Significance
MyTH4-FERM myosins (Myo7a, Myo7b, and Myo15a) regulate actin-bundle protrusion structures in various tissues, including brush border microvilli of intestines and stereocilia of inner ear hair cells. Mutations of the cargo binding MyTH4-FERM tandems of these myosins are frequently associated with human diseases, including hearing loss, vision defects, and digestive disorders, but with poorly understood mechanisms. In this work, we present the high-resolution crystal structure of Myo7b C-terminal MyTH4-FERM tandem (CMF) in complex with the USH1C PDZ3 domain. The structure, together with biochemical studies, indicates that binding to PDZ domain scaffold proteins is a general property of these myosins. The complex structure reported here also helps to explain why numerous mutations identified in Myo7a CMF can cause deafness and blindness in humans.
Keywords: myo7a, microvilli, stereocilia, Harmonin, Usher syndrome
Abstract
Unconventional myosin 7a (Myo7a), myosin 7b (Myo7b), and myosin 15a (Myo15a) all contain MyTH4-FERM domains (myosin tail homology 4-band 4.1, ezrin, radixin, moesin; MF) in their cargo binding tails and are essential for the growth and function of microvilli and stereocilia. Numerous mutations have been identified in the MyTH4-FERM tandems of these myosins in patients suffering visual and hearing impairment. Although a number of MF domain binding partners have been identified, the molecular basis of interactions with the C-terminal MF domain (CMF) of these myosins remains poorly understood. Here we report the high-resolution crystal structure of Myo7b CMF in complex with the extended PDZ3 domain of USH1C (a.k.a., Harmonin), revealing a previously uncharacterized interaction mode both for MyTH4-FERM tandems and for PDZ domains. We predicted, based on the structure of the Myo7b CMF/USH1C PDZ3 complex, and verified that Myo7a CMF also binds to USH1C PDZ3 using a similar mode. The structure of the Myo7b CMF/USH1C PDZ complex provides mechanistic explanations for >20 deafness-causing mutations in Myo7a CMF. Taken together, these findings suggest that binding to PDZ domains, such as those from USH1C, PDZD7, and Whirlin, is a common property of CMFs of Myo7a, Myo7b, and Myo15a.
Microvilli and stereocilia are both actin bundle-based, finger-like protrusions found on apical surfaces of many epithelial cells (1). Microvilli line the apical surface of epithelial cells forming a densely packed structure known as the brush border in tube-like tissues, such as intestines, kidney, and lung (2–5). The best known function of microvilli is to massively increase membrane surface area of these tissues to promote solute exchange, although these protrusions may also allow cells to communicate with their extracellular surroundings (6). Stereocilia, on the other hand, are composed of several rows of protrusions with graded height forming a staircase-like structure on the apical surface of inner ear hair cells, which collectively function to sense sound waves (7, 8). Despite clear morphological and functional differences, microvilli and stereocilia share certain features at the molecular level. For example, the packing and organization of microvilli and stereocilia both rely on homologous cadherin-based tip-link complexes that target to the distal ends of these protrusions (9, 10). Additionally, a set of homologous unconventional myosins—including Myo1, Myo3, Myo6, and Myo7—are shared by and critical for the development, maintenance, and functions of microvilli and stereocilia (1, 11, 12).
This study focuses on Myo7b, an unconventional myosin enriched in the microvilli of transporting epithelial cells (11, 13). Myo7b is characterized by a pair of MyTH4-FERM tandems in its tail cargo-binding domain. In addition to Myo7b, Myo7a, Myo10, and Myo15a also contain one or two MyTH4-FERM tandems in their tail domains (14). A common property of MyTH4-FERM myosins is their involvement in the formation or elongation/stabilization of actin-bundle supported structures, including filopodia, microvilli, and stereocilia (15). This property first appeared in ancient MyTH4-FERM myosins and is conserved over 1 billion y of evolution (14, 16, 17). Mutations of MyTH4-FERM myosins cause a broad spectrum of human diseases, including hearing loss, vision defects, digestive disorders, and cancers (18–21), often because of defects in actin bundle-supported protrusions (i.e., filopodia, microvilli, or stereocilia). The specific functions of Myo7a, Myo7b, Myo10, and Myo15a in regulating actin-bundle protrusion structures in various tissues must be a result of, at least in part, the unique cargo binding properties of their MyTH4-FERM domains. For example, the N-terminal MyTH4-FERM tandem (NMF) of Myo7a binds to the central domain (CEN) of USH1G (a.k.a., Sans) with very high affinity (22). Mutations of Myo7a or USH1G that weaken or disrupt the Myo7a/USH1G interaction can cause stereocilia deformation and hearing impairment (22–24). The Myo7b NMF binds to the central region of ANKS4B, which shares high homology with USH1G with a similar structural mechanism as the Myo7a/USH1G interaction (25, 26). The C-terminal MyTH4-FERM tandem (CMF) of Myo7b has been shown to bind to the C-terminal PDZ domains of USH1C (10, 25, 26). The direct binding of USH1C PDZ domains to Myo7b CMF is highly unexpected, as Myo7b CMF does not contain any recognizable C-terminal or internal PDZ-binding motif (PBM) (27). Additionally, binding of one or a pair of PDZ domains to a MyTH4-FERM tandem is also unique with respect to all previously characterized MyTH4-FERM/target interactions (22, 26, 28–30). Elucidating the binding mechanism for the Myo7b CMF/USH1C PDZ interaction will not only be helpful for understanding the role of Myo7b in intermicrovillar adhesion complex (IMAC) assembly but also provide valuable insights into possible interactions between MyTH4-FERM tandems from Myo7a to PDZ domains of PDZD7 and USH1C or potential interactions between Myo15a MyTH4-FERM and the PDZ domains of Whirlin.
We performed a detailed biochemical characterization of the interaction between Myo7b CMF and USH1C, and solved the high-resolution crystal structure of Myo7b CMF in complex with the extended PDZ3 domain of USH1C. The complex structure reveals previously uncharacterized interaction modes both for MyTH4-FERM tandems and for PDZ domains. The Myo7b CMF/USH1C PDZ3 complex structure, together with direct biochemical binding experiments, also helps to explain why numerous mutations in the Myo7a CMF may impair its function and thus lead to hearing and vision impairments in patients. Our findings provide a structural framework for future studies of CMFs in Myo7a, Myo7b, and Myo15a.
Results
The Overall Structure of the Myo7b CMF/USH1C PDZ3 Complex.
Our earlier study has shown that Myo7b CMF directly binds to USH1C and the interaction is mediated by Myo7b CMF and the extended PDZ3 domain of USH1C (26) (Fig. 1A). To elucidate the molecular basis governing this unexpected interaction mode, we obtained high-quality crystals of the Myo7b CMF/USH1C PDZ3 complex that diffracted to 1.8-Å resolution and solved the complex structure by a single-wavelength anomalous dispersion method using gold derivatives (Table 1).
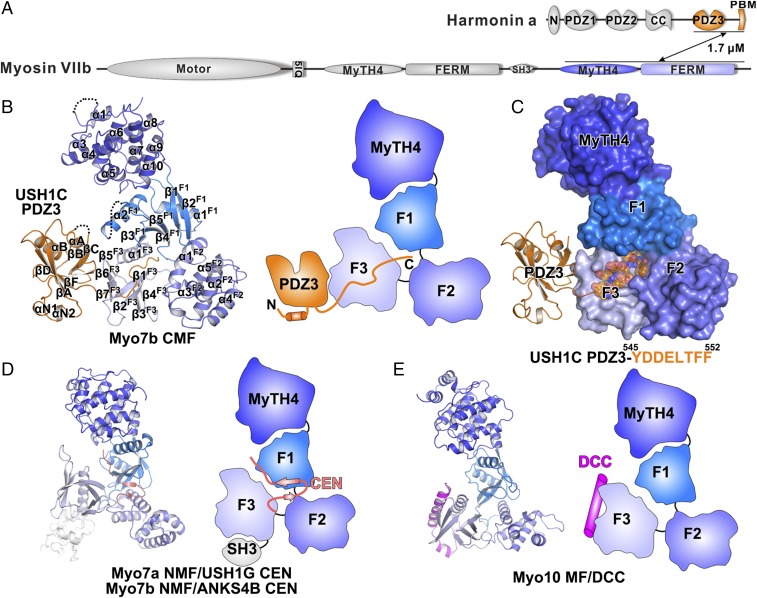
Fig. 1.
Structure of Myo7b CMF/USH1C PDZ3 complex. (A) Schematic diagrams showing the domain architectures of Myo7b and USH1C. The interaction is mediated by Myo7b CMF and USH1C PDZ3. The color coding of the domains is kept throughout the report. (B) Ribbon representation (Left) and schematic cartoon diagram (Right) of the Myo7b CMF/USH1C PDZ3 complex structure. The disordered loops are drawn as dashed lines in the ribbon representation. (C) Surface representation of the Myo7b CMF structure showing the central pocket. The USH1C PDZ3 domain is shown as ribbon, and its C-terminal tail is shown as stick and transparent spheres. The amino acid sequence of the USH1C C-terminal tail is shown below the surface diagram. (D) Ribbon representation (Left) and schematic cartoon diagram (Right) of the Myo7a NMF/USH1G CEN and Myo7b NMF/ANKS4B CEN complexes. (E) Ribbon representation (Left) and schematic cartoon diagrams (Right) of the Myo10 MF/DCC complex.
Table 1.
Statistics of X-ray crystallographic data collection and model refinement
| Data collection and refinement | Native | Au-derivative |
| Data collection | ||
| Wavelength (Å) | 0.9787 | 0.9783 |
| Space group | P21 | P21 |
| Unit cell parameters (Å) | a = 69.6, b = 42.5, c = 118.4; α = γ = 90°, β = 97.5° | a = 70.2, b = 41.9, c = 119.4; α = γ =90°, β = 98.3° |
| Resolution range (Å) | 50–1.8 (1.83–1.80) | 50–2.3 (2.34–2.30) |
| No. of unique reflections | 63,640 (3,196) | 30,149 (1,513) |
| Redundancy | 3.0 (3.0) | 12.2 (12.5) |
| I/σ | 25.9 (2.3) | 21.5 (2.5) |
| Completeness (%) | 99.1 (99.8) | 97.7 (98.4) |
| Rmerge* (%) | 5.9 (49.2) | 9.5 (75.3) |
| Structure refinement | ||
| Resolution (Å) | 50–1.8 (1.83–1.80) | |
| Rcryst†/Rfree‡ (%) | 18.71/21.07 (27.15/30.03) | |
| Rmsd bonds (Å)/angles (°) | 0.011/0.997 | |
| Average B factor§ | 37.5 | |
| No. of atoms | ||
| Protein atoms | 4,617 | |
| Ligands | 31 | |
| Water | 265 | |
| No. of reflections | ||
| Working set | 60,479 (2,956) | |
| Test set | 3,147 (174) | |
| Ramachandran plot regions§ | ||
| Favored (%) | 98.1 | |
| Allowed (%) | 1.9 | |
| Outliers (%) | 0 | |
Numbers in parentheses represent the value for the highest resolution shell.
Rmerge = Σ|Ii - <I>|/ΣIi, where Ii is the intensity of measured reflection and <I> is the mean intensity of all symmetry-related reflections.
Rcryst = Σ||Fcalc| – |Fobs||/ΣFobs, where Fobs and Fcalc are observed and calculated structure factors.
Rfree = ΣT||Fcalc| – |Fobs||/ΣFobs, where T is a test dataset of about 5% of the total unique reflections randomly chosen and set aside before refinement.
B factors and Ramachandran plot statistics are calculated using MOLPROBITY (54).
The overall structure of the Myo7b CMF/USH1C PDZ3 complex reveals previously uncharacterized target-binding modes for MyTH4-FERM tandems as well as for PDZ domains (Fig. 1B). The overall architecture of Myo7b CMF is similar to the structures of other MyTH4-FERM tandems (17, 22, 26, 28, 29), which all contain an N-terminal helical MyTH4 domain and a C-terminal clover-leaf–shaped FERM domain. The USH1C PDZ3 domain in the complex contains a canonical PDZ fold plus an N-terminal extension composed of two short α-helices and an extended C-terminal tail (Fig. 1B). Instead of using its canonical PBM binding αB/βB-groove, the USH1C PDZ3 uses an extended surface at the back of the PBM binding groove to interact with the F3 lobe of Myo7b CMF. The extended C terminus of USH1C PDZ3, which contains a putative PBM sequence (-LTFF), inserts into the central pocket formed by the three lobes of the FERM domain of Myo7b CMF with matching shape and charge complementation (Fig. 1 B and C, and see below). The USH1C PDZ3 binding mode of Myo7b CMF is significantly different from the target binding modes of other MyTH4-FERM tandems reported in the Myo7a/USH1G, Myo7b/ANKS4B, and Myo10/DCC complexes (Fig. 1 D and E) (22, 26, 28, 29).
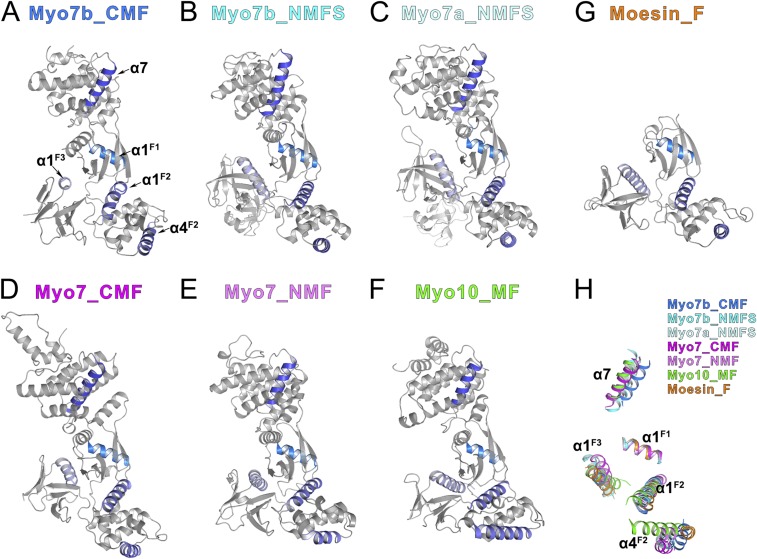
There are noticeable differences between the Myo7b CMF structure and all of the rest of MyTH4-FERM structures solved to date. Such large differences presumably prevented us from solving the Myo7b CMF structure using the molecular replacement method with currently available models. We performed a structural analysis of more than a handful of the different MyTH4-FERM structures solved to date (Fig. S1). By aligning the F1 lobes of all of the MyTH4-FERM structures (17, 22, 26, 29), as well as the FERM domain of Moesin (which is a founding member of the FERM domain) (31), it is clear that the relative orientations of the three lobes of each FERM domain are all different from each other (Fig. S1 and legend for details), indicating that the three lobes of FERM domains have considerable conformational freedoms. Such interlobe conformational differences are likely to be important for the target bindings of FERM domains, as the central interlobe interfaces are used as target binding sites of FERM domains from MyTH4-FERM tandems (22, 26).
Fig. S1.
Structural comparison of all of the MyTH4-FERM structures showing the relative rotations of the three lobes. (A–G) Ribbon diagrams showing all of the MyTH4-FERM structures solved up to date as well as the founding member Moesin FERM domain. (A) Myo7b CMF, this study; (B) Myo7b NMF, PDB ID code: 5F3Y; (C) Myo7a NMF, PDB ID code: 3PVL; (D) Dictyostelium Myo7 CMF, PDB ID code: 5EJR; (E) Dictyostelium Myo7 NMF, PDB ID code: 5EJY; (F) Myo10 MF, PDB ID code: 3PZD; (G) Moesin FERM domain, PDB ID code: 1EF1. All of the structures are shown in the same orientation by aligning the F1 lobes together. The α7 in the MyTH4 domain, α1 in the F1 lobe, α1/α4 in the F2 lobe, and α1 in the F3 lobe are highlighted in color to show the relative orientations of each domains. (H) Superposition of the five helices highlighted in A–F showing the orientation differences of the MyTH4-FERM tandems using the F1 lobe α1 (α1F1) helix as the reference.
The Detailed Interactions Between Myo7b CMF and USH1C PDZ3.
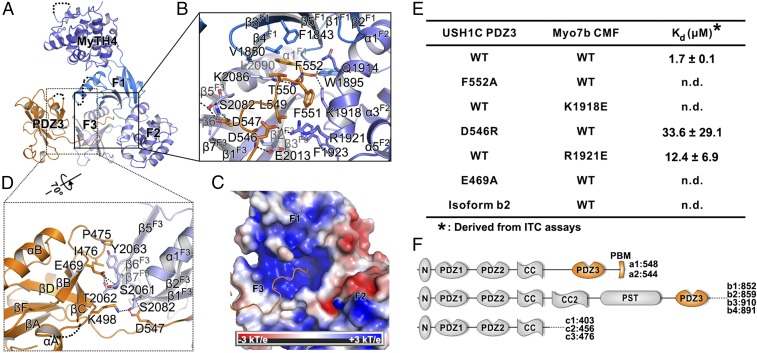
In the Myo7b CMF/USH1C PDZ3 complex, the binding interface can be divided into two parts (Fig. 2A). The C-terminal extension of USH1C PDZ3 inserts into the central pocket of the FERM clover-leaf formed by the F1_β4/β5, F2_α1, F3_β1/α1, and F1/F2_linker (Fig. 2B). The USH1C carboxyl group on the C-terminal residue Phe552 forms charge–charge interaction with Lys1918F2 as well as hydrogen bonds with Gln1914F2 and Trp1895F1/F2_linker (Fig. 2 B and C). Isothermal titration calorimetry (ITC) assays were used to evaluate the mutational effect of the critical residues (Fig. 2E). Substitution of the Lys1918F2 with Glu totally disrupted the binding (Fig. 2E). The two Phe residues upstream also make extensive hydrophobic interactions with Phe1843F1, Lys2086F3, Lys1918F2, and Phe1923F2 (Fig. 2B). Mutation of the C-terminal Phe552USH1C to Ala also abolished the binding (Fig. 2E). Additionally, a pair of salt bridges between Arg1921F2 and Asp546PDZ3 further contributes to the interaction (Fig. 2B). Reverse-charged mutations of each of these two residues, namely Arg1921Glu or Asp546Arg, significantly weakened the binding (Fig. 2E). The extensive charge–charge and hydrophobic interactions between USH1C tail and the central pocket of the FERM domain of Myo7b CMF are consistent with our previous result that the C-terminal extension of USH1C PDZ3 is essential for the Myo7b CMF and USH1C interaction (26). The canonical PDZ fold of USH1C PDZ3 uses its βB/βC-loop to interact with Myo7b CMF F3 lobe (Fig. 2A). In particular, Glu469 in the PDZ3 βB/βC-loop forms several hydrogen bonds with the sidechain of Ser2061F3 and the sidechain and backbone amide of Thr2062F3 (Fig. 2D). Fittingly, mutation of the Glu469PDZ3 to Ala also abolished the binding between USH1C PDZ3 and Myo7b CMF (Fig. 2E). Additionally, hydrophobic interactions formed by Pro475PDZ3, Ile476PDZ3 and Tyr2063F3 further stabilize the PDZ3/CMF interface (Fig. 2D). The N-terminal extension of USH1C PDZ3 is not involved in the Myo7b CMF binding. The weakening of the USH1C PDZ3 and Myo7b CMF interaction induced by truncating the PDZ3 N-terminal extension is likely a result of overall structural stability change of PDZ3. Therefore, the USH1C PDZ3 represents another example of an extended PDZ domain, the function of which requires additional sequences outside the canonical PDZ fold (32). Taken together, the above structural and biochemical results suggest that the two interfaces between USH1C PDZ3 and CMF Myo7b act synergistically to form the stable complex. The interaction between PDZ3 and F3 may help to position the PDZ3 C-terminal extension for inserting into the central pocket of the FERM domain, which in return stabilizes the PDZ3/F3 interaction mediated by only a few hydrogen bonds and a small hydrophobic interface. This analysis is consistent with our previous biochemical characterization that neither the C-terminal extension alone nor the PDZ3 without the C-terminal extension can efficiently interact with Myo7b CMF (26).
Fig. 2.
Detailed interaction between Myo7b CMF and USH1C PDZ3. (A) Ribbon representation of the Myo7b CMF/USH1C PDZ3 complex structure with the two major interfaces highlighted with the black boxes. (B) Detailed interaction between USH1C C-terminal tail and Myo7b CMF central pocket. Residues involved in binding are highlighted with stick models. Salt bridges and hydrogen bonds are indicated with dashed lines. (C) Surface electrostatic potential of the Myo7b CMF showing the positively charged central pocket. (D) Detailed interaction between USH1C PDZ3 βB/βC loop and Myo7b CMF F3 lobe. (E) Summary of dissociation constants showing that mutations of the critical residues in the interface either weakened or even abolished the binding. (F) Schematic diagrams showing domains organizations of different classes of USH1C spliced isoforms.
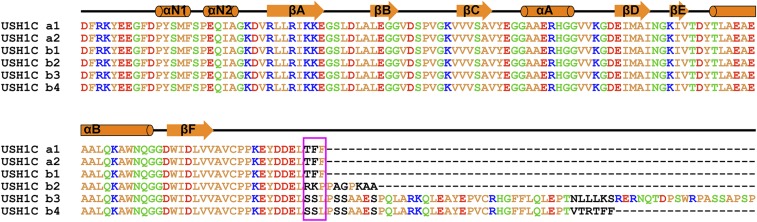
USH1C is known to undergo extensive alternative splicing (33, 34). These spliced isoforms can be divided into three classes (a–c in Fig. 2F). The c isoforms do not contain PDZ3 and thus are not expected to bind to Myo7b CMF (as well as Myo7a CMF, see below). The isoforms a1, a2, and b1 contain the extended C-terminal tail as characterized in this study, and these isoforms are expected to bind to Myo7b CMF (Fig. S2). Although the isoforms b2–b4 contain a PDZ3 with C-terminal extensions, the C-terminal extensions of these isoforms do not have the characteristic “-LTFF*” sequence required for inserting into the FERM central pocket. Therefore, these isoforms are also not expected to bind to Myo7b CMF. Indeed, replacing the C-terminal extension of isoform a1 with that of isoform b2 resulted in complete loss of binding of USH1C PDZ3 to Myo7b CMF (Fig. 2E). USH1C isoform a1 is known to be expressed in the eye, cochlea, vestibule, heart, kidney, small intestine, and testis, and isoform b1 is expressed in cochlea and vestibule. The tissues expressing the USH1C isoforms a1 and b1 are all known to express either Myo7a or Myo7b (13, 35, 36).
Fig. S2.
Sequence alignment of PDZ3 in USH1C isoforms a and b. The secondary structure elements are shown according to the USH1C structure reported in this study. Identical and highly similar residues are colored (yellow for hydrophobic residues, blue for positively charged residues, red for negatively charged residues, and green for other polar residues). The C-terminal-TFF critical for binding to Myo7b CMF are highlighted with a magenta box.
The Myo7b/USH1C Interaction Is Critical for IMAC Formation and Function in CACO-2BBE Cells.
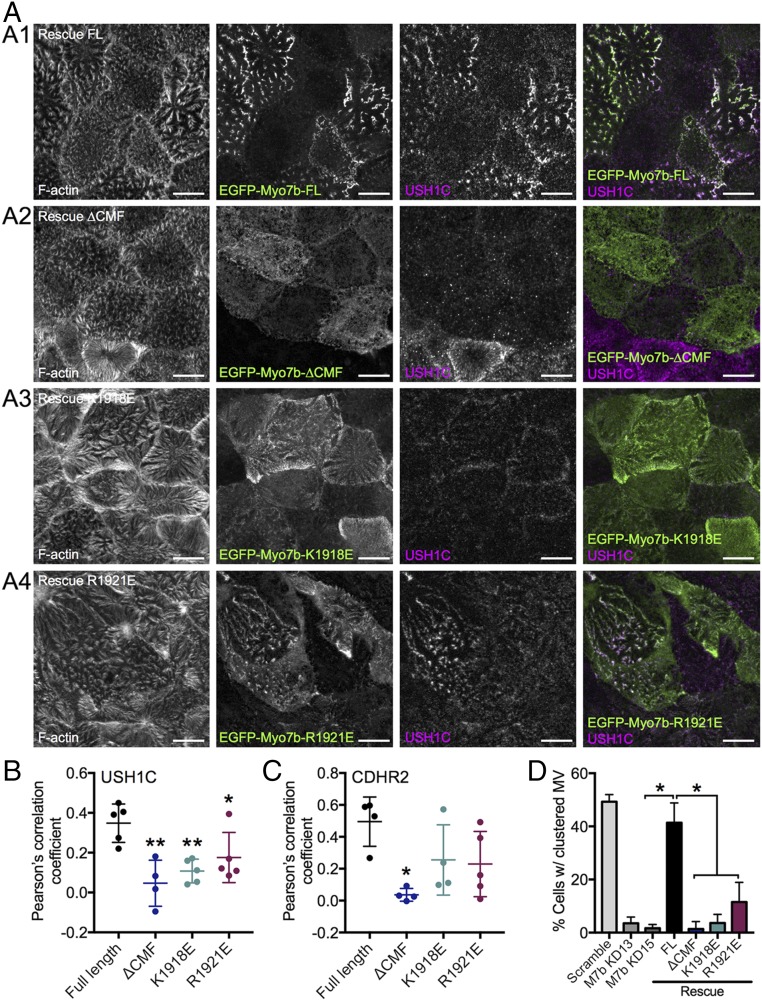
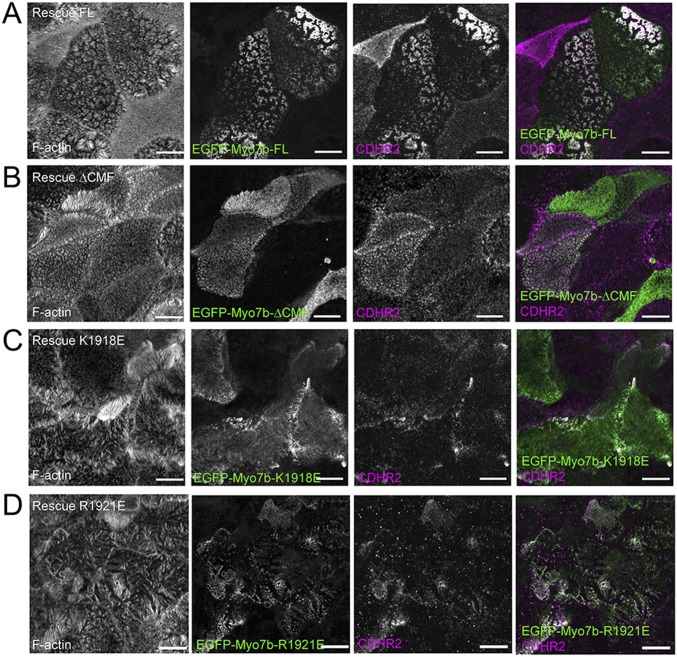
Next, we used CACO-2BBE cells to evaluate the role of Myo7b/USH1C binding in IMAC function and brush border assembly. Our previous studies showed that knockdown (KD) of Myo7b in CACO-2BBE impairs IMAC formation, as shown by loss of IMAC component (USH1C, CDHR2 and CDHR5) enrichment at microvillar tips; these phenotypes were rescued by reexpression of WT Myo7b (37). We used a similar KD/rescue approach to evaluate the function of several mutants of Myo7b defective in USH1C PDZ3 binding (K1918E and R1921E in Myo7b CMF as shown in Fig. 2 and a variant of Myo7b lacking the CMF, denoted ΔCMF). CACO-2BBE cells were subjected to Myo7b KD and then infected with lentivirus encoding Myo7b rescue constructs. After allowing cells to differentiate for 14 d, they were fixed and stained for USH1C and CDHR2 and then imaged using confocal microscopy. The resulting images were used to calculate Pearson’s correlation coefficient to examine the extent of colocalization between Myo7b constructs and USH1C or CDHR2 in microvilli. As a readout for IMAC function, we also visually scored the fraction of cells that exhibit robust microvillar clustering, as previously described (37). As a positive control, we re-expressed EGFP-tagged WT full-length Myo7b, which strongly colocalized with USH1C (Fig. 3 A, 1 and B) and CDHR2 (Fig. 3C and Fig. S3) in microvilli, and rescued microvillar clustering in KD cells (Fig. 3 A, 1 and D). In contrast, neither the point mutants nor ΔCMF fully rescued Myo7b KD-induced loss of IMAC component localization or microvillar clustering (Fig. 3 and Fig. S3). The K1918E mutant and the ΔCMF mutant, which are both unable to bind to USH1C PDZ3, completely failed to rescue the normal microvillar enrichment of USH1C and clustering of microvilli (Fig. 3 A, 2 and 3). The R1921E mutant, which is still able to bind weakly to USH1C PDZ3, showed a slightly less severe phenotype in both assays (Fig. 3), suggesting that strong Myo7b CMF/USH1C PDZ3 binding is important for stable localization and function of USH1C in microvilli.
Fig. 3.
Myo7b variants deficient in USH1C binding exhibit decreased colocalization with USH1C and impaired microvillar clustering. (A) Confocal images of 14 DPC Myo7b KD CACO-2BBE cells expressing the EGFP-tagged Myo7b rescue constructs indicated (green) and stained for USH1C (magenta) and F-actin (gray). (Scale bars, 10 μm.) (B) Colocalization analysis of Myo7b with USH1C using Pearson’s correlation coefficients. n = 5 fields, 111 cells for FL; 4 fields, 30 cells for ΔCMF; 5 fields, 66 cells for K1918E; 5 fields, 84 cells for R1921E. *P < 0.05, **P < 0.01, t test. (C) Colocalization analysis of Myo7b constructs with CDHR2 using Pearson’s correlation coefficients. n = 4 fields, 59 cells for FL; 4 fields, 37 cells for ΔCMF; 4 fields, 46 cells for K1918E; 5 fields, 61 cells for R1921E. *P < 0.01, t test. The representative images are shown in Fig. S3. (D) Quantification of cells with clustering microvilli expressed as a total percentage of cells. For KD rescue cell lines, only EGFP+ (i.e., rescue construct-expressing) cells were scored. Bars indicate mean ± SD. n = 606 cells for scramble, 573 cells of Myo7b KD13 experiment, 507 for Myo7b KD15, 271 for FL, 89 for ΔCMF, 261 for K1918E, and 264 for R1921E. *P < 0.002, t test. KD13 and KD15 represent two distinct short hairpin RNA KD lines as described previously (37).
Fig. S3.
Myo7b variants deficient in USH1C binding exhibit decreased colocalization with CDHR2. Confocal images of 14 d post confluency Myo7b KD CACO-2BBE cells expressing the EGFP-tagged Myo7b rescue constructs (A: EGFP-Myo7b-FL; B: EGFP-Myo7b-ΔCMF; C: EGFP-Myo7b-K1918E; D: EGFP-Myo7b-R1921E) indicated (green) and stained for CDHR2 (magenta) and F-actin (gray). (Scale bars, 10 μm.) The colocalization analysis of Myo7b constructs with CDHR2 using Pearson’s correlation coefficients is shown in Fig. 3C.
Myo7a CMF Also Binds to USH1C PDZ3.
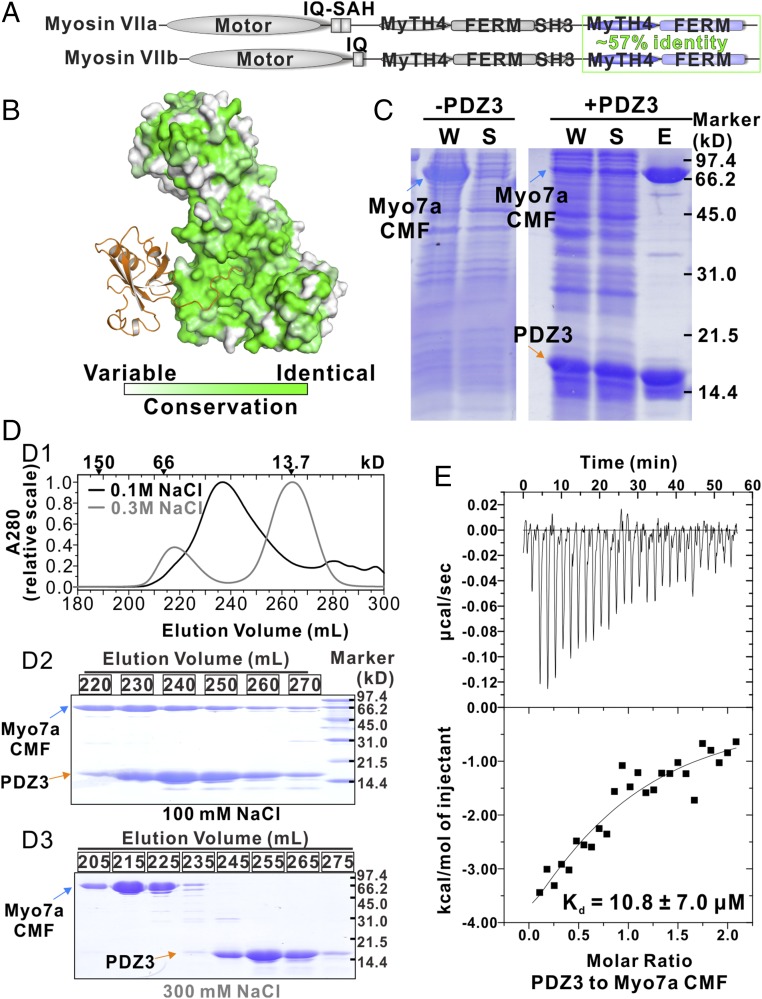
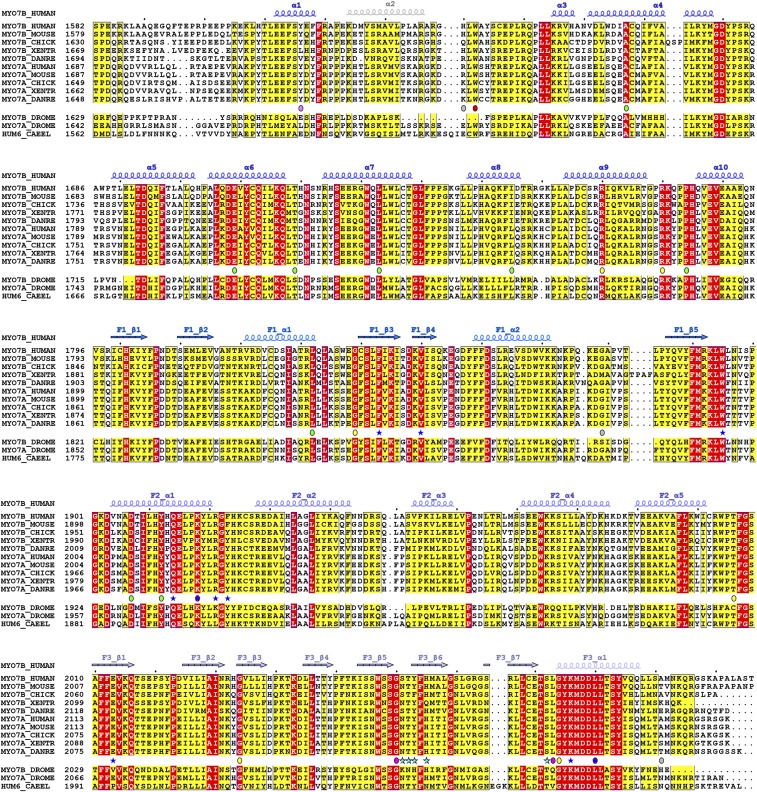
Myo7a and Myo7b are homologous and their CMFs share 57% sequence identity (Fig. 4A). Moreover, mapping of Myo7a CMF amino acid residues onto the structure of Myo7b CMF revealed that the residues responsible for USH1C PDZ3 binding (i.e., both the PDZ3 domain binding surface on F3 and the C-terminal tail binding pocket at the interface of the three FERM lobes) are nearly identical in Myo7a (Fig. 4B and Fig. S4), suggesting that USH1C PDZ3 may also interact with Myo7a CMF. We tried to test the possible interaction between Myo7a CMF and USH1C PDZ3 using purified recombinant proteins. Although isolated Myo7a CMF could not be expressed in its soluble forms in Escherichia coli, coexpression of USH1C PDZ3 with Myo7a CMF greatly enhanced the solubility of Myo7a CMF (Fig. 4C). The resulting Myo7a CMF/USH1C PDZ3 complex can be purified by immobilized metal ion-affinity chromatography followed by a size-exclusion column (Fig. 4 C, and D1 and D2), confirming the direct interaction of the two proteins. We managed to separate Myo7a CMF and USH1C PDZ3 by treating the complex with buffer containing 300 mM NaCl (Fig. 4D3). ITC-based titration experiments using separated proteins showed that USH1C PDZ3 binds to Myo7a CMF with a Kd ∼11 μM (Fig. 4E), which is a few fold weaker than that between Myo7b and USH1C (Kd ∼1.7 μM) and is consistent with the result shown in our earlier pull-down assay (26).
Fig. 4.
Interaction between Myo7a CMF and USH1C PDZ3. (A) Domain organizations of Myo7a and Myo7b. Sequence identity of their CMF is indicated. (B) The amino acid sequence conservation map of Myo7a/b CMF showing that the residues forming the USH1C PDZ3 binding surface are nearly identical. The conservation map is calculated based on amino acid sequences of mammalian Myo7a and Myo7b. (C) SDS/PAGE analysis showing that coexpression of USH1C PDZ3 can increase the solubility of Myo7a CMF. “W” means whole-cell extract; “S” denotes supernatant after centrifugation of the cell lysate; “E” stands for the fraction after elution from the Ni2+-NTA column. (D) The elution profiles of Myo7a CMF/USH1C PDZ3 complex in the buffer containing 100 mM NaCl (black) or 300 mM NaCl (gray). The elution positions of molecular size markers are indicated at the top (D1). The corresponding SDS/PAGE analysis of the eluted peaks (D2: in buffer containing 100 mM NaCl and D3: in buffer containing 300 mM NaCl). (E) ITC result showing that Myo7a CMF binds to USH1C PDZ3 with a Kd ∼11 μM.
Fig. S4.
Sequence alignment of CMF of class VII myosins from difference species. The secondary structure elements are labeled according to the Myo7b CMF structure. Residues that are identical and highly similar are indicated in red and yellow boxes, respectively. The residues involved in USH1C C-terminal tail binding and the PDZ3 domain binding are highlighted with blue and cyan stars. Missense mutations are highlighted with circles and color coded as that in Fig. 5.
Disease Mutations Impact Myo7 CMF/USH1C PDZ3 Interactions.
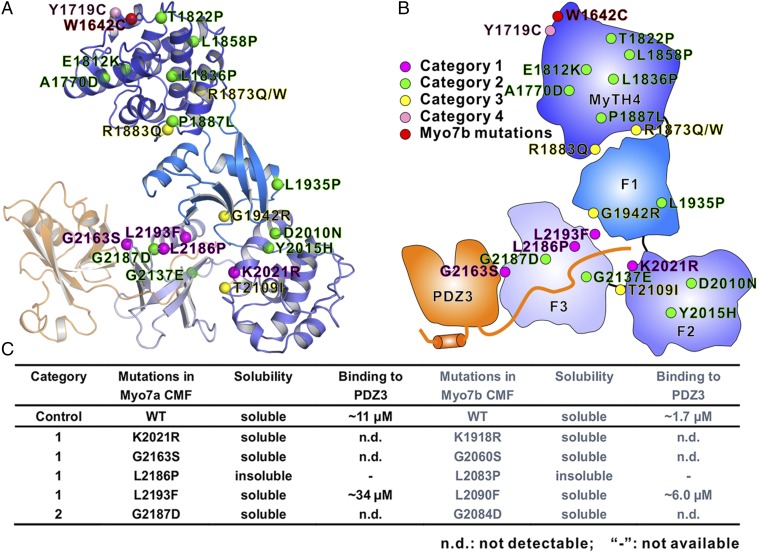
Mutations in myo7a (USH1B) are estimated to be responsible for ∼50% of all type I Usher syndrome cases, and the other half are caused by the rest of the four USH1 genes (38). A total of 25 missense mutations in 23 residues are located in Myo7a CMF based on the data extracted from ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) and HGMD database (www.hgmd.cf.ac.uk/). The high sequence identity between Myo7a and Myo7b allowed us to confidently build the Myo7a CMF structure by homology modeling (Fig. S4) (39). We then mapped the 25 mutations to the modeled structure (Fig. 5 A and B). We can divide the mutations into four categories (Fig. 5 A and B and Fig. S4). The most interesting category (category 1), which is also directly relevant to this study, contains four mutations (K2021R, G2163S, L2186P, and L2193F) located at the PDZ3 binding interface. Category 2 includes mutations within the folding core of each of the subdomains. These variants are likely to affect the folding of each individual subdomain. In category 3 are mutations at the subdomain–subdomain interfaces, and these mutations may destabilize the MyTH4-FERM supramodule formation. Interestingly, the residues in Myo7a NMF, which correspond to E1812 and P1887 in MyTH4 domain core and to R1883 at MyTH4-F1 interface of Myo7b CMF, are also found to be mutated in USH1B patients, highlighting the importance of the correct folding of the MyTH4 domain and the formation of MyTH4-FERM supramodule in the functions of Myo7a MyTH4-FERM tandems. The fourth category of mutations cannot be well explained by the current structural model, and residues in this category are located on the surface of MyTH4 (Y1719C) or in the flexible loops that cannot be visualized in our Myo7b CMF crystal structure (R1743W, R1873Q/W, and A2204P). It is noted that impact of the Y1719C mutation is ambiguous, because this mutation has been reported to be a common polymorphism in the Moroccan population (40). There is also a de novo mutation (W1642C) found in Myo7b CMF, which is proposed to be linked with autism (41). W1642 is partially exposed and the mutation may destabilize MyTH4_α1. However, the potential impact of the W1642C mutation in nervous systems requires further investigation.
Fig. 5.
Missense mutations in Myo7a CMF found in USH1B patients. (A) Ribbon representation of the Myo7a CMF structural model. The 20 missense mutation sites in USH1B patients are highlighted with spheres and colored in magenta, green, yellow, and pink, corresponding to the four categories. A de novo mutation found in Myo7b (W1642C) is shown in red. (B) Schematic cartoon diagram showing the distributions of the USH1B mutations mapped on to the structural model of Myo7a CMF. (C) Summary of impacts of some USH1B mutations on the Myo7a CMF/USH1C PDZ3 interaction. The matching mutations were also made in Myo7b CMF corresponding to each of the USH1B mutations, and their impacts on USH1C PDZ3 binding are measured.
Given the extremely high structural similarity between Myo7a and Myo7b and the easy availability of Myo7b CMF, we first tested some of these mutations using Myo7b as the surrogate. We focused on the four Myo7a CMF mutations, of which the corresponding sites are located at the PDZ3 binding interface of Myo7b CMF. Substitutions of K1918 with Arg or L2193 with Phe (corresponding to K2021R and L2193F in Myo7a CMF indicated in Fig. 5B, the two residues intimately involved in USH1C C-terminal interaction) (Fig. 2B), either abolished or weakened Myo7b CMF’s binding to PDZ3 (Fig. 5C). Although the two mutations do not dramatically change the sidechain properties (maintaining the charged or hydrophobic properties), substitutions with larger sidechains will decrease the size of the central pocket and thus impair the USH1C PDZ3 C-terminal tail binding. L2083 (corresponding to L2186 on Myo7a CMF) is also located at FERM-domain central pocket and participates the hydrophobic interaction with USH1C C-terminal extension. The substitution of L2083 with Pro led to formation of inclusion bodies in the process of protein expressions in E. coli, (Fig. 5C), which may indicate that such mutation can impair the folding of the F3 lobe of Myo7b CMF and thus prevent USH1C PDZ3 from binding. At the PDZ3/FERM F3 lobe interface, the G2060S mutation (corresponding to G2163 on Myo7a CMF) also totally abolished the interaction between Myo7b CMF and USH1C PDZ3 (Fig. 5C). We also tested another disease related mutation G2084D (corresponding to G2187 on Myo7a CMF), which is immediately following L2083 and is in the vicinity of the PDZ3 tail binding pocket. We found that the G2084D mutation also totally disrupted the interaction between Myo7b CMF and USH1C PDZ3 (Fig. 5C).
We next sought to test these disease mutations directly on Myo7a CMF. With the help of USH1C PDZ3 coexpression, we can purify four of the five mutants tested and used the ITC-based assay to assess the impact of each of the mutation on USH1C PDZ3 binding, and the result is summarized in Fig. 5C. As we have anticipated, the impact of each of the Myo7a CMF mutation on USH1C PDZ3 binding is essentially the same as what we have observed for the corresponding mutations in Myo7b CMF. The K2021R, G2163S and G2187D mutations totally abolished the interaction between Myo7a CMF and USH1C PDZ3, and the L2193F mutation weakened the interaction (Fig. 5C). As in Myo7b CMF, we were not able to express the L2186P mutant of Myo7a CMF in soluble form either.
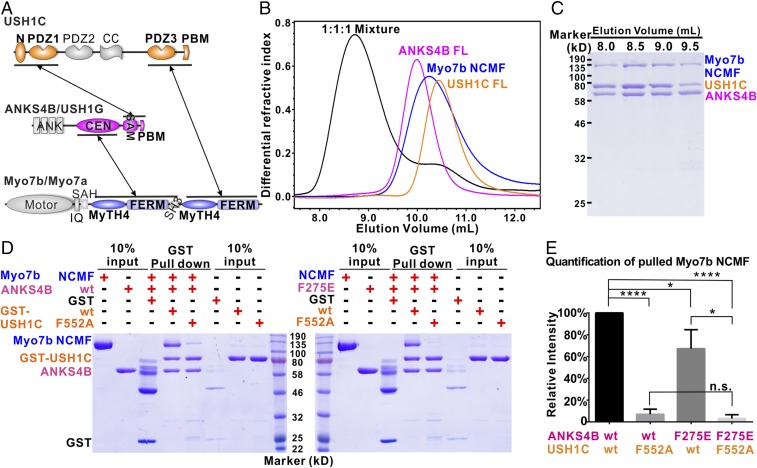
Formation of the Myo7b/ANKS4B/USH1C Tripartite Complexes Requires all Three Binary Interactions.
Up to this stage, we had quantitatively studied all binary interactions between pairs of proteins within the Myo7b/ANKS4B/USH1C tripartite complexes that are expected to form in cells (Fig. 6A). We next tried to reconstitute the tripartite complexes in vitro. After numerous trials, we successfully purified large quantities of Myo7b tail containing the two MyTH4-FERM tandems (denoted as Myo7b NCMF) with the help of coexpression of USH1C PDZ3. When loaded on a size-exclusion column, the 1:1:1 mixture of Myo7b NCMF, the full-length ANKS4B, and the full-length USH1C were eluted significantly earlier than each individual component (Fig. 6B). SDS/PAGE analysis showed that the complex consisted of all of the three proteins (Fig. 6C), indicating the formation of the Myo7b/ANKS4B/USH1C tripartite complex.
Fig. 6.
Myo7b/ANKS4B/USH1C tripartite complex formation. (A) Schematic diagram showing the detailed interaction network governing the formation of the Myo7b/ANKS4B/USH1C and Myo7a/USH1G/USH1C tripartite complexes. (B and C) Analytical gel-filtration chromatography (B) together with SDS/PAGE analysis (C) showing that the Myo7b NCMF, the full-length ANKS4B, and the full-length USH1C form a stable tripartite complex. (D) Representative pull-down experiments showing that mutations disrupting or weakening each binary interaction can affect the Myo7b/ANKS4B/USH1C tripartite complex formation (Left for ANKS4B WT and Right for ANKS4B F275E). (E) Quantification of the amount of Myo7b NCMF pulled down in the assays shown in D. The data are derived from three different batches of experiments, and the error bars are expressed as mean ± SD. *P < 0.05; ****P < 0.0001; n.s., nonsignificant; t test.
The Myo7b/ANKS4B/USH1C tripartite complex contains three pairs of binary interactions (Fig. 6A). Myo7b uses its NMF to interact with ANKS4B CEN domain and its CMF to interact with USH1C extended PDZ3 domain, each with a micromolar dissociation constant. ANKS4B also uses its SAM-PBM to interact with USH1C NPDZ1 with a Kd at nanomolar range. Next, we sought to determine how each pair of binary interactions contributes to the tripartite complex formation. Our current and earlier structural studies allowed us to design mutations to specifically disrupt or weaken each pair of binary interaction, and we used pull-down assays to evaluate each of the mutations on the Myo7b/ANKS4B/USH1C tripartite complex formation. As expected, both the full-length ANKS4B and Myo7b NCMF can be robustly pulled down by GST-USH1C, confirming that a stable tripartite complex was formed (Fig. 6D). Mutation of ANKS4B (F275E) moderately decreased the amount of pulled-down Myo7b NCMF, whereas USH1C (F552A) led to significant reduction of Myo7b NCMF being pulled down (Fig. 6 D and E). The difference between these two mutations on the tripartite complex formation is consistent with the ITC-based results on the mutation-induced binding affinity reductions. The ANKS4B (F275E) mutation decreased the ANKS4B CEN/Myo7b NMFS by fourfold, and the USH1C (F552A) mutation totally abolished the USH1C PDZ3/Myo7b CMF interaction. The data shown in Fig. 6 are also consistent with the previous cellular coimmunoprecipitation results (25) as well as the results in Fig. 3 showing that disruption of Myo7b CMF/USH1C binding resulted in lower amount of tip targeting of USH1C. As expected, when both the F275E-ANKS4B and F552A-USH1C were used in the binding experiment, essentially no Myo7b NCMF can be pulled down (Fig. 6 D and E). The above data indicate that formation of the stable Myo7b/ANKS4B/USH1C tripartite complex requires all three pairs of binary interactions illustrated in Fig. 6A.
Discussion
We have demonstrated in this study, by solving the atomic structure and performing detailed biochemical characterization, that Myo7b CMF can specifically bind to USH1C PDZ3 using a previously uncharacterized mode for MyTH4-FERM tandems, as well as for PDZ domains. We have also shown that the highly homologous Myo7a CMF can bind to USH1C PDZ3 as well. Given the domain organization similarities among USH1C, PDZD7, and Whirlin, we speculate that the recently reported Myo7a/PDZD7 complex (42, 43) and the Myo15a/Whirlin complex (44), existing in stereocilia of hair cells, may also form via a similar MyTH4-FERM/PDZ interaction observed in the Myo7b/USH1C and Myo7a/USH1C complexes. As we have illustrated in the present study, the high-resolution structure of the Myo7b CMF/USH1C PDZ3 complex structure may help explain why many of the USH1B mutations in the Myo7a CMF cause deafness and blindness in human patients. The Myo7b CMF/USH1C PDZ3 structure can also serve as a template for understanding potential impacts of mutations found in Myo15a CMF in nonsyndromic deaf patients.
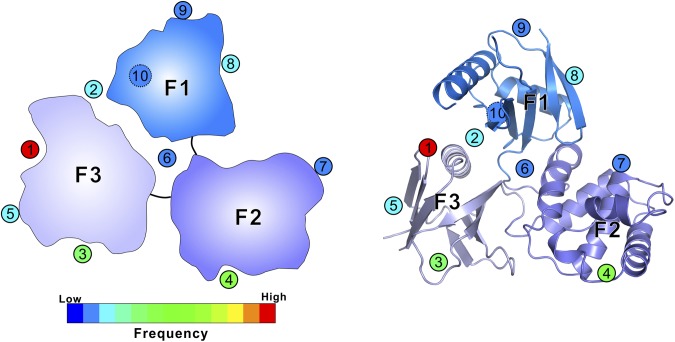
The structure of the Myo7b CMF/USH1C PDZ3 complex also extends our current knowledge about the target recognition modes of FERM and PDZ domains, both of which are highly versatile protein–protein interaction modules abundantly present in mammalian proteomes. We summarized all reported FERM/target bindings with structure coordinates deposited in PDB without considering Talin FERM because of its very different F1 lobe organization (Fig. S5 and Table S1). A total of 10 different target binding sites can be found in all three lobes of FERM domains. Additionally, the interlobe interfaces of FERM domains can also function as target binding sites. For example, the F1/F3 interface (site 2 in Fig. S5) can be protein or lipid binding sites of FERM domains (45, 46). The F1/F2/F3 interface in FERM domains of myosins binds to different protein targets with distinct mechanisms (22, 26; and present study). We also note that some targets simultaneously bind to more than one site on FERM domains, resulting in higher binding affinity and specificity. For example, we show in this study that USH1C binds to site 5 in F3 and site 6 formed by the F1/F2/F3 interface of Myo7b C-terminal FERM domain. In a number of FERM domain proteins with auto-inhibitory tails, each tail is known to simultaneously bind to sites 3 and 4 in the F3 and F2 lobes, respectively (31, 47, 48). The Crumbs tail is shown to simultaneous bind to site 1 in the F3-lobe and site 2 in the F1/F3 interface (49). Among all of the sites identified, site 1 in the α1/β5-groove of F3 is the most frequently used target binding site of FERM domains. The summary in Fig. S5 highlights the remarkable capacities of FERM domains in binding to very diverse target molecules, each with their own specificities.
Fig. S5.
Summary of the binding sites used by FERM domains to recognize targets as seen in complex structures. Schematic cartoon (Left) and ribbon diagrams (Right) of the FERM domain of Myo7b CMF are used to illustrate target binding sites of FERM domains. The 10 different sites are shown with numbers in circles. The dashed circle (site 10) indicates that the binding site is located at the back side of the F1 lobe. The circles are filled with color showing the frequencies seen in the crystal structures of FERM domains with their different targets as detailed in Table S1.
Table S1.
Diverse targets recognition modes of FERM domain
| PDB ID code | FERM domain | Target or adjacent domain | Site |
| Myo7b CMF | USH1C_PDZ3 | 5 + 6 | |
| 3BIN | DAL-1/4.1B | TSLC1 | 1 |
| 4P7I | Merlin | DCAF1 | 1 |
| 3PZD | Myo10 | DCC | 1 |
| 1J19 | Radixin | ICAM-2 | 1 |
| 2EMS | Radixin | CD43 | 1 |
| 2ZPY | Radixin | CD44 | 1 |
| 4TKN | SNX17 | Krit1 | 1 |
| 2EMT | Radixin | PSGL-1 | 1 |
| 2YVC | Radixin | NEP | 1 |
| 3U7D | Krit1 | HEG1 | 2 |
| 1GC6 | Radixin | IP3 | 2 |
| 2D10 | Radixin | NHERF-1 | 3 |
| 5IXD | JAK1 | IFNLR1 | 4 |
| 4ZRI | Merlin | Lats2 | 4 |
| 3PVL | Myo7a | Sans | 6 |
| 3X23 | Radixin | MMP14 | 8 |
| 4YL8 | Moesin | Crumbs | 1 + 2 |
| 4RM8 | Ezrin | Ezrin_CT | 3 + 4 |
| 4ZRJ | Merlin | Merlin_CT | 3 + 4 |
| 1EF1 | Moesin | Moesin_CT | 3 + 4 |
| 1SGH | Moesin | EBP50 | 3 + 4 |
| 4DXA | Krit1 | Rap1 | 7 + 8 |
| MyTH4-FERM | MyTH4 | 9 | |
| 5D68 | Krit1 | Krit1_ANK | 9 |
| 2J0J | FAK | FAK_Kinase | 7 + 8 |
| 4PO6 | TYK2 | TYK2_SH2 | 10 |
| 5IXD | JAK1 | JAK1_SH2 | 10 |
Materials and Methods
Cloning and Constructs for Recombinant Protein Expression.
The human cDNA construct of Myo7b used in this study was GI: 122937511. The protein coding sequences of Myo7b CMF (residues 1601–2116) and NCMF (residues 960–2116) were amplified by PCR from human Myo7b and cloned into a pET vector. The coding sequences of Myo7a CMF (residues 1704–2215) and NCMF (residues 965–2215) were amplified by PCR from a mouse USH1B gene (GI: U81453.1). The coding sequences of USH1C isoform b2 PDZ3 (residues 728–859) was amplified by PCR from mouse USH1C isoform b2 cDNA (a kind gift from Christine Petit, Pasteur Institute, Paris). Other constructs used have been described in our previous study (26). Point mutations were generated by PCR-based site-directed mutagenesis.
Cloning and Constructs for CACO-2BBE Transfection.
Full-length (residues 1–2116) and ΔCMF (residues 1–1587) Myo7b constructs were generated by PCR and TOPO cloned into the pCR8 Gateway entry vector (Invitrogen). Point mutations and refractory silent mutations were introduced using QuikChange site-directed mutagenesis (Agilent). All entry vectors were verified by DNA sequencing. The entry vectors were then shuttled into the destination vector pINDUCER20-EGFP-C1 (25) that was Gateway-adapted using the Gateway vector conversion kit (Invitrogen). A nontargeting scramble control shRNA (Addgene; plasmid 1864) and Myo7b KD shRNA clones were expressed in pLKO.1, corresponding to TRC clones TRCN0000247713 and TRCN0000247715 (Sigma). TRC clone TRCN0000247713 was used to KD Myo7b in the ΔCMF rescue cell line as this construct lacks the shRNA target sequence. All other Myo7b KD rescue constructs contain refractory silent mutations to TRC clone TRCN0000247715.
Protein Expression and Purification.
Recombinant proteins were expressed in BL21 (DE3) E. coli cells. The N-terminal His6-tagged proteins were purified using Ni2+-nitrilotriacetic acid agarose column followed by another step of size-exclusion chromatography (Superdex 200 column from GE Healthcare) in the final buffer of 50 mM Tris⋅HCl, 1 mM DTT, 1 mM EDTA, pH 7.8, and 100 mM NaCl. GST-fused proteins were purified by GSH-Sepharose affinity chromatography, followed by a step of size-exclusion chromatography, same as the one described above. For the separation of Myo7a CMF and USH1C PDZ3 from the copurified complex, an additional step of size-exclusion chromatography (Superdex 200 column from GE Healthcare) in the buffer of 50 mM Tris⋅HCl, 1 mM DTT, 1 mM EDTA, pH 7.8, and 300 mM NaCl was performed, and the fraction containing each of the individual components was collected.
Crystallography.
The crystals of Myo7b CMF/USH1C PDZ3 complex (∼10 mg/mL in 50 mM Tris, pH 7.8, 100 mM sodium citrate, 1 mM EDTA, and 1 mM DTT buffer) were grown in reservoir solution containing 8% tacsimate, pH 7.0, and 20% (vol/wt) PEG3350 by sitting-drop vapor diffusion methods at 16 °C. To prepare Au-derivatives, crystals were soaked in crystallization solution containing additional 2∼5 mM KAu(CN)2 for 1 to 2 d. Crystals were then soaked in reservoir solution containing extra 10% (vol/vol) glycerol for cryo-protection. Diffraction data were collected at the Shanghai Synchrotron Radiation Facility (BL17U and BL19U1) at 100 K. Data were processed and scaled using HKL2000 or HKL3000 (50).
Six gold sites were found by SHELXD (51). Subsequence sites refinement, phase calculation, density modification and initial model building were carried out with Autosol in PHENIX (52). Additional manual model building and refinement were completed iteratively using COOT (53) and PHENIX (52). The final model was validated by MolProbity (54). The final refinement statistics are summarized in Table 1. All structure figures were prepared by PyMOL (www.pymol.org).
GST Pull-Down Assay.
Formation of the tripartite complex Myo7b/ANKS4B/USH1C was assayed in 50 mM Tris buffer, pH 7.5, containing 100 mM NaCl, 1 mM EDTA, and 1 mM DTT. In each assay, purified Trx tagged Myo7b NCMF and ANKS4B FL or its mutant were first mixed with GST-tagged USH1C or its mutant at 1:1:1 ratio (∼0.28 nmol each) and then pelleted by adding 30 µL of fresh GSH-Sepharose beads. The pellet, after washing twice with the assay buffer, was analyzed by SDS/PAGE. The Coomassie Brilliant Blue-stained gel was analyzed by densitometry to determine the amount of pulled down Myo7b NCMF.
Analytical Gel-Filtration Chromatography.
Protein samples (typically 100 µL at a concentration of 20 µM pre-equilibrated with the column buffer) were injected into an AKTA FPLC system with a Superose 12 10/300 GL column (GE Healthcare) using the column buffer of 50 mM Tris⋅HCl, pH 7.8, 1 mM DTT, 1 mM EDTA, and 100 mM NaCl.
ITC Assay.
ITC measurements were carried out on a VP-ITC Microcal calorimeter (Microcal) at 25 °C. Titration buffer contained 50 mM Tris⋅HCl, pH 7.8, 1 mM DTT, 1 mM EDTA, and 100 mM NaCl. Each titration point was performed by injecting a 10-μL aliquot of a protein sample from a syringe into a protein sample in the cell at a time interval of 120 s to ensure that the titration peak returned to the baseline. The titration data were analyzed by Origin7.0 (Microcal).
Lentivirus Production and Stable Cell Line Generation.
CACO-2BBE and HEK293FT cells were cultured at 37 °C and 5% CO2 in DMEM with high glucose and 2 mM l-glutamine supplemented with 20% FBS or 10% FBS, respectively. Lentiviral particles were generated by cotransfecting HEK293FT cells with overexpression (pINDUCER20-EGFP-C1) or KD (pLKO.1) plasmids along with the packaging psPAX2 and envelope pMD2.G plasmids using FuGENE 6 (Promega). Cells were incubated for 2 d to allow for lentivirus production before collecting the media containing lentiviral particles. Media were centrifuged at 500 × g for 10 min at 4 °C to remove cells and concentrated using Lenti-X Concentrator (Clontech). For generation of stable cell lines, CACO-2BBE cells were grown to 90% confluency in T25 flasks. For lentivirus transduction of CACO-2BBE cells, the media were supplemented with 8 μg/mL polybrene and incubated with lentivirus overnight. The following day, cells were reseeded into T75 flasks and grown for 3 d. Cells were then reseeded into T182 flasks with media containing 10 μg/mL puromycin or 1 mg/mL G418 and grown to select for stable integration.
Light Microscopy and Image Analysis.
CACO-2BBE cells were washed once with warm PBS and fixed with 4% paraformaldehyde in PBS for 15 min at 37 °C. After fixation, cells were washed three times with PBS and permeabilized with 0.1% Triton X-100 in PBS for 15 min at room temperature. Cells were then washed three times with PBS and blocked overnight with 5% BSA in PBS at 4 °C. Cells were washed once with PBS and immunostaining was performed using anti-USH1C (1:70; Sigma cat#HPA027398), and anti-CDHR2 (1:75; Sigma cat#HPA012569), at room temperature for 2 h. Coverslips were then washed three times with PBS and incubated with Alexa Fluor-647 goat anti-rabbit and Alexa Fluor-568 phalloidin (1:200; Life Technologies) in PBS for 1 h at 37 °C. Cells were washed four times with PBS, and coverslips were mounted using ProLong Gold Antifade Mountant (Life Technologies). Cells were imaged using a Nikon A1R laser-scanning confocal microscope equipped with 488-, 561-, and 647-nm excitation lasers and a 100×/1.49 numerical aperture (NA) Apo total internal reflection fluorescence objective. Image acquisition was controlled with Nikon Elements software. Images were contrast enhanced and analyzed using ImageJ software (NIH). Pearson’s correlation coefficients were calculated using JACoP (Just Another Colocalization Plugin). Individual cells were blindly scored for microvillar clustering using the phalloidin signal. For all figures, error bars indicate that SD and n values are reported in the figure legends. All graphs were generated, and statistical analyses performed in Prism v.7 (GraphPad Software, Inc.). Unpaired t tests were used to determine statistical significance between reported values.
Acknowledgments
We thank the Shanghai Synchrotron Radiation Facility BL19U1 and BL17U1 for X-ray beam time. This work was supported by the Research Grants Council of Hong Kong Grants 664113, 16103614, AoE-M09-12, and T13-607/12R, and a 973 program grant from the Minister of Science and Technology of China (Grant 2014CB910204; to M.Z.); Research Grants Council of Hong Kong Grant 16149516 (to Q.L.); an American Heart Association predoctoral fellowship (to M.L.W.); National Research Service Award Predoctoral Fellowship F31DK108528 (to M.L.W.); and NIH Grants R01DK075555 and R01DK095811 (to M.J.T.). M.Z. is a Kerry Holdings Professor in Science and a Senior Fellow of the Institute for Advanced Study at the Hong Kong University of Science and Technology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5XBF).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702251114/-/DCSupplemental.
References
- 1.Nambiar R, McConnell RE, Tyska MJ. Myosin motor function: The ins and outs of actin-based membrane protrusions. Cell Mol Life Sci. 2010;67:1239–1254. doi: 10.1007/s00018-009-0254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mooseker MS. Organization, chemistry, and assembly of the cytoskeletal apparatus of the intestinal brush border. Annu Rev Cell Biol. 1985;1:209–241. doi: 10.1146/annurev.cb.01.110185.001233. [DOI] [PubMed] [Google Scholar]
- 3.Louvard D, Kedinger M, Hauri HP. The differentiating intestinal epithelial cell: Establishment and maintenance of functions through interactions between cellular structures. Annu Rev Cell Biol. 1992;8:157–195. doi: 10.1146/annurev.cb.08.110192.001105. [DOI] [PubMed] [Google Scholar]
- 4.Sauvanet C, Wayt J, Pelaseyed T, Bretscher A. Structure, regulation, and functional diversity of microvilli on the apical domain of epithelial cells. Annu Rev Cell Dev Biol. 2015;31:593–621. doi: 10.1146/annurev-cellbio-100814-125234. [DOI] [PubMed] [Google Scholar]
- 5.Delacour D, Salomon J, Robine S, Louvard D. Plasticity of the brush border—The yin and yang of intestinal homeostasis. Nat Rev Gastroenterol Hepatol. 2016;13:161–174. doi: 10.1038/nrgastro.2016.5. [DOI] [PubMed] [Google Scholar]
- 6.Helander HF, Fändriks L. Surface area of the digestive tract—Revisited. Scand J Gastroenterol. 2014;49:681–689. doi: 10.3109/00365521.2014.898326. [DOI] [PubMed] [Google Scholar]
- 7.Tilney LG, Tilney MS, DeRosier DJ. Actin filaments, stereocilia, and hair cells: How cells count and measure. Annu Rev Cell Biol. 1992;8:257–274. doi: 10.1146/annurev.cb.08.110192.001353. [DOI] [PubMed] [Google Scholar]
- 8.Schwander M, Kachar B, Müller U. Review series: The cell biology of hearing. J Cell Biol. 2010;190:9–20. doi: 10.1083/jcb.201001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kazmierczak P, et al. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature. 2007;449:87–91. doi: 10.1038/nature06091. [DOI] [PubMed] [Google Scholar]
- 10.Crawley SW, et al. Intestinal brush border assembly driven by protocadherin-based intermicrovillar adhesion. Cell. 2014;157:433–446. doi: 10.1016/j.cell.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McConnell RE, Benesh AE, Mao S, Tabb DL, Tyska MJ. Proteomic analysis of the enterocyte brush border. Am J Physiol Gastrointest Liver Physiol. 2011;300:G914–G926. doi: 10.1152/ajpgi.00005.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crawley SW, Mooseker MS, Tyska MJ. Shaping the intestinal brush border. J Cell Biol. 2014;207:441–451. doi: 10.1083/jcb.201407015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen ZY, et al. Myosin-VIIb, a novel unconventional myosin, is a constituent of microvilli in transporting epithelia. Genomics. 2001;72:285–296. doi: 10.1006/geno.2000.6456. [DOI] [PubMed] [Google Scholar]
- 14.Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. Proc Natl Acad Sci USA. 2006;103:3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weck ML, Grega-Larson NE, Tyska MJ. MyTH4-FERM myosins in the assembly and maintenance of actin-based protrusions. Curr Opin Cell Biol. November 9, 2016 doi: 10.1016/j.ceb.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petersen KJ, et al. MyTH4-FERM myosins have an ancient and conserved role in filopod formation. Proc Natl Acad Sci USA. 2016;113:E8059–E8068. doi: 10.1073/pnas.1615392113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Planelles-Herrero VJ, et al. Myosin MyTH4-FERM structures highlight important principles of convergent evolution. Proc Natl Acad Sci USA. 2016;113:E2906–E2915. doi: 10.1073/pnas.1600736113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richardson GP, de Monvel JB, Petit C. How the genetics of deafness illuminates auditory physiology. Annu Rev Physiol. 2011;73:311–334. doi: 10.1146/annurev-physiol-012110-142228. [DOI] [PubMed] [Google Scholar]
- 19.Courson DS, Cheney RE. Myosin-X and disease. Exp Cell Res. 2015;334:10–15. doi: 10.1016/j.yexcr.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peckham M. How myosin organization of the actin cytoskeleton contributes to the cancer phenotype. Biochem Soc Trans. 2016;44:1026–1034. doi: 10.1042/BST20160034. [DOI] [PubMed] [Google Scholar]
- 21.Dror AA, Avraham KB. Hearing loss: mechanisms revealed by genetics and cell biology. Annu Rev Genet. 2009;43:411–437. doi: 10.1146/annurev-genet-102108-134135. [DOI] [PubMed] [Google Scholar]
- 22.Wu L, Pan L, Wei Z, Zhang M. Structure of MyTH4-FERM domains in myosin VIIa tail bound to cargo. Science. 2011;331:757–760. doi: 10.1126/science.1198848. [DOI] [PubMed] [Google Scholar]
- 23.Grati M, Kachar B. Myosin VIIa and sans localization at stereocilia upper tip-link density implicates these Usher syndrome proteins in mechanotransduction. Proc Natl Acad Sci USA. 2011;108:11476–11481. doi: 10.1073/pnas.1104161108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan L, Zhang M. Structures of usher syndrome 1 proteins and their complexes. Physiology (Bethesda) 2012;27:25–42. doi: 10.1152/physiol.00037.2011. [DOI] [PubMed] [Google Scholar]
- 25.Crawley SW, Weck ML, Grega-Larson NE, Shifrin DA, Jr, Tyska MJ. ANKS4B is essential for intermicrovillar adhesion complex formation. Dev Cell. 2016;36:190–200. doi: 10.1016/j.devcel.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, He Y, Lu Q, Zhang M. Mechanistic basis of organization of the Harmonin/USH1C-mediated brush border microvilli tip-link complex. Dev Cell. 2016;36:179–189. doi: 10.1016/j.devcel.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 27.Ye F, Zhang M. Structures and target recognition modes of PDZ domains: Recurring themes and emerging pictures. Biochem J. 2013;455:1–14. doi: 10.1042/BJ20130783. [DOI] [PubMed] [Google Scholar]
- 28.Hirano Y, et al. Structural basis of cargo recognition by the myosin-X MyTH4-FERM domain. EMBO J. 2011;30:2734–2747. doi: 10.1038/emboj.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei Z, Yan J, Lu Q, Pan L, Zhang M. Cargo recognition mechanism of myosin X revealed by the structure of its tail MyTH4-FERM tandem in complex with the DCC P3 domain. Proc Natl Acad Sci USA. 2011;108:3572–3577. doi: 10.1073/pnas.1016567108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Q, Li J, Zhang M. Cargo recognition and cargo-mediated regulation of unconventional myosins. Acc Chem Res. 2014;47:3061–3070. doi: 10.1021/ar500216z. [DOI] [PubMed] [Google Scholar]
- 31.Pearson MA, Reczek D, Bretscher A, Karplus PA. Structure of the ERM protein Moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell. 2000;101:259–270. doi: 10.1016/s0092-8674(00)80836-3. [DOI] [PubMed] [Google Scholar]
- 32.Wang CK, Pan L, Chen J, Zhang M. Extensions of PDZ domains as important structural and functional elements. Protein Cell. 2010;1:737–751. doi: 10.1007/s13238-010-0099-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verpy E, et al. A defect in Harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat Genet. 2000;26:51–55. doi: 10.1038/79171. [DOI] [PubMed] [Google Scholar]
- 34.Reiners J, Nagel-Wolfrum K, Jürgens K, Märker T, Wolfrum U. Molecular basis of human Usher syndrome: Deciphering the meshes of the Usher protein network provides insights into the pathomechanisms of the Usher disease. Exp Eye Res. 2006;83:97–119. doi: 10.1016/j.exer.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Gibson F, et al. A type VII myosin encoded by the mouse deafness gene shaker-1. Nature. 1995;374:62–64. doi: 10.1038/374062a0. [DOI] [PubMed] [Google Scholar]
- 36.Weil D, et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374:60–61. doi: 10.1038/374060a0. [DOI] [PubMed] [Google Scholar]
- 37.Weck ML, Crawley SW, Stone CR, Tyska MJ. Myosin-7b promotes distal tip localization of the intermicrovillar adhesion complex. Curr Biol. 2016;26:2717–2728. doi: 10.1016/j.cub.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El-Amraoui A, Petit C. Usher I syndrome: Unravelling the mechanisms that underlie the cohesion of the growing hair bundle in inner ear sensory cells. J Cell Sci. 2005;118:4593–4603. doi: 10.1242/jcs.02636. [DOI] [PubMed] [Google Scholar]
- 39.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 40.Boulouiz R, et al. Analysis of MYO7A in a Moroccan family with Usher syndrome type 1B: Novel loss-of-function mutation and non-pathogenicity of p.Y1719C. Mol Vis. 2007;13:1862–1865. [PubMed] [Google Scholar]
- 41.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgan CP, et al. PDZD7-MYO7A complex identified in enriched stereocilia membranes. eLife. 2016;5:5. doi: 10.7554/eLife.18312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zou J, et al. The roles of USH1 proteins and PDZ domain-containing USH proteins in USH2 complex integrity in cochlear hair cells. Hum Mol Genet. 2017;26:624–636. doi: 10.1093/hmg/ddw421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Belyantseva IA, et al. Myosin-XVa is required for tip localization of whirlin and differential elongation of hair-cell stereocilia. Nat Cell Biol. 2005;7:148–156. doi: 10.1038/ncb1219. [DOI] [PubMed] [Google Scholar]
- 45.Hamada K, Shimizu T, Matsui T, Tsukita S, Hakoshima T. Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain. EMBO J. 2000;19:4449–4462. doi: 10.1093/emboj/19.17.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gingras AR, Liu JJ, Ginsberg MH. Structural basis of the junctional anchorage of the cerebral cavernous malformations complex. J Cell Biol. 2012;199:39–48. doi: 10.1083/jcb.201205109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phang JM, et al. Structural characterization suggests models for monomeric and dimeric forms of full-length ezrin. Biochem J. 2016;473:2763–2782. doi: 10.1042/BCJ20160541. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, et al. Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway. Cell Res. 2015;25:801–817. doi: 10.1038/cr.2015.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei Z, Li Y, Ye F, Zhang M. Structural basis for the phosphorylation-regulated interaction between the cytoplasmic tail of cell polarity protein crumbs and the actin-binding protein Moesin. J Biol Chem. 2015;290:11384–11392. doi: 10.1074/jbc.M115.643791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 52.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]