Significance
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disorder with wide clinical variability and a complex pathogenesis. One of the hallmarks of ALS is the presence of diverse inclusions formed by apparently disparate misfolded proteins, which are unrelated in sequence, structure, function, and localization. Many of these proteins, in addition, are not the usual interaction partners of the primary inclusion-forming ALS-related proteins (SOD1, TDP-43, and FUS). Here, we show that a common feature of these coaggregating proteins is their metastability to aggregation in spinal motor neurons. These results suggest that as protein homeostasis becomes progressively impaired in ALS, supersaturated proteins in an ALS-specific group fail to be maintained in their soluble states, leading to their presence in ALS-associated inclusions.
Keywords: protein aggregation, protein misfolding, protein homeostasis, supersaturation, motor neuron disease
Abstract
Amyotrophic lateral sclerosis (ALS) is a heterogeneous degenerative motor neuron disease linked to numerous genetic mutations in apparently unrelated proteins. These proteins, including SOD1, TDP-43, and FUS, are highly aggregation-prone and form a variety of intracellular inclusion bodies that are characteristic of different neuropathological subtypes of the disease. Contained within these inclusions are a variety of proteins that do not share obvious characteristics other than coaggregation. However, recent evidence from other neurodegenerative disorders suggests that disease-affected biochemical pathways can be characterized by the presence of proteins that are supersaturated, with cellular concentrations significantly greater than their solubilities. Here, we show that the proteins that form inclusions of mutant SOD1, TDP-43, and FUS are not merely a subset of the native interaction partners of these three proteins, which are themselves supersaturated. To explain the presence of coaggregating proteins in inclusions in the brain and spinal cord, we observe that they have an average supersaturation even greater than the average supersaturation of the native interaction partners in motor neurons, but not when scores are generated from an average of other human tissues. These results suggest that inclusion bodies in various forms of ALS result from a set of proteins that are metastable in motor neurons, and thus prone to aggregation upon a disease-related progressive collapse of protein homeostasis in this specific setting.
Amyotrophic lateral sclerosis (ALS) is a devastating neurodegenerative disorder in which the progressive and selective loss of upper and lower motor neurons in the motor cortex and spinal cord leads to impairment of muscle control and to muscle atrophy. Death invariably follows, generally within 3–5 y of diagnosis (1, 2). Neurodegeneration in ALS has been attributed to a multitude of processes, including glutamate excitotoxicity, oxidative stress, disruption of neurofilaments and axonal transport, mitochondrial dysfunction, endoplasmic reticulum stress, and, most recently, dysfunctional RNA metabolism (2–4). Growing evidence also indicates that protein aggregation is associated with all forms of ALS (5, 6), leading to the proposal that protein misfolding could be a common feature of the various forms of ALS (7, 8). In this respect, ALS may share similarities to other neurodegenerative disorders, such as Alzheimer’s, Parkinson’s, and Huntington’s diseases (8–10), which are characterized by the formation of aberrant protein assemblies.
A distinctive feature of ALS is its remarkable heterogeneity at the molecular level. Although most cases of ALS are sporadic [sporadic ALS (sALS)] and of unclear cause, the 5–10% of cases in which the disease is inherited [familial ALS (fALS)] can be linked to specific genetic mutations. Mutations in one or more of at least a dozen genes give rise to distinct disease neuropathological subtypes, each of which is associated with the aggregation of a variety of proteins into inclusions with histologically distinct structures. These mutations include mutations in genes encoding SOD1 (11), alsin (12), senataxin (13), FUS/TLS (14, 15), VAPB (16), angiogenin (17), TDP-43 (18, 19), dynactin (20, 21), FIG4 (22), optineurin (23), VCP (24), sequestosome-1 (25), ubiquilin-2 (26), profilin-1 (27), matrin-3 (28), and CCNF (29), as well as hexanucleotide expansions in C9ORF72 (30).
At the protein aggregate level, there are multiple forms of inclusions that have been described for ALS, including Bunina bodies; basophilic inclusions; spheroids; and ubiquitinated inclusions such as round bodies, skein-like inclusions, and hyaline inclusions (7, 31). In addition to their morphology, the inclusions in ALS can be categorized on the basis of the dominant protein involved. For example, ubiquitinated inclusions found in motor neurons in sALS and many fALS cases, including those cases associated with mutations in genes encoding TDP-43 or C9ORF72, are largely TDP-43–positive. In some fALS cases, the major constituent of ubiquitinated inclusions depends on the inherited genetic mutation, specifically FUS and SOD1 (32). Indeed, we recently observed that the aggregation processes of TDP-43, FUS, and SOD1 in cells are distinct, and that they result in inclusions with varied characteristics (33). In particular, SOD1 aggregates, which are the most intensely studied inclusions, have amorphous properties and interact transiently with a variety of other cellular proteins, including molecular chaperones, and they also bind stably with components of the ubiquitin–proteasome system and correlate with cell death (34–38). Furthermore, all three inclusion subtypes, TDP-43, FUS, and SOD1, are formed by mechanisms distinct from the highly structured amyloid aggregates of huntingtin (33).
Linking the initial events associated with the aggregation of mutant proteins in ALS with the ultimate failure of motor neurons has proved particularly challenging for a variety of reasons. One problem is the experimental challenge of characterizing in detail the array of secondary proteins associated with ALS inclusions. Although microdissection followed by mass spectrometry has been successful in identifying many of the constituent components of aggregates in other diseases (39, 40), the diversity and complexity of the aggregation processes in ALS have made this task difficult. Our current knowledge is largely due to more targeted histological approaches that have yielded sets of proteins that associate with ALS inclusions (e.g., ref. 32).
In the work reported here, we have sought to understand the reason why certain proteins are at risk for coaggregating in the inclusion bodies found in motor neurons, with the expectation that understanding this process will help explain the origins of the cellular dysfunctions associated with ALS and possible ways to ameliorate such dysfunctions therapeutically. We reasoned that two general mechanisms may be important for inclusion formation: (i) Individual aggregating proteins can pull down their normal interaction partners into deposits, and (ii) poorly soluble proteins can be recruited into the deposits. To discover the extent to which these two processes can explain the formation of inclusions in ALS, we considered the functional and biophysical characteristics of both the native interaction partners of ALS-associated proteins and coaggregating proteins in ALS. We considered in particular whether these two groups of proteins are supersaturated, because proteins associated with either the known set of dysfunctional cellular processes or the sets of proteins found in insoluble inclusions in a range of neurodegenerative diseases are known to be supersaturated (41). A protein is considered to be supersaturated, and therefore subject to a greater driving force toward aggregation (42), if its cellular concentration is high relative to its solubility (41). To examine if supersaturation could explain cell dysfunction in ALS, we assembled a comprehensive database of proteins known to coaggregate within various ALS inclusions and applied network and supersaturation analyses to them.
Our results indicate that coaggregating proteins in ALS inclusions are not simply a subset of the native interaction partners of the primary inclusion-forming proteins but that these coaggregators are significantly more supersaturated than the set of native interactors when the particular gene expression pattern of motor neurons is used to calculate the supersaturation scores. Thus, we found that the combination of a spinal motor neuron expression profile and a high supersaturation score can explain many key features of the disease-specific protein inclusion fingerprint. We have brought these results together by combining calculations of the degree of supersaturation with a network analysis of coaggregating proteins. This strategy has enabled the construction of an ALS protein network that illustrates the widespread disruption of proteome homeostasis in spinal motor neurons that contributes to the molecular heterogeneity characteristic of the ALS-associated aggregates and inclusions.
Results
TDP-43, FUS, and SOD1 Are Supersaturated.
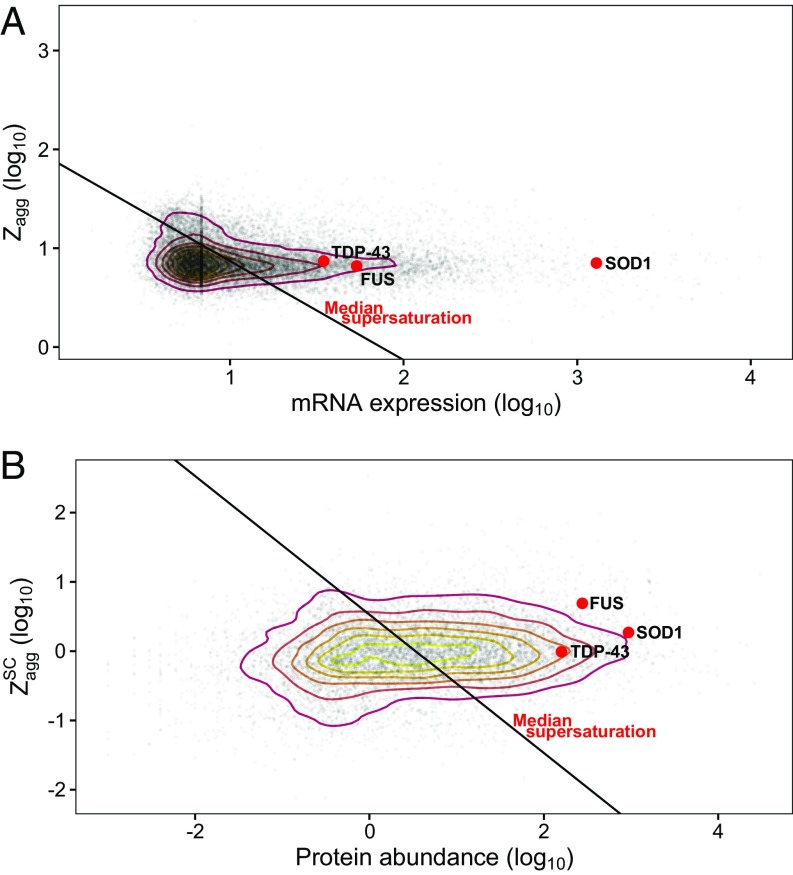
Given that all cases of ALS have either TDP-43–, FUS-, or SOD1-containing inclusions present in motor neurons, we first wanted to know if these aggregating proteins are supersaturated in the context of the proteome. To this end, we used experimental data providing mRNA and protein concentrations and the predicted aggregation propensities of each of the proteins using the Zyggregator method (43), as previously described (41). These estimates are based on wild-type sequences and wild-type expression levels averaged over a wide range of tissues. We observed that the supersaturation scores for TDP-43, FUS, and SOD1 are above the median supersaturation line, which divides the proteome into proteins whose supersaturation is relatively high and proteins whose supersaturation is low (Fig. 1). These results suggest that these proteins are metastable in their wild-type forms, and are therefore prone to become further destabilized by mutations that increase aggregation propensity. We also note that although the aggregation propensity score of SOD1 itself is close to the median aggregation propensity (Zagg) score of the proteome as a whole, this protein is predicted to be highly supersaturated due to its high abundance (Fig. 1).
Fig. 1.
Wild-type TDP-43, FUS, and SOD1 are supersaturated. Aggregation propensity (Zyggregator; Zagg) and concentration estimates are shown based on unfolded protein Zagg scores and mRNA expression (A) and structurally corrected (SC) ZaggSC scores and protein abundance estimated from mass spectrometry (B). Red points indicate TDP-43, FUS, or SOD1 as labeled; gray points indicate the rest of the proteome. Contour lines are drawn based on the density of distribution proteome values using a 2D kernel density estimator. The black line indicates the median supersaturation score for the proteome.
Native Interactors of TDP-43, FUS, and SOD1 Are Supersaturated.
A recent unbiased search of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways indicated that the biochemical pathways associated with Alzheimer’s, Parkinson’s, and Huntington’s diseases are highly enriched in supersaturated proteins (41). We thus sought to determine if ALS pathways are similarly at risk. However, in addressing this point, we realized that the annotation of the ALS KEGG pathway (www.genome.jp/kegg/pathway/hsa/hsa05014.html) is currently limited to SOD1-related pathways and, as such, does not describe very well the 98% cases of ALS not related to SOD1 mutations. To overcome this problem, because KEGG pathways are partly dependent on protein–protein interactions (44), we asked whether the network of protein–protein interactions with wild-type TDP-43, FUS, and SOD1 may be predisposed to dysfunction as a result of their supersaturation levels. Using BioGRID, we identified the native physical interaction networks of these three proteins, including only direct physical interactions observed experimentally (45) (details are provided in SI Materials and Methods).
We evaluated the supersaturation scores for both the unfolded and native states of the proteins, defining the parameters σu and σf as the unfolded and folded supersaturation scores, respectively (41). Specifically, σu values are based on mRNA expression levels as estimates of the levels of newly synthesized proteins and on predictions of aggregation propensity from the unfolded state of such proteins, whereas σf values are based on spectral counts, from label-free mass spectrometry, as an estimate of protein abundance and on predictions of aggregation propensity that account for the likelihood of a given aggregation-prone sequence being buried within the native structure (41). Scores were recentered from the scores plotted in Fig. 1 to a median supersaturation score of 0 for the proteome.
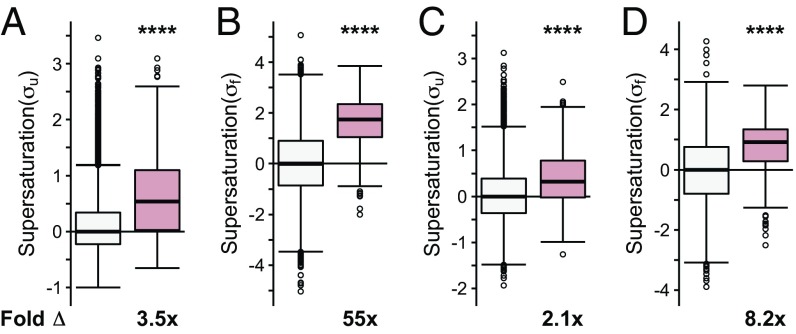
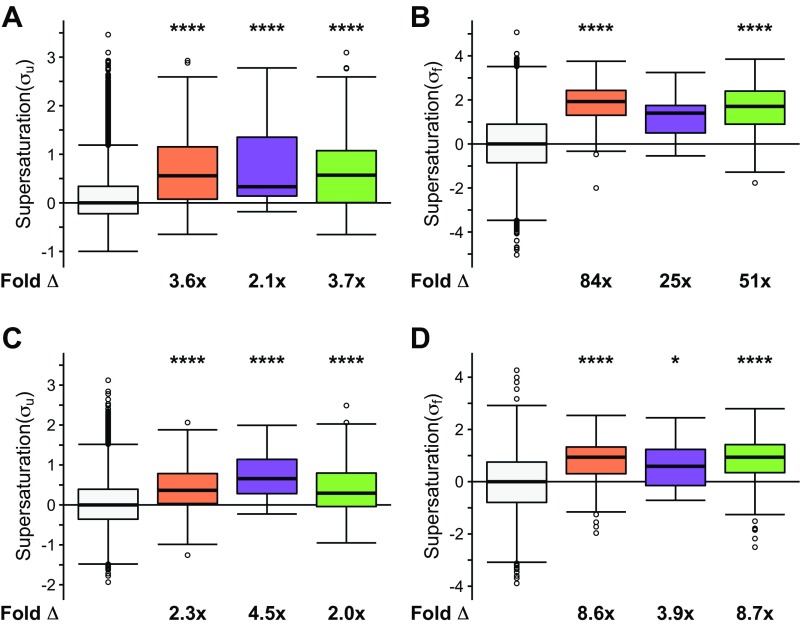
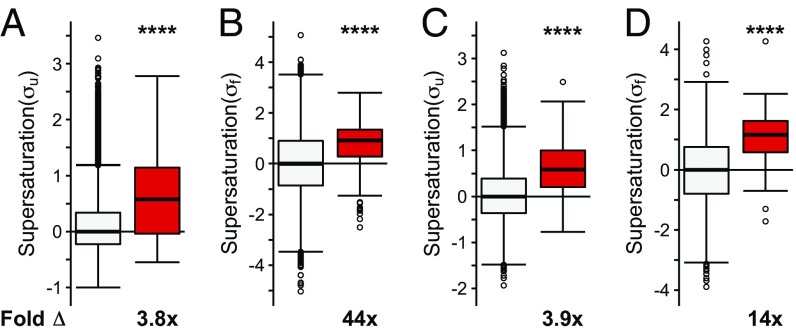
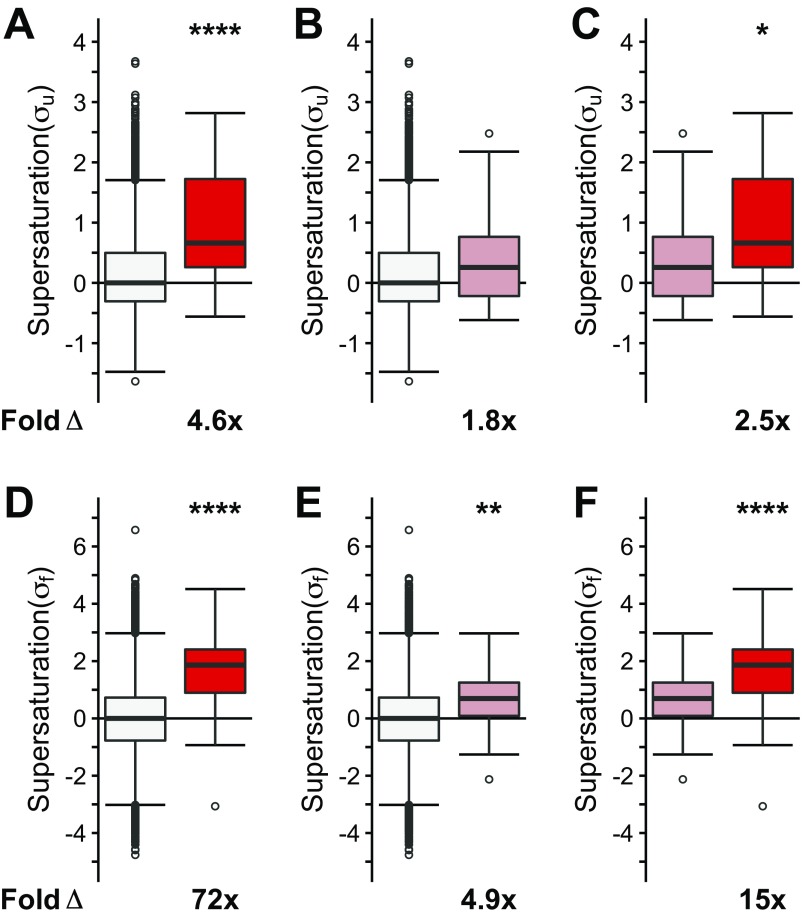
We observed that when the list of native interactors of TDP-43, FUS, and SOD1 is combined into one set, they have, on average, an increased value of both the σu (on average, 3.5-fold the proteome, P = 4.5⋅10−57) and σf (on average, 55-fold the proteome, P = 1.5⋅10−114) scores (Fig. 2 A and B). The individual sets of proteins that natively interact with SOD1 (σu: on average, 3.6-fold the proteome, P = 8.8⋅10−38; σf: on average, 84-fold the proteome, P = 4.1⋅10−74), TDP-43 (σu: on average, 2.1-fold the proteome, P = 1.1⋅10−5; σf: on average, 25-fold the proteome, P = 4.0⋅10−4), and FUS (σu: on average, 13-fold the proteome, P = 8.9⋅10−4; σf: on average, 51-fold the proteome, P = 3.8⋅10−61) are also, on average, significantly supersaturated compared with the proteome as a whole (Fig. S1 A and B). Further, using mRNA expression levels from multiple independent datasets of normal motor neurons [Genetics Selection Evolution (GSE) 20589, GSE40438] and multiple sets of protein abundance values from normal adult spinal cord, we also observed similarly elevated supersaturation levels for native interactors of TDP-43, FUS, and SOD1 in tissues related to ALS (Fig. 2 C and D and Figs. S1 C and D, S2B, and S3B).
Fig. 2.
Native interactors of TDP-43, SOD1, and FUS are supersaturated. Median supersaturation scores calculated for the unfolded (A, σu) and folded (B, σf) states are shown for native interaction partners of TDP-43, SOD1, or FUS (pink) and the total proteome (white). Supersaturation scores for the unfolded (C) and folded (D) states of proteins were also calculated using an independent set of mRNA expression levels derived from nondiseased motor neurons (GSE20589) and protein abundance values derived from nondiseased adult spinal cord. Fold Δ refers to the increase in supersaturation score from the whole proteome. Box plots extend from the lower to upper quartiles, with the internal lines referring to the median values. Whiskers range from the lowest to highest value data points within 150% of the interquartile ranges. Statistical significance was assessed by the one-sided Wilcoxon/Mann–Whitney U test with Holm–Bonferroni-corrected P values (****P < 0.0001).
Fig. S1.
Native interactors of TDP-43, SOD1, and FUS are supersaturated. Median supersaturation scores calculated for the unfolded (A, σu) and folded (B, σf) states are shown for native interaction partners of SOD1 (orange), TDP-43 (purple), or FUS (green) and the total proteome (white). Supersaturation scores for the unfolded (C) and folded (D) states of proteins were also calculated using an independent set of mRNA expression levels derived from nondiseased motor neurons (GSE20589) and protein abundance values derived from nondiseased adult spinal cord. Fold Δ refers to the increase in supersaturation score from the whole proteome. Box plots extend from the lower to upper quartiles, with the internal lines referring to the median values. Whiskers range from the lowest to highest value data points within 150% of the interquartile ranges. Statistical significance was assessed by the Wilcoxon/Mann–Whitney U test with Holm–Bonferroni-corrected P values (***P < 0.001 and ****P < 0.0001).
Fig. S2.
Supersaturation levels in an alternate dataset of spinal motor neurons. Supersaturation scores in the unfolded state (σu) were calculated based on mRNA expression levels in nondiseased spinal motor neurons (GSE40438). Comparisons of coaggregating proteins (red) with the proteome (white) (A), of native interactors (pink) with the proteome (white) (B), and of native interactors (pink) with the coaggregating proteins (red) (C) were performed. Fold Δ refers to the increase in supersaturation score from the whole proteome, except when comparing coaggregators with native interactors. Box plots extend from the lower to upper quartiles, with the internal lines referring to the median values. Whiskers range from the lowest to highest value data points within 150% of the interquartile ranges. Statistical significance was assessed by the one-sided Wilcoxon/Mann–Whitney U test with Holm–Bonferroni-corrected P values (****P < 0.0001).
Fig. S3.
Supersaturation levels based on alternative whole-organism protein abundance levels. Supersaturation scores in the folded state (σf) were calculated based on whole-organism protein abundance levels (47). Comparisons of coaggregating proteins (red) with the proteome (white) (A), of native interactors (pink) with the proteome (white) (B), and of coaggregating proteins (red) with native interactors (pink) (C) were performed. Fold Δ refers to the increase in supersaturation score from the whole proteome, except when comparing coaggregators with native interactors. Box plots extend from the lower to upper quartiles, with the internal lines referring to the median values. Whiskers range from the lowest to highest value data points within 150% of the interquartile ranges. Statistical significance was assessed by the one-sided Wilcoxon/Mann–Whitney U test with Holm–Bonferroni-corrected P values (****P < 0.0001).
Coaggregating Proteins in ALS Inclusions Are Not Merely a Subset of the Native Interactors of SOD1, TDP-43, or FUS.
Given that supersaturated proteins are prone to aggregation, the observation that the native interactors of ALS-associated proteins have high supersaturation scores raises the possibility that coaggregating proteins are a subset of the native interactors. We reasoned that native interactors may coaggregate with ALS inclusion bodies both because of their high supersaturation levels and by interacting with the soluble conformation of the inclusion-forming protein, thus increasing the likelihood of becoming trapped during the aggregation process. To test whether or not such a phenomenon could explain the coaggregation in ALS inclusions, we sought to compare the native interaction network of SOD1, TDP-43, and FUS with the proteins known to coaggregate with these proteins.
Because the amorphous nature of the inclusions formed in ALS has prevented their detailed biochemical characterization, we assembled a database of coaggregating proteins by conducting a systematic search of the literature. We carried out text mining using the terms “inclusion” + “amyotrophic” + either (“immunoreactive” or “insoluble protein” or “basophilic” or “localized” or “conglomerate” or “spheroids” or “aggregation” or “aggregate” or “hyaline” or “ubiquitin”). Proteins were only included if the published data clearly showed colocalization. This strategy yielded a collection of proteins confirmed to be colocalized with inclusions in human postmortem tissue of patients with ALS (Dataset S1).
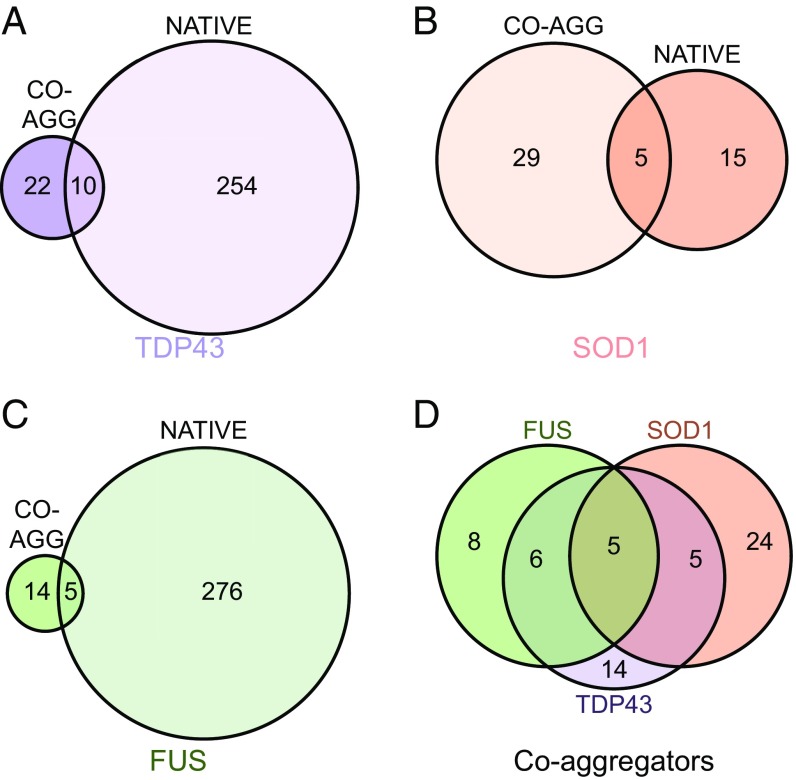
We found that a minority of proteins that coaggregate with inclusion bodies formed of TDP-43 (31%), SOD1 (15%), or FUS (26%) are also native interaction partners of these proteins (Fig. 3). Although this finding represents an enrichment of native interaction partners relative to the proteome as a whole, most proteins that coaggregate in ALS inclusions are not native interactors of the primary aggregating proteins. To examine further the differences between the native interactors (Dataset S2) and coaggregating proteins (Dataset S3), we compared them in terms of Gene Ontology (GO) biological processes (Dataset S4). Native interactors are enriched in 148 biological processes, particularly those biological processes relating to transcription and translation, whereas the coaggregators are enriched in just seven. The native interactors and coaggregators have only one enriched GO biological process in common, “regulation of mRNA stability.” It should be noted, however, that this analysis was performed on the combined interactor list and that SOD1 interactors do not contribute to the “regulation of mRNA stability” enrichment that comes entirely from TDP-43 and FUS interactors. Although limitations in experimental technique may lead to an underestimation of the overlap between coaggregators and native interactors, our results suggest that the set of proteins that aggregate in ALS are significantly different from the native interactors of the primary aggregating proteins. This result implies that the coaggregating proteins do not associate with TDP-43, SOD1, or FUS merely as a result of links through functional pathways.
Fig. 3.
Proteins that coaggregate within inclusions are largely distinct from the native interaction partners of TDP-43, SOD1, and FUS. Coaggregating (CO-AGG) proteins identified in the literature (Dataset S1) are compared with the BioGRID native physical interaction partners of TDP-43 (A), SOD1 (B), and FUS (C). (D) Overlap between the various coaggregating proteins associated with TDP-43, SOD1, and FUS.
It is reasonable to conclude that a proportion of the coaggregating proteins are present in inclusions due to their normal interactions with TDP-43, SOD1, or FUS. The results of our analysis indicate that there are 18 proteins associated with both the soluble and inclusion forms of TDP-43, SOD1, or FUS; four of these proteins are molecular chaperones (CCS, PDIA, HSPA4, and HSPA5), four are associated with degradation pathways (OPTN, RNF19A, SQSTM1, and UBQLN2), and five are genetically linked to ALS (TDP43, FUS, OPTN, SQSTM1, and UBQLN2). These findings suggest that the majority of native interaction partners that also coaggregate with TDP-43, SOD1, and FUS are either factors that contribute to protein homeostasis (molecular chaperones and components of the degradation machinery) or proteins that are themselves intrinsically at risk for aggregation (i.e., proteins in which fALS-associated mutations cause destabilization). Similarly, of the coaggregators in common between the three types of inclusions (Fig. 3D), PDIA is a molecular chaperone, whereas the rest (OPTN, SQSTM1, UBC, and UBQLN2) are associated with protein degradation. These results are consistent with the protein degradation machinery having a functional role in compartmentalization of proteins into cellular inclusions, as occurs in IPOD and JUNQ inclusions (36, 46). The common coaggregators associated with degradation are almost all native interactors with one or more of the primary aggregating proteins. This observation supports the view that a subset of those proteins that interact with both the soluble and insoluble forms of TDP-43, SOD1, and FUS plays a role in the quality control of these proteins. This observation does not, however, explain all of the coaggregators that are common between inclusion bodies.
ALS Inclusions Are Composed of Supersaturated Proteins.
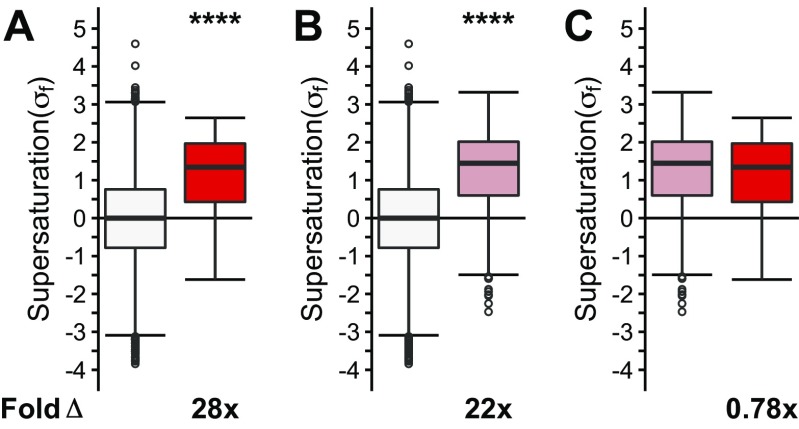
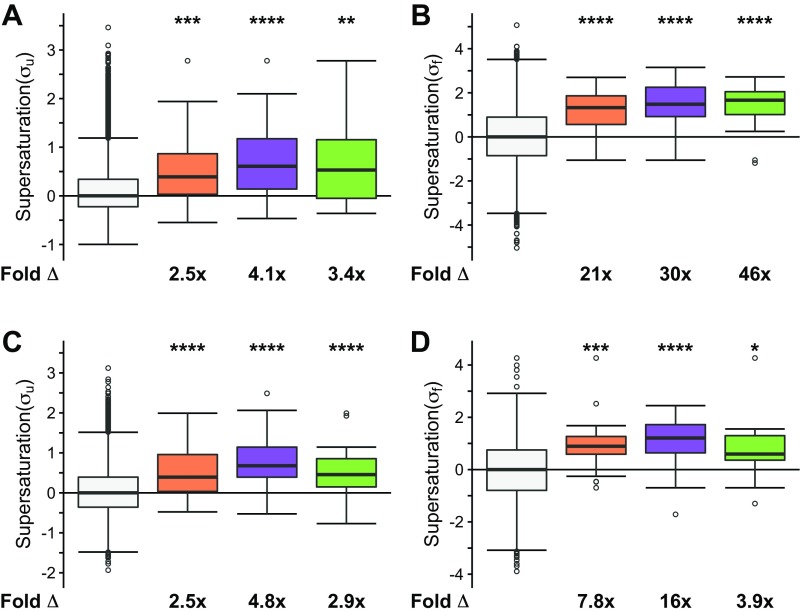
Two observations prompted us to search for an alternative explanation for the coaggregation phenomenon. First, as shown above, many coaggregating proteins are not native interactors. Second, various types of inclusion bodies share coaggregating proteins. We thus hypothesized that these proteins share biophysical features that predispose them to aggregation. Given that the proteins that coaggregate within the deposits found in neurodegenerative disorders such as Alzheimer’s and Parkinson’s diseases share supersaturation as a common biophysical property (41), we next tested if the ALS coaggregating proteins are also supersaturated. The results of this analysis indicate that the proteins present in ALS inclusions are characterized by elevated values of both the σu (on average, 3.8-fold the proteome, P = 3.4⋅10−8) and σf (on average, 44-fold the proteome, P = 8.9⋅10−13) scores (Fig. 4 A and B). To ensure that the data from specific inclusion types are not masked within the combined dataset, we analyzed the three main types of ubiquitinated inclusions separately (Fig. S4 A and B). In this case, we separated the proteins into (i) inclusions containing TDP-43 (sALS and fALS, “TDP-43–ALS inclusions,” purple box plot), (ii) SOD1-positive inclusions (“SOD1-ALS inclusions,” orange box plot), and (iii) FUS-positive inclusions (“FUS-ALS inclusions,” including basophilic inclusions, green box plot). The proteins found in SOD1-ALS inclusions (σu: 2.5-fold, P = 3.⋅10−4; σf: 21-fold, P = 3.5⋅10−6), TDP-43–ALS inclusions (σu: 4.1-fold, P = 1.7⋅10−6; σf: 30-fold, P = 3.3⋅10−7), and FUS-ALS inclusions (σu: 3.4-fold, P = 2.5⋅10−3; σf: 46-fold, P = 6.8⋅10−5) are characterized by elevated supersaturation scores in the folded and unfolded states (Fig. S4 A and B). Using independent sets of mRNA expression levels derived from normal, nondisease-associated motor neurons (GSE20589, GSE40438) and protein abundance values derived from normal adult spinal cord, we obtained similarly elevated supersaturation levels for those proteins that coaggregate with ALS inclusions (47, 48) (Fig. 4 C and D and Figs. S2A, S3A, and S4 C and D).
Fig. 4.
Proteins that coaggregate with ALS inclusions are supersaturated. Proteins identified in the literature as colocalized to all ALS inclusions are listed in Dataset S1. The median supersaturation scores calculated for the unfolded (A, σu) and folded (B, σf) states of proteins are shown for the combined set of coaggregators associated with all types of inclusions (red) and the total proteome (white). Supersaturation scores for the unfolded (C) and folded (D) states of proteins were also calculated using an independent set of mRNA expression levels derived from nondiseased motor neurons (GSE20589) and protein abundance values derived from nondiseased adult spinal cord. Fold Δ refers to the increase in supersaturation score from the whole proteome. Box plots extend from the lower to upper quartiles, with the internal lines referring to the median values. Whiskers range from the lowest to highest value data points within 150% of the interquartile ranges. Statistical significance was assessed by the one-sided Wilcoxon/Mann–Whitney U test with Holm–Bonferroni-corrected P values (****P < 0.0001).
Fig. S4.
Proteins that coaggregate with ALS inclusions are supersaturated. Proteins identified in the literature as colocalized to all ALS inclusions are listed in Dataset S1. The median supersaturation scores calculated for the unfolded (A, σu) and folded (B, σf) states of proteins are shown for coaggregators of SOD1 (orange), TDP-43 (purple), or FUS (green) and the total proteome (white). Supersaturation scores for the unfolded (C) and folded (D) states of proteins were also calculated using an independent set of mRNA expression levels derived from nondiseased motor neurons (GSE20589) and protein abundance values derived from nondiseased adult spinal cord. Fold Δ refers to the increase in supersaturation score from the whole proteome. Box plots extend from the lower to upper quartiles, with the internal lines referring to the median values. Whiskers range from the lowest to highest value data points within 150% of the interquartile ranges. Statistical significance was assessed by the Wilcoxon/Mann–Whitney U test with Holm–Bonferroni-corrected P values (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Our analysis is based on nondiseased tissue so as to determine the features of the interactors and coaggregators before disease processes altering the proteome, but we were curious as to whether disease-related changes in expression altered the supersaturation signal. We recently showed that there is a widespread transcriptional suppression of supersaturated proteins in Alzheimer’s disease (49). If a similar process were to occur in ALS, then we would predict that supersaturation scores in ALS patient tissues would be lower than in unaffected control tissue. Consistent with this idea, we find that both native interactors and coaggregators are supersaturated in SOD1-ALS–affected tissues (Fig. S5), but to a lesser extent than in healthy tissue (compare with Figs. 2 and 4).
Fig. S5.
Supersaturation levels in SOD1 ALS-affected human tissues. Supersaturation scores in the unfolded state (σu) were calculated based on mRNA expression levels in SOD1-affected tissues (GSE20589). Comparisons of coaggregating proteins (red) with the proteome (white) (A) and of native interactors (pink) with the proteome (white) (B) were performed. Fold Δ refers to the increase in supersaturation score from the whole proteome, except when comparing coaggregators with native interactors. Box plots extend from the lower to upper quartiles, with the internal lines referring to the median values. Whiskers range from the lowest to highest value data points within 150% of the interquartile ranges. Statistical significance was assessed by the one-sided Wilcoxon/Mann–Whitney U test with Holm–Bonferroni-corrected P values (****P < 0.0001).
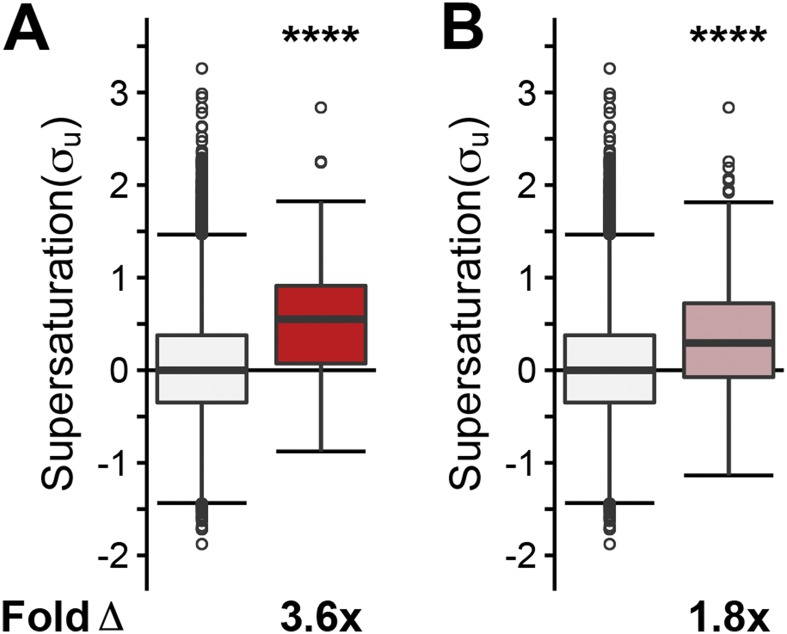
Given these results from human postmortem tissue, we next tested if these findings could be confirmed in data from a mouse model of ALS. Using a combined list of coaggregators from three independent experimental studies (50–52) (Dataset S5), we found that SOD1 coaggregators from mouse spinal cord are characterized by increased values of both σu (4.6-fold, P = 4.1⋅10−9) and σf (72-fold, P = 4.8⋅10−19) scores (Fig. S6 A and D). Intriguingly, we find that native interactors with SOD1 in the mouse also have elevated supersaturation levels, but they are not as high as the supersaturation levels for coaggregating proteins (σu: 1.8-fold, P = 5.1⋅10−2; σf: 4.9-fold, P = 2.1⋅10−3) (Fig. S6 B and E).
Fig. S6.
Proteins that coaggregate with ALS inclusions from G93A SOD1 mice are supersaturated. Proteins identified in the literature as colocalized to G93A SOD1 mouse ALS inclusions are listed in Dataset S4. The median supersaturation scores calculated for the unfolded (A–C, σu) and folded (D–F, σf) states of proteins are shown for coaggregators associated with SOD1 inclusions (red), native interactors of SOD1, and the total proteome (white). Fold Δ refers to the increase in supersaturation score from the whole proteome, except when comparing coaggregators with native interactors. Box plots extend from the lower to upper quartiles, with the internal lines referring to the median values. Whiskers range from the lowest to highest value data points. Statistical significance was assessed by the Wilcoxon/Mann–Whitney U test (*P < 0.05, **P < 0.01, ****P < 0.0001).
Coaggregators Are More Supersaturated than Native Interactors in Spinal Motor Neurons.
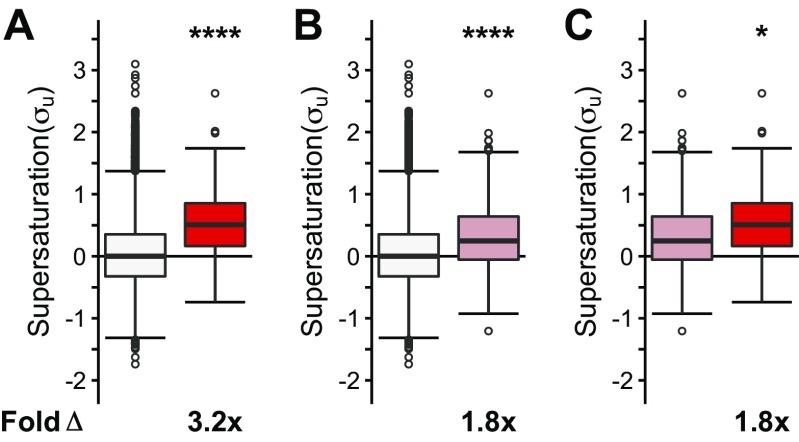
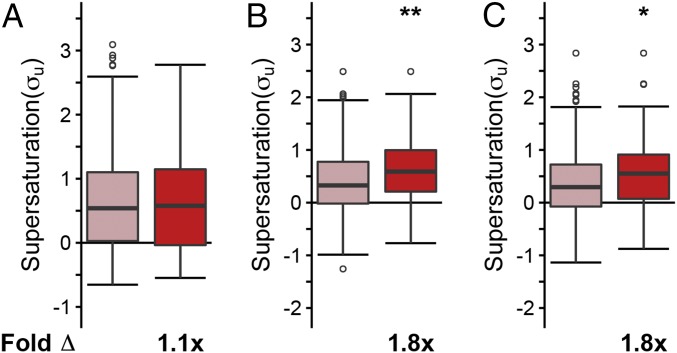
Given that coaggregators are more supersaturated than native SOD1 interactors in the mouse, (coaggregators relative to natively interacting proteins: σu: 2.5-fold, P = 1.5⋅10−2; σf: 15-fold, P = 5.5⋅10−5), we considered whether, in humans, coaggregators could have higher supersaturation scores than the supersaturation scores of the native interaction partners of TDP-43, SOD1, and FUS, most of which are not found in inclusion bodies. Given the relatively small sample sizes of each individual set of inclusion-forming proteins, we compared the combined set of coaggregators for the three types of inclusions with the combined set of native interaction partners (TDP-43 + FUS + SOD1 interactors). In an analysis where σu is calculated from data averaged over numerous tissue types, we found that the supersaturation scores of the coaggregators are not significantly different from the supersaturation scores of the native interaction partners (σu: on average, 1.1-fold relative to native interaction partners, P = 1.0; Fig. 5A).
Fig. 5.
Coaggregating proteins are significantly more supersaturated than the native interaction partners of TDP-43, SOD1, and FUS in motor neurons. (A) We found similar supersaturation scores for the combined set of coaggregating proteins (red) and for the combined set of native interaction partners of TDP-43, SOD1, and FUS (pink) across a range of tissues for the unfolded state of the proteins included. (B) By contrast, we found larger supersaturation scores for coaggregating proteins than for native interaction partners for proteins in the unfolded state in motor neurons. (C) Supersaturation scores in the unfolded state (σu) were also calculated based on mRNA expression levels in SOD1-affected tissues (GSE20589). Fold Δ refers to the increase in supersaturation score from the native interactors. Box plots extend from the lower to upper quartiles, with the internal lines referring to the median values. Whiskers range from the lowest to highest value data points within 150% of the interquartile ranges. The statistical significance was assessed by the one-sided Wilcoxon/Mann–Whitney U test with Holm–Bonferroni-corrected P values (*P < 0.05, **P < 0.01).
This finding prompted us to ask whether or not the set of proteins found in inclusion bodies is related to the specific proteome expressed in those tissues that are relevant to ALS pathology. To perform this analysis, we recalculated the supersaturation scores using mRNA expression levels from microdissected motor neurons (GSE20589) (48). Consistent with the fact that ALS inclusions are found predominantly in motor neurons, we found that the spinal motor neuron supersaturation scores of coaggregating proteins are significantly elevated relative to the supersaturation scores of the native interaction partners (σu: on average, 1.8-fold relative to native interaction partners, P = 9.1⋅10−3; Fig. 5B and Fig. S2C). We next asked whether the supersaturation signal would remain in ALS patient tissue. When using mRNA expression levels from SOD1-ALS–affected spinal motor neurons, coaggregators were significantly more supersaturated than native interactors (Fig. 5C). Therefore, we observe that the supersaturation scores of coaggregating proteins are significantly elevated relative to native interaction partners in the motor neurons affected by ALS, but we do not find these differences using supersaturation scores generated using averages of other tissue types, possibly explaining their presence in motor neuron-specific inclusions.
Definition of the ALS Network.
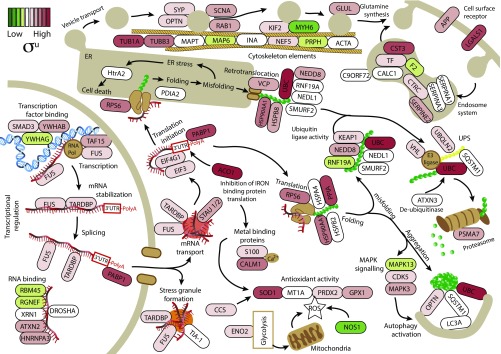
The “ALS pathway” described in the KEGG database (www.genome.jp/kegg/pathway/hsa/hsa05014.html) does not currently include recent discoveries, including key processes such as mRNA metabolism (53, 54). We therefore generated an “ALS network” by creating a list of proteins found in inclusions, data from supersaturation scores, connectivity network analysis, and existing KEGG pathways (ALS, regulation of autophagy, glutamatergic synapse, RNA transport, mRNA surveillance pathway, transcription factors, ubiquitin-mediated proteolysis, and protein processing in the endoplasmic reticulum; www.genome.jp/kegg/pathway.html), and used this list to construct a new model for functional disruption in ALS (Fig. 6). Our analysis revealed that this ALS network incorporates pathways from the KEGG ALS pathway but also includes other key processes previously identified as important in ALS pathology, such as mRNA metabolism and transport, protein misfolding and aggregation, the ubiquitin–proteasome system, autophagy, ER protein processing, and vesicle transport (2).
Fig. 6.
Schematic representation of the ALS network. The ALS network was created using the set of proteins found in ALS inclusions (Dataset S1), their known functions and pathways from the KEGG, and their calculated supersaturation values. For the purpose of this analysis, we used only supersaturation scores from the unfolded state (σu), but using the folded state (σf) gives similar results (Dataset S1). Genes are colored depending on their supersaturation scores, with red being highest and green lowest (key in upper left corner). Our analysis revealed that the ALS network covers many of the processes involved in maintaining protein homeostasis, such as transcriptional regulation, stress granule formation, translation, endoplasmic reticulum stress, vesicle transport, protein folding and degradation, and antioxidant activity. ER, endoplasmic reticulum; ROS, reactive oxygen species; UPS, ubiquitin–proteasome system.
Protein Supersaturation in the ALS Network Underlies the Protein Homeostasis Collapse in ALS.
We found that the supersaturated proteins in the ALS network are not confined to a single pathway and that many of the cellular functions represented in the ALS network contain several supersaturated proteins (Fig. 6). These results suggest that, rather than a single pathway being responsible for pathology in ALS, the physicochemical properties of the proteins in the ALS network govern their sequestration into inclusions in a widespread collapse in essentially all arms of the protein homeostasis network.
Because the proteins in the ALS network perform cellular functions that are vital for maintaining protein homeostasis, their aggregation may contribute to the cellular dysfunction observed in cellular and organismal models of ALS and in postmortem tissue from patients with ALS, including endoplasmic reticulum stress, oxidative stress, proteasome dysfunction, protein transport defects, and mRNA missplicing. However, aggregation of a protein homeostasis network component is not a requisite for its dysfunction. The functional categories identified in the ALS network described in this study can be applied to the genetic mutations in familial forms of the disease involving RNA binding (e.g., TDP-43, FUS, TAF15, MTR3), protein degradation (e.g., VCP, SQSTM1, OPTN, UBQLN2, TBK1, CCNF), cytoskeleton (PFN1 and TUBA4A), protein transport (VAPB, OPTN, and C9ORF72), and antioxidant activity (SOD1), several of which do not appear in aggregates. For example, profilin1 has so far not been found in human inclusions (27). When overexpressed, the mutant variant aggregates in cell culture (27), and structural studies show that mutations cause protein instability leading to a shorter half-life and loss of function (55). Our model predicts that destabilization of profilin1 might result in aggregate-independent proteostasis distraction by (i) production of a destabilized profilin1 that requires increased interaction from chaperones and degradation machinery and/or (ii) disruption of its function associated with cytoskeleton stability (56). Both of these possibilities may result in the aggregation of supersaturated proteins, such as TDP-43, but not do not require the aggregation of profilin1. These observations are consistent with the concept that dysfunction in any component of the protein homeostasis network may be sufficient to cause widespread and cumulative protein aggregation and associated cellular decline (57). This process may therefore eventually lead to the collapse of the entire protein homeostasis system through the aggregation of supersaturated proteins and the subsequent failure of multiple essential cellular processes (42).
SI Materials and Methods
Identification of Coaggregating Proteins.
Coaggregating proteins were identified based on text mining using the terms “inclusion” + “amyotrophic” + either (“immunoreactive” or “insoluble protein” or “basophilic” or “localized” or “conglomerate” or “spheroids” or “aggregation” or “aggregate” or “hyaline” or “ubiquitin”). Searches were conducted using PubMed (https://www.ncbi.nlm.nih.gov/pubmed). Proteins were only included if the published data clearly showed colocalization in human postmortem tissue.
Identification of Native Interactors.
Native interactors were identified based on interactions listed in BioGRID version 3.4.126. Only those interactions in which both partners had taxonomy ID 9606 (corresponding to Homo sapiens) were considered. These interactions were further filtered based on the interaction type and method. Only interactions of type “direct interaction” or “physical association” were included. This subset was further narrowed by excluding any interactions obtained by the following methods: “genetic interference,” “unspecified method,” “fluorescent resonance energy transfer,” and “imaging technique.” The goal was to obtain a list of interactors thought to be direct physical interactions as determined by relatively robust biochemical methods.
Once this final set of interactions across the proteome was compiled, BioGRID IDs were used to isolate all interactions with a particular protein of interest. The BioGRID IDs for TDP43, SOD1, and FUS are 23435, 6647, and 2521, respectively. By filtering for these IDs, a final list of interactions was obtained. Using the ID mapping database available from UniProt, the Entrez Gene or gene symbol IDs listed in BioGRID were converted to UniProt IDs. A similar approach was used to identify interactors with mouse SOD1.
Calculation of Zyggregator Scores.
Zyggregator scores were calculated as described by Ciryam et al. (41).
Calculation of Supersaturation Scores.
All tissue supersaturation scores were calculated as described by Ciryam et al. (41). The mRNA levels for the calculation of unfolded scores were derived from the study of Su et al. (84). This study was also the source of mouse mRNA expression data. For all-tissue folded scores, data were used from the whole-organism integrated set from PaxDb as well as the PaxDb-normalized data from Kim et al. (47).
For tissue-specific supersaturation scores, Zyggregator scores were the same, but different expression levels were used. For the calculation of motor neuron-specific sigma_u, microarray mRNA expression levels were obtained from Kirby et al. (48) (GSE20589) and Brockington et al. (66) (GSE40438). Mouse proteomic data were obtained from PaxDb dataset 149. In a modification from our previous analysis, we used the normalized abundance values as calculated by PaxDb (85).
Calculation of Fold Changes.
Fold change was calculated as described by Ciryam et al. (41) as the linear difference between the logarithmic medians of two sets: the linear fold difference between the medians of the supersaturation scores of the control set and experiment set being tested:
| [S1] |
Determining GO Enrichment.
UniProt GO annotations (May 2016 version) associated with the Homo sapiens taxon 9606 were downloaded from the GO website. Our analysis was restricted to those GO terms associated with UniProt IDs whose status was “reviewed.” This set was further restricted to those terms in the category “biological process,” for a total of 11,099 GO biological process terms. We considered the relative enrichment of the UniProt IDs associated with each of these terms among the coaggregators or native interactors relative to the proteome as a whole. For the purposes of this analysis, coaggregators of TDP43, SOD1, and FUS were grouped together. Separately, the native interactors of TDP43, SOD1, and FUS were grouped together as well. The human proteome list consisted of the 20,199 reviewed UniProt IDs for taxon 9606. To calculate enrichment, the one-sided Fisher exact test was used. To correct for multiple hypotheses, we adjusted these P values to the Benjamini–Hochberg-derived Q values calculated separately for the enrichment testing on the coaggregators and the native interactors.
Multiple Hypothesis Correction Family Structure.
In this analysis, numerous parallel statistical tests were performed. This multiple hypothesis testing gives rise to increasing type I error, or false-positive errors. This problem can be minimized by techniques to correct P values based on the number of parallel hypotheses corrected. Correction, however, increases the likelihood of type II error, or false-negative errors. Two basic requirements for successful multiple hypothesis correction are (i) appropriate family definitions and (ii) appropriate correction methods.
In this context, a family is a set of hypotheses thought to be parallel to each other; the P values of hypotheses in a given family are adjusted against each other. Selecting families too narrowly increases the type I error by reducing the size of the adjustment to the P values. Selecting families too broadly increases the type II error. One heuristic for defining a family is as a set of tests performed on a single experiment (86, 87). We used the following rules to organize the statistical tests in this study into families.
First, different types of questions were included in different families. The two types of questions asked in this study were (i) whether two sets of supersaturation scores were statistically different and (ii) whether sets of native interactors and coaggregators were enriched in particular GO terms. We further subdivided some of these question types.
Comparisons of sets of supersaturation scores were grouped into families according to the following rules. The first division was made between analyses of coaggregators or native interactors against the proteome and comparisons of coaggregators against native interactors. These two types of analysis were grouped in separate families. Further, the “all tissue” analysis was grouped separately from the “motor neuron/spinal cord” analysis. Comparisons of coaggregators vs. the proteome were placed in separate families from comparisons of native interactors vs. the proteome. Analyses based on σu were placed in separate families from analyses based on σf. Analyses within these groups that arose from different datasets were treated as different families. Analyses of the combined list of native interactors or coaggregators were, however, considered in the same family as analyses of the individual lists associated with TDP43, SOD1, and FUS. Although the test on the combined set is not independent of the subset tests, it was reasoned that this approach was a conservative approach.
For GO term enrichment, coaggregators and native interactors were considered as separate families.
Numerous correction methods exist to control the family-wise error rate (FWER) or the false discovery rate (FDR). Regardless of the specific method, an adjusted P value can be obtained. The most conservative method is the Bonferroni correction, in which P values are multiplied by the number of tests in the family. The Holm–Bonferroni method has greater statistical power, but it has been proven to be as effective at controlling the FWER as the Bonferroni method (88). For questions of whether two sets of supersaturation scores were statistically different, the Holm–Bonferroni method was used. For GO term enrichment, this method is likely to produce a large type II error because the number of comparisons is on the order of 10,000. The Benjamini–Hochberg method to control the FDR is a more statistically powerful method often used in datasets with a large number of comparisons (e.g., microarrays) (89). For GO enrichment analysis, this method was used.
Discussion
Spinal Motor Neuron Vulnerability to ALS Is Linked to Protein Supersaturation.
The results that we have presented indicate that (i) the network of native interactors of TDP-43, FUS, and SOD1 are, on average, supersaturated under normal physiological conditions; (ii) when mutant TDP-43, FUS, and SOD1 form inclusions, proteins in a largely distinct set coaggregate; and (iii) these coaggregators are indistinguishable from the native interactors in terms of supersaturation when averaged over numerous human tissues. To bring together these observations and rationalize them, we have shown that the supersaturation levels of coaggregators are significantly higher than the supersaturation levels of native interactors in the motor neurons specifically affected by ALS. These findings indicate that, under proteome stress, coaggregating proteins are in a metastable state and so are at a particularly high risk of misfolding, dysfunction, and aggregation in motor neurons. Strikingly, even though the specific proteins associated with TDP-43, FUS, and SOD1 in their native or aggregated state are largely structurally and functionally distinct, they share the unifying feature of being supersaturated. This remarkable observation supports the view that it is the general property of being metastable that defines the ensemble of proteins that are either disrupted or deposited with inclusion bodies in ALS and other conformational disorders (41). These results on the importance of metastable proteins prone to aggregation in relation to motor neuron vulnerability to neurodegeneration are consistent with the results recently reported for Alzheimer’s disease (58), and thus point to a common explanation of tissue-specific vulnerability in the two diseases.
The variations in the subsets of proteins associated with the different inclusion types may reflect separate primary underlying causes of proteome disruption that lead to inclusion formation in both SOD1-ALS and other subtypes of ALS. Such disruptions, however, appear to exhibit some motor neuron specificity, because supersaturation levels of coaggregating proteins are, on average, higher than supersaturation levels of native interaction partners in spinal motor neurons, but not across the human body as a whole. These results suggest that the pathological nature of the disease state in ALS involves the deposition of a set of proteins on the basis of their propensity to aggregate rather than their specific native-state binding in functional networks. Collectively, these results are consistent with a specific collapse of protein homeostasis within motor neurons that links the pathological processes of various defined subtypes of ALS. Moreover, the formation of insoluble inclusion bodies appears to redefine the interaction network of disease-causing proteins in ALS, driving a coaggregation process in motor neurons governed, in large part, by protein supersaturation.
A number of possibilities exist for the causes of motor neuron selectivity in ALS. In contrast to other types of neurons [e.g., cerebellar, cortical, pyramidal (59, 60)], motor neurons have a particularly high threshold for induction of the heat shock response (61–63), and thus may not respond as effectively as other neurons to an overload of the protein homeostasis system (64). As a result, motor neurons may be particularly vulnerable to dysregulation of protein homeostasis, a conclusion that provides a possible explanation of the highly heterogeneous nature of inclusions in ALS, where only a small set of proteins associated with protein degradation (including OPTN, SQSTM1, UBC, and UBQLN2) are present in all three categories of inclusions.
It remains to be determined what other factors, such as concentrations of protein homeostasis components, additionally contribute to the vulnerability of motor neurons to ALS. The fact that proteins central to protein degradation are found across these inclusions types points to the impairment of protein degradation as being an important aspect of the loss of protein homeostasis in ALS. The sequestration of vital protein degradation machinery into inclusions may also further exacerbate the loss of protein homeostasis (65). There is evidence that the subsets of motor neurons that are vulnerable to ALS have decreased expression of ubiquitin–proteasome system genes compared with resistant motor neurons (66). In addition to the dysfunction of individual disease-causing proteins, therefore, a cell-specific compromised protein homeostasis network may give rise to a range of cellular manifestations characteristic of ALS. Consistent with this idea, the genetic mutations associated with ALS represent a variety of processes that are all central to protein homeostasis (67). Mutations in a number of genes that cause ALS, including SOD1, FUS, and TARDBP, markedly increase the aggregation propensity of the corresponding proteins (68–70), placing an increased burden on the protein homeostasis system and amplifying the effects of its compromised function. Our observations that proteins associated with ALS inclusions have specific supersaturation patterns in motor neurons provide evidence that dysfunctional protein homeostasis may be playing an important role in ALS.
Widespread Aggregation and Supersaturation Link the Complex Pathology of ALS to Other Neurodegenerative Diseases.
A widespread and catastrophic failure to maintain the proteome in its soluble and functional state has been proposed to underlie the diverse and complex pathophysiology of neurodegenerative diseases (41). In light of this proposition, the sets of proteins found to be associated with ALS inclusions may simply reflect the specific gene expression patterns of subsets of neurons. SOD1, for example, is very highly expressed in motor neurons (71). Thus, although SOD1 is very stable once native (72), its high expression level leaves it particularly prone to misfolding and aggregation if protein homeostasis is disrupted specifically in motor neurons. Indeed, SOD1 aggregation is specific to ALS and results in large inclusions in cases of SOD1-mutant fALS (73), but smaller inclusions and misfolded forms of SOD1 can be observed in all forms of ALS (74). Disease-associated mutations decrease the solubility of SOD1, resulting in higher supersaturation levels than is the case for the wild-type protein. In addition, it is well established that misfolding of one protein can disrupt protein homeostasis and lead to the aggregation of other metastable proteins (75). Hence, the misfolding and aggregation of SOD1 are likely to lead to the destabilization and recruitment of other proteins into inclusions, either through direct interactions or indirectly through the disruption of the protein homeostasis system and aggregation of other supersaturated proteins (57).
An important example of a finding that suggests such protein homeostasis collapse in disease is the discovery that proteins whose aggregation is associated with one disease may also be found to aggregate in others. For example, TDP-43, which is found in inclusions in most forms of ALS and is considered characteristic of the disease (41), has also been found in inclusions in frontotemporal dementia (53), Machado–Joseph disease (76), spinocerebellar ataxia (77), Huntington’s disease (78), Alzheimer’s disease (79), inclusion body myositis (80), and Parkinson’s disease (81). Given the presence of disease-specific aggregation-prone proteins in these disorders, it is unlikely that TDP-43 is part of the initial aggregation phenomenon in most cases. Based on our findings, we may suggest that this protein is probably part of a group of supersaturated proteins that aggregate once protein homeostasis becomes compromised. This coincident aggregation of proteins associated with different neurodegenerative diseases may be particularly common in neurons having certain general features, such as an inefficient heat shock response and long life spans, that predispose them to increased protein aggregation (82). Compensatory mechanisms may exist once protein homeostasis machinery is compromised; our results suggest that in ALS patient tissue, coaggregators are less supersaturated consistent with a transcriptional repression similar to the transcriptional repression that we recently observed in Alzheimer’s disease (49).
Although our findings are based on the currently available data, we cannot rule out the possibility that our dataset may be incomplete and that the differences observed are a result of this incompleteness. Arguing against this possibility is the fact that the number of proteins that have been experimentally determined to be associated with inclusions in other neurodegenerative diseases is within range our ALS coaggregator list; for example, only 26 proteins are enriched in plaques compared with nonplaque tissue (39), only 72 proteins have been identified through multiple peptides in tangles (83), and, similarly, only 40 proteins are enriched in Lewy bodies compared with control material (40). Together these results suggest that regardless of the primary protein, coaggregation occurs with a similar number of other proteins. These results give us greater confidence that we have captured a major proportion of coaggregators.
Emerging evidence of widespread cellular protein aggregation in a range of disorders, including ALS, indicates that such behavior may not be limited to a small set of disease-associated proteins, and that a much larger portion of the proteome is at risk (41). Our results support the view that supersaturated proteins are particularly prone to incorporation into inclusions or to processes such as degradation (49). Thus, the various genetic modifications to the different protein homeostasis pathways and aggregation-prone proteins that give rise to different subtypes of ALS appear to converge on a more general downstream phenomenon, namely, the collapse of protein homeostasis. This collapse results in the progressive aggregation of numerous supersaturated proteins and emphasizes yet further the common or generic nature of this group of misfolding and aggregation diseases (8).
Conclusions
We have shown that ALS inclusions are formed by proteins that tend to be supersaturated under normal physiological conditions. These particular proteins are distinguished by their supersaturation levels in motor neurons from the proteins that form the functional network of normal interaction partners of the ALS-associated proteins SOD1, TDP-43, and FUS. By drawing on evidence from the supersaturation analysis of the proteins forming various ALS inclusions and from the current knowledge of biochemical pathways associated with ALS, we have defined an ALS network that connects dysfunction across clinical subtypes. We thus propose that a combination of the susceptibility to aggregation of supersaturated proteins and of the progressive failure of protein homeostasis in disease drives largely unrelated proteins to form the insoluble inclusions that underlie the cellular dysfunctions characteristic of ALS. We anticipate that the description of this ALS network and of its vulnerability to protein aggregation in motor neurons will provide a means of understanding the molecular origins of this devastating disease and may provide a target for more effective therapeutic intervention (9).
Materials and Methods
The methods of identification of coaggregating proteins and native protein interactors are described in SI Materials and Methods. The calculations of supersaturation scores and fold changes are also described in SI Materials and Methods. The multiple hypothesis correction and GO enrichment analysis are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
P.C. was supported by grants from the US-UK Fulbright Commission, St. John’s College, University of Cambridge, and NIH (Northwestern University Medical Scientist Training Program Grant T32 GM8152-28). I.A.L.-S. was supported by Rotary Health Australia. D.M.B., S.G.O., and G.F. were supported by the Wellcome Trust/Medical Research Council (Grant Code 089703/Z/09/Z). D.N.S. was supported by a National Health and Medical Research Council (NHMRC) Project grant. R.I.M. was supported by grants from the NIH (National Institute of General Medical Sciences, National Institute on Aging, and National Institute of Neurological Disorders and Stroke), Ellison Medical Foundation, Glenn Foundation, and Daniel F. and Ada L. Rice Foundation. C.M.D. and M.V. are members of the Cambridge Centre for Misfolding Diseases and were supported by the Wellcome Trust. J.J.Y. was supported by grants from the NHMRC (Grants 1095215 and 1084144), Motor Neuron Disease Research Institute of Australia, and Australian Research Council (Grant DE120102840). F.C. and G.G.T. acknowledge support from the European Research Council (RIBOMYLOME_309545) and Spanish Ministry of Economy and Competitiveness (BFU2014-55054-P).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 5065.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613854114/-/DCSupplemental.
References
- 1.Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: Disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robberecht W, Philips T. The changing scene of amyotrophic lateral sclerosis. Nat Rev Neurosci. 2013;14:248–264. doi: 10.1038/nrn3430. [DOI] [PubMed] [Google Scholar]
- 3.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: Insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 4.Turner MR, et al. Controversies and priorities in amyotrophic lateral sclerosis. Lancet Neurol. 2013;12:310–322. doi: 10.1016/S1474-4422(13)70036-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ticozzi N, Ratti A, Silani V. Protein aggregation and defective RNA metabolism as mechanisms for motor neuron damage. CNS Neurol Disord Drug Targets. 2010;9:285–296. doi: 10.2174/187152710791292585. [DOI] [PubMed] [Google Scholar]
- 6.Leigh PN, et al. Ubiquitin-immunoreactive intraneuronal inclusions in amyotrophic lateral sclerosis. Morphology, distribution, and specificity. Brain. 1991;114:775–788. doi: 10.1093/brain/114.2.775. [DOI] [PubMed] [Google Scholar]
- 7.Strong MJ, Kesavapany S, Pant HC. The pathobiology of amyotrophic lateral sclerosis: A proteinopathy? J Neuropathol Exp Neurol. 2005;64:649–664. doi: 10.1097/01.jnen.0000173889.71434.ea. [DOI] [PubMed] [Google Scholar]
- 8.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 9.Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 10.Bolognesi B, et al. ANS binding reveals common features of cytotoxic amyloid species. ACS Chem Biol. 2010;5:735–740. doi: 10.1021/cb1001203. [DOI] [PubMed] [Google Scholar]
- 11.Rosen DR, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y, et al. The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet. 2001;29:160–165. doi: 10.1038/ng1001-160. [DOI] [PubMed] [Google Scholar]
- 13.Chen YZ, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am J Hum Genet. 2004;74:1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 15.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishimura AL, et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenway MJ, et al. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat Genet. 2006;38:411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- 18.Kabashi E, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 19.Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Münch C, et al. Heterozygous R1101K mutation of the DCTN1 gene in a family with ALS and FTD. Ann Neurol. 2005;58:777–780. doi: 10.1002/ana.20631. [DOI] [PubMed] [Google Scholar]
- 21.Münch C, et al. Point mutations of the p150 subunit of dynactin (DCTN1) gene ALS. Neurology. 2004;63:724–726. doi: 10.1212/01.wnl.0000134608.83927.b1. [DOI] [PubMed] [Google Scholar]
- 22.Chow CY, et al. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet. 2009;84:85–88. doi: 10.1016/j.ajhg.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maruyama H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- 24.Johnson JO, et al. ITALSGEN Consortium Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rea SL, Majcher V, Searle MS, Layfield R. SQSTM1 mutations–bridging Paget disease of bone and ALS/FTLD. Exp Cell Res. 2014;325:27–37. doi: 10.1016/j.yexcr.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 26.Deng HX, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu CH, et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488:499–503. doi: 10.1038/nature11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson JO, et al. ITALSGEN Consortium Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci. 2014;17:664–666. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams KL, et al. CCNF mutations in amyotrophic lateral sclerosis and frontotemporal dementia. Nat Commun. 2016;7:11253. doi: 10.1038/ncomms11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki S. Neuropathology of ALS: Spheroids. Neuropathol. 1993;13:185–192. [Google Scholar]
- 32.Keller BA, et al. Co-aggregation of RNA binding proteins in ALS spinal motor neurons: Evidence of a common pathogenic mechanism. Acta Neuropathol. 2012;124:733–747. doi: 10.1007/s00401-012-1035-z. [DOI] [PubMed] [Google Scholar]
- 33.Farrawell NE, et al. Distinct partitioning of ALS associated TDP-43, FUS and SOD1 mutants into cellular inclusions. Sci Rep. 2015;5:13416. doi: 10.1038/srep13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsumoto G, Kim S, Morimoto RI. Huntingtin and mutant SOD1 form aggregate structures with distinct molecular properties in human cells. J Biol Chem. 2006;281:4477–4485. doi: 10.1074/jbc.M509201200. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto G, Stojanovic A, Holmberg CI, Kim S, Morimoto RI. Structural properties and neuronal toxicity of amyotrophic lateral sclerosis-associated Cu/Zn superoxide dismutase 1 aggregates. J Cell Biol. 2005;171:75–85. doi: 10.1083/jcb.200504050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polling S, et al. Misfolded polyglutamine, polyalanine, and superoxide dismutase 1 aggregate via distinct pathways in the cell. J Biol Chem. 2014;289:6669–6680. doi: 10.1074/jbc.M113.520189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yerbury JJ, et al. The small heat shock proteins αB-crystallin and Hsp27 suppress SOD1 aggregation in vitro. Cell Stress Chaperones. 2013;18:251–257. doi: 10.1007/s12192-012-0371-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McAlary L, Aquilina JA, Yerbury JJ. Susceptibility of mutant SOD1 to form a destabilized monomer predicts cellular aggregation and toxicity but not in vitro aggregation propensity. Front Neurosci. 2016;10:499. doi: 10.3389/fnins.2016.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao L, et al. Proteomic characterization of postmortem amyloid plaques isolated by laser capture microdissection. J Biol Chem. 2004;279:37061–37068. doi: 10.1074/jbc.M403672200. [DOI] [PubMed] [Google Scholar]
- 40.Xia Q, et al. Proteomic identification of novel proteins associated with Lewy bodies. Front Biosci. 2008;13:3850–3856. doi: 10.2741/2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ciryam P, Tartaglia GG, Morimoto RI, Dobson CM, Vendruscolo M. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Reports. 2013;5:781–790. doi: 10.1016/j.celrep.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciryam P, Kundra R, Morimoto RI, Dobson CM, Vendruscolo M. Supersaturation is a major driving force for protein aggregation in neurodegenerative diseases. Trends Pharmacol Sci. 2015;36:72–77. doi: 10.1016/j.tips.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tartaglia GG, et al. Prediction of aggregation-prone regions in structured proteins. J Mol Biol. 2008;380:425–436. doi: 10.1016/j.jmb.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 44.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chatr-Aryamontri A, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim MS, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirby J, et al. Phosphatase and tensin homologue/protein kinase B pathway linked to motor neuron survival in human superoxide dismutase 1-related amyotrophic lateral sclerosis. Brain. 2011;134:506–517. doi: 10.1093/brain/awq345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ciryam P, et al. A transcriptional signature of Alzheimer’s disease is associated with a metastable subproteome at risk for aggregation. Proc Natl Acad Sci USA. 2016;113:4753–4758. doi: 10.1073/pnas.1516604113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Basso M, et al. Characterization of detergent-insoluble proteins in ALS indicates a causal link between nitrative stress and aggregation in pathogenesis. PLoS One. 2009;4:e8130. doi: 10.1371/journal.pone.0008130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bergemalm D, et al. Superoxide dismutase-1 and other proteins in inclusions from transgenic amyotrophic lateral sclerosis model mice. J Neurochem. 2010;114:408–418. doi: 10.1111/j.1471-4159.2010.06753.x. [DOI] [PubMed] [Google Scholar]
- 52.Shaw BF, et al. Detergent-insoluble aggregates associated with amyotrophic lateral sclerosis in transgenic mice contain primarily full-length, unmodified superoxide dismutase-1. J Biol Chem. 2008;283:8340–8350. doi: 10.1074/jbc.M707751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arai T, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 54.Deng HX, et al. FUS-immunoreactive inclusions are a common feature in sporadic and non-SOD1 familial amyotrophic lateral sclerosis. Ann Neurol. 2010;67:739–748. doi: 10.1002/ana.22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boopathy S, et al. Structural basis for mutation-induced destabilization of profilin 1 in ALS. Proc Natl Acad Sci USA. 2015;112:7984–7989. doi: 10.1073/pnas.1424108112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Figley MD, Bieri G, Kolaitis RM, Taylor JP, Gitler AD. Profilin 1 associates with stress granules and ALS-linked mutations alter stress granule dynamics. J Neurosci. 2014;34:8083–8097. doi: 10.1523/JNEUROSCI.0543-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 58.Freer R, et al. A protein homeostasis signature in healthy brains recapitulates tissue vulnerability to Alzheimer’s disease. Sci Adv. 2016;2:e1600947. doi: 10.1126/sciadv.1600947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bechtold DA, Rush SJ, Brown IR. Localization of the heat-shock protein Hsp70 to the synapse following hyperthermic stress in the brain. J Neurochem. 2000;74:641–646. doi: 10.1046/j.1471-4159.2000.740641.x. [DOI] [PubMed] [Google Scholar]
- 60.Lowenstein DH, Chan PH, Miles MF. The stress protein response in cultured neurons: Characterization and evidence for a protective role in excitotoxicity. Neuron. 1991;7:1053–1060. doi: 10.1016/0896-6273(91)90349-5. [DOI] [PubMed] [Google Scholar]
- 61.Brown IR, Rush SJ. Cellular localization of the heat shock transcription factors HSF1 and HSF2 in the rat brain during postnatal development and following hyperthermia. Brain Res. 1999;821:333–340. doi: 10.1016/s0006-8993(99)01087-2. [DOI] [PubMed] [Google Scholar]
- 62.Manzerra P, Brown IR. Expression of heat shock genes (hsp70) in the rabbit spinal cord: Localization of constitutive and hyperthermia-inducible mRNA species. J Neurosci Res. 1992;31:606–615. doi: 10.1002/jnr.490310404. [DOI] [PubMed] [Google Scholar]
- 63.Batulan Z, et al. High threshold for induction of the stress response in motor neurons is associated with failure to activate HSF1. J Neurosci. 2003;23:5789–5798. doi: 10.1523/JNEUROSCI.23-13-05789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kiskinis E, et al. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell. 2014;14:781–795. doi: 10.1016/j.stem.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yerbury JJ, et al. Walking the tightrope: Proteostasis and neurodegenerative disease. J Neurochem. 2016;137:489–505. doi: 10.1111/jnc.13575. [DOI] [PubMed] [Google Scholar]
- 66.Brockington A, et al. Unravelling the enigma of selective vulnerability in neurodegeneration: Motor neurons resistant to degeneration in ALS show distinct gene expression characteristics and decreased susceptibility to excitotoxicity. Acta Neuropathol. 2013;125:95–109. doi: 10.1007/s00401-012-1058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 68.Johnson BS, et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ratovitski T, et al. Variation in the biochemical/biophysical properties of mutant superoxide dismutase 1 enzymes and the rate of disease progression in familial amyotrophic lateral sclerosis kindreds. Hum Mol Genet. 1999;8:1451–1460. doi: 10.1093/hmg/8.8.1451. [DOI] [PubMed] [Google Scholar]
- 70.Banci L, et al. Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: A possible general mechanism for familial ALS. Proc Natl Acad Sci USA. 2007;104:11263–11267. doi: 10.1073/pnas.0704307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pardo CA, et al. Superoxide dismutase is an abundant component in cell bodies, dendrites, and axons of motor neurons and in a subset of other neurons. Proc Natl Acad Sci USA. 1995;92:954–958. doi: 10.1073/pnas.92.4.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Forman HJ, Fridovich I. On the stability of bovine superoxide dismutase. The effects of metals. J Biol Chem. 1973;248:2645–2649. [PubMed] [Google Scholar]
- 73.Shibata N, et al. Intense superoxide dismutase-1 immunoreactivity in intracytoplasmic hyaline inclusions of familial amyotrophic lateral sclerosis with posterior column involvement. J Neuropathol Exp Neurol. 1996;55:481–490. doi: 10.1097/00005072-199604000-00011. [DOI] [PubMed] [Google Scholar]
- 74.Grad LI, et al. Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc Natl Acad Sci USA. 2014;111:3620–3625. doi: 10.1073/pnas.1312245111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gidalevitz T, Krupinski T, Garcia S, Morimoto RI. Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genet. 2009;5:e1000399. doi: 10.1371/journal.pgen.1000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tan CF, et al. Selective occurrence of TDP-43-immunoreactive inclusions in the lower motor neurons in Machado-Joseph disease. Acta Neuropathol. 2009;118:553–560. doi: 10.1007/s00401-009-0552-x. [DOI] [PubMed] [Google Scholar]
- 77.Toyoshima Y, et al. Spinocerebellar ataxia type 2 (SCA2) is associated with TDP-43 pathology. Acta Neuropathol. 2011;122:375–378. doi: 10.1007/s00401-011-0862-7. [DOI] [PubMed] [Google Scholar]
- 78.Schwab C, Arai T, Hasegawa M, Yu S, McGeer PL. Colocalization of transactivation-responsive DNA-binding protein 43 and huntingtin in inclusions of Huntington disease. J Neuropathol Exp Neurol. 2008;67:1159–1165. doi: 10.1097/NEN.0b013e31818e8951. [DOI] [PubMed] [Google Scholar]
- 79.Davidson YS, et al. TDP-43 pathological changes in early onset familial and sporadic Alzheimer’s disease, late onset Alzheimer’s disease and Down’s syndrome: Association with age, hippocampal sclerosis and clinical phenotype. Acta Neuropathol. 2011;122:703–713. doi: 10.1007/s00401-011-0879-y. [DOI] [PubMed] [Google Scholar]
- 80.Weihl CC, et al. TDP-43 accumulation in inclusion body myopathy muscle suggests a common pathogenic mechanism with frontotemporal dementia. J Neurol Neurosurg Psychiatry. 2008;79:1186–1189. doi: 10.1136/jnnp.2007.131334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakashima-Yasuda H, et al. Co-morbidity of TDP-43 proteinopathy in Lewy body related diseases. Acta Neuropathol. 2007;114:221–229. doi: 10.1007/s00401-007-0261-2. [DOI] [PubMed] [Google Scholar]
- 82.Wolff S, Weissman JS, Dillin A. Differential scales of protein quality control. Cell. 2014;157:52–64. doi: 10.1016/j.cell.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 83.Wang Q, et al. Proteomic analysis of neurofibrillary tangles in Alzheimer disease identifies GAPDH as a detergent-insoluble paired helical filament tau binding protein. FASEB J. 2005;19:869–871. doi: 10.1096/fj.04-3210fje. [DOI] [PubMed] [Google Scholar]
- 84.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang M, et al. PaxDb, a database of protein abundance averages across all three domains of life. Mol Cell Proteomics. 2012;11:492–500. doi: 10.1074/mcp.O111.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bender R, Lange S. Adjusting for multiple testing–when and how? J Clin Epidemiol. 2001;54:343–349. doi: 10.1016/s0895-4356(00)00314-0. [DOI] [PubMed] [Google Scholar]
- 87.Hochberg Y, Tamhane AC. Multiple Comparison Procedures. John Wiley & Sons; Hoboken, NJ: 1987. Introduction; pp. 1–16. [Google Scholar]
- 88.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- 89.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser A Stat Soc. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.