Abstract
Voltage-gated calcium ion channels are essential for numerous biological functions of excitable cells and there is wide spread appreciation of their importance as drug targets in the treatment of many disorders including those of cardiovascular and nervous systems. Each Cacna1 gene has the potential to generate a number of structurally, functionally, and in some cases pharmacologically unique CaVα1 subunits through alternative pre-mRNA splicing and the use of alternate promoters. Analyses of rapidly emerging deep sequencing data for a range of human tissue transcriptomes contain information to quantify tissue-specific and alternative exon usage patterns for Cacna1 genes. Cell-specific actions of nuclear DNA and RNA binding proteins control the use of alternate promoters and the selection of alternate exons during pre-mRNA splicing, and they determine the spectrum of protein isoforms expressed within different types of cells. Amino acid compositions within discrete protein domains can differ substantially among CaV isoforms expressed in different tissues, and such differences may be greater than those that exist across CaV channel homologs of closely related species. Here we highlight examples of CaV isoforms that have unique expression patterns and that exhibit different pharmacological sensitivities. Knowledge of expression patterns of CaV isoforms in different human tissues, cell populations, ages, and disease states should inform strategies aimed at developing the next generation of CaV channel inhibitors and agonists with improved tissue-specificity.
Keywords: Alternative splicing, Cacna1 genes, dihydropyridines, morphine, splicing factors, voltage-gated calcium channels, ziconotide
1. INTRODUCTION
The functional core of the voltage-gated calcium ion channel is the CaVα1 subunit and each may be associated with a cytoplasmic calcium channel CaVβ subunit, an extracellular CaVα2δ subunit, and other proteins such as gamma subunits, calmodulin, scaffold proteins, kinase anchoring proteins, and downstream target proteins [1]. In mammals, 10 different genes (Cacna1A-1I and 1S) encode 10 CaVα1 subunits, and in humans these reside on nine different chromosomes. Cacna1 genes encode three main classes of CaVα1 proteins grouped according to their degree of sequence homology: CaV1 (1S, 1F, 1C, 1D), CaV2 (1A, 1B, 1E) and CaV3 (1G, 1H, 1I) [2–4]. Expansion within each of the 3 main Cacna1 gene families likely occurred some 450 million years ago probably as a result of genome doublings from single ancestral vertebrate Cacna1 genes [5]. For example, phylogenic analyses suggest that Cacna1s, Cacna1c, Cacna1d, and Cacna1f genes are located within paralogous chromosome regions (spread across four different chromosomes) that contain several gene families including visual opsins [5].
As individual calcium channel genes evolved, CaV proteins acquired new functions and phenotypes that account for their unique biological activities in specific tissues and cell types. For example, of the four mammalian Cacna1 genes that encode CaV1 subunits: Cacna1s expression is limited to skeletal muscle and CaV1.1 proteins contain a unique sequence in the II–III linker region that couples the channel to the ryanodine receptor. This is the critical first step in excitation-contraction coupling in skeletal muscle [6–8]; Cacna1f is highly expressed in retina but also in other human tissues (CACNA1F, the GTEx Portal on 12/06/2014) [9], and CaV1.4 channels are important for transmission at glutamatergic ribbon synapses between photoreceptors and second-order neurons [10–13]; and Cacna1c and Cacna1d genes have relatively broad expression patterns including in brain, heart, smooth muscle, endocrine cells, and auditory hair cells [1, 14] (CACNA1C and CACNA1D, the GTEx Portal on 12/06/2014) [9]. The expression of functionally different splice isoforms of CaV1.2 and CaV1.3 in different tissues enables Cacna1c and Cacna1d genes to support a wider range of calcium-dependent cellular functions [2, 3, 15–17].
2. PHARMACOLOGICAL TOOLS THAT DISCRIMINATE AMONG MAJOR SUBTYPES OF CaV CHANNELS
Pharmacological approaches are invaluable to delineate the cellular functions of different Cacna1 genes. For example, dihydropyridine (DHP) antagonists and agonists are the molecules of choice to establish the contributions that CaV1 channels make to various cellular responses, whereas the spider toxin ω-agatoxin IVA and the cone snail toxin ω-conotoxin GVIA are highly selective inhibitors of CaV2.1 and CaV2.2 channels respectively (Table 1). As is well known, and discussed throughout this special issue, DHP antagonists are used in the clinic to lower blood pressure. All four CaV1 channels are sensitive to DHPs, but their anti-hypertensive action in vivo is dominated by the inhibitory actions of several DHPs on vascular smooth muscle, DHPs have few side effects, a surprising fact considering the relatively broad expression patterns of CaV1.2 and CaV1.3 channels in many excitable cells.
Table 1.
CACNA1 Genes, Proteins, Currents.
| Gene name: human chromosome | Protein name | Exon, splicing factor | Current | Pharmacology (influenced by exon) |
|---|---|---|---|---|
|
| ||||
| CACNA1S: chr 1 | CaV1.1 | e29, rbFox [121] | L | DHP– Isradipine and nisoldipine |
|
|
||||
| CACNA1C: chr 12 | CaV1.2 | e8a, PTB [47] e9*, rbFox [48] e21/e22, ND e33, rbFox [48] |
L | (e8/e8a; e21/22) [77,78,81,84,85] Diltiazem (e9*) [79] |
|
|
||||
| CACNA1D: chr 3 | CaV1.3 | e8b, rbFox [121] e42a, ND |
L | DHP – Nimodipine (e42a) [80] |
|
| ||||
| CACNA1A: chr 19 | CaV2.1 | e24a, Nova(+) [49] e31a, Nova (−) [49] |
P/Q | ω-Agatoxin IVA (e31a) [61] |
|
| ||||
| CACNA1B: chr 9 | CaV2.2 | e24a, Nova(+) [122] e31a, Nova(−) [49] e18a, rbFox e37a/e37b, ND |
N | μ-opioid and GABAB receptors - DAMGO, Morphine, Baclofen [107–109] |
(+) indicates enhancer of exon inclusion and (−) repressor; ND = Not determined; All genes contain numerous sites of alternative splicing but only those for which the splicing factor has been determined, or that are known to influence drug sensitivity are listed here. For a more comprehensive review of alternative splicing in Cacna1 genes see [2, 3, 15, 17, 31, 47–49, 61, 77–80, 91, 92, 108, 109, 121–124].
As shown by Bruce Bean, and by Reuter and colleagues, the efficacy of DHP antagonists is strongly voltage-dependent and these drugs are more potent inhibitors of CaV1 channels that are activated from relatively depolarized membrane potentials typical of smooth muscle cells in vivo [18, 19]. Preferential inhibition of inactivated CaV1 channels also results in reduced inhibition by DHPs of CaV1 channels activated by brief, non-inactivating stimuli applied from negative membrane potentials – such as action potential-like waveforms [20]. CaV1.3 channels have different biophysical properties and have somewhat lower intrinsic sensitivities to certain DHPs as compared to CaV1.2 channels [21, 22]; although these differences are generally insufficient to permit unambiguous functional separation of CaV1.2 and CaV1.3 currents at cellular and behavioral levels. CaV1.3 channels support low threshold calcium entry during pacemaking in some cells [23–25] and data link de novo mutations in CACNA1D to increased risk of sporadic autism spectrum disorder [26]. Risk alleles for bipolar disorder, schizophrenia and other psychiatric illnesses have also been linked to CACNA1C [27–30]. Such findings have increased efforts to develop compounds to discriminate between CaV1.2 and CaV1.3 channels, as well as among their various splice isoforms. There is also evidence discussed below, that splice isoforms of CaV1.2 have different sensitivities to DHPs.
3. CELL-SPECIFIC ALTERNATIVE PRE-mRNA PROCESSING
All mammalian Cacna1 genes studied to date contain alternatively spliced exons and many have alternative promoters; these exons combine in different patterns according to cell-specific control, with the potential to generate thousands of CaV channels [2]. We know that alternative splicing of Cacna1 pre-mRNAs is extensive in mammals, but we still have only a cursory understanding of their cell-specific functions. Recent reviews have summarized what was known of the tissue-specific expression pattern of exon usage for several Cacna1 genes and, where tested, their functional impact on CaV channel properties [2, 15, 31, 32]. More recently, the Genotype-Tissue Expression (GTEx) project provides the most comprehensive publicly available dataset on the expression pattern of human genes, including CACNA1, and the quantitative comparison of individual exon usage, genome-wide, across different tissues [9].
Multi-exon genes have the capacity to generate several – sometimes thousands – of proteins with unique structures through cell-specific selection (or elimination) of certain exons. Alternatively spliced exons often encode autonomous protein modules that introduce or eliminate specific functions or that promote or inhibit coupling to downstream signaling cascades [33–35]. Cell-specific exon choice can involve the inclusion or exclusion of a cassette exon, the choice of one of a pair or more of mutually exclusive exons, or the use of alternate splice donor or acceptor sites (Fig. 1). Cell-specific inclusion or exclusion of certain exons can also introduce premature termination codons that promote nonsense-mediated decay (NMD) thereby regulating the overall protein expression level (Fig. 1). For example, e20a in Cacna1b introduces a premature stop codon if inserted during pre-mRNA splicing and CaV2.2 mRNAs containing this exon are present in brain of mice and human (Fig. 1) (GenBank FJ609386.1; see also CACNA1B in GTEx portal [9]). Alternative splicing-linked NMD is common during early differentiation of cell-types and often involves the family of polypyrimidne tract binding protein (PTB) splicing factors, and NMD has recently been shown to involve the action of neuronal splicing factors of the neuro-oncological ventral antigen (NOVA) family, that regulate splicing of cryptic exons within introns [33, 36–38].
Fig. (1).
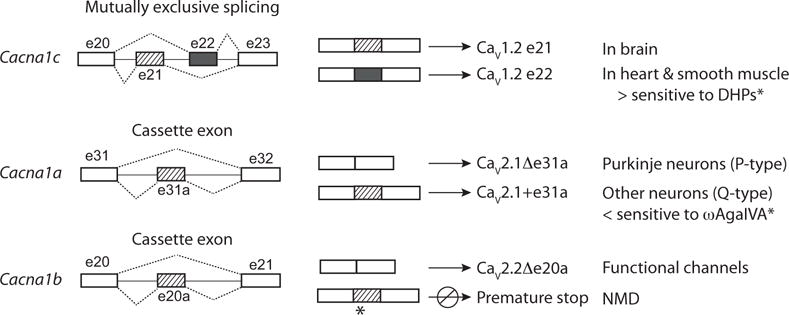
Examples of alternative pre-mRNA splicing that have been observed for Cacna1 genes. Alternative splicing can involve the selection of one of a pair of mutually exclusive exons that are the same (or similar) lengths but that encode slightly different amino acid sequences. A pair of mutually exclusive exons, e21 and e22, are present in Cacna1c. E21 is selected more frequently during pre-mRNA splicing in brain, whereas in heart and smooth muscle, e22 is selected more often (see Table 1) [85]. E21 and e22 influence the sensitivity of CaV1.2 currents to DHPs [9, 77, 78, 85]. The inclusion of one mutually exclusive exon is often required to maintain the correct reading frame, and if neither or both are included during splicing a premature stop codon is often introduced resulting in NMD. Cassette exons are included or excluded during alternative pre-mRNA splicing. E31a of Cacna1a is excluded during pre-mRNA splicing in Purkinje neurons of the cerebellum (P-type currents), whereas in other neurons including cerebellar granule cells, e31a is included (Q-type currents). The presence of e31a influences inhibition by ω-agatoxin IVA [61]. The cassette e20a in Cacna1b is repressed in most neurons but if included it introduces a premature stop codon (see GenBank FJ609386.1). Certain cells may use e20a to down regulate CaV2.2 protein expression levels.
Recent large-scale transcriptome sequencing projects suggest that alternative exon usage (pre-mRNA splicing and alternate promoters) occurs in >97% of multi-exon genes and it is extensive in mammalian brains [39–43]. Cell-specific splicing (inclusion or exclusion) of alternative exons in the nervous system is essential for development, axon targeting, neuronal excitability, circuit formation, and drug action [2, 3, 42–46]. Cell-specific splicing factors determine when and which CaV channel isoforms are expressed in a given cell type [38, 41, 47–49]. Furthermore, upstream factors regulate the expression and activity of cell-specific splicing factors including other splicing factors, miRNAs, kinases, as well as epigenetic modifiers [38, 50–52].
Cell-specific splicing factors work in concert with the ubiquitous splicing machinery to select (or skip) optional exons during pre-mRNA processing (Fig. 2) (Table 1) [33, 38, 40, 53, 54]. The expression patterns and activities of cell-specific splicing factors differ according to cell-type, age, activity, and potentially metabolic state and disease states [33]. Certain master splicing factors control the selection of exons that are essential for cell-fate determination, whereas others regulate exon choice that exert control over specific cell-functions [38]. Tremendous progress has been made in mapping splicing factor binding on a genome-wide scale, leading to the identification of splicing networks for NOVA, PTB, and rbFOX and inevitable insights into genome-wide biological significance [38, 40, 55, 56]. Splicing factors known to regulate cell-specific splicing of CaV pre-mRNAs are summarized in Table 1.
Fig. (2).
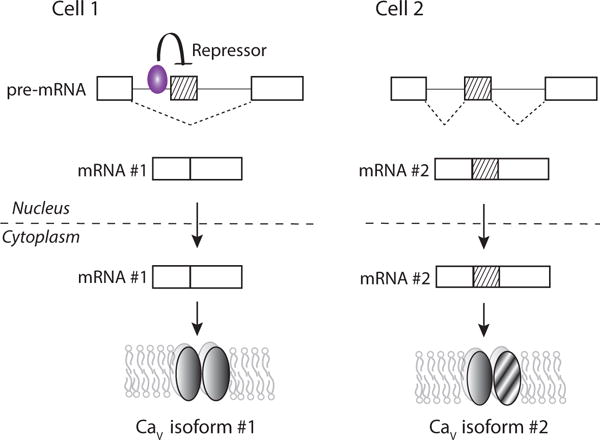
Cell-specific RNA binding proteins promote or repress exon inclusion during alternative splicing of pre-mRNAs. The example illustrates the difference in splicing patterns of the same region of pre-mRNA in Cell 1 that expresses a RNA binding splicing factor and in Cell 2 that does not. The splicing factor represses the inclusion of a cassette exon during alternative splicing of pre-mRNA. The splicing factor binds to specific nucleotide sequences upstream of the target exon in pre-mRNA. In cell 2, the absence of the splicing repressor results in exon inclusion during pre-mRNA splicing. The fully processed mRNAs are transported into the cytoplasm and are translated into different CaV splice isoforms. Certain RNA binding proteins including NOVA, rbFOX and PTB families can enhance or repress exon inclusion depending on where they bind (upstream or downstream) relative to the target exon [33, 40].
Here we focus on a few examples from three Cacna1 genes to illustrate approaches that are being employed to assess the pharmacological consequences of alternative splicing on CaV channels, in expression systems and within their native environment in vivo. Other reviews illustrate the capacity of alternative splicing to stratify CaV channel phenotypes modifying biophysical characteristics, membrane trafficking, subcellular targeting, association with downstream target proteins, and pharmacology [2, 3, 15].
4. SPLICE ISOFORMS EXPRESSED IN DIFFERENT TISSUES ARE DIFFERENT
Most, although not all, alternatively spliced cassette exons in Cacna1 genes are located in hyper-variable domains of CaV proteins that can accommodate as few as one amino acid or larger insertions comprised of several amino acids, without disrupting basic protein function. Cassette exons are either inserted or skipped and they are located in the intracellular C-termini and N-termini, intracellular I–II, II–III, and III–IV linkers, as well as S3-S4 extracellular linkers in domains III and IV of CaV channels [2, 3, 15]. Pairs of mutually exclusive exons often encode regions that do not tolerate large change in protein structure, and these have been shown to modify amino acid composition in transmembrane spanning domains including IS6, IIIS2, IVS3 and the proximal region of the C-terminus of CaV channels (Fig. 3) [2, 3, 15].
Fig. (3).
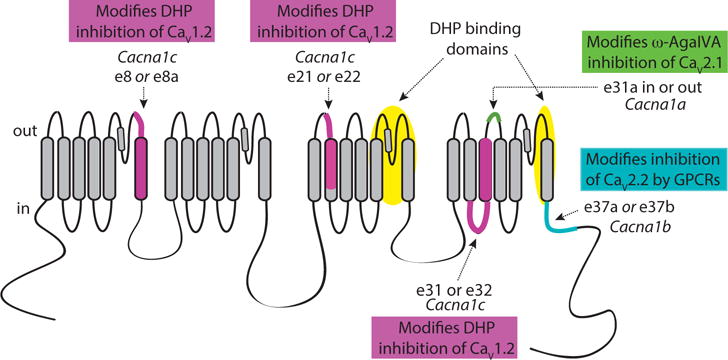
Schematic of major transmembrane spanning, intracellular, and extracellular domains of CaV channels as well as variable regions encoded by alternatively spliced exons that influence CaV channel pharmacology. Mutually exclusive exons e8a/e8, e21/e22 and e31/e32 of Cacna1c encode transmembrane spanning segments of CaV1.2 and they influence DHP inhibition by various mechanisms, including modifying voltage-dependent inhibition by DHPs [77, 78, 81, 84, 85]. Putative high-affinity DHP binding sites in CaV1.2 (shaded). Cassette e31a of Cacna1a encodes two amino acids asparagine proline (NP) in the extracellular linker IVS3–IVS4 of CaV2.1 and modifies inhibition by ω-agatoxin IVA [61]. Mutually exclusive e37a and e37b of Cacna1b encode 33 amino acids in the proximal C-terminus of CaV2.2 and they influence G protein coupled receptor mediated inhibition [107, 108].
Certain alternatively spliced exons are conserved among Cacna1 genes. For example, all but one Cacna1 gene contains an alternatively spliced cassette exon that encodes a region in the putative extracellular linker between transmembrane spanning helices S3 and S4 in domain IV of CaV channels (e.g. e29 in Cacna1s; e31a in Cacna1a; e31a in Cacna1b and e33 in Cacna1c [3] and GTEx portal [9]). The composition and length of these exons differ across genes, but all affect voltage-dependent gating of CaV channels [3]. Many other alternatively spliced exons influence channel gating including the voltage dependence of channel activation and inactivation, gating kinetics, and calcium-dependent inactivation [57–67].
Analyses of tissue-specific transcriptomes across mammals in a variety of tissues have demonstrated that alternative splicing is also frequently lineage-specific such that there is variation in the splicing of alternative exons between species [55]. This finding emphasizes the importance of study the pattern of alternative splicing in human tissue, if the ultimately goal is drug development for human use. Interestingly, transcriptome-wide analyses across several species of mammals also show that alternative splicing frequently alters the degree of protein phosphorylation implicating an important role in defining second messenger signaling cascades [55]. It is also important to point out, however, that difference in amino acid composition and protein function among CaV splice isoforms, generated from the same gene but expressed in different tissues, can also be substantial in highly variable domains including C-termini. Indeed, tissue-specific differences in amino acid compositions among CaV channel isoforms generated from one gene may be greater than those that exist across CaV channels of the same family across closely related species. This is especially dramatic for large alternative exons. For example, several Cacna1 genes have different tissue-specific promoters that generate CaV1α subunits with substantially different N-termini. Exon 1a of Cacna1c is expressed in heart and encodes 46 amino acids defining the start of the N-terminus, whereas in other tissue including brain and smooth muscle, exon 1b is expressed and it encodes a completely different 16 amino acid sequence [68–70]. By contrast, exons 1a and 1b are highly conserved across mammals and vertebrates (see multi-alignment of vertebrates in UCSC genome browser [71] and GTEx portal [9]). Similarly, exon 18a of Cacna1b, and an equivalent exon in Cacna1e, encode 21 amino acids in the II–III linkers of CaV2.2 and CaV2.3 respectively. The amino acid sequences of these alternative exons and of constitutive flanking exons, are highly conserved across species, but expression patterns of the alternative exons differ according to tissue and development [45, 71].
5. SPLICE ISOFORMS ARE TISSUE-SPECIFIC AND CELL-SPECIFIC
By definition, cell-specific processing of pre-mRNAs results in the expression of unique patterns of CaV isoforms in different cells and different tissues. This offers the opportunity to achieve a higher level of tissue targeting that might be achieved otherwise, if drugs can be identified that differentially act on splice isoforms. Indeed, the therapeutic efficacy of certain drugs can depend on their preferential action on specific splice isoforms, such as the action of non steroidal anti-inflammatory drugs that have different therapeutic profiles that depend on their differential action on isoforms of cyclooxygenase (COX-1, COX-2, and COX-3) [72–75].
Within CaV1 channels, there is interest in identifying drugs that target brain-specific CaV1.2 channel isoforms over those that dominate in cardiac and smooth muscle. The precise composition and relative abundance of CaV1.2 isoforms across different human tissue can now be analyzed from transcriptome sequencing projects (GTEx portal [9]). Certain CACNA1C exons appear to be preferentially expressed in heart including e1a (which is controlled by a cardiac tissue-specific promoter; [68, 76]), while other exons might be preferentially expressed in brain or smooth muscle [15].
6. ALTERNATIVE SPLICING OF CaV PRE-mRNAs INFLUENCE DRUG ACTION
6.1. Splice Isoforms of CaV1.2 have Different Sensitivities to DHPs
In 1995 Soldatov, Bouron, and Reuter published an important paper showing - for the first time - that different splice isoforms of CaV1.2 channels exhibit measurable differences in their biophysical as well as pharmacological properties. They suggested that the different actions of DHP antagonists on splice isoforms of CaV1.2 might account for some of their higher selectively on smooth muscle channels [77, 78] (Fig. 3). Three pairs of mutually exclusive exons in Cacna1c have been reported to modify the sensitivity of CaV1.2 calcium currents to DHPs: e8a/e8, e21/e22 and e31/e32 (>10-fold changes). While, more subtle effects of DHPs and other calcium channel blockers on inhibition of CaV1 channels (less than 10-fold changes in potency), have been reported for e9* of Cacna1c (diltiazem) [79] and for e42a of Cacna1d (nimodipine) [80].
E8a and e8 encode sequences of IS6 and part of the S5–S6 pore forming loop; photoaffinity studies indicate that the extracellular portion of this region can interact with DHPs [81–83]. Inclusion of e8 over e8a increases the sensitivity of CaV1.2 currents to DHPs [81, 84]. Interestingly, e8 and e8a have different patterns of tissue expression in mammals. E8 expression is higher in lungs and aorta than in cardiac muscle, while e8a usage is more prevalent in heart than smooth muscle of rodents and human [9, 81, 85]. This observation may also contribute to the dominant biological actions of certain DHPs as anti-hypertensive [86].
Cacna1c e21 and e22 are mutually excusive and each encodes part of the extracellular linker IIIS1–IIIS2 as well as most of the IIIS2 transmembrane helix of CaV1.2. As discussed above, e22 increases the sensitivity of CaV1.2 channels to DHP antagonists compared to e21 [77, 78, 85]. E21 and e22 are expressed throughout heart, aortic tissue and brain [9, 85, 87], although e22 is relatively more abundant in rat cardiac ventricular muscle than in aortic tissue [85]. Mutually exclusive Cacna1c exons e31 and e32 each encode most of the intracellular IVS2–IVS3 linker and the entire IVS3 transmembrane helix of CaV1.2. In heart, the selection of e31 over e32 (and vice versa) varies with development and disease state [88]. E31 dominates early in development and it reemerges in heart failure [89, 90], while e32-containing CaV1.2 mRNAs dominate in normal adult heart [89].
6.2. Splice Isoforms of CaV2.1 have Different Sensitivities to ω-Agatoxin IVA
Zamponi and colleagues showed that the actions of ω-agatoxin IVA are influenced by the presence of a two amino acid sequence, NP, encoded by e31a of the Cacna1a gene. This sequence comprises part of the putative extracellular linker between domains IVS3 and IVS4 of CaV2.1 [61]. CaV2.1 channels including NP are up to 11-fold less sensitive to ω-agatoxin IVA inhibition compared to CaV2.1 channels that lack NP. CaV2.1 mRNAs containing e31a are more abundant in Purkinje cells than in cerebellar granule cells; a result that nicely parallels the greater sensitivity of so called P-type currents, as compared to Q-type currents, to ω-agatoxin IVA [61]. The brain-specific splicing factor NOVA-2 represses the inclusion of e31a during alternative splicing of CaV2.1 pre-mRNA, and notably has the opposite action on the homologous e31a of Cacna1b [49]. NOVA-2 is known to be essential for coordinating cell-specific alternative splicing among a network of genes critical for synaptic function [91, 92].
6.3. Splice Isoforms of CaV2.2 have Different Sensitivities to G Protein Coupled Receptors
CaV2.2 channels underlie N-type currents that carry calcium entry into presynaptic terminals to control transmitter release from many synapses including primary nociceptive afferent synapses to dorsal horn neurons in spinal cord [93–98]. CaV2.2 channels are important drug targets that act directly to inhibit channel activity, as well as indirectly via G protein coupled receptors (GPCRs). Ziconotide (also known as ω-conotoxin MVIIC) is analgesic in humans and rodents when administered by intrathecal injection and it can prevent, as well as reverse symptoms of chronic pain induced by inflammation or nerve injury [99, 100]. In spinal cord, μ-opioid GPCRs inhibit presynaptic CaV2.2 channels and mediate intrathecal opioid analgesia [101–105]. We discovered that e37a, one of a pair of mutually exclusive exons the other being e37b, was expressed at higher levels in nociceptors compared to other neurons that primarily express e37b-containing CaV2.2 mRNAs [106]. We went on to show that CaV2.2 channels which contain e37a sequence exhibited different sensitivity to inhibition by GPCRs, including μ-opioid receptors, as compared to e37b-containing CaV2.2 isoforms [107]. By eliminating the contribution of e37a using either isoform-specific siRNAs or by removing e37a sequence from the Cacna1b mouse gene (replacing it with e37b; Fig. 4), we showed that e37a contributed to nociception and that it enhanced intrathecal morphine analgesia to thermal stimuli [108–110]. By contrast, ziconotide was equally effective at inhibiting e37a and e37b CaV2.2 splice isoforms and was an equally effective analgesic in vivo independent of e37a [109].
Fig. (4).
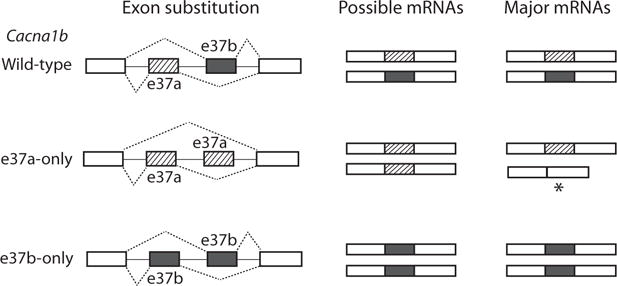
Exon substitution strategy employed to elucidate the functional and behavioral contributions of e37a and e37b of Cacna1b. Mutually exclusive e37a and e37b are the same length but about 30% of amino acids they encode differ. E37b was replaced by a second copy of e37a, in the e37a-only mouse strain, and e37a was replaced by a second copy of e37b, in the e37b-only mouse strain. E37a is repressed in most region of the nervous system but is found at higher levels in dorsal root ganglia, e37b is expressed throughout the nervous system. E37a does not substitute for e37b in the e37a-only mouse, there is increased e37 lacking CaV2.2 mRNA in neurons, and significantly reduced CaV2.2 protein levels. Whereas, in the e37b-only mouse, e37b is inserted in place of e37a and wild-type levels of CaV2.2 protein is preserved [108, 120].
7. METHODS FOR STUDYING THE BEHAVIORAL IMPORTANCE OF ALTERNATIVELY SPLICED EXONS
There has been relatively slow progress in discerning the biological and pharmacological significance of cell-specific alternative splicing of Cacna1 pre-mRNAs. This is more related to the technical challenges of separating the relative contributions of each CaV splice isoforms especially in vivo, and not the potential significance of their biological functions [2, 3]. Gene deletion and mutation studies in animal models, in conjunction with analyses of disease-causing mutations in human CACNA1 genes, have been essential in determining the biological functions of different Cacna1 genes [10, 111–117]. Interestingly, rare mutations in either one of a pair of alternatively spliced exons in CACNA1C (e8 or e8a) underlie the devastating Timothy Syndromes characterized by severe multi-organ disorders including lethal arrhythmias, webbing of fingers and toes, congenital heart disease, immune deficiency, intermittent hypoglycemia, cognitive abnormalities, and autism [86, 118, 119]. The different clinical manifestations of Type I and Type II Timothy Syndrome are correlated with the exon of CACNA1C, e8 or e8a, which carries the gain-of-function mutation [118]. Two strategies have been used to delineate the functional and behavioral consequences of different CaV2.2 splice isoforms in vivo: i) isoform-specific siRNA to achieve acute knockdown of a specific CaV protein [110]; and ii) eliminating one of a pair of alternative exons in the Cacna1b mouse gene generating novel mouse strains (Fig. 4) [108, 109]. The latter approach has the advantage of forcing inclusion of a particular exon (e37a or e37b) during splicing without disrupting intron sequence, and in the case of the e37b-only mouse, without affecting total CaV2.2 protein levels. We utilized e37b-only and wild-type mouse strains to extract the contribution of CaV2.2-e37a splice isoform in an otherwise normal background (wild-type levels of CaV2.2 current and protein expression, and no compensatory changes in non-CaV2.2 current or CaV2.1 protein) [108, 109, 120]. By comparing behavioral phenotypes we found evidence that intrathecal morphine analgesia to noxious thermal stimuli was reduced in mice lacking e37a (e37b-only) as compared to wild-type mouse strains [108, 109]. Similar analyses of other alternatively spliced exons should help establish their biological functions.
CONCLUSION
Voltage-gated calcium ion channels are essential for mediating depolarization-induced calcium entry into excitable cells, and they are important drug targets for treating several disorders including those of cardiovascular and nervous systems. Human CaV channel clones are routinely used to screening for new drugs to inhibit, or activate, calcium channels for use in humans. Even a single amino acid difference between human and non-human CaV proteins could affect drug efficacy. The amino acid composition of discrete regions of CaV channels, originating from cell-specific alternative splicing, can vary substantially across isoforms in different tissues. Intensive transcriptome sequencing projects for a range of human tissues are allowing quantification of expression patterns for each gene, including CACNA1 genes, as well as alternative exon usage according to tissue-type, age and disease state. Cell-specific alternative splicing can modify the pharmacological sensitivities of CaV channels to drugs, toxins and G protein coupled receptors. Knowledge of expression patterns of CaV isoforms in different human tissues and disease states should inform strategies aimed at developing the next generation of CaV channel inhibitors and agonists with improved tissue-specificity.
Acknowledgments
NIH grants RO1NS055251 (DL) and K99MH099405 (AA).
Biography

Diane Lipscombe
Footnotes
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
References
- 1.Catterall WA. Voltage-gated calcium channels. Cold Spring Harb Perspect Biol. 2011;3:a003947. doi: 10.1101/cshperspect.a003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lipscombe D, Allen SE, Toro CP. Control of neuronal voltage-gated calcium ion channels from RNA to protein. Trends Neurosci. 2013;36:598–609. doi: 10.1016/j.tins.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lipscombe D, Andrade A, Allen SE. Alternative splicing: functional diversity among voltage-gated calcium channels and behavioral consequences. Biochim Biophys Acta. 2013;1828:1522–1529. doi: 10.1016/j.bbamem.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catterall WA, Perez-Reyes E, Snutch TP, Striessnig J. International Union of Pharmacology. XLVIII. Nomenclature and structure-function relationships of voltage-gated calcium channels. Pharmacol Rev. 2005;57:411–425. doi: 10.1124/pr.57.4.5. [DOI] [PubMed] [Google Scholar]
- 5.Lagman D, Ocampo Daza D, Widmark J, Abalo XM, Sundstrom G, Larhammar D. The vertebrate ancestral repertoire of visual opsins, transducin alpha subunits and oxytocin/vasopressin receptors was established by duplication of their shared genomic region in the two rounds of early vertebrate genome duplications. BMC Evol Biol. 2013;13:238. doi: 10.1186/1471-2148-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beam KG, Knudson CM, Powell JA. A lethal mutation in mice eliminates the slow calcium current in skeletal muscle cells. Nature. 1986;320:168–170. doi: 10.1038/320168a0. [DOI] [PubMed] [Google Scholar]
- 7.Tanabe T, Mikami A, Numa S, Beam KG. Cardiac-type excitation-contraction coupling in dysgenic skeletal muscle injected with cardiac dihydropyridine receptor cDNA. Nature. 1990;344:451–453. doi: 10.1038/344451a0. [DOI] [PubMed] [Google Scholar]
- 8.Beam KG, Adams BA, Niidome T, Numa S, Tanabe T. Function of a truncated dihydropyridine receptor as both voltage sensor and calcium channel. Nature. 1992;360:169–171. doi: 10.1038/360169a0. [DOI] [PubMed] [Google Scholar]
- 9.The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McRory JE, Hamid J, Doering CJ, Garcia E, Parker R, Hamming K, Chen L, Hildebrand M, Beedle AM, Feldcamp L, Zamponi GW, Snutch TP. The CACNA1F gene encodes an L-type calcium channel with unique biophysical properties and tissue distribution. J Neurosci. 2004;24:1707–1718. doi: 10.1523/JNEUROSCI.4846-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Kerov V, Haeseleer F, Majumder A, Artemyev N, Baker SA, Lee A. Dysregulation of Ca(v)1.4 channels disrupts the maturation of photoreceptor synaptic ribbons in congenital stationary night blindness type 2. Channels (Austin) 2013;7:514–523. doi: 10.4161/chan.26376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doering CJ, Peloquin JB, McRory JE. The Ca(v)1.4 calcium channel: more than meets the eye. Channels (Austin) 2007;1:3–10. [PubMed] [Google Scholar]
- 13.Bech-Hansen NT, Naylor MJ, Maybaum TA, Pearce WG, Koop B, Fishman GA, Mets M, Musarella MA, Boycott KM. Loss-of-function mutations in a calcium-channel alpha1-subunit gene in Xp11.23 cause incomplete X-linked congenital stationary night blindness. Nat Genet. 1998;19:264–267. doi: 10.1038/947. [DOI] [PubMed] [Google Scholar]
- 14.Lipscombe D, Helton TD, Xu W. L-type calcium channels: the low down. J Neurophysiol. 2004;92:2633–2641. doi: 10.1152/jn.00486.2004. [DOI] [PubMed] [Google Scholar]
- 15.Liao P, Soong TW. Understanding alternative splicing of Cav1.2 calcium channels for a new approach towards individualized medicine. J Biomed Res. 2010;24:181–186. doi: 10.1016/S1674-8301(10)60027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simms BA, Zamponi GW. Neuronal voltage-gated calcium channels: structure, function, and dysfunction. Neuron. 2014;82:24–45. doi: 10.1016/j.neuron.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 17.Abernethy DR, Soldatov NM. Structure-functional diversity of human L-type Ca2+ channel: perspectives for new pharmacological targets. J Pharmacol Exp Ther. 2002;300:724–728. doi: 10.1124/jpet.300.3.724. [DOI] [PubMed] [Google Scholar]
- 18.Bean BP. Nitrendipine block of cardiac calcium channels: high-affinity binding to the inactivated state. Proc Natl Acad Sci USA. 1984;81:6388–6392. doi: 10.1073/pnas.81.20.6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kokubun S, Prod’hom B, Becker C, Porzig H, Reuter H. Studies on Ca channels in intact cardiac cells: voltage-dependent effects and cooperative interactions of dihydropyridine enantiomers. Mol Pharmacol. 1986;30:571–584. [PubMed] [Google Scholar]
- 20.Helton TD, Xu W, Lipscombe D. Neuronal L-type calcium channels open quickly and are inhibited slowly. J Neurosci. 2005;25:10247–10251. doi: 10.1523/JNEUROSCI.1089-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu W, Lipscombe D. Neuronal Ca(V)1.3alpha(1) L-type channels activate at relatively hyperpolarized membrane potentials and are incompletely inhibited by dihydropyridines. J Neurosci. 2001;21:5944–5951. doi: 10.1523/JNEUROSCI.21-16-05944.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koschak A, Reimer D, Huber I, Grabner M, Glossmann H, Engel J, Striessnig J. alpha 1D (Cav1.3) subunits can form l-type Ca2+ channels activating at negative voltages. J Biol Chem. 2001;276:22100–22106. doi: 10.1074/jbc.M101469200. [DOI] [PubMed] [Google Scholar]
- 23.Platzer J, Engel J, Schrott-Fischer A, Stephan K, Bova S, Chen H, Zheng H, Striessnig J. Congenital deafness and sinoatrial node dysfunction in mice lacking class D L-type Ca2+ channels. Cell. 2000;102:89–97. doi: 10.1016/s0092-8674(00)00013-1. [DOI] [PubMed] [Google Scholar]
- 24.Striessnig J, Pinggera A, Kaur G, Bock G, Tuluc P. L-type Ca channels in heart and brain. Wiley Interdiscip Rev Membr Transp Signal. 2014;3:15–38. doi: 10.1002/wmts.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christel CJ, Cardona N, Mesirca P, Herrmann S, Hofmann F, Striessnig J, Ludwig A, Mangoni ME, Lee A. Distinct localization and modulation of Cav1.2 and Cav1.3 L-type Ca2+ channels in mouse sinoatrial node. J Physiol. 2012;590:6327–6342. doi: 10.1113/jphysiol.2012.239954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Roak BJ, Stessman HA, Boyle EA, Witherspoon KT, Martin B, Lee C, Vives L, Baker C, Hiatt JB, Nickerson DA, Bernier R, Shendure J, Eichler EE. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nyegaard M, Demontis D, Foldager L, Hedemand A, Flint TJ, Sorensen KM, Andersen PS, Nordentoft M, Werge T, Pedersen CB, Hougaard DM, Mortensen PB, Mors O, Borglum AD. CACNA1C (rs1006737) is associated with schizophrenia. Mol Psychiatry. 2010;15:119–121. doi: 10.1038/mp.2009.69. [DOI] [PubMed] [Google Scholar]
- 28.Green EK, Hamshere M, Forty L, Gordon-Smith K, Fraser C, Russell E, Grozeva D, Kirov G, Holmans P, Moran JL, Purcell S, Sklar P, Owen MJ, O’Donovan MC, Jones L, Jones IR, Craddock N. Replication of bipolar disorder susceptibility alleles and identification of two novel genome-wide significant associations in a new bipolar disorder case-control sample. Mol Psychiatry. 2013;18:1302–1307. doi: 10.1038/mp.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, Fan J, Kirov G, Perlis RH, Green EK, Smoller JW, Grozeva D, Stone J, Nikolov I, Chambert K, Hamshere ML, Nimgaonkar VL, Moskvina V, Thase ME, Caesar S, Sachs GS, Franklin J, Gordon-Smith K, Ardlie KG, Gabriel SB, Fraser C, Blumenstiel B, Defelice M, Breen G, Gill M, Morris DW, Elkin A, Muir WJ, McGhee KA, Williamson R, MacIntyre DJ, MacLean AW, St CD, Robinson M, Van Beck M, Pereira AC, Kandaswamy R, McQuillin A, Collier DA, Bass NJ, Young AH, Lawrence J, Ferrier IN, Anjorin A, Farmer A, Curtis D, Scolnick EM, McGuffin P, Daly MJ, Corvin AP, Holmans PA, Blackwood DH, Gurling HM, Owen MJ, Purcell SM, Sklar P, Craddock N. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao P, Zhang HY, Soong TW. Alternative splicing of voltage-gated calcium channels: from molecular biology to disease. Pflugers Arch. 2009;458:481–487. doi: 10.1007/s00424-009-0635-5. [DOI] [PubMed] [Google Scholar]
- 32.Senatore A, Guan W, Spafford JD. Cav3 T-type channels: regulators for gating, membrane expression, and cation selectivity. Pflugers Arch. 2014;466:645–660. doi: 10.1007/s00424-014-1449-7. [DOI] [PubMed] [Google Scholar]
- 33.Zheng S, Black DL. Alternative pre-mRNA splicing in neurons: growing up and extending its reach. Trends Genet. 2013;29:442–448. doi: 10.1016/j.tig.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- 35.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 36.Eom T, Zhang C, Wang H, Lay K, Fak J, Noebels JL, Darnell RB. NOVA-dependent regulation of cryptic NMD exons controls synaptic protein levels after seizure. Elife. 2013;2:e00178. doi: 10.7554/eLife.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jangi M, Sharp PA. Building Robust Transcriptomes with Master Splicing Factors. Cell. 2014;159(3):487–498. doi: 10.1016/j.cell.2014.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 40.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Black DL, Grabowski PJ. Alternative pre-mRNA splicing and neuronal function. Prog Mol Subcell Biol. 2003;31:187–216. doi: 10.1007/978-3-662-09728-1_7. [DOI] [PubMed] [Google Scholar]
- 42.Grabowski PJ, Black DL. Alternative RNA splicing in the nervous system. Prog Neurobiol. 2001;65:289–308. doi: 10.1016/s0301-0082(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 43.Xie J, Black DL. A CaMK IV responsive RNA element mediates depolarization-induced alternative splicing of ion channels. Nature. 2001;410:936–939. doi: 10.1038/35073593. [DOI] [PubMed] [Google Scholar]
- 44.Asadi S, Javan M, Ahmadiani A, Sanati MH. Alternative splicing in the synaptic protein interaction site of rat Ca(v)2.2 (alpha (1B)) calcium channels: changes induced by chronic inflammatory pain. J Mol Neurosci. 2009;39:40–48. doi: 10.1007/s12031-008-9159-2. [DOI] [PubMed] [Google Scholar]
- 45.Gray AC, Raingo J, Lipscombe D. Neuronal calcium channels: splicing for optimal performance. Cell Calcium. 2007;42:409–417. doi: 10.1016/j.ceca.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jepson J, Sheldon A, Shahidullah M, Fei H, Koh K, Levitan IB. Cell-specific fine-tuning of neuronal excitability by differential expression of modulator protein isoforms. J Neurosci. 2013;33:16767–16777. doi: 10.1523/JNEUROSCI.1001-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang ZZ, Sharma S, Zheng S, Chawla G, Nikolic J, Black DL. Regulation of the mutually exclusive exons 8a and 8 in the CaV1.2 calcium channel transcript by polypyrimidine tract-binding protein. J Biol Chem. 2011;286:10007–10016. doi: 10.1074/jbc.M110.208116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang ZZ, Zheng S, Nikolic J, Black DL. Developmental control of CaV1.2 L-type calcium channel splicing by Fox proteins. Mol Cell Biol. 2009;29:4757–4765. doi: 10.1128/MCB.00608-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen SE, Darnell RB, Lipscombe D. The neuronal splicing factor Nova controls alternative splicing in N-type and P-type CaV2 calcium channels. Channels (Austin) 2010;4:483–489. doi: 10.4161/chan.4.6.12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iannone C, Valcarcel J. Chromatin’s thread to alternative splicing regulation. Chromosoma. 2013;122:465–474. doi: 10.1007/s00412-013-0425-x. [DOI] [PubMed] [Google Scholar]
- 51.Gelfman S, Cohen N, Yearim A, Ast G. DNA-methylation effect on cotranscriptional splicing is dependent on GC architecture of the exon-intron structure. Genome Res. 2013;23:789–799. doi: 10.1101/gr.143503.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gelfman S, Ast G. When epigenetics meets alternative splicing: the roles of DNA methylation and GC architecture. Epigenomics. 2013;5:351–353. doi: 10.2217/epi.13.32. [DOI] [PubMed] [Google Scholar]
- 53.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darnell JEJ. Reflections on the history of pre-mRNA processing and highlights of current knowledge: a unified picture. RNA. 2013;19:443–460. doi: 10.1261/rna.038596.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan GM, Yu D, Wang J, Soong TW. Alternative splicing at C terminus of Ca(V)1.4 calcium channel modulates calcium-dependent inactivation, activation potential, and current density. J Biol Chem. 2012;287:832–847. doi: 10.1074/jbc.M111.268722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin Z, Haus S, Edgerton J, Lipscombe D. Identification of functionally distinct isoforms of the N-type Ca2+ channel in rat sympathetic ganglia and brain. Neuron. 1997;18:153–166. doi: 10.1016/s0896-6273(01)80054-4. [DOI] [PubMed] [Google Scholar]
- 59.Soong TW, DeMaria CD, Alvania RS, Zweifel LS, Liang MC, Mittman S, Agnew WS, Yue DT. Systematic identification of splice variants in human P/Q-type channel alpha1(2.1) subunits: implications for current density and Ca2+-dependent inactivation. J Neurosci. 2002;22:10142–10152. doi: 10.1523/JNEUROSCI.22-23-10142.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soldatov NM, Zuhlke RD, Bouron A, Reuter H. Molecular structures involved in L-type calcium channel inactivation. Role of the carboxyl-terminal region encoded by exons 40–42 in alpha1C subunit in the kinetics and Ca2+ dependence of inactivation. J Biol Chem. 1997;272:3560–3566. doi: 10.1074/jbc.272.6.3560. [DOI] [PubMed] [Google Scholar]
- 61.Bourinet E, Soong TW, Sutton K, Slaymaker S, Mathews E, Monteil A, Zamponi GW, Nargeot J, Snutch TP. Splicing of alpha 1A subunit gene generates phenotypic variants of P- and Q-type calcium channels. Nat Neurosci. 1999;2:407–415. doi: 10.1038/8070. [DOI] [PubMed] [Google Scholar]
- 62.Murbartian J, Arias JM, Perez-Reyes E. Functional impact of alternative splicing of human T-type Cav3.3 calcium channels. J Neurophysiol. 2004;92:3399–3407. doi: 10.1152/jn.00498.2004. [DOI] [PubMed] [Google Scholar]
- 63.Chemin J, Monteil A, Bourinet E, Nargeot J, Lory P. Alternatively spliced alpha(1G) (Ca(V)3.1) intracellular loops promote specific T-type Ca(2+) channel gating properties. Biophys J. 2001;80:1238–1250. doi: 10.1016/S0006-3495(01)76100-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pan JQ, Lipscombe D. Alternative splicing in the cytoplasmic II–III loop of the N-type Ca channel alpha 1B subunit: functional differences are beta subunit-specific. J Neurosci. 2000;20:4769–4775. doi: 10.1523/JNEUROSCI.20-13-04769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ernst WL, Noebels JL. Expanded alternative splice isoform profiling of the mouse Cav3.1/alpha1G T-type calcium channel. BMC Mol Biol. 2009;10:53. doi: 10.1186/1471-2199-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mittman S, Guo J, Agnew WS. Structure and alternative splicing of the gene encoding alpha1G, a human brain T calcium channel alpha1 subunit. Neurosci Lett. 1999;274:143–146. doi: 10.1016/s0304-3940(99)00716-8. [DOI] [PubMed] [Google Scholar]
- 67.Zhong X, Liu JR, Kyle JW, Hanck DA, Agnew WS. A profile of alternative RNA splicing and transcript variation of CACNA1H, a human T-channel gene candidate for idiopathic generalized epilepsies. Hum Mol Genet. 2006;15:1497–1512. doi: 10.1093/hmg/ddl068. [DOI] [PubMed] [Google Scholar]
- 68.Dai B, Saada N, Echetebu C, Dettbarn C, Palade P. A new promoter for alpha1C subunit of human L-type cardiac calcium channel Ca(V)1.2. Biochem Biophys Res Commun. 2002;296:429–433. doi: 10.1016/s0006-291x(02)00894-x. [DOI] [PubMed] [Google Scholar]
- 69.Saada N, Dai B, Echetebu C, Sarna SK, Palade P. Smooth muscle uses another promoter to express primarily a form of human Cav1.2 L-type calcium channel different from the principal heart form. Biochem Biophys Res Commun. 2003;302:23–28. doi: 10.1016/s0006-291x(03)00097-4. [DOI] [PubMed] [Google Scholar]
- 70.Saada NI, Carrillo ED, Dai B, Wang WZ, Dettbarn C, Sanchez J, Palade P. Expression of multiple CaV1.2 transcripts in rat tissues mediated by different promoters. Cell Calcium. 2005;37:301–309. doi: 10.1016/j.ceca.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 71.UCSC Genome Browser Website. 2011 Available from: http://genome.ucsc.edu/goldenPath/newsarch.html [Accessed October 10, 2014]
- 72.Roos KL, Simmons DL. Cyclooxygenase variants: the role of alternative splicing. Biochem Biophys Res Commun. 2005;338:62–69. doi: 10.1016/j.bbrc.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 73.Masferrer JL, Zweifel BS, Manning PT, Hauser SD, Leahy KM, Smith WG, Isakson PC, Seibert K. Selective inhibition of inducible cyclooxygenase 2 in vivo is antiinflammatory and nonulcerogenic. Proc Natl Acad Sci USA. 1994;91:3228–3232. doi: 10.1073/pnas.91.8.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seibert K, Zhang Y, Leahy K, Hauser S, Masferrer J, Perkins W, Lee L, Isakson P. Pharmacological and biochemical demonstration of the role of cyclooxygenase 2 in inflammation and pain. Proc Natl Acad Sci USA. 1994;91:12013–12017. doi: 10.1073/pnas.91.25.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mitchell JA, Akarasereenont P, Thiemermann C, Flower RJ, Vane JR. Selectivity of nonsteroidal antiinflammatory drugs as inhibitors of constitutive and inducible cyclooxygenase. Proc Natl Acad Sci USA. 1993;90:11693–11697. doi: 10.1073/pnas.90.24.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blumenstein Y, Kanevsky N, Sahar G, Barzilai R, Ivanina T, Dascal N. A novel long N-terminal isoform of human L-type Ca2+ channel is up-regulated by protein kinase C. J Biol Chem. 2002;277:3419–3423. doi: 10.1074/jbc.C100642200. [DOI] [PubMed] [Google Scholar]
- 77.Zuhlke RD, Bouron A, Soldatov NM, Reuter H. Ca2+ channel sensitivity towards the blocker isradipine is affected by alternative splicing of the human alpha1C subunit gene. FEBS Lett. 1998;427:220–224. doi: 10.1016/s0014-5793(98)00425-6. [DOI] [PubMed] [Google Scholar]
- 78.Soldatov NM, Bouron A, Reuter H. Different voltage-dependent inhibition by dihydropyridines of human Ca2+ channel splice variants. J Biol Chem. 1995;270:10540–10543. doi: 10.1074/jbc.270.18.10540. [DOI] [PubMed] [Google Scholar]
- 79.Zhang HY, Liao P, Wang JJ, Yu de J, Soong TW. Alternative splicing modulates diltiazem sensitivity of cardiac and vascular smooth muscle Ca(v)1.2 calcium channels. Br J Pharmacol. 2010;160:1631–1640. doi: 10.1111/j.1476-5381.2010.00798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang H, Yu D, Soong TW. C-terminal alternative splicing of CaV1.3 channels distinctively modulates their dihydropyridine sensitivity. Mol Pharmacol. 2013;84:643–653. doi: 10.1124/mol.113.087155. [DOI] [PubMed] [Google Scholar]
- 81.Welling A, Ludwig A, Zimmer S, Klugbauer N, Flockerzi V, Hofmann F. Alternatively spliced IS6 segments of the alpha 1C gene determine the tissue-specific dihydropyridine sensitivity of cardiac and vascular smooth muscle L-type Ca2+ channels. Circ Res. 1997;81:526–532. doi: 10.1161/01.res.81.4.526. [DOI] [PubMed] [Google Scholar]
- 82.Kalasz H, Watanabe T, Yabana H, Itagaki K, Naito K, Nakayama H, Schwartz A, Vaghy PL. Identification of 1,4-dihydropyridine binding domains within the primary structure of the alpha 1 subunit of the skeletal muscle L-type calcium channel. FEBS Lett. 1993;331:177–181. doi: 10.1016/0014-5793(93)80321-k. [DOI] [PubMed] [Google Scholar]
- 83.Striessnig J, Murphy BJ, Catterall WA. Dihydropyridine receptor of L-type Ca2+ channels: identification of binding domains for [3H](+)-PN200-110 and [3H]azidopine within the alpha 1 subunit. Proc Natl Acad Sci USA. 1991;88:10769–10773. doi: 10.1073/pnas.88.23.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu H, Marban E. Isoform-specific inhibition of L-type calcium channels by dihydropyridines is independent of isoform-specific gating properties. Mol Pharmacol. 1998;53:902–907. [PubMed] [Google Scholar]
- 85.Tang ZZ, Hong X, Wang J, Soong TW. Signature combinatorial splicing profiles of rat cardiac- and smooth-muscle Cav1.2 channels with distinct biophysical properties. Cell Calcium. 2007;41:417–428. doi: 10.1016/j.ceca.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 86.Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, Napolitano C, Schwartz PJ, Joseph RM, Condouris K, Tager-Flusberg H, Priori SG, Sanguinetti MC, Keating MT. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 87.Tang ZZ, Liang MC, Lu S, Yu D, Yu CY, Yue DT, Soong TW. Transcript scanning reveals novel and extensive splice variations in human l-type voltage-gated calcium channel, Cav1.2 alpha1 subunit. J Biol Chem. 2004;279:44335–44343. doi: 10.1074/jbc.M407023200. [DOI] [PubMed] [Google Scholar]
- 88.Tiwari S, Zhang Y, Heller J, Abernethy DR, Soldatov NM. Atherosclerosis-related molecular alteration of the human CaV1.2 calcium channel alpha1C subunit. Proc Natl Acad Sci USA. 2006;103:17024–17029. doi: 10.1073/pnas.0606539103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Diebold RJ, Koch WJ, Ellinor PT, Wang JJ, Muthuchamy M, Wieczorek DF, Schwartz A. Mutually exclusive exon splicing of the cardiac calcium channel alpha 1 subunit gene generates developmentally regulated isoforms in the rat heart. Proc Natl Acad Sci USA. 1992;89:1497–1501. doi: 10.1073/pnas.89.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gidh-Jain M, Huang B, Jain P, Battula V, el-Sherif N. Reemergence of the fetal pattern of L-type calcium channel gene expression in non infarcted myocardium during left ventricular remodeling. Biochem Biophys Res Commun. 1995;216:892–897. doi: 10.1006/bbrc.1995.2705. [DOI] [PubMed] [Google Scholar]
- 91.Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, Darnell RB. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 92.Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, Wang H, Clark T, Fraser C, Ruggiu M, Zeeberg BR, Kane D, Weinstein JN, Blume J, Darnell RB. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- 93.Dahlhaus A, Ruscheweyh R, Sandkuhler J. Synaptic input of rat spinal lamina I projection and unidentified neurones in vitro. J Physiol. 2005;566:355–368. doi: 10.1113/jphysiol.2005.088567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heinke B, Balzer E, Sandkuhler J. Pre- and postsynaptic contributions of voltage-dependent Ca2+ channels to nociceptive transmission in rat spinal lamina I neurons. Eur J Neurosci. 2004;19:103–111. doi: 10.1046/j.1460-9568.2003.03083.x. [DOI] [PubMed] [Google Scholar]
- 95.Ikeda H, Heinke B, Ruscheweyh R, Sandkuhler J. Synaptic plasticity in spinal lamina I projection neurons that mediate hyperalgesia. Science. 2003;299:1237–1240. doi: 10.1126/science.1080659. [DOI] [PubMed] [Google Scholar]
- 96.Kerr LM, Filloux F, Olivera BM, Jackson H, Wamsley JK. Autoradiographic localization of calcium channels with [125I]omega-conotoxin in rat brain. Eur J Pharmacol. 1988;146:181–183. doi: 10.1016/0014-2999(88)90501-8. [DOI] [PubMed] [Google Scholar]
- 97.Lima D, Mendes-Ribeiro JA, Coimbra A. The spino-latero-reticular system of the rat: projections from the superficial dorsal horn and structural characterization of marginal neurons involved. Neuroscience. 1991;45:137–152. doi: 10.1016/0306-4522(91)90110-a. [DOI] [PubMed] [Google Scholar]
- 98.Maggi CA, Giuliani S, Santicioli P, Tramontana M, Meli A. Effect of omega conotoxin on reflex responses mediated by activation of capsaicin-sensitive nerves of the rat urinary bladder and peptide release from the rat spinal cord. Neuroscience. 1990;34:243–250. doi: 10.1016/0306-4522(90)90318-x. [DOI] [PubMed] [Google Scholar]
- 99.Miljanich GP. Ziconotide: neuronal calcium channel blocker for treating severe chronic pain. Curr Med Chem. 2004;11:3029–3040. doi: 10.2174/0929867043363884. [DOI] [PubMed] [Google Scholar]
- 100.Staats PS, Yearwood T, Charapata SG, Presley RW, Wallace MS, Byas-Smith M, Fisher R, Bryce DA, Mangieri EA, Luther RR, Mayo M, McGuire D, Ellis D. Intrathecal ziconotide in the treatment of refractory pain in patients with cancer or AIDS: a randomized controlled trial. JAMA. 2004;291:63–70. doi: 10.1001/jama.291.1.63. [DOI] [PubMed] [Google Scholar]
- 101.Heinke B, Gingl E, Sandkuhler J. Multiple targets of mu-opioid receptor-mediated presynaptic inhibition at primary afferent Adelta- and C-fibers. J Neurosci. 2011;31:1313–1322. doi: 10.1523/JNEUROSCI.4060-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Scholz J, Woolf CJ. Can we conquer pain? Nat Neurosci. 2002;5(Suppl):1062–1067. doi: 10.1038/nn942. [DOI] [PubMed] [Google Scholar]
- 103.Khasabova IA, Harding-Rose C, Simone DA, Seybold VS. Differential effects of CB1 and opioid agonists on two populations of adult rat dorsal root ganglion neurons. J Neurosci. 2004;24:1744–1753. doi: 10.1523/JNEUROSCI.4298-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Snutch TP. Targeting chronic and neuropathic pain: the N-type calcium channel comes of age. NeuroRx. 2005;2:662–670. doi: 10.1602/neurorx.2.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu ZZ, Chen SR, Pan HL. Differential sensitivity of N- and P/Q-type Ca2+ channel currents to a mu opioid in isolectin B4-positive and -negative dorsal root ganglion neurons. J Pharmacol Exp Ther. 2004;311:939–947. doi: 10.1124/jpet.104.073429. [DOI] [PubMed] [Google Scholar]
- 106.Bell TJ, Thaler C, Castiglioni AJ, Helton TD, Lipscombe D. Cell-specific alternative splicing increases calcium channel current density in the pain pathway. Neuron. 2004;41:127–138. doi: 10.1016/s0896-6273(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 107.Raingo J, Castiglioni AJ, Lipscombe D. Alternative splicing controls G protein-dependent inhibition of N-type calcium channels in nociceptors. Nat Neurosci. 2007;10:285–292. doi: 10.1038/nn1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Andrade A, Denome S, Jiang YQ, Marangoudakis S, Lipscombe D. Opioid inhibition of N-type Ca2+ channels and spinal analgesia couple to alternative splicing. Nat Neurosci. 2010;13:1249–1256. doi: 10.1038/nn.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jiang YQ, Andrade A, Lipscombe D. Spinal morphine but not ziconotide or gabapentin analgesia is affected by alternative splicing of voltage-gated calcium channel CaV2.2 pre-mRNA. Mol Pain. 2013;9:67. doi: 10.1186/1744-8069-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Altier C, Dale CS, Kisilevsky AE, Chapman K, Castiglioni AJ, Matthews EA, Evans RM, Dickenson AH, Lipscombe D, Vergnolle N, Zamponi GW. Differential role of N-type calcium channel splice isoforms in pain. J Neurosci. 2007;27:6363–6373. doi: 10.1523/JNEUROSCI.0307-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Singh B, Monteil A, Bidaud I, Sugimoto Y, Suzuki T, Hamano S, Oguni H, Osawa M, Alonso ME, Delgado-Escueta AV, Inoue Y, Yasui-Furukori N, Kaneko S, Lory P, Yamakawa K. Mutational analysis of CACNA1G in idiopathic generalized epilepsy. Mutation in brief #962. Online. Hum Mutat. 2007;28:524–525. doi: 10.1002/humu.9491. [DOI] [PubMed] [Google Scholar]
- 112.Baig SM, Koschak A, Lieb A, Gebhart M, Dafinger C, Nurnberg G, Ali A, Ahmad I, Sinnegger-Brauns MJ, Brandt N, Engel J, Mangoni ME, Farooq M, Khan HU, Nurnberg P, Striessnig J, Bolz HJ. Loss of Ca(v)1.3 (CACNA1D) function in a human channelopathy with bradycardia and congenital deafness. Nat Neurosci. 2011;14:77–84. doi: 10.1038/nn.2694. [DOI] [PubMed] [Google Scholar]
- 113.Chen CC, Lamping KG, Nuno DW, Barresi R, Prouty SJ, Lavoie JL, Cribbs LL, England SK, Sigmund CD, Weiss RM, Williamson RA, Hill JA, Campbell KP. Abnormal coronary function in mice deficient in alpha1H T-type Ca2+ channels. Science. 2003;302:1416–1418. doi: 10.1126/science.1089268. [DOI] [PubMed] [Google Scholar]
- 114.Groen JL, Andrade A, Ritz K, Jalalzadeh H, Haagmans M, Bradley TE, Jongejan A, Verbeek DS, Nurnberg P, Denome S, Hennekam RC, Lipscombe D, Baas F, Tijssen MA. CACNA1B mutation is linked to unique myoclonus-dystonia syndrome. Hum Mol Genet. 2015;24(4):987–993. doi: 10.1093/hmg/ddu513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pietrobon D. Calcium channels and channelopathies of the central nervous system. Mol Neurobiol. 2002;25:31–50. doi: 10.1385/MN:25:1:031. [DOI] [PubMed] [Google Scholar]
- 116.Catterall WA, Dib-Hajj S, Meisler MH, Pietrobon D. Inherited neuronal ion channelopathies: new windows on complex neurological diseases. J Neurosci. 2008;28:11768–11777. doi: 10.1523/JNEUROSCI.3901-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ophoff RA, Terwindt GM, Vergouwe MN, van Eijk R, Oefner PJ, Hoffman SM, Lamerdin JE, Mohrenweiser HW, Bulman DE, Ferrari M, Haan J, Lindhout D, van Ommen GJ, Hofker MH, Ferrari MD, Frants RR. Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell. 1996;87:543–552. doi: 10.1016/s0092-8674(00)81373-2. [DOI] [PubMed] [Google Scholar]
- 118.Splawski I, Timothy KW, Decher N, Kumar P, Sachse FB, Beggs AH, Sanguinetti MC, Keating MT. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc Natl Acad Sci USA. 2005;102:8089–96. doi: 10.1073/pnas.0502506102. discussion 8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, Dolmetsch RE. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471:230–234. doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Marangoudakis S, Andrade A, Helton TD, Denome S, Castiglioni AJ, Lipscombe D. Differential ubiquitination and proteasome regulation of Ca(V)2.2 N-type channel splice isoforms. J Neurosci. 2012;32:10365–10369. doi: 10.1523/JNEUROSCI.0851-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, Ares MJ, Mody I, Black DL. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol. 2006;16:102–110. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 123.Mermelstein PG, Foehring RC, Tkatch T, Song WJ, Baranauskas G, Surmeier DJ. Properties of Q-type calcium channels in neostriatal and cortical neurons are correlated with beta subunit expression. J Neurosci. 1999;19:7268–7277. doi: 10.1523/JNEUROSCI.19-17-07268.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Soldatov NM. Genomic structure of human L-type Ca2+ channel. Genomics. 1994;22:77–87. doi: 10.1006/geno.1994.1347. [DOI] [PubMed] [Google Scholar]


