Significance
Mutant form of p53 (mutp53) proteins are expressed at high levels in many human cancers and can promote tumor cell growth. However, their mechanisms of action have not been fully understood. Elucidation of the mechanisms may identify new therapeutic strategies for treating many cancers that contain mutp53s. We describe a role for several hotspot mutp53s in reducing the checkpoint response to replication stress through interacting with TopBP1. This finding provides a rationale for a synthetic lethality strategy to treat mutp53-harboring cancer cells with inhibitors of another ATR activator, DNA2. We also find that certain mutp53s directly promote DNA replication by bridging the interaction between TopBP1 and Treslin. These results uncover mechanisms by which mutp53 enhances DNA replication.
Keywords: ATR, Chk1, mutant p53, TopBP1, Treslin
Abstract
Accumulating evidence supports the gain-of-function of mutant forms of p53 (mutp53s). However, whether mutp53 directly perturbs the DNA replication checkpoint remains unclear. Previously, we have demonstrated that TopBP1 forms a complex with mutp53s and mediates their gain-of-function through NF-Y and p63/p73. Akt phosphorylates TopBP1 and induces its oligomerization, which inhibits its ATR-activating function. Here we show that various contact and conformational mutp53s bypass Akt to induce TopBP1 oligomerization and attenuate ATR checkpoint response during replication stress. The effect on ATR response caused by mutp53 can be exploited in a synthetic lethality strategy, as depletion of another ATR activator, DNA2, in mutp53-R273H–expressing cancer cells renders cells hypersensitive to cisplatin. Expression of mutp53-R273H also makes cancer cells more sensitive to DNA2 depletion or DNA2 inhibitors. In addition to ATR-activating function during replication stress, TopBP1 interacts with Treslin in a Cdk-dependent manner to initiate DNA replication during normal growth. We find that mutp53 also interferes with TopBP1 replication function. Several contact, but not conformational, mutp53s enhance the interaction between TopBP1 and Treslin and promote DNA replication despite the presence of a Cdk2 inhibitor. Together, these data uncover two distinct mechanisms by which mutp53 enhances DNA replication: (i) Both contact and conformational mutp53s can bind TopBP1 and attenuate the checkpoint response to replication stress, and (ii) during normal growth, contact (but not conformational) mutp53s can override the Cdk2 requirement to promote replication by facilitating the TopBP1/Treslin interaction.
Normal cells use various fundamental braking mechanisms to properly respond to environmental cues and coordinate the execution of different phases of cell cycle. Tumor suppressors p53 and pRb are the key regulators for G1 checkpoint, which are frequently lost in many types of cancer (1). Mutations of TP53 are found in half of all human cancers, including nearly all small-cell lung cancers, squamous cell lung cancers, high-grade serous ovarian cancer (2–4), and greater than 80% of glioblastoma and basal-like breast cancer (5, 6). Therefore, understanding the contribution of TP53 mutations in carcinogenesis is very important for the development of new strategies to prevent cancer progression and improve the efficacy of cancer therapy.
In addition to the loss of normal p53 function, mutant form of p53 (mutp53) proteins acquire new oncogenic properties (gain-of-function, GOF), such as promoting cancer cell proliferation, metastasis, genomic instability, resistance to chemotherapy, etc. (7–9). Among the many mechanisms of mutp53 GOF, the checkpoint activator TopBP1 (topoisomerase IIβ-binding protein) has been identified as a critical mediator for facilitating complex formation between several hotspot mutp53 proteins and either NF-Y or p63/p73 (10). TopBP1 interacts with these mutp53s and NF-Y and promotes mutp53 and p300 recruitment to NF-Y target gene promoters. TopBP1 also facilitates mutp53 interaction with p63/p73 to inhibit their transcriptional activities (10).
TopBP1 contains nine BRCA1 carboxyl-terminal (BRCT) domains with distinct functions in DNA replication initiation, ATR activation, and transcription (11). TopBP1 binds to Cdk2-phosphorylated Treslin/TICRR (TopBP1-interacitng, checkpoint, and replication regulator) to facilitate loading of Cdc45 onto replication origins (12, 13). Cdk2 phosphorylates Treslin at the Ser1000 residue during S phase and induces its association with TopBP1 (through TopBP1 first and second BRCT domains) to promote DNA replication (14). Upon DNA replication stress, TopBP1 is recruited to stalled replication forks through direct binding to the stalled forks (15, 16) or interaction of its first and second BRCT domains with the Rad9–Hus1–Rad1 (9–1–1) clamp (17). It then activates ATR through a conserved ATR-activating domain located between the sixth and seventh BRCT domains (18). It is noteworthy that in addition to TopBP1, DNA2 can also activate ATR, possibly independently of TopBP1 (19, 20). TopBP1 also regulates several transcription factors, including E2F1 (21-23), p53 (24), Miz1 (23, 25), and SPBP (26). TopBP1 is controlled by Rb/E2F and is induced when cells enter the S phase of the cell cycle (22, 27). Meanwhile, feedback regulation of E2F1 and p53 by TopBP1 is important to restrict the proapoptotic activities of both transcription factors during normal S-phase transition (22, 24).
TopBP1 is tightly controlled through different mechanisms. One of them is the regulation of its quaternary structure. Akt phosphorylates TopBP1 at the Ser1159 residue and induces its oligomerization through an intermolecular interaction between the phosphorylated Ser1159 residue (pS1159) and the seventh–eighth BRCT (BRCT7/8) domains of two individual TopBP1 molecules (23, 28). Oligomerization of TopBP1 then induces its binding to E2F1 but at the same time prevents its recruitment to chromatin and ATR binding and inhibits its checkpoint-activating functions (28). Hence, Akt switches TopBP1 function from checkpoint activation to transcriptional regulation by regulating TopBP1 quaternary structure. In cancer cells harboring high Akt activity, this mechanism is responsible for inhibition of E2F1-dependent apoptosis and ATR function (28).
Mutations of TP53 increase protein stability and lead to its accumulation in many cancer cells. As TopBP1 plays a critical role in checkpoint function and mutp53 is abundantly present in many types of cancer, the formation of the mutp53/TopBP1 complex raises intriguing questions: Do the accumulated mutp53 proteins perturb ATR/Chk1 checkpoint function? Would mutp53 affect TopBP1 function in DNA replication? Here we demonstrate that those hotspot mutp53s capable of binding TopBP1 (10) can interfere with the ATR-activating function of TopBP1 by inducing TopBP1 oligomerization independently of Akt. We also report that certain contact, but not conformational, mutp53s enhance the interaction of TopBP1 with Treslin and promote DNA replication independent of Cdk activation. Because mutp53s can perturb ATR/Chk1 checkpoint response, targeting DNA2, a TopBP1-independent ATR activator, may prove to be an effective synthetic lethality strategy to treat cancers harboring mutp53.
Results
Mutp53 Inhibits ATR/TopBP1 Interaction and Decreases the Checkpoint Response to Replicative Stress.
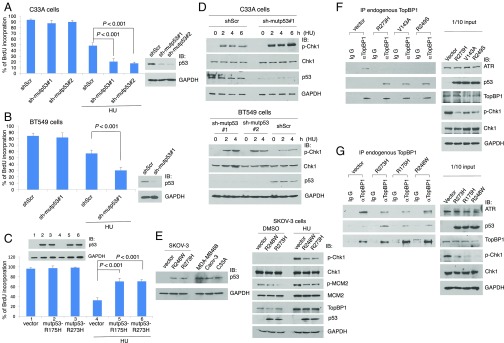
To determine whether mutp53 affects replication checkpoint response, we depleted mutp53 in C33A cervical carcinoma cells (harboring mutp53-R273C) or BT549 breast cancer cells (harboring mutp53-R249S), followed by treatment with a replication stress-inducing drug hydroxyurea (HU). BrdU incorporation assay was performed to measure DNA replication. Indeed, HU-induced S-phase checkpoint response was augmented upon depletion of mutp53 in C33A cells (Fig. 1A and SI Appendix, Fig. S1A) or BT549 cells (Fig. 1B and SI Appendix, Fig. S1B). Conversely, expression of R175H- or R273H-mutp53 in p53-null H1299 cells inhibited HU-induced S-phase checkpoint response (Fig. 1C and SI Appendix, Fig. S1 C and D). Given the pivotal role for Chk1 in the response to the replicative stress, we examined Chk1 activation by measuring the phosphorylation of Chk1. Consistent with the results obtained by BrdU incorporation assay, depletion of mutp53 enhanced HU-induced Chk1 activation in both cell lines (Fig. 1D and SI Appendix, Fig. S2). We considered the possibility that mutp53 depletion might cause DNA damage or make cells suffer from increased replicative stress and indirectly enhance Chk1 activation during HU treatment. However, in the absence of HU treatment, mutp53 depletion did not reduce BrdU incorporation (Fig. 1 A and B) nor activate Chk1 (Fig. 1D). We also examined the ATM pathway. Depletion of mutp53 in C33A cells or in BT549 cells in the absence of HU or adriamycin treatment did not cause Chk2 activation and did not induce H2AX phosphorylation, a marker for DNA damage (SI Appendix, Fig. S3 A and B). Overexpression of mutp53-R273H also did not inhibit Chk2 activation upon treatment with HU or adriamycin (SI Appendix, Fig. S4, Left). Camptothecin (CPT) can activate both Chk1 and Chk2. Although mutp53-R273H expression in H1299 cells did not have a significant effect on Chk2 activation after CPT treatment, it inhibited Chk1 activation (SI Appendix, Fig. S4, Right).
Fig. 1.
Hotspot mutp53 dampens DNA replication checkpoint response by inhibiting TopBP1 interaction with ATR. (A and B) C33A cells (A) or BT549 cells (B) stably expressing either a scrambled shRNA (shScr) or a p53 shRNA (#1 or #2) were treated with HU (2 mM) or vehicle (DMSO) for 16 h, followed by BrdU incorporation assay. At least 300 nuclei per sample were counted by fluorescence microscopy. Representative images are presented in SI Appendix, Fig. S1 A and B. Data shown represent means and SDs from six (A) or three (B) replicates. All P values are based on a two-tailed t test. Mutp53 knockdown was verified by immunoblotting. (C) H1299 cells were transfected with either an empty vector or an expression vector of mutp53 (R175H or R273H). Forty-eight hours later, cells were treated with vehicle (DMSO) or HU (2 mM) for 16 h and then subjected to BrdU incorporation assay. The data shown represent means ± SD from six replicates. Representative images are presented in SI Appendix, Fig. S1C. (D) C33A cells (Top) or BT549 cells (Bottom) were stably transfected with shScr or a p53 shRNA (sh-mutp53 #1 or #2). Cells were treated with HU (2 mM) for 2, 4, or 6 h. Whole-cell lysates were subjected to Western blot analysis as indicated. (E) Expression of mutp53-R273H or mutp53-R248W in p53-null SKOV-3 cells at a level comparable to endogenous mutp53s as seen in MDA-MB468, Caov-3, or C33A (Left) inhibits ATR activation after HU treatment. SKOV-3 cells transfected with an empty vector or the expression vector of mutp53-R248W or mutp53-R273H were treated with vehicle (DMSO) or HU (2 mM) for 20 h. The ATR activity was determined by immunoblotting of p-Chk1 and p-MCM2 (Right). (F and G) H1299 cells were transfected with an empty vector or an expression vector of mutp53 harboring R273H, V143A, R249S (F), R175H, or R248W mutation (G). Forty-eight hours later, cells were treated with HU (2 mM) for 16 h and then harvested in TNN buffer. Endogenous TopBP1 was immunoprecipitated with an anti-TopBP1 mouse monoclonal antibody or control mouse IgG, followed by immunoblotting as indicated. One tenth of the cell lysates were also subjected to Western blot analysis.
To further investigate the effect of mutp53 on checkpoint control, we expressed mutp53, either R248W or R273H, in p53-null SKOV-3 cells at a level comparable to that of endogenous mutp53 in several mutp53-bearing cancer cell lines (Fig. 1E, Left). HU-induced ATR activation was assessed by measuring the phosphorylation of ATR targets Chk1 and MCM2 (29, 30). We found that both mutp53 proteins attenuated ATR activation upon HU treatment (Fig. 1E, Right).
Our previous data showed that several hotspot mutp53 proteins could bind TopBP1 (10). Therefore, we expressed each of these hotspot mutp53s (R273H, V143A, R249S, R175H, or R248W) in p53-null H1299 cells and examined Chk1 activation and the interaction of TopBP1 with either ATR or mutp53 following HU treatment. Indeed, all of these hotspot mutp53 proteins bound to TopBP1 comparably as previously reported (10) (Fig. 1 F and G, IP) and attenuated Chk1 activation (Fig. 1 F and G, input lysates). Importantly, the interaction between TopBP1 and ATR was significantly attenuated by these mutp53s (Fig. 1 F and G, IP), consistent with the reduction of Chk1 phosphorylation. Fig. 1 F and G was performed independently. The difference in signal intensities between these two experiments was due to different exposure times of films in Western blot analysis. Taken together, these results demonstrate a role for several hotspot mutp53 proteins in perturbing the ATR/TopBP1-mediated replication checkpoint response.
Mutp53 Inhibits ATR Response in Chromatin Compaction After HU Treatment.
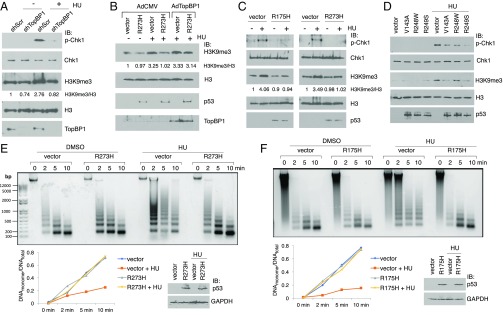
In addition to S-phase checkpoint, activation of ATR/Chk1 can elicit a global heterochromatin response and senescence during replication stress (31, 32). Therefore, we next investigated whether mutp53 could interfere with the ATR/Chk1-mediated epigenetic response through TopBP1. Previously, Di Micco et al. showed that depletion of ATR inhibited the induction of histone H3 lysine 9 trimethylation (H3K9me3), a heterochromatin marker, during oncogene-induced replication stress (31). Consistently, our data showed that in addition to Chk1 activation, HU treatment increased H3K9me3 in H1299 cells (Fig. 2 A–D). Besides, depletion of TopBP1 in H1299 cells inhibited the induction of H3K9me3 as well as Chk1 phosphorylation after HU treatment (Fig. 2A). In contrast, overexpression of TopBP1 was able to restore HU-induced H3K9me3 that was inhibited by expression of mutp53-R273H in H1299 cells (Fig. 2B). These data support a role for TopBP1 in the heterochromatin response through regulation of ATR/Chk1 and also suggest that mutp53 inhibits the HU response by restricting functional TopBP1. Because several hotspot mutp53 proteins can attenuate Chk1 response (Fig. 1 F and G), we next examined their effect on H3K9me3. Indeed, expressing these hotspot mutp53 proteins—that is, R175H, R273H, V143A, R248W, or R249S—attenuated HU-induced Chk1 phosphorylation and H3K9me3 in H1299 cells (Fig. 2 C and D). We then performed micrococcal nuclease assay to evaluate chromatin compaction (33). In vector-transfected H1299 cells, HU treatment induced chromatin compaction as evidenced by increased resistance to micrococcal nuclease digestion (Fig. 2 E and F, vector). Upon expression of either mutp53-R273H or mutp53-R175H in H1299 cells, the chromatin compaction response to HU treatment was blunted (Fig. 2 E and F). These data demonstrate a role for several hotspot mutp53 proteins in blunting ATR/Chk1 activation and heterochromatin response during replication stress.
Fig. 2.
Mutp53 blocks HU-induced chromatin compaction in cancer cells. (A) H1299 cells stably expressing either a scrambled shRNA (shScr) or TopBP1 shRNA (shTopBP1) were treated with HU (2 mM) or vehicle (DMSO) for 16 h and then harvested for Western blotting. The intensities of histone H3K9me3 and histone H3 in each lane were quantified using ImageJ software, and the signals of H3K9me3 were normalized by the corresponding H3 signals. (B) Mutp53-R273H inhibits the induction of H3K9me3 by HU; however, the effect can be rescued by overexpressing TopBP1. H1299 cells were transfected with an empty vector or mutp53-R273H. Twenty-four hours later, cells were infected with recombinant adenovirus harboring TopBP1 (AdTopBP1) or an empty vector (AdCMV) at a multiplicity of 100. Twenty-four hours later, cells were then treated with HU (2 mM) or vehicle control for 12 h. Cell lysates were harvested for Western blot analysis. (C and D) H1299 cells stably transfected with an empty vector or an expression vector of mutp53 (R175H or R273H, shown in C) or H1299 cells transiently transfected with an empty vector or an expression vector of mutp53 (V143A, R248W, or R249S, shown in D) were treated with HU for 5 h. Immunoblotting was performed to detect the indicated proteins. (E and F) H1299 cells were transfected with either an empty vector or an expression vector of mutp53 [R273H (E) or R175H (F)]. Cells were then treated with HU (2 mM) or DMSO for 16 h and subjected to the micrococcal nuclease (MNase) assay. Nuclei were isolated and digested with MNase for the indicated time. The ratio of mononucleosomal DNA versus total DNA is presented as a function of the digestion time and shown on Left Bottom. The expression of mutp53-R273H and mutp53-R175H was confirmed by immunoblotting (Right Bottom).
Mutp53 Perturbs Chromatin Recruitment of TopBP1.
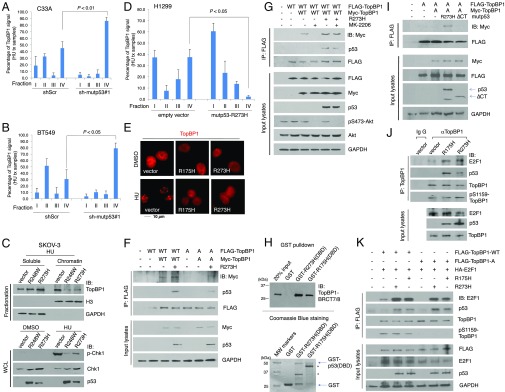
We next investigated the mechanism by which mutp53 inhibits TopBP1 checkpoint function. Because chromatin recruitment of TopBP1 is essential for TopBP1 to activate ATR upon replication stress, we performed chromatin binding assay (34) in HU-treated cells to determine the effect of mutp53 on TopBP1 chromatin recruitment. Indeed, depletion of mutp53 in C33A cells (Fig. 3A and SI Appendix, Fig. S5) and in BT549 cells (Fig. 3B and SI Appendix, Fig. S6) enhanced chromatin binding (fraction IV, which is resistant to 0.5% Nonidet P-40 extraction and represents tightly bound chromatin) (34) of TopBP1. This result is consistent with its effect on augmenting the S-phase checkpoint response (Fig. 1). Conversely, expression of mutp53-R248W or R273H in p53-null SKOV-3 cells significantly inhibited TopBP1 chromatin binding during HU treatment (Fig. 3C, Upper). Expression of mutp53 and its inhibitory effect on Chk1 phosphorylation in the same experiment were confirmed by immunoblotting of total cell lysates (Fig. 3C, Lower). Expression of mutp53-R273H in H1299 cells also inhibited TopBP1 chromatin binding during HU treatment (Fig. 3D and SI Appendix, Fig. S7). Supporting its role in preventing TopBP1 recruitment to DNA damage foci, mutp53 inhibited TopBP1 focus formation after HU treatment (Fig. 3E and SI Appendix, Fig. S8). The effect of mutp53 on inhibiting TopBP1 chromatin binding correlates with that on Chk1 activation (Fig. 1). Together, these results suggest that mutp53 binding interferes with TopBP1 recruitment to stalled replication forks and therefore inhibits TopBP1/ATR binding, which is required for ATR/Chk1 activation.
Fig. 3.
Mutp53 inhibits TopBP1 chromatin recruitment after HU treatment through induction of TopBP1 self-association. (A and B) C33A cells (A) or BT549 cells (B) expressing shScr or sh-mutp53 were treated with HU (2 mM) for 16 h, followed by chromatin fractionation. An aliquot from each fraction was subjected to immunoblotting. The intensities of TopBP1 protein levels in each fraction were quantified using ImageJ software. The graphs represent means ± SD derived from three independent experiments. Representative immunoblots are shown in SI Appendix, Figs. S5 and S6, respectively. (C) SKOV-3 cells transfected with an empty vector, mutp53-R248W, or mutp53-R273H as described in Fig. 1E were treated with HU (2 mM) for 20 h, followed by chromatin fractionation. The combined aliquots from fractions I and II (soluble fractions) and aliquots from fractions III and IV (chromatin fractions; fraction III is resistant to 0.2% Nonidet P-40 but is extracted by 0.5% Nonidet P-40; fraction IV is resistant to 0.5% Nonidet P-40 extraction and represents tightly bound chromatin) (34) for each group were separated on a 10% SDS/PAGE gel and immunoblotted with the indicated antibodies. The total cell lysates of DMSO- and HU-treated cells were also analyzed by immunoblotting (Lower; WCL, whole-cell lysate). (D) H1299 cells transfected with either an empty vector or mutp53-R273H were treated with HU (2 mM) for 16 h, followed by chromatin fractionation. An aliquot from each fraction was subjected to immunoblotting. The intensities of TopBP1 protein levels in each fraction were quantified using ImageJ software. The graphs represent means ± SD derived from three independent experiments. Representative immunoblots are shown in SI Appendix, Fig. S7. (E) H1299 cells were transfected with an empty vector, mutp53-R175H, or mutp53-R273H. After treatment with HU (2 mM) or vehicle for 16 h, cells were fixed and immunostained with anti-TopBP1 antibody, followed by Texas Red X-conjugated secondary antibody. Nuclei were stained with Hoechst 33258. Pictures shown are representative images at 100× magnification. A complete set of representative images is shown in SI Appendix, Fig. S8. (F) Mutp53-R273H was cotransfected with FLAG-TopBP1-WT (labeled as WT) and Myc-TopBP1-WT or with FLAG-TopBP1-S1159A (labeled as A) and Myc-TopBP1-S1159A into H1299 cells. Coimmunoprecipitation was performed using anti-FLAG beads, followed by immunoblotting as indicated. One tenth of the cell lysates were subjected to Western blot analysis. (G) Mutp53-R273H was cotransfected with FLAG-TopBP1-WT and Myc-TopBP1-WT into H1299 cells. After 36 h, cells were treated with vehicle or Akt inhibitor MK-2206 (5 µM) for 12 h, and then coimmunoprecipitation was performed as in F. (H) Purified TopBP1-BRCT7/8 was incubated with GST, GST-p53-R273H(DBD), or GST-p53-R175H(DBD), and GST pulldown was performed (24). Shown in Upper is immunoblotting with an antibody specific to TopBP1. In parallel, GST proteins used in the pulldown assay were resolved by SDS/PAGE and stained with Coomassie Blue (Lower). Arrow indicates GST or GST fusion protein. * indicates partially degraded proteins. (I) Mutp53-R273H or mutp53-R273H∆CT was cotransfected with FLAG-TopBP1-S1159A (labeled as A) and Myc-TopBP1-S1159A into H1299 cells. After 48 h, coimmunoprecipitation was performed using anti-FLAG beads as in F. (J) H1299 cells were transfected with an empty vector, mutp53-R175H, or mutp53-R273H. Immunoprecipitation was performed with anti-TopBP1 mouse monoclonal antibody or control mouse IgG, followed by immunoblotting as indicated. (K) HA-E2F1 was cotransfected with FLAG-TopBP1 (WT or S1159A; labeled as A) and mutp53 (R175H or R273H) into H1299 cells. Immunoprecipitation was performed using anti-FLAG beads, followed by Western blotting to detect indicated proteins.
Mutp53 Inhibits the Checkpoint Function of TopBP1 by Inducing Its Oligomerization Independent of Akt-Induced S1159 Phosphorylation.
Previously we demonstrate that Akt-mediated phosphorylation of TopBP1 at S1159 regulates its binding to BRCT7/8 domain and oligomerization, leading to a switch of its function from checkpoint activation to transcriptional regulation (28). Because p53 can form tetramers (35), we next tested whether mutp53 bound to TopBP1 and induced its oligomerization in an Akt-independent manner. Indeed, expression of mutp53-R273H in p53-null H1299 cells promoted self-association of not only wild-type (WT) TopBP1 but also S1159A mutant TopBP1 that cannot be phosphorylated by Akt (Fig. 3F). To rule out a definitive role for Akt, we used an allosteric Akt inhibitor, MK-2206 (28). As shown in Fig. 3G, MK-2206 treatment did block WT TopBP1 oligomerization in the absence of mutp53-R273H but failed to inhibit TopBP1 oligomerization when mutp53-R273H was coexpressed in H1299 cells. To further confirm that this effect is not dependent on the pS1159–BRCT7/8 interaction, we also examined the effect of mutp53 on the oligomerization of either S1273A or K1317M mutant TopBP1, which is defective in binding to pS1159 and therefore cannot form oligomers after Akt activation (28). As expected, only WT TopBP1, but not S1273A or K1317M mutant TopBP1, formed oligomers in the absence of mutp53 (SI Appendix, Fig. S9, left seven lanes). Nevertheless, expression of mutp53-R273H enhanced oligomerization of not only WT TopBP1 but also S1273A or K1317M mutant TopBP1 (SI Appendix, Fig. S9, right three lanes). These results demonstrate that mutp53 can induce TopBP1 oligomerization through a mechanism independent of Akt and the pS1159–BRCT7/8 interaction.
Our previous study showed TopBP1–BRCT7/8 domains interacted with p53-DBD (DNA-binding domain) (24). We performed GST pulldown assay and confirmed the interaction between TopBP1–BRCT7/8 and DBD of mutp53-R273H and -R175H (Fig. 3H). To investigate the role of mutp53 tetramerization, we tested the effect of a carboxyl terminus deletion, R273HΔCT (Fig. 3I), which lacks the p53 tetramerization domain but still binds TopBP1–BRCT7/8 (24). Indeed, mutp53-R273HΔCT failed to induce TopBP1 oligomerization (Fig. 3I) and therefore could not inhibit Chk1 activation after HU treatment (SI Appendix, Fig. S10).
Oligomerization of TopBP1 can also induce its binding to E2F1 (23, 28). To further provide evidence for the effect of mutp53 on TopBP1 oligomerization, we next examined the effect of mutp53 on the TopBP1 binding to E2F1. As shown in Fig. 3J, expression of mutp53-R175H or -R273H in H1299 cells greatly enhanced the interaction between endogenous TopBP1 and E2F1. Here mutp53 did not affect the levels of pS1159 TopBP1, excluding the possibility that this effect is mediated by TopBP1 phosphorylation. Consistently, mutp53-R175H or -R273H enhanced the interaction between E2F1 and TopBP1 without affecting TopBP1-S1159 phosphorylation (Fig. 3K, left four lanes). Although E2F1 did not interact with TopBP1-S1159A in H1299 cells, their binding could be induced by expression of mutp53-R175H or -R273H (Fig. 3K, right three lanes).
Taken together, these data show that hotspot mutp53 proteins use their tetramerization property to induce TopBP1 oligomerization independently of Akt and perturb replication checkpoint response. Through this mechanism, mutp53 may override the inhibitory actions of Chk1 during genotoxic or replication stress.
Mutp53 Expression and DNA2 Depletion/Inhibition Together Severely Impair ATR Function and Are Synthetic Lethal to Cancer Cells.
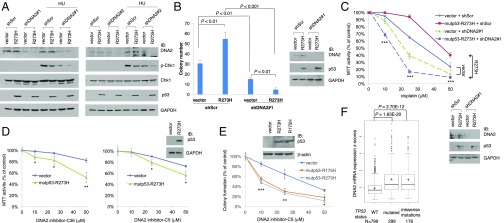
ATR mediates the S-phase checkpoint activation through two independent pathways that involve TopBP1 and DNA2 nuclease, respectively (19, 20). Although TopBP1-mediated Chk1 activation is inhibited by mutp53, ATR function is only partially attenuated in cancer cells harboring mutp53, probably due to an intact DNA2 pathway. Because a complete loss of ATR or Chk1 is not compatible with cell survival (36, 37), targeting DNA2 in mutp53-bearing cancers may severely cripple ATR function and affect their viability or cause chemosensitization. If so, this may provide an opportunity to develop novel “synthetic lethality” therapies against mutp53-bearing tumors. To test this concept, we coexpressed DNA2 shRNA (shDNA2#1 or shDNA2#2) with mutp53-R273H in H1299 cells and then evaluated HU-induced Chk1 activation to determine whether ATR/Chk1 function is further incapacitated. The result showed that HU-induced Chk1 activation was attenuated by expression of either DNA2 shRNA or mutp53-R273H, and this inhibitory effect could be further enhanced when both DNA2 shRNA and mutp53-R273H were coexpressed (Fig. 4A). The synthetic lethal interaction between DNA2 depletion and mutp53 was further demonstrated by clonogenic survival assay (Fig. 4B and SI Appendix, Fig. S11A). Consistent with our prior report (10), expression of mutp53-R273H in shScr control H1299 cells enhanced clonogenic survival. In contrast, mutp53-R273H decreased the clonogenic survival of DNA2-depleted H1299 cells. We also performed MTT assay following cisplatin treatment. As expected, depletion of DNA2 sensitized vector-transfected H1299 cells to cisplatin, whereas expression of mutp53-R273H rendered H1299 cells resistant to cisplatin (Fig. 4C). The effect of mutp53-R273H is consistent with its known GOF through inhibiting p63/p73 activity (10, 38, 39). Paradoxically, expression of mutp53-R273H in DNA2-depleted H1299 cells made cells become more sensitive to cisplatin, supporting the concept of a synthetic lethality approach. To further investigate the effect of mutp53 on the sensitivity to DNA2 inhibition, we expressed mutp53 in H1299 cells and then treated the cells with DNA2 small-molecule inhibitor C36 (NSC360177) or C5 (NSC15765) (40). Indeed, expression of mutp53-R273H or -R175H enhanced the sensitivity to DNA2 inhibitors (Fig. 4 D and E and SI Appendix, Fig. S11B). DNA2 is expressed at a higher level in breast cancer with mutated TP53 and in breast cancer containing TP53 missense mutations than WT TP53 (Fig. 4F). These data suggest that DNA2 is an ideal therapeutic target for mutp53-harboring cancers.
Fig. 4.
Combination of mutp53 expression and DNA2 depletion greatly inhibits ATR function and enhances cisplatin sensitivity. (A) An empty vector or mutp53-R273H was cotransfected with a scrambled shRNA or a DNA2 shRNA (#1, pLKO.1p shDNA2; #2, pResQ shDNA2) into H1299 cells. After treatment with HU (2 mM) or vehicle for 16 h, whole-cell lysates were subjected to Western blotting. (B) H1299 stable cell lines expressing shScr or shDNA2 were transfected with a mutp53-R273H expression vector (containing neomycin-resistance gene) or an empty vector and selected by G418 for colony formation as previously described (10). The data shown represent means ± SD (n = 3 biological replicates). Representative images are shown in SI Appendix, Fig. S11A. Some cells were harvested 2 d after transfection before G418 selection, and aliquots of the cell lysates were analyzed by immunoblotting (Right). (C) Transfected H1299 cells as described in A were seeded on 96-well plates and treated with cisplatin at the indicated concentrations for 48 h, followed by MTT assay. Data represent means ± SD (n = 3 biological replicates). The P values for the difference between any two groups are <0.005. Specifically, ***P < 0.001 when comparing “mutp53+shDNA2” with “mutp53+shScr” or with “vector+shDNA2” at the indicated concentration of cisplatin, except P < 0.005 for the difference between “mutp53+shDNA2” and “vector+shDNA2” at 50 μM cisplatin. The expression of mutp53 or DNA2 was detected by immunoblotting (Lower). (D) H1299 cells were transfected with mutp53-R273H. Next day, cells were treated with two DNA2 inhibitors, C36 and C5, at the indicated concentrations for 48 h, followed by MTT assay. Data represent means ± SD (n = 3 biological replicates). The P values for the difference between vector and mutp53 are *P < 0.05 or **P < 0.01. The expression of mutp53 was confirmed by immunoblotting (Upper). (E) H1299 cells stably transfected with an empty vector or an expression vector of mutp53 (R175H or R273H) were treated with DMSO or increasing doses of C5 as in D. Cells were then grown in fresh media without C5 until colonies formed and stained with crystal violet. Each treatment was performed in triplicate. Representative images are shown in SI Appendix, Fig. S11B. The P values for the difference between vector and mutp53 are *P < 0.05, **P < 0.01, or ***P < 0.001. Expression of mutp53 was verified by immunoblotting (Upper). (F) Box plots of DNA2 expression data in invasive breast carcinoma with WT TP53, mutated TP53, or carrying missense TP53 mutation (data analyzed from breast cancer TCGA). +, mean values.
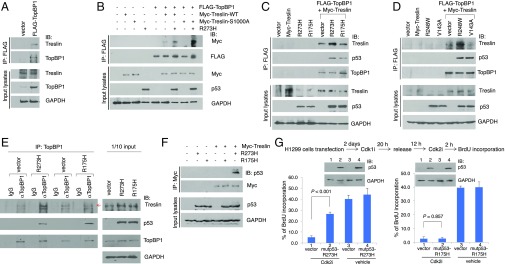
“Contact” but Not “Conformational” Mutp53s Bind Treslin and Enhance Their Interaction with TopBP1.
TopBP1 interacts with Treslin in a Cdk-dependent manner, and their interaction is critical for DNA replication initiation (14). Consistent with the report, transfected FLAG-TopBP1 associated with endogenous Treslin in actively growing HEK293 cells (Fig. 5A). We next investigated the effect of mutp53 on the interaction between TopBP1 and Treslin in H1299 cells. Indeed, mutp53-R273H enhanced their interaction (Fig. 5B, lanes 6 and 7). Previously, it has been shown that phosphorylation of Treslin by Cdk at Ser-1000 is required for its interaction with TopBP1 (14). Interestingly, although S1000A-Treslin failed to bind TopBP1 in the absence of mutp53, it bound TopBP1 very well in the presence of mutp53 (Fig. 5B, right two lanes). Moreover, two contact mutp53s, R273H and R248W, enhanced the TopBP1/Treslin interaction, whereas the other two conformational mutp53s, R175H and V143A, did not show any effect (Fig. 5 C and D). Consistently, the endogenous protein interaction between TopBP1 and Treslin was enhanced by expression of mutp53-R273H but not mutp53-R175H in H1299 cells (Fig. 5E). The differential effect of different mutp53 proteins on Treslin/TopBP1 binding is likely attributed to their ability to bind Treslin, as mutp53-R273H but not mutp53-R175H could coimmunoprecipitate with Treslin (Fig. 5F). Because WT p53 could bind Treslin like mutp53-R273H (SI Appendix, Fig. S12), it is possible that conformational mutp53 may lose a certain tertiary structure required for Treslin binding.
Fig. 5.
R273H mutp53 enhances TopBP1/Treslin interaction and bypasses the control of Cdk2 to promote G1/S phase progression. (A) HEK293 cells were transfected with an empty vector or the FLAG-TopBP1 expression vector. FLAG-TopBP1 was immunoprecipitated with anti-FLAG beads, followed by immunoblotting to detect the associated Treslin. (B) FLAG-TopBP1 was cotransfected with mutp53-R273H and either Myc-Treslin-WT or Myc-Treslin-S1000A in H1299 cells. FLAG-TopBP1 was immunoprecipitated with anti-FLAG beads, followed by immunoblotting to detect the associated Myc-Treslin. (C and D) FLAG-TopBP1 was cotransfected with Myc-Treslin and one of the mutp53 constructs in H1299 cells. Coimmunoprecipitation of FLAG-TopBP1 with Myc-Treslin was determined as described in B. (E) H1299 cells were transfected with an empty vector, mutp53-R273H, or mutp53-R175H. Endogenous TopBP1 was immunoprecipitated with anti-TopBP1 mouse monoclonal antibody or control mouse IgG, followed by immunoblotting with rabbit polyclonal antibody specific to Treslin or p53. (F) Myc-Treslin was coexpressed with either mutp53-R273H or mutp53-R175H in H1299 cells. Myc-Treslin was immunoprecipitated with anti-Myc (9E10) mouse monoclonal antibody, followed by immunoblotting with anti-p53 and anti-Myc rabbit polyclonal antibodies. (G) A scheme for cell-cycle synchronization is shown on Top. Cdk1i, Cdk1 inhibitor; Cdk2i, Cdk2 inhibitor. H1299 cells were transfected with mutp53-R273H or mutp53-R175H. Cells were synchronized by adding Cdk1i (Ro 3306, 2.5 μM) for 20 h and then were released to enter G1 phase by incubating cells in serum-containing medium without Ro 3306 for 12 h. Cells were subsequently treated with either Cdk2i (Cdk2 inhibitor II, 1 μM) or vehicle. Two hours later, BrdU incorporation assays were performed in the presence of Cdk2i or vehicle. At least 300 nuclei were counted for each sample. Representative images are presented in SI Appendix, Fig. S14. Data shown represent means ± SD from three biological replicates. Expression of mutp53 was verified by immunoblotting.
Contact but Not Conformational Mutp53 Can Promote DNA Replication in the Presence of a Cdk2 Inhibitor.
We next investigated whether mutp53 could bypass the requirement of Cdk2 in late G1 phase to enhance DNA replication. To address this, H1299 cells transfected with an empty vector, mutp53-R273H or mutp53-R175H, were synchronized at the G2/M phase border with a Cdk1 inhibitor (Cdk1i) for 20 h and then released to enter G1 phase by removing the Cdk1i. Most cells were in late G1 phase at 12 h after release and started to enter the S phase at 14 h (SI Appendix, Fig. S13). Furthermore, expression of mutp53 did not significantly affect the synchronization. After 12 h, while cells were in late G1 phase, a Cdk2 inhibitor (Cdk2i) or DMSO vehicle was added. Two hours later, BrdU incorporation was performed in the presence of a Cdk2i or DMSO vehicle for 17 h to measure the fraction of cells that transitioned through the S phase (Fig. 5G). As shown in Fig. 5G, expression of the contact mutp53-R273H (Fig. 5G, Left and SI Appendix, Fig. S14A), but not conformational mutp53-R175H (Fig. 5G, Right and SI Appendix, Fig. S14B), was able to promote DNA replication in H1299 cells despite the presence of a Cdk2i. This is not because mutp53-R273H could interfere with cell-cycle synchronization by Cdk1i, as mutp53-R273H did not affect cell-cycle arrest by Cdk1i (SI Appendix, Fig. S15). Moreover, mutp53-R273H was found to colocalize with PCNA, a marker for the replication forks, when cells entered S phase of the cell cycle (SI Appendix, Fig. S16), suggesting physical localization of mutp53-R273H to the replication forks during DNA replication. Some globular PCNA staining (in addition to punctuate staining) is likely due to its known nucleolar localization in human cancer cells during the early S phase (41–43).
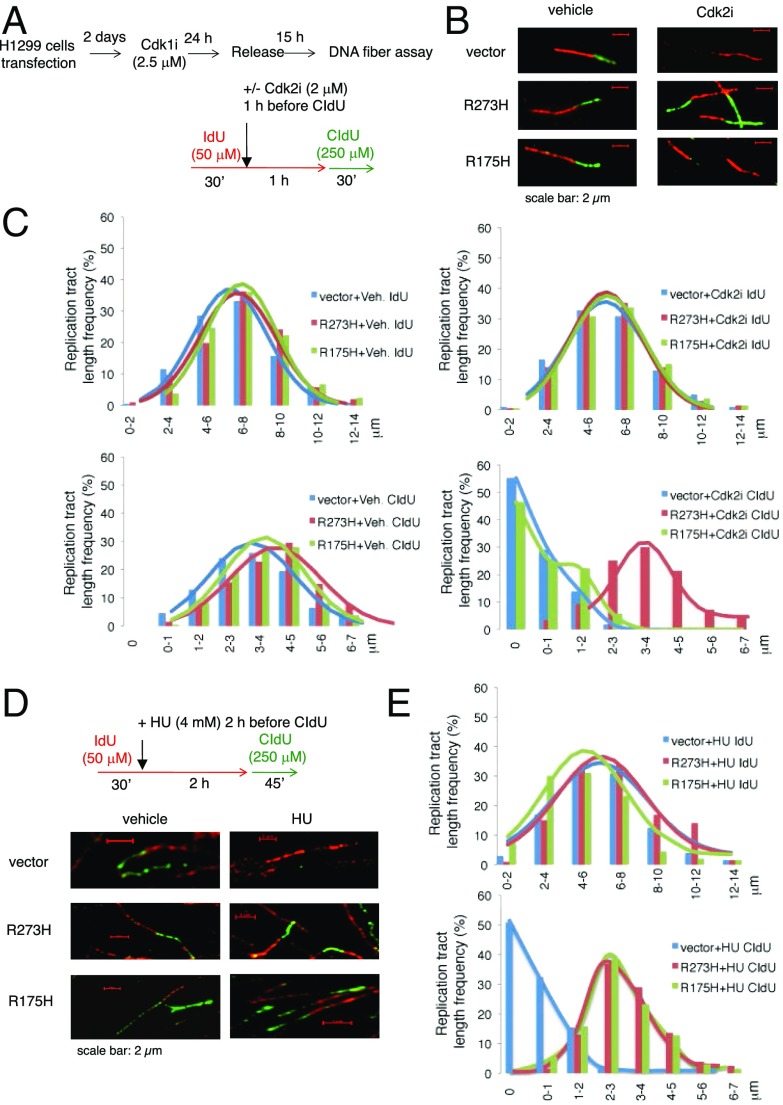
DNA Fiber Assay Confirms Perturbation of DNA Replication Checkpoint by Mutp53.
To further demonstrate the effect of mutp53 on DNA replication, we performed DNA fiber assay (44). We synchronized mutp53-transfected H1299 cells with Cdk1i, and when cells entered into the S phase, we performed sequential IdU/CIdU labeling with or without Cdk2i pretreatment before CIdU labeling (Fig. 6A). Indeed, mutp53-R273H, but not mutp53-R175H, overrode the inhibitory effect of Cdk2i and promoted CIdU incorporation (Fig. 6 B and C and SI Appendix, Fig. S17). An independent DNA fiber assay was performed in mutp53-transfected H1229 cells without synchronization that yielded the same conclusion (SI Appendix, Fig. S18). We also performed DNA fiber assay to evaluate the effect of both mutp53-R273H and mutp53-R175H on HU treatment. Consistent with the results obtained by p-Chk1 immunoblotting and BrdU incorporation assays (Fig. 1), expression of either mutp53-R273H or mutp53-R175H in H1299 cells mitigated the inhibition of HU on CIdU incorporation, as evidenced by continuous CIdU labeling on DNA fiber assay after HU treatment (Fig. 6 D and E and SI Appendix, Fig. S19).
Fig. 6.
DNA fiber analysis confirms perturbation of DNA replication control by mutp53. (A) The experimental scheme for DNA fiber analysis in B and C. H1299 cells were transfected with mutp53-R273H or mutp53-R175H. Cells were synchronized by Cdk1i and then released to enter G1 phase. At 15 h after release, cells were sequentially labeled with IdU and CIdU with treatment of Cdk2i or vehicle control 1 h preceding CIdU labeling. (B) Representative images of DNA spreading. More images are presented in SI Appendix, Fig. S17. (C) The lengths of IdU and CIdU tracts from DNA fibers were measured. Shown are tract-length distributions of IdU (Upper) and CIdU (Lower) in vehicle (Veh.) control groups (Left) and Cdk2i-treated groups (Right). More than 200 DNA fibers in each group were analyzed. P < 0.0001, comparing CIdU tract lengths between vector+Cdk2i and R273H+Cdk2i groups. (D) H1299 cells were transfected with an empty vector, mutp53-R273H, or mutp53-R175H. Two days later, cells were sequentially labeled with IdU and CIdU with HU treatment (4 mM) 2 h preceding CIdU labeling. The experimental scheme for DNA fiber analysis is shown on Top. Representative images of DNA fibers are shown on Bottom. More representative images are shown in SI Appendix, Fig. S19. (E) The lengths of IdU and CIdU tracts from DNA fibers in D were measured. Shown are tract-length distributions of IdU (Upper) and CIdU (Lower). More than 200 DNA fibers in each group were analyzed. P < 0.0001, comparing CIdU tract lengths between vector+HU and R273H+HU or R175H+HU groups.
Discussion
Mechanisms of Mutp53 GOF.
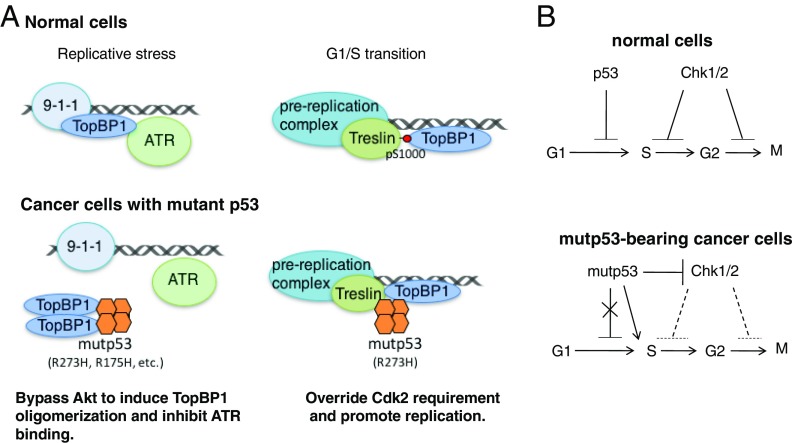
Although mutp53 GOF is well recognized, most of its known functions involve alterations in gene expression (8). Using the mutp53-R248W and mutp53-R273H knock-in mice (45), Song et al. show that these two mutp53 proteins interact with Mre11 to prevent the recruitment of Mre11–Rad50–Nbs1 (MRN) to double-stranded breaks, hence impairing ATM activation and promoting genome instability. In the present study, we elucidate two mechanisms by which mutp53 perturbs DNA replication control (Fig. 7A): (i) Many hotspot mutp53 proteins impair ATR activation by binding to TopBP1 and inducing its self-oligomerization, which is known to prevent TopBP1 checkpoint function. (ii) Some contact mutp53s may directly enhance DNA replication by facilitating the interaction of TopBP1 with Treslin, which is induced by Cdk2 under normal conditions.
Fig. 7.
A model for the mechanisms by which mutp53 perturbs TopBP1 checkpoint and replication functions in cancer cells. (A) In response to replicative stress (Left), TopBP1 plays a critical role in activating ATR in normal cells. On the contrary, in cancer cells, several mutp53s (including R273H, R175H, etc.) can bind TopBP1 and promote its oligomerization (which normally only occurs during Akt activation) to inhibit its ATR-activating function. During normal G1/S transition (Right), TopBP1 binds to Treslin in a Cdk2-dependent manner and promotes replication. However, some contact mutp53s, such as R273H, can override this Cdk2 requirement. (B) Mitigation of ATR/Chk1 activation (the current work) and ATM/Chk2 activation (45) by mutp53 and promotion of DNA replication by some contact mutp53s can lead to defects in all major cell-cycle checkpoints.
In response to genotoxic or replication stress, p53 and Chk1/2 are major mediators for halting cell-cycle progression: p53 mediates G1 checkpoint, whereas Chk1/2 are responsible for S and G2/M checkpoints (Fig. 7B, Upper). Our data suggest that mutations of p53 not only result in loss of G1 checkpoint control but also interfere with other checkpoints. Several hotspot mutp53 proteins can actively suppress the functions of Chk1 (the present study) and Chk2 (45), and some contact mutp53 proteins can directly promote S phase (Fig. 7B, Lower). Thus, mutations of TP53 can lead to the defects in all major cell-cycle checkpoints.
Many Hotspot Mutp53 Proteins Hijack the Akt-Dependent Regulatory Mechanism of TopBP1 to Inhibit ATR Checkpoint Activation.
There are several potential mechanisms by which mutp53 may inhibit TopBP1 checkpoint function. Mutp53-R175H or R273H does not affect TopBP1 phosphorylation at the Ser1159 residue (Fig. 3 J and K) but induces the oligomerization of WT TopBP1 as well as TopBP1-S1159A (Fig. 3F), even in the presence of an Akt inhibitor (Fig. 3G). Thus, mutp53 induces TopBP1 oligomerization through an Akt-independent mechanism. On the other hand, the p53-binding domains of TopBP1—that is, BRCT7/8—partly overlap with its ATR activation domain; thus, binding by mutp53 may block the ATR-activating function of TopBP1 through direct physical hindrance. However, even if this is the case, it is unlikely to be the primary mechanism, as it cannot account for our observation that mutp53 induces TopBP1 self-association and inhibits TopBP1 chromatin recruitment. Because mutp53 can form tetramers, it may serve as a bridge or a seed to promote TopBP1 oligomerization and therefore inhibit its checkpoint function. Indeed, this scenario is supported by the findings that mutp53 tetramerization is required for promoting TopBP1 oligomerization and inhibiting Chk1 activation (Fig. 3I and SI Appendix, Fig. S10).
Through inhibiting ATR/Chk1 activation in replication stress, mutp53 also blocks heterochromatin formation (Fig. 2). It should be noted that the chromatin response to HU treatment depends on the cellular p53 status. In WT p53 cancer cells, such as p53+/+ HCT116 cells and U2OS cells, H3K9me3 is reduced by HU treatment (SI Appendix, Fig. S20 A and B), probably due to either down-regulation of H3K9 methylase SUV39H1 (46, 47) or induction of H3K9 demethylase Jumonji domain 2 family demethylase (JMJD2b) (47). On the contrary, in p53-null tumor cells, such as H1299 cells and p53−/− HCT116 cells, H3K9me3 is induced by HU (Fig. 2 A–D and SI Appendix, Fig. S20A). However, the induction of H3K9me3 can be inhibited by mutp53 (Fig. 2C). Hence, in mutp53-bearing cancer cells, the H3K9me3 level remains relatively unchanged after HU treatment (Fig. 2 B–D and SI Appendix, Fig. S20 B and C) but became inducible by HU upon depletion of mutp53 (SI Appendix, Fig. S20C). Intriguingly, depletion of mutp53 reduced basal H3K9me3 in C33A and BT549 cells. High levels of H3K9me3 in cancer have been associated with poor survival (48), lymph node metastasis (49), and high tumor grade (50). Whether mutp53 directly regulates basal H3K9me3 deserves further investigation. Consistently, in p53-null H1299 cells, the level of H3K9me3 was increased by HU but was reduced by HU when expressing WT p53 and was not significantly altered by HU when expressing mutp53-R273H (SI Appendix, Fig. S20D). We note that overexpression of WT p53 to a level similar to or slightly higher than that of mutp53-R273H also attenuated HU-induced Chk1 phosphorylation, although at a lesser degree than mutp53 (SI Appendix, Fig. S20D). This is not totally surprising, as WT p53 can bind TopBP1 (24) and hence may affect TopBP1 checkpoint function as well if expressed to a high level. The accumulated p53 may contribute to tuning down Chk1 activation during the recovery phase, as suggested by a slower declining of p-Chk1 signal in p53−/− HCT116 compared with p53+/+ HCT116 cells (SI Appendix, Fig. S20E). The slight decrease of the Chk1 protein level in post-HU treatment p53+/+ HCT116 cells (but not in p53−/− cells) is consistent with the established role of p53 in down-regulating Chk1 expression (51). Further supporting our results is the observation of an inverse correlation between p53 and Chk1 phosphorylation levels in a UVB-induced mouse skin cancer model (52). The finding prompted the authors to propose that WT p53 might switch off Chk1 phosphorylation, although the mechanism was unknown. Our results suggest that stabilized WT p53 uses a similar mechanism that we elucidate for mutp53 to inhibit ATR/Chk1 and tune down checkpoint activation once stress is relieved. Mutp53 proteins make use of the existing feedback control mechanism, but due to their highly accumulated levels, they constitutively dampen ATR/Chk1 response in cancer cells. As many mutp53 proteins, including R273H and several hotspot mutp53s used in this study, can form aggregates (53–55), it will be interesting to investigate whether this property contributes to TopBP1 oligomerization in the future.
Contact Mutp53 Facilitates the Interaction Between TopBP1 and Treslin to Promote DNA Replication.
Although several hotspot mutp53 proteins bind TopBP1 (10) and inhibit ATR checkpoint function (Fig. 1), only contact, but not conformational, mutp53s can bind Treslin and induce the association of TopBP1 with Treslin (Fig. 5). As a result, only contact mutp53s (such as mutp53-R273H) but not conformational mutp53s (such as mutp53-R175H) can promote DNA replication even in the presence of Cdk2i (Figs. 5G and 6 and SI Appendix, Fig. S18). The molecular basis underlying the difference between contact and conformational mutp53s is unclear at the moment and deserves future investigation. It is tempting to suggest that contact mutp53 preserves the overall structure, which may be required for Treslin interaction (SI Appendix, Fig. S12). Alternatively, mutp53-R273H has been suggested to drive changes in chromatin conformation, as it can associate with chromatin and stabilize chromatin-associated PCNA and MCM4 (56).
The Interference of ATR Function by Mutp53 Presents an Opportunity for Synthetic Lethality Strategy to Treat Mutp53-Bearing Cancers.
Unlike the ATM–Chk2 axis, the ATR–Chk1 axis is required for cell survival due to its essential role in replication stress. Thus, a partially defective ATR function in mutp53-bearing cancer cells may present an opportunity to further cripple ATR function by inhibiting another ATR activator, DNA2. Indeed, H1299 cells with mutp53 expression and DNA2 depletion have a severely defective ATR function and are defective in clonogenic survival assay (Fig. 4 A and B). More importantly, although expression of mutp53-R273H in a p53-null cancer cell line increases chemoresistance, it paradoxically enhances the chemosensitivity when DNA2 is depleted (Fig. 4C). Moreover, expression of mutp53-R273H also renders cancer cells more sensitive to DNA2 inhibitors (Fig. 4 D and E). DNA2 is overexpressed in many human cancers (57), particularly when TP53 contains missense mutations (Fig. 4F). Hence, DNA2 may be a cancer therapeutic target, especially in mutp53-harboring cancers. Given the mechanisms of mutp53 GOF elucidated in this study, targeting DNA2 may prove to be an efficient synthetic lethality strategy to treat cancers harboring TP53 missense mutations.
Materials and Methods
Immunoprecipitation and Western Blot Analysis.
Transfected cells were harvested in TNN buffer as described previously (58). Immunoprecipitation was performed by incubating cell lysates with appropriate antibodies or anti-FLAG M2 monoclonal antibody-conjugated agarose beads (Sigma) for 3–16 h at 4 °C. After three washes, immunoprecipitates were fractionated by SDS/PAGE and electrotransferred to Imobilon-P membrane (Millipore). Equal protein loading was verified with Ponceau S staining. Immunoblotting was performed with the appropriate antibody. Sources of antibodies are presented in SI Appendix, Materials and Methods.
Chromatin Binding Assay.
Chromatin binding assay was performed as described previously (28). Briefly, transfected cells were trypsinized and incubated on ice for 5 min in 150 µL of fractionation buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM EDTA) containing 0.2% Nonidet P-40 and protease inhibitors. Following centrifugation at 1,000 g for 5 min, the supernatant was collected (fraction I), and pellets were washed with the same buffer. After centrifugation, the supernatant was collected (fraction II), and the nuclear pellets were further incubated in 150 µL of fractionation buffer containing 0.5% Nonidet P-40 on ice for 40 min. The extracts were clarified by centrifugation at 16,000 g for 15 min (fraction III). The pellets were finally lysed in 150 µL of 10% SDS/PAGE sample buffer and boiled for 5 min (fraction IV). Equal aliquots of each fraction, derived from equivalent cell numbers, were separated on 10% SDS/PAGE for Western blot analysis.
Replication Labeling and DNA Fiber Spread Assay.
H1299 growing cells in a different experimental design were labeled with 50 µM IdU for 30 min. Cells were then treated with 2 µM Cdk2 inhibitor II for 1 h (for G1/S phase cell-cycle study) or 4 mM HU for 2 h (for DNA replication stress study). Then, IdU was removed, and the cells were washed with PBS and incubated with 250 µM CIdU for 30 min or 45 min, respectively. DNA spread assay was performed as previously described (44) with some modification. A detailed description of DNA fiber spread assay is presented in SI Appendix, Materials and Methods.
Establishment of stable cell lines, antibodies for immunoprecipitation and Western blot analysis, micrococcal nuclease assay, bromodeoxyuridine incorporation assay and flow cytometry, immunofluorescence staining, GST pulldown assay, MTT assay, clonogenic survival assay, DNA fiber spread assay, and statistical analysis are discussed in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank William Dunphy for the Treslin cDNA construct, Sheila Stewart for DNA2 shRNA constructs, Bob Weinberg and Todd Waldman for p53 shRNA constructs, Bert Vogelstein for p53+/+ and p53−/− HCT116 cells, and NCI Developmental Therapeutic Program for NSC compounds. This work was supported by NIH Grants RO1CA100857 and RO1CA138641 (to W.-C.L.) and RO1CA203824 (to W.-C.L. and F.-T.L.), Department of Defense Grants W81XWH-14-1-0339 and W81XWH-14-1-0306 (to W.-C.L. and F.-T.L.), and Rivkin Center for Ovarian Cancer Pilot Award (to W.-C.L.). J.D.G. is supported by T32 Fellowship T32DK060445. We also acknowledge support from the Cytometry and Cell Sorting Core (NIAID Grants P30AI036211, NCI P30CA125123, and NCRR S10RR024574) at Baylor College of Medicine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619832114/-/DCSupplemental.
References
- 1.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous; Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous; Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.George J, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anonymous; Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anonymous; Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freed-Pastor WA, Prives C. Mutant p53: One name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 9.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2:a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu K, Ling S, Lin WC. TopBP1 mediates mutant p53 gain of function through NF-Y and p63/p73. Mol Cell Biol. 2011;31:4464–4481. doi: 10.1128/MCB.05574-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wardlaw CP, Carr AM, Oliver AW. TopBP1: A BRCT-scaffold protein functioning in multiple cellular pathways. DNA Repair (Amst) 2014;22:165–174. doi: 10.1016/j.dnarep.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 2010;140:349–359. doi: 10.1016/j.cell.2009.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sansam CL, et al. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev. 2010;24:183–194. doi: 10.1101/gad.1860310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Direct regulation of Treslin by cyclin-dependent kinase is essential for the onset of DNA replication. J Cell Biol. 2011;193:995–1007. doi: 10.1083/jcb.201102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan S, Michael WM. TopBP1 and DNA polymerase alpha-mediated recruitment of the 9-1-1 complex to stalled replication forks: Implications for a replication restart-based mechanism for ATR checkpoint activation. Cell Cycle. 2009;8:2877–2884. doi: 10.4161/cc.8.18.9485. [DOI] [PubMed] [Google Scholar]
- 16.Yan S, Michael WM. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. J Cell Biol. 2009;184:793–804. doi: 10.1083/jcb.200810185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 19.Thangavel S, et al. DNA2 drives processing and restart of reversed replication forks in human cells. J Cell Biol. 2015;208:545–562. doi: 10.1083/jcb.201406100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wanrooij PH, Burgers PM. Yet another job for Dna2: Checkpoint activation. DNA Repair (Amst) 2015;32:17–23. doi: 10.1016/j.dnarep.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu K, Lin FT, Ruppert JM, Lin WC. Regulation of E2F1 by BRCT domain-containing protein TopBP1. Mol Cell Biol. 2003;23:3287–3304. doi: 10.1128/MCB.23.9.3287-3304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu K, Luo Y, Lin FT, Lin WC. TopBP1 recruits Brg1/Brm to repress E2F1-induced apoptosis, a novel pRb-independent and E2F1-specific control for cell survival. Genes Dev. 2004;18:673–686. doi: 10.1101/gad.1180204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu K, Paik JC, Wang B, Lin FT, Lin WC. Regulation of TopBP1 oligomerization by Akt/PKB for cell survival. EMBO J. 2006;25:4795–4807. doi: 10.1038/sj.emboj.7601355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu K, et al. Regulation of p53 by TopBP1: A potential mechanism for p53 inactivation in cancer. Mol Cell Biol. 2009;29:2673–2693. doi: 10.1128/MCB.01140-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herold S, et al. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell. 2002;10:509–521. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 26.Sjøttem E, et al. The ePHD protein SPBP interacts with TopBP1 and together they co-operate to stimulate Ets1-mediated transcription. Nucleic Acids Res. 2007;35:6648–6662. doi: 10.1093/nar/gkm739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshida K, Inoue I. Expression of MCM10 and TopBP1 is regulated by cell proliferation and UV irradiation via the E2F transcription factor. Oncogene. 2004;23:6250–6260. doi: 10.1038/sj.onc.1207829. [DOI] [PubMed] [Google Scholar]
- 28.Liu K, Graves JD, Scott JD, Li R, Lin WC. Akt switches TopBP1 function from checkpoint activation to transcriptional regulation through phosphoserine binding-mediated oligomerization. Mol Cell Biol. 2013;33:4685–4700. doi: 10.1128/MCB.00373-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cortez D, Glick G, Elledge SJ. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc Natl Acad Sci USA. 2004;101:10078–10083. doi: 10.1073/pnas.0403410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoo HY, Shevchenko A, Shevchenko A, Dunphy WG. Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. J Biol Chem. 2004;279:53353–53364. doi: 10.1074/jbc.M408026200. [DOI] [PubMed] [Google Scholar]
- 31.Di Micco R, et al. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat Cell Biol. 2011;13:292–302. doi: 10.1038/ncb2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivanov A, Adams PD. A damage limitation exercise. Nat Cell Biol. 2011;13:193–195. doi: 10.1038/ncb0311-193. [DOI] [PubMed] [Google Scholar]
- 33.Carey M, Smale ST. Micrococcal nuclease-southern blot assay: I. MNase and restriction digestions. CSH Protoc. 2007;2007:pdb prot4890. doi: 10.1101/pdb.prot4890. [DOI] [PubMed] [Google Scholar]
- 34.Andegeko Y, et al. Nuclear retention of ATM at sites of DNA double strand breaks. J Biol Chem. 2001;276:38224–38230. doi: 10.1074/jbc.M102986200. [DOI] [PubMed] [Google Scholar]
- 35.Friedman PN, Chen X, Bargonetti J, Prives C. The p53 protein is an unusually shaped tetramer that binds directly to DNA. Proc Natl Acad Sci USA. 1993;90:3319–3323. doi: 10.1073/pnas.90.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Q, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 38.Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;21:1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strano S, et al. Physical interaction with human tumor-derived p53 mutants inhibits p63 activities. J Biol Chem. 2002;277:18817–18826. doi: 10.1074/jbc.M201405200. [DOI] [PubMed] [Google Scholar]
- 40.Liu W, et al. A selective small molecule DNA2 inhibitor for sensitization of human cancer cells to chemotherapy. EBioMedicine. 2016;6:73–86. doi: 10.1016/j.ebiom.2016.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andersen JS, et al. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 42.Chan PK, et al. Indirect immunofluorescence studies of proliferating cell nuclear antigen in nucleoli of human tumor and normal tissues. Cancer Res. 1983;43:3770–3777. [PubMed] [Google Scholar]
- 43.Smetana K, Gyorkey F, Chan PK, Tan E, Busch H. Proliferating cell nuclear antigen (PCNA) and human malignant tumor nucleolar antigens (HMTNA) in nucleoli of human hematological malignancies. Blut. 1983;46:133–141. doi: 10.1007/BF00320271. [DOI] [PubMed] [Google Scholar]
- 44.Nieminuszczy J, Schwab RA, Niedzwiedz W. The DNA fibre technique - Tracking helicases at work. Methods. 2016;108:92–98. doi: 10.1016/j.ymeth.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 45.Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol. 2007;9:573–580. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- 46.Mungamuri SK, et al. p53-mediated heterochromatin reorganization regulates its cell fate decisions. Nat Struct Mol Biol. 2012;19:478–484, S471. doi: 10.1038/nsmb.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng H, Chen L, Pledger WJ, Fang J, Chen J. p53 promotes repair of heterochromatin DNA by regulating JMJD2b and SUV39H1 expression. Oncogene. 2014;33:734–744. doi: 10.1038/onc.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia R, et al. High expression of H3K9me3 is a strong predictor of poor survival in patients with salivary adenoid cystic carcinoma. Arch Pathol Lab Med. 2013;137:1761–1769. doi: 10.5858/arpa.2012-0704-OA. [DOI] [PubMed] [Google Scholar]
- 49.Yokoyama Y, et al. Cancer-associated upregulation of histone H3 lysine 9 trimethylation promotes cell motility in vitro and drives tumor formation in vivo. Cancer Sci. 2013;104:889–895. doi: 10.1111/cas.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keung EZ, et al. Increased H3K9me3 drives dedifferentiated phenotype via KLF6 repression in liposarcoma. J Clin Invest. 2015;125:2965–2978. doi: 10.1172/JCI77976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gottifredi V, Karni-Schmidt O, Shieh SS, Prives C. p53 down-regulates CHK1 through p21 and the retinoblastoma protein. Mol Cell Biol. 2001;21:1066–1076. doi: 10.1128/MCB.21.4.1066-1076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bernard JJ, et al. Inverse relationship between p53 and phospho-Chk1 (Ser317) protein expression in UVB-induced skin tumors in SKH-1 mice. Exp Mol Pathol. 2014;96:126–131. doi: 10.1016/j.yexmp.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ano Bom AP, et al. Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: Implications for cancer. J Biol Chem. 2012;287:28152–28162. doi: 10.1074/jbc.M112.340638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levy CB, et al. Co-localization of mutant p53 and amyloid-like protein aggregates in breast tumors. Int J Biochem Cell Biol. 2011;43:60–64. doi: 10.1016/j.biocel.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 55.Xu J, et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol. 2011;7:285–295. doi: 10.1038/nchembio.546. [DOI] [PubMed] [Google Scholar]
- 56.Polotskaia A, et al. Proteome-wide analysis of mutant p53 targets in breast cancer identifies new levels of gain-of-function that influence PARP, PCNA, and MCM4. Proc Natl Acad Sci USA. 2015;112:E1220–E1229. doi: 10.1073/pnas.1416318112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng G, et al. Human nuclease/helicase DNA2 alleviates replication stress by promoting DNA end resection. Cancer Res. 2012;72:2802–2813. doi: 10.1158/0008-5472.CAN-11-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chowdhury P, et al. Targeting TopBP1 at a convergent point of multiple oncogenic pathways for cancer therapy. Nat Commun. 2014;5:5476. doi: 10.1038/ncomms6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.