Highlight
Sugar beet acclimation to high salinity stress involves the down-regulation of reactive oxygen species generator systems and the up-regulation of antioxidant enzymes.
Keywords: Alternative oxidase, antioxidant defense, NADPH oxidase, peroxiredoxin, RBOH, salinity stress, sugar beet, superoxide dismutase
Abstract
Fine-tuned and coordinated regulation of transport, metabolism and redox homeostasis allows plants to acclimate to osmotic and ionic stress caused by high salinity. Sugar beet is a highly salt tolerant crop plant and is therefore an interesting model to study sodium chloride (NaCl) acclimation in crops. Sugar beet plants were subjected to a final level of 300 mM NaCl for up to 14 d in hydroponics. Plants acclimated to NaCl stress by maintaining its growth rate and adjusting its cellular redox and reactive oxygen species (ROS) network. In order to understand the unusual suppression of ROS accumulation under severe salinity, the regulation of elements of the redox and ROS network was investigated at the transcript level. First, the gene families of superoxide dismutase (SOD), peroxiredoxins (Prx), alternative oxidase (AOX), plastid terminal oxidase (PTOX) and NADPH oxidase (RBOH) were identified in the sugar beet genome. Salinity induced the accumulation of Cu-Zn-SOD, Mn-SOD, Fe-SOD3, all AOX isoforms, 2-Cys-PrxB, PrxQ, and PrxIIF. In contrast, Fe-SOD1, 1-Cys-Prx, PrxIIB and PrxIIE levels decreased in response to salinity. Most importantly, RBOH transcripts of all isoforms decreased. This pattern offers a straightforward explanation for the low ROS levels under salinity. Promoters of stress responsive antioxidant genes were analyzed in silico for the enrichment of cis-elements, in order to gain insights into gene regulation. The results indicate that special cis-elements in the promoters of the antioxidant genes in sugar beet participate in adjusting the redox and ROS network and are fundamental to high salinity tolerance of sugar beet
Introduction
Sugar beet (Beta vulgaris L.) has become an important source for sugar production in temperate areas of the world. It is not only used in the food industry but also for the production of bioethanol as a source of renewable energy (Magaña et al., 2011). Sugar beet is considered to be a cash crop and requires careful agronomical practices and breeding for adaptation to biotic and abiotic stresses. It is cultivated in different climates in Europe, North America, and increasingly in Asia, South America and recently North Africa. This suggests that bred cultivars are able to cope with different environments and growth conditions. Particular traits of interest for improving sugar beet production include its ability to acclimate to biotic and abiotic stresses, such as cold in temperate climates as well as drought, heat and salinity (Vastarelli et al., 2013).
Drought and salinity are among the most serious threats for crop production and limit agricultural productivity around the world (Horie and Schroeder, 2004; Munns and Tester, 2008). Saline soils are widespread, which can in part be attributed to the common global issue of water deficiency (Turan et al., 2009). Enhanced salinity tolerance will enable more productive use of saline soil and hence mechanisms involved in this ability are important areas of plant research (Horie and Schroeder, 2004; Hussain et al., 2008; Katori et al., 2010). High salt concentrations in rooting medium induce ionic and osmotic imbalances and oxidative damage, which results in growth retardation, wilting or death (Parida et al., 2004). Successful acclimation includes physiological and biochemical changes (Taji et al., 2004). Selective ion uptake, exclusion and compartmentalization are required to maintain a proper K+/Na+-balance while synthesis of compatible solutes, such as glycine, betaine and proline, is also needed (Yeo, 1998; Hamouda et al., 2016). Analysis of the signaling pathways and regulatory mechanisms involved indicates that hormones, Ca2+, and redox cues function as central players in acclimation (Zhang et al., 2008). Sugar beet tolerates salinity of up to 500 mM sodium chloride (NaCl) for 7 d without losing viability (Yang et al., 2012). The genome sequence of sugar beet was recently reported (Dohm et al. (2014), making sugar beet an excellent model for studying plant response and tolerance to salinity stress (Yang et al., 2012).
Salinity stress affects cellular reactive oxygen species (ROS) generator systems, such as photosynthetic electron transport, photorespiratory hydrogen peroxide (H2O2) release, respiratory electron transport, and enzyme activities including glucose oxidase, xanthine oxidase and in particular, plant peroxidases and NADPH oxidase. One of the most important cellular ROS generating systems is the plasma membrane-bound NADPH oxidase RBOH (Keller et al., 1998), which decisively controls cellular redox homeostasis under salinity stress (Hossain and Dietz, 2016). ROS accumulation is a common denominator under conditions of stress (Foyer et al., 1994).
Complementary to the generator systems are enzymatic and non-enzymatic antioxidants that regulate ROS and redox homeostasis and counteract metabolic imbalances, damage to cell structures, cell death and stress adaptation (Foyer et al., 2009). Superoxide dismutases (SODs) constitute the first line of defense against superoxide (O2•−), which is produced in most cellular compartments (Elstner, 1991). SODs are therefore found in all subcellular compartments (Alscher et al., 2002). H2O2 is decomposed by thiol peroxidases, in particular glutathione peroxidases (GPXs) and peroxiredoxins (Prxs), and by type III heme peroxidases, ascorbate peroxidase (APX) and catalase (Mittler and Poulos, 2005). GPXs and Prxs decompose alkyl hydroperoxides in addition to H2O2 and employ a thiol mechanism to react with peroxide (Dietz, 2016). Following oxidation by the peroxide substrate, thiol peroxidases are regenerated by electron donors such as thioredoxins, glutaredoxins and glutathione (Navrot et al., 2006; Koh et al., 2007; Dietz, 2011). Prxs display a stable TRX-fold and are grouped into four subfamilies: 1-CysPrx, 2-CysPrx, PrxQ and PrxII-like proteins (Horling et al., 2003; Muthuramalingam et al., 2009). Prxs and GPXs play important roles as thiol antioxidants, modulators of cell signalling and redox sensors (Dietz et al., 2006; Navrot et al., 2006).
Here, we identified some of the central components of the cellular antioxidant defense system in sugar beet, and followed their transcriptional response during acclimation to 300 mmol L-1 NaCl. We tested the hypothesis that the ROS and redox network participates in the extraordinarily high salinity tolerance of sugar beet. The transcriptional response pattern was placed into the broad framework of the cellular redox state. This study addressed an additional layer of redox and ROS homeostasis, namely the activation of safety mechanisms, as there are terminal oxidases present in different subcellular compartments. Mitochondrial alternative oxidase (AOX) (Considine et al., 2002; Hossain and Dietz, 2016) and plastid terminal oxidase (PTOX) (Stepien and Johnson, 2009; Hossain and Dietz, 2016) help dissipate excess reducing power.
Materials and methods
Plant materials and NaCl treatment
Seeds of sugar beet (Beta vulgaris subsp. vulgaris), cultivar KWS2320 were sterilized with 70% (v/v) ethanol, 0.1% (w/v) HgCl2 and 0.2% (w/v) thiram, placed in germination pots in vermiculite and perlite mixture and soaked in water in darkness for one week. The tray was then placed in growth chambers with 10 h light with an intensity of 100 µmol m−2 s−1 at 21 °C and 14 h darkness at 18 °C with 55% relative humidity for another week. The growth condition was adjusted according to the temperate climate, as the cultivar is adapted to temperate regions. After 14 d, uniformly grown seedlings were transferred to hydroponic containers with Hoagland solution (Ghoulam et al., 2002). All plants grew under control conditions for 35 d and up-salting started from day 36 in the case of salinity-stressed plants. Salt concentration was gradually increased to 300 mM NaCl in 50 mM.d−1 increments (Fig. 1A). Tissue was harvested from the fully expanded third to sixth leaves and the whole root after the point of first branching, from five independent experiments at time points indicated. Tissue was then frozen in liquid nitrogen and stored at −80 °C.
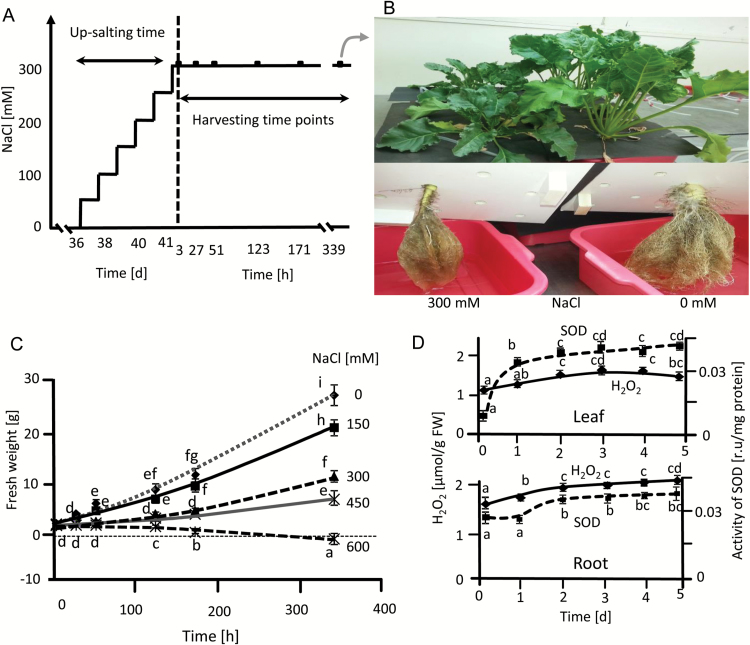
Fig. 1.
Selection of salinity conditions for sugar beet acclimation experiments. (A) The experimental design consisted of stepwise up-salting to 300 mM NaCl and harvesting after 3 h, or 339 h i.e. a 14-day period. For some analyses four additional time points were used as indicated. (B) Morphology of plants grown in hydroponics with or without 300 mM NaCl for 14 ds, with a total growth period of 55 d. (C) Growth as indicated by fresh weight increase of sugar beet at different NaCl concentrations. Data are means ±SD of n=5 experiments with three measurements each. (D) Monitoring of early responses of sugar beet during up-salting, namely H2O2 levels and SOD activity under control and salinity stress conditions. Data are means ±SD of n=5 experiments with six measurements each for H2O2 and three measurements each for SOD activity. The significant difference marked by different letters was calculated using Student’s t-test and further analyzed with Fisher LSD test, with P<0.05, using InfoStat statistical software.
Determination of photosystem II quantum yield and CO2 fixation rate
The steady-state quantum yield (FV/FM) of photosystem II (PSII) was measured using the Mini-PAM Fluorometer (Walz, Germany) under light conditions as described by Oelze et al. (2012). CO2 fixation of sugar beet leaves under stressful and control conditions was measured with a portable gas exchange system (GFS-3000, Heinz Walz GmbH, Effeltrich, Germany). The CO2 assimilation rate was measured at a light intensity of 100 µmol photons m−2 s−1, a relative humidity of 50% and at 22 °C.
Determination of osmotic potential and sodium, potassium and chloride content
Osmotic potential was measured by using an automatic cryoscopic osmometer (Knauer, Berlin, Germany), following calibration between 0 and 300 mosmol kg-1. The osmotic potential is given as mosmol kg-1. Sodium and potassium contents were determined from tissue sap using a flame photometer (Model 410; Sherwood Scientific Ltd, Cambridge, UK) calibrated between 0 and 10 ppm. Chloride content was determined in tissue sap using a Chloride Analyzer (Model 926S; Sherwood Scientific Ltd, Cambridge, UK).
Determination of antioxidant and non-protein thiols content
Ascorbate (AA) and dehydroascorbate (DHA) content were determined as described by Horling et al. (2003). Glutathione (GSH) and oxidized glutathione (GSSG) content were quantified with an enzyme cycling assay based on sequential oxidation of GSH by 5,5’-dithiobis(2-nitrobenzoate) (DTNB) and reduction by NADPH in the presence of glutathione reductase (GR) (Griffith, 1980). Tissue weighing 200 mg was extracted in 1 ml 0.1 M HCl and 0.1 mM EDTA. For total GSH content, 200 µL neutralized supernatant was incubated with 6 mM DTNB for 5 min followed by a 15 min incubation with 5 µL 2-vinylpyridine. After centrifugation, total GSH content was determined from the supernatant. The reaction was started with the addition of GR. Changes in 5’-thio-2-nitrobenzoic acid absorbance were spectrophotometrically monitored at 412 nm. To determine GSSG content, the neutralized supernatant was incubated first with 2-vinylpyridine for 15 min, then with 6 mM DTNB for 5 min and subsequently GR and NADPH were added. The difference between total glutathione and GSSG content is presented as GSH content. Non-protein thiols content was determined using 0.1 M phosphate buffer at pH 7.0, 0.5 mM EDTA and 1 mM DTNB (Del Longo et al., 1993). Absorbance was measured at 412 nm using a microplate reader (KC4, BIOTEK Instruments, Bad Friedrichshall, Germany). Values were corrected for the absorbance of a blank without extract and determined with a glutathione standard curve.
Determination of protein, malondialdehyde and hydrogen peroxide content
Total protein (mg g-1 FW) content was determined using bovine serum albumin as a standard, according to Bradford (1976). H2O2 content was quantified by chemiluminescence with luminol (Pèrez and Rubio, 2006). Malondialdehyde (MDA) content was measured colorimetrically according to Stewart and Bewley (1980) with some modification. Leaf tissue weighing 100 mg was homogenized in 0.1% trichloroacetic acid (TCA) on ice. Following centrifugation, 1.5 ml of 20% TCA containing 0.5% thiobarbituric acid (TBA) was added to 500 ml supernatant and incubated at 95 °C for 30 min. Following cooling and centrifugation, absorption was measured at 532 nm and the amount of MDA calculated, with ε=155 mM−1 cm−1.
SDS-PAGE and immunoblotting
SDS-PAGE and western blotting were performed as in Ströher et al. (2009). Binding of the first rabbit antibodies was achieved overnight in 1% skimmed milk in Tris-buffered saline with 0.1% Tween 20 (TBST) at the following dilutions: anti-At2-CysPrx, PrxQ and CuZn SOD2 at 1:3000; anti-Avena sativa D1 at 1: 5000. After secondary antibody binding, proteins were detected using chemiluminescence.
Antioxidant enzyme activities
Tissue weighing 200 mg was homogenized in 1 ml of 0.1 M phosphate buffer at pH 6.5. The supernatant was used to determine the activity of enzymes according to Urbanek et al. (1991) and protein content as above (Bradford, 1976). The reaction mixture for catalase consisted of 100 µl extract in 3 ml phosphate buffer at pH 6.8 (Urbanek et al., 1991). The detoxification of H2O2 was measured at 240 nm (Cary 300 Bio UV/VIS, Varian, Middelburg, Netherland), with ε=43.6 M cm−1. SOD activity was quantified according to Sen Gupta et al. (1993) and peroxidase according to Malik and Singh (1980), with ε=25 mM−1 cm−1. APX activity was assessed according to Yoshimura et al. (2000) by monitoring the rate of ascorbate oxidation at 290 nm, with ε=2.8 mM−1 cm−1. GR activity was measured by following the increase in absorbance in the presence of GSSG and DTNB (Sairam et al., 2002), with ε=13.6 mM−1 cm−1.
Determination of plasma membrane NADPH oxidase activity
NADPH-dependent O2− generating activity in isolated microsomal membrane vesicles was determined by following the O2−-dependent reduction of XTT (Na 3′-[1-[phenylaminocarbonyl]-3,4-tetrazolium]-bis(4-methoxy-6-nitro)benzenesulfonic acid) (Kaundal et al., 2012). The assay mixture contained 50 mM Tris-HCl at pH 7.5, 0.5 mM XTT, 0.1 mM NADPH and the membrane fraction. Tissue was ground in liquid nitrogen and 0.5 g of the powder was weighed out in empty pre-chilled Falcon tubes. 6 ml of ice-cold protein extraction buffer was added on ice and vortexed at room temperature. Homogenized tissue was filtered through four layers of cheesecloth and the flow through transferred to 2 ml microcentrifuge tubes on ice. After centrifugation at 10 000g at 4 °C for 45 min, the supernatant was transferred to ultracentrifuge tubes. Microsomal membranes were pelleted from the supernatant by centrifugation at 50 000g for 30 min. The pellet was suspended in 0.33 M sucrose, 3 mM KCl, and 5 mM potassium phosphate at pH 7.8. The plasma membrane fraction was isolated by adding the microsomal suspension to an aqueous two-phase polymer system to give a final composition of 6.2% (w/w) Dextran T500, 6.2% (w/w) polyethylene glycol (PEG) 3000, 0.33 M sucrose, 3 mM KCl, and 5 mM potassium phosphate at pH 7.8 with protease inhibitors. After three rounds of partitioning the resulting upper phase was diluted 5-fold in ice cold 10mM Tris-HCl dilution buffer at pH 7.4, containing 0.25 M sucrose, 3 mM EDTA, 1 mM DTT, 3.6 mM L-cysteine, 0.1 mM MgCl2 and protease inhibitors. The fractions were centrifuged at 120000g for 30 min. The pellets were resuspended in 1 ml of 10 mM Tris-HCl at pH 7.4 for the activity assay. All procedures were carried out at 4 °C. The reaction was initiated with NADPH. In the presence of O2−, XTT generates a yellow formazan that was quantified spectrophotometrically at 492 nm and calculated with ε=21 600 M−1 cm−1.
Transcript quantification
RNA isolation and cDNA synthesis were performed according to Wormuth et al. (2006) with a few modifications. Semi-quantitative RT-PCR analysis was carried out as previously described at the individually optimized cycle number (Finkemeier et al., 2005), using the given primer combinations (see Supplementary Table S1 at JXB online). Primers were designed with Primer3Plus software (www.bioinformatics.nl/cgi-bin/primer3plus). Loading was normalized with actin. qRT-PCR was carried out in the iCycler ™ Thermal Cycler (Bio-Rad, USA) with the iQ™SYBR Green Supermix (Bio-Rad, USA) in a final volume of 20 µl according to the manufacturer’s instructions. The iCycler was programmed to 95 °C for 1 min; 45x (95 °C for 30s, 58 °C for 40s, 72 °C for 45 s), 72 °C for 10 min, followed by a melting curve program of 55–95 °C in increasing steps of 0.5 °C. Efficiencies of each reaction were calculated using LinRegPCR software (Ruijter et al., 2009). Signal values were subsequently derived from the threshold cycles, with the average background subtracted, using the equation provided by Pfaffl (2001).
Sequence alignment and construction of phylogenetic trees
Sugar beet genes homologous to known Arabidopsis thaliana SOD, Prx, AOX and RBOH genes were searched for using FASTA and WU-BLAST2 programs. The amino acid sequences were aligned using CLUSTALW 2.1 (Larkin et al., 2007) with the default configuration. Their phylogenetic relationships were determined using the maximum likelihood (ML) algorithm incorporated in the program MEGA version 5 (Tamura et al., 2011). Bootstrap analyses with 500 replicates were performed to assess the robustness of the branches. Pairwise sequence alignment was done by blasting the isoforms of each gene group and sequence identity was retrieved (see Supplementary Table S2).
Cis-regulatory elements analysis
Promoters of stress-responsive genes in sugar beet were analyzed in silico for putative cis-elements using available genome sequences and our transcriptomic data. The Plant Promoter Analysis Navigator (PlantPAN 2.0; http://PlantPAN2.itps.ncku.edu.tw) (Chow et al., 2015) and The AthaMap (AthaMap 8; http://www.athamap.de) (Steffens et al., 2005) databases were used to identify cis-regulatory elements for 1000 bp upstream promoters of differentially regulated genes of B. vulgaris under salinity stress, with A. thaliana as reference (Table 3, Supplementary Tables S3–S5).
Table 3.
Cis-elements potentially responsible for the regulation of salinity induced transcripts in B. vulgaris but absent in the same gene from A. thaliana.
| Gene name | Cis-regulating elements | Cis-regulating elements sequence | Expression matrix |
|---|---|---|---|
| Cu-ZnSOD1; Cu-ZnSOD2; Cu-ZnSOD3; FeSOD3; MnSOD1; 2CysPrx B; Prx Q; AOX1A; AOX2; PTOX1;RBOH B | Dehydrin (LTRECOREATCOR15) | CCGAC |

|
| Cu-ZnSOD1; 2CysPrx B; AOX1A; AOX2; PTOX1 | BES1 (TFmatrixID_0494) | CACGTG* |

|
indicates details provided in Supplementary Table S5.
Statistical analysis
Pairwise comparisons were performed with Students t-test. Statistical grouping of the data was achieved by applying Fisher’s LSD, with P<0.05, using InfoStat statistical software.
Results
Effect of salinity on growth and redox state of sugar beet
The NaCl concentration in the hydroponics solution was increased stepwise to 300 mM. The plants were then kept at this level for 14 d for detailed kinetic analysis (Fig. 1A). This regime was chosen after comparing different NaCl concentrations, where growth was still measureable at 450 mM but ceased at 600 mM. Morphological changes were observed after salinity treatment (Fig. 1B–C). The high final salt level of 300 mM NaCl still enabled significant growth. The plant response was scrutinized by following the level of H2O2 and SOD activity during up-salting (Fig. 1D). H2O2 levels increased in roots and leaves after the first salt step and until the second step but did not increase further during subsequent up-salting steps. Instead levels started to decline at the highest salt concentrations in leaves. Total SOD activity roughly followed H2O2 levels indicating that sugar beet acclimated to salinity with no effects on fresh weight-related protein content, which was considered important for subsequent analyses (see Supplementary Fig. S1).
Photosynthetic quantum yield, CO2 fixation, ionic and osmotic state
The quantum yield of photosystem II (ΦPSII) was unchanged between salt-treated and control plants during up-salting and at different time points at 300 mM NaCl (Fig. 2C, Supplementary Fig. S2A, B). CO2 fixation rates decreased under salt treatment with little difference between 3 h and 14 d at 300 mM NaCl (Fig. 2C). This is despite the strong increase in osmotic potential, and sodium, chloride and potassium content (see Supplementary Fig. S2C–F).
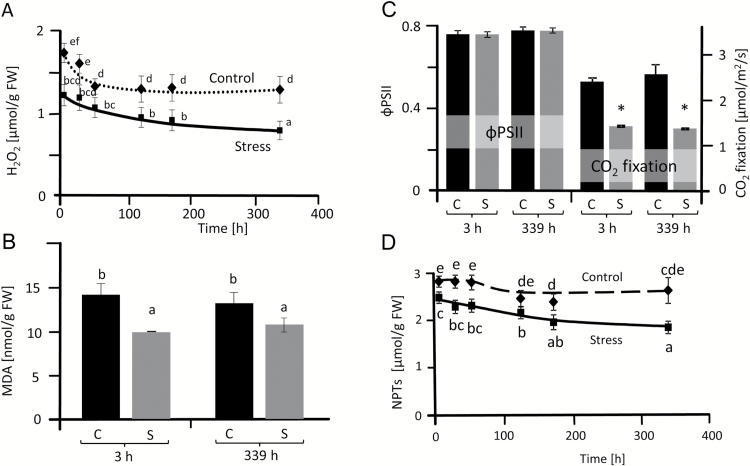
Fig. 2.
Characterization of ROS and antioxidant status of salt-stressed sugar beet. H2O2 content (A), malondialdehyde (MDA) content (B), photosynthetic quantum yield (ΦPSII) and CO2 fixation rates (C) and non-protein thiols content (D) in controls and plants stressed with 300 mM NaCl. The first analysis was carried out 3 h after reaching a concentration of 300 mM NaCl. Data are means ±SD of n=5 experiments with six measurements each for H2O2 and three for MDA and non-protein thiols. The significant difference marked by different letters was calculated using Student’s t-test and further analyzed with Fisher LSD test, with P<0.05, using InfoStat statistical software. Along the x-axis: C, control; S, salinity.
Oxidative stress and antioxidant defense
H2O2 accumulation, as a recognized marker of oxidative stress, was quantified in leaves during the 14-day period, after reaching 300 mM NaCl (Fig. 2A). The basal steady state level of H2O2 was around 1.7 and 1.2 nmol mg-1 FW in unsalted control and up-salted sugar beet, respectively. The H2O2 content in sugar beet leaves decreased during aging but importantly it was always significantly lower in salt-treated tissue compared with controls (Fig. 2A). MDA levels in salt-stressed leaves were also below control samples indicating less lipid peroxidation and membrane damage (Fig. 2B). Non-protein thiols (NPTs) were lower in stressed plants than in controls (Fig. 2D). Ascorbate and glutathione pools were analyzed as these low molecular mass antioxidants are linked to redox homeostasis and signaling and also function as reductants in the water–water cycle (Oelze et al., 2012). Glutathione levels dropped in stressed plants from 3 h to 14 d after up-salting (Table 2). Significantly lower GSH levels were observed in stressed plants at 14 d. In contrast there was no significant difference in GSSG levels for control and salinized plants. Significantly lower ascorbate levels were observed in stressed plants at 27, 123, 171 and 339 h. DHA content during stress acclimation was unchanged (Table 2). The reduction state of both metabolites was decreased at final harvest.
Table 2.
Characterization of glutathione and ascorbate status of control and salt-stressed sugar beet plants
The first analysis was done 3 h after reaching a concentration of 300 mM NaCl. Data are means ±SD of n=5 experiments with two technical replicates for glutathione and ascorbate. The significant difference was calculated using Student’s t-test and labelled with different letters after analysis with Fisher LSD test, with P<0.05, using InfoStat statistical software.
| Time (h) | GSH (µmol g-1 FW) | GSSG (µmol g-1 FW) | ASC (µmol g-1 FW) | DHA (µmol g-1 FW) | ||||
|---|---|---|---|---|---|---|---|---|
| Control | Stress | Control | Stress | Control | Stress | Control | Stress | |
| 3 | 1.83 ± 0.27 a | 1.77 ± 0.31 a | 0.39 ± 0.12 | 0.50 ± 0.12 | 1.65 ± 0.16 d | 1.57 ± 0.18 d | 0.34 ± 0.19 | 0.32 ± 0.09 |
| 27 | 1.70 ± 0.28 a | 1.69 ± 0.31 a | 0.46 ± 0.11 | 0.39 ± 0.08 | 1.74 ± 0.19 d,e | 1.42 ± 0.07 b,c | 0.25 ± 0.12 | 0.28 ± 0.09 |
| 51 | 1.59 ± 0.38 a | 1.57 ± 0.34 a | 0.38 ± 0.12 | 0.44 ± 0.11 | 1.76 ± 0.15 d,e | 1.52 ± 0.13 c,d | 0.21 ± 0.11 | 0.22 ± 0.10 |
| 123 | 1.60 ± 0.26 a | 1.41 ± 0.27 a | 0.34 ± 0.15 | 0.32 ± 0.09 | 1.73 ± 0.16 d,e | 1.32 ± 0.15 b | 0.25 ± 0.17 | 0.26 ± 0.11 |
| 171 | 1.63 ± 0.29 a | 1.33 ± 0.21 a | 0.31 ± 0.18 | 0.37 ± 0.11 | 1.57 ± 0.08 d | 1.34 ± 0.06 b | 0.18 ± 0.11 | 0.23 ± 0.10 |
| 339 | 1.65 ± 0.16 a | 1.21 ± 0.11 b | 0.29 ± 0.12 | 0.25 ± 0.09 | 1.54 ± 0.15 d | 1.25 ± 0.05 a | 0.21 ± 0.11 | 0.16 ± 0.08 |
Regulation of peroxiredoxin and superoxide dismutase proteins
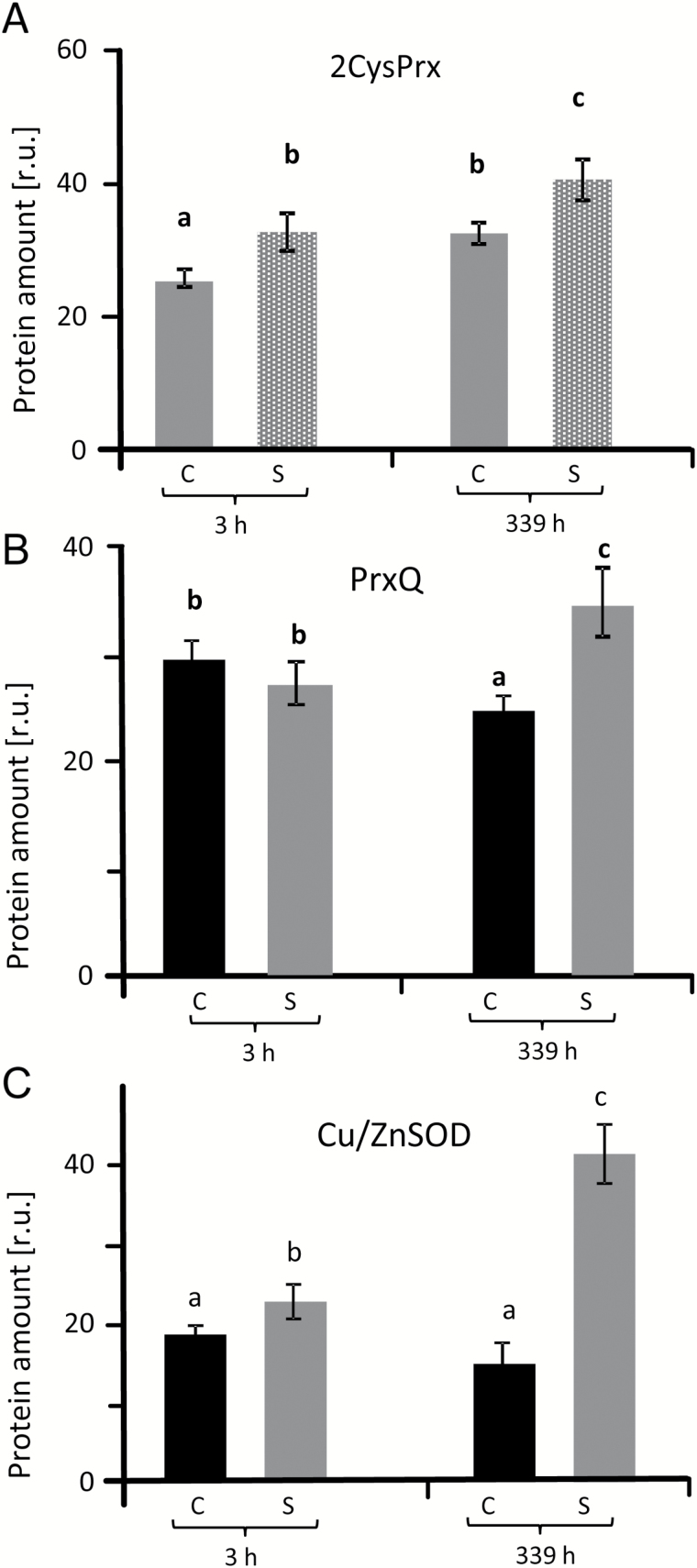
The abundance of 2-cysteine peroxiredoxin (2-CysPrx) protein was examined via western blotting using control and salt-stressed plant samples (Fig. 3A, Supplementary Fig. S3). 2-CysPrx amounts increased with aging and under salt stress. Chloroplast peroxiredoxin Q (PrxQ) levels were unchanged in salt-treated plants compared with controls at 3 h but were increased after 14 d (Fig. 3B). CuZnSOD showed the strongest response to salt stress with a 3-fold increase after 14 d (Fig. 3C, Supplementary Fig. S3A–C).
Fig. 3.
Protein amounts of 2-CysPrx (A), PrxQ (B) and CuZnSOD (C) under salinity stress and in control sugar beet. Band intensities in western blots were densitometrically quantified in three blots from independent experiments (as shown in Supplementary Fig. S3) using ImageJ software (http://imagej.nih.gov/ij/). The significant difference was calculated using Student’s t-test and labelled with different letters after analysis with Fisher LSD test, with P<0.05, using InfoStat statistical software. C, control; S, salinity.
Sugar beet SOD gene family members and their transcriptional regulation
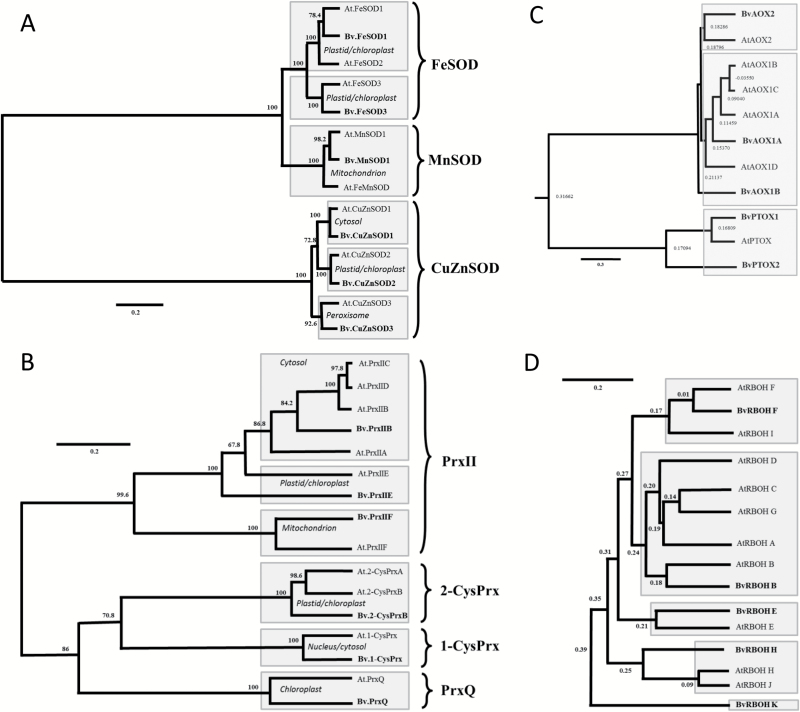
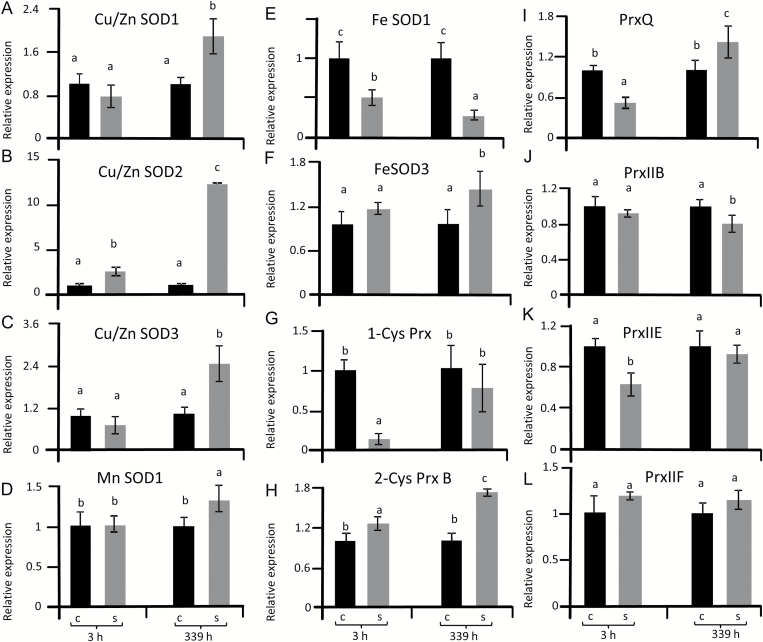
The accumulation of 2-Cys-Prx, PrxQ and CuZnSOD proteins under salinity prompted us to use bioinformatics to identify important antioxidant genes in the sugar beet genome and to quantify their transcriptional response by qPCR. Three CuZnSOD genes, one MnSOD gene and two FeSOD genes were identified (Table 1). The construction of a phylogenetic tree showed that the SOD family, with 14 available accessions in the databanks of B. vulgaris and A. thaliana, segregated into three clearly separated groups based on sequence similarity (Fig. 4A). Group I encompassed FeSOD genes that included two isoforms in sugar beet. MnSOD isoforms were related to FeSOD genes but separated into a distinct cluster. The three CuZnSOD genes in sugar beet were grouped in pairs with their cognate homologs in Arabidopsis. Relatedness was confirmed by sequence alignment (Supplementary Fig. S4). With the exception of FeSOD1, which was strongly down-regulated under salinity, all other SOD transcripts revealed significantly higher levels at 14 d of salt stress (Fig. 5A–F). Up-regulation was greatest for SOD2 with approximately a 13-fold difference between 300 mM and 0 mM NaCl (Fig. 5B). Mitochondrial MnSOD1 transcript levels were unchanged at 3 h but significantly increased at 14 d (Fig. 5D). The order of transcript up-regulation was: CuZnSOD2 > CuZnSOD3 > CuZnSOD1 > FeSOD3 > MnSOD.
Table 1.
Gene annotation of SODs, Prxs, AOXs and RBOHs in B. vulgaris subsp. vulgaris genotype KWS2320 and Arabidopsis thaliana
The table depicts the protein names and genome annotations, putative localization, protein length and calculated molecular mass. Abbreviations: cp, chloroplast; ecm, extracellular matrix, apoplast; mt, mitochondrion; per, peroxisome; vac, vacuole; cyt, cytoplasm; nuc, nucleus; pl, plastid; plast, plastoglobule; pm, plasma membrane; cw, cell wall.
| Enzyme and reaction | Protein/gene name | Reference sequence | Localization | Length (amino acids) and mol. wt (kDa) | |||
|---|---|---|---|---|---|---|---|
| B. vulgaris | A. thaliana | B. vulgaris | A. thaliana | B. vulgaris | A. thaliana | ||
| Superoxide dismutase (SOD) O2−+O2−+2H+→H2O2+O2 |
FeSOD1 | FeSOD1 | XM_010678412.1 | AT4G25100.3 | cp, mt | 324/36.26 | 212/23.78 |
| — | FeSOD2 | — | AT5G51100.1 | cp | — | 305/34.66 | |
| FeSOD3 | FeSOD3 | XM_010687302.1 | AT5G23310.1 | cp | 262/30.25 | 263/30.35 | |
| Cu/ZnSOD1 | Cu-ZnSOD1 | XM_010676650.1 | AT1G08830.1 | cyt, ecm, nuc | 152/15.25 | 152/15.10 | |
| Cu/ZnSOD2 | Cu-ZnSOD2 | XM_010690943.1 | AT2G28190.1 | cyt, cp, ecm | 228/23.13 | 216/22.24 | |
| Cu/ZnSOD3 | Cu-ZnSOD3 | XM_010695513.1 | AT5G18100.1 | cp, cyt, ecm, per, vac | 157/15.81 | 164/16.94 | |
| MnSOD1 | MnSOD1 | XM_010672327.1 | AT3G10920.1 | mt | 230/25.58 | 231/25.44 | |
| — | Fe-MnSOD | — | AT3G56350.1 | mt | — | 241/26.89 | |
| Peroxiredoxin (Prx) 2P-SH+H2O2→P-S-S-P+2H2O |
1-Cys-Prx | 1-Cys-Prx | XM_010669313.1 | AT1G48130.1 | cyt, nuc | 219/24.41 | 216/24.08 |
| — | 2-Cys-PrxA | — | AT3G11630.1 | ecm, cp | — | 266/29.09 | |
| 2-Cys-Prx | 2-Cys-PrxB | XM_010685313.1 | AT5G06290.1 | ecm, cp, mt | 271/ 29.74 | 273/29.77 | |
| 2-Cys-PrxIIF | 2-Cys-PrxIIF | XM_010696978.1 | AT3G06050.1 | mt | 174/18.66 | 201/21.44 | |
| PrxQ | PrxQ | XM_010689254.1 | AT3G26060.1 | cp, plast | 214/23.65 | 216/23.67 | |
| — | PrxIIA | — | AT1G65990.1 | cyt, nuc | — | 553/62.65 | |
| PrxIIB | PrxIIB | XM_010693075.1 | AT1G65980.1 | cp, cyt, pm | 162/17.51 | 162 /17.42 | |
| — | PrxIIC | — | AT1G65970.1 | cyt | — | 162/17.41 | |
| — | PrxIID | — | AT1G60740.1 | cyt, pm | — | 162/17.47 | |
| PrxIIE | PrxIIE | XM_010685588.1 | AT3G52960.1 | cp, pl, cw | 227/24.07 | 234/24.68 | |
| Alternative oxidase (AOX) 2e−+2H++O2→H2O |
AOX1A | AOX1A | XM_010679200.1 | AT3G22370.1 | mt | 353/39.62 | 354/39.98 |
| AOX1B | AOX1B | XP_010677502.1 | AT3G22360.1 | mt | 102/11.19 | 325/37.43 | |
| — | AOX1C | — | AT3G27620.1 | mt | — | 329/37.82 | |
| — | AOX1D | — | AT1G32350.1 | mt | — | 318/36.20 | |
| AOX2 | AOX2 | XM_010692188.1 | AT5G64210.1 | mt | 329/37.69 | 353/40.09 | |
| PTOX1 | PTOX | XM_010688453.1 | AT4G22260.1 | cp | 363/41.77 | 351/40.57 | |
| PTOX2 | — | XP_010686755.1 | — | cp | 596 /65.40 | — | |
| NADPH oxidase (RBOH) NADPH+e−+O2→NADP−+O−2+H+ |
— | RBOH A | — | AT5G07390.1 | pm | — | 902/102.94 |
| RBOH B | RBOH B | XM_010675737.1 | AT1G09090.2 | pm | 887/100.94 | 843/96.39 | |
| — | RBOH C | — | AT5G51060.1 | pm | — | 905/102.52 | |
| — | RBOH D | — | AT5G47910.1 | pm | — | 921/ 103.91 | |
| RBOH E | RBOH E | XM_010689256.1 | AT1G19230.1 | pm | 945/ 106.04 | 934/105.55 | |
| RBOH F | RBOH F | XM_010678094.1 | AT1G64060.1 | pm | 947/107.72 | 944/108.42 | |
| — | RBOH G | — | AT4G25090.1 | pm | — | 849/96.86 | |
| RBOH H | RBOH H | XM_010687568.1 | AT5G60010.1 | pm | 853/96.65 | 886/100.63 | |
| — | RBOH I | — | AT4G11230.1 | pm | — | 941/106.95 | |
| — | RBOH J | — | AT3G45810.1 | pm | — | 912/102.94 | |
| RBOH K | — | XM_010696961.1 | — | pm | 797/ 91.11 | — | |
Fig. 4.
Phylogenetic relationships among SOD, Prx, AOX, PTOX, and RBOH proteins in Arabidopsis and sugar beet. (A) Dendrogram depicting amino acid sequence-based phylogenetic relationships among 14 different SOD isoforms from B. vulgaris KWS2320 and A. thaliana available in databases. The scale bar shows the number of substitutions per amino acid. Bootstrap analysis of 500 replicates was performed. (B–D) Dendrograms depicting phylogenetic relationships among Prx isoforms (B), AOX/PTOX (C), and RBOHs (D) in sugar beet and Arabidopsis. The numbers above the branches indicate the bootstrap confidence value. The scale bar shows the number of substitutions per amino acid. Bootstrap analysis of 500 replicates was performed. Sugar beet nomenclature is derived from the Arabidopsis nomenclature, considering the closest homologs. If there was a group of related At-sequences, then the letter adopted was that which came first in the alphabet. Two AOX/PTOX and RBOH sequences, each with significant similarity to Arabidopsis genes, received numbers and letters, respectively, which were not yet used in the Arabidopsis system. For gene names and accession numbers please consult Table 1. Sequence identities between different isoforms of the same gene group of sugar beet are provided in Supplementary Table S2.
Fig. 5.
Transcript levels of SOD and Prx in sugar beet under salt stress compared to controls. Transcript amounts were quantified by qPCR at 3 h and 339 h after reaching a concentration of 300 mM NaCl and were calculated relative to actin levels. qPCR data represent the average of three independent experiments with two technical replicates each. The significant difference was calculated using Student’s t-test and labelled with different letters after analysis with Fisher LSD test, with P<0.05, using InfoStat statistical software. C, control; S, salinity.
Transcriptional regulation of Prx genes
The sugar beet genome was searched for genes encoding peroxiredoxins (Dietz, 2011). While ten Prx genes were identified in Arabidopsis, only six were identified in sugar beet and were used to construct a phylogenetic tree (Fig. 4B). Sugar beet encodes only one cytosolic Prx, PrxIIB, while Arabidopsis encodes three and one homologous pseudogene At-PrxIIA. The sugar beet genome contains a single copy of mitochondrial PrxIIF, nuclear 1-CysPrx, 2-CysPrx and PrxQ. Among these thiol peroxidases, 1-CysPrx, PrxQ and PrxIIE transcripts were lower in salt-treated plants at 3 h after up-salting (Fig. 5G–L). At the end of the experiments only transcripts encoding the two chloroplast Prxs, 2-CysPrx and PrxQ, were increased in salt-stressed plants, while cytosolic PrxIIB transcript levels were decreased and all others were similar between both treatments. Relatedness was confirmed by sequence alignment (Supplementary Fig. S5). In order to complement these data, tissue distribution was analyzed (Supplementary Fig. S6) and revealed rather ubiquitous expression, with the exception of FeSOD1 and CuZnSOD2 that were scarcely expressed in seeds. Likewise PrxQ was absent from seeds and 1-CysPrx expression was very low in leaves (Supplementary Fig. S6).
Transcript regulation of alternative and NADPH oxidases
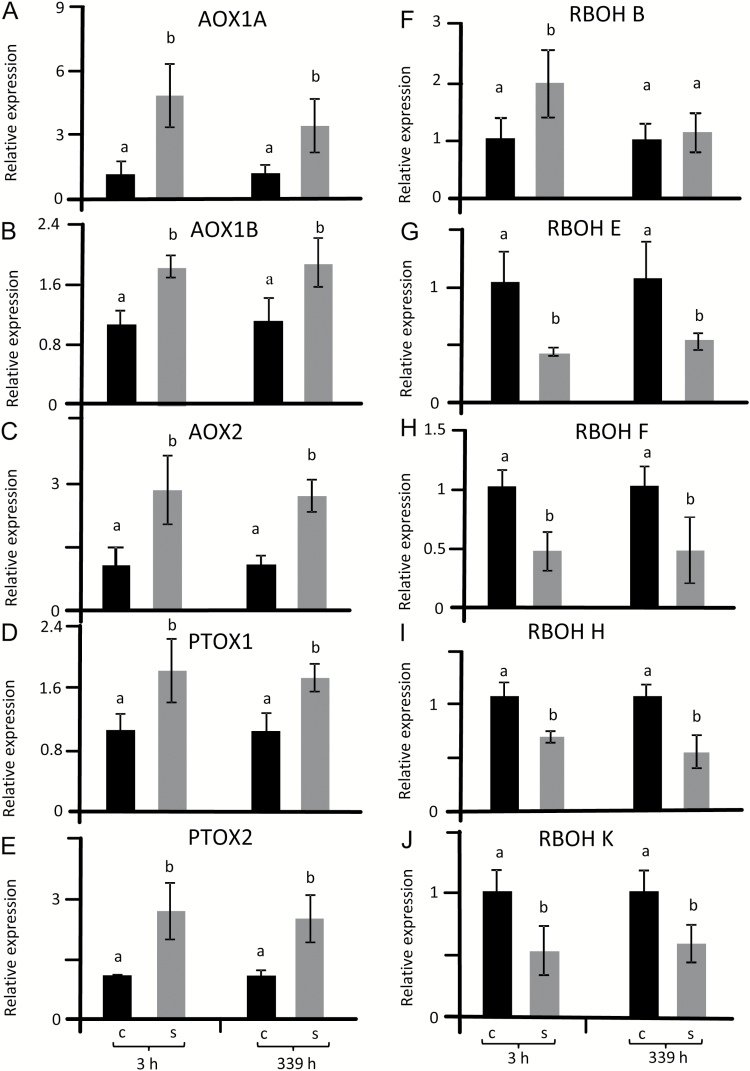
To address the regulation of redox and ROS homeostasis, the sugar beet genome was searched for homologs of important safety valves, namely alternative oxidases (AOX) in mitochondria, plastid terminal oxidase (PTOX) as well as the ROS generation systems of NADPH oxidases, including the respiratory burst oxidase homologue RBOH. Phylogenetic analysis revealed three sequences in sugar beet related to Arabidopsis AtAOX1 and AtAOX2 sequences and two with similarity to At-PTOX (Fig. 4C, Table 1, Supplementary Figs S7, S8). Each of the BvAOX and BvPTOX trancripts was significantly up-regulated in salt-treated sugar beet at 3 h and 14 d (Fig. 6A–E). The search for RBOH genes in the sugar beet genome generated seven hits. Phylogenetic analysis revealed one homolog each for AtRBOHA/B/C/D/G-, E-, F/I, H/J-like, namely BvRBOHB, BvRBOHE, BvRBOHF, and BvRBOHH (Fig. 4D). One BvRBOH isoform was particularly different and named BvRBOHK (Fig. 4D, Supplementary Figs S7, S9). With the exception of BvRBOHB that was up-regulated 3 h after salt treatment, all other BvRBOH isoforms were significantly suppressed under salinity both at 3 h and 14 d (Fig. 6F–J).
Fig. 6.
Transcript levels of AOX/PTOX and RBOH in sugar beet from stressed and non-stressed plants. Samples were taken at 3 h and 14 d after reaching a concentration of 300 mM NaCl as shown in Fig. 1. Transcript amounts were quantified by qPCR and calculated relative to actin levels. qPCR data represent the average of three independent experiments with two technical replicates each. The significant difference was calculated using Student’s t-test and labelled with different letters after analysis with Fisher LSD test, with P<0.05, using InfoStat statistical software. C, control; S, salinity.
Cis-regulatory element analysis
The promoters of the above identified stress-responsive genes of sugar beet were investigated for the presence of cis-regulatory elements. This was carried out to gain some insight into gene regulation and plant signaling under stress conditions. The occurrence of special cis-elements appears to play important roles in the differential regulation of salinity-induced transcripts in B. vulgaris as compared with A. thaliana. Over-representation or exclusive occurrence in B. vulgaris was observed for GATA, WRKY, bHLH, TCP, DOF, ZF-HD, NF-YB, TALE, TBP, dehydrin and BES1 cis-elements (Supplementary Table S3–S5). Over-represented cis-elements vary within B. vulgaris, with GATA and WRKY more abundant in promotor regions of down-regulated transcripts and bHLH, TCP, dehydrin and BES1 more abundant in up-regulated transcripts (Supplementary Tables S3, S4).
Antioxidant enzyme activities
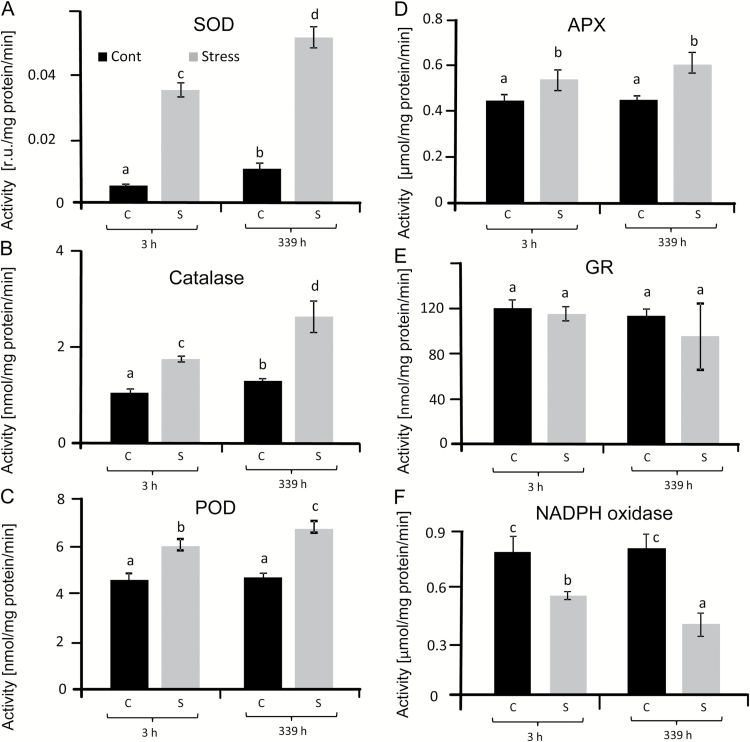
Activities of SOD, catalase, GR, APX and peroxidase (POD) were determined in leaves from salt-stressed and control plants in order to assess the state of the antioxidant systems (Fig. 7). Higher SOD activity was detected in stressed leaves, with the highest activity at 14 d with a 5-fold increase compared with controls (Fig. 7A). GR activity (Fig. 7E) was unaffected by saline growth conditions. However all other antioxidant enzyme activities were stimulated under salinity, with total APX activity showing the least increase, followed by total POD activity and catalase (Fig. 7B–D). Finally, NADPH oxidase activity was determined in plasma membrane-enriched membrane fractions of sugar beet leaves (Fig. 7F). Interestingly, NADPH oxidase was constant in control leaves at 3 h and 14 d, while it decreased by ~40% in salt-stressed plants at 3 h after up-salting up to 300 mM NaCl and by >60% at 14 d.
Fig. 7.
Activities of antioxidant enzymes in sugar beet leaves under salinity stress and control conditions: SOD (A), CAT (B), peroxidase (POD) (C), ascorbate peroxidase (APX) (D), glutathione reductase (GR) (E) and NADPH oxidase (RBOH) (F). Data are means ±SD of n=5 experiments with three measurements each. The significant difference was calculated using Student’s t-test and labelled with different letters after analysis with Fisher LSD test, with P<0.05, using InfoStat statistical software. C, control; S, salinity.
Discussion
Sugar beet is able to grow in saline soils (Ghoulam et al., 2002; Yang et al., 2012; Yang et al., 2013). In line with previous studies our results revealed efficient growth of sugar beet in medium with 300 mM NaCl. This was indicated by unchanged ΦPSII and maintained growth of ~50% of control conditions. Most notably, accumulated H2O2 levels were lower under salinity than control conditions. This response attracted our attention since salt stress usually stimulates ROS accumulation (Miller et al., 2010). Concomitantly, levels of non-protein thiols, oxidized glutathione and ascorbate were decreased in salinized sugar beet. Sugar beet therefore appears to adjust the cellular redox milieu to a lower level of oxidative stress in high NaCl conditions than under control conditions. This regulatory mechanism contrasts the response of other plant species that display salt stress-induced enhancement of ROS accumulation, such as maize (AbdElgawad et al., 2016), rice (Wutipraditkul et al., 2015) and Arabidopsis (Ben Rejeb et al., 2015).
The time-dependent increases in sodium and chloride content as well as osmotic potential (Supplementary Fig. S2) indicate the strong impact of salinity stress and precludes exclusion strategies in sugar beet. Maintenance of 50% photosynthetic CO2 fixation and unaltered ΦPSII demonstrates the high acclimation ability of sugar beet to strongly raised NaCl levels. The reduced growth rate quantitatively mirrored the reduced CO2 fixation rate. Efficient non-photochemical quenching may contribute to lowered ROS burden under salinity. Increased osmotic potential and transient ROS accumulation in halophyte roots are suggested to be instrumental in short-term activation of antioxidant defense, as well as in triggering expression of transcription factors important during long-term salinity (Ellouzi et al., 2014). ROS accumulation was found in the glycophyte Arabidopsis during short- and long-term salt stress (Ellouzi et al., 2011; Ellouzi et al., 2014). Osmotic shock triggered by salinity stress may activate initial ROS production for subsequent activation of defense mechanisms (Choudhury et al., 2013).
Efficient response of SOD in salt-stressed sugar beet
Sugar beet contains homologs of each SOD form typically present in plants such as A. thaliana supporting the assumed conserved functions. High SOD activity is needed for rapid detoxification of O2•−, for example to minimize lipid peroxidation and peroxinitrate formation, if nitric oxide is formed concomitantly. Peroxynitrite (ONOO•−) is highly reactive with many cellular constituents and reacts with various amino acid residues in proteins, in particular with Phe, Trp, Tyr, His, Met and Cys residues (reviewed by Mock and Dietz, 2016). SOD, which is present in multiple subcellular compartments, is considered a first line of defense against ROS (Alscher et al., 2002). With the exception of FeSOD1 transcript levels, which were down-regulated after 3 h and 339 h of salt stress, transcripts of CuZnSOD2, CuZnSOD3, CuZnSOD1, FeSOD3 and MnSOD1 were highly up-regulated under long-term salinity stress. CuSOD2 transcripts showed a small but significant increase after 3 h. As expected, based on the phylogenetic relatedness of SOD genes and the deduced amino acid sequences, MnSOD and FeSOD genes were highly related in sugar beet with sequence identity between both types of 93.6%. In contrast CuZnSOD genes have a very low sequence similarity with FeSOD and MnSOD genes. They separated into a unique cluster (Fig. 4a, Supplementary Fig. S4, Supplementary Table 2) and probably evolved separately in eukaryotes (Alscher et al., 2002). Alscher et al. (2002) suggested that the spatial and temporal function of SOD isozymes is defined by their subcellular location and conditional expression regulated by upstream sequences in their promoters. The pattern of salt-induced transcript responses in sugar beet differed from that reported in Arabidopsis, where CuSOD genes were scarcely responsive while FeSOD genes responded the most (Mittler et al., 2004). The focus of this study was directed towards acclimation to salinity and less to the immediate response to up-salting. Importantly, total SOD activity was up 7-fold at 3 h and 5-fold at 339 h (Fig. 7A). SOD activity determined during up-salting revealed first a parallel increase with H2O2 and then stabilized SOD activity with decreasing H2O2 at the end of the up-salting period (Fig. 1D). Up-regulation of SOD participates in adjusting the low ROS state. Produced H2O2 must be detoxified; this is facilitated by increased activities of catalase and APX (Fig. 7) and increased amounts of 2-CysPrx and PrxQ (Fig. 3). Stimulation of the ascorbate glutathione cycle is suggested to be an important mechanism of salinity tolerance (Stepien and Klobus, 2005).
The role of peroxiredoxins in sugar beet
In higher plants, the minimum set of Prx is six isoforms: one plastidal (PrxIIE), one mitochondrial (PrxIIF), one cytosolic type II Prx (PrxIIB), one cytosolic/nuclear 1-CysPrx, one plastid 2-CysPrx and one plastidal PrxQ (Dietz et al., 2006). Arabidopsis and rice express additional isoforms, ten and nine, respectively. Interestingly our search of the sugar beet genome conforms with the predicted minimum set of six Prx genes. The enzymatic activity of the different plant Prx isoforms has been well explored but the precise function within the redox regulatory network of the cell is still elusive. Functions as peroxidases, proximity peroxidases, ROS sensors, interaction partners and chaperones have all been proposed (König et al., 2012). 2-Cys-PrxB and PrxQ transcripts and protein levels were slightly increased during long-term salinity stress, while all other transcript levels decreased or were unaltered. The unresponsiveness of the single cytosolic PrxIIB gene contrasted results from Arabidopsis where AtPrxIIB, C and D showed strong responses to salinity and other stresses (Horling et al., 2002; Horling et al., 2003). This difference suggests that gene duplication in Arabidopsis allowed for diversification in stress response, while BvPrxIIB adopts a function more similar to housekeeping. These results indicate that 2-CysPrx contributes to the protection of chloroplast structures against oxidative damage by participating in detoxification processes (Baier and Dietz, 1999a,b). Furthermore, König et al. (2002) proposed that 2-Cys-Prx acts not only in the water–water cycle pathway for energy dissipation in photosynthesis but also in peroxide detoxification in plastids during the dark phase. 1-CysPrx transcripts strongly but transiently decreased during salt stress. 1-CysPrx in Arabidopsis is highly abundant in seeds compared with root tissues but was not expressed in leaf tissues or only in some cells like petiole junctions (Stacy et al., 1996; Haslekas et al., 1998; Stacy et al., 1999). 1-CysPrx in A. thaliana and B. vulgaris comprise 216 and 219 amino acids, respectively, possess a C-terminal extension with a putative nuclear signal, and as shown for Arabidopsis is localized in the nucleus and cytosol (Rouhier and Jacquot, 2002).
The AOX gene family and their transcript response to salinity
Alternative oxidases function in the dissipation of reducing power in energetic electron transport of both mitochondria (AOX) and chloroplasts (PTOX). In the sugar beet genome, three AOX genes were identified that were highly similar to AtAOX1 and AtAOX2 groups in A. thaliana and therefore named BvAOX1A, BvAOX1B and BvAOX2. Two genes coding for proteins with high homology to PTOX were identified in sugar beet and named BvPTOX1 and BvPTOX2. Efficient and coordinated up-regulation of AOX and PTOX may represent a specific feature of sugar beet during salinity stress tolerance because transcripts of all four AOX and AOX-like genes as well as PTOX were up-regulated both at 3 h and 339 h of salt stress. Up-regulation of PTOX and AOX probably stabilize the photosynthetic quantum yield of PSII under salinity despite inhibited photosynthesis. Overexpression of AtAOX1a lowers ROS formation in leaves (Smith et al., 2009). This result is in line with the hypothesis that AOX participates in cell reprogramming under salinity stress. Activation of AOX limits ROS release from the mitochondrial respiratory chain. This activation is achieved in Arabidopsis by transcriptional control and by post-translational mechanisms (Rhoads et al., 1998; Smith et al., 2009). If AOX-dependent dissipation of excess reducing power is absent, ROS accumulates and can diffuse to other cell compartments (reviewed by Dietz et al., 2016). Thus AOX contributes to mitigating oxidative stress under conditions of high salinity stress (Hanqing et al., 2010). Moreover, genetic enhancement of AOX expression decreases ROS levels in transgenic tobacco (Maxwell et al., 1999). Also, the abundance of PTOX positively correlates with salinity levels (Ivanov et al., 2012; Nawrocki et al., 2015). From these findings we conclude that transcriptional up-regulation of AOX and PTOX participates in the suppression of ROS accumulation in salt-stressed sugar beet.
The RBOH gene family and their regulation
Sugar beet expresses five RBOH and RBOH-like genes compared to ten in Arabidopsis, where AtRBOHD and AtRBOHF preferentially accumulate upon salt stress (Ma et al., 2012). Arabidopsis single mutants devoid of AtrbohD or AtrbohF are indistinguishable from wild type if stressed with 300 mM NaCl. Conversely, the double mutant AtrbohD/F reveals reduced viability in 300 mM NaCl administered for 14 d and simultaneously accumulates less H2O2 (Ma et al., 2012). Interestingly in sugar beet viability remained at 100% under salt stress despite the strong decrease in RBOH activity. Ben Rejeb et al. (2015) assigned a crucial role to RBOHs in salinity-induced regulation of the antioxidant defense in Arabidopsis under short-term treatments. The salinity-induced activation of endosomal RBOH is suppressed in the phosphatidylinositol-3-kinase mutant pi3k (Leshem et al., 2007) resulting in less oxidative stress. Post-translational control of RBOH by cellular Ca2+, ROS and phosphorylation networks participates in the early responses to salinity, particularly in glycophytes but possibly also in halophytes (Kurusu et al., 2015). Moreover, BvRBOHB levels doubled at 3 h in contrast to all other RBOH transcripts that were down-regulated both at 3 h and 339 h. Thus BvRBOHB may be involved in early acclimation responses to ionic and osmotic stress, similar to AtRBOHD and AtRBOHF in Arabidopsis (Ma et al., 2012).
Tight regulation of redox and ROS in salt-stressed sugar beet
All the data from this study lead to a consistent response pattern of sugar beet under salt stress, which ensures low ROS accumulation and maintains metabolism and growth. Up-regulation of AOX and PTOX facilitates the dissipation of excessive reducing power accumulating in stressed plants. The efficiency of this regulation is indicated by the unchanged and high ΦPSII in the growth of light-exposed plants, revealing an avoidance of over-reduction. Over-reduced electron transport chains are a prime source for the generation of superoxide, singlet oxygen and other ROS (Oelze et al., 2008). Likewise the activity of RBOH as a ROS generator system is strongly suppressed in short- and long-term salt-treated sugar beet. Up-regulation of SOD, APX, POD, catalase and some Prxs assists in lowering steady state ROS levels. Here the described efficient regulation of the ROS network in sugar beet provides a solid basis for futher investigating the underlying regulation at the level of signaling and transcription factors, including cis-elements and hormones such as abscisic acid (ABA).
ABA-dependent regulation via dehydrin in B. vulgaris may be an additional regulatory mechanism compared with A. thaliana, which coordinates up-regulation of antioxidative enzymes and lowers H2O2 levels under salt stress as compared with controls (Table 3). The cooperative action of cis-elements COR15 (dehydrin) and the promoter configuration are crucial for the regulation of ABA-induced (Busk and Pagès, 1998) and drought-regulated gene expression (Baker et al., 1994). Under salinity dehydrin1 overexpressing plants maintain lower H2O2 levels than wild type plants (Kumar et al., 2014). Dehydrin may constitute part of a general molecular mechanism used by all land plants to protect them from injury during cellular dehydration under salinity and osmotic stress (Saavedra et al., 2006).
BES1 is only present in up-regulated transcripts of antioxidant enzymes and therefore might function as a positive regulator in B. vulgaris leading to higher antioxidant defense and higher salt tolerance (Table 3). The BES1 cis-element family binds plant-specific transcription factors that cooperate with the bHLH transcription factor BIM1 to regulate brassinosteroid-induced abiotic stress responsive gene expression (Yin et al., 2005). BES1 and AtMYB30 function cooperatively to promote brassinosteroid target gene expression through a mechanism by which AtMYB30, a direct target of BES1, amplifies brassinosteroid signaling by helping BES1 activate downstream target genes (Li et al., 2009). It will be of interest to compare the genome-wide distribution and function of these elements in sugar beet and glycophytes in the future.
Supplementary Material
Acknowledgements
MSH acknowledges the support of the German Academic Exchange Service (DAAD) and AIS of the Egyptian government for approving his visiting fellowship. KJD acknowledges funding by the German Science Foundation (DI 346/14). MSH designed the study, conducted all experiments, measurements, analysis and wrote the paper. AIS helped during western blot analysis, sq-PCR and writing. MM helped in setting up the hydroponic culture, measured ΦPSII during upsalting and helped during cis-regulatory elements analysis. KJD designed and guided the study, discussed the data and wrote the paper.
Glossary
Abbreviations:
- ΦPSII
quantum yield of photosystem II
- 2-CysPrx
2-cysteine peroxiredoxin
- AA
ascorbate
- ABA
abscisic acid
- APX
ascorbate peroxidase
- AOX
mitochondrial alternative oxidase
- CO2
carbon dioxide
- DHA
dehydroascorbate
- DTNB
5,5’-dithiobis(2-nitrobenzoate)
- GPX
glutathione peroxidases
- GR
glutathione reductase
- GSH
glutathione
- GSSG
oxidized glutathione
- H2O2
hydrogen peroxide
- MDA
malondialdehyde
- ML
maximum likelihood
- NaCl
sodium chloride
- NPT
non-protein thiol
- O2•−
superoxide
- ONOO•−
peroxynitrite
- PEG
polyethylene glycol
- POD
peroxidase
- Prx
peroxiredoxins
- PrxQ
chloroplast peroxiredoxin Q
- PSII
photosystem II
- PTOX
plastid terminal oxidase
- RBOH
respiratory burst oxidase homolog
- ROS
reactive oxygen species
- SOD
superoxide dismutase
- TBA
thiobarbituric acid
- TBST
Tris-buffered saline with 0.1% Tween 20
- TCA
trichloroacetic acid
- XTT
Na 3′-[1-[phenylaminocarbonyl]-3,4-tetrazolium]-bis(4-methoxy-6-nitro)benzenesulfonic acid.
References
- AbdElgawad H, Zinta G, Hegab MM, Pandey R, Asard H, Abuelsoud W. 2016. High salinity induces different oxidative stress and antioxidant responses in maize seedlings organs. Frontiers in Plant Science 7, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alscher RG, Erturk N, Heath LS. 2002. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. Journal of Experimental Botany 53, 1331–1341. [PubMed] [Google Scholar]
- Baier M, Dietz KJ. 1999. Protective function of chloroplast 2-cysteine peroxiredoxin in photosynthesis. Evidence from transgenic Arabidopsis. Plant Physiology 119, 1407–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M, Dietz KJ. 1999. Alkyl hydroperoxide reductases: the way out of the oxidative breakdown of lipids in chloroplasts. Trends in Plant Science 4, 166–168. [DOI] [PubMed] [Google Scholar]
- Baker SS, Wilhelm KS, Thomashow MF. 1994. The 5’-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Molecular Biology 24, 701–713. [DOI] [PubMed] [Google Scholar]
- Ben Rejeb K, Benzarti M, Debez A, Bailly C, Savouré A, Abdelly C. 2015. NADPH oxidase-dependent H2O2 production is required for salt-induced antioxidant defense in Arabidopsis thaliana. Journal of Plant Physiology 174, 5–15. [DOI] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Busk PK, Pagès M. 1998. Regulation of abscisic acid-induced transcription. Plant Molecular Biology 37, 425–435. [DOI] [PubMed] [Google Scholar]
- Choudhury S, Panda P, Sahoo L, Panda SK. 2013. Reactive oxygen species signaling in plants under abiotic stress. Plant Signaling & Behavior 8, e23681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Considine MJ, Holtzapffel RC, Day DA, Whelan J, Millar AH. 2002. Molecular distinction between alternative oxidase from monocots and dicots. Plant Physiology 129, 949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CN, Zheng HQ, Wu NY, Chien CH, Huang HD, Lee TY, Chiang-Hsieh YF, Hou PF, Yang TY, Chang WC. 2015. PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Research 44, D1154–D1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DelLongo OT, Gonzalez CA, Pastori MG, Trippi VS. 1993. Antioxidant defenses under hyperoxygenic and hyperosmotic conditions in leaves of two lines of maize with differential sensitivity to drought. Plant and Cell Physiology 34, 1023–1028. [Google Scholar]
- Dietz KJ. 2011. Comprehensive review: peroxiredoxins in plants and cyanobacteria. Antioxidants and Redox Signalling 15, 1129–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ. 2016. Thiol-based peroxidases and ascorbate peroxidases: why plants rely on multiple peroxidase systems in the photosynthesizing chloroplast? Molecules and Cells 39, 20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ, Jacob S, Oelze ML, Laxa M, Tognetti V, de Miranda SM, Baier M, Finkemeier I. 2006. The function of peroxiredoxins in plant organelle redox metabolism. Journal of Experimental Botany 57, 1697–1709. [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Turkan I, Krieger-Liszkay A. 2016. Redox- and reactive oxygen species-dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiology 171, 1541–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Minoche AE, Holtgräwe D, et al. 2014. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505, 546–549. [DOI] [PubMed] [Google Scholar]
- Ellouzi H, Ben Hamed K, Hernández I, Cela J, Müller M, Magné C, Abdelly C, Munné-Bosch S. 2014. A comparative study of the early osmotic, ionic, redox and hormonal signaling response in leaves and roots of two halophytes and a glycophyte to salinity. Planta 240, 1299–1317. [DOI] [PubMed] [Google Scholar]
- Ellouzi H, Hamed KB, Cela J, Munné-Bosch S, Abdelly C. 2011. Early effects of salt stress on the physiological and oxidative status of Cakile maritima (halophyte) and Arabidopsis thaliana (glycophyte). Physiologia Plantarum 142, 128–143. [DOI] [PubMed] [Google Scholar]
- Elstner EF. 1991. Mechanisms of oxygen activation in different compartments of plant cells. In: Pell EJ, Steffen KL, eds. Active oxygen/oxidative stress and plant metabolism. Rockville, MD: American Society of Plant Physiologists, 13–25. [Google Scholar]
- Finkemeier I, Goodman M, Lamkemeyer P, Kandlbinder A, Sweetlove LJ, Dietz KJ. 2005. The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. Journal of Biological Chemistry 280, 12168–12180. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Bloom AJ, Queval G, Noctor G. 2009. Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annual Review of Plant Biology 60, 455–484. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Descourvières P, Kunert KJ. 1994. Protection against oxygen radicals: an important defence mechanism studied in transgenic plants. Plant, Cell and Environment 17, 507–523. [Google Scholar]
- Ghoulam C, Foursy A, Fares K. 2002. Effects of salinity stress on growth, inorganic ions and proline accumulation in relation to osmotic adjustment in five sugar beet cultivars. Environmental and Experimental Botany 47, 39–50. [Google Scholar]
- Griffith OW. 1980. Determination of glutathione and glutathione disulfide using glutathione reductase and 2-vinylpyridine. Analytical Biochemistry 106, 207–212. [DOI] [PubMed] [Google Scholar]
- Hamouda I, Badri M, Mejri M, Cruz C, Siddique KH, Hessini K. 2016. Salt tolerance of Beta macrocarpa is associated with efficient osmotic adjustment and increased apoplastic water content. Plant Biology 18, 369–375. [DOI] [PubMed] [Google Scholar]
- Hanqing F, Yifeng W, Hongyu L, Rongfang W, Kun S, Lingyun J. 2010. Salinity stress-induced expression of rice AOX1a is mediated through an accumulation of hydrogen peroxide. Biologia 65, 868–873. [Google Scholar]
- Haslekås C, Stacy RA, Nygaard V, Culiáñez-Macià FA, Aalen RB. 1998. The expression of a peroxiredoxin antioxidant gene, AtPer1, in Arabidopsis thaliana is seed-specific and related to dormancy. Plant Molecular Biology 36, 833–845. [DOI] [PubMed] [Google Scholar]
- Horie T, Schroeder JI. 2004. Sodium transporters in plants. Diverse genes and physiological functions. Plant Physiology 136, 2457–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horling F, König J, Dietz KJ. 2002. Type II peroxiredoxin C, a member of the peroxiredoxin family of Arabidopsis thaliana: its expression and activity in comparison with other peroxiredoxins. Plant Physiology and Biochemistry 40, 491–499. [Google Scholar]
- Horling F, Lamkemeyer P, König J, Finkemeier I, Kandlbinder A, Baier M, Dietz KJ. 2003. Divergent light-, ascorbate-, and oxidative stress-dependent regulation of expression of the peroxiredoxin gene family in Arabidopsis. Plant Physiology 131, 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MS, Dietz KJ. 2016. Tuning of redox regulatory mechanisms, reactive oxygen species and redox homeostasis under salinity stress. Frontiers in Plant Science 7, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain TM, Hazara M, Sultan Z, Saleh BK, Gopal GR. 2008. Recent advances in salinity stress biology a review. Biotechnology and Molecular Biology Reviews 3, 8–13. [Google Scholar]
- Ivanov AG, Rosso D, Savitch LV, Stachula P, Rosembert M, Oquist G, Hurry V, Hüner NP. 2012. Implications of alternative electron sinks in increased resistance of PSII and PSI photochemistry to high light stress in cold-acclimated Arabidopsis thaliana. Photosynthesis Research 113, 191–206. [DOI] [PubMed] [Google Scholar]
- Katori T, Ikeda A, Iuchi S, Kobayashi M, Shinozaki K, Maehashi K, Sakata Y, Tanaka S, Taji T. 2010. Dissecting the genetic control of natural variation in salt tolerance of Arabidopsis thaliana accessions. Journal of Experimental Botany 61, 1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaundal A, Rojas CM, Mysore KS. 2012. Measurement of NADPH oxidase activity in plants. Bio-Protocol 2, e278. [Google Scholar]
- Keller T, Damude HG, Werner D, Doerner P, Dixon RA, Lamb C. 1998. A plant homolog of the neutrophil NADPH oxidase gp91phox subunit gene encodes a plasma membrane protein with Ca2+ binding motifs. The Plant Cell 10, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CS, Didierjean C, Navrot N, Panjikar S, Mulliert G, Rouhier N, Jacquot JP, Aubry A, Shawkataly O, Corbier C. 2007. Crystal structures of a poplar thioredoxin peroxidase that exhibits the structure of glutathione peroxidases: insights into redox-driven conformational changes. Journal of Molecular Biology 370, 512–529. [DOI] [PubMed] [Google Scholar]
- König J, Baier M, Horling F, Kahmann U, Harris G, Schurmann P, Dietz KJ. 2002. The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redoxhierarchy of photosynthetic electron flux. Proceedings of National Academy of Sciences, USA 99, 5738–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Muthuramalingam M, Dietz KJ. 2012. Mechanisms and dynamics in the thiol/disulfide redox regulatory network: transmitters, sensors and targets. Current Opinion in Plant Biology 15, 261–268. [DOI] [PubMed] [Google Scholar]
- Kumar M, Lee SC, Kim JY, Kim SJ, Aye SS, Kim SR. 2014. Over-expression of dehydrin gene, OsDhn1, improves drought and salt stress tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). Journal of Plant Biology 57, 383–393. [Google Scholar]
- Kurusu T, Kuchitsu K, Tada Y. 2015. Plant signaling networks involving Ca(2+) and Rboh/Nox-mediated ROS production under salinity stress. Frontiers in Plant Science 6, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Leshem Y, Seri L, Levine A. 2007. Induction of phosphatidylinositol 3-kinase-mediated endocytosis by salinity stress leads to intracellular production of reactive oxygen species and salinity tolerance. The Plant Journal 51, 185–197. [DOI] [PubMed] [Google Scholar]
- Li L, Yu X, Thompson A, Guo M, Yoshida S, Asami T, Chory J, Yin Y. 2009. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. The Plant Journal 58, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Zhang H, Sun L, Jiao Y, Zhang G, Miao C, Hao F. 2012. NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na⁺/K⁺homeostasis in Arabidopsis under salt stress. Journal of Experimental Botany 63, 305–317. [DOI] [PubMed] [Google Scholar]
- Magaña C, Núñez-Sánchez N, Fernández-Cabanás VM, García P, Serrano A, Pérez-Marín D, Pemán JM, Alcalde E. 2011. Direct prediction of bioethanol yield in sugar beet pulp using near infrared spectroscopy. Bioresource Technology 102, 9542–9549. [DOI] [PubMed] [Google Scholar]
- Malik CP, Singh MB. 1980. Plant Enzymology and Histoenzymology. New Delhi, India: Kalyani Publishers, 53. [Google Scholar]
- Maxwell DP1, Wang Y, McIntosh L. 1999. The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proceedings of National Academy of Sciences, USA 96, 8271–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. 2010. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant, Cell & Environment 33, 453–467. [DOI] [PubMed] [Google Scholar]
- Mittler R, Poulos TL. 2005. Ascorbate peroxidase. In: Smirnoff N, ed. Antioxidants and reactive oxygen species in plants. Oxford: Blackwell Publishing, 87–100. [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. 2004. Reactive oxygen gene network of plants. Trends in Plant Science 9, 490–498. [DOI] [PubMed] [Google Scholar]
- Mock HP, Dietz KJ. 2016. Redox proteomics for the assessment of redox-related posttranslational regulation in plants. Biochimica et Biophysica Acta 1864, 967–973. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. 2008. Mechanisms of salinity tolerance. Annual Review of Plant Biology 59, 651–681. [DOI] [PubMed] [Google Scholar]
- Muthuramalingam M, Seidel T, Laxa M, Nunes de Miranda SM, Gärtner F, Ströher E, Kandlbinder A, Dietz KJ. 2009. Multiple redox and non-redox interactions define 2-Cys peroxiredoxin as a regulatory hub in the chloroplast. Molecular Plant 2, 1273–1288. [DOI] [PubMed] [Google Scholar]
- Navrot N, Collin V, Gualberto J, Gelhaye E, Hirasawa M, Rey P, Knaff DB, Issakidis E, Jacquot JP, Rouhier N. 2006. Plant glutathione peroxidases are functional peroxiredoxins distributed in several subcellular compartments and regulated during biotic and abiotic stresses. Plant Physiology 142, 1364–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki WJ, Tourasse NJ, Taly A, Rappaport F, Wollman FA. 2015. The plastid terminal oxidase: its elusive function points to multiple contributions to plastid physiology. Annual Review of Plant Biology 66, 49–74. [DOI] [PubMed] [Google Scholar]
- Oelze ML, Kandlbinder A, Dietz KJ. 2008. Redox regulation and overreduction control in the photosynthesizing cell: complexity in redox regulatory networks. Biochimica et Biophysica Acta 1780, 1261–1272. [DOI] [PubMed] [Google Scholar]
- Oelze ML, Vogel MO, Alsharafa K, Kahmann U, Viehhauser A, Maurino VG, Dietz KJ. 2012. Efficient acclimation of the chloroplast antioxidant defence of Arabidopsis thaliana leaves in response to a 10- or 100-fold light increment and the possible involvement of retrograde signals. Journal of Experimental Botany 63, 1297–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida AK, Das AB, Mittra B. 2004. Effects of salt on growth, ion accumulation, photosynthesis and leaf anatomy of the mangrove Bruguiera parviflora. Trees-Structure and Function 18, 167–174. [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pèrez FJ, Rubio S. 2006. An improved chemiluminescence method for hydrogen peroxide determination in plant tissues. Plant Growth Regulation 48, 89–95. [Google Scholar]
- Rhoads DM, Umbach AL, Sweet CR, Lennon AM, Rauch GS, Siedow JN. 1998. Regulation of the cyanide-resistant alternative oxidase of plant mitochondria. Identification of the cysteine residue involved in alpha-keto acid stimulation and intersubunit disulfide bond formation. The Journal of Biological Chemistry 273, 30750–30756. [DOI] [PubMed] [Google Scholar]
- Rouhier N, Jacquot JP. 2002. Plant peroxiredoxins: alternative hydroperoxide scavenging enzymes. Photosynthesis Research 74, 259–268. [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Research 37, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sairam RK, Rao KV, Srivastava GC. 2002. Differential response of wheat genotypes to long term salinity stress in relation to oxidative stress, antioxidant activity and osmolyte concentration. Plant Science 163, 1037–1046. [Google Scholar]
- Saavedra L, Svensson J, Carballo V, Izmendi D, Welin B, Vidal S. 2006. A dehydrin gene in Physcomitrella patens is required for salt and osmotic stress tolerance. The Plant Journal 45, 237–249. [DOI] [PubMed] [Google Scholar]
- SenGupta A, Webb RP, Holaday AS, Allen RD. 1993. Overexpression of superoxide dismutase protects plants from oxidative stress. Plant Physiology 103, 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Melino VJ, Sweetman C, Soole KL. 2009. Manipulation of alternative oxidase can influence salt tolerance in Arabidopsis thaliana. Physiologia Plantarum 137, 459–472. [DOI] [PubMed] [Google Scholar]
- Stacy RA, Munthe E, Steinum T, Sharma B, Aalen RB. 1996. A peroxiredoxin antioxidant is encoded by a dormancy-related gene, Per1, expressed during late development in the aleurone and embryo of barley grains. Plant Molecular Biology 31, 1205–1216. [DOI] [PubMed] [Google Scholar]
- Stacy RA, Nordeng TW, Culianez-Macia FA, Aalen RB. 1999. The dormancy-related peroxiredoxin anti-oxidant, PER1, is localized to the nucleus of barley embryo and aleurone cells. The Plant Journal 19, 1–8. [DOI] [PubMed] [Google Scholar]
- Steffens NO, Galuschka C, Schindler M, Bülow L, Hehl R. 2005. AthaMap web tools for database-assisted identification of combinatorial cis-regulatory elements and the display of highly conserved transcription factor binding sites in Arabidopsis thaliana. Nucleic Acids Research 33, W397–W402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien P, Johnson GN. 2009. Contrasting responses of photosynthesis to salt stress in the glycophyte Arabidopsis and the halophyte thellungiella: role of the plastid terminal oxidase as an alternative electron sink. Plant Physiology 149, 1154–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien P, Klobus GN. 2005. Antioxidant defense in the leaves of C3 and C4 plants under salinity stress. Physiologia Plantarum 125, 31–40. [Google Scholar]
- Stewart RR, Bewley JD. 1980. Lipid peroxidation associated with accelerated aging of soybean axes. Plant Physiology 65, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ströher E, Wang XJ, Roloff N, Klein P, Husemann A, Dietz KJ. 2009. Redox-dependent regulation of the stress-induced zinc-finger protein SAP12 in Arabidopsis thaliana. Molecular Plant 2, 357–367. [DOI] [PubMed] [Google Scholar]
- Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, Narusaka Y, Narusaka M, Zhu JK, Shinozaki K. 2004. Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiology 135, 1697–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turan MA, Elkarim AHA, Taban N, Taban S. 2009. Effect of salinity stress on growth, stomatal resistance, proline and chlorophyll concentrations on maize plant. African Journal of Agricultural Research 4, 893–897. [Google Scholar]
- Urbanek H, Kuzniak-Gebarowska E, Herka K. 1991. Elicitation of defense response in bean leaves by Botrytis cinerea polygalacturonase. Acta Physiologiae Plantarum 13, 43–50. [Google Scholar]
- Vastarelli P, Moschella A, Pacifico D, Mandolino G. 2013. Water stress in Beta vulgaris: osmotic adjustment response and gene expression analysis in ssp. vulgaris and maritima. American Journal of Plant Sciences 4, 11–16. [Google Scholar]
- Wormuth D, Baier M, Kandlbinder A, Scheibe R, Hartung W, Dietz KJ. 2006. Regulation of gene expression by photosynthetic signals triggered through modified CO2 availability. BMC Plant Biology 6, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutipraditkul N, Wongwean P, Buaboocha T. 2015. Alleviation of salt-induced oxidative stress in rice seedlings by proline and/or glycinebetaine. Biologia Plantarum 59, 547–553. [Google Scholar]
- Yang L, Ma C, Wang L, Chen S, Li H. 2012. Salt stress induced proteome and transcriptome changes in sugar beet monosomic addition line M14. Journal of Plant Physiology 169, 839–850. [DOI] [PubMed] [Google Scholar]
- Yang L, Zhang Y, Zhu N, Koh J, Ma C, Pan Y, Yu B, Chen S, Li H. 2013. Proteomic analysis of salt tolerance in sugar beet monosomic addition line M14. Journal of Proteome Research 12, 4931–4950. [DOI] [PubMed] [Google Scholar]
- Yeo A. 1998. Molecular biology of salinity tolerance in the context of whole-plant physiology. Journal of Experimental Botany 49, 915–929. [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. 2005. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120, 249–259. [DOI] [PubMed] [Google Scholar]
- Yoshimura K, Yabuta Y, Ishikawa T, Shigeoka S. 2000. Expression of spinach ascorbate peroxidase isoenzymes in response to oxidative stresses. Plant Physiology 123, 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lai J, Sun S, Li Y, Liu Y, Liang L, Chen M, Xie Q. 2008. Comparison analysis of transcripts from the halophyte Thellungiella halophila. Journal of Integrative Plant Biology 50, 1327–1335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.