Significance
DNMT3A is a critical epigenetic modifier and tumor suppressor in the hematopoietic system. This gene is frequently mutated in hematopoietic malignancies, including acute myeloid leukemia (AML), with Dnmt3a R878H being the most common mutant. By using a conditional knockin approach, this study shows that Dnmt3a R878H is sufficient to initiate AML and recapitulate human leukemic features in mice. The leukemia-initiating cells are enriched in hematopoietic stem/progenitor cells. Through gene expression profiling, DNA methylation and histone modification analysis, and functional tests on important regulators for cell proliferation and differentiation in an animal model, this study has not only discovered mTOR pathway activation as a key player in the disease mechanism but also revealed the potential therapeutic effects of mTOR inhibition on DNMT3A mutation-related leukemia.
Keywords: Dnmt3a R878H mutation, conditional knockin mice, single-cell RNA-seq, leukemia, mTOR inhibitor
Abstract
DNMT3A is frequently mutated in acute myeloid leukemia (AML). To explore the features of human AML with the hotspot DNMT3A R882H mutation, we generated Dnmt3a R878H conditional knockin mice, which developed AML with enlarged Lin−Sca1+cKit+ cell compartments. The transcriptome and DNA methylation profiling of bulk leukemic cells and the single-cell RNA sequencing of leukemic stem/progenitor cells revealed significant changes in gene expression and epigenetic regulatory patterns that cause differentiation arrest and growth advantage. Consistent with leukemic cell accumulation in G2/M phase, CDK1 was up-regulated due to mTOR activation associated with DNA hypomethylation. Overexpressed CDK1-mediated EZH2 phosphorylation resulted in an abnormal trimethylation of H3K27 profile. The mTOR inhibitor rapamycin elicited a significant therapeutic response in Dnmt3aR878H/WT mice.
Somatic mutations in the DNA methyltransferase DNMT3A have been identified in a subset of acute myeloid leukemia (AML), myelodysplastic syndrome (MDS), and acute lymphoblastic leukemia (ALL), with DNMT3A R882 being the hotspot (1–4). Clinical features of most AML cases with DNMT3A mutations include preferential involvement of a monocytic lineage (AML-M4 and -M5 subtypes), thrombocytosis, onset at a relatively old age, and poor prognosis (2, 5, 6). Careful genotype–phenotype correlations suggest that patients manifest DNMT3A mutations in preleukemic hematopoietic stem cells (HSCs)/multipotent progenitors (MPPs), which exhibit a competitive advantage over normal HSCs. Because these mutations occur at a very early stage among genetic abnormalities, they are likely involved in the development of leukemia (7, 8).
Functionally, the DNMT3A R882 mutation might disrupt epigenetic regulation. This kind of DNMT3A mutation confers reduced methyltransferase activity and promotes the possibility of dominant-negative consequences compared with the wild-type (WT) allele (2, 9, 10). Moreover, the DNMT3A mutation causes aberrant DNA hypomethylation and up-regulates a series of target genes involved in AML pathogenesis (11–13).
In vivo animal tests have shown that the Dnmt3a gene plays an essential role in hematopoiesis regulation. Dnmt3a−/− HSCs expand remarkably, but their differentiation is inhibited when Dnmt3a is conditionally inactivated in the murine hematopoietic system; this phenomenon is consistent with a preleukemic state (7). Moreover, all lethally irradiated mice transplanted with Dnmt3a-deleted HSCs died within 1 y and were diagnosed with a spectrum of malignancies similar to those observed in patients carrying DNMT3A mutations, including MDS, AML, primary myelofibrosis, and ALL, suggesting that Dnmt3a functions as a tumor suppressor (7). With a second hit of mutations in various genes such as N-RAS, C-KIT, or FLT3 in Dnmt3a−/− mice, Dnmt3a deletion induces leukemic transformation (14, 15). Although these results indicate a major role of Dnmt3a deletion in facilitating the development of leukemia, the in vivo roles of DNMT3A mutants in leukemogenesis still need to be addressed. In our previous work, bone marrow transplantation (BMT) of bone marrow (BM) cells retrovirally transduced with DNMT3A R882H induced chronic myelomonocytic leukemia (CMML) with mild invasive behaviors through disturbing DNA methylation and gene expression in recipient mice (12). In contrast to the aggressive manifestations of DNMT3A mutation-related AML in clinical settings, this phenotype can be ascribed to an inappropriate expression of the mutant DNMT3A gene as driven by a retroviral promoter instead of the endogenous promoter/enhancer.
To address whether the clinically significant DNMT3A mutation could really be leukemogenic, we established a conditional knockin mouse model and investigated genetic and epigenetic changes, including gene expression profiles, DNA methylation, and chromatin modification, affected by this mutation. We further explored the potential mechanisms that can explain the process by which the DNMT3A mutation hierarchically induces abnormal hematopoiesis and the manner by which specific regulators of relevant pathways in murine and human settings can be targeted for potential therapeutic applications.
Results
Dnmt3a R878H Mutation Induces AML in Conditional Knockin Mice.
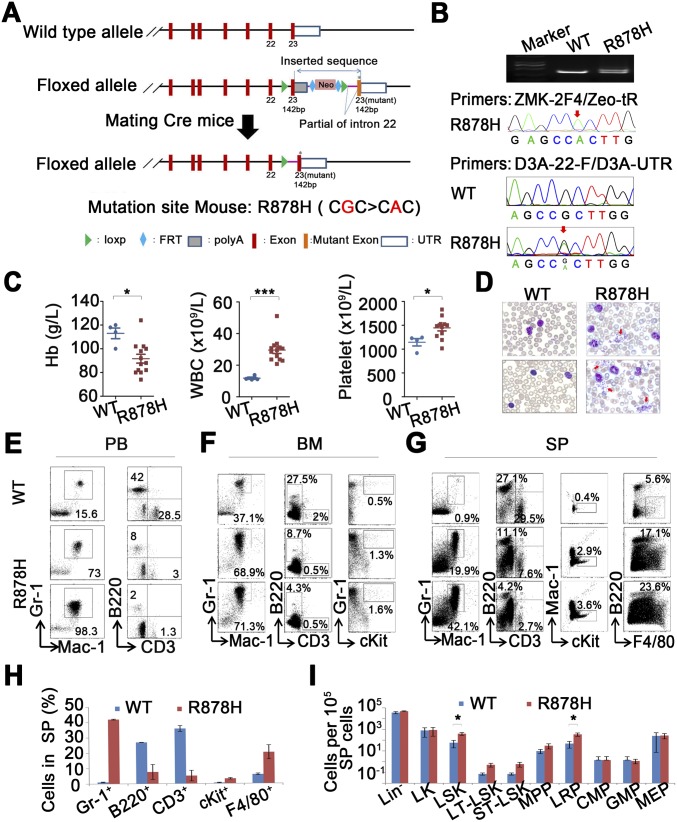
To elucidate the consequences of the DNMT3A R882 mutant in an endogenous expression environment, we generated a mouse model of Cre-mediated conditional expression of mutant Dnmt3a R878H obtained from the endogenous locus of Dnmt3a (Fig. S1A). Dnmt3a R878H knockin mice were crossed with mice carrying a Cre recombinase allele under the control of an IFN-inducible Mx1 promoter (Mx1Cre). Upon Cre induction with pIpC, the endogenous Dnmt3a exon 23 was efficiently replaced by exon 23 carrying the R878H mutation in the hematopoietic system, whereas this replacement was not found in other organs/tissues examined (Dataset S1). Conditional Dnmt3a R878H expression and activation were directed to the hematopoietic compartment by the Mx1 promoter. After obtaining Mx1Cre+Dnmt3aR878H/WT heterozygous mice (Dnmt3aR878H/WT), we confirmed the genotype at the DNA level and checked the RNA expression of the mutation by using PCR and sequence analysis (Fig. S1B).
Fig. S1.
(A) Schematic representation of the murine Dnmt3a locus, targeting vector, targeted Dnmt3a locus (LSL allele), and targeted Dnmt3a locus after Cre recombinase-mediated excision of the LSL cassette (knockin allele). (B) DNA of heterozygous mice carrying the targeted Dnmt3a locus was analyzed by PCR (primer: Loxp-tF/Loxp-tR) and sequencing (primer: ZMK-2F4/Zeo-tR). Mutant Dnmt3a of cDNA was detected in the BM cells of Dnmt3aR878H/WT mice (D3A-22-F/D3A-UTR-R). (C) Statistical analysis of hemoglobin levels and cell counts in the PB. (D) Morphological comparison of PB smears between mice. (E–G) Flow cytometric analyses of mature hematopoietic cell populations among PB (E), total nucleated BM (F), and splenic cells (G). (H and I) Flow cytometric quantification of mature hematopoietic cell populations and the indicated HSPC populations among SP cells of mice. Mean ± SEM values are shown. *P < 0.05, ***P < 0.001.
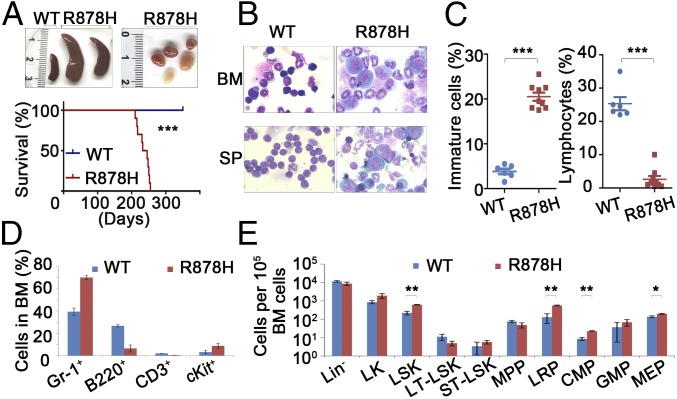
Approximately 4 to 6 mo after IFN induction, all 25 Dnmt3aR878H/WT mice were diagnosed with a myelomonocytic type of AML characterized by segmental expansion of immature cells in the BM and spleen, along with extramedullary infiltration (splenomegaly, lymphadenectasis, and cutaneous invasion). The Dnmt3aR878H/WT mice showed a relatively long median survival time of 230 d (Fig. 1A). Large leukemic cells showing a high nuclear/cytoplasmic ratio and fine chromatin were observed in the BM (23% on average) and spleen of all Dnmt3aR878H/WT mice (Fig. 1C). The nuclei of some blasts were indented, reminiscent of the identity of monoblasts (Fig. 1B). The inhibition ratio of NaF in BM cells was higher than 50%, indicating that these immature cells belonged to the monocytic lineage. Peripheral blood (PB) analysis showed significantly increased numbers of white blood cells (WBCs) and platelets and a dramatically reduced hemoglobin level (Fig. S1C). PB smears with Wright’s staining revealed a striking feature: a significant amount of early myeloid elements against a background of increased granulocytes, monocytes, and clustered platelets (Fig. S1D). In addition, the Dnmt3aR878H/WT mice displayed a reduced number of lymphocytes in the BM compared with those in Dnmt3aWT/WT controls (Fig. 1C). To determine whether cooperating mutations occurred in Dnmt3aR878H/WT leukemic mice, we performed whole-exome sequencing (WES) in BM samples and paired normal tail specimens from three diseased animals. We identified 17 potential somatic sequence changes in the coding region, including 16 single-nucleotide variations (SNVs) and 1 small deletion (Dataset S2). However, none of the SNVs was recurrent or previously reported in leukemia-related genes. Our data suggested that the Dnmt3a R878H mutant might be a driver of leukemogenesis.
Fig. 1.
Dnmt3aR878H/WT mice develop AML characterized by expansion of HSPCs. (A, Upper) Macroscopic appearance of spleen and lymphoid nodes obtained from mice. No lymphadenopathy was observed in WT animals. (Lower) Kaplan–Meier survival curves of Dnmt3aWT/WT (n = 15) and Dnmt3aR878H/WT (n = 10) mice. (B) Morphological analysis of BM and spleen (SP) obtained from mice. Wright’s staining was performed on BM and SP cytospin preparations. (C) Statistical analysis of the number of immature cells and lymphocytes in the BM. (D and E) Flow cytometric quantification of mature hematopoietic cell populations and the indicated HSPC populations in BM cells of mice. Mean ± SEM values are shown. *P < 0.05, **P < 0.01, ***P < 0.001.
Hematopoietic Progenitors Are Significantly Increased in Dnmt3aR878H/WT Mice.
In view of the significant myelomonocytosis in leukemic mice, we hypothesized that lineage-restricted progenitors (LRPs) and common myeloid progenitors (CMPs) should be affected. To further explore the unique AML phenotype induced by Dnmt3a R878H, we traced emerging hematopoietic stem/progenitor cell (HSPC) compartments for precise analysis. Flow cytometric analyses revealed an altered granulocyte/lymphocyte ratio and increased cKit+ cells in the BM, spleen, and PB of Dnmt3aR878H/WT mice (Fig. 1D and Fig. S1 E–H). Interestingly, the lineage-negative (Lin−) cells were significantly expanded in the BM, along with the majority of leukemic cells appearing in the Gr-1+ compartment. Lin−Sca1+cKit+ (LSK) cells were increased by ∼3- to 10-fold. The LSK population contains long-term HSCs (LT-HSCs), short-term HSCs (ST-HSCs), MPPs, and LRPs (16). We found that among the LSK cells the number of LT-HSCs decreased, quantity of ST-HSCs remained unchanged, and LRPs remarkably expanded from 70% up to 90% of LSKs in Dnmt3aR878H/WT mice. Moreover, the LK (Lin-Sca1-cKit+) population containing CMPs, granulocyte-macrophage progenitors (GMPs), and megakaryocyte-erythroid progenitors (MEPs) were investigated. Leukemic mice demonstrated an increase in CMPs and MEPs but not GMPs (Fig. 1E and Fig. S1I).
LSK Cells from Dnmt3aR878H/WT Mice Harbor Leukemia-Initiating Cells.
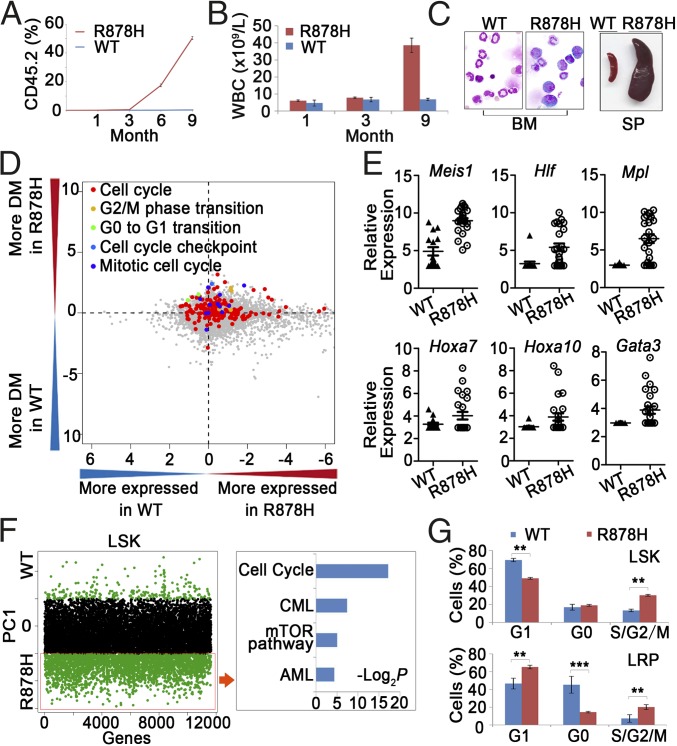
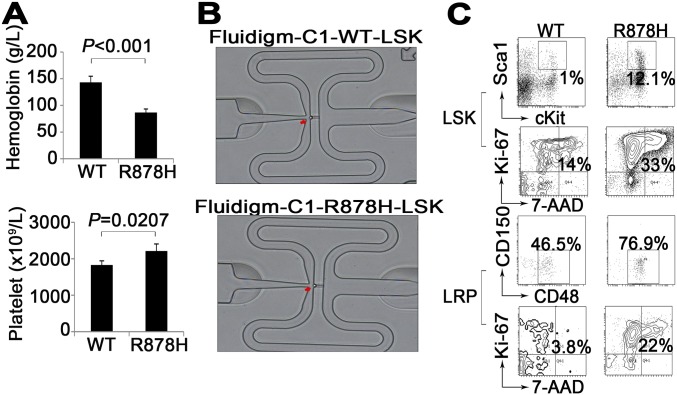
Because the LSK compartment was considerably enlarged in leukemic BM, we hypothesized that the aberrantly increased LSK cells might contain leukemia-initiating cells. Therefore, the effect of Dnmt3a R878H on leukemogenesis was further investigated through transplantation experiments by inoculating 1,000 LSK cells derived from the BM of Dnmt3aWT/WT or diseased Dnmt3aR878H/WT donor mice (with the CD45.2 marker) into sublethally irradiated (350 cGy) recipient mice (with the CD45.1 marker). As shown in Fig. 2A, almost no CD45.2-expressing elements were detectable in the PB of mice receiving Dnmt3aWT/WT LSK cells during the 9 mo after transplantation. In contrast, the percentage of CD45.2+ WBCs in the PB increased significantly to a level of 53% in recipient mice with a Dnmt3aR878H/WT transplant over the same period, suggesting that Dnmt3a R878H might provide LSK cells with an advantage in growth and survival. Detailed phenotype analysis was performed in three of the mice inoculated with Dnmt3aR878H/WT LSK cells. Similar to the observation in primary Dnmt3aR878H/WT animals, these mice manifested a high WBC count of up to 30 × 109/L, high numbers of platelets and a reduced hemoglobin level (Fig. 2B and Fig. S2A), an average of 20% immature cells in the BM, splenomegaly, and lymphadenectasis (Fig. 2C). These results indicated that the leukemia induced by knockin of Dnmt3a R878H was transplantable and that the LSK cells with the Dnmt3a mutation might harbor leukemia-initiating cells. Moreover, we also transplanted 2 × 106 spleen cells from the Dnmt3aR878H/WT leukemic mice into secondary recipient mice, which developed AML with a longer latency (12 to 18 mo) compared with the experiment using leukemic LSK cells.
Fig. 2.
Functional analysis of LSKs and single-cell transcriptome profiling of LSKs. (A) The percentage of CD45.2+ cells in the PB of recipient mice (CD45.1+ marker) after transplantation of LSKs. (B) Statistical analysis of cell counts in the PB after transplantation of LSKs. (C) Morphological analysis of BM and SP obtained from the LSK-transplanted mice. (D) Comparison between LSKs from Dnmt3aWT/WT and Dnmt3aR878H/WT mice in terms of their levels of gene expression and heterogeneity for GO categories. The logarithm (log10) of P values from two-sided paired t tests applied to mean normalized read counts (x axis) and DMs (y axis) was computed for each GO category and plotted against each other by multiplying the sign of the t statistic. (E) Expression level of genes associated with hematopoietic differentiation and leukemogenesis in LSKs. (F, Left) Distribution of genes contributing to principal component 1 (PC1) separation based on PCA. Green dots above and below the black dot zone represent down-regulated and up-regulated genes, respectively. (Right) Identification of leukemogenesis-related pathways based on KEGG enrichment analysis. (G) Flow cytometric analyses for the cell cycle of the indicated LSKs and LRPs. Mean ± SEM values are shown. **P < 0.01, ***P < 0.001.
Fig. S2.
(A) Statistical analysis of cell counts in PB from transplanted mice. Mean ± SEM values are shown. (B) A single cell was inspected using a microscope to acquire samples with only one cell captured. (C) Flow cytometric analyses for the cell cycle of the indicated LSKs and LRPs in Dnmt3aWT/WT and Dnmt3aR878H/WT mice.
Single-Cell Transcriptome Profiling of LSKs in Dnmt3aR878H/WT Leukemic Mice.
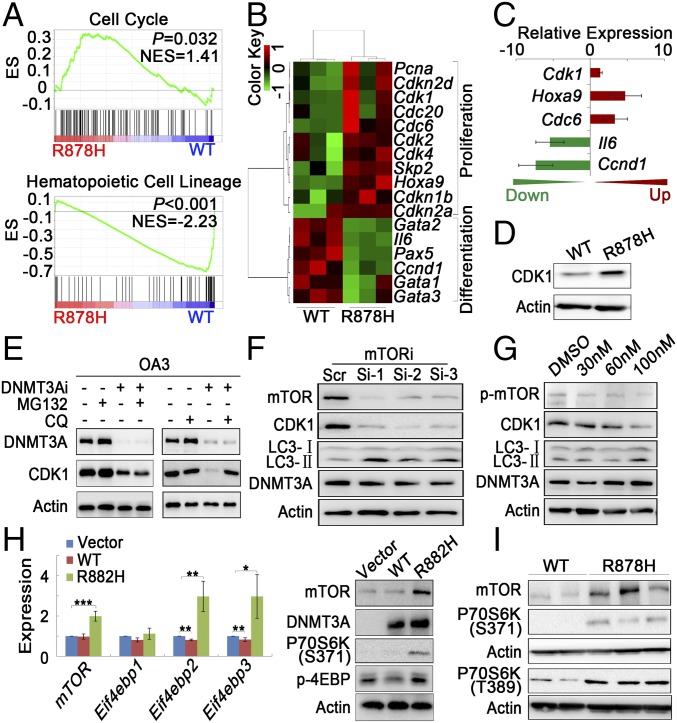
To obtain insight into the biological signature of leukemic progenitors, we compared gene expression profiles of LSKs between Dnmt3aWT/WT and Dnmt3aR878H/WT mice by using single-cell RNA sequencing (RNA-seq) (Fig. S2B). The possible differences in heterogeneity of gene expression in terms of functional categories between Dnmt3aWT/WT and Dnmt3aR878H/WT mice were further investigated (Dataset S3): We compared distance-to-the-median (DM) values of genes in mouse pairs for each Gene Ontology (GO term) as previously described (17). Altogether, 1,039 GO terms revealed significant differences in at least one pairwise comparison (P < 0.01) (Dataset S4). The expression of genes involved in cell-cycle regulation and transition were more heterogeneous in Dnmt3aR878H/WT mice than in Dnmt3aWT/WT ones (Fig. 2D). Some genes associated with leukemogenesis, such as Meis1, Hlf, Mpl, Hoxa7, Hoxa10, and Gata3 were up-regulated in LSKs of Dnmt3aR878H/WT mice (Fig. 2E). Principal component analysis (PCA) of the transcriptome demonstrated a separated LSK cluster, except for one sample, of Dnmt3aR878H/WT from LSKs derived from Dnmt3aWT/WT animals, revealing that most LSKs in diseased mice displayed distinct transcriptome identities. We then combined RNA-seq data of all the individual LSKs into Dnmt3aWT/WT and Dnmt3aR878H/WT groups to access their global expression profiles. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis (Dataset S5) showed that genes involved in the cell-cycle, chronic myeloid leukemia (CML), mTOR, and AML signaling pathways were overexpressed and responsible for the separation of leukemic and Dnmt3aWT/WT LSK clusters (Fig. 2F). These results were consistent with the cell-cycle status of LSKs as determined by flow cytometric analysis showing the transition of G0/G1 to S/G2/M (Fig. 2G and Fig. S2C). In fact, many cell-cycle regulators, such as cyclin-dependent kinase (CDK) family genes (including Cdk11b, Cdk6, Cdkn1a, and Cdk20), which are involved in this transition and presumably cause cell proliferation, were overexpressed (Dataset S3).
Transcriptome Profiling of Gr-1+ Leukemic Cells in Dnmt3aR878H/WT Mice.
The majority of leukemic cells in Dnmt3aR878H/WT mice were Gr-1+ cells, which could originate from the LSK population. Therefore, it is reasonable to hypothesize that Gr-1+ leukemic cells might possess migratory and invasive capacities resulting in the unique disease phenotype. To further address the mechanism underlying leukemogenesis, we performed transcriptome profiling through RNA-seq of Gr-1+ leukemic cells (Dataset S6). The Dnmt3a R878H allele was transcriptionally expressed. Gene set enrichment analysis (GSEA) showed a statistically significant enrichment of the cell-cycle pathway in the Gr-1+ cells of Dnmt3aR878H/WT mice compared with Dnmt3aWT/WT mice. By contrast, the enrichment of the mature hematopoietic cell-lineage pathway was prominent in Gr-1+ cells of Dnmt3aWT/WT mice (Fig. 3A). At individual gene levels, many genes associated with proliferation, such as Pcna and Cdk family members (Cdk1 and Cdk2), were overexpressed in Gr-1+ cells, which was consistent with the results in the LSKs of Dnmt3aR878H/WT mice. However, a number of hematopoietic differentiation-related genes, such as Gata1 to 3 and Pax5, were expressed at a low level in the Gr-1+ cells of Dnmt3aR878H/WT mice (Fig. 3B). A portion of the differentially expressed genes were confirmed by real-time RT-PCR (Fig. 3C).
Fig. 3.
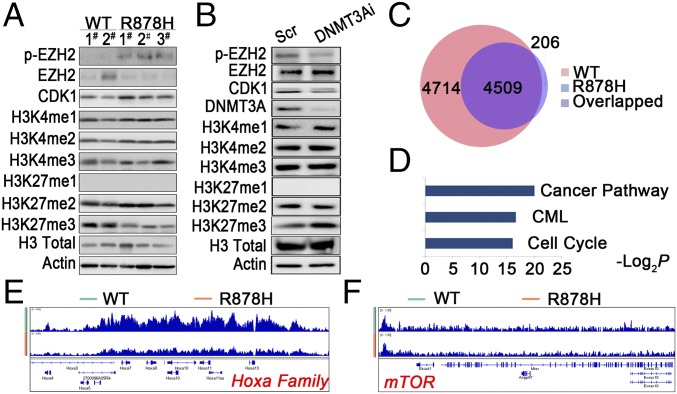
Transcriptome profiling of Gr-1+ leukemic cells and mTOR-mediated CDK1 overexpression induced by Dnmt3a mutation. (A) GSEA analysis of differentially expressed genes in Gr-1+ cells. NES, normalized enrichment score. (B) Heatmap of hematopoietic cell proliferation- and differentiation-related genes that are differentially expressed in Dnmt3aWT/WT and Dnmt3aR878H/WT mice. Red and green colors indicate up-regulated and down-regulated genes, respectively. (C) Gene expression levels were validated by RT-PCR. (D) Lysates from BM cells of mice were analyzed by Western blot with the indicated antibodies. (E) OA3 cells transfected with scrambled (DNMT3Ai−) or DNMT3A siRNA (DNMT3Ai+) were treated with MG132 or CQ, and their lysates were analyzed with Western blot. (F and G) Lysates from OA3 cells transfected with three siRNAs (1 to 3) against different mTOR regions or treated with different doses of rapamycin were analyzed with Western blot. (H) Results of RT-PCR and Western blot analysis for mTOR and its downstream genes in NIH 3T3 cells transfected with vehicle, WT, or R882H DNMT3A. (I) Lysates from BM cells of mice were analyzed with Western blot. Mean ± SEM values are shown. *P < 0.05, **P < 0.01, ***P < 0.001.
Dnmt3a Mutation Increases CDK1 Protein Level Through mTOR Activation.
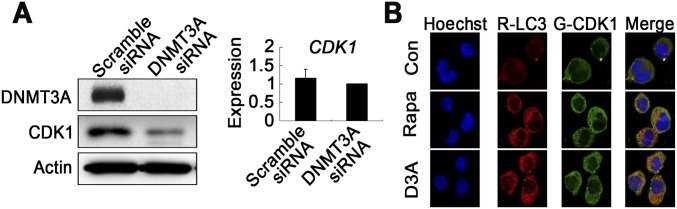
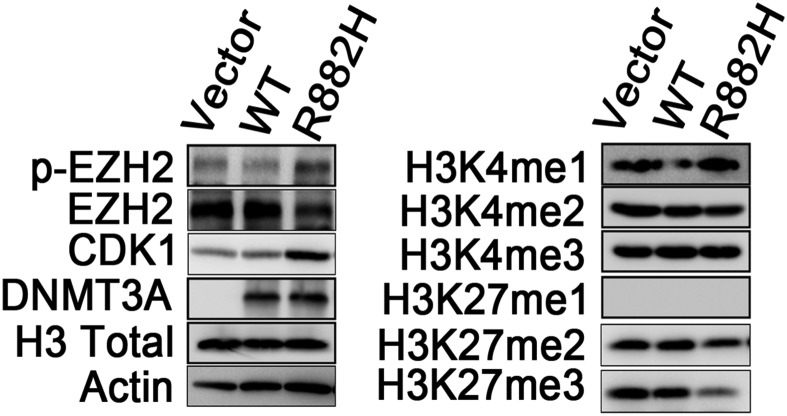
To further explore the mechanism underlying Dnmt3aR878H/WT phenotypic defects, we also tried to identify abnormal regulatory networks through protein-level analysis. To this end, CDK1 overexpression was discovered while screening proteins associated with the cell cycle in BM cells of Dnmt3aR878H/WT mice compared with Dnmt3aWT/WT animals (Fig. 3D). To facilitate detailed biochemical and molecular studies and ultimately clarify the mechanism by which the DNMT3A mutation induces CDK1 up-regulation, we used two cell models, the human cell line OCI-AML3 (OA3) harboring the DNMT3A R882C mutation and the murine fibroblast line NIH 3T3 stably expressing DNMT3A R882H. Knockdown of mutant protein expression with RNAi substantially decreased CDK1 protein, but not its mRNA, in OA3 cells, suggesting that the DNMT3A R882 mutation could affect the turnover of CDK1 protein (Fig. S3A). Furthermore, we applied the lysosome inhibitor chloroquine (CQ) or the proteasome inhibitor MG132 to OA3 cells in which knockdown of DNMT3A mutant protein was carried out. The result showed that CQ, but not MG132, reversed CDK1 expression levels, indicating that mutant DNMT3A could up-regulate CDK1 by inhibiting the autophage-lysosome pathway (Fig. 3E). Because mTOR is a key negative regulator of autophagy (18), we treated OA3 cells with specific RNAi against mTOR or the mTOR-specific inhibitor rapamycin. Indeed, CDK1 expression decreased with autophagy activation, demonstrated by an increased expression of the autophagy marker LC3-II (Fig. 3 F and G). Immunofluorescence staining revealed that CDK1 was partly colocalized with LC3. In addition, autophagy activity was further assessed by visualizing a punctate LC3 pattern using confocal microscopy in OA3 cells with knockdown of mutant DNMT3A or treatment with rapamycin (Fig. S3B). Notably, both mRNA and protein expression levels of mTOR and its downstream activated proteins, including phosphorylated P70S6K and 4EBP, were considerably higher in NIH 3T3 cells expressing DNMT3A R882H than in cells bearing vehicle or WT DNMT3A (Fig. 3H). In agreement with these data in the cell lines, the expression levels of mTOR in BM cells were much higher in Dnmt3aR878H/WT than in Dnmt3aWT/WT mice (Fig. 3I).
Fig. S3.
(A) Gene and protein expression levels of CDK1 in OA3 cells transfected with scrambled or DNMT3A siRNA. Mean ± SEM values are shown. (B) Immunofluorescence staining for CDK1 (green) and LC3 (red) in OA3 cells with DAPI staining (blue) defining the nucleus. These cells were treated with rapamycin or transfected with siRNA of DNMT3A.
Dnmt3a Mutation Promotes mTOR Expression via Hypomethylation.
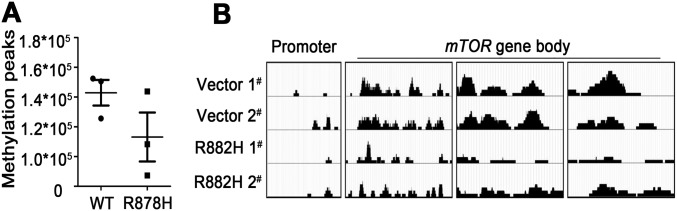
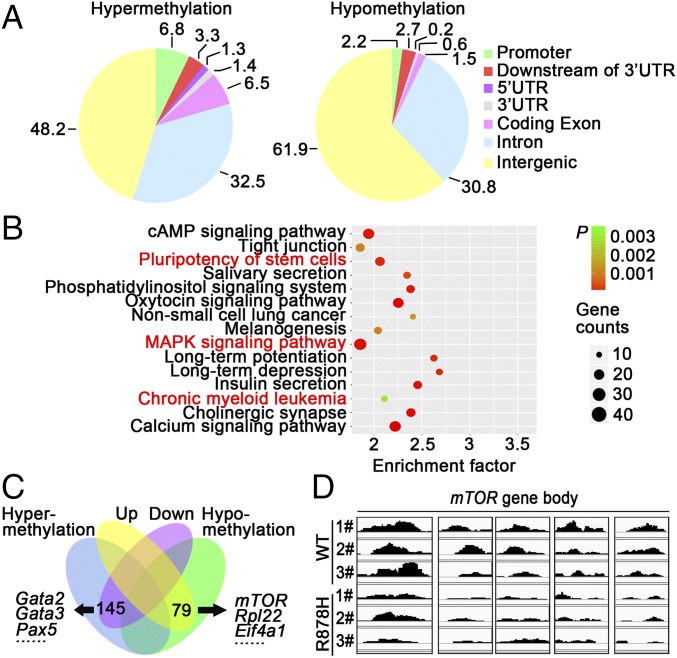
In view of the gene transcriptional changes of a large number of pathways, including mTOR in BM cells of Dnmt3aR878H/WT mice and the previous finding that DNMT3A mutations could alter DNA methyltransferase activity (2, 10), we hypothesized that the disturbance in genomic methylation might play a role in causing the aberrant transcriptome. Therefore, we performed methylated DNA immunoprecipitation (MeDIP) sequencing for Gr-1+ cells from both Dnmt3aWT/WT and Dnmt3aR878H/WT mice (Dataset S7). Although the number of DNA methylation peaks was not significantly different between the two groups, changes in hypo- and hypermethylation patterns occurred regionally throughout the genome (Fig. S4A). We then determined the DNA hypo- and hypermethylation distribution in six regions of gene structure, namely the promoter, downstream of the 3′ UTR, 5′UTR, 3′UTR, coding exon, intron, and intergenic region (Fig. 4A). Functional analysis of different methylation regions (DMRs) showed the enrichment of pathways involved in the regulation of the pluripotency of stem cells, MAPK signaling, and CML (Fig. 4B). The combination of RNA-seq and MeDIP-seq revealed 145 down-regulated and hypermethylated genes, including Gata2, Gata3, and Pax5, as well as 79 up-regulated and hypomethylated genes, such as Rpl22, Eif4a1, and mTOR (Fig. 4C and Dataset S8). Moreover, the MeDIP-seq data of leukemic cells from the same diseased mice showed hypomethylation of the mTOR gene in its body region (Fig. 4D). These results were consistent with those of a previously established DNMT3A R882H-induced CMML mouse model (Fig. S4B). Taken together, our results demonstrated that hypomethylation induced by mutant DNMT3A contributed to mTOR up-regulation, and mTOR overexpression in turn inhibited the autophagy pathway, leading to aberrantly increased CDK1 expression.
Fig. S4.
(A) Number of methylation peaks was compared between Dnmt3aWT/WT and Dnmt3aR878H/WT mice. Mean ± SEM values are shown. (B) MeDIP-seq analysis of the mTOR gene showed a local hypomethylation pattern in the gene body, whereas the promoter region exhibited no changes of methylation status in mice transduced with R882H DNMT3A compared with mice transduced with WT DNMT3A.
Fig. 4.
Dnmt3a mutation promotes mTOR expression via hypomethylation. (A) Pie charts showing the average proportions of peaks in each region defined by genomic structure. Peak ratios of hypo- and hypermethylation are shown. (B) Functional analysis of DMRs in murine Gr-1+ cells showed a series of enrichments associated with pluripotency of stem cells and CML. (C) Venn diagram of the overlapping genes enriched in DMRs and differentially expressed genes. (D) MeDIP-seq analysis of the mTOR gene showed a local hypomethylation pattern in the gene body in Gr-1+ cells from Dnmt3aR878H/WT mice compared with that of the Dnmt3aWT/WT control.
Modulation of EZH2 by CDK1 Leads to Changes of H3K27 Trimethylation Enrichment Pattern in Dnmt3aR878H/WT Mice.
It was previously reported that CDK1-mediated phosphorylation of EZH2 at threonine 487 could suppress methylation of H3K27 (19, 20). We hypothesized that a similar mechanism could exist in our leukemia model and thus bring another layer of abnormal epigenetic regulation. To explore the possible influence of histone methylation caused by a DNMT3A mutant, we compared the methylation status of histone 3 in BM cells of Dnmt3aWT/WT and Dnmt3aR878H/WT mice. H3K27 trimethylation (H3K27me3) was found decreased whereas H3K4 showed no obvious changes in BM cells of Dnmt3aR878H/WT mice. Meanwhile, EZH2 phosphorylation at threonine (p-EZH2-487) was increased in leukemic BM cells (Fig. 5A). We also examined histone 3 methylation patterns in OA3 and 293T cell models. In OA3 cells treated with DNMT3A siRNA, H3K27me3 was increased whereas p-EZH2-487 was decreased compared with cells treated with scrambled siRNA (Fig. 5B). In 293T cells transduced with R882H DNMT3A, similar patterns of H3K27 and H3K4 methylation were observed in comparison with control (Fig. S5A). These results not only provided evidence that the DNMT3A mutant inhibited H3K27me3 by inducing CDK1-mediated phosphorylation of EZH2 but also encouraged us to evaluate in a systematic way the impact of decreased H3K27me3 on gene expression. We then performed a global ChIP-seq analysis of H3K27me3 on the BM cells of Dnmt3aR882H/WT and Dnmt3aWT/WT mice. The data showed that the peaks of H3K27me3 binding were distributed mainly in the transcription start site regions in both leukemia and WT samples. Notably, the number of genes with enrichment of H3K27me3 was significantly decreased in leukemic cells compared with WT samples (Fig. 5C). Among the genes with reduced H3K27me3 enrichment levels, those related to pathways such as “cancer,” “CML,” and “cell cycle” drew our attention (Fig. 5D). For example, the Hoxa family was found to have decreased enrichment of H3K27me3, in concordance with the transcriptional activation of several Hoxa genes (Fig. 5E). However, there was no major change of H3K27me3 status in the mTOR gene (Fig. 5F). Hence, mTOR activation could more likely be caused by decreased DNA methylation (Fig. 4D).
Fig. 5.
CDK1-mediated EZH2 phosphorylation resulted in an abnormal H3K27me3 profile. (A) Lysates from BM cells of mice were analyzed with Western blot by using the indicated antibodies. (B) Lysates from OA3 cells transfected with siRNA against DNMT3A were analyzed with Western blot. (C) The number of peaks of H3K27me3 in leukemic cells compared with WT samples. (D) KEGG enrichment analysis of genes with loss of enrichment of H3K27me3 in leukemic mice. (E and F) ChIP-seq profiles of H3K27me3 at the Hoxa family genes and mTOR.
Fig. S5.
Lysates from 293T cells overexpressing vector, WT-DNMT3A, or DNMT3A R882H were analyzed by Western blot with the indicated antibodies.
Rapamycin Inhibits Proliferation of Human DNMT3A Mutation-Related Leukemia Cell Lines and Leukemic Cells in Dnmt3aR878H/WT Mice.
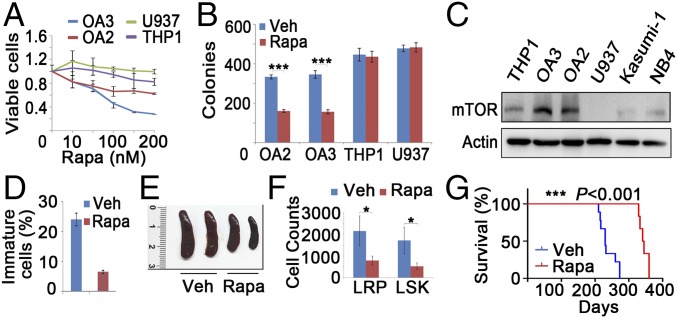
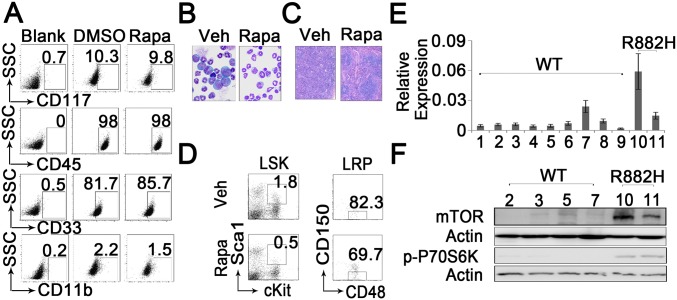
In an attempt to identify potential therapeutic agents against leukemia associated with DNMT3A mutations, we found a relatively specific inhibition of the proliferation of OA3 and OCI-AML2 (OA2) cells by rapamycin (Fig. 6A). We then performed a colony-formation assay and morphological analysis of leukemic cells with or without the DNMT3A mutation in the presence of rapamycin for 2 wk. The number of colonies carrying the DNMT3A mutation significantly decreased after treatment with rapamycin (Fig. 6B), whereas no changes of surface marker expression were found in OA3 colony cells (Fig. S6A). To determine the reason behind the high sensitivity of cells carrying the DNMT3A mutation to rapamycin, we examined the expression of mTOR in different subtypes of AML cell lines. OA2 and OA3 cells exhibited a higher mTOR expression than cells with WT DNMT3A (Fig. 6C). We further determined the in vivo effect of rapamycin (4 mg/kg for 4 mo) on BM cells in Dnmt3aR878H/WT mice. Indeed, the compound significantly reduced the percentage of immature cells in the BM (Fig. 6D and Fig. S6B) and the leukemic cell infiltration in the spleen (Fig. 6E and Fig. S6C). mTOR has been recently reported to play a role in adaptive transition of quiescent stem cells from G0 to GAlert (21). Similarly, the percentage and number of both LSK and LRP cells significantly decreased in Dnmt3aR878H/WT mice treated with rapamycin (Fig. 6F and Fig. S6D). Upon treatment with rapamycin, survival of Dnmt3aR878H/WT mice significantly improved compared with controls (Fig. 6G). Furthermore, we also examined fresh BM samples of AML patients. RT-PCR and Western blot analysis demonstrated that samples carrying DNMT3A R882H exhibited a higher expression of mTOR than those with WT DNMT3A (Fig. S6 E and F). These results suggested that rapamycin inhibits proliferation of leukemic cells carrying the DNMT3A mutation.
Fig. 6.
Rapamycin inhibits the proliferation of Dnmt3aR878H/WT and human DNMT3A mutated leukemic cells. (A) Response of OA3, OA2, THP1, and U937 cells to rapamycin (rapa). (B) Colony-formation assay of AML-related cells treated with rapamycin for 2 wk. Veh, vehicle. (C) Lysates of cells from different AML subtypes were analyzed with Western blot. (D) Statistical analysis of the number of immature cells in the BM treated with veh or rapa for 4 mo. (E) Macroscopic appearance of the spleen from Dnmt3aR878H/WT mice treated with veh or rapa for 4 mo. (F) Numbers of LSKs and LRPs in Dnmt3aR878H/WT mice treated with veh or rapa for 4 mo. (G) Kaplan–Meier survival analysis of Dnmt3aR878H/WT mice treated with veh (n = 8) or rapa (n = 6) (4 mg/kg) for 4 mo. Mean ± SEM values are shown. *P < 0.05, ***P < 0.001.
Fig. S6.
(A) Representative flow cytometric plots of AML-related cells treated with DMSO and rapamycin. (B) Morphological analysis of BM with Wright’s staining in Dnmt3aR878H/WT mice treated with vehicle (veh) and rapamycin for 4 mo. (C) Histological analysis of spleen from Dnmt3aR878H/WT mice treated with vehicle and rapamycin for 4 mo. (D) Representative flow cytometric plots and statistical analysis of BM cells obtained from Dnmt3aR878H/WT mice treated with vehicle and rapamycin for 4 mo. (E) RNA expression levels of mTOR in BM samples obtained from AML patients. Mean ± SEM values are shown. (F) Protein expression levels of mTOR in BM samples obtained from AML patients.
Discussion
The importance of DNMT3A in hematopoiesis and leukemogenesis has been demonstrated by studies using Dnmt3a knockout or DNMT3A R882-overexpressing mice (7, 12, 22). In this study, the conditional knockin mouse model allowed the inducible allelic switch from Dnmt3aWT/WT to Dnmt3aR878H/WT in the hematopoietic system of adult animals, which indeed possessed some major features of human AML associated with the DNMT3A mutation. This strategy bypassed the embryonic lethality associated with Dnmt3a R878H expression, thus providing a relevant model for studying the target genes regulated by Dnmt3a R878H. Our data therefore highlight the crucial roles of endogenous expression of mutant Dnmt3a R878H in malignant hematopoiesis. Considering that most of the reported DNMT3A mutations in human myeloid neoplasms are heterozygous (1, 2), we observed a transplantable AML phenotype in Dnmt3aR878H/WT heterozygous animals whereas homozygous mice showed growth inhibition and died soon after birth. The leukemia phenotype in mice bore some similarities to that in a Dnmt3a−/− murine model (23). Our results showed that leukemic LSKs were composed mostly of LRPs and contained leukemia-initiating cells. These data indicated that Dnmt3a mutation could promote proliferation of leukemic stem/progenitor cells by accelerating the transition of HSPCs from a quiescent status to an active one. However, it was just reported that a knockin of the same allele did not cause leukemia on its own with a 2-y follow-up (24). The generation of distinct phenotypes might be caused by different conditional knockin design, mice strain, and so forth. Moreover, we found that differentiation of erythroid progenitors was arrested, consistent with previous reports (25).
In AML patients studied with exome sequencing, 4 out of 51 cases had the DNMT3A mutation as the only known abnormal genetic event (26). In our Dnmt3aR878H/WT mice, exome sequencing and RNA-seq revealed the Dnmt3a mutation in leukemic cells without recurrent mutations of genes that are well-known coexisting events in human AML with a DNMT3A abnormality. It is worth noting that our leukemia model is a relatively moderate one, because the disease developed after a relatively long latency period (4 to 6 mo) and displayed a quite lengthy median survival time (230 d), and the average percentage of immature cells in the BM was only 23%. Hence, although our model proved the leukemogenic effect of the DNMT3A mutation, it may provide tools to study the role of cooperating mutations for a more aggressive full-blown AML to occur.
In the mechanism study, our Dnmt3a R878H mice have shown that leukemic cells have altered gene expression and epigenetic regulatory patterns contributing to the growth/survival advantage over WT mice. Single-cell RNA-seq of LSKs revealed that the gene expression heterogeneity in leukemic samples seemed to be higher than in WT controls. This, together with the significantly increased number of LSKs/LRPs in Dnmt3aR878H/WT mice, could underlie the clonal diversity and evolutionary potential of leukemia-initiating cells along with the disease course. Moreover, we found that clusters of genes, such as Hoxa family, cell-cycle, and mTOR signal pathways, which are activated in leukemic progression, were overexpressed. A recent study shed light on Dnmt3b having synergistic effects with Dnmt3a in leukemogenesis, which might contribute to the coexistence of hyper- and hypomethylated genes in Dnmt3aR878H/WT cells (27). These data have not only confirmed but also largely extended the knowledge that we previously acquired from samples obtained from AML patients.
The DNMT3A R882H mutation enhances expression of CDK1, which facilitates transition of hematopoietic cells to the S/G2/M phase. mTORC1 activity is necessary and sufficient for the transition of stem cells from a quiescent status to an active one (21). The present study found that the DNMT3A mutation contributed to hypomethylation in the gene body region of mTOR in Dnmt3aR878H/WT mice. The resultant overexpression of mTOR inhibited the autophage pathway and finally led to an aberrantly high level of CDK1. CDK1 phosphorylated EZH2 at T487, attenuated the ability of this enzyme to catalyze trimethylation of H3K27, and possibly influenced an array of genes associated with leukemic progression such as the Hoxa family.
Finally, our in vitro and in vivo data indicated a potential antitumor activity of the mTOR inhibitor in DNMT3A-mutated leukemia. AML cell lines with the DNMT3A mutation were responsive to rapamycin treatment, and the differential sensitivity displayed may be determined by the expression levels of mTOR. Consistent with in vitro results, our findings revealed that rapamycin elicited a therapeutic response in Dnmt3aR878H/WT mice. Hence, we would like to suggest that further possible preclinical and clinical studies of mTOR inhibitors be carried out in AML-M5 and -M4 subtypes with DNMT3A mutations.
Materials and Methods
Generation of Dnmt3aR878H/WT Mx1Cre+ Mice and Breeding.
Dnmt3aR878H/WT mice were obtained from the Nanjing Biomedical Research Institute of Nanjing University. Targeting vectors PL253 used to establish Dnmt3a R878H conditional knockin mice were generated through recombineering (Fig. S1A). Subsequent genotyping of offspring was performed by PCR analysis with primers specific for the Dnmt3a R878H mutation (Datasets S9 and S10). To activate transgene expression in vivo, we administered pIpC to the animals at the time of weaning (4 wk old). Details of treatment protocols are available in SI Materials and Methods. Mice were used according to animal care standards, and all protocols were approved by the Committee on Animal Use for Research at the Shanghai Jiao Tong University School of Medicine.
cDNA Library Preparation from Single Cells Using Fluidigm C1.
For each culture condition, 3,000 to 4,000 sorted LSKs were loaded onto a 10- to 17-mm Fluidigm C1 Single-Cell Auto Prep Integrated Fluidic Circuit (IFC), and cells were captured according to the manufacturer’s instructions. Libraries from one chip were pooled, and paired-end 200-bp sequencing was performed on four lanes of an Illumina MiSeq. We sequenced an average of over 4.4 million reads per cell (details are in SI Materials and Methods).
Treatment of Dnmt3aR878H/WT Mice with Rapamycin.
Five weeks after the birth of induced Dnmt3aR878H/WT mice, they were subjected to the following treatment protocols: normal saline (vehicle) or rapamycin administered at 4 mg/kg twice a wk for 4 wk, followed by the administration of the same dose once per wk for 4 mo.
The other procedures, including Western blot, flow cytometric analysis, RNA-seq, and MeDIP-seq, are described in SI Materials and Methods. All patients gave informed consent for research use. The research involving humans was approved by the institutional review board of Rui Jin Hospital.
SI Materials and Methods
Generation of Dnmt3aR878H/WT Mx1Cre+ Mice and Breeding.
Dnmt3aR878H/WT mice were obtained from the Nanjing Biomedical Research Institute of Nanjing University. Targeting vectors PL253 used to establish Dnmt3a R878H conditional knockin mice were generated through recombineering (Fig. S1A). The targeting vectors were electroporated into cultured embryonic stem cells obtained from the B6/BLU mouse strain. The correctly targeted ES clones were identified by Southern blot analysis with the restriction enzymes Apa1, Sca1, and Bcl1 and long-range PCR. The pIpC-inducible Dnmt3aR878H allele was created following the scheme shown in Fig. S1A and backcrossed into the C57BL/6 background. Subsequent genotyping of offspring was performed by PCR analysis with primers specific for the Dnmt3a R878H mutation. To activate transgene expression in vivo, we administered pIpC to the animals at the time of weaning (4 wk old). In all experiments, Mx1Cre-knockin mice were heterozygous for both the Dnmt3aR878H allele and the Mx1Cre allele (Dnmt3aR878H/WTMx1Cre+ or Dnmt3aR878H/WT). Mice were used according to animal care standards, and all protocols were approved by the Committee on Animal Use for Research at the Shanghai Jiao Tong University School of Medicine. Primers for long-range PCR and identifying the genotype of mice are listed in Dataset S9.
cDNA Library Preparation from Single Cells Using Fluidigm C1.
For each culture condition, 3,000 to 4,000 sorted LSKs were loaded onto a 10- to 17-mm Fluidigm C1 Single-Cell Auto Prep IFC, and the cells were captured according to the manufacturer’s instructions. The capture efficiency was inspected using a microscope to remove samples that captured more than one cell from the analysis (Fig. S2B). cDNA was harvested and diluted to an average of 0.2 ng/mL, and Nextera libraries were prepared using the Nextera XT DNA Sample Preparation Kit and Nextera Index Kit (Illumina) following the manufacturer’s instructions. Libraries from one chip were pooled, and paired-end 200-bp sequencing was performed on four lanes of an Illumina MiSeq. We sequenced an average of over 4.4 million reads per cell. Over 94% of reads were mapped to the Mus musculus genome (GRCm38).
Bulk RNA-Seq.
Bulk RNA-sequencing libraries were prepared and sequenced using the Wellcome Trust Sanger Institute sample preparation pipeline with Illumina’s TruSeq RNA Sample Preparation Kit, version 2. RNA was extracted from leukemic cells using TRIzol-isopropanol precipitation. The quality of the RNA was checked using Agilent RNA 6000 Nano Reagents with a 2100 Bioanalyzer. For library preparation, poly-A RNA was purified from total RNA using oligo-dT magnetic pull-down. Subsequently, mRNA was fragmented using metal ion-catalyzed hydrolysis. cDNA was synthesized using random hexamer priming, and end repair was performed to obtain blunt ends. A-tailing was done to enable subsequent ligation of Illumina paired-end sequencing adapters, and samples were multiplexed at this stage. The resulting library was amplified using 10 cycles of PCR, substituting the Kapa Hifi polymerase for the polymerase in the Illumina TruSeq Kit. Samples were checked using Agilent High Sensitivity DNA Reagent with a 2100 Bioanalyzer, and 200-bp paired-end sequencing was carried out on an Illumina MiSeq.
Reagents.
Rapamycin was obtained from Selleck (S1039). MG132 and CQ were obtained from Sigma-Aldrich (M7449 and C6628).
Western Blot Analysis.
Antibodies were purchased from Cell Signaling Technology (anti-DNMT3A, anti-CDK1, anti-FLAG, anti-GFP; anti-mTOR signaling pathway kit; Cell Signaling Technology), Abcam (anti-CDK1), and Santa Cruz (anti-actin).
Immunofluorescence.
Monolayer cells of OA3 were fixed with 4% paraformaldehyde and stained with primary antibodies specific for LC3 (Cell Signaling Technology) and CDK1 (Abcam). After washing, cells were incubated with the appropriate secondary antibodies. Nuclei were counterstained with Hoechst.
Flow Cytometric Analysis and Cell Sorting.
Cells were suspended in FACS buffer (PBS containing 1% FBS) and subsequently stained with fluorochrome-conjugated antibodies. Apoptosis was detected by the Annexin V Apoptosis Detection Kit (BD Biosciences). Flow cytometric data were collected using an LSR II flow cytometer (Becton Dickinson) and analyzed with FlowJo software (TreeStar). Cells were sorted by MoFlo (Beckman Coulter) into 1× PBS buffer.
Quantitative Real-Time RT-PCR.
The assays were performed as described using reagents according to instructions of the manufacturer (SYBR Premix Ex Taq; Takara). Reactions were performed on the ABI PRISM 7500 Fast Real-Time PCR System. Data were analyzed by using the formula 2−∆∆Ct. Primers used for RT-PCR are listed in Dataset S10.
Cell-Proliferation Analysis.
Cells were treated with inhibitors at various concentrations in a 96-well plate. After 48 h, 10 μL CCK8 was added to each well. The samples were incubated at 37 °C for 4 to 6 h and the absorbance was measured at 450 nm by spectrophotometry.
Methylation Analysis Through MeDIP-Seq.
DNA (3 μg) prepared from sorted murine Gr-1+ BM cells for each knockin mouse was used for MeDIP sequencing by Shanghai Biotechnology. The in vitro methylation assay was performed as described. After MeDIP, the remaining DNA was PCR-amplified using sequencing primers and used for sequencing (Illumina HiSeq 2500). Raw reads were preprocessed using the FASTX-Toolkit (hannonlab.cshl.edu/fastx_toolkit/index.html). MeDIP-seq peaks were calculated using MACS (Multiproject Automated Control System; liulab.dfci.harvard.edu/MACS/Download.html). The depth of genomic regions was calculated and plotted using IGVTools (software.broadinstitute.org/software/igv/download).
Whole-Exome Sequencing.
Genomic DNA of leukemic BM cells and paired normal tail specimens of Dnmt3aR878H/WT mice was extracted, and WES was performed with an average yield of 11.1 Gb per sample. Raw sequences were aligned to mouse genome build mm10 by using BWA software (bio-bwa.sourceforge.net/). SNV and indel analysis were performed using MuTect (www.broadinstitute.org/cancer/cga/mutect) and Strelka (ftp://strelka@ftp.illumina.com/).
siRNA Transfection Assay.
Cells were transfected with an siRNA oligomer of DNMT3A (12) and mTOR (28–30) by using Lipofectamine 2000 Transfection Reagent (Invitrogen).
Supplementary Material
Acknowledgments
We thank all of our colleagues at the Shanghai Institute of Hematology for constructive discussions and technical help. This work was supported by the National Key Basic Research Program of China (2013CB966800), Ministry of Health Grant 201202003, Mega-Projects of Scientific Research for the 12th Five-Year Plan (2013ZX09303302), National Natural Science Foundation of China (81123005, 81222004, and 81570151), Samuel Waxman Cancer Research Foundation, Shanghai Talent Development Fund 201458, Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant 20152507, Center for HPC Shanghai Jiao Tong University, and Innovation Foundation for Doctoral Students of Shanghai Jiao Tong University School of Medicine Grant BXJ201611.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703476114/-/DCSupplemental.
References
- 1.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan XJ, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 3.Walter MJ, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25:1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neumann M, et al. Whole-exome sequencing in adult ETP-ALL reveals a high rate of DNMT3A mutations. Blood. 2013;121:4749–4752. doi: 10.1182/blood-2012-11-465138. [DOI] [PubMed] [Google Scholar]
- 5.Shen Y, et al. Gene mutation patterns and their prognostic impact in a cohort of 1185 patients with acute myeloid leukemia. Blood. 2011;118:5593–5603. doi: 10.1182/blood-2011-03-343988. [DOI] [PubMed] [Google Scholar]
- 6.Thol F, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011;29:2889–2896. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- 7.Challen GA, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2011;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shlush LI, et al. HALT Pan-Leukemia Gene Panel Consortium Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SJ, et al. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood. 2013;122:4086–4089. doi: 10.1182/blood-2013-02-483487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russler-Germain DA, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014;25:442–454. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neri F, et al. Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell. 2013;155:121–134. doi: 10.1016/j.cell.2013.08.056. [DOI] [PubMed] [Google Scholar]
- 12.Xu J, et al. DNMT3A Arg882 mutation drives chronic myelomonocytic leukemia through disturbing gene expression/DNA methylation in hematopoietic cells. Proc Natl Acad Sci USA. 2014;111:2620–2625. doi: 10.1073/pnas.1400150111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X, et al. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Celik H, et al. Enforced differentiation of Dnmt3a-null bone marrow leads to failure with c-Kit mutations driving leukemic transformation. Blood. 2015;125:619–628. doi: 10.1182/blood-2014-08-594564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayle A, et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2015;125:629–638. doi: 10.1182/blood-2014-08-594648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiel MJ, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 17.Kolodziejczyk AA, et al. Single cell RNA-sequencing of pluripotent states unlocks modular transcriptional variation. Cell Stem Cell. 2015;17:471–485. doi: 10.1016/j.stem.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adams J. The proteasome: A suitable antineoplastic target. Nat Rev Cancer. 2004;4:349–360. doi: 10.1038/nrc1361. [DOI] [PubMed] [Google Scholar]
- 19.Wei Y, et al. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol. 2011;13:87–94. doi: 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu SC, Zhang Y. Cyclin-dependent kinase 1 (CDK1)-mediated phosphorylation of enhancer of zeste 2 (Ezh2) regulates its stability. J Biol Chem. 2011;286:28511–28519. doi: 10.1074/jbc.M111.240515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodgers JT, et al. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert) Nature. 2014;510:393–396. doi: 10.1038/nature13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang L, et al. DNMT3A loss drives enhancer hypomethylation in FLT3-ITD-associated leukemias. Cancer Cell. 2016;29:922–934. doi: 10.1016/j.ccell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guryanova OA, et al. Dnmt3a regulates myeloproliferation and liver-specific expansion of hematopoietic stem and progenitor cells. Leukemia. 2016;30:1133–1142. doi: 10.1038/leu.2015.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guryanova OA, et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat Med. 2016;22:1488–1495. doi: 10.1038/nm.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, et al. DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat Genet. 2016;48:1014–1023. doi: 10.1038/ng.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ley TJ, et al. Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Y, et al. Loss of Dnmt3b accelerates MLL-AF9 leukemia progression. Leukemia. 2016;30:2373–2384. doi: 10.1038/leu.2016.112. [DOI] [PubMed] [Google Scholar]
- 28.Wen ZH, et al. Critical role of arachidonic acid-activated mTOR signaling in breast carcinogenesis and angiogenesis. Oncogene. 2013;32:160–170. doi: 10.1038/onc.2012.47. [DOI] [PubMed] [Google Scholar]
- 29.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 30.Iwamaru A, et al. Silencing mammalian target of rapamycin signaling by small interfering RNA enhances rapamycin-induced autophagy in malignant glioma cells. Oncogene. 2007;26:1840–1851. doi: 10.1038/sj.onc.1209992. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.