Abstract
Blood for transfusion is a potential source of infection by a variety of known and unknown transmissible agents. Over the last 20 years, astounding reductions in the risk of viral infection via allogeneic blood have been achieved. As a result of this success, bacterial contamination of blood products has emerged as the greatest residual source of transfusion-transmitted disease. This paper summarizes the current status of detection, prevention, and elimination of bacteria in blood products for transfusion.
INTRODUCTION
Allogeneic blood for transfusion is a potential source of infection by a variety of known and unknown transmissible agents. Since the 1980s, with the recognition of human immunodeficiency virus contamination of the blood supply, only a blood supply with a zero risk to recipients has been politically acceptable (67). This approach has led to the concept of “creeping precautionism” in the blood supply (46). The precautionary principle has grown out of the environmental movement. Briefly, it states “…where there are threats of serious or irreversible damage to the environment, lack of full scientific certainty should not be used as a reason for postponing cost-effective measures to prevent environmental degradation…” (46). To this end, astounding reductions in the viral risk of allogeneic blood has been accomplished (27). However, a number of transfusion safety measures have been implemented to achieve this level of safety, despite relatively poor cost-effectiveness, in order to assuage the public's fears (32).
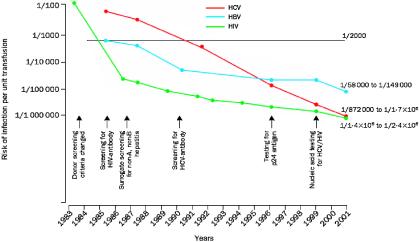
Given the reduction of viral transmission via allogeneic blood, the low but known risk of bacterial contamination has emerged as the greatest residual threat of transfusion-transmitted disease (Fig. 1).
FIG. 1.
Risk of infectious disease transmission per unit transfused by year, 1983 to 2001. Reprinted from reference 27 with permission of the publisher.
TRANSFUSION-TRANSMITTED BACTERIAL INFECTION OF RED CELLS
Each year, approximately 13,898,000 units of red cells or whole blood are transfused in the United States alone (1). This equates to one unit being transfused every 2.3 s. Despite this large number, sepsis associated with the transfusion of bacterially contaminated red cell blood components is generally regarded as a very rare event. From 1976 through September 1998, 26 fatalities thought to be secondary to contaminated whole blood or red cells were reported to the U.S. Food and Drug Administration (FDA) (36). Thus, approximately one red cell-related death per year has been reported. The majority of deaths reported to the FDA involved Yersinia enterocolitica. The highest reported incidence of Y. enterocolitica contamination was reported in New Zealand, with an incidence rate of 1 in 65,000 and a fatality rate of 1 in 104,000 red cell units transfused (59). Unrecognized cases, underreporting, and regional variation may account for observed differences in this incidence.
Interestingly, more recent passive reporting studies of bacterially contaminated red cells from the United States, France, and the United Kingdom that caused symptoms of infection show a relative paucity of Yersinia cases (Table 1) (35, 51, 57). Of the reported deaths, one was due to a coagulase-negative Staphylococcus strain and seven were due to a variety of gram-negative organisms (including Serratia liquefaciens in three cases). These organisms are all capable of growth at 1 to 6°C. Sepsis associated with the transfusion of red cells contaminated with gram-negative bacteria is typically severe and rapid in onset. Patients frequently develop high fever (temperatures as high as 109°F have been observed) and chills during or immediately following transfusion. From 1987 to February 1996, 20 recipients of Yersinia-infected red cells were reported to the Centers for Disease Control (S. T. Cookson, M. J. Arduino, S. M. Aguero, W. R. Jarvis, and the Yersinia Study Group, Program Abstr. 36th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 237, 1996). Twelve of the 20 recipients died, and the median time to death was only 25 h. Of the seven who developed disseminated intravascular coagulopathy, six died.
TABLE 1.
Organisms isolated from red cells implicated in transfusion-associated infectionsa
| Organism | No. of contaminated units in: |
Total no. | ||
|---|---|---|---|---|
| United States | United Kingdom | France (29 isolates, 25 implicated units) | ||
| Gram positive | ||||
| Coagulase-negative staphylococci | 2 (1) | 2 | 3 | 7 (1) |
| Streptococcus spp. | 4d | 4 | ||
| Staphylococcus aureus | 2b | 2 | ||
| Enteroccocus faecalis | 1c | 1 | ||
| Bacillus cereus | 2 | 2 | ||
| Propionibacterium acnes | 1 | 1 | ||
| Subtotal | 2 (1 or 50%) | 2 (0 or 0%) | 13 (0 or 0%) | 17 (1 or 6%) |
| Gram negative | ||||
| Serratia liquifaciens | 2 (2) | 1 | 2 (1) | 5 (3) |
| Serratia marcescens | 1 | 1 | ||
| Yersinia enterocolitica | 1 (1) | 1 | 2 (1) | |
| Enterobacter spp. | 1 (1) | 1 (1) | ||
| Acinetobacter spp. | 5 (1)b | 5 (1) | ||
| Pseudomonas spp. | 2 (1) | 2 (1) | ||
| Escherichia coli | 3c | 3 | ||
| Klebsiella pneumoniae | 1 | 1 | ||
| Proteus mirabilis | 1 | 1 | ||
| Subtotal | 3 (2 or 67%) | 2 (1 or 50%) | 16 (4 or 25%) | 21 (7 or 33%) |
| Total | 5 (3 or 60%) | 4 (1 or 25%) | 29 (4 or 14%) | 38 (8 or 21%) |
Summary of organisms identified in the BACON, SHOT, and BACTHEM studies (35, 51, 57). Numbers and percentages of fatalities are given in parentheses.
In one case, S. aureus and A. baumannii were both isolated from the implicated bag.
In one case, E. faecalis and E. coli were both isolated from the implicated bag.
In two cases, two isolates of Streptococcus were isolated from the implicated bag. Of the eight reported fatalities, seven (87.5%) were due to gram-negative organisms.
Prospective bacterial cultures of whole blood or red cells, however, have shown a much higher incidence of bacterial contamination (2 to 4 per 1,000 units); however, the organisms commonly cultured are Staphylococcus or Propionibacterium spp., which generally proliferate poorly during storage at 1 to 6°C (4).
TRANSFUSION-TRANSMITTED BACTERIAL INFECTION OF PLATELETS
Each year, within the United States approximately 4 million platelet units are transfused (1 × 106 single-donor apheresis platelets and 3 million whole-blood-derived platelet concentrates) (1). This equates to one platelet unit being transfused every 8 s. Unlike red cell or whole-blood components, which are stored at 1 to 6°C, platelets are stored at 20 to 24°C to preserve function and survival. Such storage makes them an excellent growth medium for a broad spectrum of bacteria. Multiple aerobic-culture surveillance studies have demonstrated that 1 in 1,000 to 2,000 platelet units are bacterially contaminated (4). Thus, 2,000 to 4,000 bacterially contaminated units would be expected to be transfused per year. Estimates of the fraction of cases that result in patient symptoms have been as low as 1 in 10 cases. However, in the only study that has prospectively cultured platelets that were transfused, eight culture-positive but Gram stain-negative platelet pools that were bacterially contaminated resulted in symptoms in three patients (37.5%) [A. Dykstra, G. Hoeltge, M. Jacobs, J. Miller, R. Domen, and R. Yomtovian, Transfusion 38(Suppl.):104S, 1998]. Notably, six Gram stain-positive pools (which would be expected to have had the highest bacterial load) were interdicted and never transfused.
Thus, clinical sepsis would be expected in at least 1 in 10 to 2 in 5 contaminated transfusions (200 to 1,600 cases). National passive-reporting studies from the United States, the United Kingdom, and France (Table 2) suggest that one-fifth to one-third would result in death (40 to 533 deaths per year) (35, 51, 57). This translates to a risk of death from a transfusion of a bacterially contaminated platelet unit of between 1 in 7,500 and 1 in 100,000. Clinical observations from university hospitals actively pursuing suspected cases of platelet-related sepsis confirm these estimates. Ness et al. from Johns Hopkins University, reported a fatality rate of 1 in 17,000 with pooled whole-blood-derived platelets and 1:61,000 with single-donor-apheresis-derived platelets (48). Similarly, University Hospitals of Cleveland observed a fatality rate of 1 in 48,000 per random platelet unit (21). The French BACTHEM study documented a fatality rate due to bacterially contaminated platelets of 7 per 106 (1 in 140,000) (51).
TABLE 2.
Organisms implicated in infections associated with platelet transfusionsa
| Organism | No. of contaminated units in: |
Total no. | ||
|---|---|---|---|---|
| United States | United Kingdom | France | ||
| Gram positive | ||||
| Bacillus cereus | 1 | 4 (1) | 2 | 7 (1) |
| Coagulase-negative staphylococci | 9 | 6 (1) | 5 | 20 (1) |
| Streptococcus spp. | 3 (1) | 2 | 5 (1) | |
| Staphylococcus aureus | 4 | 2 (1) | 6 (1) | |
| Propionibacterium acnes | 3 | 3 | ||
| Subtotal | 17 (1 or 6%) | 14 (3 or 21%) | 10 (0 or 0%) | 41 (4 or 10%) |
| Gram negative | ||||
| Klebsiella spp. | 2 (1) | 2 (1) | ||
| Serratia spp. | 2 (2) | 1 (1) | 3 (3) | |
| Escherichia coli | 5 (1) | 2 (1) | 1 | 8 (2) |
| Acinetobacter spp. | 1 | 1 | ||
| Enterobacter spp. | 2 (1) | 1 (1) | 1 | 4 (2) |
| Providencia rettgeri | 1 (1) | 1 (1) | ||
| Yersinia enterocolitica | 1 | 1 | ||
| Subtotal | 11 (5 or 45%) | 3 (2 or 67%) | 6 (2 or 33%) | 20 (9 or 45%) |
| Total | 28 (6 or 21%) | 17 (5 or 29%) | 16 (2 or 13%) | 58 (11 or 19%) |
From 1976 through September 1998, 51 fatalities thought to be secondary to contaminated platelets were reported to the U.S. FDA (36). Gram-negative organisms accounted for the majority of deaths (59.7%). Similarly, passive surveillance studies from the United Kingdom, the United States, and France show that gram-positive organisms were implicated in 41 (71%) of 58 of cases but gram-negative organisms (mostly members of the Enterobacteriaceae) account for the majority (9 [82%] of 11) of the fatalities (Table 1) (35, 51, 57).
Clinical Presentation
Clinical sequelae of the infusion of bacterially contaminated platelets are variable and may be acute or delayed (contributing to missed diagnoses). Ironically, the majority (85%) of platelets are transfused to patients in the medical oncology, hematology, pediatric hematology/oncology, and bone marrow transplantation settings, all typically immunosuppressed patients (61). A study of platelet transfusions in autologous bone marrow transplant patients found that 1 in 2,000 units of platelet concentrates associated with a febrile episode (within 24 h of transfusion) were linked to bacterially contaminated platelets (16). Of the 10 patients known to have received a bacterially contaminated unit, 4 experienced septic shock.
Because of the severity of the patient's underlying disease and the variability in presentation and timing, sepsis resulting from bacterially contaminated platelets may go unrecognized. For example, an outbreak of Salmonella enterica serovar Choleraesuis sepsis in seven patients was traced to one repeat platelet donor, subsequently found to have occult chronic osteomyelitis (52). One patient died, and two had long-term recurrences of Salmonella sepsis. The time to the onset of illness ranged from 5 to 12 days (mean, 8.6 days). In many cases, the platelet-related bacterial sepsis is discovered only in retrospect. Investigations of two Serratia liquefaciens-contaminated red cell units led to platelet products from the whole-blood donations and to the recognition of an associated septic death in one recipient and bacteremia in the other recipient (53).
TRANSFUSION-TRANSMITTED BACTERIAL INFECTION OF PLASMA AND CRYOPRECIPITATE
Cell-free products such as plasma and cryoprecipitate are stored frozen and thus are rarely associated with contamination. However, in some cases Pseudomonas cepacia and P. aeruginosa have been cultured from cryoprecipitate and plasma thawed in contaminated water baths.
SOURCES OF CONTAMINATION
Gram-positive skin commensals such as Staphylococcus epidermidis and Bacillus cereus are the organisms most often recovered from donated blood (and implicated in bacterial contamination of platelets). Such contamination is thought to occur principally during phlebotomy, as a result of incomplete disinfection and/or skin core removal (including skin appendages where the skin disinfectants may not penetrate) by the collection needle (25). These organisms typically do not grow at 1 to 6°C but survive and multiply readily at the platelet storage temperature of 20 to 24°C.
In the case of gram-negative bacterial contamination, asymptomatic donors with transient bacteremia are presumed to be responsible for most cases of contamination. For example, in the case of Y. enterocolitica contamination of red cells, implicated donors typically are found to have elevated antibody titers (immunoglobulin M or G) to Y. enterocolitica, implying a recent infection (38, 39, 60).
An outbreak of Serratia marcescens contamination of red cells in Denmark and Sweden (29, 30) was thought to involve the manufacturing process since the sterile bag sets were autoclaved and put in a clean but not sterile outer plastic package. It was thought that S. marcescens present in the dust in the factory contaminated the outside of the containers and that in the presence of moisture and a nutrient (the plasticizer diethylhexyl phthalate) the bacteria proliferated and gained entry into the bag (58).
In rare cases, unusual circumstances have been identified as the source of bacterial contamination. For example, a donor with a pet boa constrictor, who reported fever and bloody diarrhea treated with antibiotics 13 days prior to his donation, was linked to Salmonella enterica platelet sepsis in two patients (33). Scarring or dimpling of the venipuncture site (due to prior donations) can result in recessed pits that are difficult to adequately sterilize (3). A repeat single donor with a “dimpled” venipuncture site resulted in four cases of gram-positive bacterial platelet sepsis in recipients (3). Another unusual association was made between one donor in a platelet pool and the death of a leukemic recipient from septic shock. The contaminating organism was identified as C. perfringens and was thought to originate from a donor who had two young children and frequently changed their diapers (42). As stated above, an outbreak of S. enterica serovar Choleraesuis platelet sepsis in seven patients at the National Institutes of Health Clinical Center was traced to a repeat platelet donor with an occult osteomyelitis (52). However, in most cases the source of the bacterial contamination remains unidentified.
METHODS TO REDUCE THE RISK OF POST-TRANSFUSION SEPSIS
There are multiple strategies to reduce transfusion-related bacterial infection. The various approaches can be grouped into four major categories: avoidance, bacterial detection, bacterial elimination, and growth inhibition.
Avoidance
Donor screening.
Screening of donors to detect a possible risk of infection or transient bacteremia (e.g., from recent dental procedures) is routinely performed. However, questioning about symptoms suggestive of infection are often problematic; for example, gastrointestinal symptoms have not been a specific predictor (13% of donors have had gastrointestinal symptoms in the previous 30 days) (28). Such questioning also lacked sensitivity, since only 13 of 20 donors associated with Y. enterocolitica-contaminated red blood cells recalled a history of gastrointestinal symptoms (38, 39, 60; Cookson et al., Prog. Abstr. 36th ICAAC, 1996).
Skin preparation.
Skin disinfection reduces the skin bacterial load (65), but a sterile venipuncture cannot be guaranteed due to inaccessibility of organisms present in sebaceous glands and hair follicles; organisms present in skin fragments may be drawn into the component bag (25). Multiple studies have evaluated skin disinfection in the context of blood donation (25, 26, 43). A summary of the effectiveness of some of the more commonly used skin disinfection methods is given in Table 3 (43). In general, either iodine or povidone iodine are used. In donors with allergies to iodine, chlorhexidine or double isopropyl alcohol skin disinfection is employed. The difference noted in Table 3 for two different isopropyl alcohol scrubs was attributed to the abrasive nature of the sponge used in one of the preparations.
TABLE 3.
Percentage and mean count of donors with bacteria after skin disinfectiona
| No. of CFU/plate | % of contaminated donors after disinfection with: |
|||
|---|---|---|---|---|
| Povidone iodine × 2 | IPAb and iodine | IPA × 2 (sponge) | IPA × 2 (swab) | |
| 0 | 39 | 79 | 0 | 29 |
| ≤10 | 57 | 93 | 69 | 50 |
| ≤100 | 75 | 100 | 86 | 68 |
| ≤1,000 | 96 | 100 | 97 | 89 |
| Mean count | 175 | 3 | 161 | 237 |
Data are from reference 43.
IPA, isopropyl alcohol.
Diversion of the initial blood draw.
Several studies have demonstrated the partial effectiveness of diverting the first 10 to 30 ml of blood from the initial collection. A study performed with 22,000 samples of donated blood by the Red Cross in The Netherlands, in which the first 10 ml of donor blood was diverted from the primary bag, showed that 16 of the first 5-ml aliquots were bacterially contaminated while only 2 of the second 5-ml aliquots were positive after culture (19). A second study in France showed that removal of the first 10 ml of donor blood was associated with a significant decrease in bacterial contamination (18,263 collections with 0.39% contamination compared with 7,115 collections with 0.21% contamination; P < 0.05) (12).
Such methods are most effective with respect to skin contaminants (12, 19). However, it has been said that “one should not be diverted by diversion,” since most fatal cases of bacterial contamination involve gram-negative organisms and thus would not minimized by diversion. Interestingly, the addition of a diversion bag to whole-blood collections in the United States has been initially complicated by hemolysis with certain sample tubes (from Pall, East Hills, N.Y.) and dilution of the sample with anticoagulant (from Baxter, Round Lakes, Ill), requiring redesign of both diversion systems (data presented at the HHS Advisory Committee on Blood Safety and Availability, Washington, D.C.; http://www.hhs.gov/bloodsafety/pastmeetings.html#n).
Apheresis versus whole-blood-derived platelet concentrates.
Therapeutic doses of platelets can be obtained from two sources. Platelets can be collected from a donor by the use of an automated cell separator (apheresis); such collections can yield one to three therapeutic doses. Alternatively, platelets can be obtained from whole-blood donations (whole blood platelet concentrates [WBPCs]); however, a therapeutic dose requires the pooling of four to six WBPCs. Current estimates suggest that 69% of all therapeutic doses of platelets transfused in the United States are apheresis derived (1). Usage varies considerably by country.
Since bacterial contamination may result from a contaminated venipuncture or an asymptomatic bacteremic donor, it would be expected that pooled platelet products would be associated with a higher risk of contamination than a single donor product. From 1986 to 1998, the incidence of septic events related to platelets at Johns Hopkins University Hospital decreased from 1 in 4,818 to 1 in 15,098 as the hospital went from transfusing 51.7% to 99.4% single-donor apheresis units (48). The overall clinical sepsis rate was approximately 1 in 2,500 for WBPC pools and 1 in 13,400 for single-donor apheresis transfusion. The related mortality rate was approximately 1 in 17,000 for WBPC pools and 1 in 61,000 for single-donor apheresis platelets. It was concluded that the use of single-donor apheresis platelets is a simple means of reducing septic platelet transfusion reactions. Similarly, the University Hospitals of Cleveland have monitored the contamination rates of both single-donor and whole-blood-derived concentrates [A. Dykstra, M. Jacobs, and R. Yomtovian, Transfusion 38(Suppl.):104S, 1998]. The rate of bacterial contamination of apheresis-derived platelets was 1 in 2,184. While the rate for random-donor platelets was similar at 1 in 1,861, random-donor platelet concentrates in a pool of six units would be expected to result in a contamination rate of 1 in 310.
Bacterial Detection
Timing of detection.
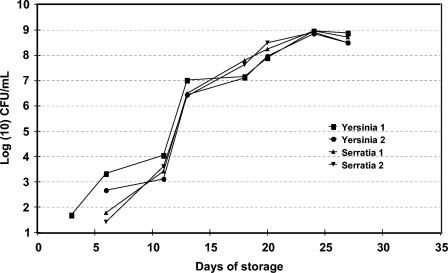
Culture studies performed both on red cell units and platelets have shown that culture on the day of collection invariably misses bacterially contaminated units that would possibly achieve dangerous levels of overgrowth during storage of the blood components [5a, 10; M. A. Blajchman, A. Ali, P. Lyn, L. Bardossy, and H. Richardson, Transfusion 7(Suppl.):7S, 1997]. This is in contrast to viral contamination of blood components, in which the virus or the immune response to the virus (antibodies) is detected from a sample obtained at the time of donation. Bacterial contamination of blood components generally requires time for the organisms to proliferate before being detectable in a small representative sample of the product. Therefore, knowledge of the growth characteristics of bacteria in blood components is essential before one considers implementation of a detection strategy (Fig. 2 and 3).
FIG. 2.
Growth curves of four units of AS-3 red blood cells inoculated with S. liqueifaciens (two units inoculated to 4.2 CFU/ml) and Y. enterocolitica (two units inoculated to 11.6 CFU/ml) (5a). Bacteria were not detectable by culture until days 3 to 6. Isolates were from strains actually implicated in post-red-blood-cell-transfusion sepsis (the isolates were kindly provided by M. Arduino, Centers for Disease Control and Prevention, Atlanta, Ga.).
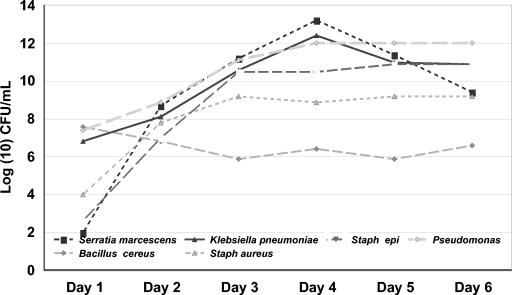
FIG. 3.
Growth curves of six bacteria species (S. marcescens, n = 7; K. pneumoniae, n = 21; S. epidermidis, n = 21; Pseudomonas sp., n = 15; B. cereus, n = 9; S. aureus, n = 22) in 95 platelet units. All bacteria were inoculated on day zero at 10-50 CFU/ml (10).
Growth characteristics of 165 platelet units inoculated (to 1 to 50 or 1,000 CFU/ml) within 1 day of collection with one of the following: B. cereus, P. aeruginosa, K. pneumoniae, S. marcescens, S. aureus, and S. epidermidis showed that after 1 day of storage all samples of B. cereus, P. aeruginosa, and K. pneumoniae had concentrations of ≥10 CFU/ml (the level of detection), while only 71.4 and 84.2% of the S. marcescens and S. aureus samples were at concentrations of ≥10 CFU/mL (10). By day 3 of storage, all units inoculated with B. cereus, P. aeruginosa, K. pneumoniae, S. marcescens, and S. aureus contained ≥105 CFU/ml. Units contaminated with S. epidermidis showed slower and more varied growth.
Bacterial culture.
Although there are a variety of automated and semi-automated systems, only two are FDA cleared for quality control of bacterial culture in platelets in the United States. The first is an automated liquid culture system (BacT/ALERT; bioMérieux, Durham, N.C.) that uses broth bottles with a colorimetric sensor which changes color as a consequence of increasing CO2 produced by bacterial proliferation. The machine monitors both the rate of change of the colorimetric sensor and the absolute color change of the sensor. Generally, one aerobic (and in some cases one aerobic and one anaerobic) bottle is inoculated with 4 ml of the platelet product. This system is not completely closed, since a needle is used to inoculate the bottles. It has been extensively validated by tests with a wide range of potentially contaminating organisms (7, 8, 11, 44). The method reliably detects contamination of platelets inoculated to 10 CFU/ml and in many cases to ≤5 CFU/ml (e.g., B. cereus, S. marcescens, C. perfringens, S. epidermidis, S. pyogenes, E. coli, K. pneumoniae, S. aureus, and viridans streptococci) in 12 to 26 h. Propionibacterium acnes (which is of questionable clinical significance in this setting) required longer incubations. Recently, our laboratory completed an 11-month pilot program in which we cultured 2,397 apheresis-derived platelets on day 2 of storage (collection day = day 0) and again at the time of issue (or following outdate: days 6 to 8) (6). This study involved the inoculation of 9,588 bottles; one “machine” false-positive bottle (negative subculture, Gram stain, and repeat culture), and one inadvertent contamination false-positive bottle (Propionibacterium sp.) were detected. Thus, the overall false-positive rate was 2 in 4,794 samplings (0.04%, or 1 in 2,397 samplings) or 2 in 9,588 bottles (0.02% or 1 in 4,794 bottles). During this period, we successfully interdicted three apheresis-derived units (one collection) contaminated with S. epidermidis but were unable to prevent the transfusion of four apheresis units contaminated with Propionibacterium sp., for which the culture bottles were reactive after 6 days of incubation. Thus, the overall true-positive rate was 7 of 2,397 apheresis units (0.29%), with a true-positive rate for aerobic organisms of 3 of 2,397 (0.13% or 1 of 799 units) and an anaerobic true-positive rate of 4 of 2,397 (0.17% or 1 of 599 units). Within our facility, transition from a research setting to a clinical microbiology laboratory has resulted in a higher false-positive (laboratory contamination) rate of 0.6% per unit (with one aerobic and one anaerobic bottle) (unpublished data). More recent experience from three large blood centers is included in Table 4.
TABLE 4.
Summary of recent experiences with culturing of apheresis-derived platelets at three large blood centersa
| Center | No. (rate) of resultsb |
||
|---|---|---|---|
| True positive | False positive | No. tested | |
| New York Blood Center | 5 (1/4,101) | 5 (1/4,101) | 20,506 |
| Florida Blood Services | 6 (1/1,790) | 5 (1/2,147) | 10,737 |
| Puget Sound Blood Center | 5 (1/1,800) | 15 (1/600) | 8,999 |
| Total | 16 (1/2,515) | 25 (1/1,610) | 40,242 |
True positive = positive bottles in which an organism was identified and a repeat sample from the bag or a retention sample also contained the same organism. False positive = positive bottles that contained no organism (machine false positive) or from which the organism could not be reisolated from the original bag or a retained sample.
The second culture system is the Pall Bacterial Detection System (BDS) (49). Sampling is obtained so as to maintain an effectively closed system. Approximately 5 to 6 ml of leukocyte-reduced platelet-rich plasma is removed from the bag and passed through a filter that removes residual white cells and platelets but allows approximately 50% of the bacteria (variable by organism) and plasma to pass into an incubation bag. The incubation bag holds 2 to 3 ml and contains sodium polyanethol sulfonate, which facilitates the growth of gram-negative organisms by inhibiting both complement and plasma lysosomal enzymes and by antagonizing the action of growth-retarding lipoproteins normally present in plasma. The incubation bag is then incubated at 35°C for at least 24 h before the oxygen concentration of the bag's headspace is measured with an oxygen analyzer. A decrease in the oxygen concentration to 19.5% or lower is an indication of bacterial growth. Thus, this system does not detect obligate anaerobic organisms. Leukocyte-reduced platelets challenged with 10 known platelet contaminants at doses of 100 to 500 CFU/ml were detected at a rate of 96.5% at 24 h and 100% at 30 h (49). External validation studies have shown lower than expected sensitivity for S. epidermidis and S. agalactiae [K.-A. T. Nguyen, T. Yamamoto, and N. Hirschler, Transfusion 43(Suppl.):S63-030K, 2003; H. C. Pleasant, G. A. Geele, and M. P. Holub, Transfusion 43(Suppl.):S62-030K, 2003 (abstract)]. Most recently, an enhanced version of this system has been cleared by the FDA, it includes modifications to the configuration (with removal of the filter, which removed some bacteria), and the incubation bag now contains a platelet-aggregating agent and tryptic soy medium to enhance the sensitivity and decrease the incubation time (http://www.pall.com/BDS.asp??noleftnav).
Less sensitive methods of detection such as Gram or Wright stains or the use of glucose and pH determination (in some case with multireagent strips) are used for rapid detection of bacteria in platelet components prior to transfusion. However, the overall sensitivity is only 106 to 107 CFU/ml (13, 62, 63, 66). Werch et al., from the M. D. Anderson Cancer Center, reported on the screening of 3,093 platelet concentrates, in which they found that multireagent strips for 30 bags had glucose levels or pH outside of the reference range (63). Two of these platelet units were culture positive for B. cereus. This screening method resulted in 1 unit per 110 being wasted but prevented two patients from receiving contaminated platelets.
Molecular biology techniques.
Because of the high sensitivity and specificity and the rapidity of obtaining results, molecular biological approaches are theoretically appealing for the detection of contaminating organisms in blood components. One such approach is the use of a nonamplified chemiluminescence-linked universal bacterial rRNA probe (5, 9). This technique was shown to detect four bacterial species in the range of 104 to 105 CFU/ml. In some cases, S. aureus was detected at levels as low as 102 to 103 CFU/ml. Other investigators have pursued the use of PCR-based amplification for a limited number of organisms (56).
However, bacterial DNA contamination of nucleotide amplification reagents (particularly the bacterially derived enzymes) and laboratory materials and the presence of bacterial DNA sequences commonly found in human blood complicate the usefulness of universal bacterial amplification techniques (22, 45, 47). It also remains to be seen if complex molecular biology techniques could be successfully deployed to a wide range of transfusion services.
Detection systems recently approved in Europe.
Recently, two bacterial detection systems were approved in Europe. The Scansystem (Hemosystem, Marseilles, France) involves a 70-min solid-phase laser cytometry. In a few studies, the Scansystem detected 11 organisms in the range of 10 to 103 CFU/ml [P. Morel, M. Deschaseaux, X. Bertrand, C. Naegelen, and D. Talon, Transfusion 43(Suppl.):SP13, 2003]. Positive samples require a technologist to perform a microscopic confirmation.
A second “rapid” method recently approved detects bacteria by using dielectrophoresis (Cell Analysis, Slough, United Kingdom). Platelet concentrates inoculated with six organisms at concentrations of 103 to 105 CFU/ml were detected after 30 min of dielectrophoresis by image analysis as they were released from the electrodes when the electric field was discontinued [C. P. McDonald, R. Smith, J. Colvin, S. Robbins, J. Barbara, W. Betts, R. Anthony, K. Chan, A. Brown, and K. Gregory, Transfusion 41(Suppl.):S119-040H, 2001].
Detection systems under development.
Currently, there are at least three other rapid-detection systems under development. Genpoint (Oslo, Norway) has developed a unique system that uses paramagnetic particles to concentrate bacteria (BUGS'n BEADS) coupled with a PCR for highly conserved DNA. The target sensitivity of this assay is 1,000 CFU/ml, and the total time is 2.5 h. Immunetics, Inc. (Cambridge, Mass.) is developing a 96-well 1-h enzyme immunoassay which employs proteins and derived peptides that bind to the peptidoglycan component of the bacterial cell wall. Preliminary results suggest a sensitivity of 103 to 104 CFU/ml (or lower) for a variety of organisms [L. A. Beausang and A. Levin, Transfusion 43(Suppl.):20A, 2003; D. Kagan, and A. E. Levin, Transfusion 41(Suppl.):S118-040H, 2001]. Verax Biomedical (Worcester, Mass.) is developing an immunoassay embodied in a lateral-flow format. Preliminary results suggest a sensitivity of 103 to 104 CFU/ml for a variety of organisms [F. Kenny, C. Woodbury, M. Pelak, J. Weinberg, and J. A. Hall, Transfusion 43(Suppl.):10A, 2003].
Implementation of detection systems: problems and considerations. (i) Platelet concentrates and prestorage pooling.
Bacterial detection of whole-blood-derived platelet concentrates presents logistical problems for systems that do not allow the prestorage pooling of platelet concentrates (e.g., United States, Canada, and Hong Kong). In Europe, whole-blood-derived buffy coat platelets are prepooled prior to storage; this allows for the culturing of platelets by the methods described above. If prepooling is not allowed, culturing of each individual platelet sample is generally not thought to be practical due to the loss of volume from each bag required per culture and the high cost of culture for each individual bag. In the absence of prepooling, many centers have chosen or are contemplating the use of less sensitive bacterial detection strategies such as Gram stains or glucose or pH measurement (or the use of multireagent strips) (2).
(ii) Anaerobic organisms.
Anaerobic organisms such as Propionibacterium sp. can be identified in platelets; however, it is not known whether anaerobic organisms such as propionibacteria are clinically significant since growth in the aerobic environment of platelet storage is slow and not favorable. Although a few cases of Propionibacterium-contaminated units have been infused into patients, no long-term sequelae have been reported (6, 40, 55). Debate continues as to the value of an anaerobic culture in this context. It is worth noting that some aerobic organisms or isolates (e.g., viridans streptococci) can be more rapidly detected in an anaerobic culture system (7, 8, 11).
(iii) Potential for reduced shelf-life.
Some centers have chosen to incubate platelet cultures for 12 to 24 h prior to release. This has in many cases led to a decrease in the available shelf-life of the platelets. Given that the current platelet products have a shelf-life of only 5 days (and that, for 1 to 2 days, such products are not available, pending completion of viral testing), any further reduction in available shelf-life can impact inventory control and hospital supply.
(iv) Use of noncleared detection systems.
Use of non-FDA-cleared detection systems (e.g., alternative automated culture systems) would at a minimum require internal validation. Examples of validation of detection systems are available in the literature (7, 8, 11, 44, 49). Patent issues currently preclude certain companies from actively pursing this market. Use of a non-FDA-cleared device for bacterial quality control of platelets, when cleared devices are available, may place an institution at higher legal risk should a patient die after receiving bacterially contaminated platelets.
Bacterial Elimination
Pathogen reduction technologies.
Continued efforts to create a “zero-risk” blood supply have led to various pathogen reduction technologies. A variety of photoactive sensitizers have been investigated for use in the reduction of viruses, bacteria, and parasites.
The best studied of these methods is the use of amotosalen HCL (S-59), a synthetic psoralen combined with a photochemical treatment process that causes permanent cross-links in DNA and RNA (INTERCEPT Platelet System; Baxter Healthcare Corp., Round Lake, Ill.) (41, 64). However, organisms capable of spore formation may be resistant to photochemical inactivation (34). The SPRINT trial, a multi-center phase III trial, has evaluated the hemostatic effectiveness of S-59 pathogen-inactivated platelets. Treated platelets were found to be as effective as conventional platelets for the prevention and treatment of bleeding; however, the treated platelets resulted in a lower post-transfusion platelet response, resulting in the need for 35% more platelets to support transfusion-dependent patients (18, 41). Concerns also remain regarding mutagenicity and teratogenicity associated with such technologies.
Growth Inhibition
The addition of antibiotics to blood components has been considered but has not been adopted due to the added risk of drug reactions (trading one infrequent reaction for another) and the potential for the development of antibiotic resistance if small amounts of antibiotics were infused with every transfusion (31).
THE PROSPECT OF SEVEN-DAY STORAGE WITH DETECTION
Currently, within the United States platelet storage is limited to 5 days due to the fear of bacterial overgrowth during storage at 20 to 24°C. In 1983, platelet storage in the United States was extended to 7 days; however, in 1986 the FDA shortened the outdate to 5 days following reports of bacterial sepsis with older units (65). With the prospect of bacterial detection, the possibility of an extended shelf-life is being reconsidered. European reports have described the use of bacterial culturing of platelets on days 1 to 3 of storage in order to extend the shelf-life of platelets to 7 days, thereby reducing the number of outdates [M. Ollgaard, I. Albjerg, and J. Georgen, Vox Sang. 74(Suppl. 1):1126, 1998; D. Vuetic, J. Taseki, B. Balint, and V. Mirovic, Vox Sang. 80(Suppl. 1):P371, 2000]. Within the United States, premarket multicenter studies (or possibly postmarket studies) are currently under discussion to provide data that may allow the extension of storage to 7 days. It would be expected that the cost of culturing would be mitigated by the savings obtained by the extension of platelet storage time (20). For example, in Denmark, when bacterial culture was implemented in 1997, it was anticipated to be cost neutral if the outdating could be reduced from 18 to 8% by extending the storage time to 7 days (24). The actual outdating rate decreased to 3 to 6%.
CONCLUSIONS
Regulatory oversight in the United States and Canada has generally concentrated on new emerging threats such as West Nile virus (leading to nationally mandated efforts to screen the blood supply with West Nile virus nucleic acid testing). However, there is no similar mandate in these countries for bacterial screening. To place these risks into some perspective, in 2002 23 transfusion-transmitted cases of West Nile virus were identified in the United States (50). Of these 23 recipients, 7 died, but only 5 of these deaths were associated with West Nile virus meningoencephalitis. During the same period, 17 deaths from bacterial contamination of blood components were reported to the FDA (14). Contaminated apheresis-derived platelets and pooled platelets were the products most often implicated. Gram-negative bacteria were the causative agents in the majority of these fatalities. It is perplexing that similar (or greater) government emphasis has not been placed on the detection of bacteria.
In blood banking and transfusion medicine, our paramount concern is to improve transfusion safety for patients, in our attempt to achieve a zero-risk blood supply. Unlike other recent threats (existing or theoretical) to blood safety, the movement for reducing bacterial contamination has come from the blood-banking and the transfusion medicine community. In this case, voluntary accrediting agencies such as the American Association of Blood Banks and the College of American Pathologists have taken the lead by developing standards to reduce the risk of bacterial contamination of blood components (17, 23). Our patients deserve no less.
REFERENCES
- 1.American Association of Blood Banks. 2003. Comprehensive report on blood collection and transfusion in the United States in 2001. National Blood Data Resource Center, Bethesda, Md.
- 2.American Association of Blood Banks. Bulletin. Guidance on implementation of new bacteria reduction and detection standard. http://www.aabb.org/members_only/archives/association_bulletins/ab03-10.htm.
- 3.Anderson, K. C., M. A. Lew, B. C. Gorgone, J. Martel, C. B. Leamy, and B. Sullivan. 1986. Transfusion-related sepsis after prolonged platelet storage. Am. J. Med. 81:405-411. [DOI] [PubMed] [Google Scholar]
- 4.Brecher, M. E. 2002. Bacterial contamination of blood products, p. 789. In T. L. Simon (ed.), Rossi's principles of transfusion medicine, 3rd ed. Lippincott, Williams and Wilkins, Baltimore, Md.
- 5.Brecher, M. E., G. Boothe, and A. Kerr. 1993. The use of a chemiluminescence-linked universal bacterial ribosomal RNA gene probe and blood gas analysis for the rapid detection of bacterial contamination in white cell reduced and nonreduced platelets. Transfusion 33:450-457. [DOI] [PubMed] [Google Scholar]
- 5a.Brecher, M. E., M. Foster, and D. Mair. 2001. Glucose and hemolysis as a rapid screen for Yersinia and Serratia RBC contamination. Vox Sang. 81:136-138. (Letter.) [DOI] [PubMed] [Google Scholar]
- 6.Brecher, M. E., S. N. Hay, and S. J. Rothenberg. 2003. Monitoring of apheresis platelet bacterial contamination with an automated liquid culture system: a university experience. Transfusion 43:974-978. [DOI] [PubMed] [Google Scholar]
- 7.Brecher, M. E., S. N. Hay, and S. J. Rothenberg. 2004. Evaluation of a new plastic culture bottle using an automated microbial detection system for 9 common contaminating organisms found in platelet components. Transfusion 44:359-363. [DOI] [PubMed] [Google Scholar]
- 8.Brecher, M. E., D. Heath, S. Hay, S. Rothenberg, and L. C. Stutzman. 2002. Evaluation of a new generation culture bottle using the BacT/ALERT 3D® Microbial Detection System on 9 common contaminating organisms found in platelet components. Transfusion 42:774-779. [DOI] [PubMed] [Google Scholar]
- 9.Brecher, M. E., J. J. Hogan, G. Boothe, A. Kerr, L. McClannan, M. R. Jacobs, R. Yomtovian, V. Chongokolwatana, G. Tegtmeier, S. Henderson, A. Pineda, V. Halling, M. Kemper, K. Kuramato, P. V. Holland, and M. Longiaru. 1994. Platelet bacterial contamination and the use of a chemiluminscence-linked universal bacterial ribosomal RNA gene probe. Transfusion 34:750-755. [DOI] [PubMed] [Google Scholar]
- 10.Brecher, M. E., P. V. Holland, A. Pineda, G. E. Tegtmeier, and R. Yomtovian. 2000. Growth of bacteria in inoculated platelets: implications for bacterial detection and the extension of platelet storage. Transfusion 40:1308-1312. [DOI] [PubMed] [Google Scholar]
- 11.Brecher, M. E., N. Means, C. S. Jere, D. Heath, S. Rothenberg, and L. C. Stutzman. 2001. Evaluation of the BacT/ALERT 3D® Microbial Detection System for platelet bacterial contamination: an analysis of 15 contaminating organisms. Transfusion 41:477-482. [DOI] [PubMed] [Google Scholar]
- 12.Bruneau, C., P. Perez, M. Chassaigne, P. Allouch, A. Audurier, C. Gulian, G. Janus, G. Boulard, P. De Micco, L. R. Salmi, and L. Noel. 2001. Efficacy of a new collection procedure for preventing bacterial contamination of whole-blood donations. Transfusion 41:74-81. [DOI] [PubMed] [Google Scholar]
- 13.Burstain, J. M., M. E. Brecher, K. Workman, M. Foster, G. H. Faber, and D. Mair. 1997. Rapid identification of bacterially contaminated platelets using reagent strips: glucose and pH analysis as markers of bacterial metabolism. Transfusion 37:255-258. [DOI] [PubMed] [Google Scholar]
- 14.CBER. 2002. Annual Report FY. 2002 (1 October 2001 through 30 September 2002). http://www.fda.gov/cber/inside/annrptpart3.htm.
- 15.Casewell, M. W., N. G. P. Slater, and J. E. Cooper. 1981. Operating theatre water-baths as a cause of Pseudomonas septicemia. J. Hosp. Infect. 2:237-240. [DOI] [PubMed] [Google Scholar]
- 15a.Centers for Disease Control and Prevention. 1997. Red blood cell transfusions contaminated with Yersinia enterocolitica—United States, 1991-1996, and initiation of a national study to detect bacteria-associated transfusion reactions. Morb. Mortal. Wkly. Rep. 46:553-555. [PubMed] [Google Scholar]
- 15b.Centers for Disease Control and Prevention. 1991. Update: Yersinia enterocolitica bacteremia and endotoxin shock associated with red blood cell transfusion—United States. 40:176-178. [PubMed]
- 16.Chiu, E. K. W., K. Y. Yuen, A. K. W. Lie, R. Liang, Y. L. Lau, A. C. Lee, Y. L. Kwong, S. Wong, M. H. Ng, and T. K. Chan. 1994. A prospective study of symptomatic bacteremia following platelet transfusion and of its management. Transfusion 34:950-954. [DOI] [PubMed] [Google Scholar]
- 17.Commission on Laboratory Accreditation. Transfusion medicine checklist. TRM.44955 phase I. College of American Pathologists, Northfield, Ill. 2003. http://www.cap.org/html/checklist_html/transfusionmedicine_1202.html. Last accessed 1 September 2004.
- 18.Corash, L. Technology for inactivation of pathogens in cellular blood products used for transfusion. Presented at the FDA/CBER Workshop on Safety and Efficacy of Methods for Reducing Pathogens in Cellular Blood Products Used in Transfusion. www.fda.gov/cber/minutes/workshop-min.htm.
- 19.DeKorte, D., J. H. Marcelis, A. J. Verhoeven, and A. M. Soeterboek. 2002. Diversion of first blood volume results in a reduction of bacterial contamination for whole-blood collections. Vox Sang. 83:13-16. [DOI] [PubMed] [Google Scholar]
- 20.Dumont, L. J., J. P. AuBuchon, P. Whitey, L. H. Herschel, A. Johnson, D. McNeil, S. Sawyer, and J. C. Roger. 2002. Seven-day storage of single-donor platelets: recovery and survival in an autologous transfusion study. Transfusion 42:847-854. [DOI] [PubMed] [Google Scholar]
- 21.Engelfriet, C. P., H. W. Reesink, M. A. Blajchman, L. Muylle, J. Kjeldsen-Kragh, R. Kekomaki, R. Yomtovian, P. Hocker, G. Stiegler, H. G. Klein, K. Soldan, Barbara J, A. Slopecki, A. Robinson, and H. Seyfried. 2000. Bacterial contamination of blood components. Vox Sang. 78:59-67. [DOI] [PubMed] [Google Scholar]
- 22.Fredricks, D. N., and D. A. Relman. 1998. Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate. J. Clin. Microbiol. 36:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fridey, J. (ed.). 2003. Standards for blood banks and transfusion services, 22nd ed. American Association of Blood Banks, Bethesda, Md.
- 24.Georgsen, J. 2003. Detection of bacterial contamination of platelet concentrates. International Forum 5. Vox Sang. 85:229-230. [DOI] [PubMed] [Google Scholar]
- 25.Gibson, T., and W. Norris. 1998. Skin fragments removed by injection needles. Lancet 2:983-985. [DOI] [PubMed] [Google Scholar]
- 26.Goldman, M., G. Roy, N. Frechette, F. Decary, L. Massicotte, and G. Delage. 1997. Evaluation of donor skin disinfection methods. Transfusion 37:309L-312L. [DOI] [PubMed] [Google Scholar]
- 27.Goodnough, L. T., A. Shander, and M. E. Brecher. 2003. Transfusion medicine: looking to the future. Lancet 361:161-169. [DOI] [PubMed] [Google Scholar]
- 28.Grossman, B. I., P. Kollins, P. M. Lau, J. L. Perreten, R. J. Bowman, S. Malcolm, and W. M. Palko. 1991. Screening blood donors for gastrointestinal illness: a strategy to eliminate carriers of Yersinia entercolitica. Transfusion 31:500-501. [DOI] [PubMed] [Google Scholar]
- 29.Heltberg, O., F. Show, P. Gerner-Smidt, H. J. Kolmos, E. Dybkjer, E. Gutschik, D. Jerne, O. B. Jepsen, M. Weischer, W. Frederiksen, and H. Sorensen. 1993. Nosocomial epidemic of Serratia marcescens septicemia ascribed to contaminated blood transfusion bags. Transfusion 33:221-227. [DOI] [PubMed] [Google Scholar]
- 30.Hogman, C. F., H. Fritz, and L. Sandberg. 1993. Post-transfusion Serratia marcescens septicemia (editorial). Transfusion 33:189-191. [DOI] [PubMed] [Google Scholar]
- 31.Jackson, B. R., M. P. Busch, S. L. Stramer, and J. P. AuBuchon. 2003. The cost-effectiveness of NAT for HIV, HCV, and HBV in whole-blood donations. Transfusion 43:721-729. [DOI] [PubMed] [Google Scholar]
- 32.Hoppe, P. A. 1992. Interim measures for detection of bacterially contaminated red cell components. Transfusion 32:199-201. (Editorial.) [DOI] [PubMed] [Google Scholar]
- 33.Jafari, M., J. Forsberg, R. O. Gilcher, J. W. Smith, J. M. Crutcher, M. McDermott, B. R. Brown, and J. N. George. 2002. Salmonella sepsis caused by a platelet transfusion from a donor with a pet snake. N. Engl. J. Med. 347:1075-1078. [DOI] [PubMed] [Google Scholar]
- 34.Knutson, F., R. Alfonso, K. Dupuis, V. Mayaudon, L. Lin, L. Corash, and C. F. Hogman. 2000. Photochemical inactivation of bacteria and HIV in buffy-coat-derived platelet concentrates under conditions that preserve in vitro platelet function. Vox Sang. 78:209-216. [DOI] [PubMed] [Google Scholar]
- 35.Kuehnert, M. J., V. R. Roth, N. R. Haley, K. R. Gregory, K. V. Elder, G. B. Schreiber, M. J. Arduino, S. C. Holt, L. A. Carson, S. N. Banerjee, and W. R. Jarvis. 2001. Transfusion-transmitted bacterial infection in the United States, 1998 through 2000. Transfusion 41:1493-1499. [DOI] [PubMed] [Google Scholar]
- 36.Lee, J. 1999. U.S. Food and Drug Administration. Presented at the FDA/CBER Workshop on Bacterial Contamination of Platelets. www.FDA.gov/CBER/minutes/bact092499.pdf.
- 37.Leiby, D. A., K. L. Kerr, J. M. Campos and R. Y. Dodd. 1997. A retrospective analysis of microbial contaminants in outdated random-donor platelets from multiple sites. Transfusion 37:259-263. [DOI] [PubMed] [Google Scholar]
- 38.Reference deleted.
- 39.Reference deleted.
- 40.Macauley, A., A. Chandrasekar, G. Geddis, K. G. Morris, and W. M. McClelland. 2003. Operational feasibility of routine bacterial monitoring of platelets. Transfusion Med. 13:189-195. [DOI] [PubMed] [Google Scholar]
- 41.McCullough, J., D. H. Vesole, R. J. Benjamin, S. J. Slichter, A. Pineda, E. Snyder, E. A. Stadtmauer, I. Lopez-Plaza, S. Coutre, R. G. Strauss, L. T. Goodnough, J. L. Fridey, T. Raife, R. Cable, S. Murphy, F. Howard IV, K. Davis, J. S. Lin, P. Metzel, L. Corash, A. Koutsoukos, L. Lin, D. H. Buchholz, and M. G. Conlan. 2004. Therapeutic efficacy and safety of platelets treated with a photochemical process for pathogen inactivation: the SPRINT Trial. Blood 104:1534-1541. [DOI] [PubMed] [Google Scholar]
- 42.McDonald, C. P., S. Hartley, K. Orchard, G. Hughes, M. M. Brett, P. E. Hewitt, and J. A. Barbara. 1998. Fatal Clostridium perfringens sepsis from a pooled platelet transfusion. Transfusion Med. 8:19-22. [DOI] [PubMed] [Google Scholar]
- 43.McDonald, C. P., P. Lowe, A. Roy, S. Robbins, S. Hartley, J. F. Harrison, A. Slopecki, N. Verlander, and J. A. Barbara. 2001. Evaluation of donor arm disinfection techniques. Vox Sang. 80:135-141. [DOI] [PubMed] [Google Scholar]
- 44.McDonald, C. P., A. Rogers, M. Cox, R. Smith, A. Roy, S. Robbins, S. Hartley, J. A. Barbara, S. Rothenberg, L. Stutzman, and G. Widders. 2002. Evaluation of the 3D BacT/ALERT automated culture system for the detection of microbial contamination of platelet concentrates. Transfusion Med. 12:303-309 [DOI] [PubMed] [Google Scholar]
- 45.Morel, P., and P. Herve. 2003. Detection of bacterial contamination of platelet concentrates. International Forum 6. Vox Sang. 85:230-232. [DOI] [PubMed] [Google Scholar]
- 46.Moreno, J. D. 2003. “Creeping precautionism” and the blood supply. Transfusion 43:840-842. [DOI] [PubMed] [Google Scholar]
- 47.Nikkari, S., I. J. McLaughin, W. Bi, D. E. Dodge, and D. A. Relman. 2001. Does blood of healthy subjects contain bacterial ribosomal DNA? J. Clin. Microbiol. 39:1956-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ness, P. M., H. G. Braine, K. King, C. Barrasso, T. Kickler, A. Fuller, and N. Blades. 2001. Single donor platelets reduce the risk of septic transfusion reactions. Transfusion 41:857-861. [DOI] [PubMed] [Google Scholar]
- 49.Ortolano, G. A., L. F. Freundlich, S. Holme, R. L. Russell, M. A. Cortus, K. Wilkins, H. Nomura, C. Chong, R. Carmen, A. Capetandes, and B. Wenz. 2003. Detection of bacteria in WBC-reduced PLT concentrates using percent oxygen as a marker for bacteria growth. Transfusion 43:1276-1285. [DOI] [PubMed] [Google Scholar]
- 50.Pealer, L. N., A. A. Marfin, L. R. Petersen, R. S. Lanciotti, P. L. Page, S. L. Stramer, M. G. Stobierski, K. Signs, B. Newman, H. Kapoor, J. L. Goodman, M. E. Chamberland, and the West Nile Virus Transmission Investigation Team. 2003. Transmission of West Nile virus through blood transfusion in the United States in 2002. N. Engl. J. Med. 349:1236-1245. [DOI] [PubMed] [Google Scholar]
- 51.Perez, P., L. R. Salmi, G. Follea, J. L. Schmit, B. de Barbeyrac, P. Sudre, R. Salamon, the BACTHEM Group, and the French Haemovigilance Network. 2001. Determinants of transfusion-associated bacterial contamination: results of the French BACTHEM case-control study. Transfusion 41:862-871. [DOI] [PubMed] [Google Scholar]
- 52.Rhame, F. S., R. K. Root, J. D. MacLowry, T. A. Dadisman, and J. V. Bennett. 1973. Salmonella septicemia from platelet transfusions. Study of an outbreak traced to a hematogenous carrier of Salmonella choleraesuis. Ann. Intern. Med. 78:633-641. [DOI] [PubMed] [Google Scholar]
- 53.Roth, V. R., M. J. Arduino, J. Nobiletti, S. C. Holt, L. A. Carson, C. F. Wolf, B. A. Lenes, P. M. Allison, and W. R. Jarvis. 2000. Transfusion-related sepsis due to Serratia liquefaciens in the United States. Transfusion 40:931-935. [DOI] [PubMed] [Google Scholar]
- 54.Rudi, K., H. K. Hoidal, T. Katla, B. K. Johansen, J. Nordal, and K. S. Jakobsen. 2004. Direct real-time PCR quantification of Campylobacter jejuni in chicken fecal and cecal samples by integrated cell concentration and DNA purification. Appl. Environ. Microbiol. 70:790-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneider, T., D. Breviere, M. F. Taillefer, A. Pujol-Rey, and J. J. Huart. 2000. Bacterial contamination of platelet concentrates by Propionibacterium acnes. Transfusion Clin. Biol. 7:540-546. [DOI] [PubMed] [Google Scholar]
- 56.Sen, K. 2000. Rapid identification of Yersinia enterocolitica in blood by the 5′ nuclease PCR assay. J Clin. Microbiol. 38:1953-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.SHOT. 2002. Serious Hazards of Transfusion. SHOT report for 2000-2001. (Cumulative data 01/10/1995-30/09/2002.) http://www.shot.demon.co.uk/toc.htm.
- 58.Szewzyk, U., R. Szewzyk, and T. A. Stenstrom. 1993. Growth and survival of Serratia marcescens under aerobic and anaerobic conditions in the presence of materials from blood bags. J. Clin. Microbiol. 31:1826-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Theakston, E. P., A. J. Morris, S. J. Streat, B. W. Baker, and D. G. Woodfield. 1997. Transfusion transmitted Yersinia enterocolitica infection in New Zealand. Aust. N. Z. J. Med. 27:62-67. [DOI] [PubMed] [Google Scholar]
- 60.Tipple, M. A., L. A. Bland, J. J. Murphy, M. J. Arduino, A. L. Panlilio, J. J. Farmer III, M. A. Tourault, C. R. Macpherson, J. E. Menitove, A. J. Grindon, P. S. Johnson, R. G. Strauss, J. A. Bufill, P. S. Ritch, J. R. Archer, O. C. Tablan, and W. R. Jarvis. 1990. Sepsis associated with transfusion of red cells contaminated with Yersinia enterocolitica. Transfusion 30:207-213. [DOI] [PubMed] [Google Scholar]
- 61.University Health System Consortium. UHC technology assessment. Platelet transfusion guidelines, 1998. http://www.uhc.org.
- 62.Wagner, S. J., and D. Robinette. 1996. Evaluation of swirling, pH, and glucose tests for the detection of bacterial contamination in platelet concentrates. Transfusion 36:989-993. [DOI] [PubMed] [Google Scholar]
- 63.Werch, J. B., P. Mhawech, C. E. Stager, E. I. Banez, and B. Lichtiger. 2002. Detecting bacteria in platelet concentrates by use of reagent strips. Transfusion 42:1027-1031. [DOI] [PubMed] [Google Scholar]
- 64.Wollowitz, S. 2001. Fundamentals of the psoralen-based Helinx technology for inactivation of infectious pathogens and leukocytes in platelets and plasma. Semin. Hematol. 38(Suppl. 11):4-11. [DOI] [PubMed] [Google Scholar]
- 65.Yomtovian, R. 2002. Bacterial contamination an overview of key issues. Presented at the FDA/CBER Safety and Efficacy of Methods for Reducing Pathogens in Cellular Blood Products Used in Transfusion Workshop. www.fda.gov/cber/minutes/workshop-min.htm.
- 66.Yomtovian, R., H. M. Lazarus, L. T. Goodnough, N. V. Hirschler, A. M. Morrissey, and M. R. Jacobs. 1993. A prospective microbiologic surveillance program to detect and prevent the transfusion of bacterially contaminated platelets. Transfusion 33:902-909. [DOI] [PubMed] [Google Scholar]
- 67.Zuck, T. F. 1987. Greetings—a final look back with comments about a policy of a zero-risk blood supply. Transfusion 27:447-448. [DOI] [PubMed] [Google Scholar]