Highlight
During Arabidopsis somatic embryogenesis phytoglobin 2 exercises its effects only when targeted to the nucleus, while when cytoplasmic it mimics the effects of its mutant form.
Keywords: Auxin, dexamethasone, phytoglobin, somatic embryogenesis.
Abstract
Mutation of phytoglobin 2 (Pgb2) increases the number of somatic embryos in Arabidopsis. To assess the effects of the cellular localization of Pgb2 on embryo formation, an inducible system expressing a fusion protein consisting of Pgb2 linked to the steroid-binding domain of the rat glucocorticoid receptor (GR) was introduced in a pgb2 mutant line lacking the ability to express Pgb2. In this transgenic system, Pgb2 remains in the cytoplasm but migrates into the nucleus upon exposure to dexamethasone (DEX). Pgb2 retention in the cytoplasm, in the absence of DEX, increased the number of somatic embryos and reduced the expression of MYC2 - an inhibitor of the synthesis of auxin, which is the inductive signal for embryogenesis. Removal of DEX also induced the expression of several genes involved in the biosynthesis of tryptophan and the auxin, indole-3-acetic acid (IAA). These genes included: tryptophan synthase-α subunit (TSA1) and tryptophan synthase-β subunit (TSB1), which are involved in the synthesis of tryptophan, cytochrome P450 CYP79B2 (CYP79B2) and amidase 1 (AMI1), which participate in the formation of IAA via indole-3-acetaldoxime, and several members of the YUCCA family, including YUC1 and 4, which are also required for IAA synthesis. Retention of Pgb2 in the cytoplasm by removal of DEX increased the staining pattern of IAA along the cotyledons of the explants generating embryogenic tissue. Staining for IAA decreased when Pgb2 translocated into the nucleus in response to the application of DEX. Collectively, these results suggest that the presence of Pgb2 in the cytoplasm, but not in the nucleus, phenocopies the effects of Pgb2 mutation in inducing somatic embryogenesis.
Introduction
Embryogenesis is a crucial event in the plant life cycle. It is initiated by the formation of the zygote, which through precise and conserved cell division and differentiation patterns, generates a mature embryo consisting of an embryonic axis separating the shoot and root apical meristems and one or more cotyledons (Jenik et al. 2007). The embryogenic program can be recapitulated in vitro through manipulating the media and culture components that induce cells to reprogram their developmental fate and embark along an embryogenic pathway. The genetic basis of this reprogramming is largely unknown (Karami et al. 2009). This process is best exemplified during somatic embryogenesis where somatic cells, i.e. cells other than gametes, can produce genetically identical somatic embryos with structural and physiological features resembling those of their zygotic counterparts (Stasolla and Yeung, 2003). These desirable characteristics, as well as the possibility to generate a large number of synchronous embryos in an enclosed environment and in a short period of time, make somatic embryogenesis a suitable model to investigate the structural, physiological and molecular events governing embryogenesis (Mordhorst et al. 2002; Su and Zhang 2009).
In Arabidopsis, somatic embryogenesis can be initiated by culturing cotyledonary zygotic embryos on an induction medium containing auxin, which is the inductive signal required for the dedifferentiation and redirection of somatic cells towards the embryogenic route (Raghavan, 2004). The initial phases on induction medium involve the expansion of the cotyledons followed by the generation of embryogenic tissue arising from the adaxial side of the cotyledons. Removal of exogenous auxin triggers the development of the somatic embryo (see Supplementary Fig. S1 available at JXB online). Formation of the embryogenic tissue during the induction phase is mainly the result of localized proliferation along the surface of the cotyledons, as the remaining regions of the zygotic embryos do not form embryogenic cells (Raghavan, 2004). Generation of auxin maxima, through PIN1-mediated polar transport, along the embryogenic tissue is essential for the establishment of stem cells and the generation of somatic embryos (Su and Zhang, 2009). Factors influencing these processes have begun to emerge (Bai et al., 2013; Elhiti et al., 2013).
Hemoglobins are present in all nucleated organisms and have been well characterized for their ability to bind and transport oxygen and other small gas ligands. In plants, proteins with hemoglobin-like features were initially found in nodules containing nitrogen-fixing bacteria and named leghemoglobins (Smagghe et al., 2009), while those later associated with organs other than nodules were referred as non-symbiotic hemoglobins (Hunt et al., 2001). The subsequent characterization of several types of plant hemoglobins with diverse functions (Garrocho-Villegas et al., 2007; Hill, 2012) has resulted in a change in nomenclature from plant hemoglobins to phytoglobins (Pgbs) (Hill et al., 2016).
Three classes of Pgbs with distinct oxygen binding characteristics and expression patterns have been described (Hunt et al., 2001), with the majority of studies centered around members of class 1 and 2. The two classes mainly differ in their oxygen binding affinities with a Km close to 2 nM for members of class 1 and 150 nM for class 2 (Hoy and Hargrove, 2008; Dordas, 2009). The Arabidopsis class 1 Pgb, Pgb1, has been mainly studied in relation to its capacity to scavenge nitric oxide (NO) in low oxygen environments (Dordas et al., 2003; Dordas et al., 2004), and its ability to protect plant cells during prolonged stress conditions (Perazzolli et al., 2004). This protective role has been further confirmed by the increase in survival and maintenance of the energy status observed in Arabidopsis roots overexpressing Pgb1 (Hunt et al., 2002). While less is known about the Arabidopsis class 2 Pgb, Pgb2, its involvement in stress responses through mechanisms characteristic of Pgb1 cannot be discounted. Like Pgb1, overexpression of Pgb2 enhances survival under hypoxic conditions through removal of cellular NO (Hebelstrup et al., 2006; Hebelstrup and Jensen, 2008; Hebelstrup et al., 2012). Distinct conditions inducing the two Pgbs, with cytokinin and cold temperatures only effective in increasing expression of Pgb2, suggest that the functions of Pgb1 and Pgb2 might not be fully redundant. This notion is further confirmed by their different localization patterns, with Pgb2 preferentially expressed in young organs including immature seeds, fruit, and leaves, as well as somatic embryos (Hendriks et al., 1998; Hunt et al., 2002; Wang et al., 2003).
Deviation in morphogenesis occurs as a result of altered Pgb2 expression. Besides affecting meristem function by accelerating the transition of vegetative meristems into inflorescence meristems (Hebelstrup and Jensen, 2008) and enhancing in vitro shoot organogenesis when ectopically expressed (Wang et al., 2011), Pgb2 regulates Arabidopsis somatic embryogenesis. Mutation of Pgb2 increases the number of Arabidopsis somatic embryos by suppressing the expression of MYC2, a repressor of auxin synthesis, and inducing transcription of several indole-3-acetic acid (IAA) biosynthetic genes (Elhiti et al., 2013). These transcriptional changes resulted in elevated levels of IAA within the embryogenic tissue generated from the adaxial sides of the cotyledons of the explants (Elhiti et al., 2013). Generation of IAA maxima, mediated by relocation of PIN1, marks the future somatic embryo initiation sites and encourages stem cell formation and production of somatic embryos (Su and Zhang, 2009; Elhiti et al., 2013). The precise transduction pathway triggered by mutation of Pgb2 and resulting in enhanced IAA production has yet to be determined and the responses controlled by Pgb2 might be influenced by its cellular localization. Phytoglobins lack specific organelle localization signals although independent reports demonstrate their presence in the cytosol and nucleus. The alfalfa Pgb protein, Mhpgb1, was localized mainly in the nucleus with only small traces detected in the cytoplasm (Seregélyes et al., 2000); an observation subsequently confirmed in Arabidopsis (Hebelstrup et al., 2008). Two mammalian hemoglobins, neuroglobin and cytoglobin, have also been detected in the nucleus (Geuens et al., 2003; Hundahl et al., 2010). Whether cellular localization i.e. cytoplasm versus nucleus, determines the Pgb response needs to be clarified.
To further understand the mode of action of Pgb2 during embryogenesis and to determine the implications of its cellular localization on the observed response, an inducible system expressing a fusion between Pgb2 and the steroid-binding domain of the rat glucocorticoid receptor (GR) was introduced in a Pgb2 mutant background. This system, in which the Pgb2-GR fusion protein remains in the cytoplasm but migrates into the nucleus upon exposure to dexamethasone (DEX), was used during Arabidopsis somatic embryogenesis to demonstrate that Pgb2 function occurs only when the protein is present in the nucleus. Retention of Pgb2 in the cytoplasm mimics pgb2 mutant phenotypes: it promotes the transcriptional activation of IAA biosynthetic genes, favors the accumulation of IAA within embryogenic tissue arising from the zygotic embryo explants, and enhances the formation of somatic embryos.
Materials and methods
Generation of the Pgb2 lines
The 35S:Pgb2-GR (A) and (B) lines were produced by introducing the Pgb2-GR fusion protein, driven by a 35S promoter, into pgb2 mutant plants. The lines were generated by subcloning Pgb2 from Arabidopsis cDNA and adding the attB1 and attB2 sites to the 5’ and 3’ ends removing the stop codon using the following primers: attb1-F adaptor, attb2-R adaptor, AtHB2-F attb1, and AtHB2-R no stop (Supplementary Table S1). By using BP clonase (Invitrogen), the fragment was then placed in the BP cloning vector pDONR Zeo to create pENTR Pgb2 zeo. The system was used to transfer the Pgb2 fragment into the R1R2 sites of R1R2 GR vector (Baud et al., 2007), using Gateway® LR Clonase® II Enzyme mix (Thermo Fisher Scientific). The resulting R1R2 GR Pgb2 vector was then employed to transform pgb2 plants containing a T-DNA insert disrupting Pgb2, (Hebelstrup et al., 2006) using GV3101 Agrobacterium tumefaciens via floral dipping (Clough and Bent, 1998). Selection was performed by germinating the seeds on 1/2 Murashige Skooge (MS) medium (Murashige and Skooge, 1962) containing 25 mg/L kanamycin. Plants were then screened to detect the presence of the construct using the 35S-F and Pgb2-R primers, as well as the presence of the T-DNA insert using the Pgb2-F and dSmp-R primers (see primers list in Supplementary Table S1). Plants from the T3 generation were utilized for this study.
Transient transformation of Arabidopsis seedlings
Transient expression of Arabidopsis seedlings was used to verify the targeting of Pgb2 into the nucleus following exposure to DEX. The YFP-Pgb2 fusion construct was cloned into the same vector R1R2 GR used for stable transformation. Generation of the YFP-Pgb2 fusion construct is briefly described below. The gBlock fragment SalI-YFP-linker-Pgb2-SalI (Supplementary Fig. S2), custom synthesized by Integrated DNA Technology, was digested with the SalI restriction enzyme. The fragment was subsequently ligated with a SalI-digested Gateway® pENTRTM1A Dual Selection Vector (Thermo Fisher Scientific) according to the manufacturer’s instructions. The orientation of the inserted cassette was verified using YFB-F and Pentr1A-R primers. The YFP-Pgb2 fusion construct was cloned into the R1R2 GR vector using the Gateway® LR Clonase® II Enzyme mix (Thermo Fisher Scientific) and then transformed into GV3101 A. tumefaciens cells. Transient transformation of Arabidopsis seedlings was conducted exactly as reported by Li et al. (2009). DEX at a concentration of 20 μM was applied 3 days prior to Agrobacterium cocultivation, which occurred seven days after germination. Pgb2 was visualized on leaf cells using confocal microscopy.
Generation of Arabidopsis somatic embryos
Arabidopsis seeds were sterilized using 70% ethanol, 0.5% Triton X100 for 15 minutes followed by 95% ethanol for 15 minutes. The seeds were then transferred on ½ strength MS medium, incubated at 4 °C in the dark for 2–3 days and then transferred to a growth cabinet at a temperature of 20–22 °C, with a 16 h light/8 h dark photoperiod. Plants were grown and green siliques were used for dissection of immature zygotic embryos.
Somatic embryogenesis was initiated according to Bassuner et al. (2007). Immature zygotic embryos were plated on induction medium containing 2,4-dichlorophenoxyacetic acid (2,4-D) for 14 days, followed by a transfer onto hormone-free development medium. Efficiency of somatic embryogenesis was estimated by counting the number of fully developed somatic embryos after 9 days on development medium. Each biological replicate consisted of one petri plate containing 20 zygotic explants and three biological replicates were used for each measurement.
Chemical treatments
The NO scavenger 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO) was applied as specified in Elhiti et al. (2013), by dispensing 10 μl of a 10 µM solution directly on the explants every other day in culture throughout the induction medium. DEX was dissolved in 20% ethanol and added to induction medium to a final concentration of 20 µM.
Analyses of transcript levels
Total RNA was extracted from day 7 and day 14 explants on induction medium using TRI Reagent (Invitrogen) followed by a treatment with DNase I (RNase-free, Promega). The RNA was then utilized for the synthesis of cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative real-time PCR was performed exactly as documented by Elhiti et al. (2013) using primers listed in Supplementary Table S1. The relative level of gene expression was analyzed by the 2−∆∆Ct method described by Livak and Schmittgen (2001), using UBQ10 (AT4G05320) as a reference (Czechowski et al., 2005; Hong et al., 2010).
Localization of IAA
Immunolocalization of endogenous IAA was performed exactly as described by Mira et al. (2016). Explants at day 7 on induction medium were pre-fixed in 4% aqueous 1-ethyl-3-(3-dimethyl-aminopropyl)-carbodiimide hydrochloride at 4 °C for 2 h, post-fixed in FAA (10% formalin, 5% acetic acid, and 50% ethanol) overnight at 4 °C, dehydrated in an ethanol series, and embedded in Paraplast Plus (Fisher). The tissue was then sectioned to a thickness of 10 µm and deparaffinised in xylene. The sections were incubated in blocking solution [1 x phosphate-buffered saline (PBS) solution pH 7, 0.1% Tween 20, 1.5% glycine, and 5% bovine serum albumin (BSA)] for 1 hour. 150 µl of monoclonal primary IAA-antibodies (1 mg/ml, Sigma) diluted 1:200 in 1x PBS containing 0.8% BSA were applied to the sections and incubated in a high humidity chamber for 4 hours at room temperature. The slides were washed first in 1x PBS containing 0.88 g/L NaCl, 0.1% Tween 20, and 0.8% BSA for 5 minutes and then in 1xPBS with 0.8% BSA for 5 minutes in order to remove any excess Tween 20. The slides were incubated in 200 µl of secondary antibody [anti-mouse IgG alkaline phosphatase conjugate (1 mg/ml), Promega, USA] overnight in a high humidity chamber, washed two times in 1x PBS containing 0.88 g/L NaCl, 0.1% Tween 20, and 0.8% BSA for 10 minutes and then incubated in water for 15 minutes to remove the excess secondary antibodies. Samples were stained using 250 µl Western Blue Stabilized Substrate for Alkaline Phosphatase (Promega) for 40 minutes.
RNA in situ hybridization
RNA in situ hybridization studies were performed according to Elhiti et al. (2010). The two full length YUC1 and YUC4 cDNAs were cloned into a pGEM-T Easy Vector System (Promega). The two cDNAs were subsequently amplified from the vector using T7 and SP6 primers and used for the preparation of digoxigenin (DIG)-labelled sense and antisense riboprobes, following the procedure outlined in the DIG Application Manual (Roche Diagnostics). Slide preparation, hybridization conditions, and color development were performed as documented previously (Elhiti et al., 2010).
Statistical analyses
The cDNA of at least three biological replicates was used for quantitative RT-PCR studies. Differences in relative transcript levels were analyzed by comparing the Least Squares (LS) Means of the interaction effect, with or without DEX treatment on genotype, using Tukey’s test. The SAS® 9.3 program (SAS Institute Inc., Cary, NC, USA) was used for all statistical analyses.
Results
Cellular localization of Pgb2 influences Arabidopsis somatic embryogenesis
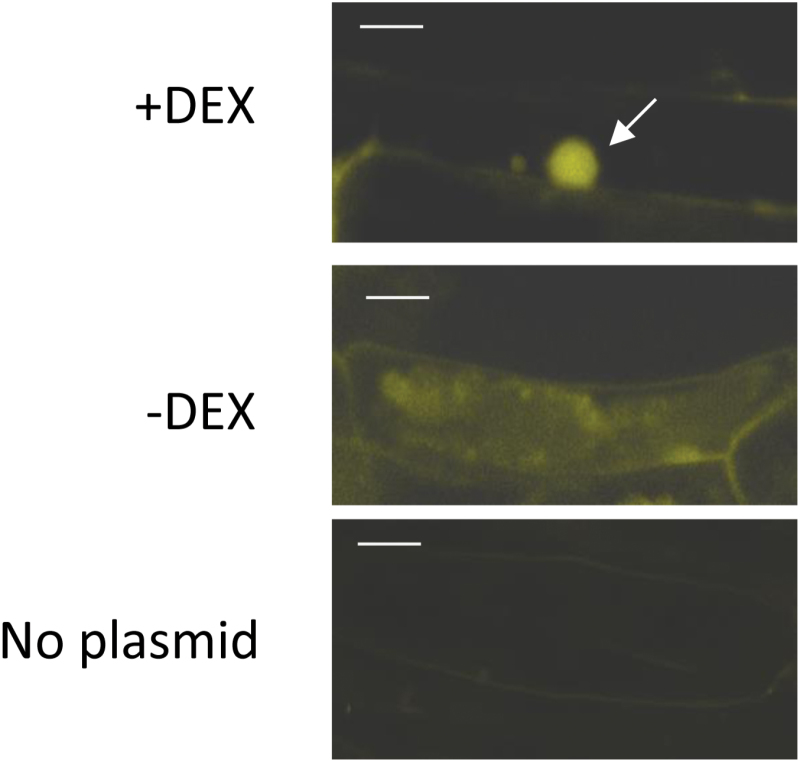
To evaluate how cellular localization of Pgb2 affects somatic embryogenesis we generated an Arabidopsis line expressing a fusion protein consisting of Pgb2 linked to the steroid-binding domain of the rat glucocorticoid receptor (GR) (Lloyd et al., 1994). The expressed PGB2-GR fusion protein remains in the cytoplasm but can be translocated into the nucleus when the steroid dexamethasone (DEX) is bound to the GR receptor site. The effectiveness of the system was tested in Arabidopsis seedlings using transient expression experiments in which the application of DEX was sufficient to translocate PGB2-YFP-GR into the nucleus (Fig. 1).
Fig. 1.
Transient transformation of Arabidopsis seedlings with 35S:Pgb2-GR-YFP. Addition of DEX targets Pgb2 to the nucleus (arrow), while in the absence of DEX Pgb2 is retained in the cytoplasm. Scale bars, 5 μm.
The 35S:Pgb2-GR construct was introduced in the pgb2 Arabidopsis mutant and two independent transformed lines, 35S:Pgb2-GR (A) and (B), were generated (Supplementary Fig. S3). The behavior of both lines during somatic embryogenesis was then compared to a line ectopically expressing Pgb2, 35S:Pgb2 line, and a line in which Pgb2 was mutated, pgb2 line. These lines have been characterized in previous studies (Hebelstrup et al., 2006; Elhiti et al., 2013).
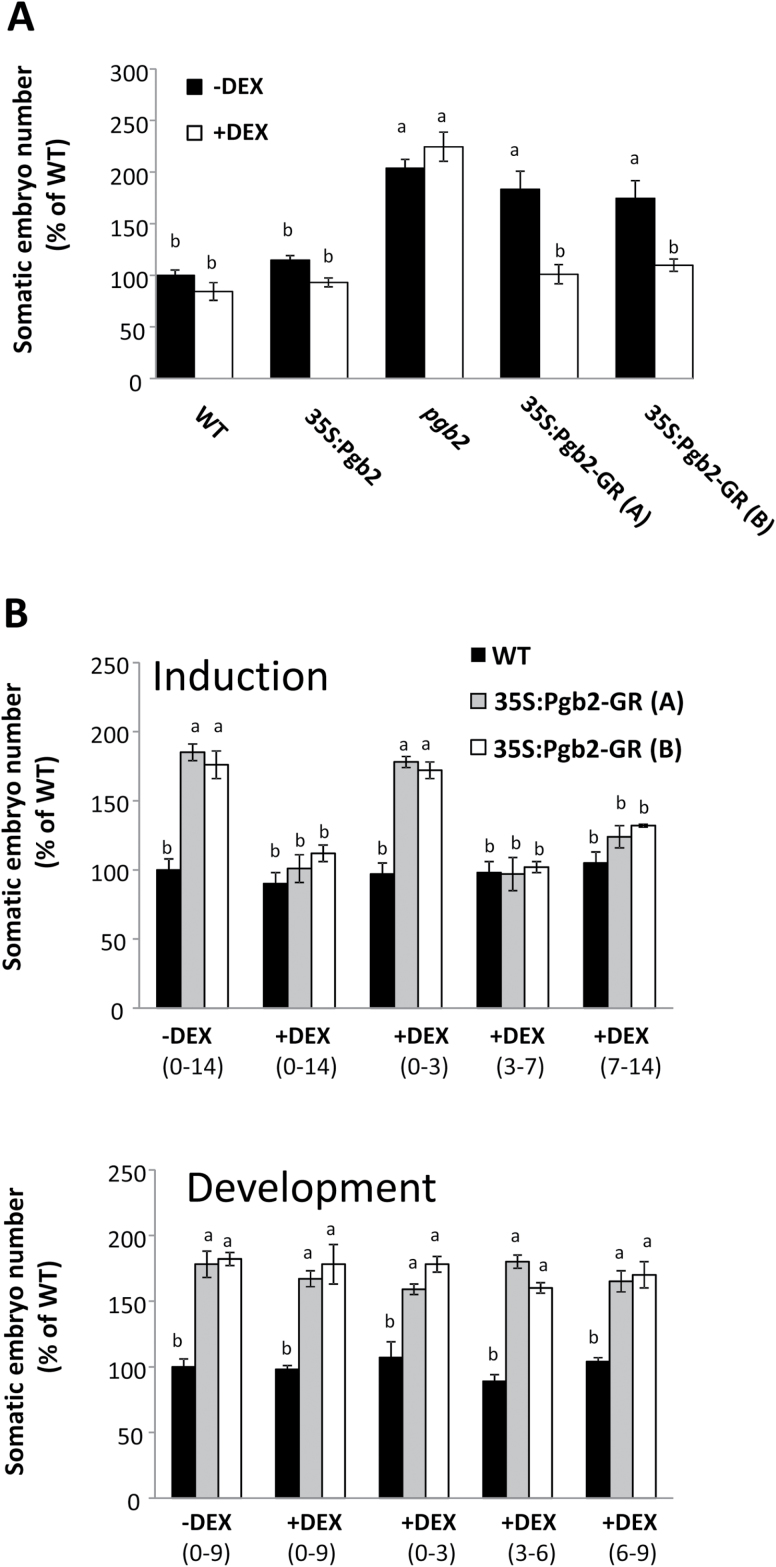
Relative to wild type, the number of embryos was not affected in the 35S:Pgb2 line while it increased in the pgb2 line, independently from DEX (Fig. 2A). An increase of similar magnitude was also observed in the 35S:Pgb2-GR (A) and (B) lines in the absence of DEX, while addition of DEX in the induction medium reduced embryo production to wild type values (Fig. 2A).
Fig. 2.
Effects of Pgb2 on somatic embryogenesis. (A) Number of Arabidopsis somatic embryos produced by the different lines. Values±SE are means of at least three biological replicates and are normalized to the value of wild type without DEX set at 100%. Letters on bars indicate statistically significant differences, P≤0.05. WT, wild type line; 35S:Pgb2, line ectopically expressing Pgb2; pgb2, line suppressing Pgb2; 35S:Pgb2-GR (A) and (B), lines in which Pgb2 tagged to the glucocorticoid receptor (GR) was overexpressed in a pgb2 background. (B) Applications of DEX at different days in culture during induction and development of the 35S:Pgb2-GR (A) and (B) lines. Values±SE are means of at least three biological replicates and are normalized to the value of wild type without DEX during days 0–14 for induction and wild type without DEX during days 0–9 for development set at 100%. Letters on bars indicate statistically significant differences, P≤0.05.
To estimate the temporal requirement of Pgb2 compartmentalization for somatic embryo formation, DEX was applied to the 35S:Pgb2-GR lines at different intervals during induction or development. In the wild type line applications of DEX at different days of induction had no significant effects on embryo yield (Fig. 2B). This was in contrast to the 35S:Pgb2-GR lines where inclusions of DEX during the first three days resulted in somatic embryo values similar to those measured in the absence of DEX. Applications of DEX after day 3 in the induction phase reduced embryo values to those of the wild type (Fig. 2B). Addition of DEX at any stage during embryo development had no effect on embryo number, producing embryo numbers similar to values in the absence of DEX.
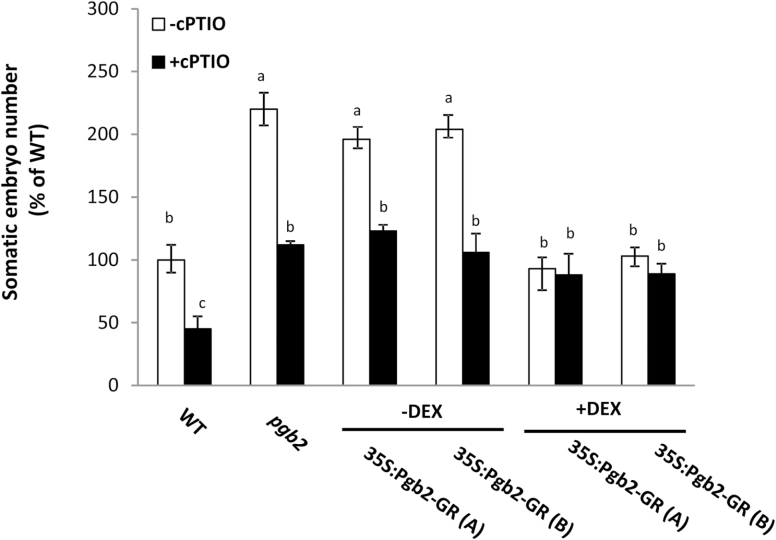
An established function of Pgbs is to scavenge nitric oxide (NO) (Hill, 2012). An experimental reduction of NO through pharmacological treatments reverses the effects of Pgb2 suppression on somatic embryogenesis (Elhiti et al., 2013). To evaluate if the observed influence of Pgb2 compartmentalization on embryo production is dependent on NO, the NO scavenger cPTIO was added to the induction medium. Consistent with previous studies (Elhiti et al., 2013), cPTIO reduced embryo production in both wild type and pgb2 lines (Fig. 3). A similar cPTIO repressive effect was also observed in the two 35S:Pgb2-GR (A) and (B) lines in the absence of DEX, but not in the same lines cultured with DEX (Fig. 3).
Fig. 3.
Number of somatic embryos produced by the same lines used in Fig. 2 in the presence or absence of the nitric oxide scavanger cPTIO added during the induction period. Values±SE are means of at least three biological replicates and are normalized to the value of wild type without cPTIO set at 100%. Letters on bars indicate statistically significant differences, P≤0.05.
Collectively, these results suggest that: 1) somatic embryo production is enhanced in the pgb2 mutant and this phenotype can be reversed by the re-introduction of Pgb2 in the nucleus but not in the cytoplasm; 2) the promotive effect on embryogenesis of suppression of Pgb2 or its restriction to the cytoplasm, occurs after day 3 on induction medium in conjunction with the formation of embryogenic tissue; and 3) this promotive effect can be attenuated by reducing the level of NO.
Transcription of MYC2 and genes of the indole acetic acid (IAA) biosynthetic pathway is affected by the cellular localization of Pgb2
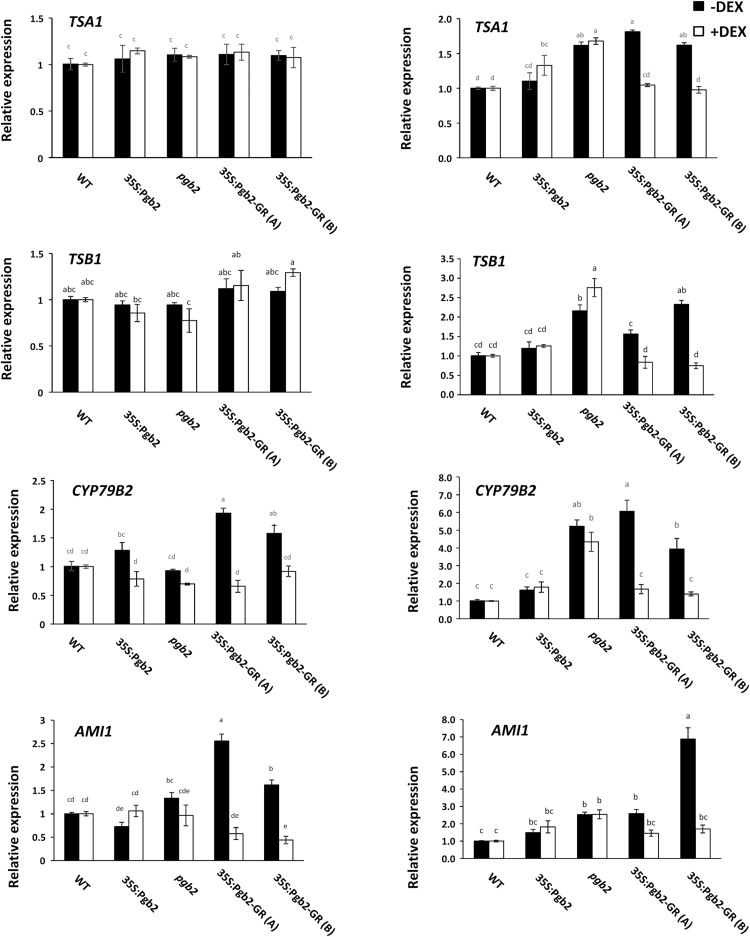
Auxin is the inductive signal required for the initiation of somatic embryogenesis (Raghavan, 2004). By suppressing MYC2, mutation of Pgb2 induces the IAA biosynthetic machinery and generates IAA maxima within the embryogenic tissue arising from the adaxial side of the cotyledons of the explants (Elhiti et al., 2013). To determine if these events are influenced by the cellular location of Pgb2, the transcript levels of MYC2 and several genes encoding auxin biosynthetic enzymes, which are upregulated by mutation of Pgb2 (Elhiti et al., 2013), were measured at day 7 and 14 on induction medium.
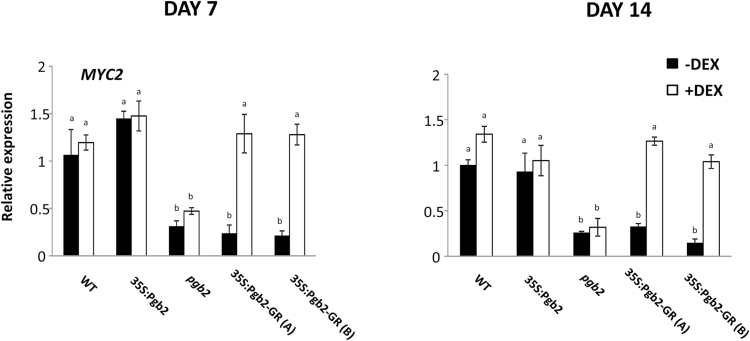
Relative to wild type, expression of MYC2 was reduced in the pgb2 mutant line and in the two inducible 35S:Pgb2-GR (A) and (B) lines cultured in the absence of DEX (Fig. 4). Applications of DEX in the inducible lines increased the transcript levels of MYC2. Genes involved in the last step of tryptophan synthesis, tryptophan synthase-α subunit (TSA1) and tryptophan synthase-β subunit (TSB1), were not affected by altered levels of Pgb2 at day 7. However, at the end of the induction period the expression of both genes increased in the pgb2 line independently from DEX, and in the 35S:Pgb2-GR (A) and (B) lines in the absence of DEX (Fig. 5).
Fig. 4.
Relative transcript levels of MYC2 at day 7 and 14 of induction. Values±SE are means of three biological replicates and are normalized to the value of wild type without DEX set at 1. Letters on bars indicate statistically significant differences, P≤0.05. Lines utilized: WT, wild type line; 35S:Pgb2, line ectopically expressing Pgb2; pgb2, line suppressing Pgb2; 35S:Pgb2-GR (A) and (B), lines in which Pgb2 tagged to the glucocorticoid receptor (GR) was overexpressed in a pgb2 background.
Fig. 5.
Relative transcript levels of genes participating in auxin biosynthesis at day 7 and 14 of induction. Values±SE are means of three biological replicates and are normalized to the respective wild type values, with or without DEX, set at 1. Letters on bars indicate statistically significant differences, P≤0.05. Genes measured included: tryptophan synthase-α subunit (TSA1), tryptophan synthase-β subunit (TSB1), cytochrome P450 CYP79B2 (CYP79B2) and amidase 1 (AMI1). Lines utilized: WT, wild type line; 35S:Pgb2, line ectopically expressing Pgb2; pgb2, line suppressing Pgb2; 35S:Pgb2-GR (A) and (B), lines in which Pgb2 tagged to the glucocorticoid receptor (GR) was overexpressed in a pgb2 background.
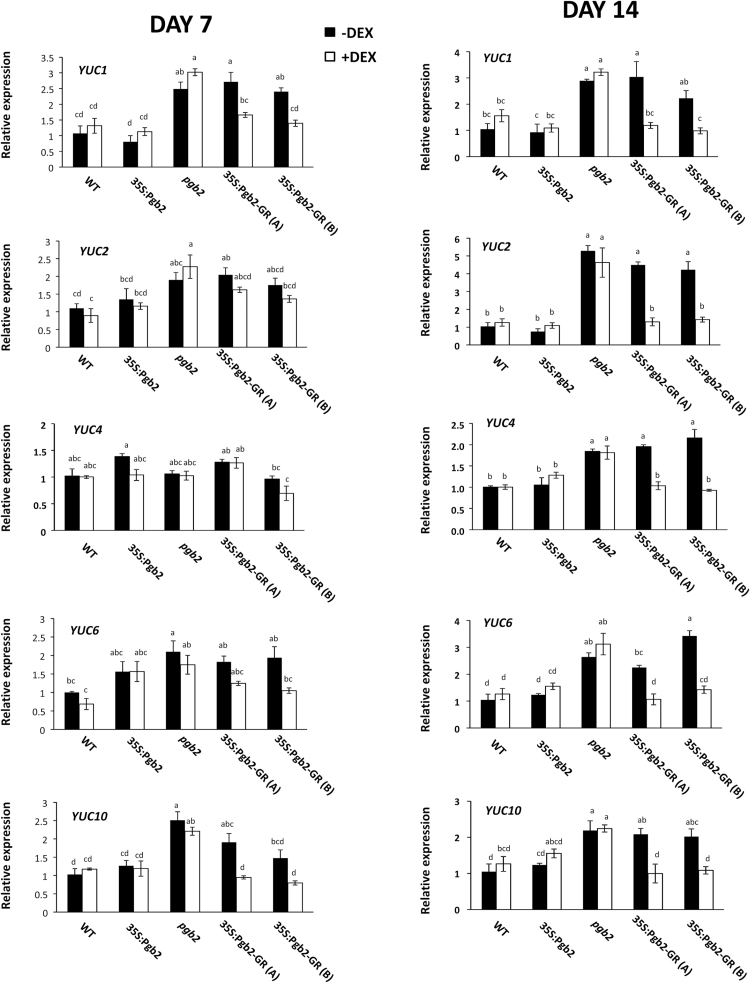
In Arabidopsis, conversion of tryptophan to IAA can occur through three distinct routes (Mashiguchi et al., 2011). The first is regulated by the sequential activity of cytochrome P450 CYP79B2 (CYP79B2) and amidase 1 (AMI1), the former producing indole-3-acetaldoxime from tryptophan and the latter converting indole-3-acetaldoxime to IAA. Conversion of tryptophan to IAA can also occur through a poorly characterized second route or through the YUCCA pathway involving members of the YUCCA family. Relative to wild type, the expression of both CYP79B2 and AMI1 increased in the two 35S:Pgb2-GR (A) and (B) lines cultured without DEX at both day 7 and 14 on induction medium (Fig. 5). In the same lines, application of DEX reduced the expression of both genes to wild type values. A DEX independent increase in CYP79B2 transcript levels was also observed in the pgb2 explants at the end of the induction period. The transcript levels of the most representative YUCCA genes operating during Arabidopsis embryogenesis, YUC1, 2, 4, 6, and 10, were also measured during the induction period. At day 7, with the exception of YUC4, all YUCs were induced in the pgb2 line and in the 35S:Pgb2-GR (A) and (B) lines cultured in the absence of DEX (Fig. 6). A similar profile was also observed for all YUCs at day 14 (Fig. 6). Previous studies showed that YUC1 and 4 can be used as markers for future somatic embryo initiation sites in explants (Bai et al., 2013). To assess if exclusion of Pgb2 from the nucleus increases the domains of these sites, the transcripts of both genes were localized in explants of the 35S:Pgb2-GR (A) and (B) lines cultured in the presence or absence of DEX. In media devoid of DEX the distribution of both YUC1 and 4 transcripts extended throughout the embryogenic tissue (arrows, Supplementary Fig. S4), while only a very faint signal was observed in the embryogenic tissue of the same lines cultured with DEX (Supplementary Fig. S4).
Fig. 6.
Relative transcript levels of YUCCA 1, 2, 4, 6 and 10 at day 7 and 14 of induction. Values±SE are means of three biological replicates and are normalized to the respective wild type values, with or without DEX, set at 1. Letters on bars indicate statistically significant differences, P≤0.05. Lines utilized: WT, wild type line; 35S:Pgb2, line ectopically expressing Pgb2; pgb2, line suppressing Pgb2; 35S:Pgb2-GR (A) and (B), lines in which Pgb2 tagged to the glucocorticoid receptor (GR) was overexpressed in a pgb2 background.
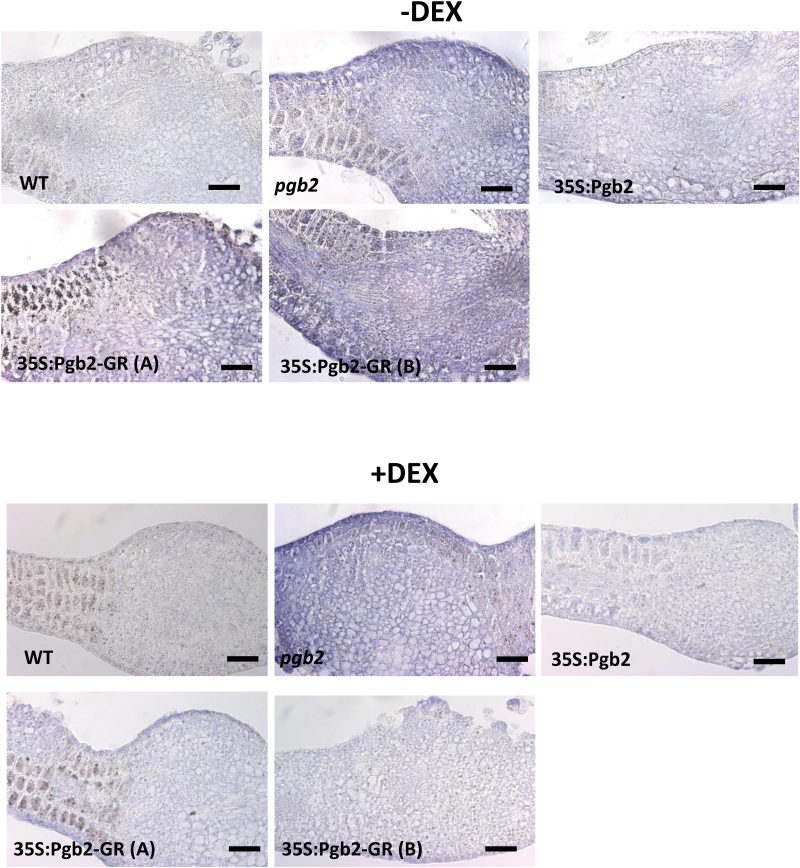
Suppression of Pgb2 or retention of Pgb2 in the cytoplasm increases IAA signal in the embryogenic tissue
Production of somatic embryos is preceded by a heavy accumulation of IAA within the embryogenic tissue arising from the cotyledons of the zygotic explants (Elhiti et al., 2013). Immunocalization studies in all Arabidopsis lines were conducted to detect IAA on the adaxial sides of the cotyledons of the zygotic embryo explants; these are the sites where embryogenic tissue forms. IAA accumulated along the cotyledons of the pgb2 explants (Fig. 7), an observation consistent with previous studies (Elhiti et al., 2013). The IAA signal was weak in wild type and 35S:Pgb2 explants independently from DEX, but intense in both 35S:Pgb2-GR (A) and (B) lines cultured in the absence of DEX. Inclusion of DEX in the same lines reduced the IAA signal (Fig. 7).
Fig. 7.
Immunolocalization of IAA at day 7 of induction in expanded cotyledons of explants of the wild type (WT) line, the 35S:Pgb2 line ectopically expressing Pgb2, the pgb2 line suppressing Pgb2, and the 35S:Pgb2-GR (A) and (B) lines in which Pgb2 tagged to the glucocorticoid receptor (GR) was overexpressed in a pgb2 background. Lines were cultured in the presence or absence of DEX. Scale bars, 75 μm.
Establishment of IAA maxima during the induction of Arabidopsis somatic embryogenesis is mediated by PIN1 (Su and Zhang, 2009), the expression of which is induced in highly embryogenic systems (Elhiti et al., 2013). Relative to wild type and 35S:Pgb2 explants, PIN1 transcript levels are induced at both day 7 and 14 in the pgb2 explants and in explants of the two 35S:Pgb2-GR (A) and (B) lines cultured without DEX (Supplementary Fig. S5).
Although the mode of action of auxin in the promotion of embryogenic tissue is debatable, accumulation of IAA increases the expression of genes directing the progression of somatic embryogenesis (Raghavan, 2004). The most representative example is somatic embryogenesis receptor kinase 1 (SERK1) (Supplementary Fig. S6), a gene sufficient to induce embryogenic competence in culture by encouraging the somatic-embryogenic cell fate transition (Hecht et al., 2001; Kwaaitaal and De Vries, 2007). Consistent with auxin, at the end of the induction period the relative levels of SERK1 transcripts are elevated in explants of the pgb2 line and in those of the 35S:Pgb2-GR lines cultured in the absence of DEX. Inclusion of DEX in the 35S:Pgb2-GR lines reduced the expression of SERK1 to levels comparable to the wild type and 35S:Pgb2 lines (Supplementary Fig. S6).
Discussion
Arabidopsis somatic embryogenesis consists of two distinct phases: an induction phase resulting in the formation of the embryogenic tissue from the adaxial side of the cotyledons of the zygotic embryo explants and an auxin-free development phase culminating with the formation of fully developed somatic embryos. It is during the induction phase that the explants start undergoing profound morphological changes, involving a cell expansion phase (days 0–3) followed by a proliferation phase (days 3–14) producing embryogenic tissue (Raghavan, 2004; Supplementary Fig. S1). These events are limited to the cotyledons of the explants as other regions of the cultured zygotic embryos do not contribute to the formation of the embryogenic tissue (Raghavan, 2004) and are tightly regulated by unknown auxin-mediated mechanisms. Besides promoting the somatic-embryogenic transition, auxin accumulates in regions of the embryogenic tissue that will give rise to somatic embryos (Su and Zhang, 2009). Genetic or pharmacological perturbations dissipating auxin accumulation inhibit the formation of the somatic embryos (Su and Zhang, 2009). The requirement of auxin for the production of somatic embryos is not restricted to Arabidopsis and is documented in a large number of species (Thorpe and Stasolla, 2001).
Auxin responses are tightly regulated by cellular NO (Pagnussat et al., 2003) and fluctuations in NO homeostasis have been implicated in many plant defense and stress response mechanisms (Delledonne et al., 1998; Mackerness et al., 2001; Gould et al., 2003), as well as morphogenic events (Leshem and Haramaty, 1996; Beligni and Lamattina, 2000; Seregelyes et al., 2003). Initially identified in the regulation of post-embryonic processes, such as root hair development, lateral and adventitious root formation, root gravitropic bending and root responses to iron deficiency (reviewed by Hill, 2012), the link between NO and auxin has been more recently extended to embryogenesis. During alfalfa somatic embryogenesis, NO is required in the induction phase where it promotes the auxin-mediated activation of cell division and embryogenic cell formation (Otvos et al., 2005). Similarly, a rise in NO levels triggered by mutation of Arabidopsis Pgb2, increases somatic embryogenesis through suppression of MYC2 (Elhiti et al., 2013). Suppression of MYC2, a repressor of IAA synthesis, induces the expression of several IAA biosynthetic enzymes and alters the pattern of IAA distribution, possibly by affecting the auxin transporter, PIN1 (Elhiti et al., 2013).
While the main role assigned to Pgbs is to scavenge NO produced as a result of using nitrite as an alternative electron acceptor, Igamberdiev and Hill (2004) did not exclude other physiological roles based on the presence of Pgbs in the nucleus, as demonstrated in alfalfa (Seregélyes et al., 2000) and Arabidopsis (Hebelstrup et al., 2008). These observations, also consistent with the localization patterns of the mamalian hemoglobins, cytoglobin and neuroglobin (Geuens et al., 2003; Hundahl et al., 2010), do not however prove whether the nuclear localization of Pgbs is required to produce an effect.
Using Arabidopsis somatic embryogenesis as a model system, we demonstrate that the Pgb2 response is triggered only when the protein is present in the nucleus. The enhanced embryo yield in the Pgb2 mutant line, pgb2 line, is phenocopied when Pgb2 is retained in the cytoplasm i.e. 35S:Pgb2-GR (A) and (B) lines cultured in the absence of DEX, and reverted to wild type values when Pgb2 is transported to the nucleus as a result of the binding of DEX to Pgb2-GR in the 35S:Pgb2-GR (A) and (B) lines (Fig. 2). This reversion is limited to a specific window of time of day, namely 3–14 on induction medium (Fig. 2B), which coincides with the formation of the embryogenic tissue (Supplementary Fig. S1). The level of NO influences the Pgb2 response (Fig. 3), in agreement with the well-established role of Pgbs as effective NO scavangers (Igamberdiev and Hill, 2004).
Formation of embryogenic tissue is closely linked to production of IAA (Raghavan, 2004; Su and Zhang, 2009), which is stimulated by suppression of MYC2 (Elhiti et al., 2013). The transcript levels of this gene are reduced by mutation of Pgb2 or when the protein is retained in the cytoplasm but are increased when Pgb2 is translocated into the nucleus (Fig. 4). Transcription of this gene influences the synthesis of IAA (Elhiti et al., 2013), which in Arabidopsis is mainly initiated by tryptophan though three distinct pathways (Mashiguchi et al., 2011). While the indole-3-acetaldoxime pathway requires the enzyme CYP79B, which converts tryptophan to indole-3-acetaldoxime, and AMI1 producing IAA from indole-3-acetaldoxime, the second pathway generates indole-3-acetaldoxime through unknown mechanisms. The third route of IAA synthesis is the YUCCA pathway that involves a variety of members of the YUCCA family (Mashiguchi et al., 2011). Confirming previous studies (Elhiti et al., 2013), suppression of Pgb2 enhances the expression of key tryptophan biosynthetic enzymes, such as TSA1 and TSB1, and this induction is only observed in those situations where Pgb2 is excluded from the nucleus i.e. pgb2 line and 35S:Pgb2-GR (A) and (B) lines cultured in the absence of DEX. A similar requirement was also observed for the induction of CYP79B2 and AMI1, regulating the indole-3-acetaldoxime pathway, as well as YUC1, 2, 4, 6, and 10, the major key players of the YUCCA pathway (Mashiguchi et al., 2011). The consistent induction of YUCs at both day 7and 14, triggered by mutation of Pgb2 or by its exclusion from the nucleus (Fig. 2A), is in agreement with the requirement of YUC genes for the formation of embryogenic tissue and the initiation of somatic embryogenesis (Bai et al., 2013). Consistent with this concept, staining of YUC1 and 4 transcripts, accurate markers of somatic embryo initiation sites (Bai et al., 2013), extended throughout the embryogenic tissue in situations when Pgb2 was excluded from the nucleus (Supplementary Fig. S4).
The transcriptional induction of tryptophan and IAA biosynthetic genes observed in the pgb2 line and 35S:Pgb2-GR (A) and (B) lines cultured in the absence of DEX coincides with the intense IAA staining pattern throughout the embryogenic tissue arising from the cotyledons of the zygotic explants (Fig. 7). We have previously demonstrated that this preferential accumulation of IAA in cotyledonary cells is also mediated by the auxin transporter PIN1, the expression of which is induced by mutation of Pgb2 (Elhiti et al., 2013) or in situations where Pgb2 is retained in the cytoplasm i.e. 35S:Pgb2-GR (A) and (B) lines in the absence of DEX (Supplementary Fig. S5).
A putative function for IAA accumulation is to activate the expression of genes necessary for the acquisition of embryogenic competence (Raghavan, 2004), including SERK1, the best characterized marker of embryogenic cells in several systems (Schmidt et al., 1997; Hecht et al., 2001; Somleva et al., 2007). The expression of this gene, sufficient to trigger the formation of apomictic embryos in vivo (Albertini et al., 2005) and somatic embryos in vitro (Hecht et al., 2001), follows a pattern similar to that of several IAA biosynthetic genes consistent with Pgb2 regulation (Supplementary Fig. S6).
In conclusion, this study demonstrates that the effects of Pgb2 on Arabidopsis somatic embryogenesis are influenced by its cellular localization. Retention of Pgb2 in the cytoplasm, where the protein retains its ability to scavenge NO (Supplementary Fig. S7), phenocopies the effects of Pgb2 mutation in inducing somatic embryogenesis through suppression of MYC2 and induction of IAA synthesis and accumulation. These effects are reversed when Pgb2 is translocated into the nucleus. Thus, we conclude that Pgb2 function during Arabidopsis somatic embryogenesis is exercised only when the protein is present in the nucleus and cannot exclude the possibility this might be a universal mechanism in dicotyledonous plants.
Supplementary Material
Acknowledgements
This work was supported by a NSERC Discovery Grant to CS and by a University of Manitoba Graduate Fellowship and a NSERC Graduate fellowship to CG. The authors thank Mr Durnin and Prof. Yeung for technical support and editing of the manuscript and Dr Kim Hebelstrup for providing the pgb2 and 35S:PGB2 Arabidopsis seeds.
Glossary
Abbreviations:
- 2,4-D
2,4-dichlorophenoxyacetic acid
- AMI1
amidase 1
- cPTIO
2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide
- CYP79B2
cytochrome P450 CYP79B2
- DEX
dexamethasone
- DIG
digoxigenin
- GR
glucocorticoid receptor
- IAA
indole-3-acetic acid
- LS
Least Squares
- MS
Murashige Skooge
- NO
nitric oxide
- Pgbs
phytoglobins
- Pgb2
phytoglobin 2
- SERK1
somatic embryogenesis receptor kinase1
- TSB1
tryptophan synthase-α subunit
- TSB2
tryptophan synthase-β subunit.
References
- Albertini E, Marconi G, Reale L, Barcaccia G, Porceddu A, Ferranti F, Falcinelli M. 2005. SERK and APOSTART. Candidate genes for apomixis in Poa pratensis. Plant Physiology 138, 2185–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai B, Su YH, Yuan J, Zhang XS. 2013. Induction of somatic embryos in Arabidopsis requires local YUCCA expression mediated by the down-regulation of ethylene biosynthesis. Molecular Plant 6, 1247–1260. [DOI] [PubMed] [Google Scholar]
- Bassuner BM, Lam R, Lukowitz W, Yeung EC. 2007. Auxin and root initiation in somatic embryos of Arabidopsis. Plant Cell Reports 26, 1–11. [DOI] [PubMed] [Google Scholar]
- Baud S, Mendoza MS, To A, Harscoët E, Lepiniec L, Dubreucq B. 2007. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. The Plant Journal 50, 825–838. [DOI] [PubMed] [Google Scholar]
- Beligni MV, Lamattina L. 2000. Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta 210, 215–221. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne M, Xia Y, Dixon RA, Lamb C. 1998. Nitric oxide functions as a signal in plant disease resistance. Nature 394, 585–588. [DOI] [PubMed] [Google Scholar]
- Dordas C. 2009. Nonsymbiotic hemoglobins and stress tolerance in plants. Plant Science 176, 433–440. [DOI] [PubMed] [Google Scholar]
- Dordas C, Hasinoff BB, Igamberdiev AU, Manac’h N, Rivoal J, Hill RD. 2003. Expression of a stress-induced hemoglobin affects NO levels produced by alfalfa root cultures under hypoxic stress. The Plant Journal 35, 763–770. [DOI] [PubMed] [Google Scholar]
- Dordas C, Hasinoff BB, Rivoal J, Hill RD. 2004. Class-1 hemoglobins, nitrate and NO levels in anoxic maize cell-suspension cultures. Planta 219, 66–72. [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. 2006. Gateway-compatible vectors for plant functional genomics and proteomics. The Plant Journal 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Elhiti M, Hebelstrup KH, Wang A, Li C, Cui Y, Hill RD, Stasolla C. 2013. Function of type-2 Arabidopsis hemoglobin in the auxin-mediated formation of embryogenic cells during morphogenesis. The Plant Journal 74, 946–958. [DOI] [PubMed] [Google Scholar]
- Elhiti M, Tahir M, Gulden RH, Khamiss K, Stasolla C. 2010. Modulation of embryo-forming capacity in culture through the expression of Brassica genes involved in the regulation of the shoot apical meristem. Journal of Experimental Botany 61, 4069–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrocho-Villegas V, Gopalasubramaniam SK, Arredondo-Peter R. 2007. Plant hemoglobins: what we know six decades after their discovery. Gene 398, 78–85. [DOI] [PubMed] [Google Scholar]
- Geuens E, Brouns I, Flamez D, Dewilde S, Timmermans JP, Moens L. 2003. A globin in the nucleus! The Journal of Biological Chemistry 278, 30417–30420. [DOI] [PubMed] [Google Scholar]
- Gould KS, Lamotte O, Klinguer A, Pugin A, Wendehenne D. 2003. Nitric oxide production in tobacco leaf cells: a generalized stress response? Plant Cell and Environment 26, 1851–1862. [Google Scholar]
- Hebelstrup KH, Hunt P, Dennis ES, Jensen SB, Jensen EØ. 2006. Hemoglobin is essential for normal growth of Arabidopsis organs. Physiologia Plantarum 127, 157–166. [Google Scholar]
- Hebelstrup KH, Jensen EO. 2008. Expression of NO scavenging hemoglobin is involved in the timing of bolting in Arabidopsis thaliana. Planta 227, 917–927. [DOI] [PubMed] [Google Scholar]
- Hebelstrup KH, Østergaard-Jensen E, Hill RD. 2008. Bioimaging techniques for subcellular localization of plant hemoglobins and measurement of hemoglobin-dependent nitric oxide scavenging in planta. Methods in Enzymology 437, 595–604. [DOI] [PubMed] [Google Scholar]
- Hebelstrup KH, van Zanten M, Mandon J, Voesenek LA, Harren FJ, Cristescu SM, Møller IM, Mur LA. 2012. Haemoglobin modulates NO emission and hyponasty under hypoxia-related stress in Arabidopsis thaliana. Journal of Experimental Botany 63, 5581–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V, Vielle-Calzada JP, Hartog MV, Schmidt ED, Boutilier K, Grossniklaus U, De Vries SC. 2001. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiology 127, 803–816. [PMC free article] [PubMed] [Google Scholar]
- Hendriks T, Scheer I, Quillet MC, Randoux B, Delbreil B, Vasseur J, Hilbert JL. 1998. A nonsymbiotic hemoglobin gene is expressed during somatic embryogenesis in Cichorium. Biochimica et Biophysica Acta 1443, 193–197. [DOI] [PubMed] [Google Scholar]
- Hill R, Hargrove M, Arredondo-Peter R. 2016. Phytoglobin: a novel nomenclature for plant globins accepted by the globin community at the 2014 XVIII conference on oxygen-binding and sensing proteins. F1000Research 5, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RD. 2012. Non-symbiotic haemoglobins—What’s happening beyond nitric oxide scavenging? AoB Plants 2012, pls004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SM, Bahn SC, Lyu A, Jung HS, Ahn JH. 2010. Identification and testing of superior reference genes for a starting pool of transcript normalization in Arabidopsis. Plant & Cell Physiology 51, 1694–1706. [DOI] [PubMed] [Google Scholar]
- Hoy JA, Hargrove MS. 2008. The structure and function of plant hemoglobins. Plant Physiology and Biochemistry 46, 371–379. [DOI] [PubMed] [Google Scholar]
- Hundahl CA, Allen GC, Hannibal J, Kjaer K, Rehfeld JF, Dewilde S, Nyengaard JR, Kelsen J, Hay-Schmidt A. 2010. Anatomical characterization of cytoglobin and neuroglobin mRNA and protein expression in the mouse brain. Brain Research 1331, 58–73. [DOI] [PubMed] [Google Scholar]
- Hunt PW, Klok EJ, Trevaskis B, Watts RA, Ellis MH, Peacock WJ, Dennis ES. 2002. Increased level of hemoglobin 1 enhances survival of hypoxic stress and promotes early growth in Arabidopsis thaliana. Proceedings of the National Academy of Science, USA 99, 17197–17202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt PW, Watts RA, Trevaskis B, Llewelyn DJ, Burnell J, Dennis ES, Peacock WJ. 2001. Expression and evolution of functionally distinct haemoglobin genes in plants. Plant Molecular Biology 47, 677–692. [DOI] [PubMed] [Google Scholar]
- Igamberdiev AU, Hill RD. 2004. Nitrate, NO and haemoglobin in plant adaptation to hypoxia: an alternative to classic fermentation pathways. Journal of Experimental Botany 55, 2473–2482. [DOI] [PubMed] [Google Scholar]
- Jenik PD, Gillmor CS, Lukowitz W. 2007. Embryonic patterning in Arabidopsis thaliana. Annual Review of Cell and Developmental Biology 23, 207–236. [DOI] [PubMed] [Google Scholar]
- Karami O, Aghavaisi B, Mahmoudi Pour A. 2009. Molecular aspects of somatic-to-embryogenic transition in plants. Journal of Chemical Biology 2, 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaaitaal MA, de Vries SC. 2007. The SERK1 gene is expressed in procambium and immature vascular cells. Journal of Experimental Botany 58, 2887–2896. [DOI] [PubMed] [Google Scholar]
- Leshem YY, Haramaty E. 1996. The characterization and contrasting effects of the nitric oxide free radical in vegetative stress and senescence of Pisum sativum Linn. Foliage. Journal of Plant Physiology 148, 258–263. [Google Scholar]
- Li JF, Park E, von Arnim AG, Nebenführ A. 2009. The FAST technique: a simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 5, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lloyd AM, Schena M, Walbot V, Davis RW. 1994. Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid-inducible regulator. Science 266, 436–439. [DOI] [PubMed] [Google Scholar]
- A-H-Mackerness S, John CF, Jordan B, Thomas B. 2001. Early signaling components in ultraviolet-B responses: distinct roles for different reactive oxygen species and nitric oxide. FEBS Letters 489, 237–242. [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, et al. 2011. The main auxin biosynthesis pathway in Arabidopsis. Proceedings of the National Academy of Science, USA 108, 18512–18517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira MM, Wally OSD, Elhiti M, El-Shanshory A, Reddy DS, Hill RD, Stasolla C. 2016. Jasmonic acid is a downstream component in the modulation of somatic embryogenesis by Arabidopsis Class 2 phytoglobin. Journal of Experimental Botany. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordhorst AP, Hartog MV, El Tamer MK, Laux T, de Vries SC. 2002. Somatic embryogenesis from Arabidopsis shoot apical meristem mutants. Planta 214, 829–836. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skooge F. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497. [Google Scholar]
- Otvös K, Pasternak TP, Miskolczi P, Domoki M, Dorjgotov D, Szucs A, Bottka S, Dudits D, Fehér A. 2005. Nitric oxide is required for, and promotes auxin-mediated activation of, cell division and embryogenic cell formation but does not influence cell cycle progression in alfalfa cell cultures. The Plant Journal 43, 849–860. [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Lanteri ML, Lamattina L. 2003. Nitric oxide and cyclic GMP are messengers in the indole acetic acid-induced adventitious rooting process. Plant Physiology 132, 1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazzolli M, Dominici P, Romero-Puertas MC, Zago E, Zeier J, Sonoda M, Lamb C, Delledonne M. 2004. Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity. The Plant Cell 16, 2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan V. 2004. Role of 2,4-dichlorophenoxyacetic acid (2,4-D) in somatic embryogenesis on cultured zygotic embryos of Arabidopsis: cell expansion, cell cycling, and morphogenesis during continuous exposure of embryos to 2,4-D. American Journal of Botany 91, 1743–1756. [DOI] [PubMed] [Google Scholar]
- Schmidt ED, Guzzo F, Toonen MA, De Vries SC. 1997. A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development 124, 2049–2062. [DOI] [PubMed] [Google Scholar]
- Seregelyes C, Barna B, Hennig J, Konopka D, Pasternak TP, Lukacs N, Feher A, Horvath GV, Dudits D. 2003. Phytoglobins can interfere with nitric oxide functions during plant growth and pathogenic responses: a transgenic approach. Plant Science 165, 541–550. [Google Scholar]
- Seregélyes C, Mustárdy L, Ayaydin F, et al. 2000. Nuclear localization of a hypoxia-inducible novel non-symbiotic hemoglobin in cultured alfalfa cells. FEBS Letters 482, 125–130. [DOI] [PubMed] [Google Scholar]
- Smagghe BJ, Hoy JA, Percifield R, et al. 2009. Review: correlations between oxygen affinity and sequence classifications of plant hemoglobins. Biopolymers 91, 1083–1096. [DOI] [PubMed] [Google Scholar]
- Somleva NM, Schmidt LED, de Vries CS. 2007. Embryogenic cells in Dactylis glomerata L. (Poaceae) explants identified by cell tracking and by SERK expression. Plant Cell Reports 19, 718–726. [DOI] [PubMed] [Google Scholar]
- Stasolla C, Yeung EC. 2003. Recent advances in conifer somatic embryogenesis: improving somatic embryo quality. Plant Cell Tissue and Organ Culture 74, 15–35. [Google Scholar]
- Su YH, Zhang XS. 2009. Auxin gradients trigger de novo formation of stem cells during somatic embryogenesis. Plant Signaling & Behavior 4, 574–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe TA, Stasolla C. 2001. Somatic embryogenesis. In: Bhojwani SS, Soh WY, eds. Current trends in the embryology of angiosperms. Dordrecht: Kluwer Academic Publishers, 279–336. [Google Scholar]
- Wang Y, Elhiti M, Hebelstrup KH, Hill RD, Stasolla C. 2011. Manipulation of hemoglobin expression affects Arabidopsis shoot organogenesis. Plant Physiology and Biochemistry 49, 1108–1116. [DOI] [PubMed] [Google Scholar]
- Wang YH, Kochian LV, Doyle JJ, Garvin DF. 2003. Two tomato non-symbiotic haemoglobin genes are differentially expressed in response to diverse changes in mineral nutrient status. Plant Cell and Environment 26, 673–680. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.