Significance
l-galactofuranose (l-Galf), the furanose form of l-galactose, is rare in nature yet plays a central role in secondary metabolite structures with medicinal potential. Importantly, the biosynthetic enzymes and mechanisms driving l-Galf biosynthesis have eluded characterization. We report here a previously unknown mutase, MtdL, that generates GDP-l-Galf from its galactopyranose analog GDP-l-Galp using unique chemistry. MtdL mediates the pyranose–furanose conversion solely by the presence of bivalent cation. The ubiquitous nature of this Galp → Galf chemistry is suggested by numerous mutase homologs that are phylogenetically widespread in various phyla of Bacteria, Eukaryota, and Archaea. Thus, a genetic marker for l-Galf-containing natural products and their producers has been discovered. Additionally, mutase homologs may constitute targets for antimicrobial drugs.
Keywords: nucleoside antibiotic A201A, biosynthesis, GDP-l-galactose mutase, methyltransferase, desaturase
Abstract
Galactose, a monosaccharide capable of assuming two possible configurational isomers (d-/l-), can exist as a six-membered ring, galactopyranose (Galp), or as a five-membered ring, galactofuranose (Galf). UDP-galactopyranose mutase (UGM) mediates the conversion of pyranose to furanose thereby providing a precursor for d-Galf. Moreover, UGM is critical to the virulence of numerous eukaryotic and prokaryotic human pathogens and thus represents an excellent antimicrobial drug target. However, the biosynthetic mechanism and relevant enzymes that drive l-Galf production have not yet been characterized. Herein we report that efforts to decipher the sugar biosynthetic pathway and tailoring steps en route to nucleoside antibiotic A201A led to the discovery of a GDP-l-galactose mutase, MtdL. Systematic inactivation of 18 of the 33 biosynthetic genes in the A201A cluster and elucidation of 10 congeners, coupled with feeding and in vitro biochemical experiments, enabled us to: (i) decipher the unique enzyme, GDP-l-galactose mutase associated with production of two unique d-mannose-derived sugars, and (ii) assign two glycosyltransferases, four methyltransferases, and one desaturase that regiospecifically tailor the A201A scaffold and display relaxed substrate specificities. Taken together, these data provide important insight into the origin of l-Galf-containing natural product biosynthetic pathways with likely ramifications in other organisms and possible antimicrobial drug targeting strategies.
Galactose (Gal), which can assume two possible stereochemical configurations (d-/l-), is able to exist in the thermodynamically favored six-membered ring form as galactopyranose (Galp), or in the less favored five-membered ring form as galactofuranose (Galf) (1, 2). d-gal in mammals is found only in the pyranose form as d-Galp, whereas in bacteria, fungi, and protozoa it exists as d-Galf; this is the form found also in many pathogenic organisms (3, 4). Indeed, in many pathogenic prokaryotes and eukaryotes, UDP-Galf, the precursor of d-Galf, is usually enzymatically generated from UDP-Galp by UDP-d-galactopyranose mutase (UGM) (3, 5, 6). Since the first identification and cloning of UGM from Escherichia coli (7), dozens of UGMs have been discovered and functionally characterized from pathogenic bacteria, fungi, parasites, and nematodes. These findings have highlighted the critical role of UGMs in Galf biosynthesis, especially as relates to virulence, cellular adhesion, parasite viability, and proper developmental processes (8–11). It has been proposed that during the formation of UDP-Galf, a flavin adenine dinucleotide (FAD)-iminium ion intermediate is formed through nucleophilic addition that facilitates galactose ring opening and contraction (5, 12–16). Three dimensional structures for members of the UGM family have been determined by X-ray crystallography in free form and in complex with various substrates, as well as in association with both oxidized and reduced forms of FAD (12, 13, 17–19). Notably, a covalent FAD-Galp intermediate involved in the UGM reaction has recently yielded to elucidation by X-ray crystallography shedding significant insight into UGM catalysis (20). Consequently, UGM is a potential antimicrobial drug target (3, 16).
Compared with the high abundance of d-Gal, l-Gal is rare in both terrestrial and marine environments (21, 22). It is worth noting that l-Gal is a key intermediate in the conversion of d-glucose to ascorbic acid in plants (23). In contrast, l-Galf, the furanose form of l-galactose, is exceptionally rare in the environment. It has been detected only in animal cells (24), plant tissues (24), early weaned piglets (25), botanical species of the Cactaceae family (26), propolis of stingless bees (27), and as a building block for the antimicrobial epipyrones from Epicoccum purpurascens (28), as well as in the oligosaccharide fraction isolated from the mycelium of Lingzhi or Reishi medicinal mushrooms Ganoderma lucidum (29). In addition, l-Galf has served as a critical intermediate in assorted carbohydrate chemical syntheses (30, 31). Despite these findings, the biosynthetic mechanisms and relevant enzymes that drive the synthesis of l-Galf have yet to be characterized.
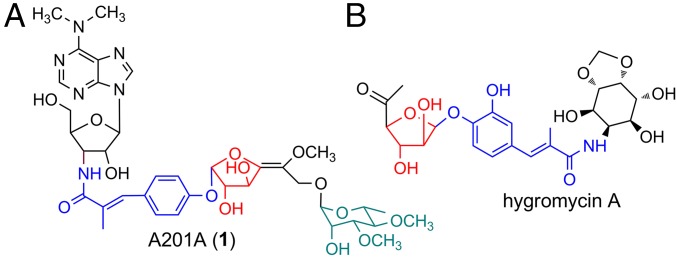
The aminonucleoside antibiotic A201A (1) was first isolated from Streptomyces capreolus NRRL 3817 (32) and then rediscovered in our laboratory from deep sea-derived Marinactinospora thermotolerans SCSIO 00652 (33) (Fig. 1). A201A is composed of a 3′-amino-3′-deoxy adenosine moiety, a p-hydroxy-α-methylcinnamic acid moiety, a hexofuranose moiety, an α-d-rhamnose moiety, and is further decorated with unusual sugar unsaturation and N- or O-methylations (Fig. 1A). The compound displays strong antibacterial activities against Gram-positive and anaerobic Gram-negative bacteria by preventing bacterial ribosomal peptide bond formation or by impairing proper tRNA–ribosome associations during protein synthesis and is only weakly toxic to mammals (32–34). Consequently, A201A possesses great potential for antiinfective drug discovery as in, for instance, the treatment of acne (32, 35, 36). The first total synthesis of A201A was achieved in 47 steps by Yu and coworkers (37). Early investigations into A201A biosynthesis by our group led to the identification of the A201A gene cluster in M. thermotolerans SCSIO 00652, demonstrating that MtdA is a negative transcriptional regulator governing A201A biosynthesis (33); efforts elsewhere identified the A201A cluster in S. capreolus (38, 39). Notably, S. capreolus failed to be genetically amenable to subsequent and more elaborate dissection. Consequently, the exact biosynthetic pathway and machineries driving A201A biosynthesis have eluded characterization over the past 10 y. To further elucidate the biosynthetic apparatus driving A201A production, especially with an eye on l-Galf incorporation, we report herein: (i) boundary determinations and heterologous expression of the A201A gene cluster in Streptomyces lividans TK64; (ii) systematic inactivation of genes coding for the biosynthesis and transfer of the two sugar units, as well as tailoring steps in M. thermotolerans enabling us to generate 10 A201A congeners from assorted mutant strains; (iii) complete elucidation of metabolite structures; and (iv) in vitro biochemical and feeding experiments enabling functional gene assignments related to biosynthesis and transfer of the unique hexofuranose and α-d-rhamnose moieties harboring a GDP-l-galactose mutase MtdL, as well as, four methyltransferases and a regiospecific tailoring desaturase showing relaxed substrate specificity.
Fig. 1.
Structures of A201A (1) and hygromycin A.
Results and Discussion
Determination of the Boundaries of the A201A Gene Cluster in M. thermotolerans SCSIO 00652 and Heterologous Expression in S. lividans TK64.
To narrow down the genes responsible for A201A biosynthesis, we first fully analyzed and annotated the A201A gene cluster (Fig. 2A and SI Appendix, Table S1). We also inactivated mtdB (encoding a peptidase S15), mtdC (encoding an unknown protein), mtdD (encoding an unknown protein), the orf1 (encoding an inositol-phosphate phosphatase), and orf2 genes using our established λ-RED-mediated PCR-targeting mutagenesis strategy in M. thermotolerans SCSIO 00652 by replacing them with an apramycin resistance gene cassette (33, 40). Genotypes of the mutants were confirmed by PCR (SI Appendix). HPLC analyses of the fermentation extracts of the resultant mutants revealed that all mutants still produced 1 in yields comparable to those of the wild-type strain (SI Appendix, Fig. S22), thereby suggesting the dispensability of these genes for A201A biosynthesis.
Fig. 2.
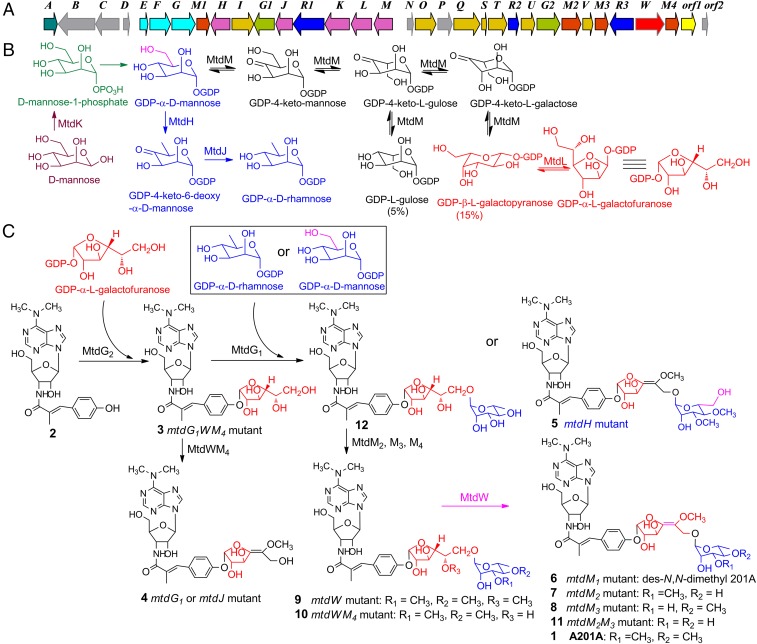
The biosynthetic gene cluster and proposed biosynthetic pathway of A201A (1) in M. thermotolerans SCSIO 00652. (A) Organization of the A201A biosynthetic genes. (B) Biosynthetic pathway of sugar moieties in A201A. (C) Tailoring steps involved in A201A assembly.
We subsequently selected cosmid 142H harboring the genes from mtdC to orf3, integrated elements for conjugation from the pSET152AB vector (41), and transferred them into S. lividans TK64 through conjugation. The recombinant S. lividans TK64/142H was fermented using the same medium as had been the case for the wild-type producer. HPLC analysis of the fermentation extract revealed that S. lividans TK64/142H indeed produced 1 (Fig. 3, trace ii), whereas the control did not (Fig. 3, trace iii), demonstrating that the narrowed gene cluster spanning mtdE to mtdM4 harbors all of the genes necessary for A201A biosynthesis. Production of 1 in S. lividans TK64/142H was ∼50% of that seen in the wild-type producer. Within the gene cluster, the gene products MtdEFGM1, homologous to those in the puromycin pathway (42), are proposed to be involved in constructing the N, N-dimethyl-3′-amino-3′-deoxyadenosine moiety (SI Appendix, Fig. S23 and Table S1). The gene products MtdINOSTUV, homologous to those in the hygromycin A (Fig. 1B) pathway (43, 44), are proposed to be involved in the biosynthesis of the p-hydroxy-α-methylcinnamic acid moiety. MtdQ, a ligase homologous to Hyg12 in the hygromycin A pathway, is proposed to converge the deoxyadenosine and cinnamic acid moieties through an amide bond to form 2 (SI Appendix, Fig. S23 and Table S1). A detailed comparison of biosynthetic proteins involved in A201A, hygromycin A, and puromycin is provided in SI Appendix, Table S1.
Fig. 3.
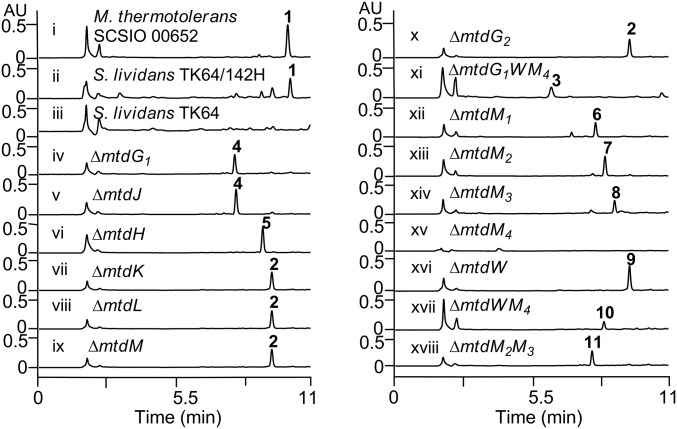
HPLC metabolite analyses for S. lividans, S. lividans TK64/142H, M. thermotolerans wild type, and mutant strains. See Fig. 2 for structures 1–11. λ = 275 nm.
Biosynthesis and Transfer of the α-d-Rhamnose Moiety Reveals Promiscuous Glycosyltransferase MtdG1 Capable of Processing both GDP-α-d-Mannose and GDP-α-d-Rhamnose.
We carried out bioinformatics analysis to identify the genes (SI Appendix, Table S1) possibly responsible for the construction and transfer of the α-d-rhamnose moiety in A201A. MtdH and MtdJ belong to members of the extended short-chain dehydrogenase-reductases (SDR) superfamily and share as low as 28% and 15% identity, respectively, with Gmd and Rmd from Aneurinibacillus thermoaerophilus L420-91T. Gmd and Rmd proteins are known to be involved in the two-step conversion of GDP-α-d-mannose to GDP-α-d-rhamnose (45). Between MtdH and MtdJ, resides a glycosyltransferase, MtdG1, as a candidate enzyme installing a sugar building block onto the A201A scaffold.
To explore the exact roles of the above-mentioned genes in 1 biosynthesis, we individually inactivated the three genes in M. thermotolerans wild type as described before. The metabolite profiles for the ∆mtdG1 and ∆mtdJ mutants both revealed that these strains do not produce 1; instead these strains both led to accumulation of product 4 (Fig. 3, traces iv and v). The ∆mtdH mutant also failed to produce 1, but yielded a product 5 (Fig. 3, trace vi). The mutant strains were then fermented on a large scale (8–16 L each) and the structures of 4 and 5 were elucidated (Fig. 2C) on the basis of comprehensive high-resolution mass spectrometry (HRMS), 1D and 2D NMR analyses (SI Appendix). The ∆mtdG1 and ∆mtdJ mutants both yielded 4 devoid of the terminal sugar, demonstrating their involvement in the transfer and synthesis of the terminal α-d-rhamnose moiety. The ∆mtdH mutant failed to produce 1 but unexpectedly accumulated product 5 in titers comparable to those of 1 in the wild-type producer. That 5 bears a d-mannose unit in place of the d-rhamnose unit found in 1 reveals that MtdH catalyzes 4, 6-dehydration of GDP-α-d-mannose to yield GDP-4-keto-6-deoxy-α-d-mannose (Fig. 2B), and that the glycosyltransferase MtdG1 shows significant substrate promiscuity because it can apparently process both GDP-α-d-mannose and GDP-α-d-rhamnose (Fig. 2C).
Biosynthesis and Transfer of the Hexofuranose Moiety as Dictated by a GDP-l-Galactose Mutase.
Three cotranscribed genes, mtdK, mtdL, mtdM (mtdKLM), were identified as candidates possibly involved in the unique hexofuranose moiety sugar unit biosynthesis. MtdK shows as low as 27–32% identity to a group of glucokinases in the GenBank database. MtdL shows the highest homology (61% identity) to Hyg20 in the hygromycin A pathway (SI Appendix, Fig. S24 and Table S1) and has been proposed to catalyze the conversion of NDP-l-fucose to NDP-l-fucofuranose (44). MtdM belongs to the extended SDR superfamily with NAD(P)+-binding sites, showing 45–57% identity to a group of GDP-mannose-3′,5′-epimerases from Arabidopsis thaliana, and rice (Oryza sativa) (SI Appendix, Fig. S25) (46, 47). Within the biosynthetic gene cluster of 1, and downstream of the mtdKLM, mtdG2 encoding another glycosyltransferase, is likely responsible for the hexofuranose transfer.
The above four genes (mtdK, mtdL, mtdM, and mtdG2) were similarly individually inactivated in the wild-type strain. LCMS analysis of the fermentation extracts revealed that ∆mtdK, ∆mtdL, ∆mtdM, and ∆mtdG2 mutants uniformly failed to produce 1 and yielded the same intermediate 2 (Fig. 3, traces vii–x) devoid of the two sugar moieties as determined by large-scale fermentation, purification, and HRMS and NMR structure elucidation (SI Appendix). These data indicate that MtdKLM are all involved in construction of the unique hexofuranose unit and that MtdG2 is responsible for transferring this unit to aglycon 2.
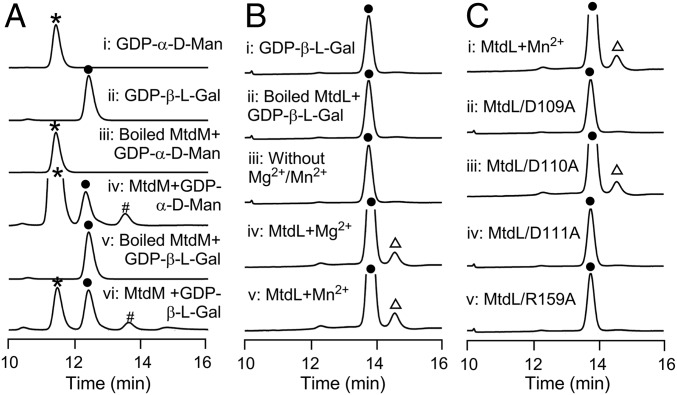
However, in the structure of 1, the hexofuranose moiety contains a double bond. Accordingly, the exact origin of this hexofuranose presents a mystery. Based on the bioinformatics data, we first asked whether or not MtdM has the in vitro GDP-mannose-3′,5′-epimerase biochemical activity. We overexpressed MtdM in E. coli BL21(DE3), and purified it to homogeneity as an N-terminus–His6-tagged protein. The enzymatic activity of MtdM was then evaluated in the presence of GDP-α-d-mannose (0.2 mM) or GDP-β-l-galactose (0.2 mM), 0.5 mM NAD+, and 50 mM Tris⋅HCl buffer (pH = 8.0). An equilibrating mixture of three products, GDP-α-d-mannose, GDP-β-l-galactose, and GDP-β-l-gulose, in a ratio of 80:15:5, was found in the enzymatic reaction (Fig. 4A). On the basis of these data it appears that the catalytic function of MtdM is similar to that of the GDP-d-mannose 3′,5′-epimerase (GME) from A. thaliana (46) (SI Appendix, Fig. S25). This result suggested that MtdL might subsequently catalyze the conversion of GDP-l-galactopyranose into GDP-l-galactofuranose (Fig. 2B).
Fig. 4.
In vitro characterization of MtdM and MtdL-catalyzed reactions. (A) HPLC analysis of in vitro MtdM assays: (i) authentic GDP-α-d-mannose (GDP-α-d-Man); (ii) authentic GDP-β-l-galactose (GDP-β-l-Gal); (iii) control assay with boiled MtdM and GDP-α-d-Man; (iv) enzymatic reaction using MtdM and GDP-α-d-Man; (v) control assay with boiled MtdM and GDP-β-l-Gal; and (vi) enzymatic reaction using MtdM and GDP-β-l-Gal. (B) HPLC analysis of in vitro MtdL assays: (i) authentic GDP-β-l-Gal; (ii) control assay with boiled MtdL and GDP-β-l-Gal; (iii) control assay without Mg2+/Mn2+; and (iv and v) enzymatic reaction with Mg2+ or Mn2+, respectively. (C) HPLC analysis of the in vitro enzymatic reaction of (i) MtdL and its mutant variants; (ii) D109A; (iii) D110A; (iv) D111A; and (v) R159A, respectively. *: GDP-α-d-Man; ●: GDP-β-l-Gal; #: GDP-l-gulose; ∆: GDP-α-l-galactofuranose. λ = 254 nm.
To validate that GDP-l-galactofuranose is incorporated as a precursor into the biosynthetic pathway for 1, we designed and successfully constructed a triple gene ∆mtdG1WM4 mutant strain (in which the mtdW and mtdM4 were in-frame deleted, and the mtdG1 was replaced with an apramycin resistance gene cassette) (SI Appendix). Fermentations of the ∆mtdG1WM4 strain, did indeed, afford important intermediate 3 (Fig. 3, trace xi). The structure of 3 (Fig. 2C), containing a furanose unit, was fully established upon isolation from a scaled-up fermentation (24 L) and following analyses of HRMS, 1D, and 2D NMR data (SI Appendix). Analyses of 1H NMR spectra and comparisons of circular dichroism (CD) data for the methanolyzed and benzoylated hydrolysate of 3 against authentic samples of methanolyzed and benzoylated l-galactose and d-galactose confirmed the presence of the l-galactose moiety in 3 (SI Appendix and SI Appendix, Figs. S26–S28).
The above results indicated that the two sugar units in 1 might be derived from a uniform precursor, GDP-d-mannose. To validate this hypothesis, we fed 1-13C-labeled d-mannose into fermentations of the wild-type M. thermotolerans strain. The corresponding A201A product was purified and subjected to 13C NMR analysis (SI Appendix, Figs. S84 and S85). As anticipated, the two anomeric carbons of the two sugar units were clearly enriched, demonstrating that the two sugars use GDP-α-d-mannose as the biosynthetic precursor.
MtdL also shows low homology (30% identity) to a rice UDP-arabinopyranose mutase (UAM) responsible for interconverting UDP-l-arabinofuranose and UDP-l-arabinopyranose in a 10:90 ratio (48, 49). Sequence alignments of MtdL with UAM, Hyg20 in the hygromycin A pathway, which may catalyze a conversion of NDP-l-fucose to NDP-l-fucofuranose, and other homologs in the GenBank with unknown functions, revealed that they all contain the conserved DxD motif usually found in the glycosyltransferase (GT)-A family (50) (SI Appendix, Fig. S24); such a motif interacts with the phosphate groups of nucleotide donor through coordination of divalent cations, typically Mn2+, and is responsible for the glycosyl transfer reaction.
To determine whether MtdL could catalyze the transformation of GDP-β-l-galactopyranose to GDP-α-l-galactofuranose in vitro, we overexpressed MtdL in E. coli BL21(DE3), and purified it to homogeneity as an N-terminus–His6-tagged protein. The enzymatic activity of MtdL was tested in a 50-μL reaction containing 0.2 mM GDP-β-l-galactose, in the absence or presence of 5 mM Mg2+ or Mn2+, 2 μM MtdL, in 50 mM phosphate buffer (pH = 8.0), at 37 °C for 20 min. HPLC analysis of the enzymatic mixture revealed that MtdL catalyzes the formation of an enzymatic product, which was found to have the identical molecular weight as the substrate by HR-LCMS analysis and the reaction absolutely requires Mg2+ or Mn2+ (Fig. 4B). The enzymatic reaction was performed in situ in an NMR tube and ultimately subjected to 1H NMR analysis following enzyme removal by centrifugal filtration. The 1H NMR spectrum of the enzymatic mixture clearly shows the appearance of the anomeric proton signal characteristic of GDP-α-l-galactofuranose (SI Appendix, Figs. S86 and S87). The ratio of the pyranose and furanose form of GDP-l-galactose is 91:9, as judged by the HPLC peak area and integration of the anomeric proton signal in the 1H NMR spectrum (SI Appendix, Figs. S86 and S87). It is noted that the coupling constant (J = 6.0 Hz) of the anomeric proton in the GDP-α-l-galactofuranose indicated an α-configuration (Fig. 2B) (51, 52), demonstrating that the stereochemistry at C-1 of the GDP-β-l-galactose was reversed during the pyranose–furanose transformation (50).
To investigate which amino acid residues of MtdL are responsible for its enzymatic activity, site-directed mutagenesis experiments were performed based on sequence alignment (SI Appendix, Fig. S24). The D109, D110, D111, and R159 residues of MtdL were each converted to alanine and the activities of purified mutant proteins were assessed in enzymatic assays. HPLC analyses of the reaction mixtures revealed that the D109A MtdL mutant (Fig. 4C, trace ii), together with the D111A (Fig. 4C, trace iv) and R159A mutants (Fig. 4C, trace v), was completely devoid of enzymatic activity, whereas the D110A mutation appeared to exert no apparent influence upon MtdL enzymatic activity (Fig. 4C, trace iii). In the glycosyltransferase A-families, the DxD motif plays a key role in the coordination of Mg2+ or Mn2+ with the phosphate groups of the nucleotide donor (50); in the UAM protein family, the arginine residue serves as a site of protein autoglycosylation (48, 49). Consequently, D109, D111, and R159 in MtdL are likely to serve as a catalytic triad for the GDP-β-l-galactopyranose and GDP-α-l-galactofuranose transformation.
A plausible chemical mechanism for MtdL-catalyzed pyranose–furanose conversion is envisioned as follows. The anomeric carbon-O-GDP bond is broken, and bicyclic intermediate 1, 4-anhydrogalactopyranose, is formed, coincident with ring contraction (SI Appendix, Fig. S90A). As indicated in SI Appendix, Fig. S90B, we envisage that Asp109 of MtdL serves as a catalytic base to deprotonate the hydroxyl group of galactose and that Asp111 interacts with the Mg2+ or Mn2+ ion as Arg159 enters into an electrostatic interaction with the phosphoryl group of GDP. A double displacement mechanism involving discrete nucleophilic catalysis and the formation of a covalently bound glycosyl-enzyme intermediate seems possible for MtdL. Central to this mechanism, in addition to the arrangement of acidic and basic amino acids within the MtdL active site, is that l-galactose is enzymatically amenable to ring contraction as noted in SI Appendix, Fig. S90 A and B. Thus, MtdL uses a flavin-independent mechanism to catalyze the GDP-l-galactose pyranose–furanose transformation. This finding contrasts sharply with the UGMs that use the flavin cofactor as a nucleophile to carry out the UDP-d-galactose pyranose–furanose exchange (SI Appendix, Fig. S90C) (11, 20).
Thus, the biosynthetic pathway for sugar moieties in A201A is initiated from the phosphorylated d-mannose catalyzed by MtdK, then converted to GDP-α-d-mannose by a GDP-sugar pyrophosphorylase, and then further processed by MtdM, a GDP-α-d-mannose 3′,5′-epimerase to yield GDP-β-l-galactose (Fig. 2B). The GDP-β-l-galactose was then transformed by MtdL, a pyranose–furanose mutase, to yield GDP-α-l-galactofuranose. GDP-α-d-mannose could also be converted to GDP-α-d-rhamnose through the agency of GDP-4-keto-6-deoxy-d-mannose by MtdH and MtdJ successively. The two sugar units were transferred to aglycon 2 by MtdG2 and MtdG1 to yield des-O-methyl desaturated A201A 10 (Fig. 2 B and C).
Phylogenetic analyses of MtdL with related proteins in the GenBank database revealed that they belong to a family of proteins with unknown functions represented across three domains: the Bacteria, Archaea, and Eukaryota (SI Appendix, Fig. S91). Homologous proteins from the Bacteria domain include proteins from actinomycetes such as Streptomyces, Micromonospora, Saccharothrix, and Frankia that have a demonstrated record of producing bioactive natural products. The Bacteria domain homologs also include proteins from Cyanobacteria, Bacteroidetes, and Proteobacteria whose secondary metabolites have only been marginally investigated. MtdL also appears to have a large number of homologous proteins from Archaea, including Halococcus, Halonotius, and Salinarchaeum from which secondary metabolites also have been largely unexplored. Notably, members of the Eukaryota including, but not limited to, O. sativa, A. thaliana, Ancylostoma ceylanicum, and Necator americanus also appear to produce MtdL homologs. Thus, it appears completely feasible that the mtdL gene represents an effective genetic marker for the mining of l-galactofuranose/l-fucofuranose/l-arabinofuranose–containing natural products in a wide variety of microorganisms.
Tailoring Steps of A201A Involving Four Regiospecific Methyltransferases and a Desaturase MtdW.
We next analyzed the genes driving the tailoring steps involved in completing the biosynthesis of 1. Structurally, A201A contains an N, N-dimethyl group, three O-methyl groups, and an unusual double bond within the hexofuranose. Installation of these moieties is achieved through a number of tailoring steps during A201A biosynthesis. Consistent with these structural considerations and aided by thorough bioinformatics analysis of the A201A gene cluster, we readily identified four methyltransferase genes, mtdM1, mtdM2, mtdM3, and mtdM4, that encode enzymes 234aa, 354aa, 242aa, and 240aa in size, respectively. These four genes were inactivated by replacement with an apramycin gene cassette as described before to yield the respective ∆mtdM1, ∆mtdM2, ∆mtdM3, and ∆mtdM4 mutants. These mutants were fermented and analyzed by HPLC-UV-MS (Fig. 3, traces xii–xv). We found that: (i) all mutants failed to produce 1; (ii) the ∆mtdM4 mutant did not yield any “unique” products; (iii) ∆mtdM1 yielded a product 6 that is produced in a 10-fold lower yield than 1 in the wild-type producer; and (iv) each of the other mutants, ∆mtdM2 and ∆mtdM3, produced analogs 7 and 8, respectively and, in each case, with titers comparable to those seen with 1 in the wild-type producer. Scaled-up fermentation of these respective mutants led to purification of compounds 6–8, whose structures were unambiguously assigned as shown in Fig. 2C upon comprehensive analysis of HRMS, 1D, and 2D NMR spectroscopic data (SI Appendix). Judging from the structures of 6–8, we conclude that: (i) MtdM1 is responsible for N, N-dimethylation of the 3′-amino-3′-deoxyadenosine moiety in 1, because the yield of 6 is significantly lower than that of 1 in the wild-type strain; (ii) MtdM2 and MtdM3 are responsible for the O-methylation of OH-C-4′ and OH-C-3′ in the α-d-rhamnose moiety of 1, respectively; and (iii) MtdM4 is responsible for the O-methylation of OH-C-5′ of the α-l-galactofuranose unit of 1.
Notably, we found mtdW residing at one far end of the A201A gene cluster encoding a 529-aa protein with as low as 37–43% identity to the glucose-methanol-choline (GMC) oxidoreductase family of proteins in GenBank whose functions have not been experimentally verified (53). Sequence alignments (SI Appendix, Fig. S29) revealed that these enzymes contain a FAD ribose moiety binding motif GxGxxG(x)18E in the N terminus and an H(x)37P motif in the C terminus presumably involved in substrate oxidation (54). The mtdW gene was similarly inactivated and the resultant ∆mtdW mutant was found to produce compound 9, as tentatively identified on the basis of HPLC-UV-MS (Fig. 3, trace xvi); 9 was generated in titers comparable to 1 produced in wild type. Scaled-up fermentation of the ∆mtdW mutant, subsequent purification, and structural elucidation on the basis of HRMS, 1D, and 2D NMR spectroscopic studies confirmed the structure of 9 as shown in Fig. 2C. Notably, the C-4/C-5 hexofuranose double bond of 1 is saturated in compound 9. Thus, MtdW represents a desaturase responsible for C-4/C-5 dehydrogenation of the α-l-galactofuranose unit of 1.
To further diversify the A201A structure, we made a ∆mtdWM4 dual mutant in which the two genes, mtdW and mtdM4, were in-frame deleted in combination, and a ∆mtdM2M3 dual mutant in which the mtdM2 was in-frame deleted and the mtdM3 gene was replaced with an apramycin gene cassette (SI Appendix). As anticipated, these mutants were fermented and found to produce two analogs 10 and 11, respectively, upon HPLC-MS analyses (Fig. 3, traces xvii and xviii). Scaled-up fermentation of these two mutants led to the purification and structure elucidation of 10 and 11 as shown in Fig. 2C by analyses of their HRMS, 1D, and 2D NMR spectroscopic data, respectively (SI Appendix). The fact that these mutant strains accumulated multiple A201A analogs reveals that the four methyltransferases: MtdM1, MtdM2, MtdM3, and MtdM4, as well as the desaturase MtdW, are highly regiospecific, yet display relaxed substrate specificities (Fig. 2C).
Antibacterial Activities of A201A and Its Analogs.
Finally, we tested the antibacterial activities of the engineered A201A analogs against a panel of Gram-positive and Gram-negative bacteria (SI Appendix, Table S11). We found that 6 displays antibacterial activity on par with that of 1. A201A analog 6 has potential as an antibacterial lead, in part, by virtue of its improved water solubility relative to 1 {solubility units: 1 as −2.95 [log(mol/L)] vs. 6 as −2.90 [log(mol/L)] at pH 7.3, as calculated by the Water Solubility Module in Percepta software of Advanced Chemistry Development, Inc}. Notably, the removal of any of the sugar-borne methyl groups or their replacement with the hydroxymethyl moiety diminished antibacterial activities by approximately two-fold. Additionally, removal of the terminal sugar or saturation of the double bond within the hexofuranose significantly decreased the bioactivity.
Conclusions
In conclusion, systematic inactivation, individually or in combination, of 18 genes located proximal to the cluster boundaries, purported to be involved in sugar unit construction/linkage, or purported to drive the tailoring steps of A201A biosynthesis in M. thermotolerans, enabled us to produce 10 A201A analogs. Gene inactivation, feeding, and in vitro biochemical experiments unveiled that d-mannose is the uniform precursor en route to the two unique sugar units involving MtdL, an enzyme involved in GDP-α-l-galactofuranose generation. Des-O-methyl desaturated A201A is decorated by four methyltransferases: MtdM1, MtdM2, MtdM3, and MtdM4, and a desaturase, MtdW. Importantly, the tailoring enzymes all appear to possess exploitable substrate promiscuities. A combinatorial biosynthetic strategy has been credibly applied to the mtd pathway leading to a number of structurally diversified natural product analogs.
Materials and Methods
Materials and methods, including heterologous expression, gene inactivations, isolation and structure elucidation of metabolites 2−11, in vitro biochemical assays, large-scale MtdL enzymatic reaction, characterization of the product, etc., are summarized in SI Appendix, SI Materials and Methods. Proposed functions of biosynthetic genes, bacteria, plasmids, primers, NMR spectral data, and antibacterial activities of the compounds are summarized in SI Appendix, Tables S1–S11. Construction and gel electrophoresis analyses of mutant strains, sequence alignments, NMR spectra of compounds 2−11, plausible chemical mechanisms for MtdL and UGM, and other supporting figures are provided in SI Appendix, Figs. S1–S91.
Supplementary Material
Acknowledgments
This work was supported in part by the National Science Foundation of China (81425022 and U1501223), the Chinese Academy of Sciences (XDA11030403), and the Guangdong Natural Science Foundation (2016A030312014).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620191114/-/DCSupplemental.
References
- 1.Richards MR, Lowary TL. Chemistry and biology of galactofuranose-containing polysaccharides. ChemBioChem. 2009;10:1920–1938. doi: 10.1002/cbic.200900208. [DOI] [PubMed] [Google Scholar]
- 2.Carlson EE, May JF, Kiessling LL. Chemical probes of UDP-galactopyranose mutase. Chem Biol. 2006;13:825–837. doi: 10.1016/j.chembiol.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Oppenheimer M, Valenciano AL, Sobrado P. Biosynthesis of galactofuranose in kinetoplastids: novel therapeutic targets for treating leishmaniasis and chagas’ disease. Enzyme Res. 2011;2011:415976. doi: 10.4061/2011/415976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tefsen B, Ram AF, van Die I, Routier FH. Galactofuranose in eukaryotes: Aspects of biosynthesis and functional impact. Glycobiology. 2012;22:456–469. doi: 10.1093/glycob/cwr144. [DOI] [PubMed] [Google Scholar]
- 5.Tanner JJ, Boechi L, Andrew McCammon J, Sobrado P. Structure, mechanism, and dynamics of UDP-galactopyranose mutase. Arch Biochem Biophys. 2014;544:128–141. doi: 10.1016/j.abb.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chlubnova I, et al. Specific and non-specific enzymes for furanosyl-containing conjugates: Biosynthesis, metabolism, and chemo-enzymatic synthesis. Carbohydr Res. 2012;356:44–61. doi: 10.1016/j.carres.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Nassau PM, et al. Galactofuranose biosynthesis in Escherichia coli K-12: Identification and cloning of UDP-galactopyranose mutase. J Bacteriol. 1996;178:1047–1052. doi: 10.1128/jb.178.4.1047-1052.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beverley SM, et al. Eukaryotic UDP-galactopyranose mutase (GLF gene) in microbial and metazoal pathogens. Eukaryot Cell. 2005;4:1147–1154. doi: 10.1128/EC.4.6.1147-1154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poulin MB, et al. Characterization of a bifunctional pyranose-furanose mutase from Campylobacter jejuni 11168. J Biol Chem. 2010;285:493–501. doi: 10.1074/jbc.M109.072157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oppenheimer M, Poulin MB, Lowary TL, Helm RF, Sobrado P. Characterization of recombinant UDP-galactopyranose mutase from Aspergillus fumigatus. Arch Biochem Biophys. 2010;502:31–38. doi: 10.1016/j.abb.2010.06.035. [DOI] [PubMed] [Google Scholar]
- 11.Wesener DA, May JF, Huffman EM, Kiessling LL. UDP-galactopyranose mutase in nematodes. Biochemistry. 2013;52:4391–4398. doi: 10.1021/bi400264d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanders DA, et al. UDP-galactopyranose mutase has a novel structure and mechanism. Nat Struct Biol. 2001;8:858–863. doi: 10.1038/nsb1001-858. [DOI] [PubMed] [Google Scholar]
- 13.Soltero-Higgin M, Carlson EE, Gruber TD, Kiessling LL. A unique catalytic mechanism for UDP-galactopyranose mutase. Nat Struct Mol Biol. 2004;11:539–543. doi: 10.1038/nsmb772. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Q, Liu H. Mechanistic investigation of UDP-galactopyranose mutase from Escherichia coli using 2- and 3-fluorinated UDP-galactofuranose as probes. J Am Chem Soc. 2001;123:6756–6766. doi: 10.1021/ja010473l. [DOI] [PubMed] [Google Scholar]
- 15.Sun HG, Ruszczycky MW, Chang WC, Thibodeaux CJ, Liu HW. Nucleophilic participation of reduced flavin coenzyme in mechanism of UDP-galactopyranose mutase. J Biol Chem. 2012;287:4602–4608. doi: 10.1074/jbc.M111.312538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kizjakina K, Tanner JJ, Sobrado P. Targeting UDP-galactopyranose mutases from eukaryotic human pathogens. Curr Pharm Des. 2013;19:2561–2573. doi: 10.2174/1381612811319140007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruber TD, Borrok MJ, Westler WM, Forest KT, Kiessling LL. Ligand binding and substrate discrimination by UDP-galactopyranose mutase. J Mol Biol. 2009;391:327–340. doi: 10.1016/j.jmb.2009.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruber TD, Westler WM, Kiessling LL, Forest KT. X-ray crystallography reveals a reduced substrate complex of UDP-galactopyranose mutase poised for covalent catalysis by flavin. Biochemistry. 2009;48:9171–9173. doi: 10.1021/bi901437v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Straaten KE, et al. Structural basis of ligand binding to UDP-galactopyranose mutase from Mycobacterium tuberculosis using substrate and tetrafluorinated substrate analogues. J Am Chem Soc. 2015;137:1230–1244. doi: 10.1021/ja511204p. [DOI] [PubMed] [Google Scholar]
- 20.Mehra-Chaudhary R, Dai Y, Sobrado P, Tanner JJ. In crystallo capture of a covalent intermediate in the UDP-galactopyranose mutase reaction. Biochemistry. 2016;55:833–836. doi: 10.1021/acs.biochem.6b00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reiter WD. Biochemical genetics of nucleotide sugar interconversion reactions. Curr Opin Plant Biol. 2008;11:236–243. doi: 10.1016/j.pbi.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Gutiérrez M, et al. l-Galactose as a natural product: Isolation from a marine octocoral of the first α-l-galactosyl saponin. Tetrahedron Lett. 2004;45:7833–7836. [Google Scholar]
- 23.Wheeler GL, Jones MA, Smirnoff N. The biosynthetic pathway of vitamin C in higher plants. Nature. 1998;393:365–369. doi: 10.1038/30728. [DOI] [PubMed] [Google Scholar]
- 24.Pabst M, et al. Nucleotide and nucleotide sugar analysis by liquid chromatography-electrospray ionization-mass spectrometry on surface-conditioned porous graphitic carbon. Anal Chem. 2010;82:9782–9788. doi: 10.1021/ac101975k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao YP, et al. Response to weaning and dietary l-glutamine supplementation: Metabolomic analysis in piglets by gas chromatography/mass spectrometry. J Zhejiang Univ Sci B. 2012;13:567–578. doi: 10.1631/jzus.B1100343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuentes VM. 1996. Compositions containing carbohydrates obtained from plants of the family cactacea. Eur Pat Appl EP 0706797.
- 27.Matienzo AC, Lamorena M. 2004. Extraction and initial characterization of propolis from stingless bees (Trigona biroi Friese). Proceeding of the Asian Apicultural Association Conference and 10 BEENET Symposium and Technoflora (University of the Philippines Los Baños, Laguna, The Philippines), pp 321–329.
- 28.Van Ginkel R, Selwood AI, Wilkins AL, Ford S. 2014. Anti-microbial compositions, US Pat 20140357580.
- 29.Tello I, et al. Anticonvulsant and neuroprotective effects of oligosaccharides from Lingzhi or Reishi medicinal mushroom, Ganoderma lucidum (Higher Basidiomycetes) Int J Med Mushrooms. 2013;15:555–568. doi: 10.1615/intjmedmushr.v15.i6.40. [DOI] [PubMed] [Google Scholar]
- 30.Hanaya T, Yasuda K, Yamamoto H, Yamamoto H. Stereoselectivity in the preparation of 5,6-dideoxy-5-dimethoxyphosphinyl-d-and-l-hexofuranoses, and an efficient synthesis of 5,6-dideoxy-5-hydroxyphosphinyl-l-galactopyranose (a P-in-the-ring l-Fucose analogue) Bull Chem Soc Jpn. 1993;66:2315–2322. [Google Scholar]
- 31.Binch H, Stangier K, Thiem J. Chemical synthesis of GDP-l-galactose and analogues. Carbohydr Res. 1998;306:409–419. [Google Scholar]
- 32.Kirst HA, et al. The structure of A201A, a novel nucleoside antibiotic. J Antibiot (Tokyo) 1985;38:575–586. doi: 10.7164/antibiotics.38.575. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Q, et al. Discovery and engineered overproduction of antimicrobial nucleoside antibiotic A201A from the deep-sea marine actinomycete Marinactinospora thermotolerans SCSIO 00652. Antimicrob Agents Chemother. 2012;56:110–114. doi: 10.1128/AAC.05278-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polikanov YS, et al. Distinct tRNA accommodation intermediates observed on the ribosome with the antibiotics hygromycin A and A201A. Mol Cell. 2015;58:832–844. doi: 10.1016/j.molcel.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamill RL, Hoehn MM. 1974. Antibiotics A201A and A201B and process for the production thereof, US Pat 3843784.
- 36.Ensminger PW, Hamill RL. 1979. Method of treating acne with antibiotic A201A, US Pat 4143141.
- 37.Nie S, Li W, Yu B. Total synthesis of nucleoside antibiotic A201A. J Am Chem Soc. 2014;136:4157–4160. doi: 10.1021/ja501460j. [DOI] [PubMed] [Google Scholar]
- 38.Saugar I, Sanz E, Rubio MA, Espinosa JC, Jiménez A. Identification of a set of genes involved in the biosynthesis of the aminonucleoside moiety of antibiotic A201A from Streptomyces capreolus. Eur J Biochem. 2002;269:5527–5535. doi: 10.1046/j.1432-1033.2002.03258.x. [DOI] [PubMed] [Google Scholar]
- 39.Saugar I, et al. Characterization of the biosynthetic gene cluster (ata) for the A201A aminonucleoside antibiotic from Saccharothrix mutabilis subsp. capreolus. J Antibiot (Tokyo) 2017;70:404–413. doi: 10.1038/ja.2016.123. [DOI] [PubMed] [Google Scholar]
- 40.Chen Q, et al. Discovery of McbB, an enzyme catalyzing the β-carboline skeleton construction in the marinacarboline biosynthetic pathway. Angew Chem Int Ed Engl. 2013;52:9980–9984. doi: 10.1002/anie.201303449. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, et al. Identification of the grincamycin gene cluster unveils divergent roles for GcnQ in different hosts, tailoring the l-rhodinose moiety. Org Lett. 2013;15:3254–3257. doi: 10.1021/ol401253p. [DOI] [PubMed] [Google Scholar]
- 42.Tercero JA, Espinosa JC, Lacalle RA, Jiménez A. The biosynthetic pathway of the aminonucleoside antibiotic puromycin, as deduced from the molecular analysis of the pur cluster of Streptomyces alboniger. J Biol Chem. 1996;271:1579–1590. doi: 10.1074/jbc.271.3.1579. [DOI] [PubMed] [Google Scholar]
- 43.Palaniappan N, Ayers S, Gupta S, Habib S, Reynolds KA. Production of hygromycin A analogs in Streptomyces hygroscopicus NRRL 2388 through identification and manipulation of the biosynthetic gene cluster. Chem Biol. 2006;13:753–764. doi: 10.1016/j.chembiol.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 44.Palaniappan N, et al. Biosynthesis of the aminocyclitol subunit of hygromycin A in Streptomyces hygroscopicus NRRL 2388. Chem Biol. 2009;16:1180–1189. doi: 10.1016/j.chembiol.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 45.Kneidinger B, et al. Identification of two GDP-6-deoxy-d-lyxo-4-hexulose reductases synthesizing GDP-d-rhamnose in Aneurinibacillus thermoaerophilus L420-91T. J Biol Chem. 2001;276:5577–5583. doi: 10.1074/jbc.M010027200. [DOI] [PubMed] [Google Scholar]
- 46.Major LL, Wolucka BA, Naismith JH. Structure and function of GDP-mannose-3′,5′-epimerase: An enzyme which performs three chemical reactions at the same active site. J Am Chem Soc. 2005;127:18309–18320. doi: 10.1021/ja056490i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolucka BA, Van Montagu M. GDP-mannose 3′,5′-epimerase forms GDP-l-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. J Biol Chem. 2003;278:47483–47490. doi: 10.1074/jbc.M309135200. [DOI] [PubMed] [Google Scholar]
- 48.Konishi T, et al. A plant mutase that interconverts UDP-arabinofuranose and UDP-arabinopyranose. Glycobiology. 2007;17:345–354. doi: 10.1093/glycob/cwl081. [DOI] [PubMed] [Google Scholar]
- 49.Rautengarten C, et al. The interconversion of UDP-arabinopyranose and UDP-arabinofuranose is indispensable for plant development in Arabidopsis. Plant Cell. 2011;23:1373–1390. doi: 10.1105/tpc.111.083931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Breton C, Snajdrová L, Jeanneau C, Koca J, Imberty A. Structures and mechanisms of glycosyltransferases. Glycobiology. 2006;16:29R–37R. doi: 10.1093/glycob/cwj016. [DOI] [PubMed] [Google Scholar]
- 51.Richards MR, Bai Y, Lowary TL. Comparison between DFT- and NMR-based conformational analysis of methyl galactofuranosides. Carbohydr Res. 2013;374:103–114. doi: 10.1016/j.carres.2013.03.030. [DOI] [PubMed] [Google Scholar]
- 52.Mangoni A. Strategies for structural assignment of marine natural products through advanced NMR-based techniques. In: Fattorusso E, Gerwick WH, Taglialatela-Scafati O, editors. Handbook of Marine Natural Products. Springer; New York: 2012. pp. 541–543. [Google Scholar]
- 53.Cavener DR. GMC oxidoreductases. A newly defined family of homologous proteins with diverse catalytic activities. J Mol Biol. 1992;223:811–814. doi: 10.1016/0022-2836(92)90992-s. [DOI] [PubMed] [Google Scholar]
- 54.Dym O, Eisenberg D. Sequence-structure analysis of FAD-containing proteins. Protein Sci. 2001;10:1712–1728. doi: 10.1110/ps.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.