Significance
Transposable elements (TEs) are important tools to study gene function in plants, more so than in animal species, because of the ease in generating large numbers of germinal transpositions. We took advantage of an engineered TE in maize to broaden this approach. We screened for a defective kernel mutant and used the green fluorescent protein encoded by the TE to locate the gene linked to the mutation. The property of the TE permitted us to generate revertants and new alleles of the tagged gene, avoiding complementation assays made cumbersome by difficult transformation protocols in maize. Based on its sequence homology to a cochaperone, the tagged gene opens a line of research on the role of cochaperones in seed development.
Keywords: Ac/Ds, Dsg, Tti2, TTT complex, PIKKs
Abstract
We have used the newly engineered transposable element Dsg to tag a gene that gives rise to a defective kernel (dek) phenotype. Dsg requires the autonomous element Ac for transposition. Upon excision, it leaves a short DNA footprint that can create in-frame and frameshift insertions in coding sequences. Therefore, we could create alleles of the tagged gene that confirmed causation of the dek phenotype by the Dsg insertion. The mutation, designated dek38-Dsg, is embryonic lethal, has a defective basal endosperm transfer (BETL) layer, and results in a smaller seed with highly underdeveloped endosperm. The maize dek38 gene encodes a TTI2 (Tel2-interacting protein 2) molecular cochaperone. In yeast and mammals, TTI2 associates with two other cochaperones, TEL2 (Telomere maintenance 2) and TTI1 (Tel2-interacting protein 1), to form the triple T complex that regulates DNA damage response. Therefore, we cloned the maize Tel2 and Tti1 homologs and showed that TEL2 can interact with both TTI1 and TTI2 in yeast two-hybrid assays. The three proteins regulate the cellular levels of phosphatidylinositol 3-kinase-related kinases (PIKKs) and localize to the cytoplasm and the nucleus, consistent with known subcellular locations of PIKKs. dek38-Dsg displays reduced pollen transmission, indicating TTI2’s importance in male reproductive cell development.
Maize has a long history of transposon genetics dating back to the discovery of transposable elements Activator (Ac) and Dissociation (Ds) by Barbara McClintock in the 1940s (1). Transposable elements comprise approximately 85% of the maize genome (2) and are mostly inert because of mutations and epigenetic silencing by DNA methylation and histone modifications (3, 4). Several of them, however, like the Ac/Ds and Mutator families, can be activated and used for forward or reverse genetics purposes (5–7). The Ac/Ds system is the method of choice when performing targeted transposon tagging, but limitations such as lack of “Ds-launching pads” near the gene of interest, and identifying germinal excisions and tracking Ds, can be a challenge. To address these major limitations, a new resource has been developed in maize by using an engineered Ds that can be tracked phenotypically for germinal excision and reinsertion through a change in seed color and green fluorescent protein (GFP) signal (8). The Ds element contains GFP (Dsg) and has the 5′ and 3′ end sequences that are recognized by Ac for transposition. As such, transpositions from these Dsg launching pads are also being made and new insertion sites being mapped to develop a collection of tagged sites for the maize genetics community (www.acdsinsertions.org). The GFP tag makes the Ds element dominant and, therefore, allows for the phenotypic selection of seeds either heterozygous or homozygous for the Dsg insertion.
We have used this system to identify recessive mutations that prevent normal seed development. Such a screen resulted in the identification of a defective kernel mutant (dek), called dek38-Dsg. Using the unique Dsg-adjacent sequences, the corresponding gene was cloned. Based on information from studies in animal and yeast systems, its gene product could be characterized as a cochaperone hypothesized to enable early seed development by controlling the action of phosphatidylinositol 3-kinase-related kinases (PIKKs) that regulate cellular signaling pathways related to gene expression, cellular growth, response to DNA damage, and nonsense-mediated mRNA decay (9). The protein, called Tel2-interacting protein 2 (TTI2), associates with Telomere maintenance 2 (TEL2) and Tel2-interacting protein 1 (TTI1), forming what is called the TTT complex (10). The TTT complex regulates the steady-state levels of PIKKs, and their assembly into functional complexes in a heat shock protein 90 (HSP90)-dependent manner (11). Here, we use the dek38-Dsg mutation to report a description of the TTT complex in plants. The mutation is lethal and results in an arrested embryo and a highly underdeveloped endosperm. The mutation also reduces protein levels of two members of maize PIKKs called Target of Rapamycin (ZmTOR) and Ataxia Telangiectasia Mutated (ZmATM), suggesting a conserved function throughout eukaryotic species.
Results
Verification and Genetic Characterization of the dek38-Dsg Mutant.
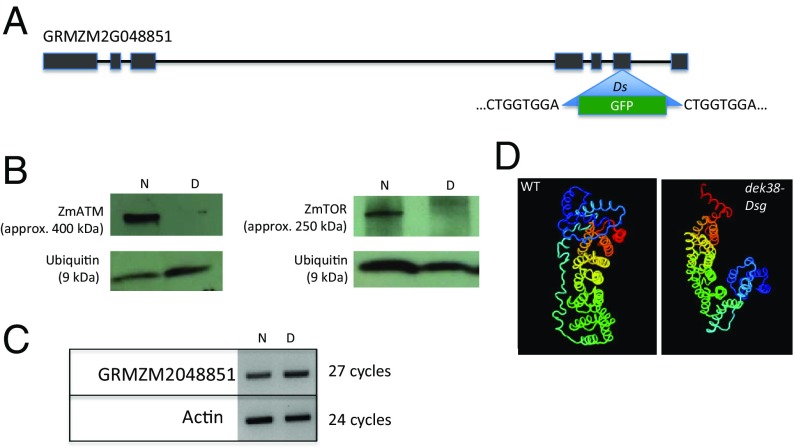
The dek38-Dsg mutant was isolated from a screen of a Dsg insertion collection for defective kernel (dek) phenotypes in a predominantly W22 genetic background (Fig. 1). Dsg-adjacent sequences derived from sequence indexing of this collection indicated that the Dsg was inserted in exon 6 of gene model GRMZM2G048851 in dek38-Dsg, based on mapping sequences to the maize inbred B73 reference genome assembly version 3 (www.acdsinsertions.org). The Dsg insertion was verified by designing PCR primer pairs on the left and right sides of the insertion, with one primer designed from the gene model, and the other from the Dsg insertion (Table S1). PCR using these two primer pairs gave amplification on the GFP containing Ds elements, but not on non-GFP containing elements. The PCR products were also sequenced and showed that left and right insertion junctions contain an 8-bp direct repeat (Fig. 2), which is characteristic of Ac/Ds transposon insertion sites (12). RT-PCR experiments showed that the Dsg insertion does not affect transcription of GRMZM2G048851. If translated, the transcript is predicted to result in a shorter protein from a premature stop codon, containing amino acid residues from parts of the Dsg sequence. Protein-folding modeling also predicts a drastic change in the 3D conformation of the protein (Fig. 2D).
Fig. 1.
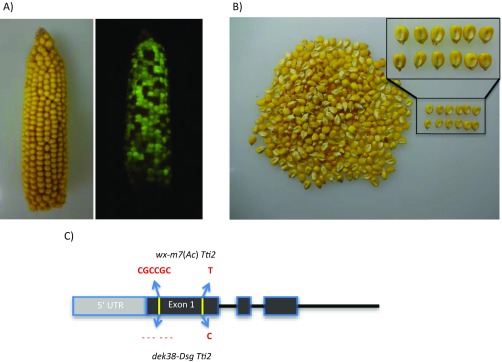
The dek38-Dsg mutant segregating in an ear (A, small kernels) and after shelling (B, Right). When viewed under a blue light (C), segregation between normal non-GFP (+/+), normal GFP (+/−), and dek GFP (−/−) is visible. The change in color of normal kernels not fluorescing GFP is because of exposure to blue light in a dark background. (D, Left) Ear from a dek38 footprint allele with with a frameshift mutation. (D, Right) Ear from the allelism test between the same footprint allele and the dek38-Dsg parent. (E) Same ears as in D, but under blue light. The frameshift allele on left lost Dsg (and fluorescence), but the kernels on the right fluoresce because of the Dsg element in the dek38-Dsg mutant. Also shown are pictures of defective kernels (embryo side up) from the footprint allele (F) and from the allelism test (G). Normal kernels are shown on top for comparison.
Table S1.
PCR primers used in this study
| Primer name | Sequence | Purpose |
| Telo2_5UTR_F | AAAGACTCGTCTATTCGAACTGTCC | Full-length CDS cloning |
| Telo2_3UTR_R | AGGACACATCCCTGCACAAC | |
| Tti1_5UTR_F | GTATTGGAAAGCAGGCAAGC | Full-length CDS cloning |
| Tti1_3UTR_R | GGGTGCTAGCATGACCAAAG | |
| Tti2_5UTR_F | ACCCCTTCAGTGCCTTGACT | Full-length CDS cloning |
| Tti2_3UTR_R | GTGACAGGATAGGCGATGGT | |
| Telo2_qPCR_F | TATTGTTTGCAGCGTCTTGC | qRT-PCR |
| Telo2_qPCR_R | TGATTTGCAGATTCCAGTGC | |
| Tti1_qPCR_F | TGATGTTCTTGCTTCCATGC | qRT-PCR |
| Tti1_qPCR_R | TCATCTGGCAAACAAGTGGA | |
| Tti2_qPCR_F | GATGCGTGCTGTCAGAACAT | qRT-PCR |
| Tti2_qPCR_R | GAGGCCCATAGCATCAAAAA | |
| Ubiquitin_qPCR_F | CGCACCCTAGCAGACTACAA | qRT-PCR |
| Ubiquitin_qPCR_R | TACGCACACACAACACAACC | |
| Dsg_left_F | TCTTCGCATTGTTGTTCCTG | Left Dsg insertion PCR |
| Dsg_left_R | TTCGCTCATGTGTTGAGCAT | |
| Dsg_right_F | CGGGATCATTCCGATTAAAA | Right Dsg insertion PCR |
| Dsg_right_R | ATTCGGTTAGGGGGATTGAG | |
| ZmMRP1_RT_F | GGCTTGTTCAGGGACAACAA | ZmMRP1 RT-PCR |
| ZmMRP1_RT_R | TCGTTGATGCTAAAGCGTTG | |
| Betl10_RT_F | CATGCATGAGCAACAAGGAC | Betl10 RT-PCR |
| Betl1_RT _R | AAATGGAGGACATGCCAAAC | |
| Pbf_RT_F1 | TGCTGAGAGGAGGGATTTTT | Pbf RT-PCR |
| Pbf_RT_F1 | GCTCTTCATGATGGCCTTG | |
| Telo2Sfi1A_F | AAGGCCGTCAAGGCCATGGCGAGCACGCGAAGC | YFP-fusion |
| Telo2Sfi1B_R | AAGGCCCATGAGGCCCATCATGTTGCCGAATGG | |
| Tti1Sfi1A_F | AAGGCCGTCAAGGCCATGGCGATGGAGGCGTCGGCG | YFP-fusion |
| Tti1Sfi1B_R | AAGGCCCATGAGGCCCACACCCTGCATCCTTCTGAA | |
| Tti2Sfi1A_F | AAGGCCGTCAAGGCCATGGCGAGCACGCGAAGC | YFP-fusion |
| Tti2Sfi1B_R | AAGGCCCATGAGGCCCATCATGTTGCCGAATGG | |
| Telo2Sfi1A_F | AAGGCCGTCAAGGCCATGGCGAGCACGCGAAGC | Yeast two-hybrid |
| Telo2Sfi1B_R_Stp | AAGGCCCATGAGGCCTCACATCATGTTGCCGAA | |
| Tti1Sfi1A_F | AAGGCCGTCAAGGCCATGGCGATGGAGGCGTCGGCG | Yeast two-hybrid |
| Tti1Sfi1B_R_Stp | AAGGCCCATGAGGCCTCACACACCCTGCATCCTTCT | |
| Tti2Sfi1A_F | AAGGCCGTCAAGGCCATGGCGAGCACGCGAAGC | Yeast two-hybrid |
| Tti2Sfi1B_R_Stp | AAGGCCCATGAGGCCTCACGGTGAACTGTTTTT |
Fig. 2.
(A) The Dsg is inserted in exon 6 of gene model GRMZM2G048851 (ZmTti2). The 8-bp target site duplication (TSD) sequence is indicated on the left and right sides of the insertion. (B) Western blots comparing ZmATM and ZmTOR protein levels between normal (N) and dek (D) kernels. (C) Gene expression of GRMZM2G048851 between WT and dek using mRNA extracted from immature seeds. (D) Predicted protein folding of ZmTTI2 in WT and dek38-Dsg alleles.
An advantage of having a Dsg-tagged gene is that it is possible to select heterozygotes among the normal seeds in the dek38-Dsg mutant by simple GFP selection. This feature made it easier to conduct a cosegregation analysis between the dek phenotype and the Dsg. Selfed progenies from 25 normal GFP seeds segregated for the dek phenotype. Conversely, none of the selfed progenies from 25 normal non-GFP seeds segregated for the dek phenotype. This result indicates a tight linkage between GFP (and thus Dsg) and the dek phenotype (0/50 recombinants). Another advantage is that it is possible to monitor the transmission of the gene (either through male or female) by counting the number of GFP and non-GFP seeds after performing a cross. To determine whether dek38 is important for male or female reproductive cell development in maize, reciprocal crosses were conducted by using dek38-Dsg either as a male or female parent. Table S2 indicates that when dek38-Dsg is used as female, the segregation of GFP and non-GFP seeds falls along the 1:1 ratio that is expected. However, when used as a male, the expected 1:1 segregation is often distorted (based on χ2 tests), indicating a defect in pollen transmission in the dek38-Dsg mutant. Based on the B73 genome reference V3, GRMZM2G048851 is located on the short arm of chromosome 5 close to another dek mutant (dek18). dek18 (accession 527A from the Maize Genetics Stock Center) also has collapsed and floury kernels like dek38-Dsg. We tested whether these two dek mutants were allelic, but they were not.
Table S2.
Pollen transmission defect in dek38-Dsg
| Cross | Observed | χ2 (P value)* | |
| GFP(+) | GFP(−) | ||
| dek38-Dsg x wx-m7(Ac) | 157 | 177 | 0.27 |
| dek38-Dsg x wx-m7(Ac) | 92 | 84 | 0.55 |
| dek38-Dsg x wx-m7(Ac) | 106 | 131 | 0.10 |
| dek38-Dsg x wx-m7(Ac) | 211 | 218 | 0.74 |
| dek38-Dsg x wx-m7(Ac) | 213 | 233 | 0.34 |
| wx-m7(Ac) x dek38-Dsg | 91 | 140 | 0.00* |
| wx-m7(Ac) x dek38-Dsg | 60 | 48 | 0.25 |
| wx-m7(Ac) x dek38-Dsg | 125 | 172 | 0.01* |
| wx-m7(Ac) x dek38-Dsg | 152 | 205 | 0.01* |
| wx-m7(Ac) x dek38-Dsg | 137 | 251 | 0.00* |
χ2, χ2 test. Note the number GFP seeds are lower than expected (bold numbers, significant χ2 values) when dek38-Dsg is used as male.
A third advantage of a marked Ds element is that it enabled us to select revertants from a lethal mutation. Revertants serve to confirm that the mutation in question is, in fact, tagged by Ds, but they are difficult to isolate from a lethal mutation (13). Putative revertants are isolated by planting a large number of normal, green fluorescent individuals, selfing them, and screening for ears that do not segregate the dek mutation, but still segregate green fluorescent kernels. These ears are revertant candidates that presumably arose from a Dsg transposition that generated a normal Dek allele. Among 500 self-pollinated ears, three putative revertants segregated GFP, but not the dek phenotype (Fig. S1 A and B). Genotyping these three plants showed that the Dsg was no longer present at the dek38 locus and that its excision had left no footprints, restoring the original wild-type sequence. To distinguish this dek38-Dsg clean excision allele from the normal allele coming from the wx-m7(Ac) parent, GFP seeds from revertants were genotyped for two polymorphisms that mark the two parental alleles: a 6-bp insertion-deletion (indel) polymorphism and a SNP present in exon 1 of GRMZM2G048851 (Fig. S1C). Sequencing several GFP seeds from each of the three putative revertant ears showed that, in fact, they were segregating both polymorphisms, indicating that the ears do not segregate the dek phenotype because the gene’s function had been restored by excision of Dsg.
Fig. S1.
(A) One of the identified revertants showing a completely normal ear (Left). The same ear is shown at Right under blue light segregating for GFP. (B) Kernels from A with a subset shown at Right. Inset shows a more close-up view of the normal kernels. (C) Sequence polymorphisms found in exon 1 of GRMZM2G048851 that enabled differentiation between the WT allele contributed by the wx-m7(Ac) parent and the WT progenitor allele of dek38-Dsg.
Besides clean Ds excision, the addition of extra nucleotides (usually variations of the TSD sequence) can cause either frameshift or in-frame mutations. The existence of these footprint alleles was first identified by PCR screening of 5,000 F2 plants grown from 2,500 GFP and 2,500 non-GFP seeds from self-pollinated F1 plants from the cross of dek38-Dsg to wx-m7(Ac). To manage the large population size, DNA were first extracted in bulks of 10 plants and genotyped for the presence of a footprint by using PCR primers that flank the insertion site (Table S1). Bulks that indicated the existence of a footprint by the presence of a larger PCR band above the wild-type band on a 3% Metaphor agarose gel were then selected for individual plant genotyping. The identified individuals from the GFP population that possessed the footprint were then genotyped with another pair of PCR primers to detect the presence or absence of the Dsg insertion (Table S1). A total of 112 individuals (102 from the GFP population and 10 from the non-GFP population) with the presence of a footprint and absence of the Dsg insertion (germinal Dsg excisions) were then tagged for self-pollination, and cross-pollination to dek38-Dsg for allelism tests.
Thirty individuals that were identified by PCR screening described above as containing germinal excision footprints were sequenced. Of these individuals, we identified four different types of frameshift insertion footprints—two types with eight extra nucleotides (fp+8), and another two with seven extra nucleotides (fp+7) (Table 1). All of the selfed ears from these individuals segregated dek kernels. Crosses with dek38-Dsg also resulted in ears segregating for the dek phenotype, indicating that the dek mutations are allelic. A notable observation is the isolation of a dek38 frameshift footprint allele from the non-GFP population, which is a result of excision and loss of Dsg that either segregated away during meiosis or did not reinsert into the genome (Fig. 1 D and E). Two in-frame mutation footprint alleles were also identified, one predicted to add an extra amino acid (fp+3) and another one adding two extra amino acids (fp+6) (Table 1). Examination of selfed ears from these individuals showed that a great majority of the kernels are normal, but occasionally dek seeds can be found randomly on the ear. A possible explanation for this observation is that the addition of extra amino acids has only a small effect on the proper function of the protein (partial reversion) (Fig. S2).
Table 1.
dek38 alleles from Dsg excision footprints
| Footprint | No. of plants | Consequence |
| CTGGTGGA GTGGTGGA | 12 | Frameshift |
| CTGGTGGA <Dsg> CTGGTGGA | ||
| CTGGTGGT CTGGTGGA | 4 | Frameshift |
| CTGGTGGA <Dsg> CTGGTGGA | ||
| CTGGTGG GTGGTGGA | 1 | Frameshift |
| CTGGTGGA <Dsg> CTGGTGGA | ||
| CTGGTGGT CGGTGGA | 1 | Frameshift |
| CTGGTGGA <Dsg> CTGGTGGA | ||
| CTGGTGG TGGA | 5 | Inframe (+1 amino acid) |
| CTGGTGGA <Dsg> CTGGTGGA | ||
| CTGGTGGT GGTGGA | 7 | Inframe (+2 amino acids) |
| CTGGTGGA <Dsg> CTGGTGGA |
The footprint sequences are in bold. The target site duplication (TSD) sequence is indicated below the footprint sequence for comparison.
Fig. S2.
(A) An ear from one of the identified partial revertants. (B) Kernels from A showing mostly normal kernels (Left) and seven dek kernels (Right). (C) Closeup of the seven dek kernels compared with the wild type.
dek38-Dsg Has Impaired Seed Development.
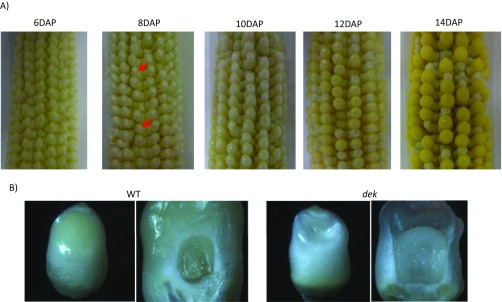
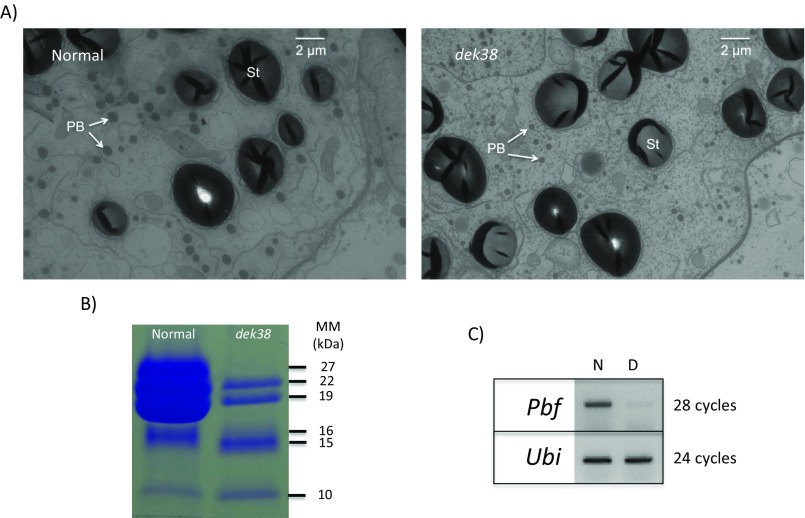
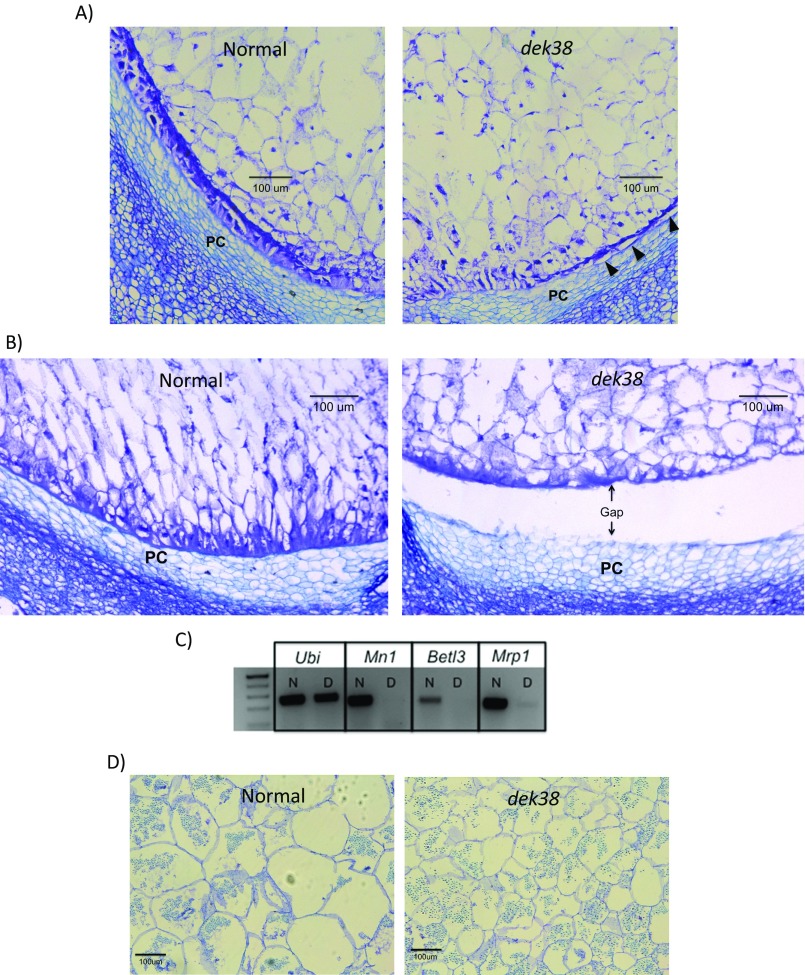
Developing ears were sampled at different days after pollination (DAP) to determine the progression of the dek phenotype. As early as 8 DAP, dek kernels can already be distinguished from WT kernels because of their smaller size (Fig. S3A). At approximately 12 DAP, the dek seeds begin to cave in at the crown because of incomplete seed filling (Fig. S3B). Proximate analysis of mature seeds reveals a lower crude protein and oil concentration, and a higher carbohydrate concentration in the dek seeds (Table S3). The lower protein concentration is reflected in the smaller protein bodies in the dek endosperm compared with normal (Fig. S4A). The increase in carbohydrate concentration in dek kernels is consistent with the unchanged size of the starch granules and the smaller overall seed size. Zein storage proteins, which make up ∼50% of the total seed protein (14), are also greatly reduced in the dek seeds. The 27-kDa gamma-zein has the largest reduction among different zeins proteins, with no detectable protein in the SDS/PAGE (Fig. S4B). Indeed, its regulator, the transcription factor Prolamin box binding factor 1 (Pbf1), whose function is conserved across cereals (15), is also reduced in the dek seeds (Fig. S4C).
Fig. S3.
(A) Heterozygote dek38-Dsg ears at different stages of development. Arrows indicate the smaller dek kernels. (B) 12DAP WT and dek kernels.
Table S3.
Proximate analysis of dek seeds compared with WT
| Component | WT, % | dek, % |
| Protein (crude) | 11.12 | 7.84 |
| Fat (crude) | 3.95 | 1.05 |
| Carbohydrates | 72.56 | 79.99 |
| Moisture | 10.60 | 9.35 |
| Ash | 1.56 | 1.77 |
Proximate analysis was done by New Jersey Food Labs Inc.
Fig. S4.
(A) Transmission electron micrograph of a normal (Left) and dek (Right) seed from the dek38-Dsg mutant (PB, protein body; St, starch granule). (B) Zein protein profile of a dek seed from the dek38-Dsg mutant compared with a normal kernel (MM, protein molecular mass in kilodaltons). (C) Gene expression (RT-PCR) of Pbf gene (N, normal; D, dek).
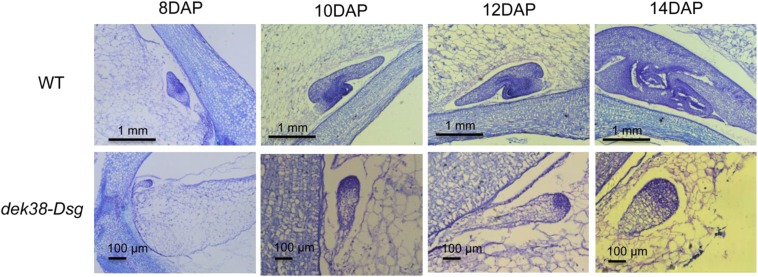
The development of the embryo and the endosperm was also studied through histological sections of seeds collected at different DAP. Dramatic differences between normal and mutant embryos are seen in Fig. 3. First is the arrest in the development of the embryo, which stopped growing at the dermatogen stage, indicating failure of further cell division and differentiation. Second are the smaller cells in the endosperm of dek kernels, which did not seem to expand as much as the normal kernels (Fig. S5D). Third is the conspicuous absence of a fully developed basal endosperm transfer layer (BETL), a darkly staining cell layer at the bottom of the developing endosperm that is specialized for nutrient transport (Fig. S5 A and B). The aberrant BETL formation is also reflected by the low gene expression of BETL-specific genes in maize (Fig. S5C). Also noticeable is the formation of a gap between the developing endosperm and the placento-chalazal cells (PC). Gap formation is visible at approximately 8 DAP and continues until the endosperm is fully dissociated from the PC layer at approximately 10 DAP (Fig. S5B).
Fig. 3.
Development of the embryo at different days after pollination. Upper, wild type; Lower, dek38-Dsg.
Fig. S5.
Defects in endosperm development in dek38-Dsg. (A) 8DAP endosperm sections showing underdeveloped BETL in dek mutant. Arrows indicate sections that appear to be separating from the base of the seed. PC, placento-chalazal layer. (B) 10DAP dek endosperm showing detachment from the base of the seed. (C) RT-PCR of BETL-specific genes (N, normal sample; D, dek sample). (D) Cell size comparison in endosperm cells between a normal and dek seed.
dek38-Dsg Is Homologous to Yeast and Mammalian Tti2.
The entire dek38-Dsg genic region is predicted to be 9,127 bp long and corresponds to the gene model GRMZM2G048851 based on current annotation (version 3). Despite its large size, the predicted transcript is only 1,263 bp long, which is divided into 7 exons. The gene contains a large LTR retrotransposon insertion in the third intron (more than 6 kb), which is also present in the W22 allelic sequence obtained from PacBio sequencing (16). Despite this intron expansion, the gene is expressed in many tissues and at different developmental stages based on available gene expression data at MaizeGDB (www.maizegdb.org). The gene annotation indicates that the predicted GRMZM2G048851 protein product contains armadillo repeats, but provides no functional data. Using primers designed from the predicted UTRs (Table S1), the full-length CDS of GRMZM2G048851 was cloned from mRNA extracted from 2-wk-old maize seedlings. From this sequence, an ORF of 1,269 bp in length was predicted to encode aprotein of 422 amino acids. A BLASTP in UniProt (www.uniprot.org) showed that GRMZM2G048851 has similarity to a protein in yeast and animals termed TTI2. Therefore, GRMZM2G048851 was designated as a putative Tti2 (ZmTti2) homolog. The ZmTTI2 protein shares 23.4% identity with its human counterpart, with the highest similarity concentrated at the TTI2 domain, which overlaps with the armadillo domain. BLAST analysis of ZmTTI2 in maize B73 reference and the W22 PacBio genome sequences (16) indicates that it is a single copy gene.
TEL2 and TTI1 Are also Present in Maize.
In animals and yeast, TTI2 partners with TEL2 and TTI1 to form the TTT complex (10, 17). Therefore, a search for TEL2 and TTI1 homologs in maize was conducted. Human TEL2 and TTI1 protein sequences were used to identify their homologous counterparts in maize via BLASTP. Both BLAST searches resulted in single hits, indicating that Tel2 and Tti1 are also single copy genes in maize. The putative Tel2 homolog corresponds to gene model GRMZM2G144166 (ZmTel2), whereas the putative Tti1 homolog corresponds to GRMZM2G056403 (ZmTti1) in maize. PCR primers were designed from the annotated 5′ and 3′ UTR regions of these genes to clone the full-length CDS (Table S1). For ZmTel2, a PCR band approximately 3 kb long was amplified, cloned, and sequenced. An ORF of 3,051 bp was identified and predicted to encode a protein 1,016 aa long. Sequence analysis of this predicted protein product at InterPro (www.ebi.ac.uk/interpro) shows the conserved TEL2 domain at amino acid residues 634–745. The TEL2 domain overlaps with an armadillo repeat, which is then flanked by two more armadillo repeats. It also shares 24.4% protein identity with human TEL2 protein. For Tti1, a 4-kb PCR product was amplified, cloned, and sequenced. This clone contains a 4,035-bp-long ORF that encodes a protein that is 1,344 aa long. Similar to ZmTEL2 and ZmTTI2, the predicted ZmTTI1 protein contains armadillo domains based on analysis with InterPro. Both ZmTEL2 and ZmTTI1 have similar structures like their respective human homologs.
Functional Characterization of the Maize TTT Complex.
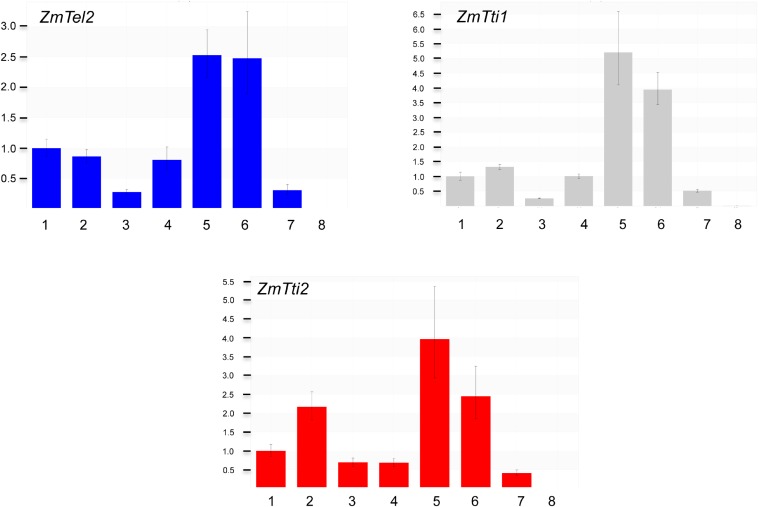
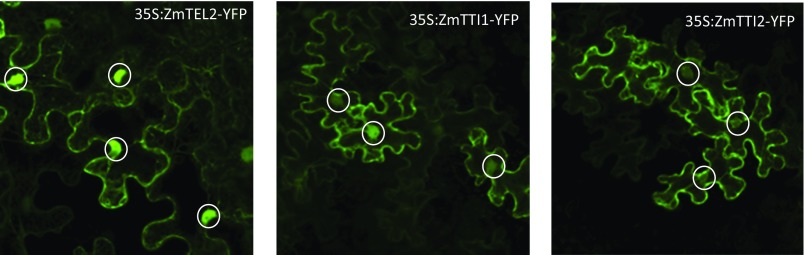
The interaction of maize TEL2, TTI1, and TTI2 was tested via yeast two-hybrid. As seen in Fig. S6, both ZmTel2-DB/ZmTti1-AD and ZmTel2-DB/ZmTti2-AD yeast diploids survived in LTH triple dropout media, indicating ZmTEL2’s interaction with both ZmTTI1 and ZmTTI2. Gene expression of ZmTel2, ZmTti1, and ZmTti2 in many different maize tissues was detected by using quantitative PCR (qPCR). Expression is particularly strong in young/developing tissues like 2-wk-old shoots and roots, as well as developing tassels and ears (Fig. S7). In contrast, gene expression was weaker in mature leaf, and almost no expression was detected in mature pollen. ZmTel2, ZmTti1, and ZmTti2 were fused to YFP and transiently expressed in tobacco leaves to determine their subcellular localization. Microscopic examinations of the leaves show YFP signals in both cytoplasm and nucleus for all of the three TTT complex members (Fig. S8). Our results are consistent with cytoplasmic and nuclear localizations of TEL2 in humans (18) and TTI2 in yeast (19). In yeast and mammals, the TTT complex is required for the stability of PIKK proteins as well as their assembly into complexes. Western blots with proteins extracted from developing mutant and wild-type seeds show that the mutant has no detectable levels of ZmATM and ZmTOR proteins (Fig. 2B).
Fig. S6.
Yeast two-hybrid between ZmTel2 and ZmTti1 (A) and between ZmTel2 and ZmTti2 (B). Growth in –LTH media indicates interaction.
Fig. S7.
Relative gene expression of ZmTel2, ZmTti1, and ZmTti2 in different maize tissues by using ubiquitin as a reference. (1) Two-week-old shoot. (2) Two-week-old root. (3) Mature leaf. (4) 14DAP seed. (5) Immature (1 cm) ear. (6) Immature (1 cm) tassel. (7) Silk. (8) Pollen.
Fig. S8.
Cellular localization of ZmTel2, ZmTti1, and ZmTti2 proteins by transient expression of YFP fusion constructs in tobacco leaves. Circles indicate YFP signal in the nucleus.
Discussion
Mutant resources are important for functional genomics. However, crop species are still notoriously hard to transform, creating an impediment toward deployment of gene editing technologies (20). Although progress in transformation has been made in some monocots (21), transformation still necessitates specialized stocks and constructs and entails tissue culture capabilities. The use of transposons in creating mutants in maize only needs a greenhouse, field space, and manual pollinations, and, therefore, can be an alternative for research groups without transformation capabilities. Also, as shown in our work, Dsg tagging has several benefits. First, it became easier to track the kernels that contain the Dsg element and identify their zygosity just by selecting for GFP fluorescence. Second, because GFP is dominant, linking the phenotype to the Dsg insertion in a lethal mutation was made easier by conducting a trait and GFP cosegregation test made possible by the phenotypic identification of seeds heterozygote for the Dsg insertion. Third, Dsg footprint sequences that can vary from different individuals provide multiple alleles that can have different phenotypic consequences, whereas clean excision events provide a clear “cause and effect” verification of the mutation. In the case of lethal mutations like the one described here, the GFP fluorescence from the Dsg enables the isolation of revertants. Some alleles that can be derived from footprint sequences have beneficial consequences such as the increased seed weight observed in a maize shrunken2 allele with 2 aa residues added as a result of a Ds footprint (22).
The dek38-Dsg, which encodes a cochaperone, is a mutant to be characterized from the Dsg insertion collection, and a member of the TTT complex characterized in plants. Because of their important roles in abiotic stress tolerance and growth regulation in response to changing environmental cues, molecular chaperones have been widely studied in plants. Most of the studies, however, have focused almost exclusively on heat shock proteins (HSPs), and little is known about their associated cochaperones. Current evidence from yeast and mammals suggests that the TTT complex associates with chaperones and various other cochaperones to regulate cellular levels of PIKK proteins. Studies in yeast and animals showing that TEL2 binds preferentially with newly synthesized PIKKs, that TEL2, TTI1, and TTI2 are required to stabilize cellular PIKK levels, and that PIKK complexes are disrupted when TEL2 or HSP90 are depleted, all point to a cochaperone activity of the TTT complex (10, 11, 18, 23).
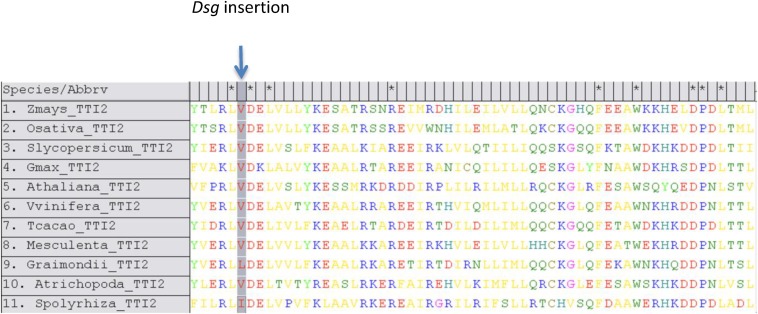
The defects in many aspects of seed development reflect the multitude of developmental pathways that are affected by the dek38 mutation, indicating that TTI2 is required in early stages of development. ZmTti2’s roles in storage protein accumulation and BETL development should be investigated further. Nevertheless, the limited nutrient transport predicted to result from a defective BETL could certainly play a role in the abnormal seed development of dek38. However, this defect alone cannot account for the embryo lethal phenotype because other mutants with defective BETL in maize like miniature seed 1 and zmsweet4c have well-differentiated and viable embryos (24, 25). Interestingly, the early embryonic lethality observed in dek38-Dsg resembles the TOR mutation in Arabidopsis. Like dek38-Dsg, the embryonic development of the AtTOR mutant is arrested at the dermatogen stage (known as transition stage in maize), which is characterized by the formation of the protoderm (26). This similarity implies that the embryonic lethality observed in dek38-Dsg might be due to aberrant ZmTOR function, which is supported by reduction of ZmTOR protein. The lethal phenotype in dek38-Dsg also implies that the C-terminal end of TTI2, where the dek38 mutations are located, is important for protein function. This hypothesis is supported by the protein sequence alignment of ZmTTI2 with its putative plant homologs showing high degree of conservation at several amino acid residues at the C-terminal end (Fig. S9). In addition, the dek38-Dsg revertants that were isolated in our screen all show complete restoration of the WT sequence. This result is also consistent with findings in yeast wherein C-terminal end truncations of TTI2 are also lethal (27).
Fig. S9.
Multiple sequence alignment of maize TTI2 protein with other putative plant TTI2 proteins showing high degree of conservation at the C-terminal end. The Dsg insertion point is indicated. The accession numbers of the sequences are as follows: (1) Zea mays, GRMZM2G048851; (2) Oryza sativa, LOC_Os06g47900; (3) Solanum lycopersicum, Solyc06g009690; (4) Glycine max, Glyma.18G031000; (5) Arabidopsis thaliana, AT2G39910; (6) Vitis vinifera, GSVIVG01016380001; (7) Theobroma cacao, Thecc1EG042614; (8) Manihot esculenta, Manes.10G018500; (9) Gossypium raimondii, Gorai.010G199300; (10) Amborella trichopoda, evm_27.TU.AmTr; (11) Spirodela polyrhiza, Spipo12G0055600.
The abnormal segregation of GFP and non-GFP seeds from the expected 1:1 ratio when dek38-Dsg is used as a pollen donor indicates ZmTti2 is important for male reproductive cell development. This observation is likely due to ZmTti2’s role in stabilizing the ATM protein. In humans, the ATM gene is expressed four times higher in testes than in somatic cells (28) and mutations in mice result either in embryonic lethality (29) or complete infertility (30). In Arabidopsis, ATM mutants are only partially sterile, although defects in meiosis have also been observed in both male and female reproductive cell development (31). If the pollen transmission defect in dek38-Dsg is actually due to reduced ATM function caused by the ZmTti2 mutation, then ATM may not be as critical for female reproductive cell development in maize compared with Arabidopsis and mammals. It is also possible that ZmTTI2 functions independently of ATM in reproductive cell development.
The conserved interaction of TEL2, TTI1, and TTI2 in yeast, mice, humans, and now maize, provides strong evidence of the importance of this complex in organismal development. In a large-scale mutagenesis experiment, the homolog of Tel2 in Arabidopsis has been identified as one of the many genes that affect embryo development and has been named embryo defective 2423, which corresponds to gene model AT3G48470 (32). The mutant, however, remains uncharacterized. No Tti1 mutants have been reported in the literature outside of yeast, but it is likely that null mutants will also be lethal. A recent finding that a “chaperome” network that includes TTT complex members facilitates tumor survival in humans (33) raises the possibility of their potential use to genetically engineer plants with enhanced stress tolerance. Our analysis now provides a foundation for new lines of investigation in maize in terms of embryo and endosperm development, BETL formation, and DNA damage repair among others. Because additional mutants in TTT function are likely to be lethal, weaker mutant alleles or conditional mutant alleles would be preferred. The partial dek38 revertant alleles isolated in this study can be used in the future to study other aspects of TTT complex function that were not possible with lethal alleles, such as DNA damage response. In the case of PIKKs (or other proteins with multiple domains), Dsg can be useful for targeting specific domains of these proteins for mutagenesis via intragenic transposition.
Materials and Methods
Plant Materials.
dek38-Dsg was identified in 2013 from the available Dsg insertion collection after a screen of ears segregating for defective kernel phenotypes. It has been referred to as dek34-Dsg (MaizeGDB.org). Seeds were deposited in the Maize Genetics Cooperative Stock Center and can be obtained under accession no. tdsgR07A07. The dek18 mutant used for allelism test was obtained from the Maize Genetics Stock Center. The wx-m7(Ac) stock that was used as the transposase source for the reversion and Dsg excision footprint experiments is in a predominant W22 background.
Dsg Excision Footprint and Reversion Analysis.
Revertants were isolated by first crossing dek38-Dsg to wx-m7(Ac). Five hundred GFP-fluorescent seeds from this cross were then planted in the field and self-pollinated. The ears from these plants were then evaluated for GFP and defective kernel segregation, and ears that segregated for GFP but not defective kernels were identified. GFP kernels from these putative revertants were then planted and genotyped for two polymorphisms that distinguish the WT allele from wx-m7(Ac) and the Dek38 revertant allele from dek38-Dsg. These individuals were also genotyped for Dsg presence/absence by using PCR primers from Table S1.
For isolating Dsg footprint alleles, the dek38-Dsg mutant was also crossed to wx-m7(Ac). The F1 were then self-pollinated to produce F2 seeds. From these seeds, 2,500 GFP and 2,500 non-GFP seeds were planted in the field. At the seedling stage, DNA was extracted from leaf samples bulked from 10 individuals and assayed for Dsg excision footprints by PCR with primers that amplify 100-bp regions flanking the Dsg insertion (Table S1). PCR products were electrophoresed in 3% Metaphor agarose (Lonza) to enable resolution of the excision footprint PCR band that appears as a slightly slower band above the wild-type band. To identify the actual plants within the bulk that possessed the footprint, DNA from individual plants were extracted and genotyped for the presence of the footprint. Individual plants from the GFP population identified to contain footprints were then genotyped for the presence or absence of Dsg by using PCR primers designed to amplify the insertion junctions (Table S1). Individual plants containing a footprint were then tagged, self-pollinated, and crossed to dek38-Dsg for allelism test.
Histological Analysis.
Developing kernels were fixed in 4% paraformaldehyde in PBS and processed according to a described protocol (34). Sections were then mounted on slides, stained with Toluidine blue, and documented by using a Leica DM5500B light microscope system. A published method was also used for transmission electron microscopy of developing kernels (35).
Transient Expression in Tobacco Leaves.
Maize Tel2, Tti1, and Tti2 C-terminal YFP fusions were created by first PCR amplifying the coding sequences (without stop codon) using primers with Sfi1A and Sfi1B restriction sites (Table S1) from pGEM-Teasy full-length CDS clones of Tel2, Tti1, and Tti2, respectively. The PCR products were then directionally cloned into pENTRSfi-223.1 plasmid by standard cloning protocol. The pENTR vectors containing Tel2, Tti1, and Tti2 were then Gateway-recombined into pEARLEYGATE101 (36) by using LR clonase. The resulting gene-YFP fusion constructs were then transformed into Agrobacterium and used for transient expression by leaf injection. Leaf punches were then imaged 3–5 d after injection in Leica SP8 confocal microscope by using 514 nm excitation and 520–550 nm emission ranges.
Yeast Two-Hybrid.
Maize Tel2, Tti1, and Tti2 full-length CDS were cloned into pENTRSfi-223.1 as described above. The pENTR-Tel2 construct was then Gateway-recombined into pDestDB (ZmTel2-DB), whereas pENTR-Tti1 and pENTR-Tti2 were Gateway-recombined into pDestAD (ZmTti1-AD and ZmTti2-AD). The DB and AD constructs were then transformed into yeast strain Y8930 and Y8800, respectively, by using the lithium acetate method (37). Autoactivation of pDestDB-Tel2 was then tested by growing in a medium lacking histidine with 1 mM 3AT. Transformed DB and AD constructs were mated by plating on top of each other in YPDA medium overnight. Diploids (ZmTel2-DB/ZmTti1-AD and ZmTel2-DB/ZmTti2-AD) were then selected and grown on media lacking leucine, tryptophan, and histidine (−LTH) with 1 mM 3AT for 3 d to test for reporter gene activation.
Western Blotting.
To extract total buffer-soluble proteins, seed samples were powdered in liquid nitrogen and lysed in NETT buffer (20 mM Tris⋅HCl, 5 mM EDTA, 100 mM NaCl, 1 mM DTT, 0.5% Triton X-100) supplemented with 1 mM NaF, 1 mM β-glycerophosphate, 1 mM Na2VO4, and cOmplete Ultra Tablet Mini protease inhibitor mixture (Roche). Samples were then centrifuged at 15,000 × g for 15 min at 4 °C, and the cleared supernatant was saved to another clean tube. To ensure equal loading, the protein extracts were quantified by using the DC Protein Quantification kit (Bio-Rad). Samples were then loaded in SDS/PAGE for electrophoresis and blotted on PVDF membrane (Bio-Rad). The blots were then further processed according to membrane manufacturer’s protocol. The ZmATM antibody development was outsourced to GenScript by using the carboxyl terminal 200 aa as the antigen, the same part that was used for previous development of ATM antibody in Arabidopsis (31). Maize and Arabidopsis ATM proteins have 83% sequence identity on this part. The antibody against TOR is commercially available and was purchased from AgriSera.
Gene Expression Analysis.
Total RNA was extracted by using Qiagen RNEasy Mini Kit, treated with DNase (Invitrogen), and reverse-transcribed to cDNA with oligo-DT primers using qScript Supermix (Quanta Biosciences). The first-strand cDNA was then used as template for quantitative RT-PCR (qRT-PCR). The PerfeCTa SYBR Green Fastmix (Quanta Biosciences) was used for qPCR in an Illumina Eco Real-Time PCR System. Three biological and technical replicates were used. Relative gene expression was calculated by using the ΔΔCT (threshold cycle) method as implemented in the Illumina EcoStudy software using ubiquitin gene expression as a reference. The primers used are listed in Table S1.
Protein Folding Prediction.
The translated protein sequences of ZmTTI2 (WT and with Dsg insertion) were submitted to Phyre (38) for de novo protein folding prediction. Although such methods do not substitute for crystal structure analysis of proteins, they still can provide information on protein truncations, as it is the case here. The top hit models were then visualized in the free visualization software RasMol.
Acknowledgments
We thank Marc Probasco for field and greenhouse management and plant care and the Gallavotti laboratory at Rutgers University for providing the yeast strains and some of the plasmids used in this study. This research was supported by the Selman A. Waksman Chair in Molecular Genetics of Rutgers University (to J.M.) and National Science Foundation Plant Genome Program Grant IOS 13-39238 (to H.K.D.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KY513721, KY513722, and KY513723).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703498114/-/DCSupplemental.
References
- 1.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 2.Schnable PS, et al. The B73 maize genome: Complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 3.Almeida R, Allshire RC. RNA silencing and genome regulation. Trends Cell Biol. 2005;15:251–258. doi: 10.1016/j.tcb.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 5.Brutnell TP. Transposon tagging in maize. Funct Integr Genomics. 2002;2:4–12. doi: 10.1007/s10142-001-0044-0. [DOI] [PubMed] [Google Scholar]
- 6.Cowperthwaite M, et al. Use of the transposon Ac as a gene-searching engine in the maize genome. Plant Cell. 2002;14:713–726. doi: 10.1105/tpc.010468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCarty DR, et al. Steady-state transposon mutagenesis in inbred maize. Plant J. 2005;44:52–61. doi: 10.1111/j.1365-313X.2005.02509.x. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Segal G, Wang Q, Dooner HK. Gene tagging with engineered Ds elements in maize. In: Peterson T, editor. Methods in Molecular Biology: Plant Transposable Elements. Humana; New York: 2013. pp. 83–99. [DOI] [PubMed] [Google Scholar]
- 9.Abraham RT. PI 3-kinase related kinases: ‘Big’ players in stress-induced signaling pathways. DNA Repair (Amst) 2004;3:883–887. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Hurov KE, Cotta-Ramusino C, Elledge SJ. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev. 2010;24:1939–1950. doi: 10.1101/gad.1934210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takai H, Xie Y, de Lange T, Pavletich NP. Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes Dev. 2010;24:2019–2030. doi: 10.1101/gad.1956410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pohlman RF, Fedoroff NV, Messing J. The nucleotide sequence of the maize controlling element Activator. Cell. 1984;37:635–643. doi: 10.1016/0092-8674(84)90395-7. [DOI] [PubMed] [Google Scholar]
- 13.Ma Z, Dooner HK. A mutation in the nuclear-encoded plastid ribosomal protein S9 leads to early embryo lethality in maize. Plant J. 2004;37:92–103. doi: 10.1046/j.1365-313x.2003.01942.x. [DOI] [PubMed] [Google Scholar]
- 14.Lending CR, Larkins BA. Changes in the zein composition of protein bodies during maize endosperm development. Plant Cell. 1989;1:1011–1023. doi: 10.1105/tpc.1.10.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia N, Zhang W, Wu Y, Messing J. Evolution of gene expression after gene amplification. Genome Biol Evol. 2015;7:1303–1312. doi: 10.1093/gbe/evv075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong J, et al. Analysis of tandem gene copies in maize chromosomal regions reconstructed from long sequence reads. Proc Natl Acad Sci USA. 2016;113:7949–7956. doi: 10.1073/pnas.1608775113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi T, et al. Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits. Genes Cells. 2007;12:1357–1370. doi: 10.1111/j.1365-2443.2007.01141.x. [DOI] [PubMed] [Google Scholar]
- 18.Horejsí Z, et al. CK2 phospho-dependent binding of R2TP complex to TEL2 is essential for mTOR and SMG1 stability. Mol Cell. 2010;39:839–850. doi: 10.1016/j.molcel.2010.08.037. [DOI] [PubMed] [Google Scholar]
- 19.Genereaux J, et al. Genetic evidence links the ASTRA protein chaperone component Tti2 to the SAGA transcription factor Tra1. Genetics. 2012;191:765–780. doi: 10.1534/genetics.112.140459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altpeter F, et al. Advancing crop transformation in the era of genome editing. Plant Cell. 2016;28:1510–1520. doi: 10.1105/tpc.16.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowe K, et al. Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell. September 6, 2016 doi: 10.1105/tpc.16.00124. tpc.00124.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giroux MJ, et al. A single mutation that increases maize seed weight. Proc Natl Acad Sci USA. 1996;93:5824–5829. doi: 10.1073/pnas.93.12.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaizuka T, et al. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J Biol Chem. 2010;285:20109–20116. doi: 10.1074/jbc.M110.121699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng WH, Taliercio EW, Chourey PS. The Miniature1 seed locus of maize encodes a cell wall invertase required for normal development of endosperm and maternal cells in the pedicel. Plant Cell. 1996;8:971–983. doi: 10.1105/tpc.8.6.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sosso D, et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat Genet. 2015;47:1489–1493. doi: 10.1038/ng.3422. [DOI] [PubMed] [Google Scholar]
- 26.Menand B, et al. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA. 2002;99:6422–6427. doi: 10.1073/pnas.092141899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffman KS, et al. Saccharomyces cerevisiae Tti2 regulates PIKK proteins and stress response. G3 (Bethesda) 2016;6:1649–1659. doi: 10.1534/g3.116.029520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galetzka D, et al. Expression of somatic DNA repair genes in human testes. J Cell Biochem. 2007;100:1232–1239. doi: 10.1002/jcb.21113. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto K, et al. Kinase-dead ATM protein causes genomic instability and early embryonic lethality in mice. J Cell Biol. 2012;198:305–313. doi: 10.1083/jcb.201204098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barlow C, et al. Atm deficiency results in severe meiotic disruption as early as leptonema of prophase I. Development. 1998;125:4007–4017. doi: 10.1242/dev.125.20.4007. [DOI] [PubMed] [Google Scholar]
- 31.Garcia V, et al. AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell. 2003;15:119–132. doi: 10.1105/tpc.006577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzafrir I, et al. Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 2004;135:1206–1220. doi: 10.1104/pp.104.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodina A, et al. The epichaperome is an integrated chaperome network that facilitates tumour survival. Nature. 2016;538:397–401. doi: 10.1038/nature19807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallavotti A, et al. The control of axillary meristem fate in the maize ramosa pathway. Development. 2010;137:2849–2856. doi: 10.1242/dev.051748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y, Holding DR, Messing J. Gamma-zeins are essential for endosperm modification in quality protein maize. Proc Natl Acad Sci USA. 2010;107:12810–12815. doi: 10.1073/pnas.1004721107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 37.Consortium AIM. Arabidopsis Interactome Mapping Consortium Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]