Abstract
The nocturnal increase in circulating melatonin in vertebrates is regulated by the activity of arylalkylamine N-acetyltransferase (AANAT), the penultimate enzyme in the melatonin pathway (serotonin → N-acetylserotonin → melatonin). Large changes in activity are linked to cyclic AMP-dependent protein kinase-mediated phosphorylation of AANAT T31. Phosphorylation of T31 promotes binding of AANAT to the dimeric 14-3-3 protein, which activates AANAT by increasing arylalkylamine affinity. In the current study, a putative second AANAT cyclic AMP-dependent protein kinase phosphorylation site, S205, was found to be ≈55% phosphorylated at night, when T31 is ≈40% phosphorylated. These findings indicate that ovine AANAT is dual-phosphorylated. Moreover, light exposure at night decreases T31 and S205 phosphorylation, consistent with a regulatory role of both sites. AANAT peptides containing either T31 or S205 associate with 14-3-3ζ in a phosphorylation-dependent manner; binding through phosphorylated (p)T31 is stronger than that through pS205, consistent with the location of only pT31 in a mode I binding motif, one of two recognized high-affinity 14-3-3-binding motifs AANAT protein binds to 14-3-3ζ through pT31 or pS205. Two-site binding lowers the Km for arylalkylamine substrate to ≈30 μM. In contrast, single-site pS205 binding increases the Km to ≈1,200 μM. Accordingly, the switch from dual to single pS205 binding of AANAT to 14-3-3 changes the Km for substrates by ≈40-fold. pS205 seems to be part of a previously unrecognized 14-3-3-binding motif-pS/pT (X1–2)-COOH, referred to here as mode III.
Keywords: pineal, circadian, cAMP, kinase
The daily rhythm in circulating melatonin is a highly conserved feature of vertebrate physiology; in all cases, high levels occur at night. Day/night differences in circulating melatonin levels provide a hormonal analog signal of environmental lighting, which is used in a variety of ways to optimize circadian and circannual rhythms in physiology (1). The melatonin rhythm is controlled by large changes in the rate of melatonin production in the pineal gland; these changes reflect changes in the penultimate enzyme in melatonin synthesis, arylalkylamine N-acetyltransferase (AANAT) (2, 3). Changes in the activity of this enzyme are strongly influenced by cAMP-dependent binding to the bowl-shaped, dimeric 14-3-3 protein (4, 5). Binding is thought to protect the enzyme against proteasomal proteolysis (6, 7); moreover, binding to 14-3-3 increases the affinity of AANAT by ≈10-fold for arylalkylamine substrates, e.g., serotonin and tryptamine (4, 5).
Pinealocyte cAMP is controlled in mammals through a photoneural system, which includes the eyes and the suprachiasmatic nucleus, the site of the master circadian clock (1). In lower vertebrates, cAMP is regulated by light acting directly on pinealocytes (8). It is likely that cAMP-dependent binding to 14-3-3 occurs in all vertebrates, based on the ubiquitous presence of 14-3-3 proteins and the presence of two putative cyclic AMP-dependent protein kinase (PKA) sites centered on T31 and S205 (3).∥ This cAMP-dependent association is thought to be a dominant regulatory mechanism controlling AANAT protein and activity in animals in which AANAT mRNA levels do not change on a rhythmic basis and also to be essential in those with a rhythm in AANAT mRNA (3, 9–12).
The T31 PKA site of AANAT is nested within a nascent 14-3-3-binding motif (13–15). It is physiologically phosphorylated and phosphorylated (p)T31 mediates binding of AANAT to 14-3-3 by using an amphipathic groove in one member of the 14-3-3 dimer (4, 5). In contrast, little is known about the S205 PKA site other than high conservation and that the S205G mutation of human AANAT (hAANAT) seems to prevent intracellular cAMP activation of hAANAT (4).** Physiological phosphorylation of S205 has not been demonstrated, nor has it been determined that pS205 is important for binding to 14-3-3 in conjunction with pT31, although dual binding is theoretically possible (5). Furthermore, there also is reason to doubt the importance of pS205, because it was absent from the truncated form of AANAT (oAANAT1–201) used to crystallize the AANAT/14-3-3 dimer complex (5) and it is not nested within a consensus 14-3-3-binding motif (13–15).
Here we studied pS205 and discovered that it plays a dual role in mediating 14-3-3-dependent activation and inhibition.
Materials and Methods
Tissues. Pineal glands used for Fig. 1 A–D and for tissue in an antisera screen (J.L.W. and D.C.K., unpublished data) were from adult sheep (outbred Dorsett × Hampshire mix, mostly females with some castrated males; National Institutes of Health Animal Center, Poolesville, MD). They were housed in covered outdoor pens with natural lighting during the day and dim white light during the night; feeding was at 0700 and 1430 hours; before killing at night (dark), animals were blindfolded at 1800 hours. Animals were killed in October and November (≈11:13 light/ dark cycle) by ketamine injection into the carotid. Tissues were removed and placed on solid CO2 immediately thereafter.
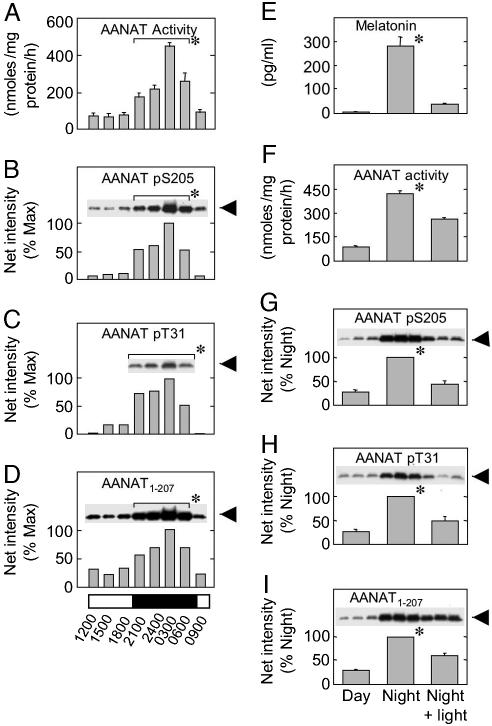
Fig. 1.
Daily and light-induced changes in AANAT pS205 and related parameters. (A–D) Pineal glands were removed at the times indicated, and tissue extracts (1 mg/10 μl) were prepared. Representative results from a single study are presented. Similar results were obtained in two additional experiments. (E–I) Blood was obtained, and glands were removed during the day (≈1100 hours), at night (≈0200 hours), and at night after exposure to 30 min of light (≈0230 hours). At each time point, three pineal glands were removed under each lighting condition (1 pineal per supernatant, 75 μg per lane; proteins were resolved by SDS/PAGE). Blots were prepared in triplicate and immunodetected. The experiment was repeated three times, and the results were pooled; each bar represents the mean ± SE (n = 9). (A and F) AANAT activity in a gland was assayed in triplicate (1 mM tryptamine/0.5 mM [3H]acetyl-CoA; final specific activity, 4.2 Ci/mol; 20 min, 37°C). (B–D and G–I) Samples of supernatant (75 μg) were resolved by SDS/PAGE. Epitopes were detected with the following: AANAT pS205 and anti-oAANAT194–207 (As 5901); AANAT pT31 and anti-rAANAT22–37 pT31 (As 3352); and AANAT1–207 and anti-oAANAT1–207 (As 2819). (G–I) The three immunosignals that appear above each bar are representative of the nine samples used for quantitation of the corresponding bar. Immunosignals were captured on a Kodak Image Station 2000 mm and analyzed with Kodak 1d image-analysis software. The percent maximum value was calculated based on net intensity by using the highest night value as 100%. (E) Melatonin was measured by RIA. Data are presented as the mean ± SE (A and E–I) or the mean of two determinations (B–D). *, Statistical analysis: for the data shown in A–D, night (2100–0600 hours) samples were pooled, compared with a pool of day (0900–1800 hours) samples, and found to be statistically different (n = 8, P < 0.01); for the data shown in E–I, night values were found to be significantly (P < 0.01) greater than the day and the night + light groups. For additional details, see Materials and Methods.
Pineal glands used for Figs. 1 E–I and 2 and Table 1 were obtained from sheep in a field-maintained flock [male and female, 3–4 months old, 35–40 kg (authorization for animal experimentation: French Ministry of Agriculture no. A37801); Ile-de France and Romanov, Institut National de la Recherche Agronomique, Research Center of Nouzilly, France]; ≥3 weeks before slaughter they were transferred to and maintained thereafter in light-controlled compartments (12:12 light/dark cycle). Care, blood collection, and slaughter have been detailed (12). Times of killing and lighting schedules are given in the figures and figure legends.
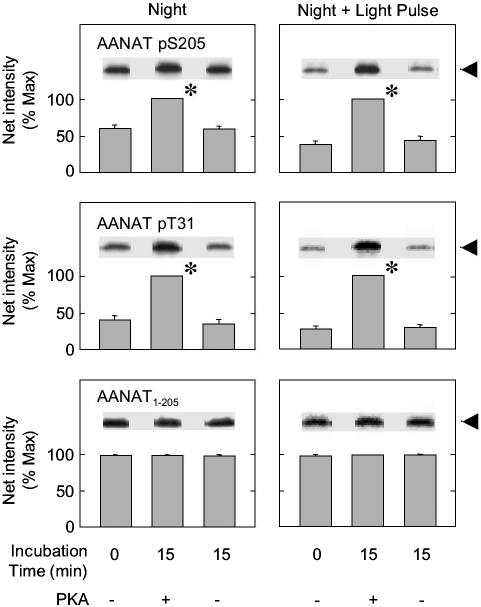
Fig. 2.
Light exposure reduces the degree of phosphorylation of T31 and S205. The percentage of phosphorylated AANAT pT31 and AANAT pS205 in ovine pineal glands at night (Left) and after exposure to 30 min of light at night (Right) was determined. AANAT was maximally phosphorylated by incubation with ATP and PKA. Each sample was analyzed in duplicate on a 15% gel containing the three treatment groups indicated; there was no decrease in phosphorylation during incubation in the absence of PKA and ATP. The data on the left were not normalized to the data on the right. Each digital image is an example of one sample representative of the experimental group. Each bar represents the mean ± SE of the results of analysis of three samples (two glands per sample). For additional details, see Materials and Methods and the Fig. 1 legend. *, Incubation with PKA and ATP increased the degree of phosphorylation (P < 0.01).
Table 1. Effect of light exposure at night on degree of phosphorylation of T31 and S205.
| AANAT residue | Night (% 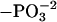 ) ) |
Night + 30′ light pulse (% 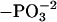 ) ) |
Light-induced change |
|---|---|---|---|
| T31 | 37.0 ± 2.4 | 25.4 ± 3.0 | -11.6 ± 3.8* |
| S205 | 55.1 ± 2.9 | 35.2 ± 2.3 | -19.9 ± 3.7* |
Pineal glands were obtained during the night (0200 hours) or after a 30-min exposure to light at night (0230 hours); for each time, pools of two glands each were used to prepare supernatants. Samples of each supernatant were incubated with ATP and the catalytic subunit of PKA for 15 min at 20°C or without ATP and PKA for 0 min at 4°C. Proteins were resolved by SDS/PAGE; each gel contained a complete set of samples of unphosphorylated and phosphorylated night and night + light supernatants; results were normalized within a single blot and are presented as percent of T31 and S205 that is phosphorylated in AANAT obtained from pineal glands at night and at night after 30-min pulse of light.
The data are expressed as a mean ± SE (n = 5, P < 0.05). For further details see Materials and Methods and the Fig. 2 legend.
Tissue Preparation. Unless otherwise indicated, tissues were homogenized in 7 vol of buffer [100 mM sodium phosphate buffer, pH 6.8, containing the following protease/phosphatase “inhibitor mixture”: Mixture EDTA-free, 1 tablet per 50 ml (Roche, Indianapolis); 50 nM microcystin-LR (Calbiochem); and 50 mM NaF and 0.2 mM Na3VO4 (Sigma)]. Homogenates were centrifuged (15,000 × g, 8 min), and samples of the supernatant were used for analysis.
Phosphorylation of Expressed and Native Proteins. Expressed proteins were phosphorylated by incubation for either 30 min (20°C) or 16 h (4°C) in 20 mM sodium phosphate buffer, pH 6.8 (10 μg of protein/500-μl reaction), containing inhibitor mixture, 15 mM MgCl2, 5 mM DTT, 50 μg of thyroglobulin, and 200 units of the catalytic subunit of PKA (Promega) to which a 10× stock solution of ATP (pH 7.0) was added to a final concentration of 2 mM. These conditions produce ≈100% phosphorylation of expressed AANAT with a stoichiometry of 2 mol of phosphate/mol of AANAT, as determined by using 0.1 mM ATP and tracer [γ-32P]ATP [final specific activity = 1μCi/nmol (1 Ci = 37 GBq)]; there was no phosphorylation using AANAT T31A S205G, indicating that this treatment only phosphorylates N- and C-terminal PKA sites. Maximal phosphorylation of the naturally occurring AANAT was achieved by incubation at 20°C for 15 min. Incubation (15 min, 20°C) with 2 mM ATP and PKA (160 units) completely phosphorylates the C- and N-terminal PKA sites of naturally occurring AANAT based on standard curves generated with bacterially expressed AANAT1–207 pT31 pS205, as detected by using anti-AANAT pT31, anti-AANAT pS205, anti-AANAT1–207, and anti-AANAT1–25. Phosphorylation does not increase if the concentration of ATP is increased to 10 mM, if the concentration of PKA is doubled, or if PKA treatment is increased to 30 min or with PKA added at zero time and 15 min. Controls were prepared without the addition of PKA or ATP.
Proteins in homogenates were phosphorylated by incubating (15 min, 20°C) a sample (150 μl of a homogenate of two glands) in a final volume of 200 μl containing 2 mM MgCl2, 5mMDTT, 2 mM ATP, and 160 units of the catalytic subunit of PKA. These conditions were found to produce 100% phosphorylation based on immunological detection using phosphorylated expressed AANAT as standards. Controls were 0 min at 4°C (–PKA and –ATP; Fig. 2 and Table 1) and 15 min at 20°C (–PKA and –ATP; Fig. 2). Reactions were ended by the addition of 4 μl of 500 mM EDTA, 200 μl of SDS/PAGE sample buffer (see Supporting Methods and Materials, which is published as supporting information on the PNAS web site), and boiling (7 min).
Bacterially Expressed Proteins. Recombinant oA ANAT, oAANAT1–201, and 14-3-3ζ were prepared as described (4, 5). GST-fused oAANAT mutant constructs (GST-oAANAT T31A and GST-oAANAT S205A) were made by PCR-based site-directed mutagenesis (QuikChange kit, Stratagene) using the wild-type pGEX-4T1-oAANAT as the template according to manufacturer instructions. GST-oAANAT T31A S205G was made with the same technique by using the pGEX-4T1-oAANAT T31A template. cDNA encoding AANAT32–207 was amplified by PCR using the template pGEX-oAANAT and was ligated into pGEX-4T1 plasmid by using BamHI and XhoI sites. Sequences were verified (Veritas Laboratory, Rockville, MD). Thrombin (5 units/mg protein; Roche) digestion of GST-AANAT fusion proteins resulted in two additional residues (GS-) at the N terminus of the recombinant proteins (16).
Antisera. Anti-rat AANAT22–37 pT31 (As 3352), anti-oAANAT1–25 (As 3343), and anti-oAANAT1–207 (As 2819, also referred to as anti-AANAT1–205) have been described (4, 17, 18). Antisera were immunopurified (19) by using synthetic phosphopeptide (As 3352, As 5901), unphosphorylated peptide (As 3343), or unphosphorylated protein (As 2819). Immunopurified anti-oAANAT194–207 pS205 serum selectively detects oAANAT T31A pS205 (see Fig. 5, which is published as supporting information on the PNAS web site) and does not detect unphosphorylated AANAT, oAANAT T31A S205G, or AANAAT pT31 S205A. Immunopurified anti-rat AANAT22–37 pT31 detects oAANAT pT31 S205A but not oAANAT T31A pS205. Details are in Supporting Methods and Materials.
Immunodetection of AANAT. Images of AANAT immunosignals were captured by using a Kodak Image Station 2000 mm and quantitated by using Kodak 1d image-analysis software. Images were captured also by using Bio-MAX MR or X-Omat AR film (Eastman Kodak) (see Supporting Methods and Materials for details).
Assays. Published assays were used for determination of protein (protein assay kit, Bio-Rad; ref. 17), AANAT activity in tissue homogenates (Figs. 1 and 2 and ref. 12), and melatonin (18). Additional details appear in the figure legends.
Solid-Phase 14-3-3-Binding Assay. GST-14-3-3 was incubated with glutathione Sepharose beads (2 mg; Amersham Pharmacia) with GST-14-3-3 (0.5 mg) in binding buffer (150 mM NaCl in 20 mM PBS, pH 7.4); unbound 14-3-3 was removed by washing three times in binding buffer. The beads (0.2 mg of GST-14-3-3/mg of beads) were suspended (4 ml of binding buffer) and stored at 4°C for <1 week. This preparation is also referred to as “immobilized” 14-3-3.
For an assay, beads in 40 μl of the immobilized 14-3-3 suspension were resuspended in 40 μl of buffer A (100 mM NH4Ac buffer, pH 6.8, containing 5 mM DTT, 5% glycerol, and 0.1 mg/ml BSA). Binding of 32PO4-labeled peptides (specific activity, 1,200–1,400 dpm/pmol; 2.5 nM to 6.25 μM) to immobilized 14-3-3 was measured after incubation (90 min, 24°C) in 0.5 ml of buffer A; the beads were collected by centrifugation (300 × g, 3 min), washed twice with 0.5 ml of buffer A, and transferred to glass fiber discs (GF/C, 24 mm, Whatman; presaturated with 5% trichloroacetic acid) on a vacuum manifold. The filters were washed with 1 ml of 5% trichloroacetic acid and 0.5 ml of ethanol, dried, and analyzed for radioactivity. Maximum binding of [32PO4]oAANAT24–39 was ≈0.8 mol of peptide/mol of 14-3-3. Thirty-five percent maximal binding was achieved at a concentration of 50 nM.
The relative affinity of selected peptides and proteins was estimated by incubating a range of concentrations with 25 pmol of 32P-labeled AANAT24–39 peptide (1,200 dpm/pmol peptide; 50 nM) in 0.5 ml of buffer A (90 min, 24°C) and determining bound radioactivity.
Analysis of Effects of 14-3-3 on AANAT Activity. Kinetic analysis (Table 2) was as described (4). The influence of 14-3-3 on AANAT activity at physiological concentrations of substrates was estimated by incubating (30°C, 3 min) AANATs and 14-3-3 with [14C]serotonin (3 μM) and AcCoA (10 μM) in 40 μl of 100 mM NH4Ac buffer, pH 6.8, containing 5 mM DTT. The reaction was stopped by adding 10 μl of ethyl alcohol containing 1 mM N-acetylserotonin. Serotonin and N-acetylserotonin were resolved by TLC [silica-coated aluminum sheets; Whatman 4420 222; CHCl3/MeOH/HAc (glacial, 90:10:1)]. Migrating radioactivity (detected by Storm phosphorimager, Amersham Pharmacia) was associated with authentic N-acetylserotonin (detected by UV quenching).
Table 2. Kinetic analysis of the effects of T31 and S205 mutations on AANAT activity and effect of 14-3-3ζ.
| AANAT proteins | 14-3-3ζ protein | Km (μM) | Vmax (nmol/h per ng) |
|---|---|---|---|
| T31 S205 | - | 180 ± 3.5 | 2.9 ± 0.40 |
| pT31 pS205 | - | 170 ± 6.0 | 3.3 ± 0.27 |
| + | 24 ± 0.7* | 0.64 ± 0.09 | |
| T31A S205 | - | 310 ± 2.6 | 3.7 ± 0.56 |
| T31A pS205 | - | 300 ± 9.0 | 4.5 ± 0.28 |
| + | 1,200 ± 11.0* | 2.5 ± 0.12 | |
| T31 S205A | - | 280 ± 3.0 | 3.4 ± 0.42 |
| pT31 S205A | - | 250 ± 1.0 | 2.9 ± 0.36 |
| + | 170 ± 1.1* | 2.3 ± 0.18 |
Km of phosphorylated or unphosphorylated oAANAT and its mutants (≈15 ng AANAT enzyme/100-μl assay mixture; 30°C; 20-min incubation) for tryptamine using a saturating concentration of [3H]AcCoA (0.5 mM) in the presence or absence of 14-3-3ζ isoform (1:5 molar ratio of pAANAT/AANAT/14-3-3ζ monomer). Km was calculated by using nonlinear regression analysis (kaleidagraph, Synergy Software, Reading, PA), and the changes in the values were found to be significant.
P < 0.01. Km and Vmax values are represented as mean ± SE (n = 4). For further details, see Materials and Methods.
Statistics. Replicates and sample sizes are given in the figure legends. Error bars represent the standard error of the mean. Statistical analysis was by Student's t test using sigmaplot 2000 (Systat, Chicago).
Results
Daily Rhythm in Pineal AANAT pS205. AANAT pS205 was detected at low levels during the day and at 4- to 10-fold higher levels at night (Fig. 1 B and G). The 24-hour profile of AANAT pS205 (Fig. 1B) was generally similar to those of AANAT activity, protein, and pT31 in these glands (Fig. 1 A, C, and D); the latter observations confirm previous reports (3, 4, 11).
The degree to which the C- and N-terminal sites were phosphorylated was estimated by comparison of apparent to maximal phosphorylation achieved by PKA treatment (Fig. 2 and Table 1), which revealed that ≈40% of T31 sites and ≈55% of S205 sites are phosphorylated at night.
Photic Control of Phosphorylation of S205. Light exposure at night leads to a decrease in oAANAT activity, AANAT and protein, and circulating melatonin (3, 4, 11, 20), which was confirmed here (Fig. 1 E–I); the decrease in melatonin was approximately twice that in other parameters. In addition, light exposure decreased the percentage of each PKA site in AANAT that was phosphorylated (Fig. 2 and Table 1).
AANAT192–207 pS205 Binds to Immobilized 14-3-3ζ. The functional importance of pS205 in binding to 14-3-3ζ was examined by using a solid-phase binding assay in which GST-14-3-3ζ was immobilized. Previous studies had suggested that AANAT24–39 pT31 could bind to 14-3-3ζ because it competed with AANAT for binding to 14-3-3ζ (4). Binding of the pT31 peptide to 14-3-3ζ was confirmed here (Fig. 3A); it was found also that AANAT192–207 pS205 associates with 14-3-3ζ with >10-fold lower affinity.
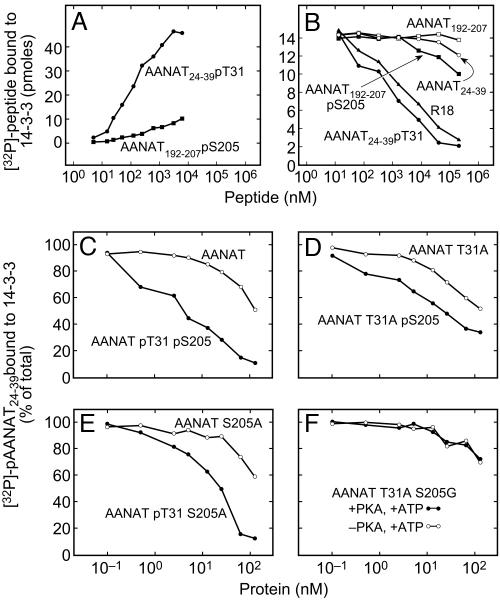
Fig. 3.
Binding of oAANAT-derived peptides, oAANAT, and mutated oAANAT proteins to immobilized 14-3-3ζ.(A) Binding of 32P-labeled peptides, [32P]AANAT24–39 pT31and [32P]AANAT192–207 pS205, to GST-14-3-3ζ immobilized on glutathione-Sepharose beads. (B) Inhibition of [32P]AANAT24–39 pT31 binding to immobilized 14-3-3 by phosphorylated and unphosphorylated AANAT-derived peptides and by R-18 (21). In the case of AANAT24–39 pT31, the decrease in specific activity was accounted for by determining binding of the ligand. (C–F) Inhibition of [32P]AANAT24–39 pT31 binding to immobilized 14-3-3ζ by phosphorylated and unphosphorylated AANATs: AANAT and AANAT pT31 pS205 (C), AANAT T31A and AANAT T31A pS205 (D), AANAT S205A and AANAT pT31 S205A (E), and AANAT T31A S205G and a preparation subjected to phosphorylation (ATP/PKA) (F). The plots represent a typical binding experiment in which each value is a mean of duplicates and each such experiment was repeated at least three times. The inhibition constant (Ki) for ligands that were measurable is indicated in Results and was calculated from their corresponding IC50 values by using prism (GraphPad, San Diego) and as described by Cheng and Prusosoff (27). Ki values are calculated from three independent experiments as mean ± SE (n = 3, P < 0.01). For additional details, see Materials and Methods.
R-18 is a peptide that selectively binds to the amphipathic binding groove of 14-3-3ζ proteins through phosphorylation-independent mechanisms (21). It inhibited binding of [32PO4]AANAT24–39 pT31 (Fig. 3B), indicating that the solid-phase competitive binding assay used provides a valid indication of binding by means of the amphipathic binding groove of 14-3-3ζ. Binding was also suppressed by AANAT24–39 pT31 > AANAT24–39 and AANAT192–207 pS205 > AANAT192–207 (Fig. 3B), demonstrating that these peptides bind to the amphipathic binding groove of AANAT in a phosphorylation-dependent manner. This study also revealed that intrinsic affinity of AANAT24–39 pT31 for 14-3-3 was >10-fold greater than that of AANAT192–207 pS205, confirming the results shown in Fig. 3A.
Phosphorylation of AANAT S205 or T31 Promotes Binding to 14-3-3ζ. 14-3-3 binding to full-length AANAT, AANAT T31A, AANAT S205A, and AANAT T31A S205G was studied as a function of PKA phosphorylation (Fig. 3 C–F). The unphosphorylated proteins bound to 14-3-3ζ >100-fold more strongly than the unphosphorylated T31 and S205 peptides (Fig. 3B), probably because of more numerous protein–protein interactions (5).
PKA phosphorylation caused an ≈30-fold increase in the affinity of AANAT for 14-3-3ζ (Fig. 3C). In contrast, the same treatment of the double-PKA site AANAT mutant, AANAT T31A S205G, had no effect (Fig. 3F), revealing that effects of PKA treatment are only caused by phosphorylation of T31 and S205.
PKA-dependent phosphorylation of the AANAT T31A and AANAT S205A increased their affinity for 14-3-3ζ by ≈7-fold (Ki = 226 ± 4.6 versus 31.8 ± 1.2 nM) and ≈10-fold (Ki = 232 ± 3.6 versus 21.7 ± 3.5 nM), respectively (Fig. 3 D and E). The PKA-dependent increase in affinity of AANAT for 14-3-3 was ≈30-fold (Ki = 224 ± 2.2 versus 7.2 ± 0.34 nM; Fig. 3C); the apparent affinity of the dual-phosphorylated protein was significantly greater (P < 0.01) than that due to single phosphorylation. These findings are consistent with a model in which 14-3-3 affinity for AANAT is highest when mediated by both pT31 and pS205.
Phosphorylation of S205 Is Required for 14-3-3-Dependent Regulation of AANAT Activity. It is known that phosphorylation of AANAT increases arylalkylamine affinity by ≈10-fold (4) in a 14-3-3ζ-dependent manner to <30 μM, as confirmed here (Table 2). In contrast, 14-3-3 reduces arylalkylamine affinity of single-phosphorylated AANAT T31A pS205 by ≈4-fold to 1,200 μM (Table 2). In contrast, substrate affinity of single-phosphorylated AANAT pT31 S205A did not show this 14-3-3-dependent change.
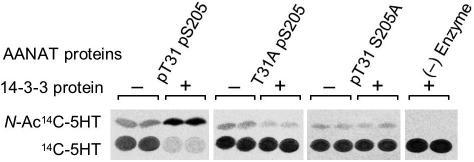
These effects of 14-3-3 binding were confirmed by using an assay that used nonsaturating concentrations of substrates that approximated nighttime concentrations of substrates (Fig. 4). 14-3-3ζ increased the conversion of [14C]serotonin to [14C]N-acetylserotonin by AANAT pT31 pS205 but inhibited AANAT T31A pS205-mediated conversion. AANAT pT31 S205A activity was not affected by 14-3-3. The findings shown in Table 2 and Fig. 4 establish that phosphorylation of both T31 and S205 of AANAT is essential for 14-3-3ζ-dependent activation, and AANAT activity is inhibited markedly if AANAT binds to 14-3-3 only by pS205.
Fig. 4.
Effect of 14-3-3ζ on the activity of phosphorylated AANAT at physiological concentrations of substrates (4, 28). A sample containing 15 ng of phosphorylated AANAT proteins oAANAT pT31 pS205, oAANAT S205A pT31, and oAANAT T31A pS205 was incubated with an ≈10-fold molar excess of 14-3-3ζ (125 ng). The addition of 14-3-3ζ increased 14C-labeled N-acetylserotonin (N-Ac-[14C]5HT) formation by AANAT pT31 pS205 from 22.6 to 109.0 pmol (average of two assays in which the results were essentially identical); there was no increase with the AANAT T31A pS205 or pT31A S205. For additional details, see Materials and Methods.
Discussion
The results of these studies are of interest in understanding the control of AANAT; of even broader interest is that they add to a growing body of evidence indicating that functionally relevant binding of proteins to 14-3-3 can occur through multiple sites in a protein and this interaction can involve a sequence that is not a commonly recognized and accepted consensus 14-3-3 binding motif (22). These points are discussed below.
Physiological Phosphorylation of AANAT. It is now clear that both the N- and C-terminal PKA sites in AANAT are physiologically phosphorylated (Fig. 1). The degree of phosphorylation of T31 and S205 seems to be ≈40% and ≈55%, respectively. The finding that major fractions of T31 and S205 are not phosphorylated may reflect the existence of a dynamic equilibrium between production, phosphorylation, dephosphorylation, and destruction of AANAT that maintains large pools of unphosphorylated and phosphorylated forms. However, it is possible also that pT31 and pS205 are dephosphorylated during the process of tissue removal and homogenate preparation and that only a minor pool of dephosphorylated AANAT exists in the cell.
It has been argued that a key step in the sequence of events leading to a light-induced decrease in AANAT protein is a decrease in cAMP and that this reduction in cAMP results in a decrease in PKA-mediated phosphorylation of AANAT. As a result, there is a shift in AANAT from the phosphorylated/14-3-3-bound form to an unbound form that is destroyed by proteasomal proteolysis. Our finding here that light exposure decreases the fractions of T31 and S205 that are phosphorylated is consistent with this scheme. However, this result does not rule out possible additional indirect effects of cAMP on the turnover of AANAT, i.e., through effects on proteasomal proteolysis or translation.
14-3-3 Binding of AANAT. The results of binding analysis establish that both pT31 and pS205 can interact with 14-3-3 and promote complex formation and that each site can act independently of each other. In the case of pT31, this finding is not surprising, because it is part of a consensus 14-3-3-binding motif, characterized by a proline plus two of the pT/pS residues in mode I (RSx[pS/pT]xP) and mode II (Rx Φx[pS/pT]xP).†† However, the binding of pS205 to 14-3-3 is not predictable because it is not located in such a site (13–15). Single-site binding of AANAT is of interest, because it also seems sufficient to slow proteasomal destruction of AANAT, as demonstrated by using semisynthetic forms of AANAT containing nonhydrolyzable pT31 or pS205 mimetics (ref. 7 and W. Zheng, D. Schwarzer, A. LeBeau, J.L.W., D.C.K., and P. A. Cole, unpublished data).
It seems clear to us that under physiological conditions, a single molecule of AANAT binds to a 14-3-3 dimer by virtue of contacts between pT31 and the amphipathic groove of one 14-3-3 monomer and also between pS205 with the other monomer. In addition, important protein–protein interactions occur at multiple sites (5), which contribute to the higher affinity of phosphorylated AANATs as compared to AANAT-based phosphopeptides (Fig. 3). This 1:1 AANAT/14-3-3 dimer model is consistent with the results of solution analysis (4) and a theoretical model (5). The 2:1 structure obtained by physical studies reflects the use of a truncated form of AANAT that lacked pS205 (5), thereby permitting two molecules of AANAT to bind to 14-3-3, each by means of the pT31 site.
Effects of 14-3-3 Binding on AANAT Kinetics. The importance of dual phosphorylation of AANAT is evident from the finding that 14-3-3-dependent activation occurs only if both sites are phosphorylated (Table 2 and Fig. 4). Although it is apparent that AANAT can weakly associate to 14-3-3 by multiple interactions in addition to the two PKA sites, these interactions alone do not change arylalkylamine kinetics. The two-site AANAT/14-3-3-binding model fits well with a two-site 14-3-3-binding model proposed by Yaffe (23) in which one high-affinity site, the “gatekeeper,” is required for binding; in the case of AANAT, the gatekeeper is pT31. Binding of the gatekeeper leads to binding of a second low-affinity site, in this case pS205, which reflects both the intrinsic affinity of that site and the “high local concentration induced by its proximity” (23). Yaffe also proposes that as a relatively firm structure, 14-3-3 acts as an “anvil” to induce conformational changes in the bound protein. In the case of AANAT, these changes are in the conformation of loop 1, a floppy element of AANAT that is part of the arylalkylamine-binding pocket (5, 16). Dual-site binding of AANAT to 14-3-3 seems to produce optimal configuration of this loop for high-affinity binding (≈30 μM).
The results of our study indicate that this model can be extended to account for the unexpected finding that single-site pS205 binding to 14-3-3 inhibits the enzyme by increasing the Km to 1.2 mM (Fig. 4 and Table 2), which may reflect binding of AANAT to 14-3-3 in a configuration that deconstructs the active site.
It is reasonable to consider that there may be an ordered association and dissociation in which the gatekeeper pT31 binds first, followed by binding of pS205. Subsequent shift to a single-site pT31-binding complex would reverse this activation; shift to pS205 binding would inhibit. The shift from dual to single binding could be directed toward the pS205-bound form by contacts formed in the complex that preferentially stabilize binding by means of pS205 relative to that through pT31. These contacts could involve the unstructured N-terminal portion of AANAT, which can interact with C-terminal sequence; the somewhat flexible C-terminal element of 14-3-3 (24); or both.
The dramatic inactivation of AANAT mediated by pS205 is physiologically relevant, because it would explain why exposure to light at night decreases circulating melatonin (Fig. 1E) more rapidly than total AANAT protein (Fig. 1 D and I).
Nonconsensus Site Binding to 14-3-3. As discussed above, it is widely recognized that most proteins bind to 14-3-3 through phosphorylated motifs with a proline plus two of the pT/pS. The binding of AANAT through a nonproline-containing sequence adds to the growing list of proteins that bind through nonconsensus 14-3-3-binding motifs (23). A striking similarity exists between atypical C-terminal 14-3-3-binding sequences of AANAT (–RRNpSDR-COOH), plant plasma membrane H+-ATPase (–QQYpTV-COOH; ref. 25), and Ibα subunit of glycoprotein Ib-IX-V (–RYSGHSL-COOH; ref. 26): all are C-terminal sequences. Accordingly, it seems that the terminal -COOH functionally substitutes for the kink that the proline provides by enhancing binding (22). The AANAT and ATPase sequences are of special interest because they can mediate phosphorylation-dependent binding. Based on this common feature, it is possible to propose a mode III 14-3-3-binding motif: pS/pT(X1–2)-COOH. The search for other mode III sequences may reveal that they are generally important for two-site binding to 14-3-3.
Two-site binding not only improves positioning but also allows more complex regulation to occur, i.e., regulation of each site by different kinases and phosphatases. For example, it is possible that AANAT is selectively dephosphorylated at the pT31 site by a phosphatase that has low activity toward pS205, which could confer ordered dissociation. Differential regulation of pT31 and of pS205 is consistent with the finding that the degree of phosphorylation of these sites is different (Fig. 2 and Table 1).
Supplementary Material
Acknowledgments
We express appreciation to Christian Schwartz, Mark Rollag, and Steven L. Coon for permitting inclusion of their results from unpublished studies; Weiping Zheng, Dirk Schwarzer, Aaron LeBeau, and Philip A. Cole for allowing us to cite their unpublished work; and Didier Chesneau, Christian Moussu, and Jean-Philippe Dubois for the melatonin assay and management of experimental animals. We thank Prof. A. Aitken for reading the manuscript and for helpful suggestions.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AANAT, arylalkylamine N-acetyltransferase; PKA, cyclic AMP-dependent protein kinase; oAANAT, ovine AANAT; p-, phosphorylated; hAANAT, human AANAT.
Footnotes
Numbering is based on the ovine AANAT (oAANAT) sequence.
The ≈10-fold forskolin-induced increase in intracellular AANAT activity that occurs in COS7 cells transfected with hAANAT but not with hAANAT pT31A (4) does not occur if cells are transfected with the hAANAT S205A mutant (C. Schwartz, S. L. Coon, M. Rollag, and D.C.K., unpublished data).
Uppercase indicates a highly conserved position; Φ, an aromatic or aliphatic amino acid; x, any amino acid.
References
- 1.Arendt, J. (1995) Melatonin and the Mammalian Pineal Gland (Chapman & Hall, London), pp. 201–285.
- 2.Klein, D. C. & Weller, J. L. (1970) Science 169, 1093–1095. [DOI] [PubMed] [Google Scholar]
- 3.Klein, D. C., Coon, S. L., Roseboom, P. H., Weller, J. L., Bernard, M., Gastel, J. A., Zatz, M., Iuvone, P. M., Rodriguez, I. R., Begay, V., et al. (1997) Recent Prog. Horm. Res. 52, 307–358. [PubMed] [Google Scholar]
- 4.Ganguly, S., Gastel, J. A., Weller, J. L., Schwartz, C., Jaffe, H., Namboodiri, M. A., Coon, S. L., Hickman, A. B., Rollag, M., Obsil, T., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 8083–8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Obsil, T., Ghirlando, R., Klein, D. C., Ganguly, S. & Dyda, F. (2001) Cell 105, 257–267. [DOI] [PubMed] [Google Scholar]
- 6.Gastel, J. A., Roseboom, P. H., Rinaldi, P. A., Weller, J. L. & Klein, D. C. (1998) Science 279, 1358–1360. [DOI] [PubMed] [Google Scholar]
- 7.Zheng., W., Zhang, Z., Ganguly, S., Weller, J. L., Klein, D. C. & Cole, P. A. (2003) Nat. Struct. Biol. 10, 1054–1057. [DOI] [PubMed] [Google Scholar]
- 8.Klein, D. C., Baler, R., Roseboom, P. H., Weller, J. L., Bernard, M., Gastel, J. A., Zatz, M., Iuvone, P. M., Begay, V., Falcon, J., et al. (1998) in Handbook of Behavioral State Control: Cellular and Molecular Mechanisms, eds. Lydic, R. & Baghdoyan, H. (CRC, Boca Raton, FL), Vol. 4, pp. 45–59. [Google Scholar]
- 9.Coon, S. L., Roseboom, P. H., Baler, R., Weller, J. L., Namboodiri, M. A., Koonin, E. V. & Klein, D. C. (1995) Science 270, 1681–1683. [DOI] [PubMed] [Google Scholar]
- 10.Klein, D. C., Ganguly, S., Coon, S., Weller, J. L., Obsil, T., Hickman, A. & Dyda, F. (2002) Biochem. Soc. Trans. 30, 365–373. [DOI] [PubMed] [Google Scholar]
- 11.Klein, D. C., Ganguly S, Coon, S. L., Shi, Q., Gaildrat, P., Morin, F., Weller, J. L., Obsil, T., Hickman, A. & Dyda, F. (2003) J. Neuroendocrinol. 15, 370–377. [DOI] [PubMed] [Google Scholar]
- 12.Coon, S. L., Del Olmo, E., Young, W. S., III, & Klein, D. C. (2002) J. Clin. Endocrinol. Metab. 87, 4699–4706. [DOI] [PubMed] [Google Scholar]
- 13.Aitken, A. (2003) Methods Mol. Biol. 211, 465–485. [DOI] [PubMed] [Google Scholar]
- 14.Yaffe, M. B. (2004) Biochem. J. 379, 395–408.14744259 [Google Scholar]
- 15.Fu, H., Subramanian, R. R. & Masters, S.C. (2000) Annu. Rev. Pharmacol. Toxicol. 40, 617–647. [DOI] [PubMed] [Google Scholar]
- 16.Hickman, A. B., Namboodiri, M. A. A., Klein, D. C. & Dyda, F. (1999) Cell 97, 361–369. [DOI] [PubMed] [Google Scholar]
- 17.Coon, S. L., Weller, J. L., Korf, H. W., Namboodiri, M. A. A., Rollag, M. & Klein, D. C. (2000) J. Biol. Chem. 275, 24097–24107. [DOI] [PubMed] [Google Scholar]
- 18.Coon, S. L., Zarazaga, L. A., Malpaux, B., Ravault, J. P., Bodin, L., Voisin, P., Weller, J. L. Klein, D. C. & Chemineau, P. (1999) Am. J. Physiol. 277, E792–E797. [DOI] [PubMed] [Google Scholar]
- 19.Smith, D. E. & Fisher, P. A. (1984) J. Cell Biol. 99, 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Namboodiri, M. A., Sugden, D., Klein, D. C., Grady, R., Jr., & Mefford, I. N. (1985) J. Neurochem. 45, 832–835. [DOI] [PubMed] [Google Scholar]
- 21.Wang, B., Yang, H., Liu, Y. C., Jelinek, T., Zhang, L., Ruoslahti, E. & Fu, H. (1999) Biochemistry 38, 12499–12504. [DOI] [PubMed] [Google Scholar]
- 22.Aitken, A. (2002) Plant Mol. Biol. 50, 993–1010. [DOI] [PubMed] [Google Scholar]
- 23.Yaffe, M. B. (2002) FEBS Lett. 513, 53–57. [DOI] [PubMed] [Google Scholar]
- 24.Silhan, J., Obsilova, V., Vecer, J., Herman, P., Sulc, M., Teisinger, J. & Obsil, T. (2004) J. Biol. Chem. 279, 49113–49119. [DOI] [PubMed] [Google Scholar]
- 25.Svennelid, F., Olsson, A., Piotrowski, M., Rosenquist, M., Ottman, C., Larsson, C., Oecking, C. & Sommarin, M. (1999) Plant Cell 11, 2379–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews, R. K., Harris, S. J., McNally, T. & Berndt, M. C. (1998) Biochemistry 37, 638–647. [DOI] [PubMed] [Google Scholar]
- 27.Cheng, Y. & Prusosoff, W. H (1973) Biochem. Pharmacol. 22, 3099–3108. [DOI] [PubMed] [Google Scholar]
- 28.Namboodiri, M. A., Sugden, D., Klein, D. C., Tamarkin, L. & Mefford, I. N. (1985) Comp. Biochem. Physiol. 80, 731–736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.