot start of 10 min at 94°C. A melting curve was calculated at the end of the reaction to confirm a single product. A series of 10-fold dilutions of the cDNA was used in both the control and the experimental reactions. The induction data were taken from dilutions that came up 3.3 cycles apart, indicating that the reaction was in the linear range. The data were normalized to actin cDNA expression amplified in the set of PCRs.
Generation of laccase deletion constructs.
The lac1Δ and tsa1Δ mutants were described previously (16, 17). Overlap PCR gene deletion technology (3) was used to generate gene-specific deletion cassettes of LAC1, LAC2, and TSA1 that included a nourseothricin, hygromycin, or G418 resistance cassette (8, 14) and resulted in the deletion of the entire coding regions of the appropriate genes. Mutant strains were reconstituted by fusing a ∼4-kb fragment that contained the LAC1 or LAC2 coding sequence and ∼900 bp of the putative promoter sequence to a nourseothricin-, phleomycin-, or G418-selectable marker (8, 14), and the entire construct was transformed into the appropriate mutant strains.
Transformation of C. neoformans.
H99 and mutant strains were transformed by using biolistic techniques (8, 22). Cells were grown in YPD to late log phase, concentrated, and plated onto YPD agar for transformation. The cells were bombarded with 0.6-μm gold beads (Bio-Rad, Richmond, Calif.) which were coated with DNA of the target construct according to the manufacturer's recommendations. Following the transformation, the cells were incubated at 30°C for 4 h on nonselective medium to allow for recovery and then transferred with 0.8 ml of sterile phosphate-buffered saline to the appropriate selective medium. Transformants were observed in 3 to 5 days.
Analysis of transformants.
To isolate stable transformants, all transformants were passaged three times on nonselective YPD medium and then tested for resistance to the appropriate selective marker. Only those transformants that grew equally well on selective medium and nonselective medium were used as stable transformants. A three-primer PCR screen was used to prove homologous integration on both the 5′ and 3′ ends of the deletion construct (17). An example of this screen is the 5′ end of the lac2 deletion construct. Three PCR primers that anneal to genomic sequence outside the deletion construct (A), genomic sequence from the deleted portion of the gene (B), and the selectable marker cassette (C) are used in a PCR together. If the wild-type gene is present, a 948-bp PCR product is made from the A and B primers, and if a homologous recombination event has occurred, a 1,334-bp PCR product from the A and C primers will be synthesized. In this manner, homologous recombinants can be distinguished from the wild type. Integrations at the 3′ end of the lac2 deletions were identified in the same manner. A PCR screen using primers outside the deletion construct amplified the entire gene region; the wild-type gene was 4.4 kb, and the homologous recombinants were 3.4 kb, demonstrating that a single copy of the transforming DNA had been inserted at the LAC2 locus. Southern blotting was performed to screen for single integration. There were single bands observed on a Southern blot when probed with a selectable marker-specific probe. All deletion strains generated for this work had a single deletion construct homologously integrated at the appropriate locus and no other insertions in the genome (data not shown).
Genomic DNA preparation.
Genomic DNA was prepared by a modification of the glass bead DNA extraction protocol described by Fujimura and Sakuma (5). C. neoformans cells were suspended in a microfuge tube in 500 μl of lysis buffer (50 mM Tris [pH 7.5], 20 mM EDTA, 1% sodium dodecyl sulfate [SDS]) with 400 mg of glass beads (425 to 600 μm; G-9268; Sigma). Cells were disrupted by alternating 1 min of vortexing and 1 min on ice five times, followed by a 10-min incubation at 70°C. After brief vortexing, 200 μl of 5 M potassium acetate and 150 μl of 5 M NaCl were added. The tubes were placed on ice for 20 min and centrifuged at 14,000 rpm in an Eppendorf 5417C microcentrifuge for 20 min. The supernatant was mixed with 500 μl of phenol-chloroform and spun for 2 min at 14,000 rpm. The aqueous phase was then mixed with 450 μl of chloroform and spun for 2 min at 14,000 rpm. The DNA was then precipitated by addition of 200 μl of ethanol, washed with 70% ethanol, dried, and resuspended in 50 μl of deionized water.
Southern hybridizations.
Approximately 10 μg of genomic DNA from each strain was digested with various restriction endonucleases according to the manufacturer's recommendations. Restriction fragments were separated on a 1% agarose gel and transferred to nylon membranes using a Turbo-Blot apparatus (Schleicher & Schuell) and 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) as a transfer buffer. Probes for Southern analysis were prepared by random priming (random priming kit; Roche) using 50 μCi of [α-32P]dCTP (AA0005; Amersham) according to the manufacturer's instructions. The blots were incubated in 10 ml of a 6× SSC, 0.1% SDS, and 5% nonfat dry milk (Carnation) solution for 1 h at 65°C, and then probe was added to this solution and the blots were hybridized at 65°C overnight. The blots were washed twice in 2× SSC-0.1% SDS at room temperature for 10 min and once for 10 min in 0.2× SSC-0.1% SDS that had been prewarmed to 65°C.
Phenolic substrate oxidation activity assay.
Fifty milliliters of YPD cultures were pelleted and washed five times with sterile water. Cells were then plated on asparagine plates and incubated at 30°C for two nights. Cells were scraped and resuspended in 0.1 M sodium acetate (pH 5) or 0.05 M sodium phosphate (pH 6.5) for either the 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) or epinephrine oxidation assay, respectively. Cells were diluted to 2.2 × 108 cells/ml. One hundred eighty microliters of diluted cells were combined with 20 μl of the appropriate substrate (10 mM ABTS or 100 mM epinephrine). The mixture was incubated at 37°C for 30 min, and the reaction was stopped with 22 μl of 100 mM NaCN. The optical densities of cells stopped at time zero and at 30 min were read at 405 nm.
Oxidative and nitrosative stress plates.
Solid minimal medium was made with designated amounts of H2O2 or NaNO2. C. neoformans strains were grown to mid-log phase in asparagine media plus 1 mM L-Dopa, and 10-fold dilutions were made. Five microliters (each) of the undiluted and diluted cultures for each strain were spotted onto the solid minimal medium and grown at 30°C for two nights.
Macrophage assay.
Resident peritoneal exudate cells were harvested from 129 [129Sv(ev)] mice by peritoneal lavage with 10 ml of balanced salt solution, washed twice with Connaught’s Medical Research Laboratory (CMRL) medium supplemented with l-glutamine (2 mM) and penicillin-streptomycin (100 U/ml), and adjusted to a final concentration of 105 cells/ml in CMRL. Macrophages represent the major constituent of isolated peritoneal cells (>80%) (1, 13). One hundred microliters (104 macrophages) was plated into each well of a pretreated microtiter dish and incubated at 37°C and 5% CO2 for 24 h. C. neoformans cells were incubated in asparagine medium plus 1 mM L-Dopa for 12 h and diluted in CMRL to 105 cells/ml. The cryptococcal cells were added to the macrophages at a multiplicity of infection of 1 and incubated at 37°C in 5% CO2 for 24 h. One hundred microliters of 5% SDS was added to each well, and the mixture was incubated at RT for 5 min to lyse the macrophages. Serial dilutions were plated on YPD agar and incubated at 30°C for 2 days. Control wells without macrophages were used for each strain to control for growth of cryptococcal cells in CMRL media.
RESULTS
The first two introns of the LAC2 gene are inefficiently spliced.
A nucleotide BLAST search of LAC1 against the C. neoformans genomes resulted in two hits 8 kb apart on the same chromosome. In addition to the first hit, LAC1, we designated the second hit the LAC2 gene. LAC1 and LAC2 have very similar gene structures, sharing 76% nucleotide and 72% amino acid identity among their 14 exons (Fig. 1). The homology of these two genes is much greater in the coding regions than in the regulatory regions upstream and downstream of the genes. This suggests possible differences in the expression and regulation of the two laccases. Analysis of the sequences of the two genes revealed that there is an early termination codon in the LAC2 gene, resulting in a 14th exon that is smaller than that of LAC1. The LAC2 gene also has an insertion in the second intron, making it an unusually large, 255-bp intron. Large introns have been shown to be inefficiently spliced in other organisms, including Arabidopsis thaliana (10). RT-PCR using primers to the predicted full-length transcript revealed two bands, one of the predicted size (∼1,400 bp) and one that was ∼300 bp larger (data not shown). By sequencing clones from both bands, we show that transcripts exist with and without the first two introns, suggesting that these two introns are inefficiently spliced (GenBank accession AY817107 and AY817108). Independent RT-PCR using different sets of primers confirmed that this incompletely spliced transcript was present in multiple samples. Complete read-through of these introns from the first ATG is not possible due to the presence of stop codons, but a start codon exists in the beginning of the third exon, and therefore an alternative 55.7-kDa, rather than the 65.1-kDa, protein product is possible with the larger, alternatively spliced transcript.
FIG. 1.
Schematic alignment of the two laccase genes showing the unusually large second intron and truncated 14th exon in LAC2. Blocks represent exon sequences determined by alignment of cDNA sequence with genomic sequence. It is also noted that the first two introns (marked by arrows) on LAC2 are inefficiently spliced, potentially resulting in two different protein products.
Both laccases contribute to melanin production.
Deletion mutants of each and both of the laccase genes were generated. The lac1Δ and lac2Δ mutants were each reconstituted with the respective wild-type gene, while the lac1Δ lac2Δ double mutant was reconstituted with the LAC1 gene. As shown in Fig. 2, the laccase mutants all showed reduced melanin production when grown in the presence of the phenolic substrate L-Dopa, and these phenotypes were all reconstituted as expected. Similarly, all the laccase mutants had reduced ability to oxidize other phenolic substrates, including epinephrine and ABTS (data not shown). In addition, the tsa1Δ mutant (16) showed reduced epinephrine and ABTS oxidation, though it melanized in L-Dopa culture similarly to the wild type (data not shown). It is clear from this functional analysis that Lac1 is the primary contributor to melanin production and that Lac2 plays a minor role. Since the two laccase enzymes are homologous, it seems possible that both enzymes can perform this function and that there are differences in level of expression, enzyme activity, or localization of the two laccases.
FIG. 2.
Melanin production in wild-type strain H99 and in lac1Δ, lac2Δ, lac1Δ lac2Δ, and reconstituted strains when grown in asparagine media with 1 mM L-Dopa for 12 h at 30°C.
Laccases are differentially localized in the cell.
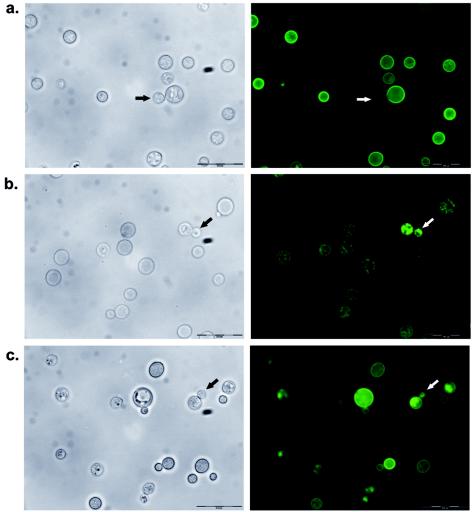
To determine the localization of both laccases, translational fusions of the laccases with GFP were engineered. The coding sequence for GFP was fused in frame to the 3′ end of the genomic sequence for each of the laccase genes. The constructs included 1 kb of untranslated 5′ sequence so that each fusion would be under the control of the appropriate laccase promoter. The Lac1-GFP and Lac2-GFP constructs were transformed into the lac1Δ and lac2Δ strains, respectively, and stable transformants were examined for complementation of the melanin defects and for GFP localization. Lac1-GFP was localized to the cell wall in a lac1Δ mutant background (Fig. 3a), confirming results of previous localization studies (31). In budding cells, Lac1-GFP was expressed only in the cell wall of mother cells but not in daughter cells (Fig. 3). Lac2-GFP was not localized to the cell wall but appeared to be cytoplasmic in a lac2Δ mutant background (Fig. 3b). Note that these fusion proteins are functional, since they reconstitute the melanin phenotypes in mutant strains (data not shown). These data suggest that the differences in melanin production of the laccase mutants could be due to a difference in localization. Enzymes localized to the cell wall would be more accessible to their substrates in the media. Interestingly, when we examined the expression of Lac2-GFP in a lac1Δlac2Δ mutant background, we saw partial localization to the cell wall in addition to its cytoplasmic localization (Fig. 3c). This suggests that the localization of Lac2 is altered in the absence of Lac1.
FIG. 3.
GFP fusion protein localization of laccases. Left panels, phase contrast; right panels, GFP fluorescence. (a) Lac1-GFP expressed in the lac1Δ mutant; (b) Lac2-GFP expressed in the lac2Δ mutant; and (c) Lac2-GFP expressed in the lac1Δ lac2Δ mutant. Arrows point to daughter cells in the process of budding, showing differential localization described in the text. Scale bar = 200 μM.
LAC2 is induced in response to nitrosative stress in the tsa1Δ mutant.
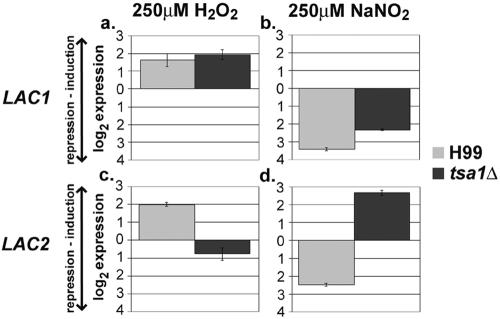
Using real-time PCR with specific primers for each laccase gene, we examined the expression of LAC1 and LAC2 in response to oxidative and nitrosative stress in C. neoformans (Fig. 4). LAC1 and LAC2 were similarly regulated in response to oxidative and nitrosative stress in the wild-type strain, H99. Both laccases were induced during hydrogen peroxide stress (Fig. 4a and 4c) and greatly repressed during nitric oxide stress in the wild type, H99 (Fig. 4b and d). Interestingly, in response to stress in the tsa1Δ mutant, LAC2 was regulated differently than it is in H99, while LAC1 expression was similar in both strains. LAC2 was no longer induced during peroxide stress in the tsa1Δ mutant (Fig. 4c), while LAC2 was induced rather than repressed in the tsa1Δ mutant during nitrosative stress (Fig. 4d). This suggested a possible compensation of LAC2 in the absence of TSA1 or a regulation pathway involving the expression of both these genes.
FIG. 4.
Laccase gene expression in response to stress in H99 and the tsa1Δ strain. LAC1 expression in response to (a) oxidative and (b) nitrosative stress and LAC2 expression in response to (c) oxidative and (d) nitrosative stress are shown. Note the altered expression of LAC2 in the tsa1Δ mutant with both oxidative and nitrosative stresses.
Lac2 is important for resistance to nitrosative stress in the absence of Tsa1.
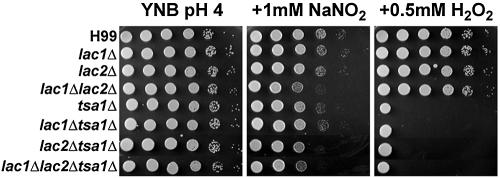
To determine if these differences in expression relate to phenotypic analysis, we generated combination mutants by deleting the TSA1 gene from the single and double laccase mutants. All the laccase mutants and combination mutants with the deletion in TSA1 were tested for sensitivities to various oxidative and nitrosative stresses to understand the importance of the laccases to the stress resistance of C. neoformans. Because it has been shown that LAC1 is derepressed in the absence of glucose (29) and that melanization helps protect cryptococcal cells from reactive oxygen species (27), we pretreated the cells in low-glucose medium and in the presence of 1 mM L-Dopa. Only the lac1Δ lac2Δ mutant sensitivity to nitric oxide was accentuated under these conditions. Interestingly, the single-laccase mutants did not show increased sensitivities to either peroxide or nitric oxide, while the double lac1Δ lac2Δ mutant was slightly sensitive to nitric oxide (Fig. 5). Though the lac2Δ tsa1Δ mutant was slightly sensitive to peroxide stress in comparison to the tsa1Δ mutant, this double mutant was more sensitive to nitric oxide stress. The triple lac1Δ lac2Δ tsa1Δ mutant did not show any additional sensitivity. We have reconstituted the laccase mutants and have rescued the oxidative and nitrosative stress phenotypes back to the parental strain phenotypes as expected (data not shown). Thus, Lac2 is the more important laccase for resistance to nitrosative stress in the absence of the Tsa1 thiol peroxidase.
FIG. 5.
Mutant phenotypes on nitric oxide or hydrogen peroxide. Tenfold dilutions of cultures incubated in asparagine media plus 1 mM L-Dopa for 16 h were plated on YNB (pH 4) plates and subjected to the indicated stress. Plates were incubated at 30°C for 48 h. Wild-type H99, laccase mutants, and laccase mutants with the additional deletion in TSA1 are shown.
Laccases are important for macrophage survival.
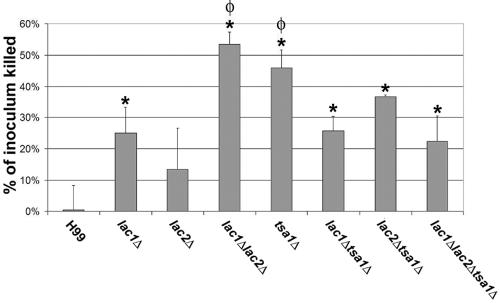
In order to determine the role of laccases in evasion of the innate immune response during infection, we tested the ability of the mutant strains to survive the oxidative attack of primary macrophages (Fig. 6). We confirmed the work of Liu et al. (12), showing a significant difference in killing of lac1Δ cells from that of wild-type cells by primary macrophages (P = 0.0190). Though the effects on survival in macrophages of the lac2Δ mutant were not significant (P = 0.2009), the effects from both laccase mutants appeared to be additive in the lac1Δ lac2Δ mutant, since it was killed significantly more than the wild type (P = 0.005) or the lac1Δ mutant (P = 0.0056). These macrophage data are consistent with the in vivo data from Pukkila-Worley et al. (19) using independently generated lac1Δ and lac2Δ mutants in a mouse inhalation model. The tsa1Δ mutant, which is avirulent in our mouse model (16), showed survival similar to that of the lac1Δ lac2Δ mutant. Interestingly, the additional deletion of the TSA1 gene in the lac1Δ strain appeared to partially rescue survival in macrophages, showing sensitivity similar to that of the lac1Δ single mutant (P = 0.8949) but significantly less than that of the tsa1Δ mutant (P = 0.0095). The lac1Δ lac2Δ tsa1Δ mutant also shows survival similar to that of the lac1Δ mutant (P = 0.7303) and significantly different from that of the lac1Δ lac2Δ or tsa1Δ mutant (P = 0.0039 or P = 0.0155, respectively). This suggests that regulation of the TSA1 and laccase genes is complex and is affected not only during nitrosative stress but during the stresses encountered by C. neoformans in the more elaborate environment of phagocytic cells.
FIG. 6.
Killing of C. neoformans strains by primary peritoneal macrophages. Strains were premelanized in asparagine media plus 1 mM L-Dopa for 12 h and incubated with macrophages at a multiplicity of infection of 1. The percentage of original inoculum that is killed within 24 h is presented. The asterisk denotes significant difference in killing compared to that for H99, and φ denotes significant difference compared to lac1Δ (P < 0.03).
DISCUSSION
C. neoformans encodes two laccases which are both regulated by oxidative and nitrosative stresses. These two genes, oriented in tandem in the C. neoformans genome, are very similar in structure. We found differences in expression of these two highly homologous genes with a tsa1Δ mutant background. This divergence in regulation and expression has been shown to be common in duplicated genes in many organisms, including two xanthine dehydrogenases from A. thaliana (6, 7, 11). We also report the existence of two forms of the LAC2 transcript. If these alternatively spliced products are both functional and result in different protein products, future studies should determine whether the splicing of LAC2 is regulated under different conditions and whether the two possible Lac2 isoforms perform different functions.
By generating deletion mutants of the laccase genes, we show that both Lac1 and Lac2 are capable of contributing to melanin production using L-Dopa as a substrate, an observation made independently by Pukkila-Worley et al. (19). It is possible that under the conditions used to induce melanin production, the majority of the LAC2 transcript is fully spliced, resulting in an enzyme extremely similar to Lac1. We do, however, observe a difference in melanin production between the lac1Δ and lac2Δ mutants, allowing us to conclude that Lac1 is more efficient at producing melanin in C. neoformans. This difference could result from a distinction in expression or localization of the two enzymes. An expression difference could be due to either a difference in transcription of these two genes, as suggested by Pukkila-Worley et al. (19), differences in message stability, or the inefficient splicing of LAC2, resulting in fewer molecules with functional Lac2 enzymatic activity.
Distinct localization of different isoforms has been shown to alter functional contribution of other enzymes (25), and we show that the laccases are differentially localized in C. neoformans. Lac1 is localized to the cell wall (Fig. 3a)(31), while Lac2 is cytoplasmic. This difference may in part account for the greater ability of Lac1 to produce melanin, since a substrate in the medium will be more accessible to the cell wall-localized enzyme and the deposition of melanin in the cell wall would not require additional transport. This differential location may contribute to the differences in substrate specificity observed by Pukkila-Worley et al. (19) in cells. We also find a difference in the localization of Lac2 in the absence of Lac1, since Lac2 appears to partially localize to the cell wall in addition to its cytoplasmic location. These two localizations could be due either to a change in expression of Lac2, to different protein products that have differential intracellular localization, or to a factor required for transport to the wall that is much more efficient for Lac1 and only transports Lac2 in the absence of Lac1. Further studies may uncover changes in regulation of LAC2 transcription or splicing that may account for this modification of localization.
Our expression studies demonstrate the importance of the laccases to oxidative stress defense in C. neoformans. We also show that the expression in response to oxidative or nitrosative stress in the wild type, H99, is similar for both LAC1 and LAC2. Interestingly, we observe a distinct regulation of each gene in response to stress in the absence of the TSA1 gene. This divergence in regulation results in the induced expression of LAC2 in the absence of Tsa1 during nitrosative stress, suggesting the down-regulation of a LAC2 repressor in the tsa1Δ mutant. Our functional analysis is consistent with these data, showing that one laccase enzyme is important for survival in the oxidative environment of macrophages and again suggesting the importance of Lac2 during nitrosative stress only in the absence of Tsa1. It is possible that this second laccase in C. neoformans is up-regulated in the absence of Tsa1 as a compensation mechanism to protect the cell from nitric oxide-induced damage, since laccases and other copper-containing oxidases have been shown to react with nitric oxide and metabolize it to the nontoxic nitrite (23, 24). This rapid reaction, which results in a peroxide intermediate of the laccase, also holds implications for the regulation of copper enzymes by nitric oxide (24, 30).
The down-regulation of LAC2 in response to nitrosative stress implies the presence of a repressor while the induction of LAC2 during this stress in the absence of TSA1 suggests the additional presence of an activator. Both activators and repressors are implicated in transcriptional responses to stress, including those mediated in S. cerevisiae by the Hog1 pathway involving Sko1 as a repressor of some target genes involved in oxidative stress defense (18, 20). It is possible that Tsa1 is involved in activating a stress-activated protein kinase pathway like a peroxiredoxin in S. pombe (26), which may in turn activate a repressor similar to Sko1 to repress LAC2 during nitrosative stress. In the absence of Tsa1, the repressor would not be activated and LAC2 would no longer be repressed. We observe an induction, and not merely an absence of repression, of LAC2 in the tsa1Δ mutant in response to NO. This could be explained by the presence of an activator that competes with the repressor to regulate the stress response as seen in the Sko1/Gcn4 system in S. cerevisiae (18). A homolog of Sko1 is not found in the C. neoformans genome, but a similar mechanism of regulation for LAC2 and other stress-regulated genes may exist. In addition, the proposed positive transcription factor for LAC2 could regulate other genes required to protect cells from this stress, since the lac2Δ mutant is not sensitive to nitric oxide.
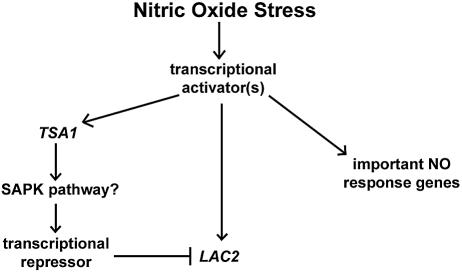
We propose a model based on our findings and stress-regulation results presented for other systems (Fig. 7). The nitric oxide stress response involves the up-regulation of one or more transcriptional activators regulating important stress response genes, including TSA1 and LAC2. The TSA1 protein product then activates a transcriptional repressor either directly or indirectly through a stress-activated protein kinase pathway. This transcriptional repressor acts competitively on LAC2, resulting in the down-regulation of LAC2 during nitrosative stress. In the absence of TSA1, the repressor is not expressed and therefore the activator is able to bind and initiate transcription from the LAC2 promoter, resulting in the induction of LAC2 during nitrosative stress in a tsa1Δ mutant.
FIG. 7.
Proposed model for nitric oxide stress regulation in C. neoformans.
Further studies need to be done to validate this model with C. neoformans. We have shown that a novel regulatory mechanism connects what were thought to be two separate defenses against oxidative and nitrosative stresses in this fungal pathogen. Since both are known to be important to virulence, understanding the interactions of these two interrelated systems may provide insights into the mechanisms by which C. neoformans establishes infection.
Acknowledgments
We gratefully thank Joel Eisenberg, Emi Torigoi, and Drew Walton for review of the manuscript prior to submission as well as John Perfect for providing plasmids. We also thank the C. neoformans H99 sequencing project, Duke Center for Genome Technology (http://cgt.genetics.duke.edu), the Broad Institute (www.broad.mit.edu/annotation/fungi/cryptococcus_neoformans), and the Genome Sequence Centre, BC Cancer Research Centre (http://www.bcgsc.bc.ca/), as well as the C. neoformans serotype D Genome Project, Stanford Genome Technology Center, funded by the NIAID/NIH under cooperative agreement U01 AI47087, and The Institute for Genomic Research, funded by the NIAID/NIH under cooperative agreement U01 AI48594. We thank the C. neoformans cDNA sequencing project at the University of Oklahoma (http://www.genome.ou.edu/cneo.html), funded by the NIH/NIAID AI147079.
This work was supported by an AHA fellowship to T.A.M., by NIH-NIAID grant RO1-AI44458 to J.A.C., and by NIH-NIAID grants RO1-AI051209 and RO1-AI50184 to J.K.L.
REFERENCES
- 1.Campbell, P. A., B. P. Canono, and J. L. Cook. 1988. Mouse macrophages stimulated by recombinant gamma interferon to kill tumor cells are not bactericidal for the facultative intracellular bacterium Listeria monocytogenes. Infect. Immun. 56:1371-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox, G. M., T. S. Harrison, H. C. McDade, C. P. Taborda, G. Heinrich, A. Casadevall, and J. R. Perfect. 2003. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect. Immun. 71:1538-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davidson, R. C., J. R. Blankenship, P. R. Kraus, M. de Jesus Berrios, C. M. Hull, C. D'Souza, P. Wang, and J. Heitman. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607-2615. [DOI] [PubMed] [Google Scholar]
- 4.de Jesus-Berrios, M., L. Liu, J. C. Nussbaum, G. M. Cox, J. S. Stamler, and J. Heitman. 2003. Enzymes that counteract nitrosative stress promote fungal virulence. Curr. Biol. 13:1963-1968. [DOI] [PubMed] [Google Scholar]
- 5.Fujimura, H., and Y. Sakuma. 1993. Simplified isolation of chromosomal and plasmid DNA from yeasts. BioTechniques 14:538-540. [PubMed] [Google Scholar]
- 6.Gu, Z., S. A. Rifkin, K. P. White, and W. H. Li. 2004. Duplicate genes increase gene expression diversity within and between species. Nat. Genet. 36:577-579. [DOI] [PubMed] [Google Scholar]
- 7.Hesberg, C., R. Hansch, R. R. Mendel, and F. Bittner. 2004. Tandem orientation of duplicated xanthine dehydrogenase genes from Arabidopsis thaliana: differential gene expression and enzyme activities. J. Biol. Chem. 279:13547-13554. [DOI] [PubMed] [Google Scholar]
- 8.Hua, J. H., J. D. Meyer, and J. K. Lodge. 2000. Development of positive selectable markers for the fungal pathogen Cryptococcus neoformans. Clin. Diagn. Lab. Immunol. 7:125-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobson, E. S., and S. B. Tinnell. 1993. Antioxidant function of fungal melanin. J. Bacteriol. 175:7102-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarvis, P., F. Belzile, and C. Dean. 1997. Inefficient and incorrect processing of the Ac transposase transcript in iae I and wild-type Arabidopsis thaliana. Plant J. 11:921-931. [DOI] [PubMed] [Google Scholar]
- 11.Langkjær, R. B., P. F. Cliften, M. Johnston, and J. Piskur. 2004. Yeast genome duplication was followed by asynchronous differentiation of duplicated genes. Nature 421:848-852. [DOI] [PubMed] [Google Scholar]
- 12.Liu, L., R. P. Tewari, and P. R. Williamson. 1999. Laccase protects Cryptococcus neoformans from antifungal activity of alveolar macrophages. Infect. Immun. 67:6034-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maggi, L. B., J. M. Moran, R. M. Buller, and J. A. Corbett. 2003. ERK activation is required for double-stranded RNA- and virus-induced interleukin-1 expression by macrophages. J. Biol. Chem. 278:16683-16689. [DOI] [PubMed] [Google Scholar]
- 14.McDade, H. C., and G. M. Cox. 2001. A new dominant selectable marker for use in Cryptococcus neoformans. Med. Mycol. 39:151-154. [DOI] [PubMed] [Google Scholar]
- 15.Missall, T. A., J. K. Lodge, and J. E. McEwen. 2004. Mechanisms of resistance to oxidative and nitrosative stress: implications for fungal survival in mammalian hosts. Eukaryot. Cell 3:835-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Missall, T. A., M. E. Pusateri, and J. K. Lodge. 2004. Thiol peroxidase is critical for virulence and resistance to nitric oxide and peroxide in the fungal pathogen, Cryptococcus neoformans. Mol. Microbiol. 51:1447-1458. [DOI] [PubMed] [Google Scholar]
- 17.Nelson, R. T., B. A. Pryor, and J. K. Lodge. 2003. Sequence length required for homologous recombination in Cryptococcus neoformans. Fungal Genet. Biol. 38:1-9. [DOI] [PubMed] [Google Scholar]
- 18.Pascual-Ahuir, A., R. Serrano, and M. Proft. 2001. The Sko1p repressor and Gcn4p activator antagonistically modulate stress-regulated transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:16-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pukkila-Worley, R., Q. D. Gerrald, P. R. Kraus, M.-J. Boily, M. J. Davis, S. S. Giles, G. M. Cox, J. Heitman, and J. A. Alspaugh. 2005. Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. Eukaryot. Cell 4:190-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rep, M., M. Proft, F. Remize, M. Tamas, R. Serrano, J. M. Thevelein, and S. Hohmann. 2001. The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol. Microbiol. 40:1067-1083. [DOI] [PubMed] [Google Scholar]
- 21.Salas, S. D., J. E. Bennett, K. J. Kwon-Chung, J. R. Perfect, and P. R. Williamson. 1996. Effect of the laccase gene, CNLAC1, on virulence of Cryptococcus neoformans. J. Exp. Med. 184:377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torres, J., D. Svistunenko, B. Karisson, C. E. Cooper, and M. T. Wilson. 2002. Fast reduction of a copper center in laccase by nitric oxide and formation of a peroxide intermediate. J. Am. Chem. Soc. 124:963-967. [DOI] [PubMed] [Google Scholar]
- 24.Torres, J., and M. T. Wilson. 1999. The reactions of copper proteins with nitric oxide. Biochim. Biophys. Acta 1411:310-322. [DOI] [PubMed] [Google Scholar]
- 25.Valadi, A., K. Granath, L. Gustafsson, and L. Adler. 2004. Distinct intracellular localization of Gpd1p and Gpd2p, the two yeast isoforms of NAD+-dependent glycerol-3-phosphate dehydrogenase, explains their different contributions to redox-driven glycerol production. J. Biol. Chem. 279:39677-39685. [DOI] [PubMed] [Google Scholar]
- 26.Veal, E. A., V. J. Findlay, A. M. Day, S. M. Bozonet, J. M. Evans, J. Quinn, and B. A. Morgan. 2004. A 2-Cys peroxiredoxin regulates peroxide-induced oxidation and activation of a stress-activated MAP kinase. Mol. Cell 15:129-139. [DOI] [PubMed] [Google Scholar]
- 27.Wang, Y., P. Aisen, and A. Casadevall. 1995. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect. Immun. 63:3131-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, Y., and A. Casadevall. 1994. Susceptibility of melanized and nonmelanized Cryptococcus neoformans to nitrogen- and oxygen-derived oxidants. Infect. Immun. 62:3004-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williamson, P. R. 1994. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J. Bacteriol. 176:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson, M. T., and J. Torres. 2004. Reactions of nitric oxide with copper containing oxidases; cytochrome c oxidase and laccase. IUBMB Life 56:7-11. [DOI] [PubMed] [Google Scholar]
- 31.Zhu, X., J. Gibbons, J. Garcia-Rivera, A. Casadevall, and P. R. Williamson. 2001. Laccase of Cryptococcus neoformans is a cell wall-associated virulence factor. Infect. Immun. 69:5589-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]