Significance
Structured illumination microscopy (SIM) is a superresolution technique that illuminates the specimen with a sine-modulated pattern. The frequency of the illumination pattern, i.e., the inverse of the resolution, is limited by the angular aperture of the objective lens. SIM in combination with light-sheet-based fluorescence microscopy (csiLSFM) uses a pair of illumination objective lenses to form two counterpropagating light sheets that interfere in the fluorescent specimen at an angle π. The frequency of the illumination pattern becomes the physical limit of a linear interference pattern. Whereas SIM is restricted to flat specimens and surfaces, csiLSFM operates deep inside three-dimensional specimens. Our implementation is able to generate different spatial frequencies and thus sample the frequency space intelligently.
Keywords: light sheet, structured illumination, SPIM, SIM, LSFM
Abstract
Light-sheet-based fluorescence microscopy (LSFM) features optical sectioning in the excitation process. It minimizes fluorophore bleaching as well as phototoxic effects and provides a true axial resolution. The detection path resembles properties of conventional fluorescence microscopy. Structured illumination microscopy (SIM) is attractive for superresolution because of its moderate excitation intensity, high acquisition speed, and compatibility with all fluorophores. We introduce SIM to LSFM because the combination pushes the lateral resolution to the physical limit of linear SIM. The instrument requires three objective lenses and relies on methods to control two counterpropagating coherent light sheets that generate excitation patterns in the focal plane of the detection lens. SIM patterns with the finest line spacing in the far field become available along multiple orientations. Flexible control of rotation, frequency, and phase shift of the perfectly modulated light sheet are demonstrated. Images of beads prove a near-isotropic lateral resolution of sub-100 nm. Images of yeast endoplasmic reticulum show that coherent structured illumination (csi) LSFM performs with physiologically relevant specimens.
The prime reason to use fluorescence light microscopy is the observation of live, thick, multicellular, and 3D specimens under close to natural conditions. Thus, one has to work with specimens that are mounted in an aqueous medium and not attached to or established close to a coverslip. In addition, the main issues in fluorescence microscopy, fluorophore bleaching, endogenous organic molecule phototoxicity, and solar-level intensities have to be addressed (1).
Light-sheet-based fluorescence microscopy (LSFM) has emerged as one of the most valuable novel tools in developmental biology (1–5), plant biology (6), and 3D cell biology (7). In contrast to an epifluorescence arrangement, LSFM uses at least two independently operated microscope objective lenses. The lenses used in the excitation of the fluorophores are arranged at an angle of 90° relative to those used for the detection of the 3D fluorophore density distribution. In addition, special optical arrangements ensure that only a thin planar section centered on the focal planes of the detection lenses receives light. Hence, optical sectioning (8, 9), which is not available in conventional fluorescence microscopy, as well as no phototoxicity and no fluorophore bleaching outside a small volume close to the focal plane, are intrinsic properties of LSFM. The energy required to excite the fluorophores when recording a 3D stack of images can be reduced by two to four orders of magnitude relative to wide-field, confocal, and two-photon fluorescence microscopy (10, 11). Modern cameras are used to record millions of pixels in parallel, i.e., tens to hundreds of images with subcellular resolution within a few seconds.
Because the lateral resolution of LSFM is similar to that of a conventional epifluorescence microscope, superresolution techniques have been combined with LSFM. A promising complementary contrast could be photoactivated localization microscopy (PALM), but the acquisition speed of localization microscopy (12) is slow. The combination with stimulated emission depletion (STED) microscopy improves the axial resolution (13) but suffers from a very low signal. The most reasonable approach combines structured illumination fluorescence microscopy (SIM) with LSFM. One advantage of SIM over localization microscopies is the high acquisition speed (14–17), another one is the applicability of SIM for live imaging (18, 19). In combination with TIRF, a 2D SIM achieves a frame rate in excess of 10 Hz (18). However, SIM images are usually recorded close to the surface of the specimen whereas biological samples usually feature a 3D structure and hence a certain thickness.
Because the detection path in LSFM is identical to that of conventional fluorescence microscopy, essentially all known fluorescence-based methods can be implemented, e.g., FCS (20), fluorescence lifetime imaging/fluorescence resonance energy transfer (FLIM/FRET) (21, 22), Raman imaging (23), and certainly SIM. However, very few setups combining LSFM and SIM have been presented. Most impressive are the concepts of Eric Betzig, which combine parts of the SIM concept with LSFM by scanning Bessel beams (24) and by implementing an optical lattice (25) to create a periodic illumination pattern. However, the pattern cannot be rotated in the lattice light-sheet microscope (LLSM), thus the resolution is only improved in one axis in the lateral plane. The frequency of the lattice pattern is also limited and the resolution improvement is only by a factor between 1.3 and 1.5.
We used two coherent counterpropagating light sheets to generate a 100% modulated structured illumination pattern. We refer to it explicitly as coherent structured illumination (csi) light-sheet-based fluorescence microscopy (csiLSFM). The structured illumination pattern can be rotated, and the spatial frequency as well as the spatial phase shift of the pattern are precisely controlled. We measured a resolution of 89 nm with 40-nm fluorescent beads. Such a resolution is similar to that presented in a TIRF-SIM close to the surface of a glass plate (18, 26, 27). We applied this instrument to generate images of endoplasmic reticulum (ER) in yeast. In addition, our method to control the path of light is different from that in a conventional LSFM and will become the basis for further versions of both LSFM and SIM.
Results
Ideal Instrument.
In SIM, the achievable resolution improvement is proportional to the spatial period of the pattern. A structured pattern is created by two interfering laser beams, which intersect at an angle α in the focal plane of the detection system (14, 17, 18). The spatial frequency (f) of the pattern depends on the angular aperture (AA, also known as ) of the objective lens and is described by
| [1] |
where λ is the vacuum wavelength of the laser light, n is the refractive index of the medium, and α is the half-intersection angle of the two laser beams. The maximal frequency is achieved with two counterpropagating beams. In this case, the spatial period of the interference pattern becomes , and the resolution reaches the linear physical optimum (SI Appendix, Fig. S2). Because conventional SIM uses the same lens for illumination as well as detection, the achievable frequency is limited by the AA of the objective lens. However, in LSFM the illumination and detection paths are operated independently, which provides further degrees of freedom. LSFM also improves the axial resolution, which results in less out-of-focus light, lower photobleaching, and fewer phototoxicity side effects.
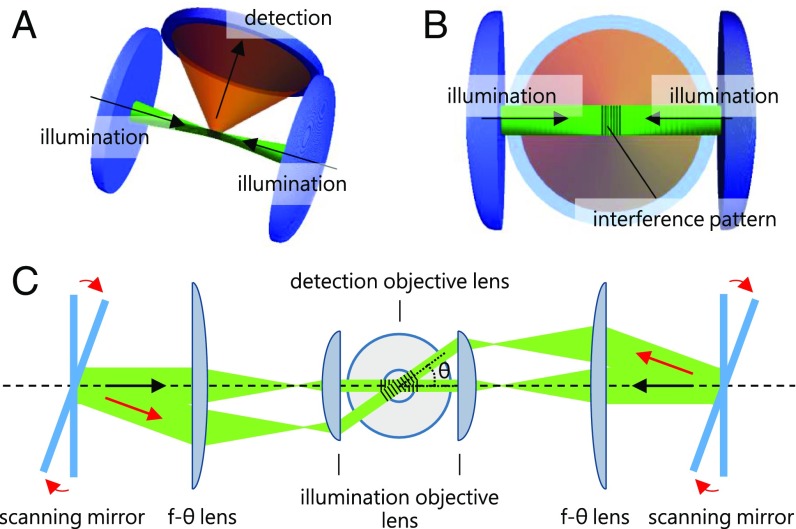
The basic concept of our implementation is illustrated in Fig. 1. The structured illumination pattern generated by the interference of two counterpropagating light sheets overlaps with the focal plane of the detection objective lens. Two scanning mirrors and two f-theta (f-θ) lenses are mounted in two separately and independently operated but coherent illumination light paths. The scanning mirrors tilt the light sheets, i.e., rotate them around the optical axis of the detection system and, consequently, generate interference patterns of two counterpropagating light sheets along different orientations.
Fig. 1.
csi of a fluorescent specimen with counterpropagating light sheets. (A) Two microscope objective lenses are used to illuminate the specimen and at least one microscope objective lens collects the fluorescence light. (B) The two coherent counterpropagating light sheets interfere in the specimen and generate the structured illumination pattern in the focal plane of the detection lens. (C) The interference of two complementarily tilted counterpropagating light sheets generates a structured illumination pattern of the same spatial frequency that is rotated around the optical axis of the detection system.
Ideally, the orientations of the interference pattern are spaced evenly in a 360° circle for an isotropic high-resolution image, for example, 0°, 60°, and 120° for three orientations and 0°, 45°, 90°, and 135° for four orientations (17). However, in an optical system that consists of two opposing illumination lenses the maximal angle is determined by their AA. A structured pattern with an orientation of 60° requires an AA of 0.866 and an orientation of 45° requires an AA of 0.707. The detection objective lens should also have a large AA for high-resolution imaging.
Real Instrument.
Whereas Fig.1 illustrates the layout of an ideal system, a real system has to take the free working distances and the diameters of the lenses into account and hence operate with space in between at least three objective lenses. Consequently, a long-working-distance objective lens is highly desirable. In practice, the working distance is inversely proportional to the AA. The optimal objective lens we could obtain recently is a 63× water-immersion objective lens with a working distance of 2.1 mm and a numerical aperture of 1.0 (Carl Zeiss, 63×/N.A. 1.0, W Plan-Apochromat M27, 421480–9900-000, Carl Zeiss). This lens provides a maximum angle of 48.75° for rotating the pattern. The high-resolution image in SIM with only 0°, 45°, and 135° reconstruction may not be perfectly isotropic but suffices for initial experiments that are merely limited by technical issues. We cannot achieve the ideal frontal arrangement of three objective lenses, due to the extents of the objective lenses. Therefore, the illumination objective lenses are tilted by about 25° horizontally in an arrangement shown in Fig. 2C. We achieve an intersection angle of 130° for the two light sheets that enter the center of the objective lenses. More objective lenses can be combined to either to generate the interference pattern with other orientations or to simultaneously detect an image along further directions. However, with the currently available objective lenses, we can only use a setup with three lenses. An appropriate chamber for the three objective lenses arrangement is shown in SI Appendix, Fig. S3.
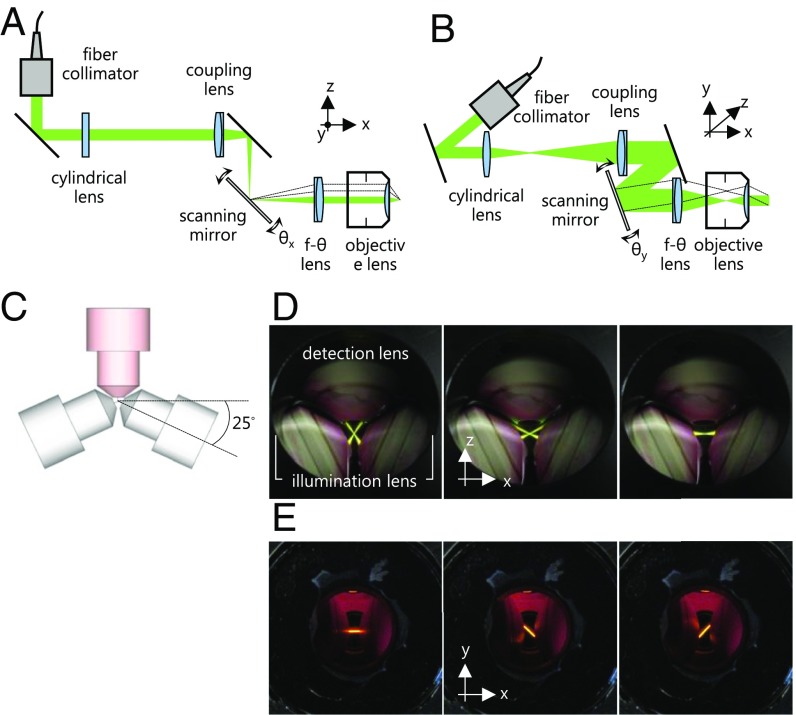
Fig. 2.
Flexible control of light sheets. (A and B) Top and side views of one arm of the illumination system. The scanning mirror is used to direct the light sheet along two axes as indicated by the dashed lines. (C) Top view of three objective lenses, which lie in a single plane. The two illumination lenses are centered on the common focal point of three lenses and rotated by an angle of ∼25° relative to the focal plane of the detection lens. (D) Top view of the two light sheets. The two scanning mirrors are used complementarily to guide the two light sheets along different angles (52°, 25°, and 0°). (E) Side view of two light sheets recorded through the front window of the specimen chamber. The chamber is filled with 50 μM Rose Bengal in PBS buffer to visualize the fluorescence emission generated by the light sheets with a 543-nm HeNe laser (25-LGP-193–230, Melles Griot). Two light sheets are rotated by an angle of 45° relative to the normal axis of the focal plane. The top view of the directions of two light sheets in E is identical to the image in the middle of D.
Flexible Control of the Interference Pattern.
A particular advantage of our system is the flexible control of the two light sheets. One cylindrical lens, one coupling lens, one two-axis scanning mirror, one f-θ lens (also known as scanning lens), and one objective lens are mounted in both of the identical illumination arms. The cylindrical lens and the coupling lens create a line-shaped beam on the two-axis scanning mirror. The f-θ lens and the objective lens form a telecentric optical configuration and transform the line-shaped beam into a light sheet in the focal plane of the detection system (28). Therefore, tilting the scanning mirror in the x axis (θx) influences the intersection angle of the two light sheets, as shown in Fig. 2A. The vertical tilt of the scanning mirror (θy) results in the rotation of the light sheet in the lateral plane of the detection objective lens, as shown in Fig. 2B. In short, this setup allows a manipulation of the light sheet in three dimensions. Some examples of the possible interference conditions are shown in SI Appendix, Fig. S6. Fig. 2 D and E shows the macroscopic movements of the light sheets as described above.
Controlling Frequency and Orientation of the Interference Pattern.
The N.A. 1.0 objective lens has a half-aperture angle α of 48.75°. The smallest period of the interference pattern that can be observed with this objective lens is 244 nm (, , and ). The period of the interference pattern of two counterpropagating light sheets (T = 183.5 nm) or two light sheets with an intersection angle of 130° (T = 202.4 nm) is beyond the diffraction limit of the detection lens and, therefore, too fine to be visualized directly. To observe the interference pattern and characterize as well as demonstrate its properties, e.g., the period, the rotation, and the phase shift, we position the scanning mirrors to let the light sheets enter both the illumination and detection objective lenses, as shown in the leftmost image in Fig. 2D. Initially, we demonstrate the interference of two collimated beams in Fig. 3 A and B. The cylindrical lenses and the coupling lenses were removed and two collimated beams covering the entire field of view make it easier to visualize the modulation of the interference pattern. The control of the period of the interference pattern is shown in Fig. 3A. A series of interference patterns with different periods is acquired by varying the intersection angle between two collimated beams. Next, we fix the period of the interference pattern and simultaneously tilt the two scanning mirrors vertically, thus rotating the interference pattern (Fig. 3B). The phase of the interference pattern must be controlled precisely and should change rapidly. In our system, we use a piezo-driven device to move one of the collimators as shown in SI Appendix, Fig. S5A. Another means to control the phase of the interference pattern using only the scanning mirrors is illustrated in SI Appendix, Fig. S4.
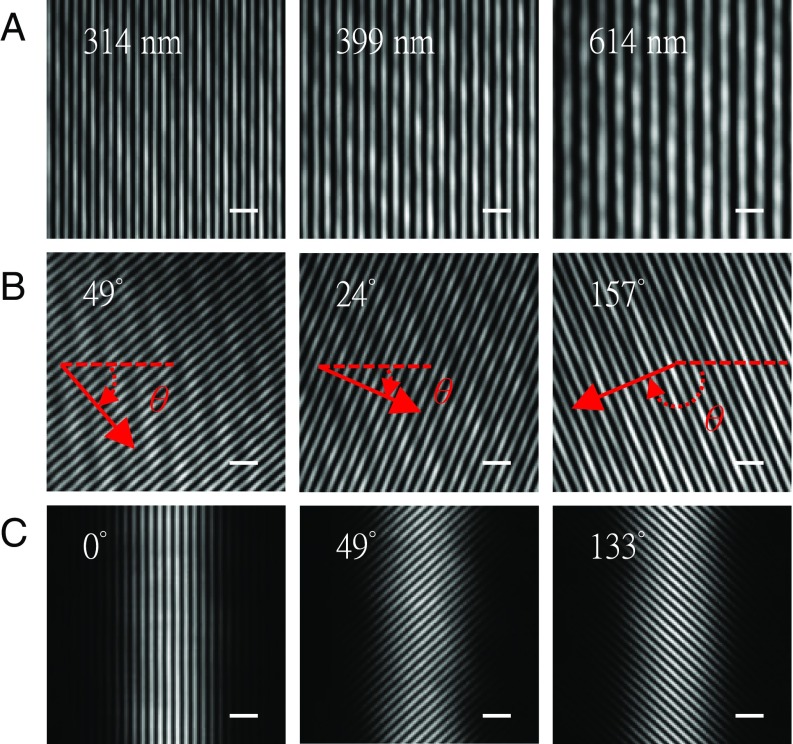
Fig. 3.
Flexible control of interference pattern. For demonstration purposes, the scanning mirrors are tilted to direct the coherent beams into the detection objective lens and thus on the camera. (A) Three images demonstrate the generation of different spatial periods in the illumination pattern by changing the relative angle θx (defined in Fig. 2) of the scanning mirrors. The interference pattern is observable within a certain angular range, i.e., as long as the beams pass both the illumination and detection objective lenses. (B) Three images show the effect of changing the angle θy (defined in Fig. 2) and thus the pattern orientation angles of 49°, 24°, and 157°. (C) Three images show the interference pattern of two light sheets with pattern orientation angles of 0°, 49°, and 133°. The spatial period of the interference pattern is 301 nm. (Scale bars, 1 μm wide.)
Next, the cylindrical lenses and the coupling lenses were reinserted and the line-shaped laser beams in the back focal plane of the illumination objective lenses and the equivalent light sheets were formed. The interference pattern of the two light sheets follows a straight line within the Rayleigh range (Fig. 3 and SI Appendix, Fig. S7A). The interference patterns of two light sheets with three orientations are shown in Fig. 3C. The entire line-shaped beam still enters the illumination objective lens when the maximum rotary angle of the interference pattern is set to 49° (SI Appendix, Fig. S7). The diameter of the entire line-shaped beam or a portion exceeds the diameter of the aperture of the objective lens if a larger rotary angle is applied. We validated the period, the orientation, and the phase shift of the interference pattern with the Fourier transform of the pattern image.
Superresolution Image at 89 nm.
To demonstrate the resolution improvement, we embedded 40-nm (yellow-green, F8795, Invitrogen) or 100-nm fluorescent latex beads (yellow-green, F8803, Invitrogen) in 1.5% low-melt agarose (6351.5, Carl Roth). We commenced to image with a small intersection angle of two light sheets and gradually increased the angle. We also increased the rotation angle of the interference pattern at a small intersection angle and inspected the overlap of the two light sheets. They overlapped perfectly when the positions of the scanning mirrors were slightly adjusted with the motorized linear actuators (SI Appendix, Fig. S5A). Thus, we were able to document the raw images of SIM (SI Appendix, Fig. S8), the resolution improvement (SI Appendix, Fig. S9), and the maximum rotation angle of the pattern (SI Appendix, Fig. S10).
Finally, we successfully overlapped the counterpropagating light sheets, which created the finest interference pattern. The striking improvement of the resolution is shown in Fig. 4. With the interference pattern created by counterpropagating light sheets, we are able to reach the optimal resolution in a linear SIM. An average resolution of 114.1 ± 10.43 nm is obtained. The Richardson–Lucy deconvolution replaces the Wiener deconvolution commonly used in SIM and provides a final resolution of 89.3 ± 13.47 nm (29)(SI Appendix, SIM reconstruction algorithm in Note 1, Fig. S14, and Table S1). We stress the further improvement of the resolution with the deconvolution is a windfall gain. The final resolution is a 2.4× and a 3.2× improvement compared with a deconvolved wide-field (204.9 ± 4.75 nm) and a conventional wide-field (283.0 ± 12.18 nm) image, respectively (SI Appendix, Table S1). In the linear regime such a resolution has only been reported for TIRF-SIM (18, 26, 27).
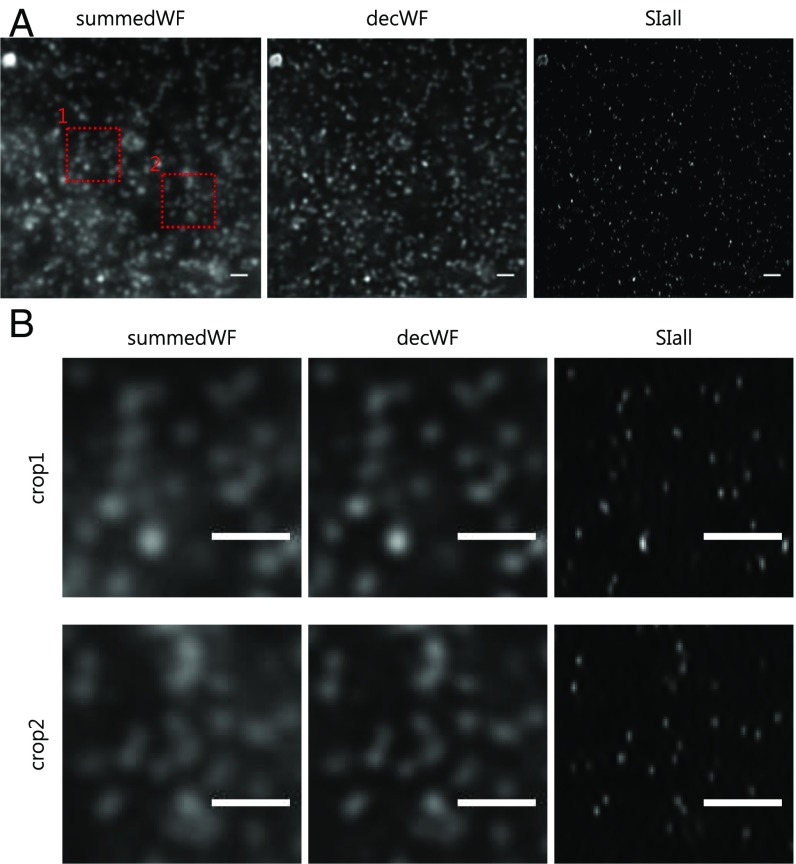
Fig. 4.
High-resolution image with counterpropagating light sheets. (A) Conventional wide-field (summedWF), deconvolved wide-field (decWF), and SIM (SIall) images. The beads are embedded in 1.5% low-melt agarose. (B) Magnified images of the red dashed boxes in A clearly show the resolution improvement in the SIM (SIall) image. (Scale bars, 1 µm wide.)
Near-Isotropic High-Resolution Image at Sub-100 nm.
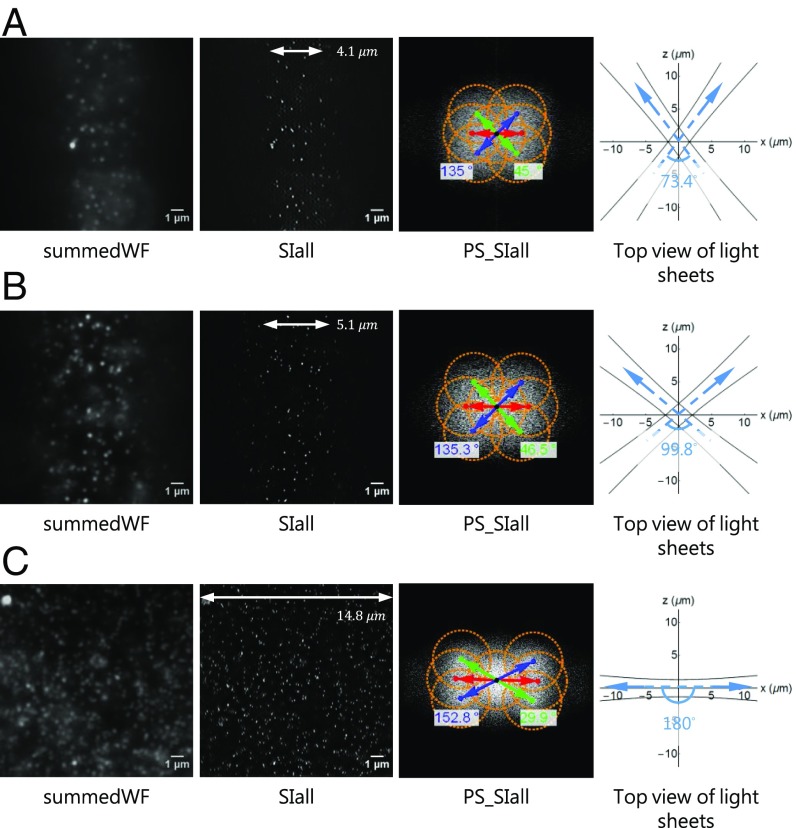
With counterpropagating illumination the resolution is anisotropic. This is due to the spatial constraints of objective lenses (Fig. 2C and SI Appendix, Fig. S6). However, we can intersect two light sheets at different angles. The corresponding resolution, image quality, maximum rotation angle of the interference pattern, and illuminated area are shown in Fig. 5. The detailed quantifications of the patterns and the resolutions are summarized in SI Appendix, Table S2. The resolution improves as the intersection angle and hence the pattern frequency increase (from Fig. 5A to Fig. 5C). The illumination by counterpropagating light sheets confirms less background and a larger illuminated area of the image because the light sheets are orthogonal to the detection axis whereas the rotation angle of its interference pattern is smaller, i.e., restricted to ∼27° due to the available objective lens (Fig. 5C). However, the rotation angle of the interference pattern can be increased (∼45° in Fig. 5B) with a smaller intersection angle between two light sheets. The resolution is ∼100 nm [SI Appendix, Table 2, 97.8 ± 11.47 nm (x) and 117.4 ± 10.92 nm (y)], the background is slightly higher, and the lateral illuminated area is smaller (Fig. 5 and SI Appendix, Fig. S16). The illumination configuration shown in Fig. 5B is considered a good compromise between resolution, image quality, and isotropy of the resolution improvement. When imaging biological specimens, we mostly use this illumination configuration because of a more isotropic image and a reasonable background quality. Finally, a high-resolution 3D image stack can be acquired by simply scanning the specimen along the detection axis (SI Appendix, Figs. S12 and S14). The bead images shown in SI Appendix, Figs. S12 and S14 essentially represent the 3D point-spread function of our system. The axial resolution in csiLSFM is similar to that in a conventional LSFM, which is 880.29 ± 72.84 nm in our measurements.
Fig. 5.
Resolution, image quality, and illuminated area at different intersection angles. Conventional wide-field image (summedWF), SIM image (SIall), and the power spectra of the SIM image (PS_SIall). The drawings overlaying the spectra indicate the extended frequency information in the SIall images and the achievable rotation angle of the pattern. The orange circles represent the optical transfer function (OTF) region, whose radius is 2 N.A.∕λ (N.A. = 1.0, λ = 515 nm). The white arrows represent the corresponding illuminated areas. The top view illustrates the configuration of two light sheets at 0° pattern orientation. The intersection angles of the 0° pattern in A, B, and C are 73.4°, 99.8°, and 180°, respectively, which result in pattern periods of 307, 240, and 183 nm, respectively.
Applications to Biology.
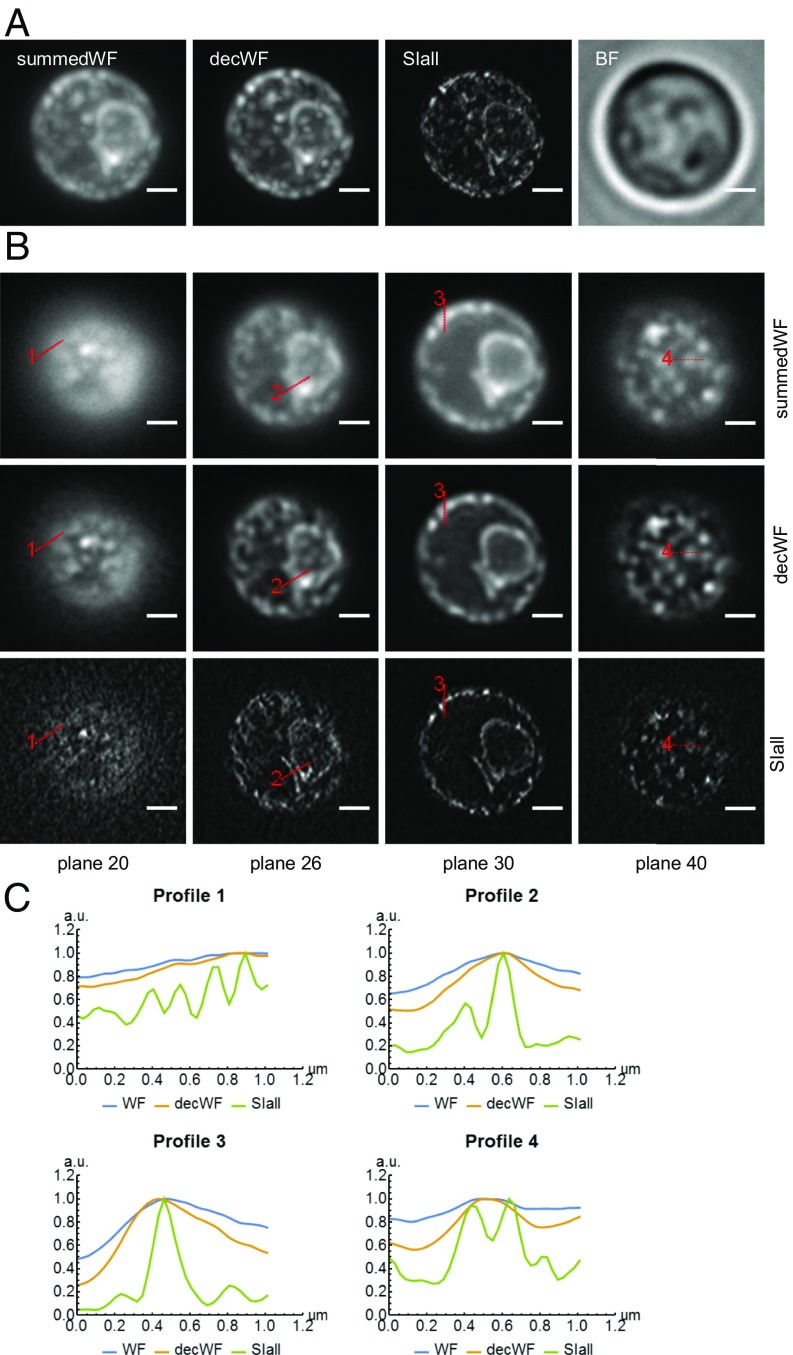
Because csiLSFM is designed to work with fluorescently labeled biological specimens, we recorded images of GFP-tagged endoplasmic reticulum in wild-type yeast (BY4741, transformed with pRS415-ER-sfGFP-HDEL) (30). We directly embedded the live yeast cells in 1.5% low-melt agarose. The cells remain alive for many hours with this preparation method. One example of the high-resolution images of yeast is shown in Fig. 6. A 3D rendering of the same image stack is shown in Movie S1. The images were recorded with an exposure time of 100 ms for each image, three images (phases) per orientation, and three orientations in each plane. Both the maximum-intensity projections and several single planes show the striking improvement of the image with our csiLSFM. We also profiled ER structures and showed that the SIM images reveal many details that cannot be observed in either wide-field or deconvolved wide-field images. More examples of high-resolution yeast images are shown in SI Appendix, Figs. S17 and S18. We also imaged fixed yeast cells; however, the ER structures appear different from the ones in a live yeast cell (SI Appendix, Fig. S19). We conclude that our fixation process damages the cell and suggest to maintain the biological specimen in a state that allows it to live long and prosper. The setup also allows to mount the sample on a fluorinated ethylene propylene (FEP) film, which assists future applications of csiLSFM (SI Appendix, Fig. S20).
Fig. 6.
High-resolution images of GFP-tagged ER structure within live yeast cells embedded in 1.5% low-melt agarose. (A) Maximum-intensity projections of conventional wide-field (summedWF), deconvolved wide-field (decWF), and SIM (SIall) image stacks as well as the bright-field (BF) image of a live yeast cell. (B) Four planes were chosen to allow a more detailed comparison between the summedWF, decWF, and the SIall image. The detailed structure of the ER, which appears to be quite blurry in both summedWF and decWF images, is resolved in the SIM images. The image stack consists of 50 planes with an axial spacing of 200 nm. (Scale bars, 1 μm wide.) (C) The line profiles of the SIM (SIall) image show the ER structure, which cannot be observed in either the summedWF or the decWF image.
Discussion
We developed a versatile approach to beam-scanning LSFM, which is based on a single mirror that is tilted along two orthogonal axes. The beam is focused either into the entrance aperture or into the image plane of an objective lens. This means the beam can be rotated around well-defined locations and can be moved to any location in the focal plane of the objective lens. Two such well-controlled counterpropagating beams or light sheets generate the pattern required for csi. The incoherent structured illumination (isi) (31) in LSFMs has been implemented by inserting a grating in a complementary image plane of a single-plane illumination fluorescence microscope (32) and by systematically modulating the illumination beam in a digital scanned light-sheet-based fluorescence microscope (33). However, the isi does not improve the resolution but rejects out-of-focus light to improve image quality similar to HiLo background rejection, which uses a modified reconstruction algorithm (34). The modulation depth of the isi pattern decreases, essentially linearly, with the frequency of the pattern until it becomes zero. Hence, its cutoff frequency is low. In contrast, the intensities and the collinear polarization state (s polarization) of two beams are well controlled in csiLSFM, enabling a 100% modulation depth of the pattern, as well as a frequency that can be up to a factor of 4 higher than the cutoff frequency of the isi pattern (18, 35). Whereas the axial resolution is not enhanced in the current implementation, csiLSFM improves two weaknesses of LLSM. The lateral resolution of csiLSFM is better (sub-100 nm vs. 150 nm) and closer to isotropic because the pattern can be rotated in csiLSFM.
High spatial and temporal (more than one stack per second) resolution, together with low photodamage and low phototoxicity for long time-lapse imaging in live 3D specimens are the most obvious direction for further developments of advanced light microscopy (11, 24, 25). csiLSFM, which combines advantages of SIM and LSFM, is certainly one option to approach this goal (25). The resolution improvement in SIM relies on the illumination pattern frequency. Whereas a TIRF-SIM only observes the surface of a specimen close to the coverslip (18), csiLSFM realizes the maximum frequency by interfering two counterpropagating beams and the superresolution is not limited to the surface. Thus, csiLSFM is a suitable approach for the investigation of biological specimens which are cultivated in a 3D environment, i.e., the specimens are not collapsed to a flat structure. csiLSFM reduces the out-of-focus fluorescence. It uses 2D-SIM rather than 3D-SIM, which increases the acquisition speed by a factor of 5/3. csiLSFM is applicable to any small biological specimen. Following the imaging of ER in wild-type yeast, the dynamics of the ER membrane in the context of ER stress will be further investigated (36). The number of autophagosome can be measured more precisely and the autophagy in fungi can be studied further (37). Spatial organization of RNA polymerase and the dynamics of nucleoids in Escherichia coli were studied with superresolution techniques. csiLSFM definitely fits such topics very well (38, 39).
The resolution anisotropy is due to the technical specifications of the lenses. Longer working distances and different AAs are necessary to simplify the current optical arrangement. One option to address the resolution anisotropy is to add more illumination lenses and space them evenly in a 360° circle on the plane perpendicular to the detection objective lens. Improving axial resolution and achieving isotropic superresolution are parts of our ongoing developments. I5S microscopy achieves impressive 100-nm resolution in both lateral and the axial directions because it applies the interference pattern from six beams and collects interference images from two objective lenses (40). However, the alignment to create image interference (I2M) is challenging. In contrast, csiLSFM applies an obvious but stable method to create the interference pattern reaching the finest limit. Additionally, csiLSFM is capable of rotating the specimen and implementing multiple-views imaging to improve the axial resolution (41, 42). Lower wavelengths can be used that work well with smaller molecules. In this manner, isotropic sub-100-nm resolution can be achieved. csiLSFM can also be combined with the oblique illumination used, e.g., in LLSM and increases the field of view (25).
As pointed out, we used a fairly relaxed approach to LSFM. In fact, it comprises many of the approaches that have been discussed and implemented over the past 15 y and does not discard the advantages, for which LSFM is known. We used this approach to implement a few aspects of csiLSFM and thus demonstrate the feasibility of superresolution in an LSFM. Many more approaches that are currently not possible with SIM can now be attempted. For example, a specimen can be observed with multiple different periods and with more than three phases to fill the k space. Alternatively, our system allows the observation of specimens along multiple directions by generating patterns that intersect the focal plane of the detection lens from different angles. Finally, technical developments such as new lenses will allow us to further relax the optical layout of the system.
Materials and Methods
Details of experimental setup, method of embedding samples in gel, SIM reconstruction algorithm, image presentation, and our resolution measurement are presented in SI Appendix.
Supplementary Material
Acknowledgments
We thank Prof. Dr. Andreas Reichert and Matthias Müller for the yeast cell line in the preliminary test and helpful remarks; the Goethe Universität’s workshop for custom hardware; Frederic Strobl for suggesting the phytagel preparation and the design of the sample holder; and Prof. Dr. Rainer Heintzmann and Aurélie Jost (IPC, Friedrich-Schiller-Universität Jena) for discussions of the reconstruction algorithm. We especially thank Prof. Dr. Robert Ernst and Stephanie Ballweg [Buchmann Institute for Molecular Life Sciences (BMLS), Goethe Universität Frankfurt am Main] for the yeast cell line used in this article and valuable comments and Eric Snapp for kindly providing the plasmid pRS415-ER-sfGFP-HDEL used in the yeast cell. We also thank Sven Plath for substantial assistance concerning the hardware and Alexander Schmitz for help with the Mathematica-based image processing. The research was funded by the Cluster of Excellence for Macromolecular Complexes granted to the Goethe Universität Frankfurt am Main by the Deutsche Forschungsgemeinschaft (DFG Grant EXC-115). The research was also supported by the National Science Council (NSC Grant 100-2917-I-564-030) in Taiwan, Republic of China and European Molecular Biology Organization (EMBO) (Fellowship ASTF 404-2012).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609278114/-/DCSupplemental.
References
- 1.Stelzer EHK. Light-sheet fluorescence microscopy for quantitative biology. Nat Methods. 2015;12:23–26. doi: 10.1038/nmeth.3219. [DOI] [PubMed] [Google Scholar]
- 2.Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EHK. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 3.Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EHK. Digital scanned laser light-sheet fluorescence microscopy (DSLM) of zebrafish and Drosophila embryonic development. Cold Spring Harb Protoc. 2011;2011:1235–1243. doi: 10.1101/pdb.prot065839. [DOI] [PubMed] [Google Scholar]
- 4.Strobl F, Stelzer EHK. Non-invasive long-term fluorescence live imaging of Tribolium castaneum embryos. Development. 2014;141:2331–2338. doi: 10.1242/dev.108795. [DOI] [PubMed] [Google Scholar]
- 5.Strobl F, Schmitz A, Stelzer EHK. Live imaging of Tribolium castaneum embryonic development using light-sheet-based fluorescence microscopy. Nat Protoc. 2015;10:1486–1507. doi: 10.1038/nprot.2015.093. [DOI] [PubMed] [Google Scholar]
- 6.Maizel A, von Wangenheim D, Federici F, Haseloff J, Stelzer EHK. High-resolution live imaging of plant growth in near physiological bright conditions using light sheet fluorescence microscopy. Plant J. 2011;68:377–385. doi: 10.1111/j.1365-313X.2011.04692.x. [DOI] [PubMed] [Google Scholar]
- 7.Pampaloni F, Ansari N, Stelzer EHK. High-resolution deep imaging of live cellular spheroids with light-sheet-based fluorescence microscopy. Cell Tissue Res. 2013;352:161–177. doi: 10.1007/s00441-013-1589-7. [DOI] [PubMed] [Google Scholar]
- 8.Cox IJ. Scanning optical fluorescence microscopy. J Microsc. 1984;133:149–154. doi: 10.1111/j.1365-2818.1984.tb00480.x. [DOI] [PubMed] [Google Scholar]
- 9.Engelbrecht CJ, Stelzer EH. Resolution enhancement in a light-sheet-based microscope (SPIM) Opt Lett. 2006;31:1477–1479. doi: 10.1364/ol.31.001477. [DOI] [PubMed] [Google Scholar]
- 10.Reynaud EG, Krzic U, Greger K, Stelzer EHK. Light sheet-based fluorescence microscopy: More dimensions, more photons, and less photodamage. HFSP J. 2008;2:266–275. doi: 10.2976/1.2974980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Planchon TA, et al. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat Methods. 2011;8:417–423. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cella Zanacchi F, et al. Live-cell 3D super-resolution imaging in thick biological samples. Nat Methods. 2011;8:1047–1049. doi: 10.1038/nmeth.1744. [DOI] [PubMed] [Google Scholar]
- 13.Friedrich M, Gan Q, Ermolayev V, Harms GS. STED-SPIM: Stimulated emission depletion improves sheet illumination microscopy resolution. Biophys J. 2011;100:L43–L45. doi: 10.1016/j.bpj.2010.12.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gustafsson MGL, Agard DA, Sedat JW. Doubling the lateral resolution of wide-field fluorescence microscopy using structured illumination. In: Conchello J-A, Cogswell CJ, Tescher AG, Wilson T, editors. BiOS 2000 The International Symposium on Biomedical Optics. International Society for Optics and Photonics; Bellingham, WA: 2000. pp. 141–150. [Google Scholar]
- 15.Fedosseev R, Belyaev Y, Frohn J, Stemmer A. Structured light illumination for extended resolution in fluorescence microscopy. Opt Lasers Eng. 2005;43:403–414. [Google Scholar]
- 16.Heintzmann R, Cremer CG. Laterally modulated excitation microscopy: Improvement of resolution by using a diffraction grating. In: Bigio IJ, Schneckenburger H, Slavik J, Svanberg K, Viallet PM, editors. BiOS Europe ’98. International Society for Optics and Photonics; Bellingham, WA: 1999. pp. 185–196. [Google Scholar]
- 17.Chang B-J, Chou L-J, Chang Y-C, Chiang S-Y. Isotropic image in structured illumination microscopy patterned with a spatial light modulator. Opt Express. 2009;17:14710–14721. doi: 10.1364/oe.17.014710. [DOI] [PubMed] [Google Scholar]
- 18.Kner P, Chhun BB, Griffis ER, Winoto L, Gustafsson MGL. Super-resolution video microscopy of live cells by structured illumination. Nat Methods. 2009;6:339–342. doi: 10.1038/nmeth.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirvonen LM, Wicker K, Mandula O, Heintzmann R. Structured illumination microscopy of a living cell. Eur Biophys J. 2009;38:807–812. doi: 10.1007/s00249-009-0501-6. [DOI] [PubMed] [Google Scholar]
- 20.Wohland T, Shi X, Sankaran J, Stelzer EHK. Single plane illumination fluorescence correlation spectroscopy (SPIM-FCS) probes inhomogeneous three-dimensional environments. Opt Express. 2010;18:10627–10641. doi: 10.1364/OE.18.010627. [DOI] [PubMed] [Google Scholar]
- 21.Greger K, Neetz MJ, Reynaud EG, Stelzer EHK. Three-dimensional fluorescence lifetime imaging with a single plane illumination microscope provides an improved signal to noise ratio. Opt Express. 2011;19:20743–20750. doi: 10.1364/OE.19.020743. [DOI] [PubMed] [Google Scholar]
- 22.Costa A, Candeo A, Fieramonti L, Valentini G, Bassi A. Calcium dynamics in root cells of Arabidopsis thaliana visualized with selective plane illumination microscopy. PLoS One. 2013;8:e75646. doi: 10.1371/journal.pone.0075646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oshima Y, et al. Light sheet-excited spontaneous Raman imaging of a living fish by optical sectioning in a wide field Raman microscope. Opt Express. 2012;20:16195. [Google Scholar]
- 24.Gao L, et al. Noninvasive imaging beyond the diffraction limit of 3D dynamics in thickly fluorescent specimens. Cell. 2012;151:1370–1385. doi: 10.1016/j.cell.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen B-C, et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beck M, Aschwanden M, Stemmer A. Sub-100-nanometre resolution in total internal reflection fluorescence microscopy. J Microsc. 2008;232:99–105. doi: 10.1111/j.1365-2818.2008.02075.x. [DOI] [PubMed] [Google Scholar]
- 27.Fiolka R, Beck M, Stemmer A. Structured illumination in total internal reflection fluorescence microscopy using a spatial light modulator. Opt Lett. 2008;33:1629–1631. doi: 10.1364/ol.33.001629. [DOI] [PubMed] [Google Scholar]
- 28.Greger K, Swoger J, Stelzer EHK. Basic building units and properties of a fluorescence single plane illumination microscope. Rev Sci Instrum. 2007;78:023705. doi: 10.1063/1.2428277. [DOI] [PubMed] [Google Scholar]
- 29.Perez V, Chang BJ, Stelzer EH. Optimal 2D-SIM reconstruction by two filtering steps with Richardson-Lucy deconvolution. Sci Rep. 2016;6:37149. doi: 10.1038/srep37149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lajoie P, Moir RD, Willis IM, Snapp EL. Kar2p availability defines distinct forms of endoplasmic reticulum stress in living cells. Mol Biol Cell. 2012;23:955–964. doi: 10.1091/mbc.E11-12-0995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neil MAA, Juškaitis R, Wilson T. Method of obtaining optical sectioning by using structured light in a conventional microscope. Opt Lett. 1997;22:1905–1907. doi: 10.1364/ol.22.001905. [DOI] [PubMed] [Google Scholar]
- 32.Breuninger T, Greger K, Stelzer EHK. Lateral modulation boosts image quality in single plane illumination fluorescence microscopy. Opt Lett. 2007;32:1938–1940. doi: 10.1364/ol.32.001938. [DOI] [PubMed] [Google Scholar]
- 33.Keller PJ, et al. Fast, high-contrast imaging of animal development with scanned light sheet-based structured-illumination microscopy. Nat Methods. 2010;7:637–642. doi: 10.1038/nmeth.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mertz J, Kim J. Scanning light-sheet microscopy in the whole mouse brain with HiLo background rejection. J Biomed Opt. 2010;15:016027. doi: 10.1117/1.3324890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang H-C, Chang B-J, Chou L-J, Chiang S-Y. Three-beam interference with circular polarization for structured illumination microscopy. Opt Express. 2013;21:23963–23977. doi: 10.1364/OE.21.023963. [DOI] [PubMed] [Google Scholar]
- 36.Westrate LM, Lee JE, Prinz WA, Voeltz GK. Form follows function: the importance of endoplasmic reticulum shape. Annu Rev Biochem. 2015;84:791–811. doi: 10.1146/annurev-biochem-072711-163501. [DOI] [PubMed] [Google Scholar]
- 37.Knuppertz L, Hamann A, Pampaloni F, Stelzer E, Osiewacz HD. 2014 Identification of autophagy as a longevity-assurance mechanism in the aging model Podospora anserina. Autophagy. Available at www.tandfonline.com/doi/full/10.4161/auto.28148. Accessed October 12, 2015.
- 38.Endesfelder U, et al. Multiscale spatial organization of RNA polymerase in Escherichia coli. Biophys J. 2013;105:172–181. doi: 10.1016/j.bpj.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spahn C, Endesfelder U, Heilemann M. Super-resolution imaging of Escherichia coli nucleoids reveals highly structured and asymmetric segregation during fast growth. J Struct Biol. 2014;185:243–249. doi: 10.1016/j.jsb.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 40.Shao L, et al. I5S: Wide-field light microscopy with 100-nm-scale resolution in three dimensions. Biophys J. 2008;94:4971–4983. doi: 10.1529/biophysj.107.120352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verveer PJ, et al. High-resolution three-dimensional imaging of large specimens with light sheet-based microscopy. Nat Methods. 2007;4:311–313. doi: 10.1038/nmeth1017. [DOI] [PubMed] [Google Scholar]
- 42.Swoger J, Verveer P, Greger K, Huisken J, Stelzer EHK. Multi-view image fusion improves resolution in three-dimensional microscopy. Opt Express. 2007;15:8029–8042. doi: 10.1364/oe.15.008029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.