Significance
Exogenous cannabinoids such as marijuana exert their influence through cannabinoid receptors. Endogenous cannabinoids such as anandamide (AEA) function through the same receptors, and their physiological roles are a subject of intense study. Here, we show that AEA plays a pivotal role in maintaining immunological health in the gut. The immune system in the gut actively tolerates the foreign antigens present in the gut through mechanisms that are only partially understood. We show that AEA contributes to this critical process by promoting the presence of CX3CR1hi macrophages, which are immunosuppressive. These results uncover a major conversation between the immune and nervous systems. In addition, with the increasing prevalence of ingestion of exogenous marijuana, our study has significant implications for public health.
Keywords: mucosal immunity, T-regulatory cells, cannabis, diabetes, CX3CR1 macrophage
Abstract
Endogenous cannabinoids (endocannabinoids) are small molecules biosynthesized from membrane glycerophospholipid. Anandamide (AEA) is an endogenous intestinal cannabinoid that controls appetite and energy balance by engagement of the enteric nervous system through cannabinoid receptors. Here, we uncover a role for AEA and its receptor, cannabinoid receptor 2 (CB2), in the regulation of immune tolerance in the gut and the pancreas. This work demonstrates a major immunological role for an endocannabinoid. The pungent molecule capsaicin (CP) has a similar effect as AEA; however, CP acts by engagement of the vanilloid receptor TRPV1, causing local production of AEA, which acts through CB2. We show that the engagement of the cannabinoid/vanilloid receptors augments the number and immune suppressive function of the regulatory CX3CR1hi macrophages (Mϕ), which express the highest levels of such receptors among the gut immune cells. Additionally, TRPV1−/− or CB2−/− mice have fewer CX3CR1hi Mϕ in the gut. Treatment of mice with CP also leads to differentiation of a regulatory subset of CD4+ cells, the Tr1 cells, in an IL-27–dependent manner in vitro and in vivo. In a functional demonstration, tolerance elicited by engagement of TRPV1 can be transferred to naïve nonobese diabetic (NOD) mice [model of type 1 diabetes (T1D)] by transfer of CD4+ T cells. Further, oral administration of AEA to NOD mice provides protection from T1D. Our study unveils a role for the endocannabinoid system in maintaining immune homeostasis in the gut/pancreas and reveals a conversation between the nervous and immune systems using distinct receptors.
The endocannabinoid system (ECS) is highly conserved in evolution dating back to at least 600 million years (1). It consists of (i) lipid endocannabinoids; (ii) their receptors such as the G protein-coupled receptors, cannabinoid receptor 1 (CB1), cannabinoid receptor 2 (CB2), and a ligand-gated cation channel vanilloid receptor 1 (i.e., TRPV1); and (iii) the enzymes such as fatty acid amide hydrolyase (FAAH) that regulate the levels of endocannabinoids in vivo (2). The ECS impacts several aspects of mammalian physiology, particularly in the gut. Endogenous cannabinoids such as anandamide (AEA) belong to the N-acylethanolamine family and are synthesized from membrane glycerophospholipids (3, 4). AEA is an intestinal endocannabinoid, which engages its cognate receptors on the enteric nervous system and contributes to control of appetite and energy balance (5). Here, we have unraveled a role of the ECS in regulating immune homeostasis in the gut–pancreas axis.
The intestinal immune system is continuously exposed to a variety of antigens. An effective immune response must be launched against pathogenic insults; however, it must maintain tolerance to the vast amount of antigens such as those in commensal flora, endogenous metabolites, and food components. Mononuclear phagocytes (MNPs) such as CX3CR1hi macrophage (Mϕ) and CD103+ dendritic cells (DCs), which are present abundantly in the small intestinal lamina propria (siLP), play an instrumental role in maintaining this exquisite balance between inflammation and tolerance (6).
On the basis of expression level of the chemokine receptor CX3CR1, two functionally and phenotypically distinct populations of MNPs in the gut have been identified. Although both populations differentiate from blood derived monocytes, CX3CR1hi Mϕ are regulatory in nature, whereas CX3CR1lo cells are perpetuators of inflammation (7). Adoptive transfer of CX3CR1hi Mϕ provide protection from inflammatory diseases like colitis in mouse models (8). However, the microenvironment signal that “educates” the monocytes and promotes differentiation into CX3CR1hi (as opposed to CX3CR1lo) Mϕ still remains elusive. It is increasingly appreciated that the MNPs play a crucial role in gut equilibrium. Identifying key elements that condition the siLP MNPs to become tolerogenic will enable exploration of strategies that can be helpful in providing protection from inflammatory diseases.
The studies described here connect the nervous system (responsible for sensing exogenous cannabinoids such as marijuana, as well as endogenous ones) to one of the most fundamental properties of the immune system, i.e., maintenance of an immune tolerant environment in the gut. These studies have a bearing on marijuana use and abuse, particularly with respect to the formulations meant for ingestion, and very significantly, for new avenues of understanding and treating human diseases.
Results
CX3CR1hi Mϕ Express the Highest Levels of ECS Receptors CB2 and TRPV1.
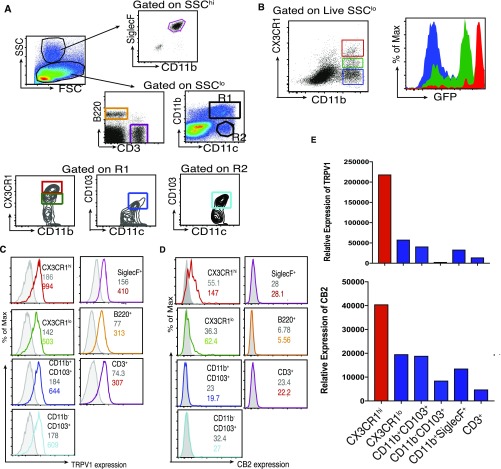
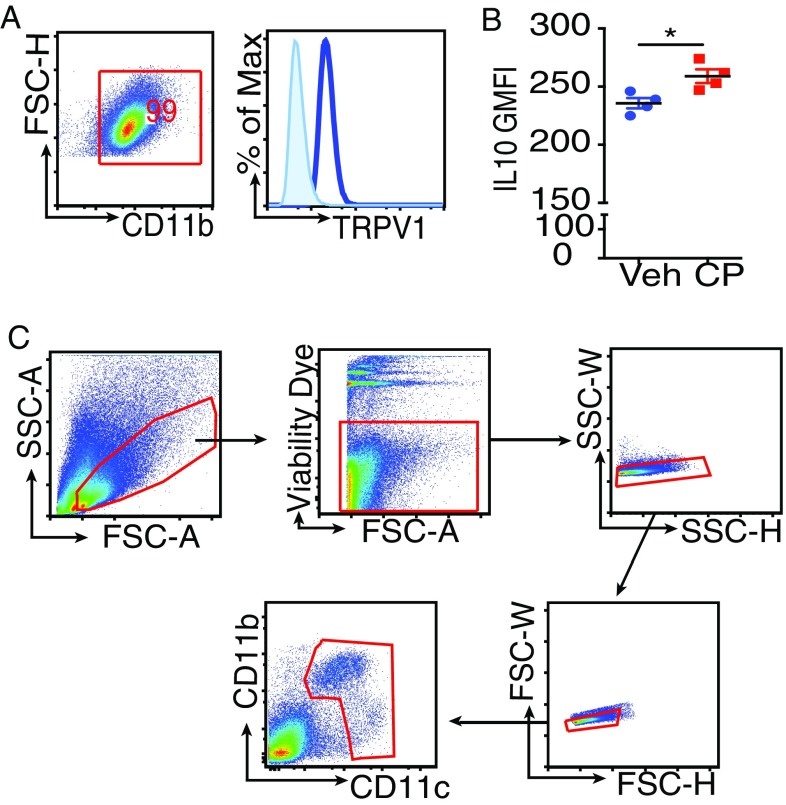
siLP cells were isolated after removing the Peyer’s patches (PPs), and cell suspensions obtained were analyzed. Fig. S1A depicts the gating strategy to identify the different cell populations. The SSChi cells were gated for CD11b+SiglecF+ cells (i.e., eosinophils) and the SSClo cells for lymphocytes (B220+ and CD3+) and the MNPs. Phenotypic characterization of the MNPs was based on the expression of integrins CD11b (αM), CD11c (αX), CD103 (αE), and the chemokine receptor CX3CR1. Special attention was paid to the CX3CR1 phenotype because CX3CR1hi Mϕ but not the CX3CR1lo cells play a significant role in immune homeostasis (7). To facilitate the identification of CX3CR1hi and CX3CR1lo cells, we used the CX3CR1+/gfp reporter mice in which GFP expression is a marker for CX3CR1 expression (9). As a quality-control measure, cell suspensions derived from siLP of these mice were stained with antibody against CX3CR1 and also checked for GFP by flow cytometry. The two patterns were consistent (Fig. S1B). We classified the MNPs as R1 (CD11b+CD11c+) and R2 (CD11b−CD11c+). R1 was further subclassified into CD11b+CD11c+CX3CR1hi, CD11b+CD11c+CX3CR1lo, and CD11b+CD11c+CD103+, and, in the R2, we looked for CD11b−CD11chiCD103+ (Fig. S1A, Bottom). Selected populations were analyzed for the expression level of TRPV1 (Fig. S1C) and CB2 (Fig. S1D). Of note, the MNPs expressed higher levels of TRPV1 and CB2 compared with the lymphocytes and eosinophils. Strikingly, CX3CR1hi cells expressed higher levels of TRPV1 and CB2 compared with the other cells analyzed (Fig. 1A). This finding was corroborated by quantitative PCR (qPCR) analysis of mRNA levels of TRPV1 and Cnr2 (gene encoding CB2) on the various gut immune populations (Fig. S1E). Because of the substantial overlap in surface phenotype of intestinal DCs and Mϕ, designation of CX3CR1hi cells as DCs or Mϕ is debatable. However, Tamoutounour et al. have shown that expression of CD64 (the high-affinity FcγR1) and absence of CD103 can be used in combination with CD11c and MHCII to definitively define functionally distinct Mϕ in intestine of mice (10). The population predominantly expressing TRPV1 and CB2 was found to meet these criteria (Fig. 1B, Top). Consistent with Tamoutounour et al., we show that CX3CR1lo cells are a heterogeneous population comprising of Mϕ (CD64+) and DCs (CD64−; Fig. S2A). Previous reports demonstrate that CX3CR1hi Mϕ make IL-10 ex vivo without stimulation (7). The IL-10–producing capacity of the CX3CR1hi Mϕ was tested ex vivo without any stimulation by using the IL-10–GFP reporter mice (11); it was observed that CX3CR1hi Mϕ express high levels of IL-10 (Fig. 1B, Bottom). Thus, TRPV1 and CB2, which are integral components of the ECS, are preferentially expressed by the CX3CR1hi regulatory Mϕ.
Fig. S1.
(A) Gating strategy for siLP cells of CX3CR1gfp/+ mice. SSChi and SSClo cells were gated. CD11b+SiglecF+ cells were analyzed in the SSChi population. SSClo cells were gated for CD3+ and B220+ cells and mononuclear phagocytes comprising R1 (CD11b+CD11c+) and R2 (CD11b−CD11c+) cells. R1 and R2 were further analyzed for expression of CX3CR1 and/or CD103. (B) CX3CR1-antibody staining correlates with CX3CR1-GFP expression in CX3CR1gfp/+ mice. Dot plot represents three different populations shown by gates in distinct colors: CD11b+CX3CR1hi Mφ (red), CD11b+CX3CR1lo (green), and CD11b+CX3CR1− cells (blue) among live SSClo siLP cells of CX3CR1gfp/+ mice gated based on staining with CX3CR1 antibody (Left). Histograms represent expression of CX3CR1-GFP by each of the gated populations in distinct colors (Right), showing the correlation between CX3CR1− antibody staining and CX3CR1-GFP among live SSCloCD11b+ siLP cells of CX3CR1gfp/+. (C and D) Histogram with the normal serum (control) is shown in gray, and the histogram of expression of TRPV1 (C) or CB2 (D) by the selected population from A is shown in distinct colors. The number inside each box represents the GMFI, numbers in gray represent the GMFI of control, and the numbers in distinct colors represent GMFI of anti-TRPV1 or anti-CB2 antibody for the given population. (E) Total RNA was extracted from various immune cell populations sorted from siLP of mice, and expression levels of TRPV1 and CB2 (encoded by Cnr2) were evaluated by qPCR.
Fig. 1.
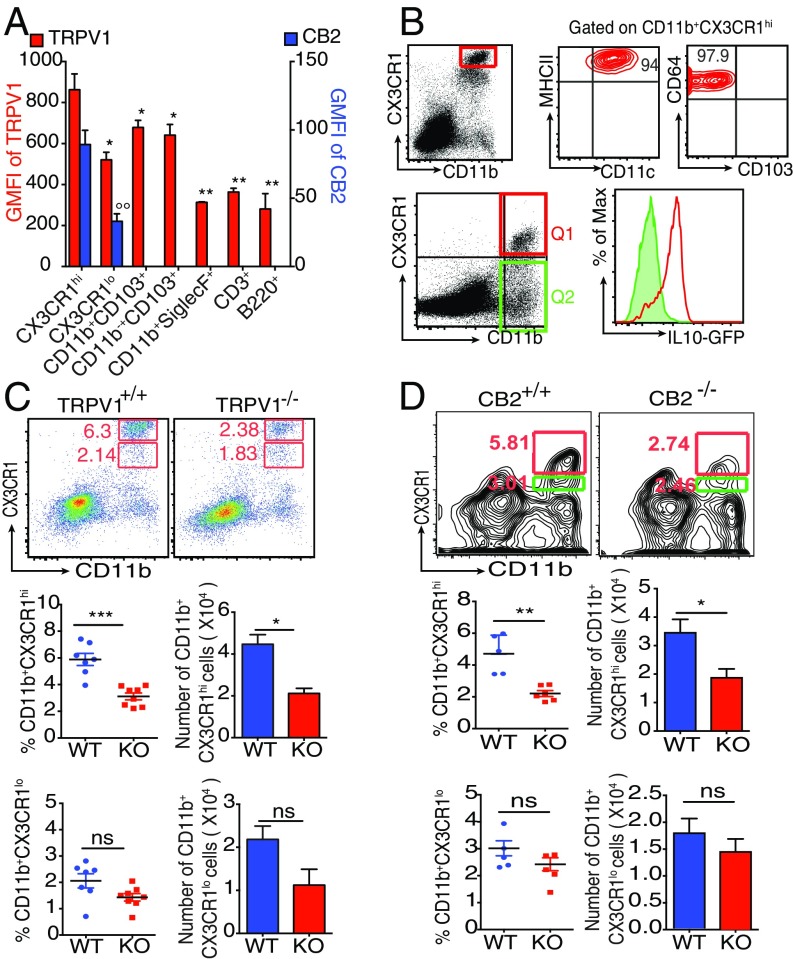
The ECS influences the siLP CX3CR1hi Mϕ at steady state. (A) Bar graph represents geometric mean fluorescence intensity (GMFI) of TRPV1 and CB2 on indicated siLP cells (asterisk and degree symbol represent statistical comparison of TRPV1 and CB2 expression, respectively, of each indicated cell type with expression of these receptors by CD11b+CX3CR1hi cells). (B) Phenotype of CD11b+CX3CR1hi cells (Top). Expression of IL-10 by CD11b+CX3CR1hi Mϕ (Q1, red) and CD11b+CX3CR1lo and CD11b+CX3CR1− cells (Q2, green; Bottom). (C) Representative FACS plots and column scatter plots represent the frequency (n = 7–8 mice per group), and bar graphs represent the absolute number (n = 5 mice per group) of CD11b+CX3CR1hi Mϕ (Middle) and CD11b+CX3CR1lo cells (Bottom) in the siLP of CX3CR1gfp/+ TRPV1+/+ and CX3CR1gfp/+ TRPV1−/− mice. (D) Representative FACS plots and column scatter plots represent the frequency, and bar graphs represent the absolute number of CD11b+CX3CR1hi Mϕ (Middle) and CD11b+CX3CR1lo cells (Bottom), in the siLP cells of CB2+/+ and CB2−/− mice (n = 5 mice per group). (ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, and °°P < 0.01, unpaired Student’s t test; data represent mean ± SEM).
Fig. S2.
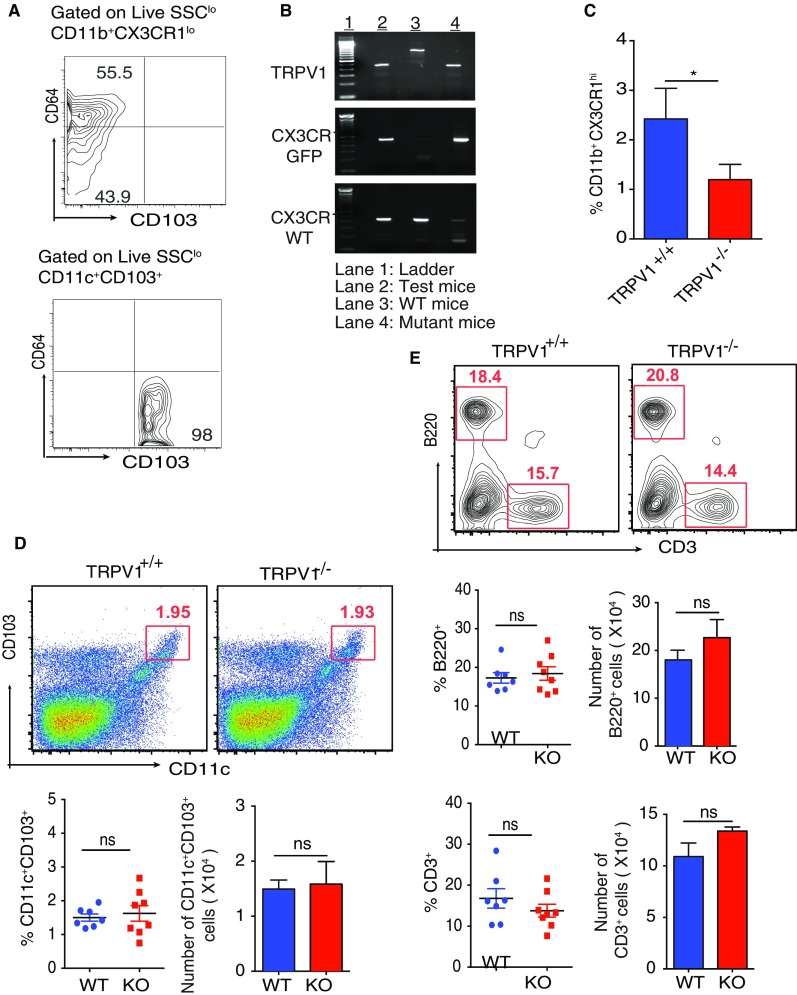
(A) Contour plot shows the expression of CD64 and CD103 by CD11b+CX3CR1lo cells (Top) and CD11c+CD103+ DCs (Bottom). (B) Genotype of the CX3CR1gfp/+TRPV1−/− mice constructed (C) siLP cells were purified from TRPV1+/+ and TRPV1−/− mice and were analyzed by flow cytometry by using antibody staining for CX3CR1. Bar graph represents the frequency of CX3CR1hi Mφ among live SSClo siLP cells of the TRPV1+/+ and TRPV1−/− mice. (D) Frequency and absolute number of CD11c+CD103+ DCs in the siLP of TRPV1+/+ and TRPV1−/− mice (n = 7–8 mice per group). (E) Frequency and absolute number of B220+ and CD3+ in the siLP of TRPV1+/+ and TRPV1−/− mice (n = 7–8 mice per group). (ns, not significant; n = 3; *P < 0.05, unpaired Student’s t test; data represent mean ± SEM).
ECS Receptors Regulate CX3CR1hi Mϕ Homeostasis in the Gut.
Given the high expression of TRPV1 and CB2 on CX3CR1hi Mϕ, we analyzed the role of each receptor. For TRPV1, we back-crossed TRPV1−/− mice (12) with CX3CR1gfp/gfp reporter mice to obtain CX3CR1gfp/+ TRPV1−/− mice. We validated the expression of functional (i.e., WT) allele and GFP allele for CX3CR1 and the absence of TRPV1 by PCR (Fig. S2B). Introduction of GFP disrupts the expression of endogenous CX3CR1 such that a homozygous mice does not express functional CX3CR1 protein whereas heterozygous mice do, and the expression of CX3CR1 in them can be followed through GFP. By analyzing the siLP cells of the CX3CR1gfp/+TRPV1−/− mice, we observed that the absence of TRPV1 influences the frequency and number of CX3CR1+ cells differently: the frequency and number of CX3CR1hi Mϕ are reduced significantly (Fig. 1C, Middle), whereas, in case of the CX3CR1lo cells, there is a trend of decreasing number and frequency but the difference is not statistically significant (Fig. 1C, Bottom). These results in CX3CR1gfp/+TRPV1−/− mice were also reproduced in TRPV1−/− mice in which CX3CR1 was identified with antibody staining (Fig. S2C). The frequency and number of the other cells such as CD103+ DCs and CD3+ and B220+ cells remain unaffected in the absence of TRPV1 (Fig. S2 D and E).
Next, we tested the importance of CB2 on the homeostasis of CX3CR1hi Mϕ by using mice genetically lacking CB2: the frequency and absolute number of CX3CR1hi Mϕ was significantly reduced in CB2-deficient mice (Fig. 1D, Middle). As in TRPV1−/− mice, there is a trend of decreasing number and frequency among the CX3CR1lo cells in CB2−/− mice as well, but the difference is not statistically significant (Fig. 1D, Bottom).
Stimulation of the ECS Leads to Expansion of CX3CR1hi Mϕ Population.
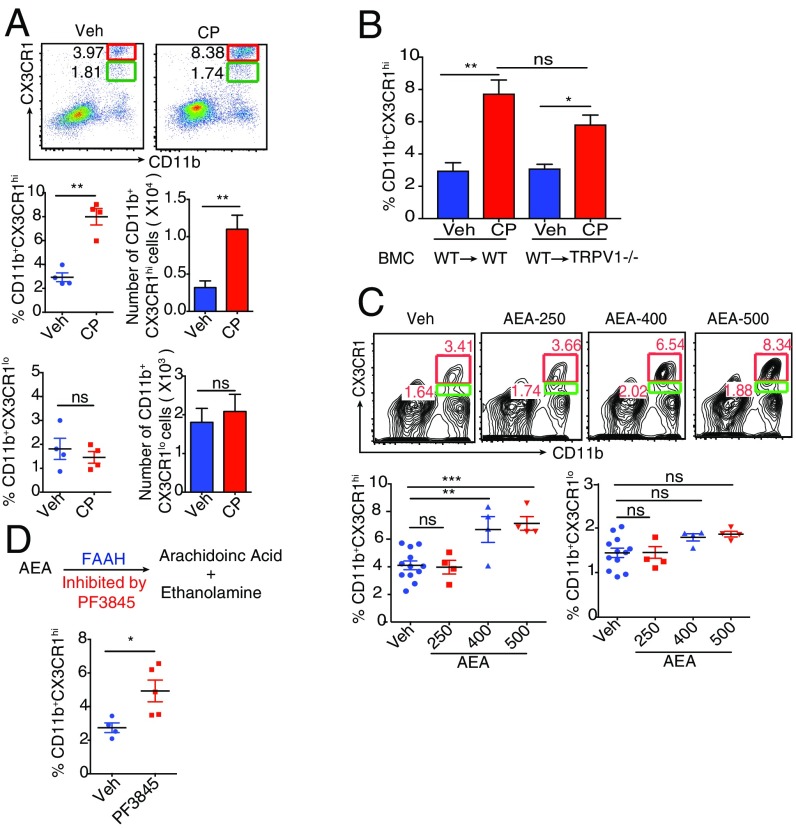
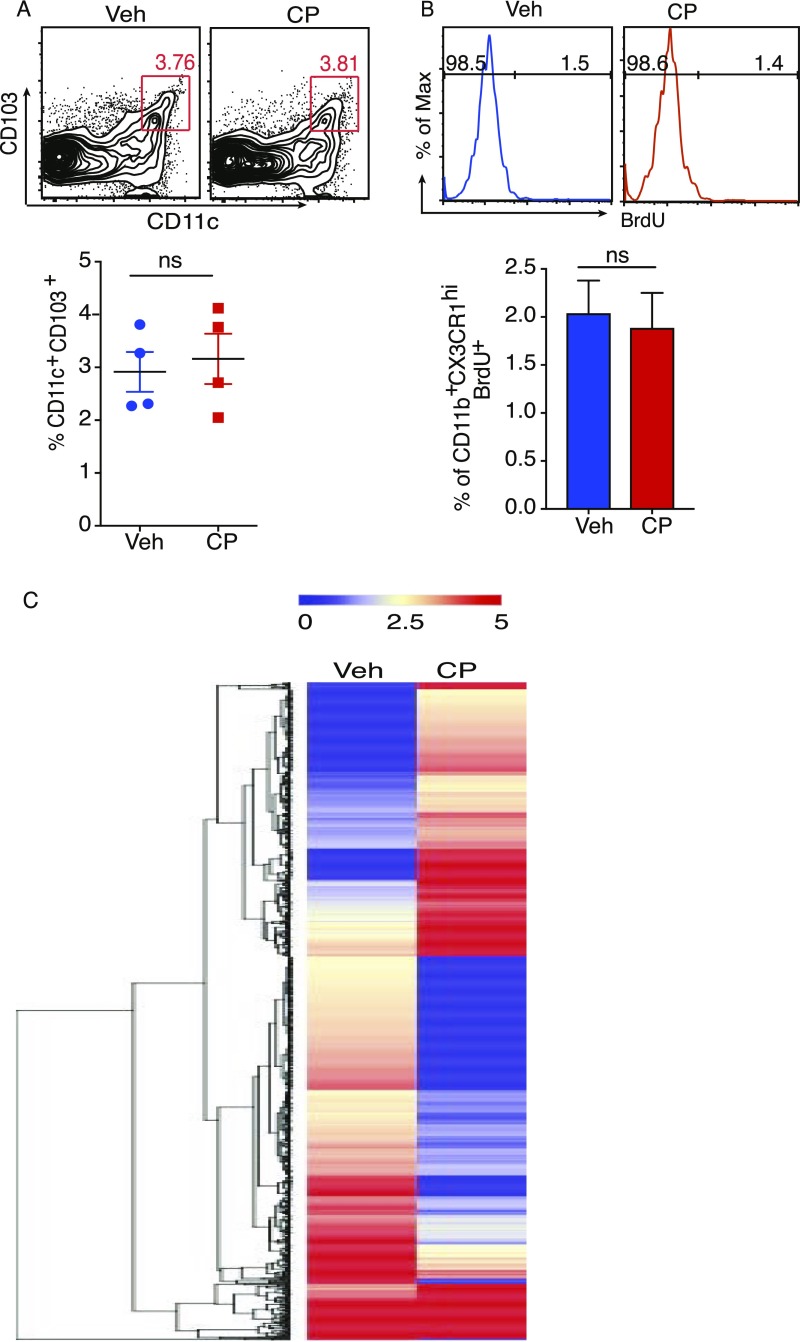
CX3CR1gfp/+TRPV1−/− mice underwent oral gavage with 10 μg capsaicin (CP) or vehicle, and the siLP cells were isolated 24 h after treatment. CP-elicited changes in the MNPs were analyzed. CP significantly increased the frequency of the regulatory CX3CR1hi Mϕ but not the frequency and number of the CX3CR1lo cells (Fig. 2A). CP did not affect the frequency of CD11c+CD103+ DCs (Fig. S3A). To test if CP-mediated increase in CX3CR1hi Mϕ frequency was caused by enhanced proliferation, an in vivo BrdU incorporation assay was performed in mice that had undergone oral gavage with CP: feeding of CP did not result in incorporation of BrdU in CX3CR1hi Mϕ (Fig. S3B). We performed RNA sequencing and IsoEM/IsoDE analysis (13, 14) (as described in SI Materials and Methods) on the total MNPs of siLP to study the changes mediated by treatment with CP; the heat map for hierarchical clustering of the top 1,200 differentially expressed genes (DEGs) shows a clear presence of genes that are significantly up- or down-regulated by CP treatment (Fig. S3C). We used Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatic Resource, version 6.8, to study the functional-related gene groups and pathways. We found that CP treatment significantly enhances the cytokine/chemokine-mediated signaling pathways such as CCL2, CXCL12, CSF1, and IL-10 (Table S1). Thus, the increase in the frequency of CX3CR1hi Mϕ must derive from augmented migration or differentiation or a combination of both processes.
Fig. 2.
CP and AEA expand the CX3CR1hi Mϕ population in vivo. (A) CP-elicited changes (24 h after feeding) in the frequency (Middle and Bottom Left) and absolute numbers (Middle and Bottom Right) of CX3CR1hi Mϕ and CX3CR1lo cells in the siLP of CX3CR1gfp/+ mice (n = 4 mice per group). Veh, vehicle-treated mice. (B) TRPV1−/− mice (n = 4 mice per group) were lethally irradiated and, 24 h later, received CD45.1 C57BL/6 BM as described in Materials and Methods. After reconstitution (6 wk), TRPV1−/− BM chimeras were orally gavaged with CP or vehicle. Graph indicates CP-mediated changes in the frequency of CX3CR1hi Mϕ. (C) FACS plots and column scatter plots represent changes in the frequency (24 h after feeding) of siLP CX3CR1hi Mϕ elicited by AEA (data are pooled from three independent experiments testing different doses of AEA; n = 4–5 mice per group). (D) Changes in the frequency (24 h after feeding) of siLP CX3CR1hi Mϕ elicited by PF3845 (n = 4–5 mice per group). (ns, not significant; *P < 0.05, **P < 0.01, and ***P < 0.001, unpaired Student’s t test or one-way ANOVA; data represent mean ± SEM).
Fig. S3.
(A) Contour plots and column scatter plots represent frequency of CD11c+CD103+ DCs among live siLP cells 24 h after oral administration of CP (n = 4 mice per group). (B) Histogram and bar graph represent frequency of CX3CR1hi Mφ among live siLP cells that incorporated BrdU after oral administration of CP. (C) Hierarchical clustering heat map displaying the statistical overrepresentation of the top 1,200 up- or down-regulated DEGs in siLP MNPs after CP treatment (Left). (ns, not significant; data represent mean ± SEM).
Table S1.
RNA-Seq of MNPs sorted from the siLP of CP- or vehicle-treated mice
| Annotation category | Term | Count | P value | Up-regulated genes |
| GOTERM_BP_DIRECT | GO: 0006955∼immune response | 47 | 0.0087 | CXCL1, CCL2, CXCL2, SUSD2, IL-15, etc. |
| UP_KEYWORDS | Cytokine | 35 | 0.0010 | IL-10, CXCL1, CCL2, CSF1, CXCL2, IL-15, etc. |
| GOTERM_MF_DIRECT | GO: 0005125∼cytokine activity | 35 | 0.0037 | IL-10, CXCL1, CCL2, CSF1, CXCL2, CCL7, CXCL10, etc. |
| KEGG_ PATHWAY | mmu04060: cytokine-cytokine receptor interaction | 35 | 0.0615 | IL-10, CCL2, CSF1, GDF5, CXCL10, etc. |
| UP_KEYWORDS | Chemotaxis | 21 | 7.79 × 10−4 | CXCL1, CCL2, CXCL2, CXCL9, CCL4, CCL7, CXCL10, etc. |
| GOTERM_BP_DIRECT | GO: 0006935∼chemotaxis | 21 | 0.0140 | RARRES2, CCL2, CXCL2, CXCL9, CCL4, CCL7, CXCL10, etc. |
| GOTERM_BP_DIRECT | GO: 0070098∼chemokine mediated signaling pathway | 15 | 0.0013 | CXCL1, CCL2, GPR75, CXCL2, CCL7, CXCL10, etc. |
| SMART | SM00199:SCY | 14 | 2.52 × 10−4 | CXCL1, CCL2, CXCL2, etc. |
| INTERPRO | IPR001811: Chemokine Interleukine-8 like domain | 14 | 0.0012 | CXCL1, CCL2, CXCL2, etc. |
| GOTERM_MF_DIRECT | GO: 0008009∼chemokine activity | 13 | 0.0029 | CXCL1, CCL2, CXCL2, etc. |
| GOTERM_BP_DIRECT | GO: 0071346∼cellular response to IFN gamma | 12 | 0.0769 | CCL24, CCL11, CCL4, CCL7, etc. |
| GOTERM_BP_DIRECT | GO: 0048247∼lymphocyte chemotaxis | 8 | 0.0570 | CCL2, CCL21B, CCL4, CCL7, etc. |
| GOTERM_BP_DIRECT | GO: 0002548∼monocyte chemotaxis | 8 | 0.1249 | CCL12, CCL25, CCL2, CCL4, CCL7, etc. |
| INTERPRO | IPR000827: CC chemokine, conserved site | 6 | 0.0394 | CCL2, CCL24, CCL11, CCL12, CCL4, CCL7, etc. |
| INTERPRO | IPR018048: CC chemokine, conserved site | 5 | 0.170 | CXCL1, CXCL2, etc. |
| INTERPRO | IPR001098: CXC chemokine | 5 | 0.0117 | CXCL1, CXCL2, etc. |
| GOTERM_MF_DIRECT | GO: 0045236∼CXCR chemokine receptor binding | 5 | 0.0177 | CXCL1, CXCL2, CXCL9, PF4, CXCL10, etc. |
| GOTERM_MF_DIRECT | GO: 0048248∼CXCR3 chemokine receptor binding | 3 | 0.0796 | CXCL9, PF4, CXCL10, etc. |
| GOTERM_MF_DIRECT | GO: 0031727∼CCR2 chemokine receptor binding | 3 | 0.0796 | CCL2, CCL12, CCL7, etc. |
DAVID Bioinformatic Resource 6.8 was used to study the functional-related gene groups and pathways. Table represents CP up-regulated pathways.
To distinguish between the contribution of TRPV1-expressing neurons vs. immune cells to the phenomenon shown in Fig. 2A, TRPV1−/− mice reconstituted with bone marrow (BM) from TRPV1+/+ mice were treated with CP. The siLP CX3CR1hi Mϕ populations in all groups were monitored: CP-fed BM chimeric (BMC) mice behaved like the WT mice, indicating that TRPV1 expression on the hematopoietic cells was sufficient for activity. There was a notable, albeit statistically insignificant, difference between the expansion observed in the WT mice compared with the BMC mice, suggesting a possible, though not definitive, contribution from the neuronal TRPV1 in the process (Fig. 2B).
The effect of oral administration of AEA, an endogenous compound capable of activating vanilloid and cannabinoid receptors, on CX3CR1hi Mϕ of siLP was examined. Oral administration of AEA led to a significant increase in the frequency of CX3CR1hi Mϕ (Fig. 2C, Bottom Left). As expected, we did not observe a difference in the frequency of CX3CR1lo cells after treatment with AEA (Fig. 2C, Bottom Right). We reasoned that, if administering exogenous AEA can affect the CX3CR1hi Mϕ, increasing the endogenous levels of AEA by inhibiting the enzyme that catabolizes it should yield similar results. Endogenous levels of AEA are controlled by the enzyme FAAH, which catabolizes AEA into arachidonic acid and ethanolamine (Fig. 2D, Top). Inhibition of FAAH, the enzyme that catabolizes AEA, increases the levels of AEA in the brain and peripheral tissues (15). Massa et al. demonstrated that mice genetically deficient in FAAH are protected from 2,4-dinitrobenzene sulfonic acid-induced colitis, highlighting the antiinflammatory role of AEA in the gut (16).
Mice underwent gavage with an orally viable, irreversible FAAH inhibitor, PF3845, and the effect of this treatment on CX3CR1hi Mϕ in LP was examined 24 h after feeding. Treatment of mice with PF3845 increased the frequency of the regulatory Mϕ demonstrating that endogenous AEA can profoundly influence the homeostasis of CX3CR1hi Mϕ (Fig. 2D, Bottom).
CP Induces AEA Production in Myeloid Cells.
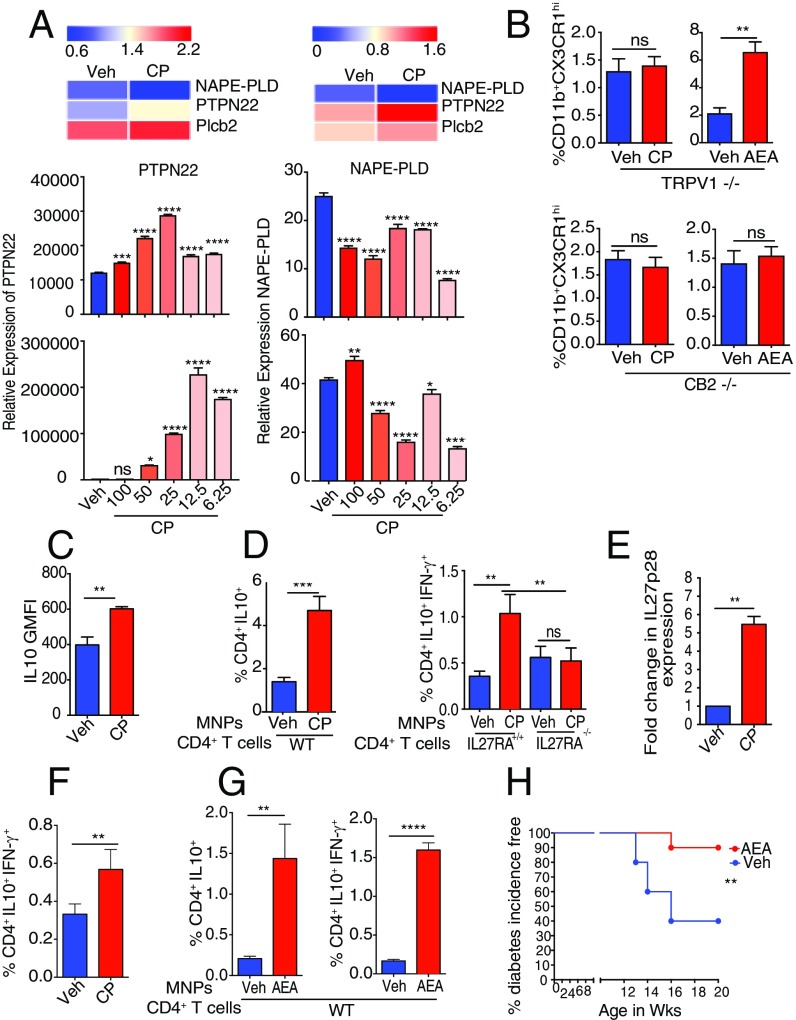
As shown here earlier, CP and AEA influence CX3CR1hi Mϕ in a similar manner. The potential of cross-talk between the two was investigated. AEA biosynthesis involves hydrolysis of a membrane phospholipid precursor N-arachidonoyl phosphatidylethanolamine (NAPE) by a phospholipase C to yield phospho-AEA (pAEA), which is then dephosphorylated by phosphatases such as the tyrosine phosphatase PTPN22; this process in Mϕ is associated with an increase of the level of PTPN22 (17) and a decrease in the level of phospholipase D NAPE-PLD (17). Consistent with this pattern, analysis of the transcriptome of siLP MNPs from CP-treated mice revealed that CP leads to increase in the expression of PTPN22 with a concomitant decrease in the level of NAPE-PLD (Fig. 3A, Top Left).
Fig. 3.
The relationship between CP, AEA, and the tolerogenic properties of siLP MNPs. (A) Heat map of RNA sequencing shows changes in enzymes involved in AEA biosynthesis after CP treatment in siLP MNPs (Top Left) and in RAW 264.7 cells (Top Right). Bar graph shows qPCR of enzymes involved in AEA biosynthesis in RAW 264.7 cells 2 h (Middle) and 4 h (Bottom) after CP treatment in vitro. (B) CP- or AEA-elicited changes (24 h after feeding) in the frequency of CX3CR1hi Mϕ in the siLP of TRPV1−/− mice (Top) and CB2−/− mice (Bottom; n = 4 mice per group). (C) CP-elicited changes in the expression (GMFI) of IL-10 (n = 4 mice per group). (D–G) Mice underwent oral gavage with vehicle (Veh) or CP (10 μg) or AEA (500 μg), and siLP MNPs (CD11b+CD11c+ and CD11b−CD11c+) were sorted after 24 h (D and G) and 48 h (E). (D) MNPs were sorted from CP-treated or vehicle-treated mice and cocultured (1:1) with naïve splenic CD4+CD25−CD62L+ T cells derived from WT (IL-27RA+/+) or IL-27RA−/− mice for 4 d. Expression of IL-10 and IFN-γ among the CD4+ cells was analyzed by flow cytometry. Bar graphs represent the frequency of CD4+IL-10+ and CD4+IL-10+IFN-γ+ (Tr1) cells. (Data are representative of two independent experiments and show mean values ± SEM of duplicate or triplicate determinations in which MNPs were sorted from 12 pooled mice per group). (E) Total RNA was extracted from MNPs sorted from CP-treated or vehicle-treated mice, and expression levels of IL-27-p28 were evaluated by qPCR. (Data represent duplicate determinations from 12 pooled mice per group). Bar graph represents fold increase in expression of IL-27–p28 in CP-treated samples with respect to vehicle-treated samples. (F) Frequency of CP-elicited CD4+IL-10+IFN-γ+ (Tr1) cells among live SSClo siLP cells 2 wk after treatment (n = 4 mice per group). (G) MNPs were sorted from AEA-treated or vehicle-treated mice and cocultured with WT T cells as described in D. Bar graphs represent the frequency of CD4+IL-10+ and CD4+IL-10+IFN-γ+ (Tr1) cells. (Data show mean values ± SEM of triplicate determinations in which MNPs were sorted from 10 pooled mice per group.) (H) Female NOD mice were orally administered AEA or vehicle at 9th and 10th weeks of age, and urine glucose was monitored to study disease progression. (ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, unpaired Student’s t test; data represent mean ± SEM; Mantel–Cox test was used for survival curve).
To explore this phenomenon mechanistically in a pure myeloid population (as opposed to in vivo, in which the effects could be indirect), the Mϕ cell line RAW264.7, which expresses TRPV1 (Fig. S4A), was used. RNA sequencing (RNA-Seq) analysis of CP-treated cells shows that treatment with CP activates the AEA biosynthesis pathway as described earlier (Fig. 3A, Top Right). The changes in the expression levels of PTPN22 and NAPE-PLD in CP-treated RAW264.7 cells were validated by a qPCR analysis (Fig. 3A, Middle and Bottom).
Fig. S4.
(A) Histogram showing TRPV1 expression in RAW 264.7 cells. (B) IL-10–GFP reporter mice underwent oral gavage with vehicle or CP (10 μg). siLP cells were isolated after 24 h, and CP-elicited changes in IL-10 expression among siLP CD11c+CD103+ DCs were analyzed by flow cytometry. Column scatter graphs represent GMFI of IL-10 among siLP CD11c+CD103+ DCs (n = 4 mice per group). (C) Gating strategy for sorting siLP MNPs. siLP cells were harvested, and SSClo live cells were sorted on the basis of the expression of CD11b and CD11c after exclusion of doublets. (*P < 0.05, unpaired Student’s t test; data represent mean ± SEM).
AEA Is the Central Molecule in Expansion of CX3CR1hi Mϕ Population.
As shown in Fig. 2 A and C, oral administration of CP or AEA leads to expansion of the CX3CR1hi population in the siLP. This phenomenon was now tested in TRPV1−/− and CB2−/− mice. In TRPV1−/− mice, CP failed to mediate expansion of CX3CR1hi cells, but AEA still did so (Fig. 3B, Top). As AEA has a higher binding affinity for CB2 than for TRPV1 (18), the same phenomenon was tested in CB2−/− mice as well. Neither CP nor AEA mediated expansion of the CX3CR1hi population in the CB2−/− mice (Fig. 3B, Bottom). These results (Fig. 3 A and B) indicate that AEA, through its interaction with CB2, is the final mediator of the expansion of the CX3CR1hi population in siLP upon oral administration of CP or AEA. The effect of CP is mediated through two steps: one, generation of AEA upon its interaction with TRPV1; and two, interaction of AEA with CB2.
Stimulation of ECS by AEA or CP Enhances the Tolerogenic Properties of MNPs and Provides Protection from Autoimmune Diabetes.
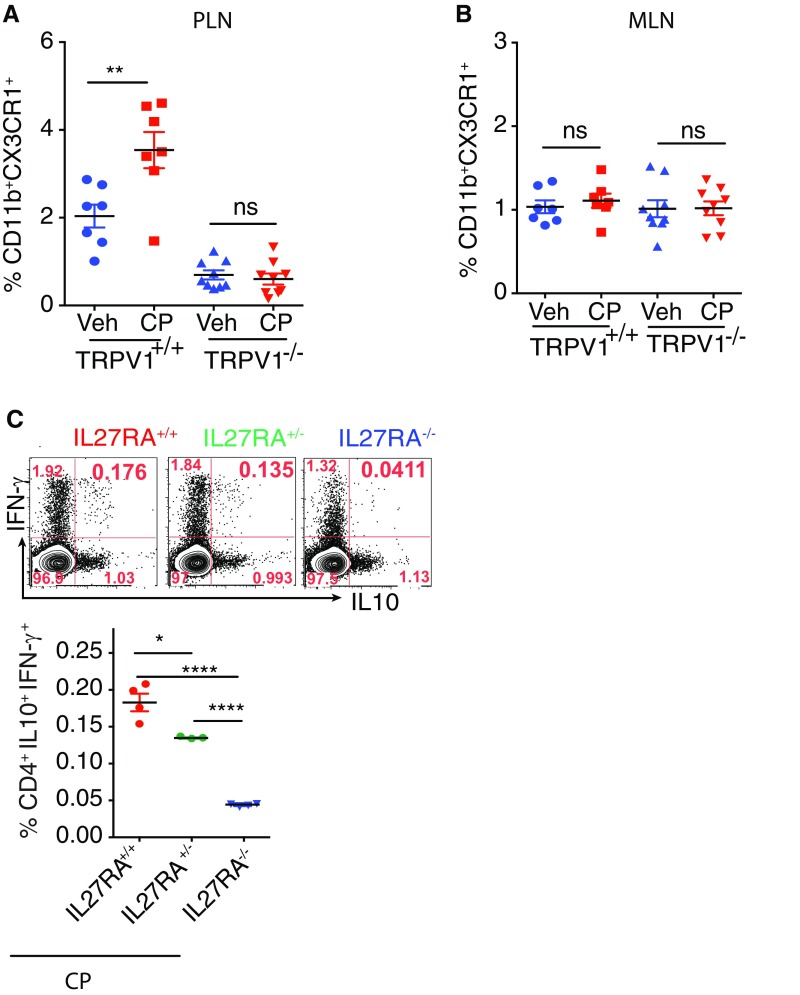
IL-10 is a cytokine with potent tolerogenic and antiinflammatory properties (19, 20). It is particularly important in the gut, as it has been demonstrated that mice deficient in IL-10 develop spontaneous enterocolitis (21). We looked at IL-10 production as a measure of tolerogenicity in the CX3CR1hi Mϕ and CD103+ DCs 24 h after treatment with CP by using the IL-10 GFP-reporter mice. Compared with vehicle-treated mice, the CX3CR1hi Mϕ of CP-fed mice showed a significant increase in the production of IL-10 (Fig. 3C). CP treatment also resulted in a modest but statistically significant increase in IL-10 production in CD103+ DCs (Fig. S4B). The capacity of CP to alter the ability of the siLP MNPs to induce IL-10–producing regulatory T cells was tested. The CX3CR1hi Mϕ and CD103+ DCs constitute the overwhelming majority of siLP MNPs; therefore, the total MNPs (CD11b+CD11c+ and CD11b−CD11c+ cells; Fig. S4C) were sorted from mice treated with vehicle or CP 24 h earlier and cocultured with naïve splenic CD4+ T cells (1:1) for 4 d, and IL-10– and IFN-γ–producing CD4+ T cells were analyzed. CP-elicited MNPs led to a significant increase in the expression of IL-10 in the CD4+ T cells (Fig. 3D, Left). Additionally, CP-elicited MNPs led to significant increase in the induction of a population of CD4+ T cells that express IL-10 and IFN-γ (Fig. 3D, Right). CD4+ T cells, which produce IL-10 and IFN-γ, have been previously described as a potent regulatory subset called Tr1 cells (22). IL-27, a member of the IL-12 family, plays a dominant role in the induction of Tr1 cells (23). The requirement of IL-27 for the induction of Tr1 cells by CP-elicited MNPs was tested. Functional IL-27 receptor has two subunits, IL-27RA (WSX1) and gp130, gp130 being a common receptor shared with IL-6R family, whereas IL-27RA is specific to IL-27 signaling (24). The sorted MNPs were cocultured with naïve CD4+ T cells from IL-27RA+/+ (WT) or IL-27RA−/− mice: the increase in the frequency of CD4+IL-10+IFN-γ+ T cells seen in the presence of CP-elicited MNPs is completely abrogated in the T cells derived from the IL-27RA−/− mice (Fig. 3D, Right). The possibility that CP treatment induces IL-27 production in the MNPs was tested. IL-27 (p28) levels were quantified by qPCR: at 2 d after CP treatment, there was a fivefold increase in the level of IL-27p28 in the MNPs (Fig. 3E). Altogether, these data suggest that (i) CP-elicited MNPs have a superior ability to induce a subset of regulatory T cells called Tr1 cells, (ii) this phenomenon is dependent on IL-27–IL-27R interaction, and (iii) CP treatment makes the milieu in the gut more amiable for induction of IL-27 in the MNPs. We then tested the ability of CP to elicit such regulatory T cells in vivo. Two weeks after CP treatment, the frequency of CD4+IL-10+IFN-γ+ Tr1 cells in siLP was measured. Consistent with the differentiation of Tr1 cells in vitro, CP-fed mice showed expansion of Tr1 cells in siLP (Fig. 3F).
We also tested the ability of AEA-elicited MNPs to induce Tr1 cells in vitro and observed that AEA-elicited MNPs did lead to a significant increase in the differentiation of Tr1 cells compared with vehicle-elicited MNPs (Fig. 3G). Given that the ECS promotes several aspects of tolerance-enhancing mechanisms, we tested the protective effect of AEA in context of type 1 diabetes (T1D). We observed (Fig. 3H) that orally gavaged AEA provided significant protection from T1D in a mouse model, as seen by us previously in CP-fed mice (25).
Lymph Node Specificity of Increase in the Number of Regulatory Mϕ by Stimulation of ECS.
The pancreatic lymph node (PLN) is the first site in which initiation and priming of autoreactive T cells occur in context of T1D (26). Besides draining the pancreas, the PLN has been shown to drain the gastrointestinal tract (27). Hence, the PLN is located at the crux of the pancreas and the gut and can sample the environment of both. Turley et al. showed that perturbations in the gut can affect the quality of immune response in the PLN that in turn can alter the course of T1D (27). Based on this observation that establishes a strong immunological axis between the gut and the pancreas, we tested the possibility that ECS-mediated tolerance was evident in the PLN as well.
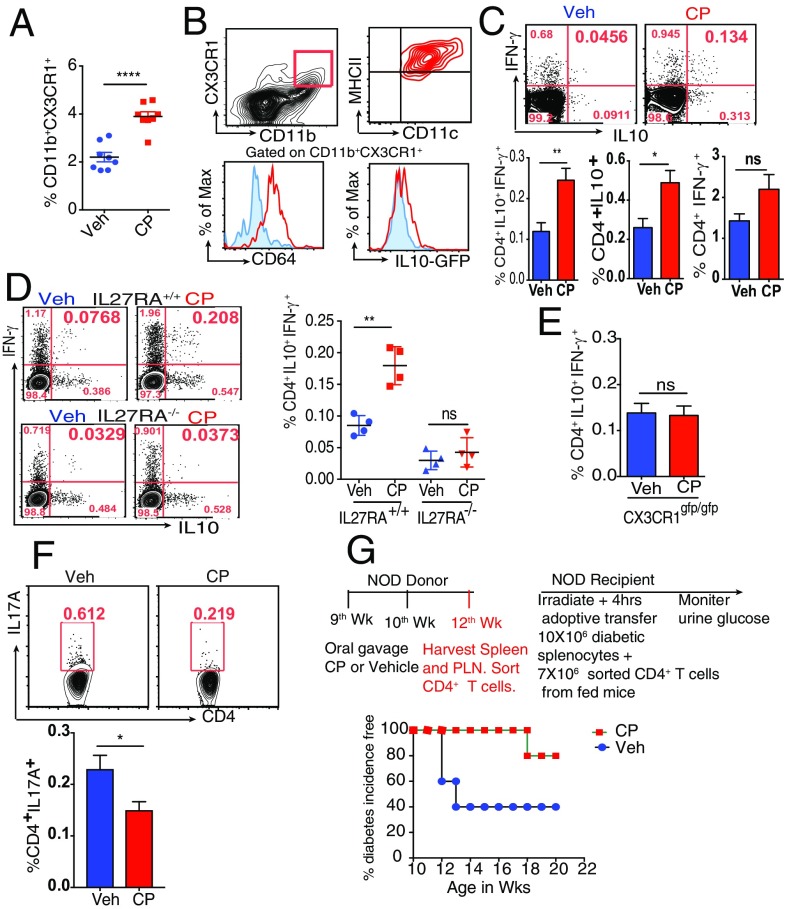
The effect of oral administration of CP on the PLN Mϕ of nonobese diabetic (NOD) mice was tested. Engagement of TRPV1 led to a significant increase in the frequency of CD11b+CX3CR1+ cells (Fig. 4A). Given that the PLN of NOD mice is inflamed, we wanted to determine if CP-mediated effect on the PLN Mϕ is a universal phenomenon and whether it is affected by the absence of TRPV1. To answer these questions, TRPV1+/+ and TRPV1−/− C57BL/6 mice underwent oral gavage with CP. Three days after CP administration, there was a significant increase in the frequency of CX3CR1+ Mϕ in the PLN of TRPV1+/+ mice but not in TRPV1−/− mice (Fig. S5A). These data demonstrate that the ECS-mediated regulation of the PLN Mϕ is not restricted to NOD mice and that this phenomenon requires the presence of functional TRPV1. Although the PLNs and mesenteric lymph node (MLNs) drain the gut (27), the CP-elicited increase in the frequency of Mϕ was evident in the PLN but not in the MLN (Fig. S5B).
Fig. 4.
Feeding CP enhances tolerance in PLN and mediates protection against T1D. (A) Frequency of PLN CD11b+CX3CR1+ cells of NOD mice 3 d after oral gavage with CP (10 μg; n = 8 mice per group). (B) Contour plot (Top Left) shows the gate for PLN CD11b+CX3CR1+ cells (red gate). Contour plot (Top Right) shows the expression of MHCII and CD11c on the gated cells. Left histogram shows the expression of CD64 (red) and isotype control (blue) by the gated cells; right histogram shows IL-10 expression by gated cells from IL-10–GFP reporter mouse (red) and C57BL/6 mouse with no GFP (blue). (C) Frequency of CP-elicited CD4+IL-10+IFN-γ+ (Tr1) cells, CD4+IL-10+ cells, and CD4+IFN-γ+ among live PLN cells of NOD mice (n = 4 mice per group). (D and E) Frequency of CP-elicited Tr1 cells among live PLN cells in (D) IL-27RA+/+ and IL-27RA−/− mice (n = 4 per group) and (E) CX3CR1gfp/gfp mice (n = 4 mice per group). (F) Frequency of CP-elicited CD4+IL-17A+ cells among live CD4+ PLN cells of NOD mice (n = 4 mice per group). (G) Schematic representation of the experimental design (Top). Survival curve represents difference in diabetes development between the two recipient groups (Bottom; n = 15 mice per group, P < 0.01). (ns, not significant; *P < 0.05, **P < 0.01, and ****P < 0.0001, unpaired Student’s t test or Mantel–Cox test; error bars indicate ± SEM).
Fig. S5.
(A and B) Frequency of CD11b+CX3CR1+ cells among live FSChi cells of TPRV1+/+ or TRPV1−/− mice in the PLN (A) and MLN cells (B) 3 d after oral gavage with CP (10 μg; n = 7–9 mice per group). (C) Frequency of CP-elicited Tr1 cells among live PLN cells in IL-27RA+/+, IL-27RA−/− and IL-27RA+/− mice (n = 3–4 mice per group). (*P < 0.05 and ****P < 0.0001, unpaired Student’s t test; data represent mean ± SEM).
The phenotype of the PLN Mϕ was examined and was observed to be akin to that of the siLP Mϕ (Fig. 1B) in terms of expression of CX3CR1, CD11c, MHCII, and CD64 and spontaneous production of IL-10 (Fig. 4B). The phenotypic similarity of the siLP and PLN Mϕ and the kinetics of the changes (changes in the siLP Mϕ were seen as early as 24 h after feeding with CP, whereas they were seen only after 3 d in the PLN) suggest that these Mϕ migrate from the gut to the PLN.
Engagement of ECS Expands a Tolerogenic Subset of CD4+ T Cells in the PLN in an IL-27–Dependent Manner.
Two weeks after CP treatment, the PLNs were harvested, and the frequency of Tr1 cells in the PLN was measured. Consistent with the CP-mediated expansion of Tr1 cells in siLP (Fig. 3F), CP-fed mice showed an expansion of Tr1 cells in the PLN (Fig. 4C). Closer analysis revealed that CP leads to a significant increase in the frequency of CD4+IL-10+ but not CD4+IFN-γ+ cells. The mechanism of Tr1 induction was investigated. Experiments were carried out in IL-27RA+/+ and IL-27RA−/− mice and, consistent with our study in vitro, we observed that CP-elicited Tr1 cells depend on the presence of functional IL-27–IL-27R signaling (Fig. 4D). Surprisingly, when the effects of IL-27 signaling were tested in CP-fed mice that lacked IL-27RA (i.e., −/−), mice that are hemizygous (i.e., +/−) and mice that are homozygous (i.e., +/+), a clear gene dose-dependence of Tr1 cell elicitation was observed (Fig. S5C). Based on these data, it would be important to study the development of T1D in IL-27RA−/− and IL-27RA+/− mice in an NOD background. Given that the CX3CR1hi Mϕ are critical to TRPV1-induced tolerance, we asked if functional CX3CR1 is required for CP-elicited expansion of Tr1 cells. CX3CR1gfp/gfp mice underwent oral gavage with CP or vehicle as a control, and Tr1 cells were analyzed in the PLNs. It was observed that, in the absence of functional CX3CR1 protein, CP does not elicit Tr1 cells (Fig. 4E). Of note, we found that oral administration of CP also leads to a significant decrease in the frequency of CD4+IL-17A+ T cells in the PLN (Fig. 4F). We note the low frequency of cells in these experiments in Fig. 4, but the clear consistency and very high reproducibility of these modest numbers persuade us as to their significance. Further testing of these results in IL-27RA−/−IL-10–GFP and IL-17A–GFP reporter mice in NOD background is necessary for a more definitive proof of our conclusions in this regard. The physiological consequence of the phenomenon seen in experiments described here earlier was partially tested in NOD mice; CD4+ T cells were isolated from the PLN and spleen of CP- or vehicle-treated female NOD mice. Naïve female NOD mice received the CD4+ T cells that were derived from the CP- or vehicle-treated NOD donors. Disease was monitored by checking the urine glucose levels twice per week. It was observed that CP-elicited CD4+ T cells could provide significantly higher protection from T1D compared with the vehicle-elicited CD4+ T cells (Fig. 4G). However, we have not tested the possibility that the transfer of IL-10–producing Mϕ may also recapitulate the phenomenon partly or wholly.
SI Materials and Methods
Mice.
All animals were housed in the Center for Laboratory Animal Care at the University of Connecticut Health Center (UCHC). All experiments were conducted under the guidelines approved by the UCHC Animal Care Committee.
Cell Isolation.
The siLP cells were isolated per the protocol previously established with some modifications (30). In brief, small intestines were removed from the mice, luminal contents were flushed with cold HBSS (Invitrogen) with 5% FBS, and small intestines were opened longitudinally after removing PPs and cut into small pieces. The epithelial cells were removed by incubation with HBSS containing 1.3 mM EDTA (Invitrogen) at 37 °C with shaking at 350 rpm (Thermo Scientific, Variomag Poly 15, part no. 50094595). The intestinal pieces were then washed with PBS solution (Invitrogen) and digested in 100 U/mL collagenase IV (Sigma) at 37 °C with shaking at 550 rpm (Thermo Scientific, Variomag Poly 15, part no. 50094595). Leukocytes were enriched by using Percoll (GE Healthcare) gradient. The PLN, MLN, and splenic cells were isolated by gently crushing the organs through a 70-μm cell strainer (BD Falcon). Splenocytes were incubated with ACK lysing buffer (Invitrogen) to eliminate red blood cells for 3 min at 37 °C and washed twice with PBS solution.
Flow Cytometry.
Fluorescently labeled antibodies used for surface staining included CD11b (M1/70), CD11c (N418), CX3CR1 (polyclonal), CD103 (2E7), CD64 (X54-5/7.1), MHCII (M5/114.15.2), B220 (RA3-6B2), CD3 (145-2C11), CD4 (RM4-5), CD25 (PC61), and CD62L (ME2-14), and those used for intracellular staining included TRPV1 (polyclonal), IL-10 (JES5-16E3), TNF-α (MP6-XT22), IFN-γ (XMG1.2), and IL-17A (TC11-18H10.1). For detection of CB2, primary antibody against CB2 (polyclonal) was used, followed by fluorescently labeled secondary antibody. All antibodies and isotype controls were purchased from Biolegend except for TNF-α (Becton Dickinson), CX3CR1 (R&D), TRPV1 (Santa Cruz Biotechnology), IL-10 (eBioscience), and CB2 (Thermo Scientific). Normal rabbit serum and goat serum were obtained from Sigma. Cells were acquired with an LSRII flow cytometer (Becton Dickinson) or FACSCalibur or sorted with a FACSAria machine (Becton Dickinson). Flow cytometry analysis was done with FlowJo version 9.7.6 (Tree Star).
Intracellular Staining for TRPV1.
After surface staining and fixing, cells were incubated in permeabilization buffer (BD Biosciences). Cells were then incubated in 4% blocking serum for 30 min to prevent nonspecific binding of the secondary antibody. After washing, cells were incubated with primary antibody against TRPV1 (sc-12498 or sc-28759; Santa Cruz Biotechnology) or unconjugated normal goat or rabbit IgG as a control for 30 min. Next, the cells were washed two times and stained with fluorochrome-labeled anti-goat or anti-rabbit secondary antibody for 30 min. After washing two times, cells were analyzed by flow cytometry.
Intracellular Cytokine Staining.
Cells were incubated with PMA (50 ng/mL; Sigma) plus ionomycin (1 μg/mL; Sigma) in the presence of a Golgi plug (BD Biosciences) for 5 h at 37 °C, followed by surface staining with fluorochrome-conjugated antibodies, and then intracellular staining of IL-10, TNF-α, IFN-γ, and IL-17A was performed by fixation and permeabilization according to manufacturer’s protocol (BD Cytofix/Cytoperm kit).
In Vitro Coculture Assays.
For in vitro coculture assays, siLP MNPs were sorted as described before from C57BL/6 or CX3CR1gfp/+ mice. MNPs (10 × 104) were cocultured for 4 d with splenic naïve CD25−CD62Lhi CD4+ T cells sorted from untreated C57BL/6 or IL-27RA−/− mice at 1:1 ratios in the presence of soluble αCD3 mAb (145-2C11; 2 μg/mL) in U-bottomed 96-well plates. The cultured cells were maintained in RPMI 1640 medium supplemented with 5% FBS, 1 mM sodium pyruvate, 1 mM nonessential amino acids, 2 mM l-glutamine, 50 U/mL streptomycin, and 50 μM β-mercaptoethanol, all purchased from Gibco/Invitrogen. After 4 d, the cultured cells were washed with PBS solution and surface-stained with CD4, and intracellular staining was performed for IL-10 and IFN-γ.
Oral Administration of CP, AEA, and PF3845.
Mice were anesthetized with i.p. injection of ketamine (2 mg/kg) and xylazine (0.1 mg/kg). Vehicle control [Tween 80 (5%), ethanol (85%) in PBS solution] or CP (10 μg; Enzo Life Sciences) or AEA (250 μg, 400 μg or 500 μg; Cayman Chemical) or PF3845 (5 mg/kg; Cayman Chemical) were gavaged in 100 μL volume by using an 18-gauge feeding needle (Popper & Sons).
BrdU Incorporation Assay.
Mice underwent oral gavage with CP or vehicle, and 20 h later, mice were injected i.p. with 100 μL (1 mg) of BrdU solution. Four hours later, the mice were killed, and siLP cells were analyzed for BrdU incorporation by flow cytometry.
RNA-Seq Analysis.
RNA samples from siLP MNPs from CP- or vehicle-treated mice or from RAW264.7 cells treated with CP or vehicle were sequenced on the ION Torrent PGM and were profiled. RNA-Seq single-end reads were aligned to the CCDS Release 16 mouse transcriptome reference by using TMAP (ioncommunity.iontorrent.com). Updated version of IsoEM (13), an expectation-maximization algorithm for inference of isoform- and gene-specific expression levels from RNA-Seq data, was used to estimate gene expression levels. Gene expression is reported as transcript per million mapped reads (TPM) units. Each gene was assigned the value of log2 (TPM + 1). An updated version of IsoDE (14) was run for gene differential expression. A twofold expression change cutoff combined with exclusion of those with a P value greater than 0.05 was used to define DEGs. The Web-based tool Morpheus (https://software.broadinstitute.org/morpheus/) was used to generate heat maps of DEGs. The DAVID Bioinformatic Resource (version 6.8) was used for gene function categories and pathways (31, 32).
RAW 264.7 Cell Treatment with CP in Vitro.
RAW 264.7 cells (American Type Culture Collection) were maintained in RPMI 1640 medium supplemented with 5% FBS, 1 mM sodium pyruvate, 1 mM nonessential amino acids, 2 mM l-glutamine, 50 U/mL streptomycin, and 50 μM β-mercaptoethanol, all purchased from Gibco/Invitrogen, in a CO2 incubator at 5% CO2. Cells were washed with OPTI-Mem media before treatment with CP. CP was dissolved in ethanol at a concentration of 100 mM. Cells in Opti-MEM media were then treated with various doses of CP (100 μM, 50 μM, 25 μM, 12.5 μM, 6.25 μM, or 3.125 μM) or ethanol as control for 2 h and 4 h. Cells were harvested after the completion of the treatment period.
qPCR.
Total RNA was isolated from RAW 264.7 cells or from sorted siLP MNP by using the RNeasy Mini Plus kit (Qiagen). cDNA was made from total RNA by using iSCRIPT cDNA synthesis kit (Bio-Rad). Expression of the following genes were studied by using TaqMan Gene Expression Assay (Life Technologies) and CFX96 Real-Time PCR detection system (Bio-Rad): PTPN-22, NAPE-PLD, Cnr2 (gene encoding Cannabinoid receptor 2), and IL-27. TRPV1 expression was studied by using SsoFast EvaGreen Supermix (Bio-Rad) and the following primers: forward, CGAGGATGGGAAGAATAACTC; reverse, GGATGATGAAGACAGCCTTGA. Gene expression was normalized to GAPDH.
Adoptive Transfer.
Female NOD mice underwent oral gavage with CP 10 μg or vehicle at 9 and 10 wk of age as described earlier (25). Two weeks later, CD4+ T cells were sorted from the PLN and spleen of the treated mice. All recipient female NOD mice (11 wk of age) were irradiated, and, 4 h later, they received 10 × 1010 diabetogenic splenocytes i.p. followed by 7 × 106 sorted CD4+ T cells from the donors via retroorbital injection. Development of diabetes was monitored by measuring urine glucose by using Diastix reagent strips (Bayer). Mice were considered diabetic after two consecutive readings of >2,000 mg/dL of urine glucose.
Discussion
The studies reported here show that the endogenous cannabinoid AEA is a major participant in maintaining tolerance in the gut. It does so through two nonoverlapping and mutually reenforcing pathways, i.e., through maintenance/differentiation of the well-known immune regulatory CX3CR1hi Mϕ population and by mediating expansion of the regulatory T cells called Tr1 cells. With respect to the former, we show that the CX3CR1hi Mϕ express the highest levels of the cannabinoid receptors CB2 and TRPV1. Additionally, oral administration of CP (ligand of TRPV1) or AEA (ligand of TRPV1 and CB2) led to a significant increase in the number of functionally active CX3CR1hi Mϕ. Consistent with this observation, CB2−/− as well as TRPV1−/− mice have a significantly decreased frequency of the CX3CR1hi Mϕ. In these same mice, the frequency of CX3CR1lo cells also showed a decreasing trend, although this decrease was not statistically significant.
The divergence between CX3CR1hi and CX3CR1lo cells with respect to their behavior in the presence or absence of ECS ligands or receptors suggests two interesting ideas. One, this observation is important because CX3CR1hi and CX3CR1lo cells are ontogenically related and arise from Ly6Chi blood-derived monocytes. Hence, the decrease in the frequency of the CX3CR1+ cells in the absence of CB2 or TRPV1 suggests that the ECS may be playing a role in the recruitment of the monocyte precursors. Second, the observation that the absence of TRPV1 or CB2 powerfully influences the CX3CR1hi Mϕ in particular suggests that some mechanism other than recruitment governs the homeostasis of these cells. It is conceivable that the ECS may be the erstwhile esoteric microenvironmental signal that plays a critical role in the conditioning and maturation of CX3CR1hi Mϕ.
Our results clarify the relationship between the roles of CP, AEA, TRPV1, and CB2. Both CP and AEA, administered orally, lead to an expansion of the CX3CR1hi Mϕ population, and protection from T1D in NOD mice. However, AEA, through its engagement with CB2, is the actual and final mediator of expansion of CX3CR1hi Mϕ. CP simply mediates release of endogenous AEA through its interaction with TRPV1, and the AEA in turn acts through CB2.
Engagement of ECS also leads to a significant increase in the frequency of the regulatory Tr1 cells in the siLP and PLN, although it does not, as reported earlier, increase the number and/or function of Foxp3+Tregs (25). Induction of Tr1 cells in vivo occurs through the established IL-27–IL-27R axis (28, 29). In our study as well, the requirement of IL-27 for TRPV1-mediated Tr1 expansion is seen in IL-27RA−/− mice. Importantly, CP-mediated Tr1 expansion in mice that are IL-27RA(−/−), hemizygous (+/−), or homozygous (+/+) demonstrated a gene dose-dependence of Tr1 cells on IL-27RA. Such dose-dependency of IL-27 signaling is an observation that suggests that the quantity of IL-27RA is the limiting factor in physiological conditions. Additionally, engagement of TRPV1 causes a modest but significant decrease in the frequency of IL-17A CD4+ T cells in the PLN. Thus, CP can skew the CD4+ T-cell compartment toward tolerance promotion by inhibiting Th17 cells and augmenting the Tr1 cells. Although modest, the cytokine changes in the CD4+ T-cell compartment were significant and consistent. However, the use of IL-27RA−/−IL-10–GFP reporter mice and IL-17A–GFP reporter mice in NOD background would be necessary for a final confirmation of our conclusion. Functional relevance of this modulation in the cytokine production is exemplified by the study that transfer of CP-elicited CD4+ T cells could transfer tolerance to naïve NOD mice. Further, as IL-27RA−/− mice are not available in the NOD background, the involvement of IL-27 in ECS-mediated protection from diabetes has not been directly proven, even though it is supported by several key observations. Study of the course of T1D development in IL-27RA−/− mice in an NOD background is required for a firm delineation of the role of IL-27 in this autoimmune disease.
Our studies report an example of a neurologically active endocannabinoid playing a substantial immunological role, and suggests interesting possibilities of concordant regulation of immune tolerance and energy balance through one ligand and two receptors.
Materials and Methods
Mice.
C57BL/6, NOD/Lt, TRPV1−/−, CB2−/−, IL-27RA−/− (also known as WSX-1−/−), CX3CR1gfp/gfp, and IL-10–GFP reporter mice were obtained from the Jackson Laboratory. All animals except NOD/Lt were on a C57BL/6 background. CX3CR1gfp/gfp were crossed with C57BL/6 mice to obtain CX3CR1gfp/+ mice. CX3CR1gfp/gfp mice were backcrossed with TRPV1−/− to obtain CX3CR1gfp/+TRPV1−/− mice.
Generation of BMCs.
Eight-week-old TRPV1−/− mice received 1,000 rads in a single dose of γ-irradiation. Two hours later, via retroorbital injection, irradiated hosts were adoptively transferred with 1.5 × 106 BM cells harvested from femurs and tibias of CD45.1+C57BL/6 WT mice. After 7 wk, BM chimeric mice were bled via tail bleeding to check for BM reconstitution by FACS.
Statistical Analyses.
Data were analyzed by Student’s t test (unpaired, one-tailed) or one-way ANOVA except for the survival curves, which were analyzed by Mantel–Cox test (GraphPad). P < 0.05 was considered significant.
Acknowledgments
The authors acknowledge Anupinder Kaur and Joshua Tagore for technical assistance, Dr. Kepeng Wang (University of Connecticut) for assistance with qPCR analysis, and Drs. Robert Clark and Vijay Rathinam (University of Connecticut) for critically reading the manuscript. This work was supported by a Mucosal Immunology Study Team Junior Faculty award (to S.B.) and the Neag Cancer Immunology Translational Program (P.K.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.A. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612177114/-/DCSupplemental.
References
- 1.McPartland JM, Matias I, Di Marzo V, Glass M. Evolutionary origins of the endocannabinoid system. Gene. 2006;370:64–74. doi: 10.1016/j.gene.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Di Marzo V, Bifulco M, De Petrocellis L. The endocannabinoid system and its therapeutic exploitation. Nat Rev Drug Discov. 2004;3:771–784. doi: 10.1038/nrd1495. [DOI] [PubMed] [Google Scholar]
- 3.Schmid HH, Schmid PC, Natarajan V. N-acylated glycerophospholipids and their derivatives. Prog Lipid Res. 1990;29:1–43. doi: 10.1016/0163-7827(90)90004-5. [DOI] [PubMed] [Google Scholar]
- 4.Schmid HH. Pathways and mechanisms of N-acylethanolamine biosynthesis: Can anandamide be generated selectively? Chem Phys Lipids. 2000;108:71–87. doi: 10.1016/s0009-3084(00)00188-2. [DOI] [PubMed] [Google Scholar]
- 5.Di Marzo V, Matias I. Endocannabinoid control of food intake and energy balance. Nat Neurosci. 2005;8:585–589. doi: 10.1038/nn1457. [DOI] [PubMed] [Google Scholar]
- 6.Varol C, Zigmond E, Jung S. Securing the immune tightrope: Mononuclear phagocytes in the intestinal lamina propria. Nat Rev Immunol. 2010;10:415–426. doi: 10.1038/nri2778. [DOI] [PubMed] [Google Scholar]
- 7.Bain CC, et al. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 2013;6:498–510. doi: 10.1038/mi.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kayama H, et al. Intestinal CX3C chemokine receptor 1(high) (CX3CR1(high)) myeloid cells prevent T-cell-dependent colitis. Proc Natl Acad Sci USA. 2012;109:5010–5015. doi: 10.1073/pnas.1114931109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung S, et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamoutounour S, et al. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur J Immunol. 2012;42:3150–3166. doi: 10.1002/eji.201242847. [DOI] [PubMed] [Google Scholar]
- 11.Madan R, et al. Nonredundant roles for B cell-derived IL-10 in immune counter-regulation. J Immunol. 2009;183:2312–2320. doi: 10.4049/jimmunol.0900185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis JB, et al. Vanilloid receptor-1 is essential for inflammatory thermal hyperalgesia. Nature. 2000;405:183–187. doi: 10.1038/35012076. [DOI] [PubMed] [Google Scholar]
- 13.Nicolae M, Mangul S, Măndoiu II, Zelikovsky A. Estimation of alternative splicing isoform frequencies from RNA-Seq data. Algorithms Mol Bio l. 2011;6:9. doi: 10.1186/1748-7188-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al Seesi S, Tiagueu YT, Zelikovsky A, Măndoiu II. Bootstrap-based differential gene expression analysis for RNA-Seq data with and without replicates. BMC Genomics. 2014;15:S2. doi: 10.1186/1471-2164-15-S8-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cravatt BF, Lichtman AH. The endogenous cannabinoid system and its role in nociceptive behavior. J Neurobiol. 2004;61:149–160. doi: 10.1002/neu.20080. [DOI] [PubMed] [Google Scholar]
- 16.Massa F, et al. The endogenous cannabinoid system protects against colonic inflammation. J Clin Invest. 2004;113:1202–1209. doi: 10.1172/JCI19465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, et al. A biosynthetic pathway for anandamide. Proc Natl Acad Sci USA. 2006;103:13345–13350. doi: 10.1073/pnas.0601832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ross RA. Anandamide and vanilloid TRPV1 receptors. Br J Pharmacol. 2003;140:790–801. doi: 10.1038/sj.bjp.0705467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bogdan C, Vodovotz Y, Nathan C. Macrophage deactivation by interleukin 10. J Exp Med. 1991;174:1549–1555. doi: 10.1084/jem.174.6.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 21.Kühn R, Löhler J, Rennick D, Rajewsky K, Müller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 22.Groux H, et al. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature. 1997;389:737–742. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 23.Awasthi A, et al. A dominant function for interleukin 27 in generating interleukin 10-producing anti-inflammatory T cells. Nat Immunol. 2007;8:1380–1389. doi: 10.1038/ni1541. [DOI] [PubMed] [Google Scholar]
- 24.Pflanz S, et al. WSX-1 and glycoprotein 130 constitute a signal-transducing receptor for IL-27. J Immunol. 2004;172:2225–2231. doi: 10.4049/jimmunol.172.4.2225. [DOI] [PubMed] [Google Scholar]
- 25.Nevius E, Srivastava PK, Basu S. Oral ingestion of capsaicin, the pungent component of chili pepper, enhances a discreet population of macrophages and confers protection from autoimmune diabetes. Mucosal Immunol. 2012;5:76–86. doi: 10.1038/mi.2011.50. [DOI] [PubMed] [Google Scholar]
- 26.Höglund P, et al. Initiation of autoimmune diabetes by developmentally regulated presentation of islet cell antigens in the pancreatic lymph nodes. J Exp Med. 1999;189:331–339. doi: 10.1084/jem.189.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turley SJ, Lee JW, Dutton-Swain N, Mathis D, Benoist C. Endocrine self and gut non-self intersect in the pancreatic lymph nodes. Proc Natl Acad Sci USA. 2005;102:17729–17733. doi: 10.1073/pnas.0509006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Apetoh L, et al. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat Immunol. 2010;11:854–861. doi: 10.1038/ni.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pot C, et al. Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. J Immunol. 2009;183:797–801. doi: 10.4049/jimmunol.0901233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medina-Contreras O, et al. CX3CR1 regulates intestinal macrophage homeostasis, bacterial translocation, and colitogenic Th17 responses in mice. J Clin Invest. 2011;121:4787–4795. doi: 10.1172/JCI59150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]