Significance
Stage-specific and gene-specific molecular machines are recruited to elongating RNA polymerase II (Pol II) through reversible phosphorylation of its carboxyl-terminal domain. This unusual domain is composed of a tandemly repeating Y1S2P3T4S5P6S7 motif. Here, we identify a class of noncoding RNA that relies on phospho-Thr4 for effective termination. We also identify protein complexes that rely on Thr4 to associate with Pol II. Rtt103, one of the proteins that engages phospho-Thr4, also binds phospho-Ser2 and facilitates transcription termination of protein-coding genes. Using NMR, we show that Rtt103 binds with phospho-Thr4 in a nearly identical manner as phospho-Ser2. Our genomic, proteomic, and structural data suggest that phospho-Ser2 and phospho-Thr4 enable the recruitment of Rtt103 to different gene classes.
Keywords: CTD code, CTD interactome, noncoding RNA, NMR, phosphothreonine
Abstract
The carboxyl-terminal domain (CTD) of the largest subunit of RNA polymerase II (Pol II) orchestrates dynamic recruitment of specific cellular machines during different stages of transcription. Signature phosphorylation patterns of Y1S2P3T4S5P6S7 heptapeptide repeats of the CTD engage specific “readers.” Whereas phospho-Ser5 and phospho-Ser2 marks are ubiquitous, phospho-Thr4 is reported to only impact specific genes. Here, we identify a role for phospho-Thr4 in transcription termination at noncoding small nucleolar RNA (snoRNA) genes. Quantitative proteomics reveals an interactome of known readers as well as protein complexes that were not known to rely on Thr4 for association with Pol II. The data indicate a key role for Thr4 in engaging the machinery used for transcription elongation and termination. We focus on Rtt103, a protein that binds phospho-Ser2 and phospho-Thr4 marks and facilitates transcription termination at protein-coding genes. To elucidate how Rtt103 engages two distinct CTD modifications that are differentially enriched at noncoding genes, we relied on NMR analysis of Rtt103 in complex with phospho-Thr4– or phospho-Ser2–bearing CTD peptides. The structural data reveal that Rtt103 interacts with phospho-Thr4 in a manner analogous to its interaction with phospho-Ser2–modified CTD. The same set of hydrogen bonds involving either the oxygen on phospho-Thr4 and the hydroxyl on Ser2, or the phosphate on Ser2 and the Thr4 hydroxyl, can be formed by rotation of an arginine side chain, leaving the intermolecular interface otherwise unperturbed. This economy of design enables Rtt103 to engage Pol II at distinct sets of genes with differentially enriched CTD marks.
Each stage of transcription relies on ordered recruitment and exchange of specific protein complexes that act on RNA polymerase II, its nascent transcripts, and the underlying chromatin. This dynamic process is orchestrated via patterned posttranslational modifications of the carboxyl-terminal domain (CTD). This unusual and essential domain of Rpb1, the largest component of the 12-subunit polymerase, consists of repeating Y1S2P3T4S5P6S7 heptapeptides (26 repeats in budding yeast and 52 in humans) (1). The mechanistic consequences of phosphorylating Ser5 and Ser2 have been well documented (2–11). However, the role of Thr4 phosphorylation (pThr4), and even the necessity of Thr4 for cellular survival, appears to differ among closely related species and between growth conditions within a given species (12–15). Recent mass spectrometric analysis of an extensively engineered CTD revealed a paucity of pThr4, raising questions about the importance of this mark (16). In contrast, similar studies found pThr4 marks at levels comparable to or greater than the ubiquitously placed pSer2 mark in both yeast and human cells (17). Thus, much remains to be understood about the natural abundance and functional role of pThr4 marks on the endogenous, unmodified CTD.
Previous studies suggest that pThr4 has roles in transcriptional elongation, 3ʹ-end processing, and termination (12–14, 18). Our data, as well as other recent studies, suggest that CTD bearing pThr4 is bound by Rtt103 (16, 18), a well-known component of the Rat1 exosome that is thought to play a role in transcription termination of protein-coding genes. However, the bulk of previous genetic, biochemical, and structural studies show Rtt103 binds pSer2 and may cooperatively recruit Pcf11 to the 3ʹ ends of protein-coding genes (19, 20). Very few CTD-interacting proteins recognize multiple forms of phosphorylated CTD (3, 21–25), and the structures of even fewer are resolved. One intriguing example is Ssu72, which binds and dephosphorylates pSer5-CTD or pSer5-pThr4 peptides in the opposite orientation as pSer7-CTD peptides (22, 24, 26–28). How does Rtt103 discriminate between pSer2 and pThr4 CTD? Does Rtt103 use the same interface to engage pThr4 or pSer2 marks? Does pThr4-bound Rtt103 impact similar genes as Rtt103 recruited by pSer2? Here, we aim to answer these questions and further elucidate the role of pThr4 in Pol II function.
We report that substituting all threonine 4 residues in the Saccharomyces cerevisiae CTD with alanines (T4A) results in defective transcription of a coherent set of small nucleolar RNA (snoRNA) genes. Genome-wide chromatin immunoprecipitation (ChIP) studies show high pThr4 and low pSer2 levels at snoRNA genes that display defective termination. Consistent with this observation, snoRNA that are unaffected by T4A substitution show high pSer2 and low pThr4 levels. Quantitative mass spectrometry with T4A bearing Pol II revealed dramatically reduced binding of discrete protein complexes involved in transcription elongation and termination. Among the many partners, Rtt103 displayed the greatest sensitivity to the removal of the Thr4 side chain. Remarkably, our NMR studies show that Rtt103 binds analogously to pThr4 and pSer2 peptides and that a key Arg (R108) within Rtt103 mediates key intermolecular interactions with both phosphorylated peptides. We propose that, whereas pSer2 functions as a generic termination mark at most Pol II-transcribed genes, pThr4 engages the transcriptional termination machinery in a gene class-specific manner.
Results
Thr4 Is Required for Transcription Termination.
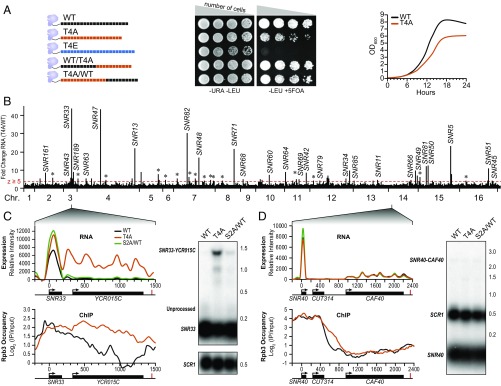
To further characterize the role of pThr4 in budding yeast, we examined the effect of Rpb1 alleles in which Thr4 was substituted with an alanine (T4A) or glutamate (T4E). Alanine substitution is commonly used to map binding interfaces because it is considered a benign perturbation, and this residue occurs in structured and unstructured protein folds (29). Moreover, alanine cannot be phosphorylated and does not replace the polar hydroxyl moiety of threonine with a hydrophobic methyl group (as is the case with a valine substitution). Alanine substitutions were made across all heptad repeats or in the proximal or distal halves of the CTD. We also created a strain in which Thr4 was substituted with the phosphomimic glutamate (T4E) in each repeat (Fig. 1A, Left). Whereas the T4E strain was inviable, the T4A strain displayed a slow growth phenotype (Fig. 1A, Middle Right).
Fig. 1.
pThr4 levels correlate with snoRNA termination in S. cerevisiae. (A) Rpb1 with mutant CTDs are shown with black (WT), orange (T4A), and blue (T4E) CTD repeats. Plasmids bearing mutant Rpb1 and a LEU2 auxotrophic marker were transformed into yeast. Shown are serial dilutions of cells in the presence of 5-FOA, which counterselects against yeast retaining the URA3-bearing plasmid containing WT Rpb1. −LEU, agar plates lacked leucine; OD, optical density; −URA, agar plates lacked uracil. (B) T4A expression (from genome tiling microarrays) normalized to WT is plotted across the whole yeast genome with chromosomes (Chr.) listed below. Transcripts above the red line denote regions with z ≥ 5 (*protein-coding genes). (C) RNA expression (Top) and Rpb3 ChIP analysis (Bottom) from WT (black), T4A (orange), or S2A/WT (green) are shown at SNR33. Arrows denote the transcription start site, black bars indicate gene bodies, and red lines indicate cleavage and polyadenylation sites. Northern blot shows probing SNR33 (Right). IP, immunoprecipiate. SCR1 is a loading control. (D) Same as C at the SNR40 locus.
Under exponential growth conditions in rich medium, the transcriptomes of WT and T4A cells displayed few global differences when examined with high-density genome tiling microarrays (30, 31). Avian histone transcripts require pThr4 to process 3ʹ ends and are not polyadenylated (12); however, in budding yeast, histone genes are polyadenylated (32). Consistent with this species-specific difference, no defects in 3ʹ-end processing of histone genes were observed in T4A-bearing yeast strains (SI Appendix, Fig. S1). Interestingly, we observed a minor but significant correlation between T4A and a strain containing a shortened CTD (33) (SI Appendix, Fig. S2A and Dataset S1).
Plotting the fold change in expression between T4A and WT, ∼50 striking peaks were observed (Fig. 1B). Using a stringent z-score cutoff of ≥5, we found significant enrichment at 3ʹ ends of snoRNAs. In the T4A strain, 37 of 77 snoRNA transcripts extended well past the mature 3ʹ end (Fig. 1C, Top and SI Appendix, Fig. S3). Whereas many formed chimeric transcripts that used the polyA signals of downstream protein-coding genes to terminate transcription (e.g., SNR33) (Fig. 1C) (34–36), others were read through and terminated even in the absence of a downstream gene (e.g., SNR48) (SI Appendix, Fig. S3). In contrast, 40 snoRNAs displayed no 3ʹ-read-through defect (e.g., SNR40) (Fig. 1D, Top and SI Appendix, Fig. S3). We further performed genome-wide transcription analysis on a strain in which Ser2 was substituted with an alanine only in the first half of the CTD (S2A/WT) and observed no significant read-through at any snoRNA gene (Fig. 1 C and D and SI Appendix, Table S1).
Northern blots were performed to confirm that these 3ʹ extensions were bona fide read-through events and not spurious initiation of new transcripts. RNA from WT, T4A, or S2A/WT cells was analyzed with a probe targeting SNR33 or SNR40 (Fig. 1 C and D, Right). A strong band at ∼1.5 kb corresponding to the chimeric SNR33-YCR015C transcript was observed in T4A but not in WT. Evidence of minor read-through was observed in S2A/WT, consistent with previous results (37). As evident, no chimeric SNR40-CAF40 transcript was observed in any strain tested (Fig. 1D, Right).
In addition to Northern blot analysis of specific transcripts, genome-wide chromatin immunoprecipitation of the Rpb3 subunit of Pol II was performed to further distinguish 3ʹ-processing defects from bona fide transcription termination defects (Fig. 1 C and D, Bottom). Rpb3 was strongly retained downstream of SNR33, further suggesting a role for pThr4 in snoRNA termination.
Thr4-Dependent and -Independent snoRNAs.
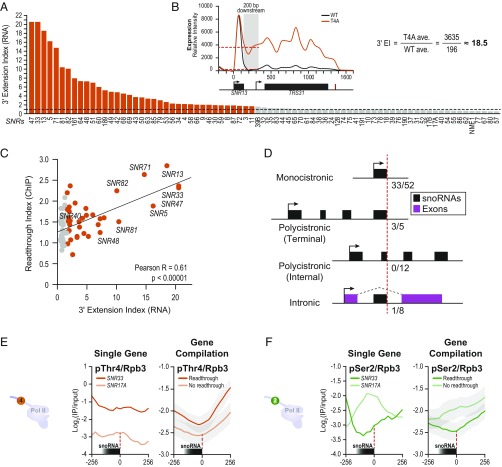
We quantified read-through at each snoRNA, as previously described (35) (Fig. 2B), and identified 37 snoRNAs with a 3ʹ extension (Fig. 2A and SI Appendix, Table S1). The Pol II read-through index (from ChIP of the Rpb3 subunit) in the region downstream of the 3ʹ end of processed snoRNAs was significantly correlated (P < 0.00001) with the 3ʹ-RNA extension index (from RNA expression) (Fig. 2C). However, no correlation was observed between the read-through defect and the length (P = 0.44) or the abundance of the snoRNA transcripts (P = 0.67) (SI Appendix, Fig. S2 B and C).
Fig. 2.
Thr4-dependent and -independent snoRNAs. (A) The 3′-extension index (EI) for each snoRNA in T4A is plotted from highest to lowest. Orange bars indicate an EI of >1.7. (B) Example gene (SNR13) and equation used to calculate EI, defined as fold change in the average expression 200 bp downstream of the 3′ end of snoRNAs. (C) Comparison of RNA extension index to Rpb3 ChIP read-through, defined as fold change in the average Rpb3 ChIP occupancy in the 1,000-bp window downstream of the 3′ end of snoRNAs. (D) Four snoRNA classes are illustrated with black boxes representing snoRNAs and purple representing exons. The fractions of snoRNAs in each class with 3′ extensions in T4A are noted next to the red dotted line depicting the mature 3′ end of respective snoRNAs. (E) pThr4 occupancy (normalized to Rpb3) centered at the 3′ end of SNR33 or SNR17A (Left) or a gene compilation of pThr4 occupancy across all snoRNAs read-through in T4A (dark) or no read-through (light). Shaded regions indicate 95% confidence interval. (F) Similar traces for pSer2.
To determine why only a subset of snoRNA genes is read through in the T4A mutant, we examined each of the four classes of snoRNA processing sites. Of the 37 snoRNAs with significant read-through, 36 occurred at terminal processing sites of monocistronic or polycistronic transcripts (P = 3.3 × 10−6, Fisher’s exact test) (Fig. 2D and SI Appendix, SI Materials and Methods). We also observed that several snoRNAs, even those with minimal 3ʹ-end extension, displayed high levels of Pol II downstream of the termination site when assayed by genome-wide ChIP analysis (Figs. 1D and 2C). This result indicates that certain snoRNA transcripts are more defective for transcription termination rather than 3ʹ-transcript processing.
Previous work suggests a role for pSer2 in snoRNA termination (37–39). Therefore, we examined the levels of pSer2 as well as pThr4, normalized to Rpb3 levels, across snoRNA genes in a WT strain. Remarkably, SNR33, a transcript with a termination defect and high read-through in T4A, displayed significantly more pThr4 downstream of the 3ʹ end than SNR17A, a snoRNA transcript that displays little, if any, read-through (Fig. 2E, Left). This pattern was observed across gene compilations of all snoRNAs (Fig. 2E, Right). Concomitant with an increased pThr4 level downstream of the 3ʹ end, lower pSer2 levels were observed downstream of snoRNA genes that displayed defective termination in the T4A strain. Consistent with this pattern, significantly higher levels of pSer2 were observed downstream of the compilation of snoRNAs that do not show a termination defect in T4A strain (Fig. 2F). Striking correlations between 3ʹ-extension index, high pThr4, and low pSer2 levels implicate pThr4 in snoRNA termination. As α-pThr4 antibodies yield modest ChIP signals, we examined pThr4 signals in other genome-wide studies of this mark (15, 40). Similar reciprocal patterns of pThr4 and pSer2 profiles were evident in pThr4 ChIP data obtained with a different (6D7) α-pThr4 antibody (40, 41) (SI Appendix, Fig. S4A).
Thr4 Is Required for Association of Elongation and Termination Factors.
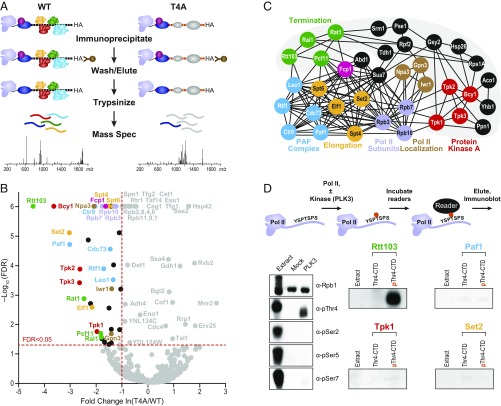
Previous efforts to identify factors that bind pThr4 used pThr4-specific antibodies to affinity purify RNA polymerase II bearing this mark (18). Because pThr4-bound factors may be competitively displaced by the pThr4 antibody, we sought to identify binding partners in a CTD-antibody–independent manner. We affinity enriched HA-tagged Pol II from strains bearing WT, T4A, and T4E CTDs. A control strain (Z26) lacking the HA-tagged Rpb1 was subjected to identical affinity enrichment procedure. Label-free quantitative proteomics (22, 42–44) was used to identify and quantify the relative abundance of affinity-enriched complexes (Fig. 3A). For Pol II enriched from T4A extracts, dramatic reduction was observed for multiple members of the Pol II termination complex (Rtt103, Rat1, Rai1, and Pcf11), cAMP-dependent PKA kinase holoenzyme and its regulatory subunit (Tpk2, Tpk3, and Tpk1, and Bcy1), PAF1c complex that bridges elongation and termination (Paf1, Ctr9, Cdc73, Rtf1, and Leo1), elongation factors (Set2, Spt4, Spt6, and Elf1), Pol II nuclear import proteins (Npa3, Iwr1, and Gpn3), as well as Fcp1, a phosphatase that acts on pThr4 (45) (Fig. 3B and Dataset S2). Many of these proteins were not known to rely on Thr4 for association with Pol II. Furthermore, the loss of all members of a given protein complex (e.g., Rtt103, PKA, and PAF1c) strongly supports the role of the Thr4 in association with the entire complex with Pol II, and it also serves as an internal control for label-free quantitative proteomic analysis.
Fig. 3.
Thr4 is required for association of elongation and termination factors. (A) Flowchart of the method to identify factors whose binding is compromised in T4A. (B) Volcano plot illustrating the fold change ln(T4A/WT) and the significance –Log10[false discovery rate (FDR)] of differential binding. Proteins depleted at least one natural log in T4A with an FDR of <0.05 are color coded and labeled as in C. (C) Physical interconnectivity (46) between the significantly depleted proteins identified in B. Gray background indicates low density connectivity. (D) Flowchart of the experimental design to assay binding of factors to human CTD (Top). Phosphorylation state of HeLa extract, unphosphorylated human Rpb1, or Rpb1 phosphorylated with human PLK3 (Left). Enriched hypophosphorylated Pol II (Thr4-CTD) and PLK3-treated CTD (pThr4-CTD) was incubated with the indicated TAP-tagged proteins, expressed in yeast. Indicated proteins were assayed for binding to pThr4-bearing Rpb1 and probed with α-TAP (Right).
Next, we charted the connectivity between Thr4-dependent partner proteins (Thr4 readers) using the publicly available STRING database of physical contacts (46). Substantial interconnectivity between Thr4 readers was evident, as were two groups of high- and low-density networks (Fig. 3C). Rtt103 termination complex, PKA, and nuclear localization factors form the outer set of interacting nodes, whereas PAF1c, elongation factors, Fcp1, and Pol II subunits form an intricate high-density network. The factors in the high-density network interact with Pol II subunits, suggesting that their interaction with Pol II may be less CTD dependent than the low-density network containing the Rtt103 termination complex and PKA. This interpretation is consistent with our unbiased and unsupervised quantitative mass spectrometry data that show greatest depletion of the Rtt103 and PKA complexes in the interactome of Pol II bearing the T4A substitution. Thus, our interactome data and the network analysis with known protein–protein interaction studies are internally consistent and mutually reinforcing.
Rtt103 Binds to CTD Phosphorylated by PLK3.
Our mass spectrometry-based approach relies on quantifying differential binding to Rpb1 that lacks the Thr4 side chain. Thus, we cannot exclude the possibility that the factors identified in our study necessarily bind to phospho-Thr4. Therefore, we biochemically assayed for binding of these factors to pThr4. To examine binding to the CTD, we immunopurified (1C7) hypophosphorylated Pol II and phosphorylated the CTD with PLK3, a highly selective human Thr4 kinase (Fig. 3D, Top) (13). Significant phosphorylation of Thr4 was observed with negligible phosphorylation at other residues of the CTD (Fig. 3D, Left). Tandem affinity purification (TAP)-tagged Rtt103, a representative of the termination complex; Paf1-TAP of the eponymous PAF1 complex; Tpk1-TAP, the PKA subunit that associates with Pol II across the transcribed gene (47); and Set2-TAP, an H3K36 methyltransferase, were expressed and purified from budding yeast and assayed for binding to pThr4-bearing Pol II. Rtt103 showed a dramatic increase in binding over the unphosphorylated Pol II, further suggesting an interaction between the termination factor and the pThr4-bearing CTD (Fig. 3D, Right). Purified Paf1, Tpk1, and Set2 did not bind appreciably to Thr4-phosphorylated CTD (Fig. 3D, Right and SI Appendix, Fig. S5B). To examine the role of pThr4 in recruiting Rtt103 in vivo, we performed genome-wide ChIP analysis of Rtt103 in WT and T4A cells. In T4A, we observed delayed recruitment of Rtt103 at read-through snoRNAs (∼160 bp 3ʹ), confirming the role of pThr4 levels in engaging the termination machinery in vivo (SI Appendix, Fig. S4B).
Rtt103 Forms a Structurally Analogous Interface with Different Phosphoisoforms of the CTD.
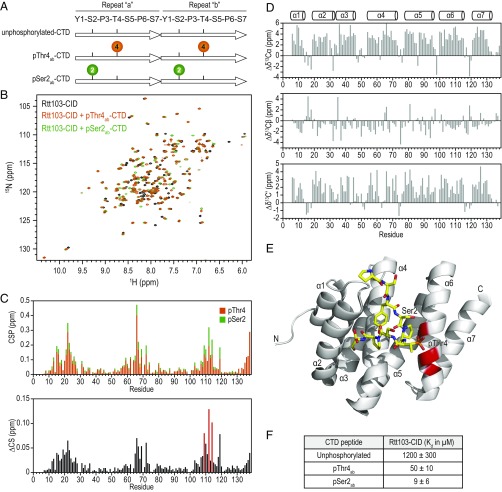
To elucidate the molecular basis of pThr4-CTD binding to Rtt103, we performed NMR titrations with a diheptad phosphorylated at Thr4 on repeat “a” and repeat “b” (denoted as pThr4ab-CTD) and compared it to a pSer2ab-CTD diheptad peptide studied by us in the past (20) (Fig. 4A). The first 131 residues of Rtt103, comprising the CTD interacting domain (CID), was expressed and purified in bacteria. Peptide binding to the CID of Rtt103 was examined using 2D 1H-15N heteronuclear single quantum coherences (HSQCs) to monitor chemical shift perturbations (CSPs) of amide resonances upon addition of the respective peptide. The 1H-15N HSQCs of the CID bound to the pThr4-marked peptide overlays well with the 1H-15N HSQC of the CID bound to the pSer2-marked peptide, indicating that the CID forms a structurally analogous interface with both phosphoisoforms (Fig. 4B). By comparing the chemical shift perturbation upon addition of each peptide (Fig. 4C), we observed that the two peptides affect the chemical shifts of Rtt103-CID in the same way, even though they are phosphorylated at different positions. The chemical shift deviations from random coil for backbone carbons confirm that the secondary structure of Rtt103 is unchanged (Fig. 4D). The chemical shift differences between the two bound Rtt103-CIDs are very small, and the most significant changes occur in close proximity to the region where Ser2 and Thr4 are located in our structure of pSer2 bound to Rtt103 (Fig. 4E) (20).
Fig. 4.
The CID domain of Rtt103 binds analogously to pThr4ab-CTD and pSer2ab-CTD peptides. (A) Schematic of diheptad CTD peptides used in NMR titrations. Each CTD repeat (arrow) is labeled as repeat “a” or repeat “b.” Sequences of the peptides are shown on the Top. Phosphothreonines and phosphoserines are indicated by orange and green circles, respectively. (B) Overlay of 1H-15N HSQC spectra of free Rtt103-CID (black) and of the same domain in complex with pThr4ab-CTD (orange) and pSer2ab-CTD (green). (C) Chemical shift perturbations (CSPs) of Rtt103-CID with diheptad peptides phosphorylated at Thr4 (orange) or Ser2 (green). Very similar trends are observed for both peptides, with generally smaller overall amplitude changes for pThr4ab-CTD (Top). (Bottom) Chemical shift difference for Rtt103-CID in complex with the two peptides. Three residues V109, I112, and K114 (red) have the largest chemical shift differences (>0.08 ppm) in the two complexes. (D) The secondary structure is unchanged for Rtt103-CID in complex with pThr4ab-CTD. Chemical shift deviations from random coil shifts for 13Cα (Top), 13Cβ (Middle), and 13C′ (Bottom) versus residue number are shown. (E) Model of Rtt103-CID in complex with pThr4ab-CTD, generated from the existing structure of Rtt103-pSer2ab-CTD, by substituting pSer2 with Ser2 and Thr4 with pThr4 and performing energy minimization within PyMOL. The three amino acids experiencing the largest changes between the two phosphorylated peptides, V109, I112, and K114 (red), are located in the immediate proximity of the sites of phosphorylation (Ser2 or Thr4). (F) Affinities of Rtt103-CID for diheptad repeat CTD peptides, as measured by NMR titrations. Values are averaged over multiple amino acids (>10) used for extracting independent Kd values by fitting of chemical shift changes vs. the ratio of peptide to Rtt103. Data are represented as mean ± SD.
This result is in good agreement with the previous finding by us and others that the structures of CIDs bound to CTD peptides generally overlap quite well. Specifically, the Rtt103-CID bound to pSer2-CTD (20) overlaps well with the other CIDs bound to CTD peptides with different phosphorylated states, with a 5.5-Å Cα rmsd to Nrd1-CID bound to pSer5 (48), 5.2-Å Cα rms deviation to the SCAF8-CID bound to pSer2 and pSer5 (49), 5.5-Å Cα rms deviation to the SCAF8-CID bound to pSer2 and pSer7 (49), 3.9-Å Cα rms deviation to Pcf11-CID bound to pSer2 (25), and 1.1-Å Cα rmsd to the RPRD1A-CID bound to pSer7 (50) over the entire length of the polypeptide chain (SI Appendix, Fig. S8). Whereas CID is a widely conserved protein fold, it is important to note that other proteins can interact with the CTD using different folds, including Pin1 (WW domain) (51), Tdrd3 (Tudor domain) (52), and Ssu72 (PTP domain) (26–28).
We measured binding affinities for each peptide by fitting the chemical shift changes versus the ratio of added CTD peptide to CID protein for every resonance exhibiting significant changes upon peptide addition (Fig. 4F). pThr4ab-CTD binds to Rtt103 CID fivefold weaker than pSer2ab-CTD, but still 25-fold stronger than the unphosphorylated CTD peptide. Recent structural and mutational studies suggest that the functional CTD unit is not defined by the YSPTSPS heptad motif and may well extend to a longer sequence (53–55). Therefore, we examined binding of Rtt103-CID to longer CTD peptides (SI Appendix, Figs. S6 and S7). Consistent with the binding to two-repeat peptides, similar structural interface was observed between Rtt103 CID and a four-repeat CTD peptide bearing pThr4 residues at the internal two-repeats (SI Appendix, Figs. S6 and S7). These results provide a framework for understanding how two different phosphoisoforms interact with a common interaction surface of Rtt103.
Structural Basis for pThr4–Rtt103 Interaction.
The solution-state NMR ensemble of structures of Rtt103 revealed a hydrogen bond between Arg108 of Rtt103 and the phosphate on pSer2 of the CTD (20). The data reveal that the Arg108 side chain can adopt many conformations when bound to the pSer2-CTD peptide (SI Appendix, Fig. S5D). To examine whether pThr4 could be accommodated at the same binding interface of Rtt103, we modeled the CTD with this mark on to our NMR structure and performed energy minimization in PyMOL (56) (Fig. 5A). The triad hydrogen bond/salt bridge interaction involving pSer2-Thr4-Arg108 in our previous structure of Rtt103 bound to the pSer2 peptide (20) can be replaced by a new triad Ser2-pThr4-Arg108 in pThr4ab-CTD bound complex (Fig. 5 A and B) without altering the interface significantly, a result consistent with the conservative pattern of chemical shift changes (Fig. 4 B and C). Further, these modeling results are consistent with our biochemical experiments that show strong binding between Rtt103 and pThr4 marks (Fig. 3D).
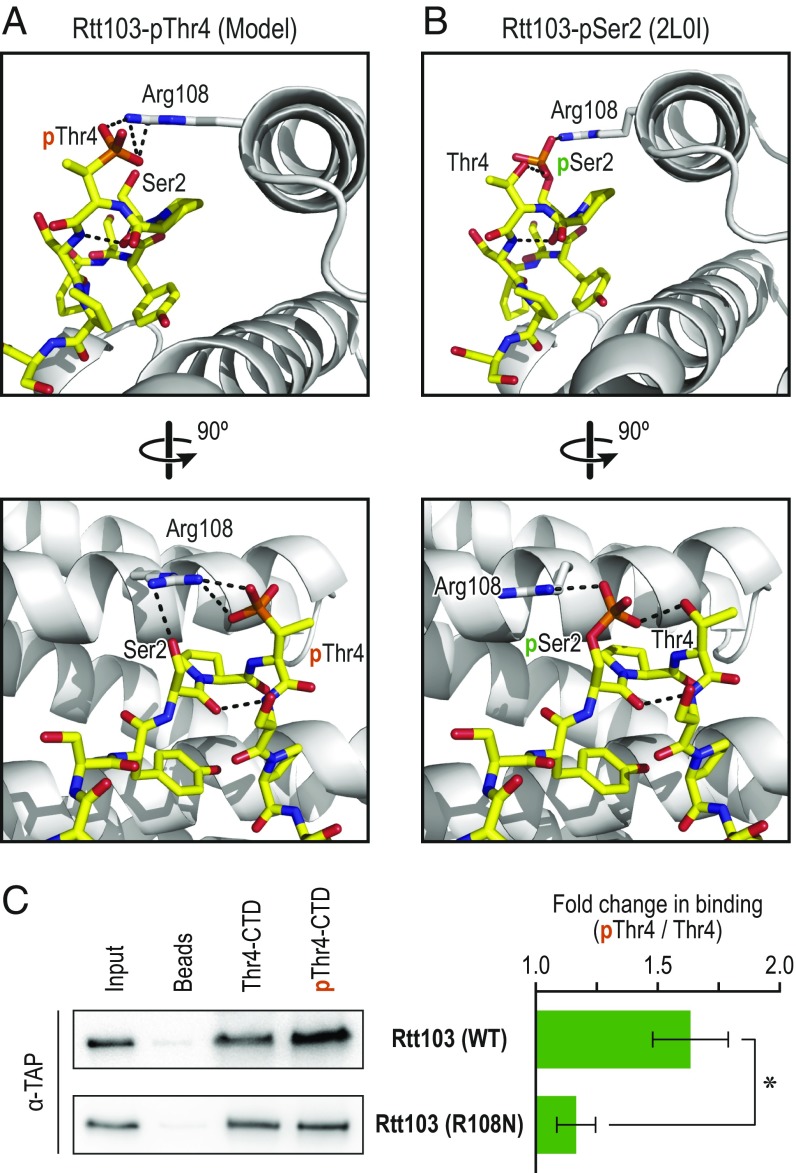
Fig. 5.
Model of Rtt103 in complex with pThr4-CTD. (A) Two orthogonal views of the model of Rtt103 in complex with pThr4-CTD and (B) representative solution state structures of Rtt103 in complex with pSer2-CTD are shown. All structural figures were generated with PyMOL (56). (C) Biochemical validation of the model is shown. Purified Rtt103 (WT) or the mutant Rtt103 (R108N) was incubated with a bead only control, or beads bound to GST-CTD that were either in their unphosphorylated state (Thr4-CTD) or phosphorylated state (pThr4-CTD). Bar graph shows fold change in binding to the phosphorylated CTD vs. unphosphorylated (*P < 0.01).
To validate this model, we purified a mutant variant of Rtt103 in which the positively charged Arg108 was substituted with a neutral Asn (R108N). Previous studies confirm that this mutant is deficient in binding to pSer2 peptides (20), and we hypothesized that it would also be incapable of binding to pThr4. Unphosphorylated or Thr4-phosphorylated GST-CTD bearing 14 repeats was bound to glutathione beads and incubated with WT Rtt103 or the R108N mutant. GST-CTD was used to definitively show that Rtt103 directly binds to pThr4-CTD, because it was purified in bacteria that lack eukaryotic CTD-binding proteins. Consistent with the model, Rtt103 (WT) bound significantly better to pThr4 than to the unmodified CTD. No pThr4-dependent increase in binding was observed with the mutant Rtt103, confirming that R108 plays a crucial role in recognizing both pThr4 and pSer2 marks (Fig. 5C). Whereas we did not observe binding of Rtt103 to beads, both WT and mutant Rtt103 display low levels of binding to unphosphorylated CTD. This low level binding of Rtt103 to CTD is not observed with human Pol II (Fig. 3D). However, immunoprecipitated human Pol II is likely to co-enrich factors that bind unmodified CTD and prevent Rtt103 from interacting with unmodified CTD. On the other hand, the recombinant GST-CTD fusion does not contain eukaryotic CTD binding proteins and is therefore accessible for low levels of Rtt103 association.
Discussion
The CTD undergoes sequential waves of posttranslational modifications during the transcription cycle. These modifications facilitate engagement of cellular machines that process nascent transcripts, place posttranslational marks on histones, and facilitate each stage of the transcription cycle. Unlike other ubiquitous phosphorylation marks on the CTD, pThr4 plays a critical role in regulating transcription at specific gene classes. Convergence of results from independent experimental approaches have revealed (i) two classes of noncoding RNAs that rely on either pThr4 or pSer2 marks to recruit the transcriptional termination machinery, (ii) an interactome that includes protein complexes not known to rely on Thr4 to associate with the CTD, and (iii) the structural basis of the interaction between pThr4 and Rtt103. That Rtt103 binds analogously to pThr4-CTD and pSer2-CTD suggests the remarkable economy of design where two different CTD marks, which are enriched on different sets of genes, are recognized by the cellular machinery that affects a common function at both sets of genes.
More specifically, we find that substitution of Thr4 with Ala in all heptad repeats of the CTD results in defective transcription termination at a subset of snoRNAs in vivo (Figs. 1C and 2A and SI Appendix, Table S1). The genome-wide transcriptome data from the T4A strain reveal two classes of snoRNA genes with termination at one class dependent primarily on pThr4 and termination at the other class primarily dependent on pSer2 marks. We hypothesize that a yet-to-be-identified yeast kinase may selectively place pThr4 marks on the CTD at this subset of snoRNAs. Remarkably, ChIP studies as well as single transcript analysis by Northern blots, show that in the absence of the pThr4 mark, Pol II transcribes through the SNR33 termination site and continues to transcribe the entire downstream protein-coding gene, YCR015C, using the accruing pSer2 marks to terminate synthesis of the chimeric SNR33-YCR015C transcript (Fig. 1C).
Previous studies defined a role for pSer5 in recruiting the Nrd1-Nab3-Sen1 complex, which facilitates termination of many noncoding RNA transcripts (36, 57–59). This finding prompted us to ask whether the T4A substitution inherently prevents other CTD residues from being phosphorylated. Western blots of WT, S2A/WT, and T4A cells confirm that pSer5 levels (H14 antibody) were equally abundant among all strains. Further, we only observed a partial reduction in pSer2 in the T4A strain (SI Appendix, Fig. S9). This partial reduction in pSer2 levels in the T4A strain may contribute to reduction in binding of known pSer2-CTD “readers” such as Set2 (60–63) and Paf1C (64, 65).
The genomic and proteomic data indicate that a balance between pThr4 and pSer2 levels leads to engagement of common components of the termination machinery at different genes. Rtt103 is known to bind pSer2-marked CTD and facilitate termination at protein-coding genes (19, 20); however, our results suggest that Rtt103 may also participate in termination at noncoding snoRNA genes. A minor reduction in pSer2 in the T4A strain (SI Appendix, Fig. S9) may explain the reduction in Rtt103 occupancy at snoRNAs; there remains sufficient signal to efficiently terminate at genes that are not dependent on the Thr4 side chain (SI Appendix, Fig. S4B). Whereas the precise role of Rtt103 in termination is being actively investigated by several groups, the ability of termination factors, such as Pcf11, that function at both protein-coding and -noncoding genes has been well documented. Interestingly, NMR studies had previously shown that Rtt103 interacts with pSer2 marks and cooperatively engages Pcf11 at protein-coding genes (20). However, a strain harboring pcf11-13, an allele that disrupts interaction with the CTD, displays defective termination at a set of snoRNA genes (38). These pcf11-13-sensitive snoRNAs coincide with snoRNAs that display termination defects in our T4A strain (P = 0.011) (Fig. 2A and SI Appendix, Table S1). This functional overlap is consistent with our quantitative proteomics analysis that shows reduced association of Pcf11 with T4A-bearing Pol II (Fig. 3). It is interesting to note that structural studies of Pcf11 with pSer2 peptides do not show any direct interaction with this mark but may rely on structure of CTD stabilized by this modification (25, 66, 67). Whereas pThr4 may stabilize a similar conformation in the CTD repeats, it remains to be determined whether such repeat structure or cooperative assembly with direct pThr4 readers such as Rtt103 engages Pcf11 at noncoding snoRNA genes.
In addition to termination factors such as Pcf11 and Rat1 exosome, we also observe reduced association of all subunits of the PAF1 complex with T4A bearing Pol II (Fig. 3B). Consistent with the role of the PAF1 complex in snoRNA termination (35), the read-through defects in T4A strain overlapped remarkably well with those observed in paf1Δ cells (P = 0.00014) (SI Appendix, Table S1). In the context of the chromatin landscape, PAF1c plays a key role in snoRNA termination by recruiting the machinery that modifies histone H3 (35). In agreement with this finding, significantly lower levels of histone H3 modifications were observed across snoRNA genes that display termination defects in T4A strains (GSE72802) (SI Appendix, Fig. S4C). Further, a genetic link between Thr4 and chromatin remodeling through the histone variant Htz1 has been reported (15). Consistent with a termination defect, Htz1, a histone H2A variant that yields more labile nucleosomes (68, 69), was highly enriched downstream of snoRNAs that display read-through in T4A (70) (SI Appendix, Fig. S4D).
The exquisite dependence of Rtt103 on pThr4 led us to investigate the molecular basis for the ability of this factor to engage two distinct phosphoisoforms of the CTD. As noted above, pSer2 and pThr4 marks are reciprocally enriched at the two classes of snoRNA genes. Moreover, pSer2 plays a key role in engaging the transcription termination machinery at protein-coding genes. NMR-based structural models of the CTD interacting domain of Rtt103 indicate that the same surface of the protein interacts with the both pSer2- and pThr4-modified CTD peptides. A highly conserved arginine residue on Rtt103 can readily engage either phospho-CTD mark with minimal perturbation of the remaining interface. This structurally analogous mode of engagement contrasts with those observed for Ssu72 binding to pSer5- and pSer7-modified CTD peptides. However, the preference for one modification over the other is also observed in the interactions of Rtt103 with pSer2 or pThr4 bearing CTD peptides. The NMR titration data indicate that Rtt103 prefers pSer2 over pThr4 by fivefold and lead us to hypothesize that Rtt103 might have a more direct role at pSer2-enriched genes, whereas it may function more cooperatively with other components of the transcriptional machinery at pThr4-enriched genes.
Taken together, the model that emerges from our results posits that pThr4 and pSer2 play overlapping roles in engaging the transcriptional termination machinery at distinct sets of genes. Unlike the near ubiquitous role for pSer2 in transcription termination, phosphorylation of Thr4 provides a ready means to selectively engage the termination machinery at specific genes. This model is consistent with variable levels of pThr4 marks observed at different gene classes and the differing sets of genes that display the dependence on pThr4 in response to different environmental and physiological cues.
Materials and Methods
Mutant CTD Construct and Plasmid Shuffle.
Mutant Rpb1 CTDs were constructed and tested for viability in vivo as previously described with some modifications (71). Briefly, the CTD repeats were constructed by annealing and ligating 5ʹ-phosphorylated oligonucleotides containing WT or mutant codons at position 4 (SI Appendix, Table S3). WT and mutant Rpb1 CTDs were cut with AvaI and ligated into similarly cut pSB0, which was transformed into DH5α and screened. pSB0 vectors were cut with KpnI and SnaBI and ligated into a similarly cut pY1 vector, a modified pRS315 vector containing full-length Rpb1. pY1 constructs were transformed into Z26, which contains a URA3-linked WT Rpb1 gene. Transformants were plated on synthetic complete (SC) media lacking uracil and leucine. Single colonies were grown overnight in YPD, and plated on SC −Leu +5-FOA (1 mg/mL) (Toronto Research Chemicals) to counterselect against any colonies maintaining the WT Rpb1.
RNA Expression Analysis.
Cell collection, RNA preparation, cDNA synthesis and labeling, array hybridization, and normalization were as previously described (72), using cyanine dyes (Flownamics) and SuperScript III (Life Technologies). Samples were hybridized to whole-genome tiled DNA microarrays (Roche NimbleGen). Biological triplicates were averaged, and significant read-through was quantified as the fold change (mutant/WT) in the region 200 bp downstream of the 3ʹ end of processed snoRNAs as previously described (35). All expression data are deposited in the Gene Expression Omnibus (accession no. GSE95419). Additional details on Fisher’s exact test used in Fig. 2D are described in SI Appendix, SI Materials and Methods.
Northern Blot Analysis.
RNA was extracted from 25-mL cultures at OD600 0.5 using hot phenol. A total of 20 μg of RNA was run on 1× MOPS [(3-(N-morpholino)propanesulfonic acid], 6% formaldehyde, and 1.5% agarose gel and transferred to Hybond-N membrane. 32P-UTP was incorporated into probes (SI Appendix, Table S3) complementary to SNR33, SNR40, or SCR1 and was hybridized overnight to the blots. Blots were exposed to a phosphorimaging screen and scanned on a Typhoon FLA9000 (GE Healthcare). Additional details are in SI Appendix, SI Materials and Methods.
Chromatin Immunoprecipitation.
ChIP was done as previously described with minor modifications (9, 31). Chromatin was sonicated on a Misonix 4000 sonicator. Rpb3 (NeoClone, W0012, currently BioLegend), pThr4 (1G7), or the FLAG-tag on Rtt103 (M2, Sigma) was immunoprecipitated. Samples were labeled, hybridized, and processed as previously described (9, 31). All ChIP data are deposited in the Gene Expression Omnibus (accession no. GSE95419). Additional details are in SI Appendix, SI Materials and Methods.
Coimmunoprecipitation and Label-Free Quantitative Proteomics.
WT, T4A, T4E (unshuffled), and Z26 were grown to OD600 ∼1.0 in 2 L of selective media. Cells were harvested and lysed via milling with a Retsch Mixer Mill MM 400. Lysed yeast was added to 1× NET-seq buffer (73) with protease and phosphatase inhibitors and 2,000 units DNase I (Sigma). Debris was pelleted, and Pol II was immunoprecipitated with α-HA Dynabeads (Life Technologies). Following washes, samples were eluted three times with 2 mg/mL HA peptide in TBS. Fractions were combined, and proteins were precipitated with TCA. Triplicate samples were analyzed by label-free quantitative proteomics analysis as previously described (42). Significant proteins were identified using QSpec (74). Raw data are available at the ProteomeXchange (accession no. PXD005967). Additional details are described in SI Appendix, SI Materials and Methods.
Validation of Binding to pThr4.
Endogenous Pol II from HeLa whole-cell extract was immunoprecipitated with the antibody 1C7 (13) to enrich hypophosphorylated CTD (Pol IIA). Pol IIA was treated with PLK3 for 60 min at 28 °C in kinase buffer A [20 mM Tris⋅HCl (pH 7.4), 20 mM NaCl, 10 mM MgCl2, 1 μM DTT, and 2 μM ATP] to selectively phosphorylate Thr4. TAP-tagged proteins were expressed in yeast (SI Appendix, SI Materials and Methods) and incubated with phosphorylated CTD substrate for 60 min at 30 °C. Beads were washed, and proteins were eluted with Laemmli sample buffer before Western blot analysis using antibodies targeting CTD modifications, Pol II, or the TAP-tag.
NMR Experiments.
The CID domain of Rtt103 (Rtt103-CID) used for NMR titration and chemical shift assignments was prepared as described previously (20) and 15N or 15N, 13C labeled. The diheptad peptides were purchased from AnaSpec or VCPBio Lab. All of the NMR experiments were recorded at 25 °C on a Bruker Avance 600 MHz spectrometer equipped with cryocooled 1H/13C/15N triple-resonance probes and pulse-field gradients. NMR data were processed with NMRPipe (75) and analyzed within CcpNmr (76). The complex of Rtt103-CID with diheptad peptides was formed by titrating the peptide into the CID and monitoring the change of 15N-edited HSQC spectra until saturation was observed. The chemical shifts of backbone and side-chain resonances were assigned using standard triple resonance NMR experiments including 3D HNCO, HNCACB, and CBCA(CO)NH (77). The composite chemical shift changes for each HN cross-peak in 15N-edited HSQC are quantified using empirical equation , where ΔδH and ΔδN are the chemical shift changes for 1H and 15N, respectively. For binding affinities, a series of 15N-edited HSQC spectra were collected with increasing molar ratio of peptide to protein ranging from 0:1 to 3:1. The Kds were calculated using the module “follow shift changes” within CcpNmr by fitting the fractional chemical shift changes against the ratios of ligand to protein.
Supplementary Material
Acknowledgments
We thank Prof. Jeffry Corden for providing the S2A/WT CTD mutant strain and Prof. Steve Buratowski for providing the Rtt103(R108N)-TAP mutant strain. This work was funded by National Science Foundation (NSF) Grant MCB1413547, NIH Grant GM117362, and W. M. Keck Foundation awards (to A.Z.A.); NIH Grant GM64440 (to G.V.); Stowers Institute for Medical Research (M.P.W.); Deutsche Forschungsgemeinschaft Grant SFB1064; Chromatin Dynamics (D.E.); NIH Grant GM083989 (to A.P.G.); NIH Chemistry–Biology Interface Training Program (Grant T32GM008505) (to C.M.N.); NIH Molecular Biosciences Training Grant T32GM007215 (to S.C.T.); and NSF Graduate Research Fellowship DGE-1256259 (to C.M.N. and S.C.T.). Support for this research was also provided by the University of Wisconsin–Madison, Office of the Vice Chancellor for Research and Graduate Education, with funding from the Wisconsin Alumni Research Foundation.
Footnotes
Conflict of interest statement: A.Z.A. is the sole member of VistaMotif, LLC and founder of the nonprofit WINStep Forward.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE95419). Raw data are available at the ProteomeXchange (accession no. PXD005967).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700128114/-/DCSupplemental.
References
- 1.Corden JL, Cadena DL, Ahearn JM, Jr, Dahmus ME. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci USA. 1985;82:7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang DW, Rodríguez-Molina JB, Tietjen JR, Nemec CM, Ansari AZ. Emerging views on the CTD code. Genet Res Int. 2012;2012:347214. doi: 10.1155/2012/347214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eick D, Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev. 2013;113:8456–8490. doi: 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- 4.Zaborowska J, Egloff S, Murphy S. The pol II CTD: New twists in the tail. Nat Struct Mol Biol. 2016;23:771–777. doi: 10.1038/nsmb.3285. [DOI] [PubMed] [Google Scholar]
- 5.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 6.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodríguez-Molina JB, Tseng SC, Simonett SP, Taunton J, Ansari AZ. Engineered covalent inactivation of TFIIH-kinase reveals an elongation checkpoint and results in widespread mRNA stabilization. Mol Cell. 2016;63:433–444. doi: 10.1016/j.molcel.2016.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanin EI, et al. Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc Natl Acad Sci USA. 2007;104:5812–5817. doi: 10.1073/pnas.0611505104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akhtar MS, et al. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell. 2009;34:387–393. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsin JP, Sheth A, Manley JL. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science. 2011;334:683–686. doi: 10.1126/science.1206034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hintermair C, et al. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J. 2012;31:2784–2797. doi: 10.1038/emboj.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwer B, Bitton DA, Sanchez AM, Bähler J, Shuman S. Individual letters of the RNA polymerase II CTD code govern distinct gene expression programs in fission yeast. Proc Natl Acad Sci USA. 2014;111:4185–4190. doi: 10.1073/pnas.1321842111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosonina E, et al. Threonine-4 of the budding yeast RNAP II CTD couples transcription with Htz1-mediated chromatin remodeling. Proc Natl Acad Sci USA. 2014;111:11924–11931. doi: 10.1073/pnas.1412802111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suh H, et al. Direct analysis of phosphorylation sites on the Rpb1 C-terminal domain of RNA polymerase II. Mol Cell. 2016;61:297–304. doi: 10.1016/j.molcel.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schüller R, et al. Heptad-specific phosphorylation of RNA polymerase II CTD. Mol Cell. 2016;61:305–314. doi: 10.1016/j.molcel.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Harlen KM, et al. Comprehensive RNA polymerase II interactomes reveal distinct and varied roles for each phospho-CTD residue. Cell Reports. 2016;15:2147–2158. doi: 10.1016/j.celrep.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim M, et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 20.Lunde BM, et al. Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2010;17:1195–1201. doi: 10.1038/nsmb.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeronimo C, Bataille AR, Robert F. The writers, readers, and functions of the RNA polymerase II C-terminal domain code. Chem Rev. 2013;113:8491–8522. doi: 10.1021/cr4001397. [DOI] [PubMed] [Google Scholar]
- 22.Zhang DW, et al. Ssu72 phosphatase-dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J Biol Chem. 2012;287:8541–8551. doi: 10.1074/jbc.M111.335687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu H, Hu C, Hinnebusch AG. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol Cell. 2009;33:752–762. doi: 10.1016/j.molcel.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo Y, et al. Novel modifications on C-terminal domain of RNA polymerase II can fine-tune the phosphatase activity of Ssu72. ACS Chem Biol. 2013;8:2042–2052. doi: 10.1021/cb400229c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- 26.Xiang K, Manley JL, Tong L. An unexpected binding mode for a Pol II CTD peptide phosphorylated at Ser7 in the active site of the CTD phosphatase Ssu72. Genes Dev. 2012;26:2265–2270. doi: 10.1101/gad.198853.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Werner-Allen JW, et al. cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J Biol Chem. 2011;286:5717–5726. doi: 10.1074/jbc.M110.197129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang K, et al. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature. 2010;467:729–733. doi: 10.1038/nature09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 30.Steinmetz EJ, et al. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 31.Tietjen JR, et al. Chemical-genomic dissection of the CTD code. Nat Struct Mol Biol. 2010;17:1154–1161. doi: 10.1038/nsmb.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fahrner K, Yarger J, Hereford L. Yeast histone mRNA is polyadenylated. Nucleic Acids Res. 1980;8:5725–5737. doi: 10.1093/nar/8.23.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aristizabal MJ, et al. High-throughput genetic and gene expression analysis of the RNAPII-CTD reveals unexpected connections to SRB10/CDK8. PLoS Genet. 2013;9:e1003758. doi: 10.1371/journal.pgen.1003758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheldon KE, Mauger DM, Arndt KM. A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol Cell. 2005;20:225–236. doi: 10.1016/j.molcel.2005.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomson BN, et al. Effects of the Paf1 complex and histone modifications on snoRNA 3′-end formation reveal broad and locus-specific regulation. Mol Cell Biol. 2013;33:170–182. doi: 10.1128/MCB.01233-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim M, et al. Distinct pathways for snoRNA and mRNA termination. Mol Cell. 2006;24:723–734. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 37.Lenstra TL, et al. The role of Ctk1 kinase in termination of small non-coding RNAs. PLoS One. 2013;8:e80495. doi: 10.1371/journal.pone.0080495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grzechnik P, Gdula MR, Proudfoot NJ. Pcf11 orchestrates transcription termination pathways in yeast. Genes Dev. 2015;29:849–861. doi: 10.1101/gad.251470.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chinchilla K, et al. Interactions of Sen1, Nrd1, and Nab3 with multiple phosphorylated forms of the Rpb1 C-terminal domain in Saccharomyces cerevisiae. Eukaryot Cell. 2012;11:417–429. doi: 10.1128/EC.05320-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayer A, et al. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 41.Mayer A, et al. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336:1723–1725. doi: 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to label free proteome quantitation: How to deal with peptides shared by multiple proteins. Anal Chem. 2010;82:2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- 43.Mosley AL, et al. Quantitative proteomics demonstrates that the RNA polymerase II subunits Rpb4 and Rpb7 dissociate during transcriptional elongation. Mol Cell Proteomics. 2013;12:1530–1538. doi: 10.1074/mcp.M112.024034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang K, et al. Characterization of human cyclin-dependent kinase 12 (CDK12) and CDK13 complexes in C-terminal domain phosphorylation, gene transcription, and RNA processing. Mol Cell Biol. 2015;35:928–938. doi: 10.1128/MCB.01426-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsin JP, Xiang K, Manley JL. Function and control of RNA polymerase II C-terminal domain phosphorylation in vertebrate transcription and RNA processing. Mol Cell Biol. 2014;34:2488–2498. doi: 10.1128/MCB.00181-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jensen LJ, et al. STRING 8: A global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- 48.Kubicek K, et al. Serine phosphorylation and proline isomerization in RNAP II CTD control recruitment of Nrd1. Genes Dev. 2012;26:1891–1896. doi: 10.1101/gad.192781.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Becker R, Loll B, Meinhart A. Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2008;283:22659–22669. doi: 10.1074/jbc.M803540200. [DOI] [PubMed] [Google Scholar]
- 50.Ni Z, et al. RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation. Nat Struct Mol Biol. 2014;21:686–695. doi: 10.1038/nsmb.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 52.Sikorsky T, et al. Recognition of asymmetrically dimethylated arginine by TDRD3. Nucleic Acids Res. 2012;40:11748–11755. doi: 10.1093/nar/gks929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwer B, Sanchez AM, Shuman S. Punctuation and syntax of the RNA polymerase II CTD code in fission yeast. Proc Natl Acad Sci USA. 2012;109:18024–18029. doi: 10.1073/pnas.1208995109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu P, Greenleaf AL, Stiller JW. The essential sequence elements required for RNAP II carboxyl-terminal domain function in yeast and their evolutionary conservation. Mol Biol Evol. 2008;25:719–727. doi: 10.1093/molbev/msn017. [DOI] [PubMed] [Google Scholar]
- 55.Liu P, Kenney JM, Stiller JW, Greenleaf AL. Genetic organization, length conservation, and evolution of RNA polymerase II carboxyl-terminal domain. Mol Biol Evol. 2010;27:2628–2641. doi: 10.1093/molbev/msq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schrödinger LLC. 2010. The PyMOL Molecular Graphics System, Version 1.3 (Schrödinger LLC, New York)
- 57.Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- 59.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 60.Xiao T, et al. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krogan NJ, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 63.Li J, Moazed D, Gygi SP. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem. 2002;277:49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- 64.Qiu H, Hu C, Gaur NA, Hinnebusch AG. Pol II CTD kinases Bur1 and Kin28 promote Spt5 CTR-independent recruitment of Paf1 complex. EMBO J. 2012;31:3494–3505. doi: 10.1038/emboj.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rozenblatt-Rosen O, et al. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol Cell Biol. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Noble CG, et al. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat Struct Mol Biol. 2005;12:144–151. doi: 10.1038/nsmb887. [DOI] [PubMed] [Google Scholar]
- 67.Hollingworth D, Noble CG, Taylor IA, Ramos A. RNA polymerase II CTD phosphopeptides compete with RNA for the interaction with Pcf11. RNA. 2006;12:555–560. doi: 10.1261/rna.2304506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li B, et al. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci USA. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Santisteban MS, Hang M, Smith MM. Histone variant H2A.Z and RNA polymerase II transcription elongation. Mol Cell Biol. 2011;31:1848–1860. doi: 10.1128/MCB.01346-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gu M, Naiyachit Y, Wood TJ, Millar CB. H2A.Z marks antisense promoters and has positive effects on antisense transcript levels in budding yeast. BMC Genomics. 2015;16:99. doi: 10.1186/s12864-015-1247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.West ML, Corden JL. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140:1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berry DB, Gasch AP. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol Biol Cell. 2008;19:4580–4587. doi: 10.1091/mbc.E07-07-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choi H, Fermin D, Nesvizhskii AI. Significance analysis of spectral count data in label-free shotgun proteomics. Mol Cell Proteomics. 2008;7:2373–2385. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 76.Vranken WF, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 77.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Mag Res Sp. 1999;34:93–158. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.