Significance
The yellow, orange, and red colors of birds are produced through the deposition of carotenoid pigments into feathers and skin, and often function as signals in aggressive interactions and mate choice. These colors are hypothesized to communicate information about individual quality because their expression is linked to vital cellular processes through the mechanisms of carotenoid metabolism. To elucidate these mechanisms, we carried out genomic and biochemical analyses of the white recessive canary breed, which carries a heritable defect in carotenoid uptake. We identified a mutation in the SCARB1 gene in this breed that disrupts carotenoid transport function. Our study implicates SCARB1 as a key mediator of carotenoid-based coloration and suggests a link between carotenoid coloration and lipid metabolism.
Keywords: coloration, carotenoids, lipid metabolism, Serinus canaria
Abstract
Yellow, orange, and red coloration is a fundamental aspect of avian diversity and serves as an important signal in mate choice and aggressive interactions. This coloration is often produced through the deposition of diet-derived carotenoid pigments, yet the mechanisms of carotenoid uptake and transport are not well-understood. The white recessive breed of the common canary (Serinus canaria), which carries an autosomal recessive mutation that renders its plumage pure white, provides a unique opportunity to investigate mechanisms of carotenoid coloration. We carried out detailed genomic and biochemical analyses comparing the white recessive with yellow and red breeds of canaries. Biochemical analysis revealed that carotenoids are absent or at very low concentrations in feathers and several tissues of white recessive canaries, consistent with a genetic defect in carotenoid uptake. Using a combination of genetic mapping approaches, we show that the white recessive allele is due to a splice donor site mutation in the scavenger receptor B1 (SCARB1; also known as SR-B1) gene. This mutation results in abnormal splicing, with the most abundant transcript lacking exon 4. Through functional assays, we further demonstrate that wild-type SCARB1 promotes cellular uptake of carotenoids but that this function is lost in the predominant mutant isoform in white recessive canaries. Our results indicate that SCARB1 is an essential mediator of the expression of carotenoid-based coloration in birds, and suggest a potential link between visual displays and lipid metabolism.
The yellow, orange, and red coloration of the feathers, skin, and beaks of birds is most commonly produced through the deposition of carotenoid pigments (1). Carotenoid coloration of birds has been a focus of study in the fields of behavior, evolution, and physiology because it plays a key role in mate assessment in many species. In addition, it is frequently an indicator of individual quality, and can signal species identity (2–4). Birds cannot synthesize carotenoids de novo and must acquire them through their diet (1), potentially linking coloration to the acquisition of pigments from the environment (3). Thus, key hypotheses related to honest signaling and sexual selection have been shaped by and are currently being tested in carotenoid-ornament systems (5, 6). Ultimately, the information content and evolutionary trajectories of carotenoid ornaments are a function of the physiological mechanisms underlying color expression, yet our understanding of these mechanisms is limited.
The expression of carotenoid coloration in birds involves four distinct physiological steps: uptake in the gut, transport in circulatory and lymphatic systems, metabolism either at the site of deposition or in the liver, and deposition in the integument (7). Recent progress has been made in understanding how carotenoids are metabolized to novel forms. In 2016, two studies independently identified a key carotenoid metabolism enzyme, CYP2J19, that mediates the oxidation of yellow carotenoids into red ketocarotenoids and is used by many bird species to produce red feathers and bare parts (8, 9). In other studies, expression of the carotenoid-cleaving enzyme β-carotene-9′,10′-dioxygenase (BCO2) was found to be associated with loss of yellow leg coloration in chickens (10), and sequence variation around this same gene was associated with yellow versus white breast plumage in two sister species of wood warblers (4). The discoveries of the roles that CYP2J19 and BCO2 play in the carotenoid pigmentation of birds provide important insights into the metabolic mechanisms that underlie carotenoid coloration.
Progress has also been made in understanding the mechanism whereby carotenoids are transported via the blood to target tissues. It has long been known that birds transport carotenoids in the circulatory and lymphatic systems via lipoproteins (11, 12), and recent experimental evidence confirms a key role for lipoproteins in carotenoid transport in birds. For example, the Wisconsin hypoalpha mutant chicken has a mutation in the ABCA1 transporter, which results in markedly reduced levels of circulating high-density lipoprotein (HDL) and a consequent reduction in the level of carotenoids in the blood and peripheral tissues (13, 14). Despite these insights, many aspects of carotenoid transport and delivery remain to be discovered.
Domesticated canaries (Serinus canaria) provide an ideal system in which to explore the mechanisms of avian carotenoid physiology because these birds have been selectively crossed for centuries to produce breeds with fixed expression of distinct carotenoid-based plumage color phenotypes (15, 16). Wild canaries and typical domesticated varieties pigment their feathers with yellow carotenoids (Fig. 1A) (17, 18). The white recessive canary has entirely white plumage (Fig. 1B) as well as congenital vitamin A deficiency that is inherited as an autosomal recessive trait (19, 20). It is hypothesized that the white recessive phenotype is the result of a mutation affecting carotenoid uptake (19, 20), but the specific gene(s) involved remains unknown. Thus, the white recessive canary presents an opportunity to discover mechanisms of carotenoid uptake that are necessary for carotenoid coloration in birds.
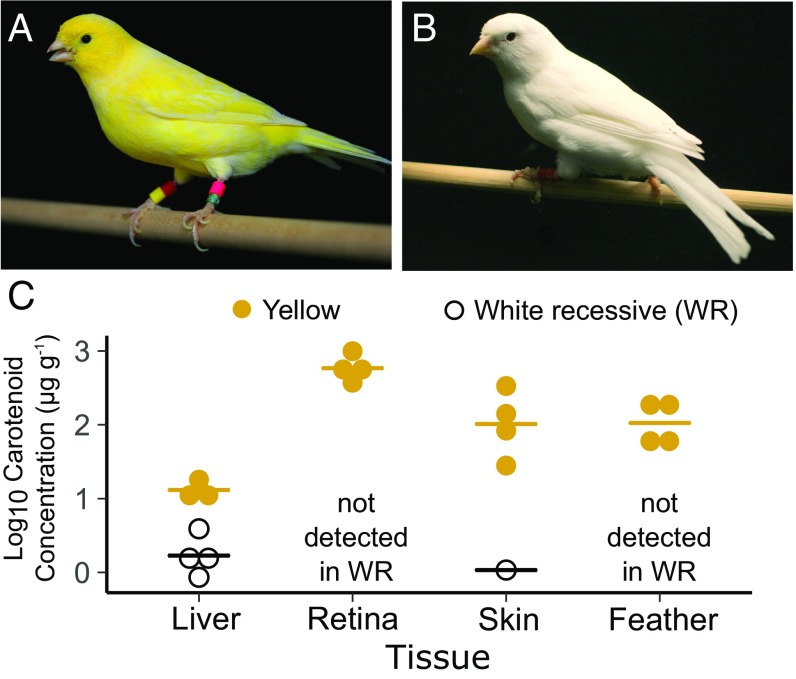
Fig. 1.
White recessive canary has white plumage and very low carotenoid levels in its tissues. (A and B) Representative images of the (A) lipochrome domestic canary and (B) white recessive canary. (C) Carotenoid concentrations of the liver, retina, skin, and feathers of typical yellow (yellow points) and white recessive (WR; open points) canaries. The lines represent the means for each breed and tissue. Carotenoid concentration was calculated relative to protein content in liver, retina, and skin samples, or a dry mass of feathers. All carotenoid types within a given sample were summed to give the total concentration.
Results
Carotenoids Are Absent or at Very Low Concentrations in the Tissues of White Recessive Canaries.
The white recessive canary is characterized by completely white plumage that appears to lack the yellow carotenoid pigmentation typical of wild-type canaries (Fig. 1 A and B). To better understand the carotenoid physiology of the white recessive canary breed, we used high-performance liquid chromatography (HPLC) to identify and quantify carotenoids in the feathers, skin, liver, and retina, tissues that are important sites of carotenoid accumulation in birds (1, 7, 21) (Fig. 1C). Carotenoids were not detectable in the feathers of white recessive canaries. In contrast, feathers collected from the same tracts of typical yellow canaries contained a high concentration of carotenoids (mean ± SE; 128.89 ± 37.78 µg⋅g−1 feather mass; Fig. 1C), primarily canary xanthophylls (SI Appendix, Fig. S1A and Table S1). We did not detect carotenoids in the skin of three white recessive canaries, and only trace amounts in the skin of a fourth individual (1.08 µg⋅g−1 protein; Fig. 1C). Skin from comparable regions of typical yellow canaries contained >100-fold higher carotenoid concentrations (146.85 ± 66.84 µg⋅g−1 protein), primarily lutein (SI Appendix, Fig. S1B and Table S1). The livers of both white recessive and typical yellow canaries contained low, but detectable, levels of carotenoids, with lutein being the dominant type. However, concentrations were significantly lower in the white recessives (white recessive: 1.97 ± 0.66 µg⋅g−1 protein; wild-type: 13.39 ± 1.70 µg⋅g−1 protein; t = −6.26, df = 3.89, P = 0.0036; Fig. 1C and SI Appendix, Table S1). Carotenoids were not detectable in the retinas of white recessive canaries but were present at very high concentrations in the retinas of typical yellow birds (622.57 ± 131.73 µg⋅g−1 protein; Fig. 1C and SI Appendix, Table S1). Together, these results indicate that the levels of carotenoids are greatly reduced across multiple tissues in white recessive canaries.
Identification of the Genomic Region Associated with White Recessive Coloration.
The white recessive breed was created through fixation of a spontaneous mutation by artificial selection (19, 22, 23). In crosses with typical yellow canaries, this white phenotype is transmitted in a manner consistent with the existence of a single recessive allele. It is thus expected that the genomic region underlying white feather coloration will show elevated differentiation in allele frequency between white and colored canaries compared with the remainder of the genome. To search for such regions in the white recessive genome, we used whole-genome sequencing data of pooled DNA samples (detailed in SI Appendix, Table S2). We compared white recessive canaries with other domestic breeds and one wild population, all characterized by yellow or red coloration. Because the white recessive phenotype is explained by an autosomal mutation (23), we restricted our analysis to the autosomal chromosomes.
We summarized allele frequency differentiation across the genome using the fixation index (FST) and a sliding window approach (Fig. 2A). Average levels of genetic differentiation between white recessive canaries and the remaining populations were moderate (FST ∼ 0.15). This level of genetic differentiation is conducive to identifying regions of unusually high genetic differentiation, consistent with directional selection. One region clearly stood out that contains the top nine values of the empirical distribution of FST, providing a strong candidate for the sequence harboring the white recessive gene. We extended the boundaries of this region by merging all windows having FST estimates above the 95th percentile of the empirical distribution (FST ≥ 0.23), if not separated by more than five windows below this threshold. The resulting region spanned ∼1.4 Mb on scaffold NW_007931140 (11,350,000 to 12,790,000 bp), which is homologous to zebra finch chromosome 15 (∼970,000 to 2,620,000 bp).
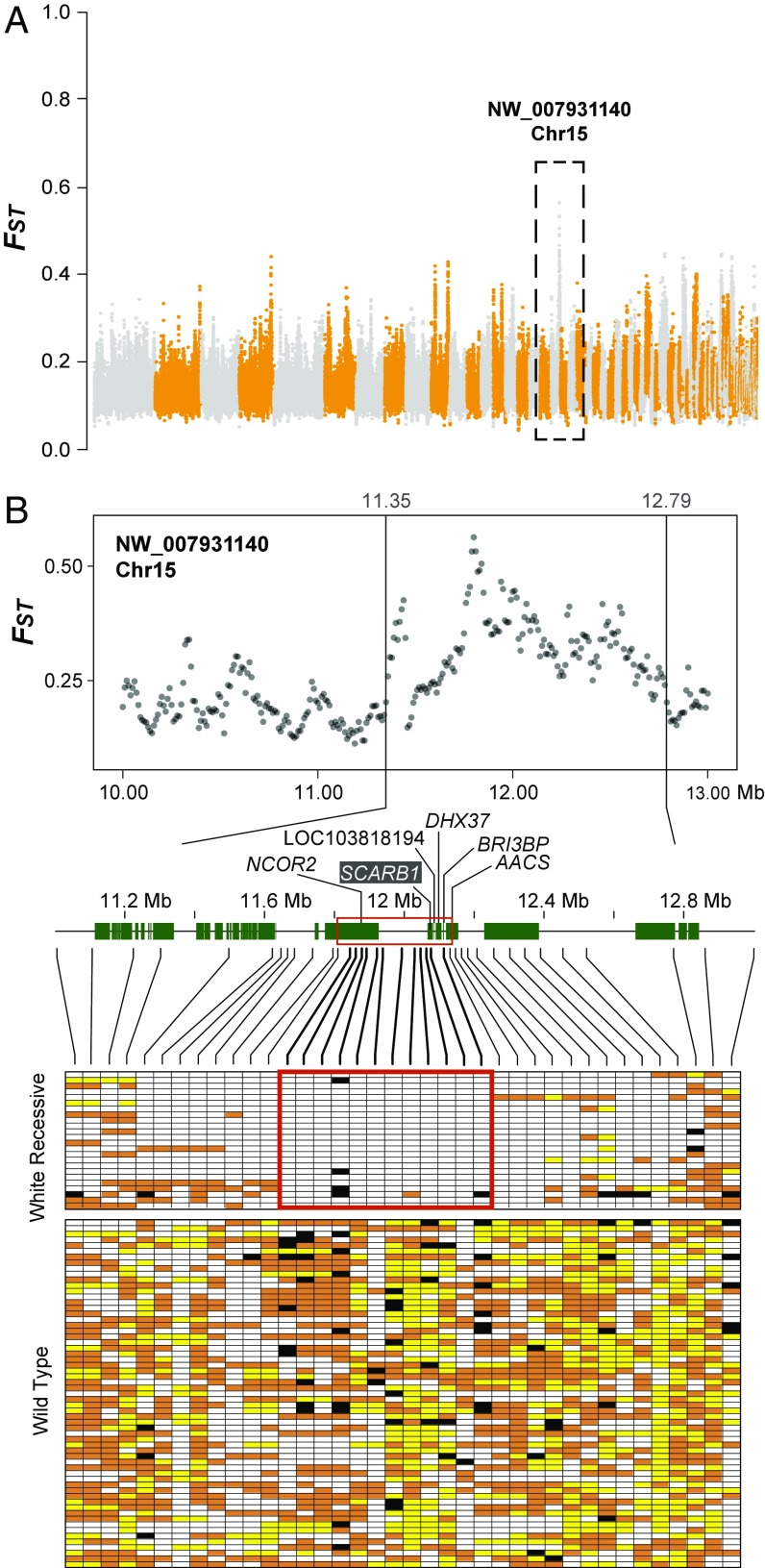
Fig. 2.
Mapping of the white recessive mutation. (A) Selective sweep mapping. FST between white recessive and nonwhite breeds/populations across the autosomal scaffolds. Each dot represents FST in 20-kb windows iterated every 10 kb. The different scaffolds are presented along the x axis in the same order as they appear in the canary reference genome assembly. (B) FST zoom-in and IBD mapping. (Top) FST in 20-kb windows iterated every 10 kb across the outlier region (delineated by vertical lines). (Bottom) The protein-coding genes found within this region are indicated by green boxes, and black lines point to the position of the genotyped SNPs in the IBD analysis. For the IBD analysis, 38 SNPs were genotyped for 24 white recessive canaries and 61 individuals belonging to 8 breeds with yellow or red coloration. Alleles more common in white recessive canaries are represented by white boxes, alternative alleles are in yellow, and heterozygotes are in orange. The red-outlined boxes indicate a segment of high homozygosity in white recessive canaries and black boxes indicate missing data.
Patterns of molecular variation do not provide direct information about the underlying phenotypes. To confirm the association between our candidate region and the white recessive phenotype, we generated a backcross mapping population segregating for white and yellow coloration. Briefly, F1 individuals derived from a cross between white recessive and yellow parental individuals were backcrossed to white recessive individuals (Materials and Methods). We found a perfect association between our candidate region and white coloration: All 24 yellow backcrossed birds were heterozygous for a SNP diagnostic between parental individuals, and all 14 white birds were homozygous. Overall, the combination of selective sweep mapping and linkage analysis allows us to conclude that our candidate region contains the gene that explains white recessive coloration in canaries.
Fine Mapping Within the Candidate Region.
To achieve better resolution at the causative locus, we used identical-by-descent (IBD) mapping (Fig. 2B). The assumption behind this approach is that the white recessive mutation appeared once and, therefore, the causal mutation should be located within the minimum shared haplotype present in white recessive individuals. Variants were selected from the whole-genome resequencing data, and chosen randomly with respect to allele frequency differences between white recessive and other breeds. We genotyped these variants in a larger cohort of birds, including white recessive individuals (n = 24) as well as individuals belonging to several breeds with red or yellow coloration (n = 61). Compared with other breeds, white recessive canaries were nearly all homozygous for a segment defined by multiple consecutive variants—with the exception of a single polymorphic position in a single individual that might be explained by a double recombination event or a de novo mutation. Assuming that this region represents a single IBD segment, the minimum haplotype spans ∼321 kb between nucleotide positions 11,808,700 and 12,130,178. An examination of the annotation of the canary reference genome (22) revealed six protein-coding genes within this chromosomal interval: NCOR2, SCARB1, LOC103818194 (polyubiquitin-like), DHX37, BRI3BP, and AACS (Fig. 2B; further gene information is provided in SI Appendix, Table S3).
A Splice-Site Mutation in SCARB1 Explains the White Recessive Phenotype.
We next searched within the IBD interval for potential causative mutations, including single-base changes, indels, and structural changes (i.e., inversions, duplications, translocations). We identified a single point mutation in this interval at nucleotide position 12,075,002 that was unique to white recessive canaries compared with the remaining pooled samples. This variant was predicted to be a mutation in the splice-donor site immediately downstream of exon 4 of the SCARB1 gene, which could potentially disrupt splicing of the transcript (see below). SCARB1 is an excellent candidate gene for white recessive coloration in canaries. It encodes scavenger receptor class B, member 1, which is known to serve as an HDL receptor and to mediate carotenoid uptake in mammals (24–28).
Next, we genotyped the splice variant in the cohort of samples used for the IBD analysis. Consistent with an autosomal recessive mode of inheritance, we found that all white recessive individuals were homozygous for the splice mutation (“G”), whereas all yellow and red individuals across several breeds were either homozygous (n = 60) or heterozygous (n = 1) for the wild-type allele (“T”). This mutation occurs in a genomic position that shows universal evolutionary conservation in whole-genome alignments across a wide diversity of bird species (SI Appendix, Fig. S2). These results suggest that the splice-site mutation detected in SCARB1 is likely to cause the white recessive phenotype.
The Splice Mutation Leads to the Production of Alternative Isoforms of SCARB1.
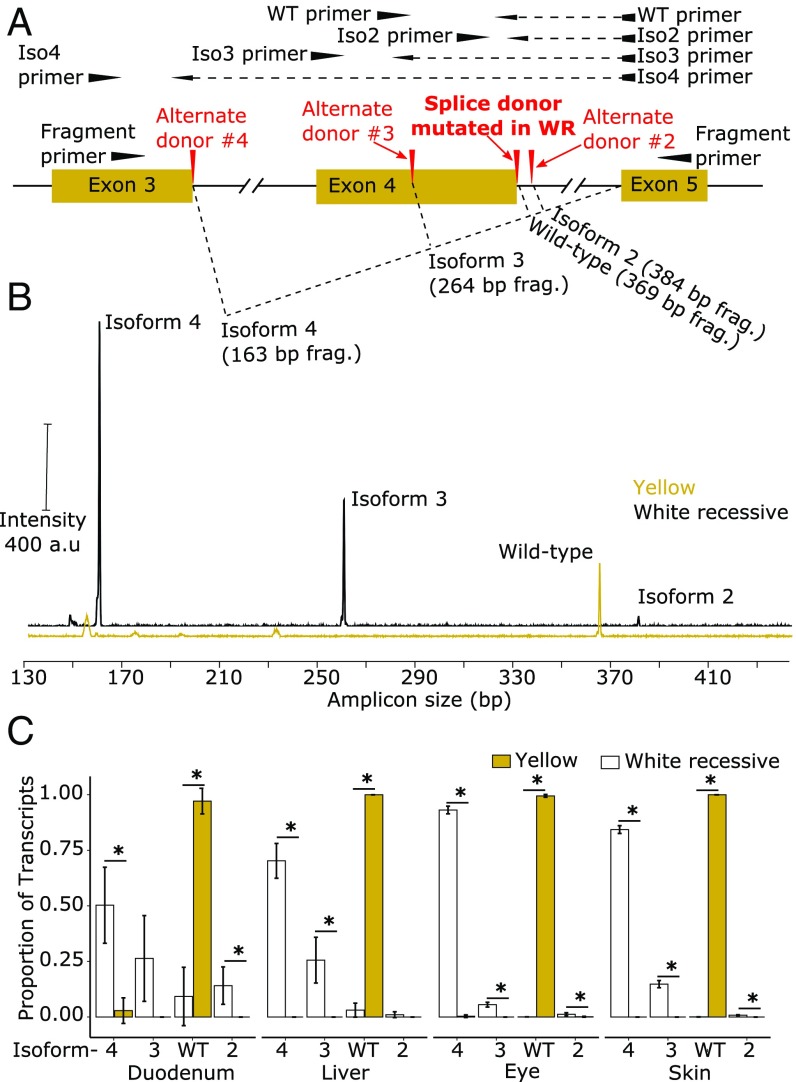
Between exons 3 and 5 of SCARB1, there are several potential splice donor sites that may be used as alternatives to the exon 4 splice donor mutated in white recessive canaries (Fig. 3A). To evaluate the use of these alternative splice donors, we reverse-transcribed RNA extracted from skin samples of adult white recessive and yellow canaries and PCR-amplified SCARB1 transcript across these sites (fragment primers, Fig. 3A). We then used capillary electrophoresis fragment analysis and Sanger sequencing to characterize the amplicons and found that white recessive canaries lack wild-type SCARB1 but express three different splice isoforms (Fig. 3B and SI Appendix, Fig. S3). Isoform 2 is spliced at NW_007931140 (12,074,986 to 12,074,987 bp), includes 15 bp of the intron, and maintains the reading frame but adds 5 amino acids between exon 4 and 5 (Fig. 3A and SI Appendix, Fig. S3). Isoform 3 is spliced at NW_007931140 (12,075,092 to 12,075,093 bp) within exon 4 and maintains the reading frame but omits 35 amino acids encoded by exon 4 (Fig. 3A and SI Appendix, Fig. S3). In isoform 4, exons 3 and 5 are spliced at their canonical donor and acceptor sites, which maintains the reading frame of the transcript but entirely skips exon 4, omitting 68 amino acids from the protein (Fig. 3A and SI Appendix, Fig. S3). Next, we used quantitative PCR (qPCR) to measure the overall expression of SCARB1 and the relative expression of each splice isoform in the duodenum, liver, eye, and skin of white recessive and yellow canaries. First, we targeted a portion of the transcript common to all of the isoforms (exons 7/8) and measured expression relative to a housekeeping gene, GAPDH. Overall expression of SCARB1 differed significantly among tissue types, with the highest levels of expression in the skin (F3,24 = 70.5, P = 4.8 × 10−12; SI Appendix, Fig. S4). There was a significant interaction between breed and tissue type (F3,24 = 8.2, P = 0.0006; SI Appendix, Fig. S4), and white recessive canaries had twofold lower levels of SCARB1 in the duodenum compared with wild-type canaries (Tukey's post hoc test, P = 0.022; SI Appendix, Fig. S4). However, expression in the other tissue types did not differ significantly between the breeds (Tukey's post hoc test, P ≥ 0.083; SI Appendix, Fig. S4). Next, we designed primers that spanned the unique splice junctions of each SCARB1 isoform (Fig. 3A) and measured the expression of each isoform relative to exon 7/8. In yellow canaries, we found that the wild-type isoform of SCARB1 constituted between 97.1 and 100% of all transcripts derived from this gene (Fig. 3C). In contrast, in the tissues of the white recessive canaries, the wild-type isoform made up less than 9.3% of all SCARB1 transcripts (Fig. 3C). Isoform 4 (50.3 to 93.1% of all transcripts) was most abundant in the white recessive tissues, and the remainder consisted of varying levels of the two other alternative transcripts (Fig. 3C). Taken together, these results indicate that the candidate splice-site mutation results in the production of alternative isoforms of SCARB1, and that the predominant transcripts in white recessive birds exclude a large portion or the entirety of exon 4.
Fig. 3.
Splicing variants and expression levels of SCARB1 in white recessive and wild-type individuals. (A) SCARB1 contains alternative splice donor sites that potentially yield different transcript isoforms. To investigate these alternatives, we designed primers (fragment) to amplify across this region, yielding different amplicon sizes from each isoform (B). To quantify the abundance of each alternative isoform (C), we designed qPCR primers spanning each potential splice junction (dashed lines). (B) Capillary electrophoresis fragment analyses of amplicons across the exon 4 splice junctions, generated from skin cDNA of white recessive or yellow canaries, indicate that three alternative SCARB1 transcript isoforms are present in the skin of the white recessive canary. This multitemplate amplification is biased toward short amplicons, and therefore the intensity of the peaks does not necessarily reflect transcript abundance. a.u., arbitrary units. (C) The mean ± SD of relative expression of SCARB1 transcript isoforms in the duodenum, liver, eye, and skin of yellow (n = 4; yellow bars) and white recessive (n = 4; white bars) canaries. For each sample, expression was measured by qPCR and calculated relative to a transcript region 3′ of the splice sites (exon 7/8) and then normalized to the sum of the four isoforms. The asterisk indicates a significant difference between yellow and white recessive (P ≤ 0.045).
Absence of Exon 4 Results in a Nonfunctional Form of SCARB1.
Isoform 4 of SCARB1, the most abundant transcript isoform in the white recessive canary, is predicted to lack 68 amino acids, a deficiency that is likely to have a strong impact on SCARB1 protein function. To explore the effects of this deficiency, we aligned the amino acid sequence of canary SCARB1 to the SCARB1 proteins from representatives of four different vertebrate classes and found that 15 of the amino acids encoded by exon 4 are identical across this broad phylogenetic sample (SI Appendix, Fig. S5A). Next, we generated a structural model of SCARB1 using a recently published crystal structure of a related receptor, human CD36 (29) (SI Appendix, Fig. S5B). Members of the CD36 superfamily of scavenger receptors (including SCARB1 and CD36) are integral membrane proteins that contain a large hydrophobic tunnel that extends through the length of the central vertical axis of the protein (SI Appendix, Fig. S2B). This tunnel is hypothesized to form a conduit for the passage of lipophilic molecules into cells (30). In human SCARB1, this tunnel is surrounded by extracellular subdomains that bind specific lipid carrier proteins and mediate selective uptake (31–34). In the canary, portions of SCARB1 encoded by exon 4 are homologous to the ligand-binding domain identified in other members of the CD36 family (SI Appendix, Fig. S5C). Thus, the exclusion of exon 4 from SCARB1 in white recessive canaries has the potential to disrupt essential ligand binding and impair transport function.
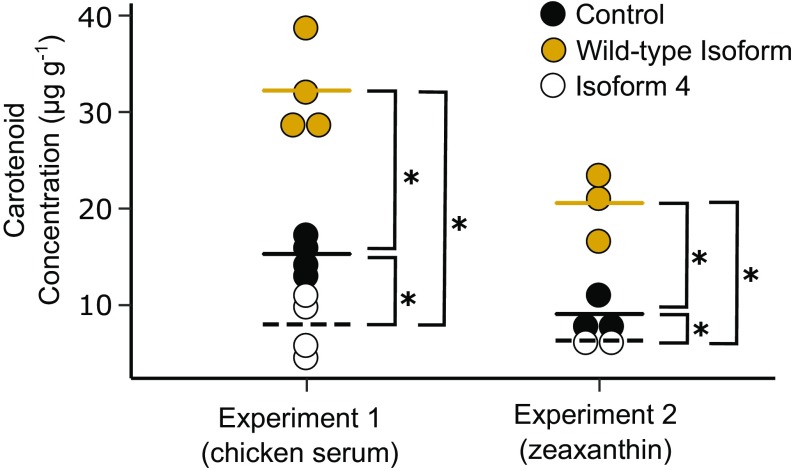
To directly test the functional consequences of the exclusion of exon 4, we compared the activity of the wild-type isoform of SCARB1 with that of isoform 4. We cloned the isoforms of SCARB1 into a eukaryotic expression vector that coexpresses green fluorescent protein (GFP), transfected these constructs into an avian fibroblast cell line, and confirmed transfection by monitoring GFP expression. We then delivered carotenoids to these cells in two different ways. In the first experiment, to ensure that the carotenoids were presented to the cells by the appropriate lipoprotein carriers (11, 12), we supplemented the cells with 20% whole-chicken serum, which is rich in lipoproteins containing carotenoids (lutein: 2.65 μg⋅mL−1 serum; zeaxanthin: 1.40 μg⋅mL−1 serum). In the second experiment, we followed a previously established protocol (26) to solubilize purified zeaxanthin in cell media with a low-concentration detergent (Tween 40). We incubated the cells with these carotenoid substrates overnight, harvested the cells, and measured carotenoid uptake by HPLC. In both experiments, the expression of wild-type SCARB1 significantly enhanced the uptake of carotenoids into the cells compared with controls (experiment 1: F2,9 = 51.83, P = 1.2 × 10−5, Tukey’s post hoc, P = 1.8 × 10−4; experiment 2: F2,5 = 24.69, P = 0.0026, Tukey’s post hoc, P = 0.0056; Fig. 4). In contrast, the expression of isoform 4 of SCARB1 resulted in a small but significant reduction in the uptake of carotenoids relative to the controls (experiment 1: Tukey’s post hoc, P = 1.0 × 10−5; experiment 2: Tukey’s post hoc, P = 0.0035; Fig. 4). These results support the hypothesis that the predominant form of SCARB1 expressed in white recessive canaries is nonfunctional with respect to carotenoid transport.
Fig. 4.
Analysis of the carotenoid transport function of SCARB1 splice isoforms. Here are presented the total carotenoid concentration (µg carotenoid per g of protein) of avian fibroblast cells expressing wild-type SCARB1 (yellow points), the exon 4-deficient isoform (isoform 4) of SCARB1 (open points), or a fluorescent protein-only control (black points). The cells were supplemented with carotenoid-containing whole-chicken serum to a final concentration of 0.81 μg⋅mL−1 of carotenoid in media (experiment 1) or 1 μg⋅mL−1 of pure zeaxanthin solubilized with Tween 40 (experiment 2). The lines represent the mean for each condition within each experiment. Significant differences (P ≤ 0.006) between conditions are denoted with an asterisk.
Discussion
Through genomic and biochemical analyses of the white recessive canary breed, we identified a single-nucleotide substitution that results in the near-total absence of carotenoid-based plumage coloration. This mutation is in a splice donor site of the gene encoding the integral membrane protein SCARB1 and results in the production of alternative transcript isoforms. Our in vitro analyses indicate that, whereas wild-type SCARB1 facilitates the cellular uptake of carotenoids, the most abundant isoform in the white recessive canary (isoform 4) has lost this function. These findings implicate SCARB1 as an important mediator of carotenoid uptake in birds and suggest a link between carotenoid-based coloration and lipid metabolism.
Our observations add to a growing body of evidence implicating scavenger receptors as key mediators of carotenoid uptake across a diversity of animals. SCARB1 was first characterized in humans as a high-density lipoprotein receptor that mediates the selective uptake of cholesterol, and was subsequently demonstrated to have a role in the cellular uptake of a variety of lipids, including phospholipids, products of triglycerol hydrolysis, and the lipophilic vitamins A and D (28, 35, 36). SCARB1 homologs have now been implicated as mediators of carotenoid uptake in fruit flies (Drosophila melanogaster), silkworms (Bombyx mori), salmon (Salmo salar), mice, and humans (24, 26, 37–41). These findings suggest that SCARB1 is an ancient and conserved mechanism of carotenoid uptake in animals.
Although the white recessive mutation in SCARB1 has a profound impact on plumage coloration and the accumulation of carotenoids in feathers, this mutation does not completely abrogate carotenoid uptake in all tissues. We observed very low concentrations of carotenoids in the liver and skin of some white recessive canaries, suggesting that the white recessive mutation may not completely eliminate the function of SCARB1. It is possible that other isoforms (i.e., isoform 2 or 3) maintain some carotenoid transport function. This would not be surprising, because isoform 2 retains the entire coding sequence of SCARB1 with a small in-frame addition of five amino acids to the mature protein. Another possibility is that there exist SCARB1-independent mechanisms of carotenoid uptake in birds. In silkworms and humans, other members of the CD36 family have been identified as mediators of carotenoid uptake, and passive diffusion is a possible mechanism of uptake when carotenoids are present at high concentrations (37, 40, 42, 43). These considerations notwithstanding, the dramatic phenotype of the white recessive canary indicates that SCARB1 plays a central role in carotenoid uptake in birds.
The confirmation of SCARB1 as an essential mediator of carotenoid uptake in birds presents new avenues of investigation into the proximate basis of carotenoid-based color expression and the information content of these color ornaments. In mammals, SCARB1 expression and carotenoid uptake are regulated by retinoid (vitamin A) status via the transcription factor ISX (44–46). Such a mechanism is consistent with the vitamin A-redox hypothesis, which proposes that carotenoid coloration reveals information through interdependencies in the uptake, transport, and metabolism of vitamin A and carotenoids (7). SCARB1 may also play a central role in avian reproduction. In birds, oogenesis involves the massive mobilization of cholesterol and other lipids, including carotenoids, from the body into the egg, and these lipid resources are essential for the growth and development of the embryo (47). Thus, the broad roles of SCARB1 in avian carotenoid, retinoid, and lipid physiology potentially link the expression of carotenoid-based colors to an individual’s redox status, lipid physiology, and fecundity.
In humans, SCARB1 is an important mediator of cholesterol homeostasis, and mutations in SCARB1 are associated with the accelerated development of coronary artery disease [e.g., (48)]. The pathophysiology of the white recessive mutation has not been fully characterized, but further study of this canary breed could offer broad insights into the role of SCARB1 in disease.
Materials and Methods
Analysis of Tissue Carotenoid Content.
The carotenoid content of white recessive and yellow canary tissues was analyzed by HPLC using methodology adapted from previous studies (21). All experiments were conducted in accordance with Directive 2010/63/EU on the protection of animals, and protocols were examined by the Órgão Responsável pelo Bem-Estar Animal of CIBIO.
Whole-Genome Resequencing, Read Mapping, and SNP Calling.
Whole-genome resequencing data from DNA pools of four canary breeds were obtained in the course of a different study (8). Each pool was sequenced using an Illumina instrument to an effective coverage of ∼17 to 23× (SI Appendix, Table S1). Sequencing reads were mapped to the canary reference genome assembly (49) using BWA-MEM (50), and SNP calling was performed using the Bayesian haplotype-based method implemented in FreeBayes (51).
Genetic Mapping.
Genetic differentiation between white recessive and other breeds was summarized in windows of 20 kb moved in steps of 10 kb using the FST as implemented in the PoPoolation2 package (52). To confirm the association between our candidate region and the white recessive phenotype, a family consisting of 38 backcrosses segregating for the white recessive allele and wild-type yellow coloration was used for linkage analysis. An SNP diagnostic between parental individuals used in the cross was genotyped in all backcross individuals using Sanger sequencing IBD mapping was performed by genotyping 85 individuals (24 white recessive and 61 from other breeds) for 38 SNPs within and flanking the candidate region using Sequenom’s iPLEX technology.
Search for Causative Mutations.
SNP and indel variants within the candidate region were functionally annotated using the genetic variant annotation and effect prediction toolbox SnpEff (53). The identification of structural variants was performed using three approaches: BreakDancer (54), DELLY (55), and LUMPY (56). The splice-site mutation was genotyped using Sanger sequencing for the same 85 individuals used for IBD.
Whole-Genome Alignment of Bird Species.
To evaluate conservation of the candidate causative mutation in SCARB1 across the avian phylogeny, we obtained whole-genome alignments of available avian genomes using LASTZ (57), chainNet (58), and MULTIZ (59).
Analysis of SCARB1 Transcript Isoform Expression.
To identify alternative splice isoforms, we PCR-amplified from exons 3 to 5 of the SCARB1 transcript from cDNA derived from the skin of wild-type and white recessive canaries, and examined the amplicons by capillary electrophoresis fragment analysis and Sanger sequencing. We measured the relative expression of the SCARB1 transcript isoforms in white recessive and yellow canaries by qPCR with primers that targeted the splice junctions unique to each isoform. RNA extraction, cDNA production, and qPCR analyses were carried out as described previously (8).
Functional Analysis of SCARB1 Variants.
The wild-type and isoform 4 of SCARB1 were cloned from canary cDNA (above) into eukaryotic expression vectors and expressed in DF1 cells. These cells were cultured overnight in carotenoid-enriched media, and cellular carotenoid uptake was analyzed by HPLC. All methods and materials are further detailed in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank the multiple breeders who contributed samples for this project. The work was supported by Fundação para a Ciência e Tecnologia (FCT) through POPH-QREN funds from the European Social Fund and Portuguese MCTES [FCT Investigator grant to M.C. (IF/00283/2014/CP1256/CT0012) and a postdoctoral fellowship to R.J.L. (SFRH/BPD/84141/2012)]; a research fellowship to M.A.G. in the scope of the Biodiversity, Genetics, and Evolution (BIODIV) PhD program at CIBIO/InBIO and University of Porto (PD/BD/114042/2015); the projects “Genomics and Evolutionary Biology” and “Genomics Applied to Genetic Resources” cofinanced by North Portugal Regional Operational Programme 2007/2013 (ON.2–O Novo Norte) under the National Strategic Reference Framework and European Regional Development Fund; and an EU FP7 REGPOT grant (CIBIO-New-Gen) (286431). M.B.T. and J.C.C. were funded in part by a Human Frontiers Science Program grant (RGP0017/2011) and National Institutes of Health grants (R01EY026672 and RO1EY024958). J.C.C. was also supported by a grant from Research to Prevent Blindness. M.B.T. received fellowship support from the National Science Foundation (Award 1202776), National Institutes of Health (T32EY013360), and McDonnell Center for Cellular and Molecular Neurobiology at Washington University. G.E.H. was funded by the Department of Biological Sciences and College of Science and Mathematics at Auburn University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank (BioProject PRJNA300534) and Dryad (doi:10.5061/dryad.bt816) databases.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700751114/-/DCSupplemental.
References
- 1.McGraw KJ. Mechanics of carotenoid-based coloration. In: Hill GE, McGraw KJ, editors. Bird Coloration. I. Mechanisms and Measurements. Harvard Univ Press; Cambridge, MA: 2006. pp. 177–242. [Google Scholar]
- 2.Hill GE. Female mate choice for ornamental coloration in birds. In: Hill GE, McGraw KJ, editors. Bird Coloration. II. Function and Evolution. Harvard Univ Press; Cambridge, MA: 2006. pp. 137–200. [Google Scholar]
- 3.Hill GE. Environmental regulation of ornamental coloration. In: Hill GE, McGraw KJ, editors. Bird Coloration. I. Mechanisms and Measurements. Harvard Univ Press; Cambridge, MA: 2006. pp. 507–560. [Google Scholar]
- 4.Toews DP, et al. Plumage genes and little else distinguish the genomes of hybridizing warblers. Curr Biol. 2016;26:2313–2318. doi: 10.1016/j.cub.2016.06.034. [DOI] [PubMed] [Google Scholar]
- 5.Hill GE. A Red Bird in a Brown Bag: The Function and Evolution of Colorful Plumage in the House Finch. Oxford Univ Press; New York: 2002. [Google Scholar]
- 6.Svensson PA, Wong BBM. Carotenoid-based signals in behavioural ecology: A review. Behaviour. 2011;148:131–189. [Google Scholar]
- 7.Hill GE, Johnson JD. The vitamin A-redox hypothesis: A biochemical basis for honest signaling via carotenoid pigmentation. Am Nat. 2012;180:E127–E150. doi: 10.1086/667861. [DOI] [PubMed] [Google Scholar]
- 8.Lopes RJ, et al. Genetic basis for red coloration in birds. Curr Biol. 2016;26:1427–1434. doi: 10.1016/j.cub.2016.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mundy NI, et al. Red carotenoid coloration in the zebra finch is controlled by a cytochrome P450 gene cluster. Curr Biol. 2016;26:1435–1440. doi: 10.1016/j.cub.2016.04.047. [DOI] [PubMed] [Google Scholar]
- 10.Eriksson J, et al. Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 2008;4:e1000010. doi: 10.1371/journal.pgen.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trams EG. Carotenoid transport in the plasma of the scarlet ibis (Eudocimus ruber) Comp Biochem Physiol. 1969;28:1177–1184. doi: 10.1016/0010-406x(69)90558-1. [DOI] [PubMed] [Google Scholar]
- 12.Parker RS. Absorption, metabolism, and transport of carotenoids. FASEB J. 1996;10:542–551. [PubMed] [Google Scholar]
- 13.Attie AD, et al. Identification and functional analysis of a naturally occurring E89K mutation in the ABCA1 gene of the WHAM chicken. J Lipid Res. 2002;43:1610–1617. doi: 10.1194/jlr.m200223-jlr200. [DOI] [PubMed] [Google Scholar]
- 14.Connor WE, Duell PB, Kean R, Wang Y. The prime role of HDL to transport lutein into the retina: Evidence from HDL-deficient WHAM chicks having a mutant ABCA1 transporter. Invest Ophthalmol Vis Sci. 2007;48:4226–4231. doi: 10.1167/iovs.06-1275. [DOI] [PubMed] [Google Scholar]
- 15.Walker GBR, Avon D. Coloured, Type, and Song Canaries: A Complete Guide. Sterling; New York: 1993. [Google Scholar]
- 16.Birkhead T. A Brand New Bird: How Two Amateur Scientists Created The First Genetically Engineered Animal. Basic Books; New York: 2003. [Google Scholar]
- 17.Brockmann H, Völker O. Der gelbe Federfarbstoff des Kanarienvogels [Serinus canaria canaria (L.)] und das Vorkommen von Carotinoiden bei Vögeln. Mit 5 Figuren auf Tafel II. Hoppe Seylers Z Physiol Chem. 1934;224:193–215. German. [Google Scholar]
- 18.Koch RE, McGraw KJ, Hill GE. Effects of diet on plumage coloration and carotenoid deposition in red and yellow domestic canaries (Serinus canaria) Wilson J Ornithol. 2016;128:328–333. [Google Scholar]
- 19.Wolf P, Bartels T, Sallmann HP, Heisler K, Kamphues J. Vitamin A metabolism in recessive white canaries. Anim Welf. 2000;9:153–165. [Google Scholar]
- 20.Preuss SE, Bartels T, Schmidt V, Krautwald-Junghanns ME. Vitamin A requirements of alipochromatic (‘recessive-white’) and coloured canaries (Serinus canaria) during the breeding season. Vet Rec. 2007;160:14–19. doi: 10.1136/vr.160.1.14. [DOI] [PubMed] [Google Scholar]
- 21.Toomey MB, et al. A complex carotenoid palette tunes avian colour vision. J R Soc Interface. 2015;12:20150563. doi: 10.1098/rsif.2015.0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorrestein GM, Schrijver J. [A genetic disorder of vitamin A metabolism is recessively white canaries] Tijdschr Diergeneeskd. 1982;107:795–799. Dutch. [PubMed] [Google Scholar]
- 23.Perez-Beato O. Fundamentals of Color Genetics in Canaries: Reproduction and Control. RoseDog Books; Pittsburgh: 2008. [Google Scholar]
- 24.Kiefer C, Sumser E, Wernet MF, Von Lintig J. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc Natl Acad Sci USA. 2002;99:10581–10586. doi: 10.1073/pnas.162182899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukasawa M, et al. SRB1, a class B scavenger receptor, recognizes both negatively charged liposomes and apoptotic cells. Exp Cell Res. 1996;222:246–250. doi: 10.1006/excr.1996.0030. [DOI] [PubMed] [Google Scholar]
- 26.During A, Doraiswamy S, Harrison EH. Xanthophylls are preferentially taken up compared with beta-carotene by retinal cells via a SRBI-dependent mechanism. J Lipid Res. 2008;49:1715–1724. doi: 10.1194/jlr.M700580-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Bennekum A, et al. Class B scavenger receptor-mediated intestinal absorption of dietary beta-carotene and cholesterol. Biochemistry. 2005;44:4517–4525. doi: 10.1021/bi0484320. [DOI] [PubMed] [Google Scholar]
- 28.Reboul E, Borel P. Proteins involved in uptake, intracellular transport and basolateral secretion of fat-soluble vitamins and carotenoids by mammalian enterocytes. Prog Lipid Res. 2011;50:388–402. doi: 10.1016/j.plipres.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Hsieh FL, et al. The structural basis for CD36 binding by the malaria parasite. Nat Commun. 2016;7:12837. doi: 10.1038/ncomms12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodrigueza WV, et al. Mechanism of scavenger receptor class B type I-mediated selective uptake of cholesteryl esters from high density lipoprotein to adrenal cells. J Biol Chem. 1999;274:20344–20350. doi: 10.1074/jbc.274.29.20344. [DOI] [PubMed] [Google Scholar]
- 31.Kartz GA, Holme RL, Nicholson K, Sahoo D. SR-BI/CD36 chimeric receptors define extracellular subdomains of SR-BI critical for cholesterol transport. Biochemistry. 2014;53:6173–6182. doi: 10.1021/bi500706x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu X, Kozarsky K, Krieger M. Scavenger receptor class B, type I-mediated [3H]cholesterol efflux to high and low density lipoproteins is dependent on lipoprotein binding to the receptor. J Biol Chem. 2000;275:29993–30001. doi: 10.1074/jbc.275.39.29993. [DOI] [PubMed] [Google Scholar]
- 33.Chadwick AC, Sahoo D. Functional characterization of newly-discovered mutations in human SR-BI. PLoS One. 2012;7:e45660. doi: 10.1371/journal.pone.0045660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neculai D, et al. Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36. Nature. 2013;504:172–176. doi: 10.1038/nature12684. [DOI] [PubMed] [Google Scholar]
- 35.Acton S, et al. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 1996;271:518–520. doi: 10.1126/science.271.5248.518. [DOI] [PubMed] [Google Scholar]
- 36.Rigotti A, Acton SL, Krieger M. The class B scavenger receptors SR-BI and CD36 are receptors for anionic phospholipids. J Biol Chem. 1995;270:16221–16224. doi: 10.1074/jbc.270.27.16221. [DOI] [PubMed] [Google Scholar]
- 37.Tsuchida K, Sakudoh T. Recent progress in molecular genetic studies on the carotenoid transport system using cocoon-color mutants of the silkworm. Arch Biochem Biophys. 2015;572:151–157. doi: 10.1016/j.abb.2014.12.029. [DOI] [PubMed] [Google Scholar]
- 38.Sundvold H, Helgeland H, Baranski M, Omholt SW, Våge DI. Characterisation of a novel paralog of scavenger receptor class B member I (SCARB1) in Atlantic salmon (Salmo salar) BMC Genet. 2011;12:52. doi: 10.1186/1471-2156-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.During A, Harrison EH. Mechanisms of provitamin A (carotenoid) and vitamin A (retinol) transport into and out of intestinal Caco-2 cells. J Lipid Res. 2007;48:2283–2294. doi: 10.1194/jlr.M700263-JLR200. [DOI] [PubMed] [Google Scholar]
- 40.Borel P, et al. CD36 and SR-BI are involved in cellular uptake of provitamin A carotenoids by Caco-2 and HEK cells, and some of their genetic variants are associated with plasma concentrations of these micronutrients in humans. J Nutr. 2013;143:448–456. doi: 10.3945/jn.112.172734. [DOI] [PubMed] [Google Scholar]
- 41.Voolstra O, et al. The Drosophila class B scavenger receptor NinaD-I is a cell surface receptor mediating carotenoid transport for visual chromophore synthesis. Biochemistry. 2006;45:13429–13437. doi: 10.1021/bi060701u. [DOI] [PubMed] [Google Scholar]
- 42.Sakudoh T, et al. CD36 homolog divergence is responsible for the selectivity of carotenoid species migration to the silk gland of the silkworm Bombyx mori. J Lipid Res. 2013;54:482–495. doi: 10.1194/jlr.M032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Lintig J. Colors with functions: Elucidating the biochemical and molecular basis of carotenoid metabolism. Annu Rev Nutr. 2010;30:35–56. doi: 10.1146/annurev-nutr-080508-141027. [DOI] [PubMed] [Google Scholar]
- 44.Lobo GP, et al. ISX is a retinoic acid-sensitive gatekeeper that controls intestinal beta,beta-carotene absorption and vitamin A production. FASEB J. 2010;24:1656–1666. doi: 10.1096/fj.09-150995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi MY, et al. A dynamic expression survey identifies transcription factors relevant in mouse digestive tract development. Development. 2006;133:4119–4129. doi: 10.1242/dev.02537. [DOI] [PubMed] [Google Scholar]
- 46.Widjaja-Adhi MA, Lobo GP, Golczak M, Von Lintig J. A genetic dissection of intestinal fat-soluble vitamin and carotenoid absorption. Hum Mol Genet. 2015;24:3206–3219. doi: 10.1093/hmg/ddv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Surai PF. Natural Antioxidants in Avian Nutrition and Reproduction. Nottingham Univ Press; Nottingham, UK: 2002. [Google Scholar]
- 48.Zanoni P, et al. CHD Exome+ Consortium CARDIoGRAM Exome Consortium Global Lipids Genetics Consortium Rare variant in scavenger receptor BI raises HDL cholesterol and increases risk of coronary heart disease. Science. 2016;351:1166–1171. doi: 10.1126/science.aad3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frankl-Vilches C, et al. Using the canary genome to decipher the evolution of hormone-sensitive gene regulation in seasonal singing birds. Genome Biol. 2015;16:19. doi: 10.1186/s13059-014-0578-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. 2012;arXiv:1207.3907. [Google Scholar]
- 52.Kofler R, Pandey RV, Schlötterer C. PoPoolation2: Identifying differentiation between populations using sequencing of pooled DNA samples (pool-seq) Bioinformatics. 2011;27:3435–3436. doi: 10.1093/bioinformatics/btr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen K, et al. BreakDancer: An algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rausch T, et al. DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: A probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harris RS. 2007. Improved pairwise alignment of genomic DNA. PhD thesis (Pennsylvania State University, State College, PA)
- 58.Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution’s cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci USA. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blanchette M, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.