Significance
This article describes the characterization of the antigenic determinant of the capsular polysaccharide from the clinically relevant serotype III of group B Streptococcus (GBS). NMR and X-ray crystallography have been applied to elucidate the interaction of type III GBS oligosaccharides obtained by synthetic and depolymerization procedures of the bacterial polysaccharide with a functional monoclonal antibody. A Fab–GBS oligosaccharide complex structure has been solved at high resolution (2.7 Å). The results demonstrate the existence of a sialic acid-dependent functional epitope of GBS that is fully contained within four consecutive sugars deriving from the type III GBS polysaccharide backbone and one branched disaccharide present in this sequence. This finding has implications for the development of vaccines against GBS infection.
Keywords: group B Streptococcus, capsular polysaccharide, antigen
Abstract
Despite substantial progress in the prevention of group B Streptococcus (GBS) disease with the introduction of intrapartum antibiotic prophylaxis, this pathogen remains a leading cause of neonatal infection. Capsular polysaccharide conjugate vaccines have been tested in phase I/II clinical studies, showing promise for further development. Mapping of epitopes recognized by protective antibodies is crucial for understanding the mechanism of action of vaccines and for enabling antigen design. In this study, we report the structure of the epitope recognized by a monoclonal antibody with opsonophagocytic activity and representative of the protective response against type III GBS polysaccharide. The structure and the atomic-level interactions were determined by saturation transfer difference (STD)-NMR and X-ray crystallography using oligosaccharides obtained by synthetic and depolymerization procedures. The GBS PSIII epitope is made by six sugars. Four of them derive from two adjacent repeating units of the PSIII backbone and two of them from the branched galactose–sialic acid disaccharide contained in this sequence. The sialic acid residue establishes direct binding interactions with the functional antibody. The crystal structure provides insight into the molecular basis of antibody–carbohydrate interactions and confirms that the conformational epitope is not required for antigen recognition. Understanding the structural basis of immune recognition of capsular polysaccharide epitopes can aid in the design of novel glycoconjugate vaccines.
Bacterial cell surface carbohydrates are the interface of multiple host interactions and have been targeted to develop highly efficacious glycoconjugate vaccines against severe infections caused by Streptococcus pneumoniae, Haemophilus influenzae type b, and Neisseria meningitidis (1, 2). Glycoconjugate vaccines against other important pathogens are under clinical or preclinical development (2).
The mapping of polysaccharide (PS) epitopes recognized by functional antibodies mediating protection from infection is crucial for understanding the mechanism of action of this type of vaccine. In many cases, the antigenic determinants of the immunological properties of PS and the structural details of the minimal epitope targeted by specific functional antibodies are unknown. Structural biology has been commonly practiced in the last decade for the characterization of protein antigen–antibody interactions (3). However, it has been less applied to carbohydrate antigens, in part because of the well-known difficulty of crystallizing carbohydrates.
Minimal epitopes can be composed of short, defined glycans comprising 2–3 monosaccharides, as for the β-(1→2) mannans of the Candida albicans cell wall (4), Vibrio cholerae O1 (5), Shigella flexneri variant Y (6), and Salmonella (7) O-antigens, or a tetrasaccharide, as for the repeating unit (RU) of S. pneumoniae type 14 PS (Pn14) (8, 9), and even six sugar residues, as in the case of S. flexneri serotype 2a O-antigen (10). In contrast, the type III PS of Streptococcus agalactiae (group B Streptococcus, GBS) has been proposed as a prototype of a unique length-dependent conformational epitope (11).
GBS is an encapsulated Gram-positive β-hemolytic pathogen causing neonatal sepsis and meningitis, particularly in infants born to mothers carrying the bacteria (12). The GBS capsular PS is constituted by multiple RUs (from ∼50 up to 300 per polymer) of four to seven monosaccharides shaped to form a backbone and one or two side chains. Ten serotypes presenting a unique pattern of glycosidic linkages have been identified and their primary structures elucidated (13). Three monosaccharides (β-d-glucopyranose, β-d-Glc; β-d-galactopyranose, β-d-Gal; and β-d-N-acetylglucosamine, β-d-GlcNAc) are present in all of the described serotypes, and sialic acid (α-N-acetyl-neuraminic acid, NeuNAc) is always found at the terminus of one chain (13). Maternal concentrations of IgG directed to the different GBS capsular serotypes inversely correlate with the risk of newborn infection (14), suggesting that antibodies able to cross the placenta can confer serotype-specific infant protection (15). PS conjugates of different serotypes elicit antibodies that mediate GBS type-specific opsonophagocytic killing (OPK) in functional assays and protection against GBS challenge in neonate mice (16–20). Furthermore, Ia, Ib, II, III, and V monovalent vaccines, as well as a trivalent combination (Ia, Ib, and III), have been proven to be safe and immunogenic in nonpregnant and pregnant women, and elicited maternal antibodies were efficiently transferred to neonates (21–23).
GBS strains belonging to the serotype III (GBSIII) are epidemiologically the most relevant in neonatal infections (24). Pioneering immunological studies aimed at elucidating type III-specific antigenic determinants indicated the presence of the terminal NeuNAc residue as essential for the elicitation of protective antibodies (25). Subsequent studies revealed that sera from rabbits immunized with GBSIII bacteria and from humans vaccinated with PSIII conjugates contained two types of anticarbohydrate antibodies—that is, a major population recognizing the native PS but not its incomplete core antigen derivative lacking sialic acid (corresponding to the S. pneumoniae PS group 14, Pn14), and a minor variable population reacting with both the native PSIII and the core antigen (26, 27). Remarkably, both types of human PSIII-induced antibodies were shown to mediate GBSIII OPK, whereas antibodies elicited by Pn14 (desialylated PSIII) did not recognize GBSIII bacteria and therefore did not mediate GBS OPK (27).
Studies using 13C NMR spectroscopy highlighted ring-linkage signal displacements in the core versus the native PS, suggesting that NeuNAc residues exert a specific control over the conformation of the native PS (26, 28). Molecular dynamics simulations confirmed a more flexible and disordered structure for desialylated PSIII and suggested that native PSIII could form extended helical structures where each turn was made by more than four RUs (29–32). PSIII fragments smaller than four RUs appeared as weak inhibitors of the binding of native PSIII to its specific antibodies and failed to elicit an efficient immune response following conjugation (33, 34). Based on the above observations, it was concluded that the native PSIII forms a sialic acid-dependent conformational epitope that is essential for the elicitation and recognition of functional antibodies, and the length dependency of this conformational epitope was ascribed to its localization on extended helices within a random coil structure (33). According to the proposed model, the side chain NeuNAc moiety would exercise a remote control over immunological determinants of the PS backbone, without being directly involved in molecular interactions as part of the epitope (34).
Structural glycobiology studies using bacterial oligosaccharides in complex with functional monoclonal antibodies (mAbs) able to protect against the target pathogen could represent the most direct methodology to gain information on PS immunological determinants at the atomic level. However, due to the high polarity of the hydroxyl groups and flexibility of the carbohydrate structures, crystallization of carbohydrate–protein complexes is a challenging task (35). To date, only a very limited number of carbohydrate antigen–Fab complexes, in comparison with a large number of protein antigen–Fab complexes, have been resolved by X-ray crystallography (5–7, 10). As an alternative, a combination of techniques [NMR, surface plasmon resonance (SPR), ELISA, etc.] is typically used to gain insights into carbohydrate–protein interactions (36, 37).
In the present study, we used saturation transfer difference–NMR (STD-NMR) in conjunction with X-ray crystallography to investigate the interaction of GBSIII oligosaccharides obtained by synthetic and depolymerization procedures with a protective antibody.
Results
Selection and Immunochemical Characterization of a Functional Anti-PSIII Rabbit mAb.
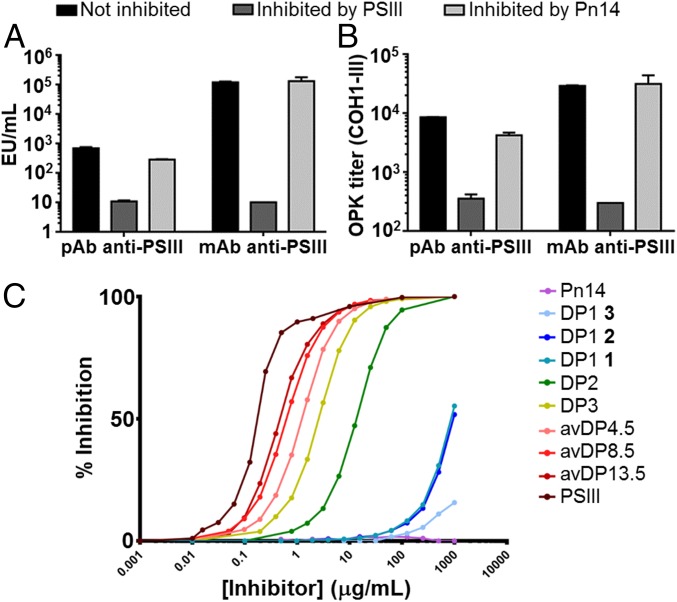
To investigate the binding of GBS PSIII to functional antibodies at the atomic level and further decipher the role of the NeuNAc moiety, we first generated a rabbit anti-PSIII mAb capable of mediating GBS OPK. Similar to a previous report (26), rabbits immunized with native PSIII conjugated to genetically detoxified diphtheria toxin (CRM197) developed two types of antibodies recognizing the native PS but differing in their capacity to bind the PSIII desialylated core antigen. Indeed, inhibition ELISA experiments showed that binding of IgG from rabbit sera to immobilized PSIII unconjugated (SI Appendix, Fig. S1) or conjugated to human serum albumin (HSA) (38) (Fig. 1A) could be partially blocked by preincubation with soluble Pn14 or desialylated PSIII. Pn14 also partially inhibited antibody-mediated GBSIII OPK (Fig. 1B), confirming functional activity of both types of NeuNAc-dependent and -independent rabbit antibodies. Similar results were described previously in the case of human responses to GBSIII (27).
Fig. 1.
PSIII antibody specificity and functional activity toward PSIII. Inhibition experiments with PSIII and Pn14 of rabbit anti-PSIII polyclonal sera (pAb) and mAb in (A) ELISA, binding to immobilized PSIII conjugated to HSA, and (B) OPK. (C) Competitive SPR of the binding between the rabbit Fab and PSIII. Different length fragments were used as inhibitors. PSIII and the Pn14 were used as positive and negative controls, respectively.
An IgG1 mAb was selected by hybridoma cell line technology (NVS-1-19-5), and its activity was ascertained by OPKA (OPK titer, 1.03 μg/mL). The selected monoclonal was representative of the typical response to type III PS and had the properties described in the literature (33, 34). Complete inhibition by native PSIII and absence of inhibition by Pn14 in ELISA and OPKA experiments (Fig. 1 A and B) confirmed that the epitope recognized by this mAb was NeuNAc-dependent. A Fab fragment of mAb NVS-1-19-5 was prepared by papain proteolytic cleavage, and its high-affinity binding toward the GBS PSIII was measured by SPR (KD estimated with the Fab 4 × 10−8 M).
The NeuNAc-Gal covalent link in the lateral chain of the PSIII RU is the most chemically labile glycosidic bond within the PS, and complete loss of NeuNAc residues or reduction of their carboxylic groups results in PS structures incapable of inducing functional antibodies (25, 26). To investigate the potential of the obtained mAb for probing the integrity of the PSIII antigen, we subjected PSIII-CRM197 to mild acid hydrolysis for different incubation times and obtained conjugates with decreasing sialylation levels of 100, 60–70, 20–30 and <5%, as estimated by NMR analysis (SI Appendix, Fig. S2 and Table S1). The resulting glycoconjugates were characterized for their structural integrity and saccharide/protein content (SI Appendix, Fig. S3 and Table S2). ELISA experiments using immobilized native PSIII as the antigen and soluble native or partially desialylated PSIII-CRM197 as the competitor showed decreasing binding of the mAb to conjugates with increasing desialylation levels (SI Appendix, Fig. S4A). The critical role of the NeuNAc in elicitation of functional antibodies was confirmed by immunizing female mice with three doses of the different glycoconjugates, followed by sera analysis by ELISA for quantification of anti-PSIII antibodies, and by OPKA to assess antibody functional activity. Lower levels of sialylation resulted in diminished recognition of HSA-conjugated native PSIII and decreasing in vitro killing of GBS bacteria (SI Appendix, Fig. S4 B and C).
Selection of PSIII Glycan Fragments for Structural Studies.
Antibodies elicited in mice by PSIII have been shown to bind oligosaccharide fragments in a length-dependent manner (33, 34, 39). Using a competitive ELISA, it was demonstrated that at least two unmodified RUs were necessary for suboptimal binding to a GBSIII-specific mouse mAb. Inhibition with 3–7 RU PSIII fragments showed a moderate increase and became much higher above this number of RUs (29). SPR experiments using a Fab of the same mAb confirmed that the affinity (KD) was comparable between 2 and 6–7 RUs, it increased by threefold from 6–7 RUs to 20 RUs, and it remained constant beyond this point (33).
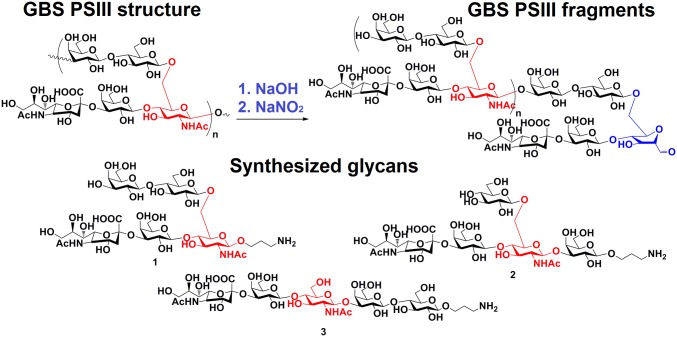
To investigate more in detail the epitope recognized by the selected functional mAb, we generated a set of PSIII oligosaccharide fragments by partial de–N-acetylation followed by nitrosation (40). Improvements in the purification process of the PSIII fragments by using an anionic exchange HPLC in place of size exclusion HPLC, as described by Paoletti et al. (41), allowed for the isolation of fragments with a degree of polymerization (DP) in the range of 2–15 with a more defined composition (SI Appendix, Fig. S5). These fragments were composed of a modified RU and a variable number of unmodified RUs (Fig. 2). The identity of these oligosaccharides was established by 1H NMR, and the ratio between the signals of the generated hydrated aldehyde group and the H-3e (i.e., equatorial) position of NeuNAc or the H-2 position of Glc was used to determine the oligosaccharide length (SI Appendix, Fig. S6). The relative ratio of monosaccharide components in the obtained oligosaccharides was ascertained by high-performance anion-exchange chromatography coupled with pulsed amperometric detection (HPAEC-PAD) (SI Appendix, Table S3). MALDI-TOF MS spectra in negative mode confirmed (SI Appendix, Fig. S7) the structure of the first two peaks eluted on the analytical MonoQ anionic exchange column as DP2 and DP3. Both were composed of homogeneous glycans, whereas longer oligosaccharides contained mixtures of glycans and are hereafter indicated as average DP (avDP). A single PSIII RU was chemically synthesized by conventional carbohydrate chemistry procedures (42). Given that the five residues composing the RU can be arranged according to three different sequences, three alternative glycans were assembled with an end-terminal sugar bearing a linker for possible conjugation (Fig. 2). After deprotection, the defined glycans were purified on size exclusion chromatography and characterized by NMR and MS (procedures for synthesis and characterization data are reported elsewhere) (42).
Fig. 2.
Chemical structure of GBS PSIII and glycan probes used in this study. The red highlighted GlcNAc residue is used as the reference monosaccharide to outline the three possible sugar sequences related to PSIII RU. The 2,5-anhydri-d-mannose residue obtained by chemical depolymerization of PSIII is indicated in blue.
Length-dependent recognition of the different fragments was confirmed by competitive ELISA with the selected rabbit mAb (SI Appendix, Fig. S8) (33). Inhibition of mAb binding increased two logs between DP1 and DP2 and then slightly increased another 1.5 log, changing the PS size from DP2 to DP13, and became five-log higher when PSIII was used as an inhibitor (Fig. 1C and SI Appendix, Table S4). To exclude the effect of the bivalent IgG interaction on the avidity, we performed a competitive SPR assay where different oligosaccharide fragments (DP1–13 range) were tested as competitors for the binding of soluble Fab fragment to PSIII conjugated to HSA immobilized on the chip. Overall, two major populations of inhibitors, DP ≥ 2 and DP < 2, were differentiated. DP ≥ 2 oligosaccharides showed asymptotically increasing affinity up to the native PSIII, with only a 2-log difference between native PSIII and DP2. Concerning the single RUs, a 2-log difference was observed between DP2 and the synthetic DP1 RU 1 and 2 had a β-Glc-(1→6)–β-GlcNAc branching; conversely, a very weak inhibition was detected for the linear structure 3, suggesting that the arrangement of sugars in the RU impacts Ab recognition and the ramification point is relevant for binding.
The binding affinities of the native PSIII and the DP2 fragment to the rabbit Fab were compared by conjugating the two PS structures to CRM197 and immobilizing them on an SPR chip. The two binding constants determined according to a 1:1 fitting model differed by less than 10-fold (SI Appendix, Fig. S9 and Table S5) (33). Overall, this analysis confirmed a length-dependent affinity of anti-PSIII GBS antibody but also indicated that DP2 contains the PSIII portion necessary for high-affinity antibody binding and could be used for further structural analysis.
Identification of the PSIII Antigenic Determinant by STD-NMR.
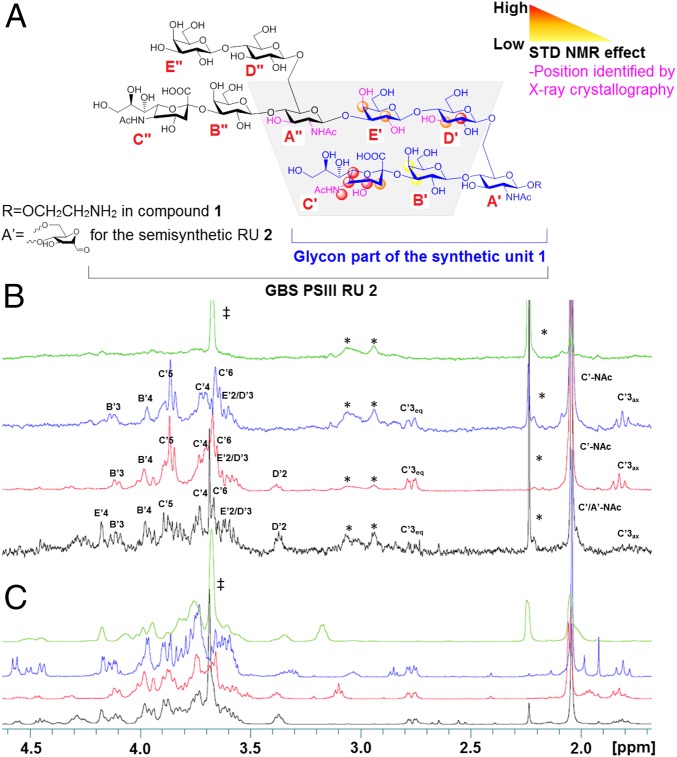
To map the interactions of PSIII oligosaccharides with the protective mAb, STD-NMR studies were undertaken (43). STD difference (STDD)-NMR spectra were derived by subtracting the STD-NMR spectrum of the glycan in the bound state with the mAb (ligand/protein 15–50:1 molar ratio) to the reference spectrum in the unbound state. The DP3 STDD spectrum was superimposable to the one obtained for the DP2–mAb complex (SI Appendix, Fig. S10), highlighting that the antibody was recognizing identical regions in the two fragments. Due to their higher affinity constants and larger size, experiments carried out on longer fragments (5.5 and 8 RUs) resulted in lower signal intensities that did not allow unequivocal detection of saturation transfer effects.
Notably, most of the 1H NMR resonances that could unequivocally be assigned to the DP2–mAb complex were related to the α-NeuNAc-(2→3)–β-GalB-(1→4) residues (Fig. 3A), indicating a direct involvement of this branch in antibody binding, in addition to the Glc and Gal residues of the backbone. In the DP2 oligosaccharide, it was not possible to differentiate the NAc group of NeuNAc and GlcNAc. To better discriminate the exact positions engaged in antibody interaction, the synthesized DP1 was complexed with the mAb and analyzed by STDD-NMR. As shown in Fig. 3 B and C, well-recognizable resonances of the branched RU 1 were recovered in the STDD spectrum with medium–high saturation intensity (>50%). The highest transfer of saturation was observed for H-5 (3.85 ppm), H-4 (3.70 ppm), H-3 (2.76 and 1.82 ppm for equatorial and axial, respectively), and H-6 (3.65 ppm) of NeuNAc (Fig. 3B), confirming that NeuNAc is the major anchoring site for mAb interaction. A similar pattern of signals related to the NeuNAc was obtained with units 3 (Fig. 3) and 2 (SI Appendix, Fig. S11). Further, the involvement of the NeuNAc N-acetyl group, not clearly distinguishable from the one of the GlcNAc residue in the DP2, was unveiled in the synthetic probes at 2.0 ppm. When the desialylated tetrasaccharide [β-Gal-(1→4)–β-GlcNAc-(1→3)–β-Gal-(1→4)–α/β-Glc] was analyzed in the presence of the mAb, no transfer of saturation was observed. A high STD signal was also observed for the Glc H-2 position at 3.37 ppm in the DP2 and DP1 RUs 1 and 2 (SI Appendix, Fig. S11) in complex with the mAb, whereas no recovery was seen for this position in the linear RU 3, where the Glc moiety is located far from the NeuNAc (SI Appendix and Fig. 3 B and C). Previous conformational studies revealed that the orientation of the Glc backbone residue was affected by removal of sialic acid, suggesting spatial proximity of the two residues (32). The transfer of saturation to the H-2 of Glc indicated that mAb binding of this residue is determined by its intrinsic location in the sugar sequence.
Fig. 3.
GBS PSIII epitope mapping. (A) Protons interacting with the mAb identified by STD-NMR and X-ray crystallography. (B) STDD and (C) related 1H NMR spectra of the desialylated tetrasaccharide [β-Gal-(1→4)–β-GlcNAc-(1→3)–β-Gal-(1→4)–α/β-Glc in green], the linear 3 (blue) and branched 1 (red) RUs, and the DP2 fragment (black). Proton positions receiving saturation after irradiation of the protein are indicated. ‡ indicates the CH2 signal of the Tris buffer; * refers to signals related to the protein.
Owing to the better resolution of the STD-NMR spectra of the synthetic glycans, saturation transfer was more precisely quantified for some of the protons involved in binding. Experiments at increasing saturation times from 0.5 up to 5.0 s were performed to avoid possible bias in the calculation of STD effects due to different proton longitudinal relaxation times (T1) or intramolecular spin diffusion within the bound state (SI Appendix, Fig. S12). In the synthetic glycan 1, an STD relative effect of 100%, 96%, and 70% was measured for H-5, H-3eq signals of NeuNAc, and H-2 of Glc, respectively.
Taken together, these results unambiguously showed that the side chain NeuNAc and Gal residues, in conjunction with the Gal and Glc backbone sugars of the RU, directly interact with the mAb.
Structural Studies of the PSIII–Fab Complex by X-Ray Crystallography.
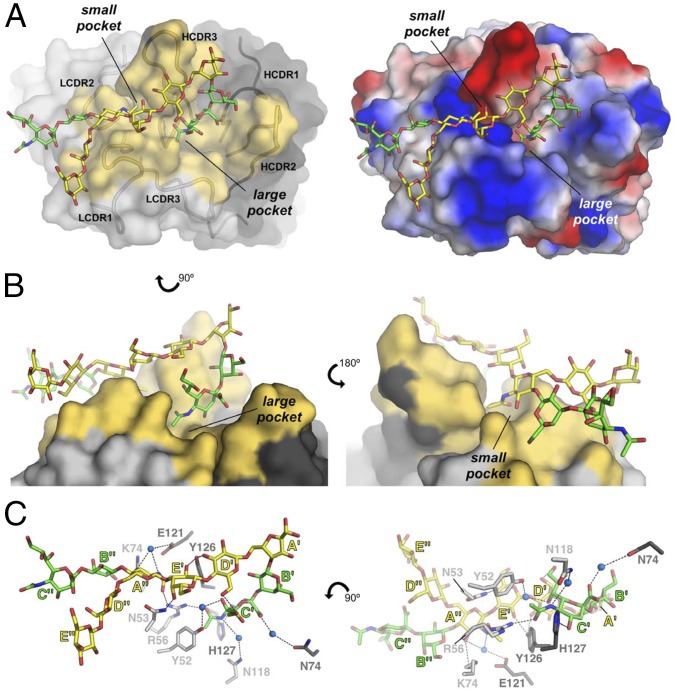
To further elucidate the basis for the PSIII–mAb interaction at the atomic level, we used X-ray crystallography to determine the 3D structure of DP2 in complex with the Fab NVS-1-19-5. Crystals of the complex belong to space group C2221, contain two copies of a 1:1 DP2–Fab complex in the asymmetric unit (ASU), and diffract to a resolution of 2.7 Å. The structure was solved by molecular replacement, using as template input model the coordinates of a rabbit Fab [Protein Data Bank (PDB) ID code 4JO1] with which Fab NVS-1-19-5 shares 79% sequence identity. Excellent electron densities were observed for the Fab portions and allowed the modeling of residues Gln20–Ser245 of the heavy (H) chain and Val24–Cys238 of the light (L) chain. Additional difference densities were observed in front of the Fab complementarity determining regions (CDRs), and these were modeled with the DP2 oligosaccharide (SI Appendix, Fig. S13). Of the total 10 monosaccharide residues of DP2, 6 could be modeled with very good fitting in clear electron density maps and in both copies of the complex, whereas the terminal saccharides residues (B″, C″, D″, and E″) had low sigma electron densities for one of the copies of the complex only. For completeness, the full DP2 was included in the final refined model, although the terminal residues had low sigma electron density signals and subsequently high B-factors, both of which reflect uncertainty in their exact position. Importantly, the stereochemistry and geometry of final refined DP2 molecules were validated using Privateer (SI Appendix, Table S6). The coordinates of the complex were refined to final Rwork/Rfree values of 23.6%/28.2% (SI Appendix, Table S7). Structural superimposition of the two copies of the complex present in the ASU resulted in very low carbon alpha root mean square deviation (Cα rmsd) values (∼0.5 Ǻ; the two structures are essentially identical), and their bound DP2 conformations were also highly similar; therefore, one copy will be used for the description of the structure.
The Fab fragment binds the glycan with an extended interface (577 Å2) formed by two adjacent pockets that are located between the L and H chain of the Fab (Fig. 4 A and B and SI Appendix, Fig. S14). Most of the observed contacts with the Fab involve a single PS RU, whereas the sugar residues of the consecutive RU depart from the binding pockets, with the exception of GlcNAc-A″, which is directly bound to Fab amino acid residues. The larger pocket hosts the NeuNAc of the RU branch that interacts with multiple hydrophilic residues of the CDRs from both the H and L chains. Specifically, Arg56 of the L chain forms a hydrogen bond with the N-acetyl group of NeuNAc-C′, and His127 interacts with its O-4 (Fig. 4C). Additional weaker interactions of NeuNAc-C′ occur through a water molecule bridge between the carboxylic group of the sugar and Asn74 of the H chain and the CH-π interaction between Tyr52 of the L chain and the sialic NAc methyl group (Fig. 4C). The GlcNAc-A″ residue appears involved in the binding to the smaller anchoring pocket of the Fab through at least two hydrogen bonds formed by the L chain Arg56 and Lys74 with the NAc group and the O-3 position of the sugar, respectively. The NAc group is also stabilized by a CH-π interaction with Tyr126 (Fig. 4C). Interestingly, the guanidine group of Arg56 is located within H-bond distance from the NAc carbonyl groups of both NeuNAc-C″ and GlcNAc-A″, whose methyl groups are at the same time favorably oriented to make CH-π interactions with the phenyl rings of Tyr52 of the L chain and Tyr126 of the H chain, respectively. This arrangement allows for a peculiar symmetric network of interactions involving the NeuNAc of one RU and the GlcNAc of a consecutive unit (Fig. 4C). Complete de–N-acetylation of these residues resulted in loss of Fab recognition as demonstrated by competitive SPR (SI Appendix, Fig. S15). Inspection of the glycan structure revealed how Gal-E′, although positioned outside of the two binding pockets, interacts with side chain atoms of Asn53 of the L chain and of Tyr126 of the H chain. The side chain of Tyr126 seems also favorably positioned to engage H-bond–mediated interactions with the O-3 position of Glc-D′. Interestingly, GlcNAc-A′ that was modified into 2,5-anhydro-d-mannose during the depolymerization reaction appears to be far from the anchoring pocket; hence, its chemical manipulation during depolymerization did not interfere with Fab binding. The DP2–Fab complex crystal structure also revealed how NeuNAc-C′ and Glc-D′ are in close contact, with a calculated distance between the O-9 atom of NeuNAc and the O-6 of Glc of 2.8 Å (Fig. 4C). This suggests that hydrogen bonds between these two positions might be further stabilized upon binding to the Fab. O-acetylation of the NeuNAc glycerol chain was noticed in PSIII from GBS clinical isolates, and it affects inhibition of neutrophil suppression and virulence (44, 45). We observed that the mAb interacts with the O-7 of NeuNAc through an H2O molecule, and dimensionally, this space could also host an acetyl group. This could explain the data from the literature reporting that O-acetylation does not interfere with protection (46).
Fig. 4.
Crystal structure of DP2 bound to Fab NVS-1-19-5. (A, Left) The Fab is depicted with surfaces and the DP2 with sticks. Fab residues involved in direct binding with DP2 (the paratope) are colored in yellow, and the binding pockets are labeled. (A, Right) Surface electrostatic potential distribution of the Fab, oriented as on Left. (B) Two views (Left and Right) of the large and small pockets where the DP2 binds. (C) Details of the interactions between DP2 and Fab. Carbon atoms of the DP2 backbone are colored in yellow, and those belonging to the branches are in green, whereas carbon atoms of the Fab are colored in light and dark gray for the L and H chain, respectively. Nitrogen and oxygen atoms are colored in blue and red, respectively, and water molecules are shown as blue spheres.
In summary, our structure clearly showed that the side chain NeuNAc and the backbone Gal and Glc of the first RU, as well as the backbone GlcNAc of the consecutive RU, interact with multiple hydrophilic residues of the CDRs from both the H and the L chain, whereas the NeuNAc glycerol moiety is involved in internal interactions rather than in direct binding to the Fab.
Discussion
Conjugate vaccines have been one of the major developments of the last 40 y. The nature of the repetitive protective epitopes present in the PS has been difficult to define, mostly because of the difficulty in obtaining crystals from PSs. In many instances, it has been speculated that the protective epitopes would be formed from distant residues that would become close in a helical structure assumed by the PS (28, 29).
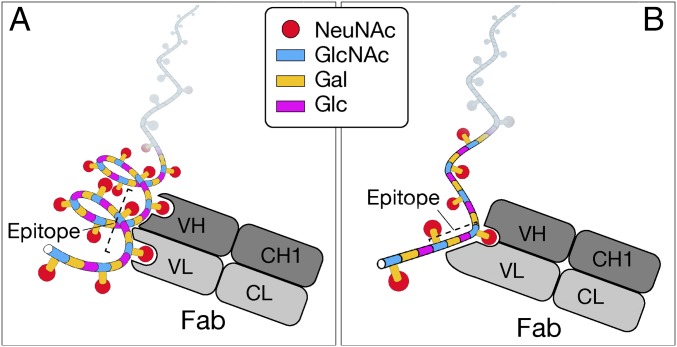
In this study, we determined the molecular structure of a protective epitope of GBS PSIII. Based on NMR simulation studies indicating the formation of several potential helical conformations (29, 32), where sialylated side chains are arranged on the exterior surface of the helix, the protective epitope dependent on a conformational structure has been proposed for GBS PSIII. In marked contrast, the data reported in this paper show that the protective epitope is linear and overlaps two RUs of the PS.
In the proposed conformational epitope structure, the PSIII helix structure was supposed to be stabilized by the presence of specific interactions between the side chain of the sialic acid and the backbone glucosyl and galactosyl residues, influencing the orientation of the side chain and stabilizing the conformation of the backbone, and required a carbohydrate chain length of at least four pentasaccharide RUs to form a conformational epitope equivalent to one present in the native PSIII (33, 34). Shorter carbohydrate chains were thought to be subjected to conformational changes so that in the absence of specific interactions the overall helical structure would collapse (29). The need for a helical epitope for antibody recognition was hypothesized considering that GBS PSIII showed a length-dependent binding of specific monoclonal IgGs (33). Zou et al. (33) observed that KD measured by SPR for monovalent binding remained virtually constant from two to seven RUs, which demonstrated that the epitope optimization was occurring from two to seven RUs, whereas beyond 20 RUs this phenomenon would overlap with the multivalent exposition of stabilized epitopes. Our competitive ELISA and SPR data indicate that the functional mAb characterized here shares similar features to the ones used by Zou et al. (33) in that it exhibits a similar length-dependent affinity. However, this length-dependent affinity could also be explained by a multivalency effect instead of the need for a helical conformation.
Johnson et al. showed by STD-NMR experiments that NeuNAc was not involved in binding to a monoclonal IgM (47), indicating the existence of antibodies recognizing the PS backbone. Molecular dynamics simulations confirmed that sialic acid plays a role in maintaining the helical structure and led to the hypothesis that this PS conformation was pivotal for antibody binding (29).
Our results demonstrate the existence of a sialic acid-dependent functional epitope of GBS that is fully contained within four consecutive sugars deriving from the PSIII backbone and one branched disaccharide present in this sequence. The nature of the mapped epitope fully explains the well-known critical role of the sialic acid and, as schematically shown in Fig. 5, does not require a conformational epitope (29, 33, 34). However, our study does not rule out the possibility that mAbs like those described by Johnson et al. (47) may recognize different regions of GBS PSIII.
Fig. 5.
Schematic representation of GBS PSIII–Fab interaction according to (A) the helical model previously proposed (29–34) and (B) the model that fully explains the well-known critical role of NeuNAc.
Our findings have profound implications in understanding the protective immunity against type III GBS and in the design of conjugate vaccines and suggest that the nature of protective epitopes should be investigated in all PS vaccines, especially those where the helical structure has been proposed (29). Carbohydrates present a higher level of structural complexity compared with other classes on natural biopolymers such as polypeptides or polynucleotides, because of the presence of α- or β-glycosidic bonds and the occurrence of connections at different positions of the ring that create ramifications. These unique features may result in spatial motifs relevant for antibody recognition and specificity even in the presence of a limited number of sugar residues, as it has been demonstrated here for GBS PSIII. In this manner, the hypothesized need for the human immune system to recognize GBS PS by means of length-dependent helical structures, thus avoiding recognition of self-antigens containing sialic acid, would need to be reconsidered (34). The existence of helical conformational epitopes, which would imply the necessity of using a long PS antigen, has been considered a challenge when applying these technologies to certain pathogens (2). The elucidation and understanding of the structural base for the immune recognition of carbohydrate epitopes reported here paves the way for designing modern glycoconjugate vaccines with short oligosaccharides obtained by synthetic, chemoenzymatic, or bioengineering methods (48).
Materials and Methods
SI Appendix, Materials and Methods feature additional information to that provided here.
The clone producing the rabbit mAb anti-PSIII NVS-1-19-5 was obtained using hybridoma technology by EPITOMICS Inc., and the relative Fab was prepared by using a “Fab Preparation Kit” (Pierce). The GBS PSIII fragments were prepared by deamination (40), and different chain-length oligosaccharides were separated by anionic exchange chromatography. NMR experiments were carried out on a Bruker 500 MHz NMR instrument, and the X-ray diffraction data were collected at the European Synchrotron Radiation Facility (ESRF).
Supplementary Material
Acknowledgments
We thank Matthew James Bottomley of the GSK group of companies for useful discussions and critical reading of the manuscript. This study was sponsored by Novartis Vaccines, now part of the GSK group of companies, which was involved in all stages of the study conduct and analysis.
Footnotes
Conflict of interest statement: All authors are employees of GSK Vaccines (known as Novartis Vaccines at the time of the study).
Data deposition: The crystal structure has been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5M63) (deposition ID D_1200001992).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701885114/-/DCSupplemental.
References
- 1.Avci FY, Kasper DL. How bacterial carbohydrates influence the adaptive immune system. Annu Rev Immunol. 2010;28:107–130. doi: 10.1146/annurev-immunol-030409-101159. [DOI] [PubMed] [Google Scholar]
- 2.Costantino P, Rappuoli R, Berti F. The design of semi-synthetic and synthetic glycoconjugate vaccines. Expert Opin Drug Discov. 2011;6:1045–1066. doi: 10.1517/17460441.2011.609554. [DOI] [PubMed] [Google Scholar]
- 3.Anish C, Schumann B, Pereira CL, Seeberger PH. Chemical biology approaches to designing defined carbohydrate vaccines. Chem Biol. 2014;21:38–50. doi: 10.1016/j.chembiol.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Lipinski T, et al. A β-mannan trisaccharide conjugate vaccine aids clearance of Candida albicans in immunocompromised rabbits. Vaccine. 2012;30:6263–6269. doi: 10.1016/j.vaccine.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Villeneuve S, et al. Crystal structure of an anti-carbohydrate antibody directed against Vibrio cholerae O1 in complex with antigen: Molecular basis for serotype specificity. Proc Natl Acad Sci USA. 2000;97:8433–8438. doi: 10.1073/pnas.060022997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vyas NK, et al. Molecular recognition of oligosaccharide epitopes by a monoclonal Fab specific for Shigella flexneri Y lipopolysaccharide: X-ray structures and thermodynamics. Biochemistry. 2002;41:13575–13586. doi: 10.1021/bi0261387. [DOI] [PubMed] [Google Scholar]
- 7.Cygler M, Wu S, Zdanov A, Bundle DR, Rose DR. Recognition of a carbohydrate antigenic determinant of Salmonella by an antibody. Biochem Soc Trans. 1993;21:437–441. doi: 10.1042/bst0210437. [DOI] [PubMed] [Google Scholar]
- 8.Safari D, et al. Identification of the smallest structure capable of evoking opsonophagocytic antibodies against Streptococcus pneumoniae type 14. Infect Immun. 2008;76:4615–4623. doi: 10.1128/IAI.00472-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Safari D, et al. The immune response to group B streptococcus type III capsular polysaccharide is directed to the -Glc-GlcNAc-Gal- backbone epitope. Glycoconj J. 2011;28:557–562. doi: 10.1007/s10719-011-9354-1. [DOI] [PubMed] [Google Scholar]
- 10.Vulliez-Le Normand B, et al. Structures of synthetic O-antigen fragments from serotype 2a Shigella flexneri in complex with a protective monoclonal antibody. Proc Natl Acad Sci USA. 2008;105:9976–9981. doi: 10.1073/pnas.0801711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jennings HJ. The role of sialic acid in the formation of protective conformational bacterial polysaccharide epitopes. In: Kosma P, Muller-Loennies S, editors. Anticarbohydrate Antibodies. Springer; Vienna: 2012. pp. 55–73. [Google Scholar]
- 12.Nuccitelli A, Rinaudo CD, Maione D. Group B Streptococcus vaccine: State of the art. Ther Adv Vaccines. 2015;3:76–90. doi: 10.1177/2051013615579869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berti F, et al. Structure of the type IX group B Streptococcus capsular polysaccharide and its evolutionary relationship with types V and VII. J Biol Chem. 2014;289:23437–23448. doi: 10.1074/jbc.M114.567974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker CJ, Kasper DL. Correlation of maternal antibody deficiency with susceptibility to neonatal group B streptococcal infection. N Engl J Med. 1976;294:753–756. doi: 10.1056/NEJM197604012941404. [DOI] [PubMed] [Google Scholar]
- 15.Munoz FM, Ferrieri P. Group B Streptococcus vaccination in pregnancy: Moving toward a global maternal immunization program. Vaccine. 2013;31:D46–D51. doi: 10.1016/j.vaccine.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 16.Paoletti LC, et al. Effects of chain length on the immunogenicity in rabbits of group B Streptococcus type III oligosaccharide-tetanus toxoid conjugates. J Clin Invest. 1992;89:203–209. doi: 10.1172/JCI115564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paoletti LC, Kennedy RC, Chanh TC, Kasper DL. Immunogenicity of group B Streptococcus type III polysaccharide-tetanus toxoid vaccine in baboons. Infect Immun. 1996;64:677–679. doi: 10.1128/iai.64.2.677-679.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wessels MR, et al. Immunogenicity in animals of a polysaccharide-protein conjugate vaccine against type III group B Streptococcus. J Clin Invest. 1990;86:1428–1433. doi: 10.1172/JCI114858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasper DL, Baker CJ, Galdes B, Katzenellenbogen E, Jennings HJ. Immunochemical analysis and immunogenicity of the type II group B streptococcal capsular polysaccharide. J Clin Invest. 1983;72:260–269. doi: 10.1172/JCI110965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker CJ, Rench MA, Paoletti LC, Edwards MS. Dose-response to type V group B streptococcal polysaccharide-tetanus toxoid conjugate vaccine in healthy adults. Vaccine. 2007;25:55–63. doi: 10.1016/j.vaccine.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 21.Edwards MS. Group B streptococcal conjugate vaccine: A timely concept for which the time has come. Hum Vaccin. 2008;4:444–448. doi: 10.4161/hv.4.6.6507. [DOI] [PubMed] [Google Scholar]
- 22.Donders GG, et al. Maternal immunization with an investigational trivalent group B streptococcal vaccine: A randomized controlled trial. Obstet Gynecol. 2016;127:213–221. doi: 10.1097/AOG.0000000000001190. [DOI] [PubMed] [Google Scholar]
- 23.Clarke ET, et al. Polysaccharide-specific memory B cells generated by conjugate vaccines in humans conform to the CD27+IgG+ isotype-switched memory B cell phenotype and require contact-dependent signals from bystander T cells activated by bacterial proteins to differentiate into plasma cells. J Immunol. 2013;191:6071–6083. doi: 10.4049/jimmunol.1203254. [DOI] [PubMed] [Google Scholar]
- 24.Edmond KM, et al. Group B streptococcal disease in infants aged younger than 3 months: Systematic review and meta-analysis. Lancet. 2012;379:547–556. doi: 10.1016/S0140-6736(11)61651-6. [DOI] [PubMed] [Google Scholar]
- 25.Kasper DL, et al. Immunodeterminant specificity of human immunity to type III group B streptococcus. J Exp Med. 1979;149:327–339. doi: 10.1084/jem.149.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jennings HJ, Lugowski C, Kasper DL. Conformational aspects critical to the immunospecificity of the type III group B streptococcal polysaccharide. Biochemistry. 1981;20:4511–4518. doi: 10.1021/bi00519a001. [DOI] [PubMed] [Google Scholar]
- 27.Guttormsen H-K, et al. Type III group B streptococcal polysaccharide induces antibodies that cross-react with Streptococcus pneumoniae type 14. Infect Immun. 2002;70:1724–1738. doi: 10.1128/IAI.70.4.1724-1738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jennings HJ, et al. Structure, conformation and immunology of sialic acid containing polysaccharides of human pathogenic bacteria. Pure Appl Chem. 1984;56:893–905. [Google Scholar]
- 29.González-Outeiriño J, Kadirvelraj R, Woods RJ. Structural elucidation of type III group B Streptococcus capsular polysaccharide using molecular dynamics simulations: The role of sialic acid. Carbohydr Res. 2005;340:1007–1018. doi: 10.1016/j.carres.2004.12.034. [DOI] [PubMed] [Google Scholar]
- 30.Kadirvelraj R, et al. Understanding the bacterial polysaccharide antigenicity of Streptococcus agalactiae versus Streptococcus pneumoniae. Proc Natl Acad Sci USA. 2006;103:8149–8154. doi: 10.1073/pnas.0602815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woods R, Yongye A. Computational techniques applied to defining carbohydrate antigenicity. In: Kosma P, Müller-Loennies S, editors. Anticarbohydrate Antibodies. Springer; Vienna: 2012. pp. 361–383. [Google Scholar]
- 32.Brisson JR, et al. NMR and molecular dynamics studies of the conformational epitope of the type III group B Streptococcus capsular polysaccharide and derivatives. Biochemistry. 1997;36:3278–3292. doi: 10.1021/bi961819l. [DOI] [PubMed] [Google Scholar]
- 33.Zou W, et al. Conformational epitope of the type III group B Streptococcus capsular polysaccharide. J Immunol. 1999;163:820–825. [PubMed] [Google Scholar]
- 34.Zou W, Jennings HJ. The conformational epitope of type III group B Streptococcus capsular polysaccharide. Adv Exp Med Biol. 2001;491:473–484. doi: 10.1007/978-1-4615-1267-7_31. [DOI] [PubMed] [Google Scholar]
- 35.Bush CA, Martin-Pastor M, Imberty A. Structure and conformation of complex carbohydrates of glycoproteins, glycolipids, and bacterial polysaccharides. Annu Rev Biophys Biomol Struct. 1999;28:269–293. doi: 10.1146/annurev.biophys.28.1.269. [DOI] [PubMed] [Google Scholar]
- 36.Oberli MA, et al. Molecular analysis of carbohydrate-antibody interactions: Case study using a Bacillus anthracis tetrasaccharide. J Am Chem Soc. 2010;132:10239–10241. doi: 10.1021/ja104027w. [DOI] [PubMed] [Google Scholar]
- 37.Coelho H, et al. The quest for anticancer vaccines: Deciphering the fine-epitope specificity of cancer-related monoclonal antibodies by combining microarray screening and Saturation Transfer Difference NMR. J Am Chem Soc. 2015;137:12438–12441. doi: 10.1021/jacs.5b06787. [DOI] [PubMed] [Google Scholar]
- 38.Bhushan R, Anthony BF, Frasch CE. Estimation of group B streptococcus type III polysaccharide-specific antibody concentrations in human sera is antigen dependent. Infect Immun. 1998;66:5848–5853. doi: 10.1128/iai.66.12.5848-5853.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wessels MR, Muñoz A, Kasper DL. A model of high-affinity antibody binding to type III group B Streptococcus capsular polysaccharide. Proc Natl Acad Sci USA. 1987;84:9170–9174. doi: 10.1073/pnas.84.24.9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michon F, et al. Group B streptococcal type II and III conjugate vaccines: Physicochemical properties that influence immunogenicity. Clin Vaccine Immunol. 2006;13:936–943. doi: 10.1128/CVI.00122-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paoletti LC, et al. An oligosaccharide-tetanus toxoid conjugate vaccine against type III group B Streptococcus. J Biol Chem. 1990;265:18278–18283. [PubMed] [Google Scholar]
- 42.Cattaneo V, et al. 2016 Synthesis of group B Streptococcus type III polysaccharide fragments for evaluation of their interactions with monoclonal antibodies. Pure & Appl Chem, www.degruyter.com/view/j/pac.ahead-of-print/pac-2016-0918/pac-2016-0918.xml.
- 43.Meyer B, Peters T. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew Chem Int Ed Engl. 2003;42:864–890. doi: 10.1002/anie.200390233. [DOI] [PubMed] [Google Scholar]
- 44.Lewis AL, Nizet V, Varki A. Discovery and characterization of sialic acid O-acetylation in group B Streptococcus. Proc Natl Acad Sci USA. 2004;101:11123–11128. doi: 10.1073/pnas.0403010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiman S, et al. O-Acetylation of sialic acid on group B Streptococcus inhibits neutrophil suppression and virulence. Biochem J. 2010;428:163–168. doi: 10.1042/BJ20100232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pannaraj PS, et al. Group B streptococcal conjugate vaccines elicit functional antibodies independent of strain O-acetylation. Vaccine. 2009;27:4452–4456. doi: 10.1016/j.vaccine.2009.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson MA, et al. NMR studies of carbohydrates and carbohydrate-mimetic peptides recognized by an anti-group B Streptococcus antibody. J Biol Chem. 2003;278:24740–24752. doi: 10.1074/jbc.M301846200. [DOI] [PubMed] [Google Scholar]
- 48.Berti F, Adamo R. Recent mechanistic insights on glycoconjugate vaccines and future perspectives. ACS Chem Biol. 2013;8:1653–1663. doi: 10.1021/cb400423g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.