Highlight
Loss-of-function of the rice eEF1A-like protein SPL33 causes a lesion-mimic phenotype by triggering programmed cell death, and increases disease resistance, likely through activating defense pathways.
Keywords: Defense responses, eukaryotic translation elongation factor 1 alpha (eEF1A), lesion-mimic mutant, Oryza sativa, programed cell death, SPL33.
Abstract
Lesion-mimic mutants are useful to dissect programmed cell death and defense-related pathways in plants. Here we identified a new rice lesion-mimic mutant, spotted leaf 33 (spl33) and cloned the causal gene by a map-based cloning strategy. SPL33 encodes a eukaryotic translation elongation factor 1 alpha (eEF1A)-like protein consisting of a non-functional zinc finger domain and three functional EF-Tu domains. spl33 exhibited programmed cell death-mediated cell death and early leaf senescence, as evidenced by analyses of four histochemical markers, namely H2O2 accumulation, cell death, callose accumulation and TUNEL-positive nuclei, and by four indicators, namely loss of chlorophyll, breakdown of chloroplasts, down-regulation of photosynthesis-related genes, and up-regulation of senescence-associated genes. Defense responses were induced in the spl33 mutant, as shown by enhanced resistance to both the fungal pathogen Magnaporthe oryzae and the bacterial pathogen Xanthomonas oryzae pv. oryzae and by up-regulation of defense response genes. Transcriptome analysis of the spl33 mutant and its wild type provided further evidence for the biological effects of loss of SPL33 function in cell death, leaf senescence and defense responses in rice. Detailed analyses showed that reactive oxygen species accumulation may be the cause of cell death in the spl33 mutant, whereas uncontrolled activation of multiple innate immunity-related receptor genes and signaling molecules may be responsible for the enhanced disease resistance observed in spl33. Thus, we have demonstrated involvement of an eEF1A-like protein in programmed cell death and provided a link to defense responses in rice.
Introduction
Plants have developed elaborate mechanisms of protection from attack by pathogens, the most common of which is the hypersensitive response (HR), which triggers rapid programmed cell death (PCD) to inhibit further invasion or proliferation of many pathogens in host plant tissues (Heath, 2000). Typical physiological processes associated with the HR include systemic signals such as bursts of reactive oxygen species (ROS) and production of free radicals, induction of pathogenesis-related (PR) genes, accumulation of antimicrobial compounds, and cell wall fortification through callose deposition (Qiao et al., 2010; Shirsekar et al., 2014). Although the HR plays an important role in host resistance to pathogens, its underlying molecular mechanisms are not fully understood.
As a means of dissecting HR-mediated PCD, researchers have identified and characterized a number of lesion-mimic mutants (LMMs) in a range of plant species, including Arabidopsis (Lorrain et al., 2004), maize (Johal et al., 1995) and rice (Shirsekar et al., 2014; Fekih et al., 2015; Wang et al., 2015a). These mutants spontaneously develop localized cell death lesions resembling those caused by HR in the absence of pathogen infection, abiotic stress or mechanical damage (Xu et al., 2014). Many LMMs display significantly enhanced resistance to disease (Wang et al., 2015a; Wang et al., 2015b) and it is believed that such mutants are ideal tools for deciphering the signal pathways of PCD and defense responses in plants. Therefore, cloning and characterization of the genes involved in LMMs should aid the elucidation of molecular mechanisms underlying plant disease resistance, possibly opening a way to development of broad-spectrum disease resistance (Wang et al., 2015b).
Previous studies have shown that the functional alleles of LMM genes encode various types of different proteins, including membrane-associated proteins (Lorrain et al., 2004; Noutoshi et al., 2006), ion channel proteins (Rostoks et al., 2006; Mosher et al., 2010), zinc-finger proteins (Wang et al., 2005), transcription factors (Yamanouchi et al., 2002; Li et al., 2004), E3 ubiqutin ligase (Zeng et al., 2004), porphyrin (Ishikawa et al., 2001), oxidoreductases (Tanaka et al., 2003; Yang et al., 2004), protein kinases (Liang et al., 2003; Wang et al., 2015c), clathrin-associated adaptor protein (Qiao et al., 2010), nucleotide-binding site leucine-rich repeat (NBS-LRR) type proteins (Tang et al., 2011), splicing factors (Chen et al., 2012), UDP-N-acetylglucosamine pyrophosphorylase (Wang et al., 2015b), and AAA-type ATPase (Fekih et al., 2015), as well as proteins involved in biosynthesis or metabolism of fatty acids/lipids, porphyrin and phenolic compounds (Undan et al., 2012). Thus the molecular mechanisms regulating lesion-mimic cell death and defense response in plants seem to be very complicated, and the study of LMMs not only identifies the mutated gene loci but also indicates how they might be involved in various physiological and defense-related signals (Undan et al., 2012).
Eukaryotic translation elongation factor 1 alpha (eEF1A) has a pivotal role in protein synthesis by catalysing GTP-dependent binding of aminoacyl-tRNA to the acceptor site of the ribosome, and is the second most abundant protein in eukaryote cells after actin (Mateyak and Kinzy, 2010). Besides its canonical role in translation, eEF1A has been implicated in PCD (apoptosis) in higher vertebrates (Mateyak and Kinzy, 2010). In mammals, eEF1A has two isoforms, eEF1A1 and eEF1A2, which are encoded by different genes in the eEF1A gene family. eEF1A1 is expressed in all tissues and has pro-apoptotic properties and a possible anti-apoptotic behavior; eEF1A2 is present only in brain, heart, and skeletal muscle and has anti-apoptotic properties in ovarian, breast, pancreatic, liver, and lung cancer (Abbas et al., 2015; Migliaccio et al., 2015). It was suggested that oncogenes encoding eEF1As could be used as potential prognostic markers for certain cancers, and perhaps research on those genes could lead to identification of novel therapeutic targets (Lee, 2003). Although a handful of genes encoding eEF1As have been cloned and characterized in plants (Xu et al., 2007), the biological phenotypes resulting from eEF1A gene mutations have not been reported, and thus the question of whether eEF1A genes are involved in plant PCD and defense responses remains to be answered.
In order to further elucidate the molecular mechanism of PCD and defense response in plants, we isolated and characterized a new rice lesion-mimic mutant named spotted leaf 33 (spl33). This mutant exhibits spotted leaves and enhances resistance to both the rice blast and bacterial blight diseases. The SPL33 allele was identified through a map-based cloning strategy combined with sequencing, and was predicted to encode an eEF1A-like protein. This gene is constitutively expressed in all tissues and developmental stages examined, and its encoded protein, SPL33, is localized in the endoplasmic reticulum (ER). Cell death, early leaf senescence and defense responses were observed in the spl33 mutant. These results indicated that loss of SPL33 function was the cause of regulated cell death, early senescence and enhanced defense responses.
Materials and methods
Plant materials and growth conditions
The rice spotted leaf mutant spl33 was isolated from an ethyl methane sulfonate mutant pool of the japonica rice cv. Nipponbare. For light treatments the middle part of leaf blades of seedlings in a growth chamber (12 h of light at 30 °C/12 h of darkness at 20 °C) was wrapped with aluminum foil to block light entry. Reciprocal crosses were made between spl33 and wild type (WT) for preliminary genetic analysis, and detailed genetic analysis was performed on an F2 population from a cross between spl33 and indica cv. Dular. All F2 individuals and corresponding parents were grown in a paddy field at the Changping Experimental Station of the Institute of Crop Science from April to October.
DNA extraction and molecular marker development
Genomic DNA was extracted from frozen young leaves using the cetyltrimethylammonium bromide (CTAB) method (Murray and Thompson, 1980). Insertion and deletion (InDel) markers were developed as described by Ma et al. (2015).
Linkage analysis and mapping of spl33
Two hundred and twelve InDel markers evenly distributed across all the 12 chromosomes (Lei et al., 2013) were screened for polymorphisms between the parents used. Twenty individuals with the spl33 phenotype and 20 with WT phenotype from an F2 population of spl33/Dular were initially used for linkage analysis; 476 F2 individuals with the spl33 phenotype were used for fine-mapping. InDel marker development and PCR amplifications were performed as described previously (Ma et al., 2015). Markers used in this study are listed in Supplementary Table S1 at JXB online.
Complementation and overexpression of SPL33 in spl33 mutant plants
For complementation of the spl33 mutation, a 9881 bp WT genomic DNA fragment containing the entire SPL33 coding region along with 2732 bp upstream and 2851 bp downstream sequences was amplified by PCR using the primers pP1305F and pP1305R (see Supplementary Table S2). The PCR product was inserted into the binary vector pCAMBIA1305.2 to generate the transformation plasmid pGSPL33. To construct an overexpression vector, the 1968 bp cDNA sequence of SPL33 was amplified by PCR using the primers pC1390F and pC1390R (Supplementary Table S2), inserted downstream of the ubiquitin promoter, and then fused in the vector pCUbi1390, resulting in the plasmid pUbi::SPL33. To determine the function of domains in SPL33, the cDNA fragments of 1–669 bp (spanning the amino acids 1–223) and 670–1968 bp (spanning the amino acids 224–655) were amplified using the primers p1390-SPL331–223F/R and p1390-SPL33224–655F/R, respectively, inserted downstream of the ubiquitin promoter in pCUbi1390, resulting in pUbi::SPL331–223 and pUbi::SPL33224–655. All the constructs were verified by sequencing and subsequently introduced into spl33 by Agrobacterium tumefaciens-mediated transformation as described previously (Jeon et al., 2000).
Multiple sequence alignment
Gene prediction was performed using the Rice Genome Annotation Project database (RGAP, http://rice.plantbiology.msu.edu/). The major functional domains of SPL33 were predicted by the Simple Modular Architecture Research Tools (SMART) program (http://smart.embl-heidelberg.de/). Homologous sequences of SPL33 were identified using the NCBI Blastp search program (http://www.ncbi.nlm.nih.gov/) and Phytozome (http://www.phytozome.net/). Multiple sequence alignments were conducted using the software MEGA v4.1 (http://www.megasoftware.net/) and DNAMAN v6.0 (http://www.lynnon.com/).
Quantitative real-time PCR analysis
RNA was extracted from flag leaves, leaf sheaths, culms, and young panicles at the booting stage, and from seedling roots. For expression analysis of anti-oxidative enzyme-, photosynthesis-, and senescence-related genes, RNA was extracted from leaves of spl33 and WT at 28 days after sowing (DAS). The total RNA extraction, reverse transcription and Real-time PCR were performed as described previously (Ma et al., 2015). Primer pairs designed using GenScript (https://www.genscript.com/ssl-bin/app/primer) are listed in Supplementary Table S3. The rice Ubiquitin gene (LOC_Os03g13170) was used as a reference (primer pair Ubi).
Histochemical β-glucuronidase assay
A 2724 bp promoter fragment upstream of the ATG start of SPL33 was amplified by PCR using primers Pro-SPL33F and Pro-SPL33R (see Supplementary Table S2), and the amplicon was cloned into the BamHI/BglII site of pCAMBIA1305.1. This resulting construct, pSPL33 promoter–GUS, was introduced into WT by the Agrobacterium-mediated method as described previously (Jeon et al., 2000). β-Glucuronidase (GUS) signals were observed as described previously (Jefferson, 1987).
Subcellular localization
A 1965 bp SPL33 cDNA fragment was amplified by the primers SPL33-GFP-F/R and 1305GFP-SPL33F/R (see Supplementary Table S2). The PCR product was cloned into the N-terminus of the green fluorescent protein (GFP) coding region in pAN580 and pCAMBIA1305.1-GFP vectors, to generate an SPL33–GFP fusion expression vector under the control of the CaMV 35S promoter using the Clonetech in-fusion PCR cloning system (TaKaRa). Transient expression constructs were co-transformed into rice protoplasts with the marker plasmid mCherry–HDEL (Nelson et al., 2007), and transfected protoplasts were incubated as described previously (Chen et al., 2006). GFP fluorescence was detected using a laser confocal scanning microscope (Leica TCS SP5). For transient expression in intact leaves, A. tumefaciens strain AH109 carrying the GFP constructs together with the p19 strain (Voinnet et al., 2003) and ER marker mCherry–HDEL were infiltrated into ~5–6-week-old Nicotiana benthamiana leaves as described previously (Lin et al., 2012).
Pigment determination and transmission electron microscopy analysis
Equal weights of freshly collected leaves from spl33 and WT at 28 DAS were used to determine contents of chlorophyll and carotenoids following the method of Arnon (1949). Transverse sections of leaves from plants at 28 DAS grown in paddy conditions were used for transmission electron microscopy observation as described previously (Dong et al., 2013).
Histochemical marker staining assay
Leaves of spl33 and WT at 28 DAS were harvested for histochemical assay. Trypan blue staining for dead cells and 3,3′-diaminobenzidine (DAB) staining for H2O2 accumulation were tested as described previously (Dietrich et al., 1994; Thordal-Christensen et al., 1997). Callose accumulation in leaves was examined as described previously (Wolter et al., 1993).
Determination of resistance to rice blast and bacterial blight in spl33
Isolates of 12 Magnaporthe oryzae (M. oryzae) pathotypes virulent on WT were used to infect spl33 plants. Inoculum preparation and seedling inoculation with the isolates followed a previously described procedure (Li et al., 2008). Disease reactions were scored 7 days post-inoculation on a scale of 0–5. The disease index was calculated as described previously (Wang et al., 2013).
Isolates of 11 Xanthomonas oryzae pv. oryzae (Xoo) pathotypes virulent on WT were used to evaluate resistance to bacterial blight in spl33 plants. The isolates were prepared and inoculated as described by Wang et al. (2014). Lesion lengths were measured 2 weeks after inoculation.
Transcriptome sequencing and data analysis
The spl33 and WT leaves of 4-week-old plants in a growth chamber with 16 h of light at 28 °C and 8 h of darkness at 25 °C were used for total RNA extraction. RNAs were checked using an Agilent Bioanalyzer 2100 (Agilent Technologies, Waldbronn, Germany). High-quality samples with total RNA concentration ≥40 ng μl–1, and RNA integrity (RIN) value ≥7.0 were used for construction of cDNA libraries. The cDNA libraries were constructed using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (New England Biolabs, USA) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. High-throughput sequencing was performed on an Illumina HiSeq 2500 (version 4.0) genome analyser (Illumina, San Diego, CA, USA). The 150 bp paired-end reads generated from spl33 and WT plants were processed as described previously (Hiremath et al., 2011). In total 27 892 108 and 27 909 005 clean reads were obtained for spl33 and WT, respectively. Clean reads were aligned to the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) using Bowtie2 (Langdon, 2015). Differentially expressed genes (DEGs) and transcript expression analyses were performed using Cufflinks (Trapnell et al., 2012), DEseq2 (Love et al., 2014) and EdgeR (Robinson et al., 2010). DEGs were determined by at least two packages, with an absolute log2-fold change value ≥1 and a false discovery rate ≤0.001. Gene Ontology (GO) annotations of DEGs were obtained from the Rice Genome Annotation Project. GO functional enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed as described previously (Tang et al. 2013).
Results
Characterization of the spl33 mutant
Small, reddish-brown lesions started to appear on the leaves of three-leaf-stage spl33 mutant seedlings (about 20 DAS) and continued to form throughout ripening growth stages (Fig. 1A–C). Notably, newly emerging leaves showed no lesions and were indistinguishable from wild-type (WT) Nipponbare (Nip), but gradually developed lesions as they expanded (Fig. 1A). The spl33 phenotype was seen under both field conditions in summer (25–35 °C) and greenhouse conditions in winter (25–30 °C) (Fig. 1A). Young seedlings displayed fewer and relatively small reddish-brown lesions on fully expanded leaves; the lesions increased in both number and size at the early flowering stage, followed by leaf chlorosis and rapid senescence of the whole plant at the late flowering stage, while the leaves of WT plants stayed green (Fig. 1B, C). In addition, tiller number, plant height, panicle length, total grain number per plant, spikelet fertility and 1000-grain weight of the spl33 mutant were all significantly decreased relative to WT plants (see Supplementary Fig. S1).
Fig. 1.
Comparison of wild type (Nip) and spl33 plants. (A) Lesion mimic phenotype in spl33. a, clean leaf blade in the Nipponbare wild type (WT); b and c, lesion mimics at seedling and tillering stages of greenhouse-grown spl33 in winter; d and e, lesion mimics at seedling and tillering stages of field-grown spl33 in summer. (B) spl33 plant at tillering stage showing early senescence in spl33. (C) WT and spl33 plants at heading stage. Note that spl33 was full of lesion mimics. Nip, Nipponbare. Scale bar: 1cm in (A), 10 cm in (B, C).
To determine whether the lesion phenotype of the spl33 mutant was affected by environmental factors, such as light, we performed light avoidance assays on the spl33 mutant and WT. In the light avoidance experiment, part of the newly emerging leaves was covered with aluminum foil to block light while the uncovered leaf areas remained exposed to light. The leaf area without exposure to light did not develop lesions of any size in contrast to the neighboring uncovered areas, which developed scattered lesions (see Supplementary Fig. S2), indicating that lesion formation in spl33 is light-dependent and that light-induced lesion-related signals are not transported across the leaf. Thus, we identified an HR-mimicking mutant, spl33, that develops lesions in a light-dependent manner.
Map-based cloning of the SPL33 locus
Reciprocal crosses were made between the spl33 mutant and WT. The F1 plants exhibited WT phenotype and the segregation of F2 populations fitted a phenotype ratio of 3 WT:1 mutant indicating that the spl33 phenotype was caused by a single recessive nuclear gene mutation (see Supplementary Table S4).
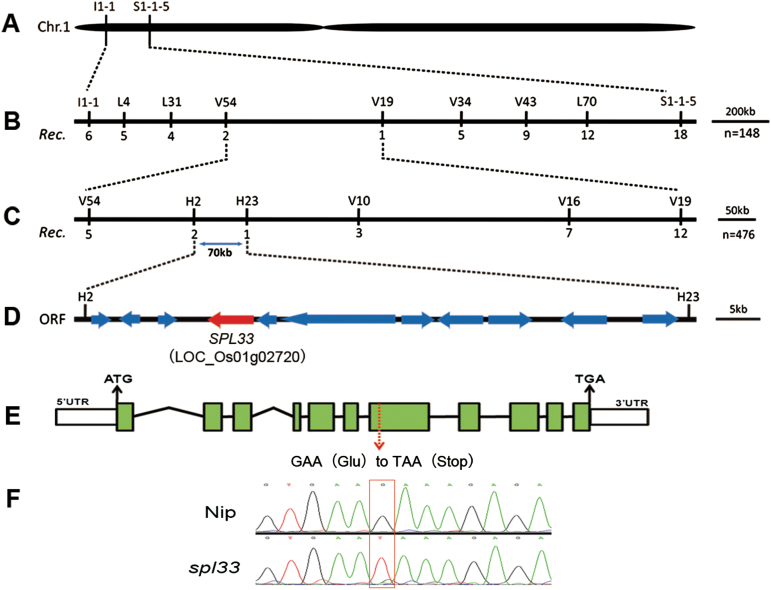
For mapping of the spl33 locus, an F2 population from the cross of spl33 × indica cv. Dular was used. When 20 F2 individuals with mutant phenotype and 20 with WT phenotype were genotyped using polymorphic markers selected from 212 insertion and deletion (InDel) markers (Lei et al., 2013), we localized the mutation locus between the InDel markers I1-1 and S1-1-5 on chromosome 1 (Fig. 2A). With further genotyping on 148 mutant F2 individuals derived from the same cross with seven additional polymorphic markers, the spl33 locus was delimited to the V54–V19 interval (Fig. 2B). Using an additional 476 F2 mutant individuals and four newly developed polymorphic markers, we finally anchored spl33 to a 70 kb region between InDel markers H2 and H23 (Fig. 2C). Eleven open reading frames (ORFs) were predicted in this region (Fig. 2D and Supplementary Table S5) (http://rice.plantbiology.msu.edu/). Sequencing and comparison of those ORFs cloned from spl33 and WT revealed that the fourth ORF (LOC_Os01g02720) had a single-base substitution (G2493→T2493) in its seventh exon, leading to a premature stop (Fig. 2E, F), thus placing LOC_Os01g02720 as a candidate position for SPL33.
Fig. 2.
Genetic and physical maps of the SPL33 gene. (A) The SPL33 gene was located on chromosome 1 between InDel markers I1-1 and S1-1-5. (B) The SPL33 gene was delimited to the V54-V19 interval using 148 F2 mutant individuals; marker names and number of recombinants are shown. (C) Fine genetic mapping of the SPL33 gene based on 476 mutant F2 individuals. (D) Eleven putative ORFs were located in an ~70-kb region. (E) Gene structure LOC_Os01g02720. Eleven exons and ten introns are indicated by green rectangles and black lines, respectively; a G to T point mutation was identified in the seventh exon (red arrow) generating a premature termination codon. (F) Sequence analysis of the G-to-T mutation site in plants of wild type and spl33.
Functional complementation of the spl33 mutant with LOC_Os01g02720
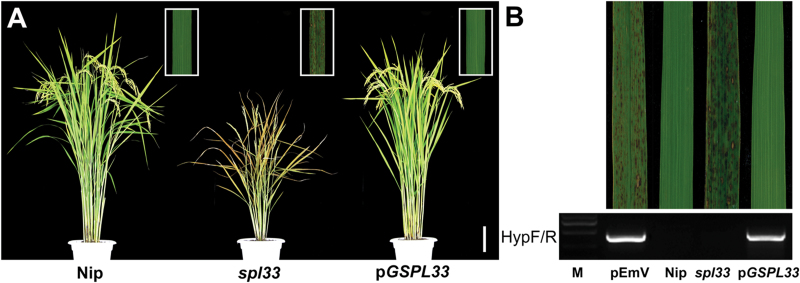
To verify whether the single base substitution in LOC_Os01g02720 was responsible for the spl33 phenotype, we constructed the vector pGSPL33 that contains a WT-derived 9881 bp genomic DNA fragment consisting of the entire SPL33 coding region, 2732 bp upstream and 2851 bp downstream sequences, and introduced it into spl33 by A. tumefaciens-mediated transformation. The corresponding empty vector pEmV was also transformed as a control. Of 72 regenerated T0 plants, 64 were positive transformants, and all of them exhibited a complete rescue of the mutant phenotype (Fig. 3A), whereas none of the plants transformed with the control vector displayed the WT phenotype (Fig. 3B). The rescued phenotype was seen again in T1 plants positive for the transgene (see Supplementary Fig. S3). Meanwhile, we also introduced an overexpression vector (pUbi::SPL33) containing the entire cDNA sequence of SPL33 into the spl33 mutant. Normal green phenotypes were also completely recovered in 32 independent pUbi::SPL33 transgenic lines (see Supplementary Fig. S4). Taken together, those results demonstrate that LOC_Os01g02720 is SPL33 and that the single-base substitution G2493→T2493 in spl33 causes the lesion-mimic phenotype.
Fig. 3.
Genetic complementation of spl33. (A) spl33 plant transformed with the genomic sequence of SPL33 (pGSPL33) was completely recovered to the wild type Nipponbare (Nip) phenotype. The insert indicates enlargement of leaf section with lesion spots. (B) Transgenic plants were verified by the presence of the hygromycin selectable marker gene. M, molecular markers; pEmV, the empty vector. Scale bar: 10 cm in (A).
SPL33 encodes an eEF1A-like protein
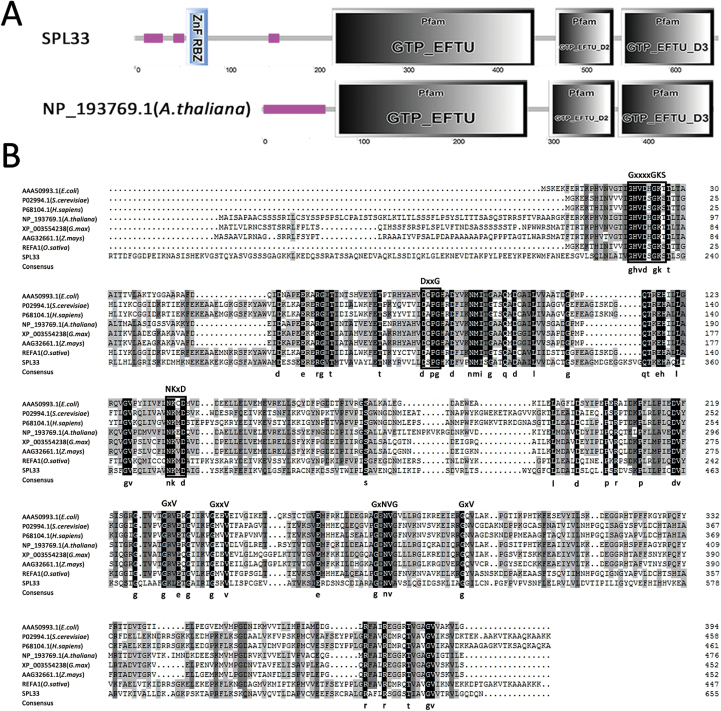
Sequence comparison between genomic DNA and cDNA showed that SPL33 is composed of 11 exons separated by 10 introns (Fig. 2E). The coding sequence (CDS) of SPL33 consists of 1968 nucleotides, and encodes a putative 655 amino acid protein with a molecular mass of 71 kDa. SPL33 was predicted to encode an eEF1A-like protein, containing a zinc finger domain, a GTP-binding domain (GTP_EFTU), and two oligonucleotide binding domains referred to as domain 2 (GTP_EFTU_D2) and domain 3 (GTP_EFTU_D3) (Fig. 4A). GTP_EFTU_D2 adopts a β-barrel structure, and is involved in binding to an aminoacyl-tRNA. GTP_EFTU_D3 is involved in binding to both aminoacyl-tRNA and eukaryotic factor 1 beta (eEF1B). The N-terminal of SPL33 also has a zinc finger domain that is not present in other eEF1As, such as Arabidopsis eEF1A (Fig. 4A). Comparison of the amino acid sequences of SPL33 with those orthologs from other organisms showed that the three regions significant for binding of GTP and hydrolysis of GTP to GDP (Bourne et al., 1991), and some other active amino acid sites, are highly conserved in all aligned eEF1A or EF-Tu (Fig. 4B).
Fig. 4.
SPL33 encodes a eukaryotic translation elongation factor1A (eEF1A)-like protein. (A) Predicted domains of SPL33 by Simple Modular Architecture Research Tools (SMART). SPL33 has a zinc finger domain (blue rectangle) at the N-terminus. (B) Alignment of the conserved motifs of eukaryotic translation elongation factor 1 alpha (eEF1A) and prokaryotic elongation factor (EF-Tu) from multiple organisms. The amino acid sequence alignment indicated that eEF1A is a highly conserved domain. The region corresponding to the GTP/GDP binding domain of GTP-binding proteins is indicated by white boxes, the conserved amino acid sequences are shown at the top, and the consensus amino acid residues are shown at the bottom.
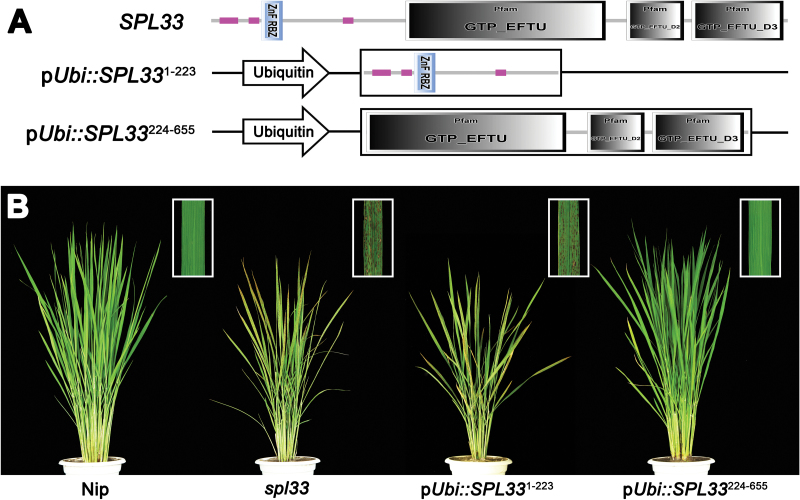
To further confirm the functions of different domains of SPL33, we performed complementation tests by separately transforming the 1–669 bp sequence of SPL33 (SPL331–223, encoding the first 223 amino acids) containing the zinc finger domain and the 670–1968 bp sequence of SPL33 (SPL33224–655, encoding the remaining 432 amino acids) containing the three EF-Tu domains, both under control of the ubiquitin promoter, into the spl33 mutant. The mutant phenotype was completely rescued in the pUbi::SPL33224–655 positive plants, whereas the pUbi::SPL331–223 vector failed to complement the spl33 mutant phenotype (Fig. 5). These results indicated that the C-terminal region of SPL33 containing the three EF-Tu domains is enough for functioning, whereas the N-terminal zinc finger domain is redundant, at least for preventing rice leaves from showing lesion mimics.
Fig. 5.
Dissection of SPL33 by transformation. (A) Schematic diagram of the overexpression vectors. pUbi::SPL331–223: the 1–669 bp sequence of SPL33 (SPL331–223, encoding amino acids 1–223 of SPL33) that contains the zinc finger domain under the control of the ubiquitin promoter; pUbi::SPL33224–655: the 670–1968 bp sequence of SPL33 (SPL33224–655, encoding amino acids 224–655 of SPL33) that contains three EF-Tu structural domains under the control of the ubiquitin promoter. (B) Phenotypes of wild-type (Nip), spl33 mutant, and two transgenic T0 plants carrying pUbi::SPL331–223 and T0 pUbi::SPL33224–655, respectively. The inset indicates enlargement of leaf section with lesion spots.
Expression pattern analysis of SPL33
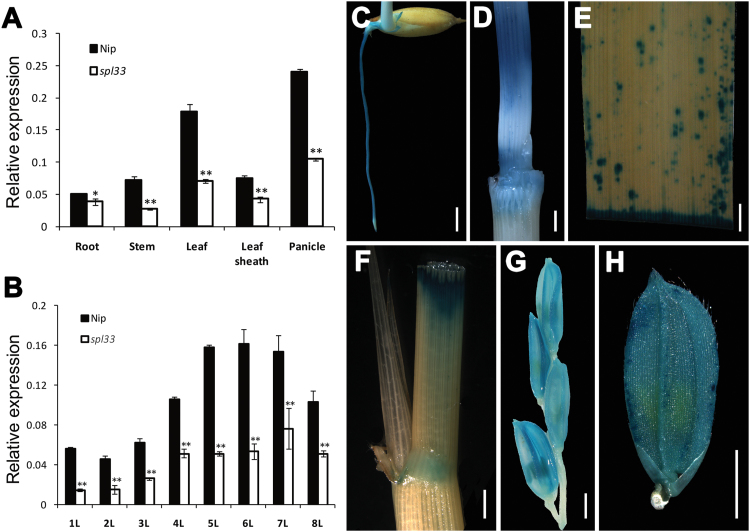
Quantitative real-time PCR (qRT-PCR) analysis of SPL33 in various organs showed that SPL33 was ubiquitously expressed in all organs examined (Fig. 6A). The strongest expression was detected in panicles and leaves, with relatively weak expression in roots, stems and leaf sheaths. Expression levels in all these organs in the spl33 mutant were significantly decreased relative to those of the wild-type (Fig. 6A). In addition, the expression level of SPL33 differed in leaves at different growth stages, and peaked in the sixth leaf (Fig. 6B). We generated transgenic plants of pSPL33::GUS expressing the GUS reporter gene driven by the native promoter of SPL33 to more precisely examine the spatial and temporal expression patterns of SPL33. Consistent with the qRT-PCR results, we observed GUS signals in all tissues of transgenic plants (Fig. 6C–H). These results indicated that SPL33 is broadly expressed in all organs and at all developmental stages examined.
Fig. 6.
Expression analysis of SPL33. (A) Expression of SPL33 in roots of 7-day-old seedling, stem, flag leaf blade and sheath, and young panicle at booting stage of wild type Nipponbare (Nip) and mutant spl33 analysed by quantitative RT-PCR. (B) Developmental expression pattern of SPL33 in different leaves along plant growth. 1L–8L represent the first to eighth leaf, respectively. (C–H) Histochemical signals in plants carrying the SPL33 promoter–GUS reporter gene. GUS signals were detected in the root (C), stem (D), leaf (E), leaf sheath (F), panicle (G), and spikelet (H). Scale bar: 2 mm in (C–H). Error bars in (A, B) indicate standard deviations of three independent samples. Data are means±SD of three biological replicates (Student’s t-test: *P<0.05; **P<0.01).
Subcellular localization of SPL33 protein
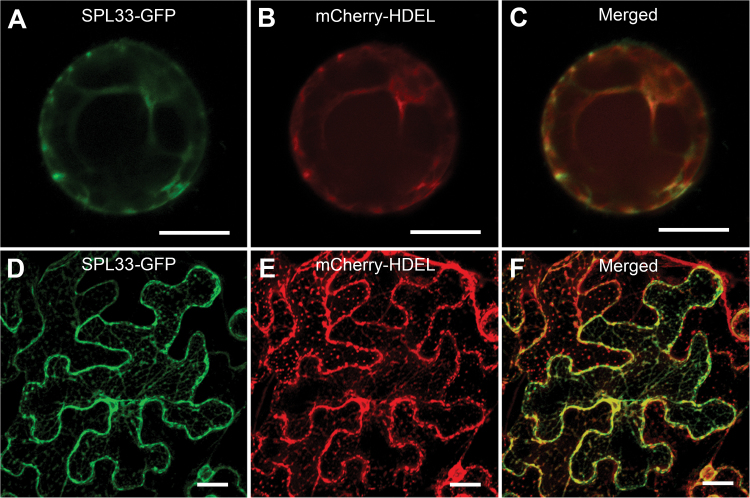
To determine the subcellular localization of SPL33, the full-length coding sequence of SPL33 was fused to the N-terminus of green fluorescent protein (GFP). When transiently expressed in rice protoplasts, the GFP signal was co-localized with the ER marker mCherry–HDEL (Nelson et al., 2007) (Fig. 7A–C). To validate this observation, we transformed the plasmid containing SPL33–GFP fusion into N. benthamiana leaves. SPL33–GFP protein was exclusively detected in the ER (Fig. 7D, E). These results indicated that the SPL33 protein is localized to the ER.
Fig. 7.
Subcellular localization of SPL33 protein. (A–C) Rice protoplast transient assay. (D–F) N. benthamiana leaf assay. SPL33–GFP, SPL33 fused to GFP; mCherry–HDEL, endoplasmic reticulum (ER) marker. Scale bar: 20 μm.
The SPL33 regulates ROS accumulation and cell death in rice
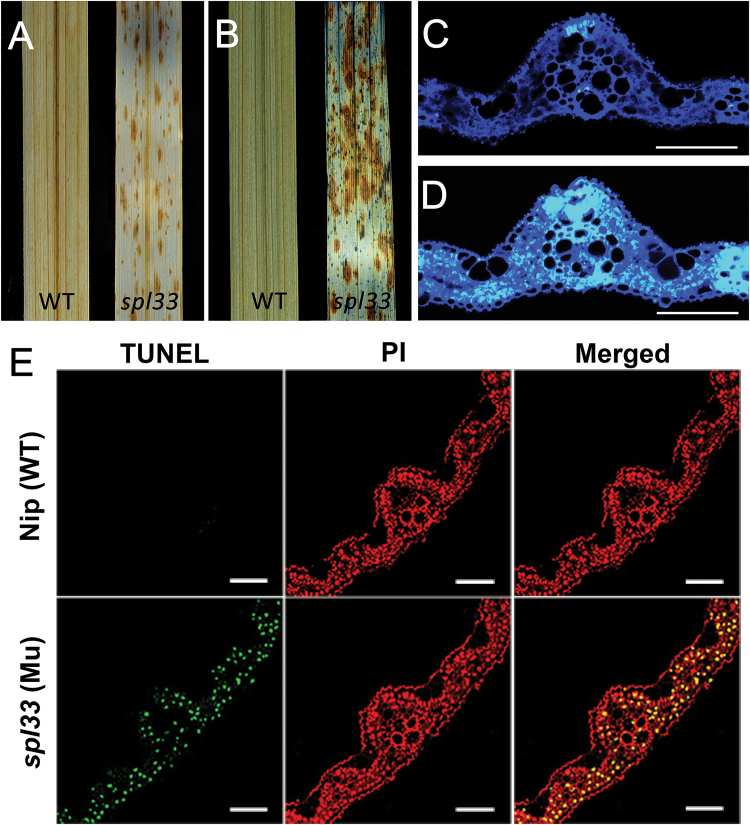
Reactive oxygen species (ROS) are major signaling molecules in plant PCD, and high concentrations of ROS, including superoxide and hyperoxide, may cause cellular damage or trigger PCD (Khanna-Chopra, 2012). DAB is an indicator of H2O2 accumulation (Wang et al., 2015a). Using DAB staining, we found that the spl33 mutant had much stronger H2O2 accumulation than WT (Fig. 8A). Typan blue staining, a traditional method for selective staining of dead tissues or cells (Qiao et al., 2010), consistently showed that the spl33 mutant had a large number of blue spots after staining, whereas the wild-type did not (Fig. 8B). In addition, callose, a plant polysaccharide component of primary cell walls induced by wounding or pathogen infection (Qiao et al., 2010), accumulated at a much higher level in vascular bundles, sclerenchyma and mesophyll cells of spl33 leaves than in WT (Fig. 8C, D). Together, our results suggested that the reddish-brown lesions in the spl33 mutant were caused by ROS accumulation and irreversible membrane damage. Further, we performed a terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) assay for detection of DNA fragmentation, a hallmark of programmed cell death (Kim et al., 2011). TUNEL signals in nuclei were strong and randomly distributed in the spl33 mutant, whereas no TUNEL signals were detected in WT (Fig. 8E). These results suggested that loss of function of SPL33 triggers the PCD pathway, ultimately leading to development of the spl33 lesion-mimic phenotype.
Fig. 8.
H2O2 accumulation and PCD detection in spl33. (A) DAB staining for H2O2 accumulation. (B) Trypan blue staining for cell death. (C, D) Aniline blue staining for callose accumulation under UV light; fluorescent regions indicate callose accumulation. (C) WT; (D) spl33. Scale bar: 100 μm. (E) DNA fragmentation detection in mesophyll cells by TUNEL assay. Red signal represents staining with propidium iodide, and yellow and green signals indicate TUNEL-positive nuclei of dead cells resulting from PCD. Scale bar: 100 μm. WT and spl33 leaf samples in (A–E) were analysed at 28 DAS.
To further understand the biochemical mechanism involved in ROS accumulation in the spl33 mutant we undertook expression analyses of the genes encoding anti-oxidative enzymes superoxide dismutase (SOD), catalase (CAT) and peroxidase (POD) in spl33 mutant and WT. These three enzymes are normally activated to remove ROS under oxidative stress (Miller et al., 2010). However, the expressions of both CAT and POD in the spl33 mutant were not obviously changed, even though the expression of SOD in the spl33 was elevated by almost two-fold (see Supplementary Fig. S5). Those results suggested that loss-of-function of SPL33 activated SOD, which might remove some superoxide, but not CAT and POD, which might result in insufficient capability for scavenging excessive H2O2.
SPL33 regulates early leaf senescence in rice
Early leaf senescence is a consequence of uncontrolled PCD, and has been described as a characteristic of some LMMs (Wang et al., 2015b). To examine early leaf senescence in the spl33 mutant, we measured four indicators of senescence, namely chloroplast structure (Wang et al., 2015b), chlorophyll content (Jiao et al., 2012), photosynthesis-related gene expression (Lim et al., 2007), and senescence-related gene expression (Park et al., 2007).
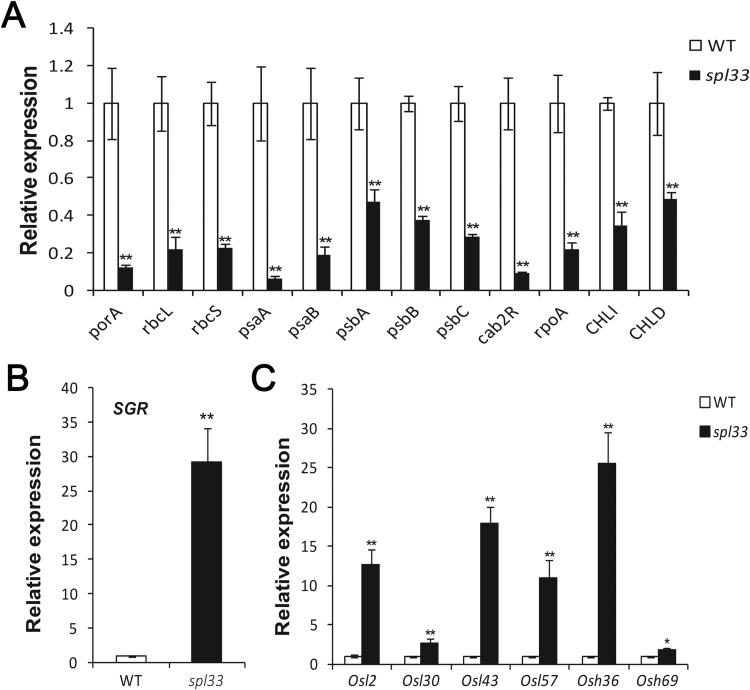
Examination of ultra-structures of spl33 and WT mesophyll cells in leaves of 28-day-old seedlings by transmission electron microscopy showed that chloroplasts in mesophyll cells of WT leaves were fully developed with intact membranes, whereas mesophyll cells surrounding the spotted areas of spl33 leaves were completely disrupted, leaving membrane-bound bodies in the cytoplasm (see Supplementary Fig. S6A–D. Coincidently, chlorophyll (a, b) and carotenoid contents were significantly decreased in spl33 leaves (Supplementary Fig. S6E). These results implied that irreversible degradation of chloroplasts and decreased photosynthesis in spl33 leaves could be responsible for the early leaf senescence. On the other hand, qRT-PCR of 12 photosynthesis-related genes revealed that the expression levels of porA, rbcL, rbcS, psaA, psaB, psbA, psbB, psbC, cab2R, rpoA, CHLI, and CHLD in spl33 were significantly down-regulated to 0.12-, 0.22-, 0.23-, 0.07-, 0.19-, 0.47-, 0.38-, 0.29-, 0.09-, 0.22-, 0.35-, and 0.49-fold, respectively, of those in WT (Fig. 9A). qRT-PCR of the senescence-induced STAYGREEN (SGR) gene and six senescence-associated genes revealed that the expression levels of SGR, Osl2, Osl30, Osl43, Osl57, Osh36, and Osh69 were dramatically up-regulated, being 29.2-,12.6-, 2.8-, 17.9-, 10.9-, 25.6-, and 1.8-fold higher than in comparative WT leaves (Fig. 9B, C). These results provided molecular evidence for the early leaf senescence in spl33 plants. We concluded that SPL33 may play an important role in regulating leaf senescence, and that its loss-of-function may accelerate early leaf senescence.
Fig. 9.
Identification of early leaf senescence in spl33 at the molecular level. Relative expression of photosynthesis-related genes (A), STAY GREEN (SGR) gene (B), and senescence-associated genes (C). Wild-type (WT) and spl33 leaves were collected from 28-day-old seedling; the expression level of each gene in WT was normalized to 1. Data are means±SD of three biological replicates (Student’s t-test: *P<0.05; **P<0.01).
SPL33 regulates defense responses in rice
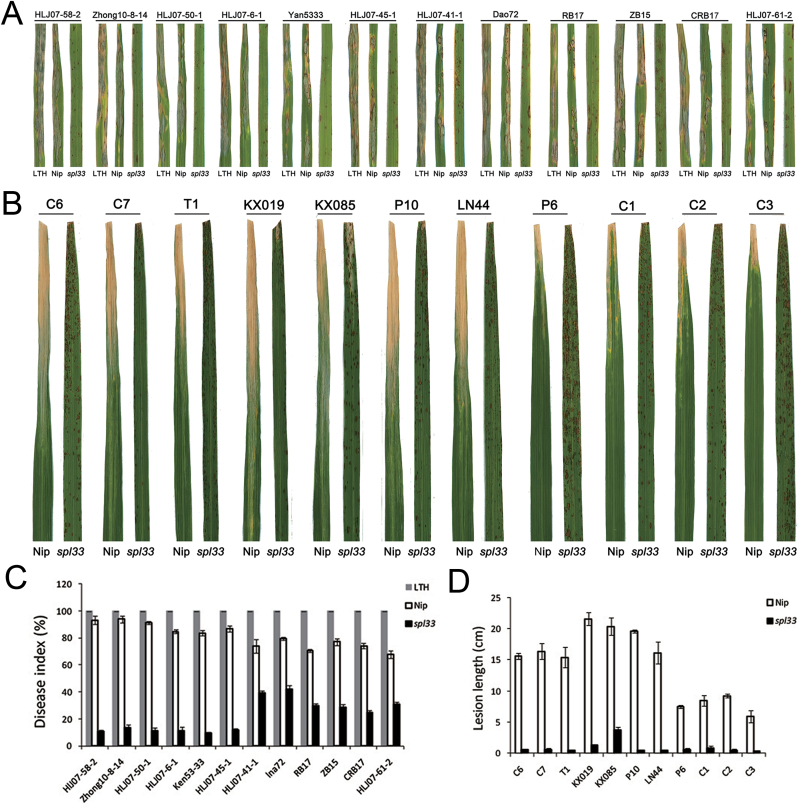
The lesion-mimic phenotype in spl33 plants resembles the HR that occurs in plants following infection by many pathogens. Some LMMs show enhanced resistance to fungal and bacterial pathogens (Fekih et al., 2015; Wang et al., 2015b). To examine whether the spl33 mutant also gains disease resistance, we inoculated the mutant and the wild-type plants with isolates of 12 M. oryzae pathotypes and 11 Xoo pathotypes virulent against the WT. The spl33 plants exhibited significantly enhanced resistance to all tested isolates of both pathogens (Fig. 10).
Fig. 10.
Enhanced resistance in the spl33 mutant to Magnaporthe oryzae and Xanthomonas oryzae pv. oryzae isolates. (A) Reactions to 12 M. oryzae isolates. The variety LTH was used as a susceptible control. (B) Reactions to 11 Xoo isolates. (C) Disease indices of WT and spl33 to M. oryzae isolates. (D) Lesion lengths of wild-type Nipponbare (Nip) and spl33 to 11 Xoo isolates. Data are means±SD of 15 plants.
Defense-response genes were activated during lesion development in some rice LMMs (Fekih et al., 2015; Wang et al., 2015b). To determine whether and when defense-response genes were activated in the spl33 mutant, we examined the expression of defense-response marker genes PR1a, PBZ1 and PO-C1 (Shirsekar et al., 2014; Wang et al., 2015b) in both the mutant and WT plants by qRT-PCR at 14 DAS (before appearance of visible lesions in spl33 plants), 21 DAS (lesions just visible) and 28 DAS (lesions clearly visible). At 14 DAS expression levels of PR1a, PBZ1 and PO-C1 in the spl33 mutant were 2.14-, 1.11-, and 6.66-fold, respectively, of those in WT plants; at 21 DAS expression of the three genes in spl33 was 199.17-, 86.47-, and 296.52-fold higher, respectively, than WT; and at 28 DAS their expression dropped but was still higher than WT by 5.44-, 8.57-, and 19.83-fold, respectively (see Supplementary Fig. S7). Together, these results suggest that lack of the eEF1A-like protein triggers the defense response, which leads to enhanced disease resistance associated with lesion formation in spl33.
Transcriptome sequencing suggests that SPL33 may have multiple functions
SPL33 encodes an eEF1A-like protein, known to be involved in PCD, nuclear export, proteasome-mediated degradation of damaged protein, and actin-binding and bundling besides a central role in translation (Migliaccio et al., 2015). However, almost all those known functions were documented from studying Saccharomycetes and mammals, with scant knowledge of their role in plants (Gao et al., 2013). In order to investigate the functions of the eEF1A-like protein encoded by SPL33 in rice, we performed transcriptome sequencing of both the spl33 mutant and WT at 28 DAS when mutant plants were exhibiting multiple lesions.
Based on the RNA-seq data, there were 4792 DEGs between the spl33 mutant and WT, of which 3867 were up-regulated and 925 down-regulated (see Supplementary Dataset S1). All the reliable DEGs were then analysed for GO functional enrichment. Genes involving translation (GO:0006412), structural molecule activity (GO:0005198) and ribonucleoprotein complex (GO:0030529) were significantly enriched in spl33, suggesting that SPL33 may be involved in protein biosynthesis. Genes involved in response to biotic stimulus (GO:0009607), response to stress (GO:0006950), and cell death (GO:0008219) were significantly enriched among the up-regulated group (Supplementary Dataset S2, Up-regulated). GO terms associated with photosynthesis (GO:0015979) and signal transduction (GO:0007165) were among the down-regulated genes (Supplementary Dataset S2, Down-regulated). These results of GO enrichment were consistent with the earlier results, suggesting that SPL33 functions in disease response and photosynthesis in addition to protein translation.
To further explore the biological pathways in which SPL33 may be involved, we performed KEGG enrichment analysis for the DEGs between the spl33 mutant and WT. Seven pathways were significantly (P<0.01) enriched for up-regulated genes and four for down-regulated ones (see Supplementary Dataset S3). The highly enriched up-regulated pathways were mainly related to protein synthesis (ribosome, biosynthesis of amino acids and ribosome biogenesis in eukaryotes), PCD and defense response (Supplementary Dataset S3, Up-regulated). The highly enriched pathways with down-regulated genes were related to photosynthesis (photosynthesis–antenna proteins, photosynthesis–porphyrin and chlorophyll metabolism) (Supplementary Dataset S3, Down-regulated). These results suggest that SPL33 is involved in protein translation, and regulation of cell death, leaf senescence, and defense response, consistent with the results of GO enrichment.
The enriched plant–pathogen interaction pathway involved 45 genes, among which 43 were significantly up-regulated and two were significantly down-regulated in spl33 (see Supplementary Dataset S4). Up-regulation involved genes that participate in multiple signaling-related events in innate immunity (Supplementary Fig. S8 and Supplementary Dataset S4). Examples include the pattern recognition receptor (PPR) OsCEBiP involved in recognition of pathogen- or microbe-associated molecular patterns; MEKK1 and two MKKs involved in phosphorylation of kinase cascades; CNGC, three CPKs, three CAMs and seven CMLs involved in calcium signaling; Rboh involved in generation of ROS as an NADPH oxidase; rice homologs of RPM1 and RPS2 involved in recognition of avirulence effectors produced by pathogens; and two PR1s that serve as antimicrobial proteins. The up-regulated receptor genes and signaling molecules in the spl33 mutant may induce innate immunity, cell wall reinforcement, antimicrobial proteins and phytoalexin accumulation, all of which have been implicated in disease response.
Further analysis on the 4792 DEGs identified eight salicylic acid (SA) biosynthesis-related PAL genes, 21 jasmonic acid (JA) biosynthesis-associated genes, and 3 ethylene (ETH) biosynthesis-associated genes. Interestingly, all but 2 of them were up-regulated in spl33 (see Supplementary Table S6). Those results indicate that the eEF1A-like gene plays a negative role in both SA and JA/ETH biosynthesis and that its loss-of-function may result in higher SA and JA/ETH accumulation, thus enhancing the plant defense network.
Discussion
SPL33 encodes an eEF1A-like protein containing a zinc finger domain in addition to three conserved EF-Tu domains
LMMs are important for understanding PCD and defense responses in plants. In the present study, we isolated an LMM, spl33, from a tissue culture-derived population of cv. Nipponbare. The mutant displayed small reddish brown lesions on the leaves from the seedling stage, early leaf senescence at the late flowering stage, semi-dwarfness, fewer tillers and reduced spikelet fertility (Fig. 1). The SPL33 gene was identified as LOC_Os01g02720, encoding an eEF1A-like protein. Functional complementation with wild-type LOC_Os01g02720 rescued all mutant phenotypes observed in spl33 (Fig. 3 and Supplementary Fig. S4). It is worth noting that the SPL33 protein contains a zinc finger domain in addition to three highly conserved EF-Tu domains of typical eEF1As present in a wide range of organisms (Fig. 4). Complementation tests by expressing truncated SPL33 in the spl33 mutant confirmed that only the three EF-Tu domains were necessary for SPL33 function, whereas the zinc finger domain was dispensable (Fig. 5). This suggests that SPL33 could fulfill the functions of typical eEF1As. Transcriptome sequencing of spl33 and WT showed that both protein translation-related GO terms and protein synthesis-related KEGG pathways were highly enriched (Supplementary Datasets S2 and S3), indicating that SPL33 was involved in protein translation, the canonical role of typical eEF1As.
There are at least ten other distinct rice genes that are predicted to encode putative eEF1As (see Supplementary Fig. S9). The SPL33 protein shares 97.26% amino acid identity with LOC_Os04g50870, which is located on chromosome 4, which was also predicted to contain a zinc finger domain, and has 17.58–49.24% amino acid identity with the other nine putative eEF1A proteins (Supplementary Fig. S9). There was no sequence variation of LOC_Os04g50870 between WT Nipponbare and the spl33 mutant (data not shown). Expression analysis showed that LOC_Os04g50870 was expressed at relatively low levels in both spl33 mutant (about 1/7 of SPL33) and WT plants (about 1/31 of SPL33) compared with SPL33 (Supplementary Fig. S10). In addition, the expression level of LOC_Os04g50870 in WT plants was only half of that in the mutant plants, which is contrary to that the expression level of SPL33 in the wild-type plants was 2.1-fold of that in the mutant plants (Supplementary Fig. S10). These results suggest that the two homolog genes, SPL33 and LOC_Os04g50870, seem to have no functional redundancy in regulating the mutant phenotypes. Further investigation is needed to determine the relationship between SPL33 and LOC_Os04g50870.
The spl33 allele in the mutant has a single-base G2493→T2493 substitution that causes a premature stop of the SPL33 protein (Fig. 2), and it is loss-of-function of SPL33 that induces the mutant phenotypes including spotted leaves, early leaf senescence and enhanced resistance to M. oryzae and Xoo. To our knowledge, this is the first report of an eEF1A-like mutant in plants, and also the first report that an eEF1A-like protein is involved in cell death and the defense response in plants.
Cell death and early leaf senescence are caused by loss-of-function of SPL33
The spl33 mutant exhibited spontaneous leaf spotting starting from the young seedling stage, mimicking the localized HR that occurs following infection by many pathogens. Our study revealed that mutation of the eEF1A-like protein encoded by SPL33 was responsible for the mutant phenotype. Even though eEF1As have been reported to function in PCD in higher vertebrates (Migliaccio et al., 2015), there were no reports describing a similar function in plants. In order to determine whether the eEF1A-like protein encoded by SPL33 controls PCD in plants, we performed expression analyses of several histochemical markers using staining methods, including DAB staining for H2O2 accumulation, trypan blue staining for cell death, aniline blue staining for callose accumulation and propidium iodide staining for TUNEL-positive nuclei of PCD cells (Qiao et al., 2010; Kim et al., 2011; Fekih et al., 2015; Wang et al., 2015a), and the transcriptome sequencing in spl33 and wild-type plants, respectively. All the results obtained (Fig. 8 and Supplementary Datasets S2 and S3) provided evidence that the eEF1A-like protein controls PCD in plant. Considering that the spl33 mutation is characterized by spontaneous cell death in leaves, we thus concluded that SPL33 serves as a negative regulator of PCD-mediated cell death in rice.
Early leaf senescence also occurred in the spl33 mutant, and this phenotype was obviously accelerated at the tillering and flowering stages (Fig. 1). To date, only a few studies have described senescence as a phenotype of LMM plants, and the senescence syndromes were not well analysed in those mutants (Qiao et al., 2010; Wang et al., 2015b). Therefore, it was necessary to determine that early leaf senescence does occur in the spl33 mutant. For this purpose, we selected four indicators for leaf senescence, namely loss of chlorophyll (Jiao et al., 2012), breakdown of chloroplasts (Wang et al., 2015b), down-regulation of photosynthesis-related genes (Wang et al., 2015b), and up-regulation of senescence-associated genes (Park et al., 2007), and performed relevant senescence assays. All results (Supplementary Fig. S6 and Fig. 9) suggest that early leaf senescence happens in spl33 plants, and this was further verified by the transcriptome sequencing of the spl33 mutant and WT (Supplementary Datasets S2 and S3). Since loss-of-function of SPL33 resulted in early leaf senescence we deduced that the eEF1A-like protein encoded by SPL33 may function in regulation of leaf senescence.
The PCD-mediated leaf senescence is generally regarded to have a more complex physiological basis than cell death in that the senescence is normally characterized by nutrient remobilization between organs and is an integral part of normal plant development (Rogers, 2015). It is worth noting that the PCD-mediated cell death and early leaf senescence seem to occur simultaneously in spl33 plants (Fig. 1A–C). In order to better dissect the regulatory mechanisms for cell death and leaf senescence, it is necessary to investigate whether cell death and early leaf senescence are a continuum or distinct processes, or regulated separately in spl33 plants.
Defense responses are induced by loss-of-function of SPL33
Defense response genes may be activated during the development of symptoms in lesion-mimic mutants, and may contribute to enhanced resistance to pathogens (Wang et al., 2015b). The loss-of-function of SPL33 results in induced HR-like lesions. Enhanced resistances to blast and bacterial blight were observed in the spl33 mutant (Fig. 10). Defense responses can also involve activation of PR genes and PR1a, PBZ1 and PO-C1 have been used as molecular markers for monitoring defense responses in rice. These three PR genes were all significantly up-regulated in spl33 plants during the development of lesion mimics (see Supplementary Fig. S7), and may have been involved in enhancing the resistances. The induced resistance in spl33 appears to be broad-spectrum and non-specific (Fig. 10).
KEGG enrichment analysis suggested that the plant–pathogen interaction pathway was constitutively activated in the spl33 mutant (Supplementary Fig. S8 and Supplementary Dataset S4). A number of genes involved in pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) were significantly up-regulated, including pattern recognition receptors, e.g. OsCEBiP (Akamatsu et al., 2013); MAP kinase kinases, e.g. OsMPK3, OsMKK4, and OsMPK6 (Kishi-Kaboshi et al., 2010; Hu et al., 2015); MEK kinase 1, e.g. OsMKK1 (Kumar et al., 2008); cyclicnucleotide-gated channels, e.g. CNGC10 (Guo et al., 2008); calmodulin-like proteins, e.g. OsMSR2 (Xu et al., 2013); calcium-dependent protein kinases, e.g. OsCPK10 (Fu et al., 2013); WRKYs, e.g. OsWRKY53 (Chujo et al., 2007); and respiratory burst oxidase homologs, e.g. OsrbohA and OsrbohE (Yoshie et al., 2005). A variety of genes involved in effector-triggered immunity (ETI) were also significantly up-regulated in the spl33 mutant, including resistance (R) proteins, e.g. RPM1 and RPS2 (Mackey et al., 2002, 2003); heat shock proteins, e.g. HSP90 (Shirasu, 2009); suppressors of the G2 allele of skp1, e.g. SGT1 (Shirasu, 2009); and PRs, e.g. OsPR1a and OsPR1b (Agrawal et al., 2001). Collectively, both the PTI- and ETI-related receptor genes and signaling molecules were significantly up-regulated in spl33, suggesting that the loss-of-function of SPL33 leads to negative regulation of innate immunity, including both PTI and ETI, reinforcement of cell walls, induction of anti-microbial proteins and accumulation of phytoalexins, thus resulting in broad-spectrum resistance of spl33 to pathogens.
No previous report has suggested that an eEF1A-like protein functions in cell death and defense responses in plants. The present study identified a novel rice LMM caused by functional loss of an eEF1A-like protein. We characterized the roles of this eEF1A-like protein, and confirmed that lost function of eEF1A-like protein leads to cell death and induces defense responses in rice.
Supplementary Material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant numbers 31471758), the Agricultural Science and Technology Innovation Program of CAAS ‘Crop Functional Genomics’, and the Special Fund for Agro-scientific Research in the Public Interest Program of China (201203014).
References
- Abbas W, Kumar A, Herbein G. 2015. The eEF1A proteins: at the crossroads of oncogenesis, apoptosis, and viral infections. Frontiers in Oncology 5, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal GK, Rakwal R, Jwa NS, Agrawal VP. 2001. Signaling molecules and blast pathogen attack activates rice OsPR1a and OsPR1b genes: a model illustrating components participating during defense/stress response. Plant Physiology and Biochemistry 39, 1095–1103. [Google Scholar]
- Akamatsu A, Wong HL, Fujiwara M, et al. 2013. An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host & Microbe 13, 465–476. [DOI] [PubMed] [Google Scholar]
- Arnon DI. 1949. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiology 24, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, McCormick F. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349, 117–127. [DOI] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL. 2006. A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Molecular Plant Pathology 7, 417–427. [DOI] [PubMed] [Google Scholar]
- Chen X, Hao L, Pan J, Zheng X, Jiang G, Jin Y, Gu Z, Qian Q, Zhai W, Ma B. 2012. SPL5, a cell death and defense-related gene, encodes a putative splicing factor 3b subunit3 (SF3b3) in rice. Molecular Breeding 30, 939–949. [Google Scholar]
- Chujo T, Takai R, Akimoto-Tomiyama C, et al. 2007. Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochimica et Biophysica Acta 1769, 497–505. [DOI] [PubMed] [Google Scholar]
- Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL. 1994. Arabidopsis mutants simulating disease resistance response. Cell 77, 565–577. [DOI] [PubMed] [Google Scholar]
- Dong H, Fei GL, Wu CY, et al. 2013. A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiology 162, 1867–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekih R, Tamiru M, Kanzaki H, et al. 2015. The rice (Oryza sativa L.) LESION MIMIC RESEMBLING, which encodes an AAA-type ATPase, is implicated in defense response. Molecular Genetics and Genomics 290, 611–622. [DOI] [PubMed] [Google Scholar]
- Fu L, Yu X, An C. 2013. Overexpression of constitutively active OsCPK10 increases Arabidopsis resistance against Pseudomonas syringae pv. tomato and rice resistance against Magnaporthe grisea. Plant Physiology and Biochemistry 73, 202–210. [DOI] [PubMed] [Google Scholar]
- Gao S, Zha X, Pan J. 2013. Research advances on biological function of eukaryotic translation elongation factor. Journal of Zhejiang Normal University Natural Science 36, 444–449 (in Chinese with English abstract). [Google Scholar]
- Guo KM, Babourina O, Christopher DA, Borsics T, Rengel Z. 2008. The cyclic nucleotide-gated channel, AtCNGC10, influences salt tolerance in Arabidopsis. Physiologia Plantarum 134, 499–507. [DOI] [PubMed] [Google Scholar]
- Heath MC. 2000. Hypersensitive response-related death. Plant Molecular Biology 44, 321–334. [DOI] [PubMed] [Google Scholar]
- Hiremath PJ, Farmer A, Cannon SB, et al. 2011. Large-scale transcriptome analysis in chickpea (Cicer arietinum L.), an orphan legume crop of the semi-arid tropics of Asia and Africa. Plant Biotechnology Journal 9, 922–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Ye M, Li R, Zhang T, Zhou G, Wang Q, Lu J, Lou Y. 2015. The rice transcription factor WRKY53 suppresses herbivore-induced defenses by acting as a negative feedback modulator of mitogen-activated protein kinase activity. Plant Physiology 169, 2907–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa A, Okamoto H, Iwasaki Y, Asahi T. 2001. A deficiency of coproporphyrinogen III oxidase causes lesion formation in Arabidopsis. The Plant Journal 27, 89–99. [DOI] [PubMed] [Google Scholar]
- Jefferson RA. 1987. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Molecular Biology Reporter 5, 387–405. [Google Scholar]
- Jeon JS, Lee S, Jung KH, et al. 2000. T-DNA insertional mutagenesis for functional genomics in rice. The Plant Journal 22, 561–570. [DOI] [PubMed] [Google Scholar]
- Jiao BB, Wang JJ, Zhu XD, Zeng LJ, Li Q, He ZH. 2012. A novel protein RLS1 with NB-ARM domains is involved in chloroplast degradation during leaf senescence in rice. Molecular Plant 5, 205–217. [DOI] [PubMed] [Google Scholar]
- Johal GS, Hulbert S, Briggs SP. 1995. Disease lesion mimics of maize: a model for cell death in plants. BioEssays 17, 685–692. [Google Scholar]
- Khanna-Chopra R. 2012. Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation. Protoplasma 249, 469–481. [DOI] [PubMed] [Google Scholar]
- Kim SG, Kim ST, Wang Y, Yu S, Choi IS, Kim YC, Kim WT, Agrawal GK, Rakwal R, Kang KY. 2011. The RNase activity of rice probenazole-induced protein1 (PBZ1) plays a key role in cell death in plants. Molecules and Cells 31, 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi-Kaboshi M, Okada K, Kurimoto L, et al. 2010. A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. The Plant Journal 63, 599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K, Rao KP, Sharma P, Sinha AK. 2008. Differential regulation of rice mitogen activated protein kinase kinase (MKK) by abiotic stress. Plant Physiology and Biochemistry 46, 891–897. [DOI] [PubMed] [Google Scholar]
- Langdon WB. 2015. Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Mining 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM. 2003. The role of protein elongation factor eEF1A2 in ovarian cancer. Reproductive Biology and Endocrinology 1, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei C, Hao K, Yang Y, et al. 2013. Identification and fine mapping of two blast resistance genes in rice cultivar 93-11. The Crop Journal 1, 2–14. [Google Scholar]
- Li J, Brader G, Palva ET. 2004. The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. The Plant Cell 16, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Lei C, Cheng Z, et al. 2008. Identification of SSR markers for a broad-spectrum blast resistance gene Pi20(t) for marker-assisted breeding. Molecular Breeding 22, 141–149. [Google Scholar]
- Liang H, Yao N, Song JT, Luo S, Lu H, Greenberg JT. 2003. Ceramides modulate programmed cell death in plants. Genes & Development 17, 2636–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. 2007. Leaf senescence. Annual Review of Plant Biology 58, 115–136. [DOI] [PubMed] [Google Scholar]
- Lin Q, Wang D, Dong H, et al. 2012. Rice APC/C(TE) controls tillering by mediating the degradation of MONOCULM 1. Nature Communications 3, 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Lin B, Auriac MC, Kroj T, Saindrenan P, Nicole M, Balagué C, Roby D. 2004. Vascular associated death1, a novel GRAM domain-containing protein, is a regulator of cell death and defense responses in vascular tissues. The Plant Cell 16, 2217–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Lei C, Xu X, et al. 2015. Pi64, encoding a novel CC-NBS-LRR protein, confers resistance to leaf and neck blast in rice. Molecular Plant-Microbe Interactions 28, 558–568. [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. 2003. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389. [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF, 3rd, Wiig A, Dangl JL. 2002. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754. [DOI] [PubMed] [Google Scholar]
- Mateyak MK, Kinzy TG. 2010. eEF1A: thinking outside the ribosome. The Journal of Biological Chemistry 285, 21209–21213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliaccio N, Ruggiero I, Martucci NM, Sanges C, Arbucci S, Tatè R, Rippa E, Arcari P, Lamberti A. 2015. New insights on the interaction between the isoforms 1 and 2 of human translation elongation factor 1A. Biochimie 118, 1–7. [DOI] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. 2010. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant, Cell & Environment 33, 453–467. [DOI] [PubMed] [Google Scholar]
- Mosher S, Moeder W, Nishimura N, Jikumaru Y, Joo SH, Urquhart W, Klessig DF, Kim SK, Nambara E, Yoshioka K. 2010. The lesion-mimic mutant cpr22 shows alterations in abscisic acid signaling and abscisic acid insensitivity in a salicylic acid-dependent manner. Plant Physiology 152, 1901–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research 8, 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Noutoshi Y, Kuromori T, Wada T, Hirayama T, Kamiya A, Imura Y, Yasuda M, Nakashita H, Shirasu K, Shinozaki K. 2006. Loss of NECROTIC SPOTTED LESIONS 1 associates with cell death and defense responses in Arabidopsis thaliana. Plant Molecular Biology 62, 29–42. [DOI] [PubMed] [Google Scholar]
- Park SY, Yu JW, Park JS, et al. 2007. The senescence-induced staygreen protein regulates chlorophyll degradation. The Plant Cell 19, 1649–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Jiang W, Lee J, et al. 2010. SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit micro 1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). The New Phytologist 185, 258–274. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HJ. 2015. Senescence-associated programmed cell death. In: Gunawardena AN, McCabe PF, eds. Plant Programmed Cell Death. Heidelberg: Springer, 203–233. [Google Scholar]
- Rostoks N, Schmierer D, Mudie S, Drader T, Brueggeman R, Caldwell DG, Waugh R, Kleinhofs A. 2006. Barley necrotic locus nec1 encodes the cyclic nucleotide-gated ion channel 4 homologous to the Arabidopsis HLM1. Molecular Genetics and Genomics 275, 159–168. [DOI] [PubMed] [Google Scholar]
- Shirasu K. 2009. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annual Review of Plant Biology 60, 139–164. [DOI] [PubMed] [Google Scholar]
- Shirsekar GS, Vega-Sanchez ME, Bordeos A, Baraoidan M, Swisshelm A, Fan J, Park CH, Leung H, Wang GL. 2014. Identification and characterization of suppressor mutants of spl11-mediated cell death in rice. Molecular Plant-Microbe Interactions 27, 528–536. [DOI] [PubMed] [Google Scholar]
- Tanaka R, Hirashima M, Satoh S, Tanaka A. 2003. The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a: inhibition of the pheophorbide a oxygenase activity does not lead to the “stay-green” phenotype in Arabidopsis. Plant & Cell Physiology 44, 1266–1274. [DOI] [PubMed] [Google Scholar]
- Tang J, Zhu X, Wang Y, Liu L, Xu B, Li F, Fang J, Chu C. 2011. Semi-dominant mutations in the CC-NB-LRR-type R gene, NLS1, lead to constitutive activation of defense responses in rice. The Plant Journal 66, 996–1007. [DOI] [PubMed] [Google Scholar]
- Tang S, Liang H, Yan D, Zhao Y, Han X, Carlson JE, Xia X, Yin W. 2013. Populus euphratica: the transcriptomic response to drought stress. Plant Molecular Biology 83, 539–557. [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. 1997. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. The Plant Journal 11, 1187–1194. [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Undan JR, Tamiru M, Abe A, et al. 2012. Mutation in OsLMS, a gene encoding a protein with two double-stranded RNA binding motifs, causes lesion mimic phenotype and early senescence in rice (Oryza sativa L.). Genes & Genetic Systems 87, 169–179. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. The Plant Journal 33, 949–956. [DOI] [PubMed] [Google Scholar]
- Wang C, Fan Y, Zheng C, Qin T, Zhang X, Zhao K. 2014. High-resolution genetic mapping of rice bacterial blight resistance gene Xa23. Molecular Genetics and Genomics 289, 745–753. [DOI] [PubMed] [Google Scholar]
- Wang J, Ye B, Yin J, et al. 2015a Characterization and fine mapping of a light-dependent leaf lesion mimic mutant 1 in rice. Plant Physiology and Biochemistry 97, 44–51. [DOI] [PubMed] [Google Scholar]
- Wang L, Pei Z, Tian Y, He C. 2005. OsLSD1, a rice zinc finger protein, regulates programmed cell death and callus differentiation. Molecular Plant-Microbe Interactions 18, 375–384. [DOI] [PubMed] [Google Scholar]
- Wang SH, Lim JH, Kim SS, Cho SH, Yoo SC, Koh HJ, Sakuraba Y, Paek NC. 2015c Mutation of SPOTTED LEAF3 (SPL3) impairs abscisic acid-responsive signalling and delays leaf senescence in rice. Journal of Experimental Botany 66, 7045–7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Liu SY, Mao XQ, et al. 2013. Identification and characterization of rhizosphere fungal strain MF-91 antagonistic to rice blast and sheath blight pathogens. Journal of Applied Microbiology 114, 1480–1490. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang Y, Hong X, et al. 2015b Functional inactivation of UDP-N-acetylglucosamine pyrophosphorylase 1 (UAP1) induces early leaf senescence and defence responses in rice. Journal of Experimental Botany 66, 973–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter M, Hollricher K, Salamini F, Schulze-Lefert P. 1993. The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Molecular and General Genetics 239, 122–128. [DOI] [PubMed] [Google Scholar]
- Xu GY, Cui YC, Li MJ, Wang ML, Zhang B, Huang L, Xia XJ. 2013. OsMSR2, a novel rice calmodulin-like gene, confers enhanced salt tolerance in rice (Oryza sativa L.). Australian Journal of Crop Science 7, 368–373. [Google Scholar]
- Xu WL, Wang XL, Wang H, Li XB. 2007. Molecular characterization and expression analysis of nine cotton GhEF1A genes encoding translation elongation factor 1A. Gene 389, 27–35. [DOI] [PubMed] [Google Scholar]
- Xu X, Zhang L, Liu B, Ye Y, Wu Y. 2014. Characterization and mapping of a spotted leaf mutant in rice (Oryza sativa). Genetics and Molecular Biology 37, 406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanouchi U, Yano M, Lin H, Ashikari M, Yamada K. 2002. A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proceedings of the National Academy of Sciences of the United States of America 99, 7530–7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Wardzala E, Johal GS, Gray J. 2004. The wound-inducible Lls1 gene from maize is an orthologue of the Arabidopsis Acd1 gene, and the LLS1 protein is present in non-photosynthetic tissues. Plant Molecular Biology 54, 175–191. [DOI] [PubMed] [Google Scholar]
- Yoshie Y, Goto K, Takai R, Iwano M, Takayama S, Isogai A, Che FS. 2005. Function of the rice gp91phox homologs OsrbohA and OsrbohE genes in ROS-dependent plant immune responses. Plant Biotechnology 22, 127–135. [Google Scholar]
- Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang GL. 2004. Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. The Plant Cell 16, 2795–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.