Highlight
Mitochondrial AtTrxo1 has a role in redox homeostasis during seed germination under salt conditions, and it could act as a possible sensor of saline stress and an inducer of H2O2 accumulation, independently of other ROS parameters
Keywords: Arabidopsis thaliana, AtTrxo gene family, AtTrxo1 gene expression, germination, ROS homeostasis, saline conditions, transcriptional regulation
Abstract
Mitochondrial thioredoxin-o (AtTrxo1) was characterized and its expression examined in different organs of Arabidopsis thaliana. AtTrxo1 transcript levels were particularly high in dry seeds and cotyledons where they reached a maximum 36 h after imbibition with water, coinciding with 50% germination. Expression was lower in seeds germinating in 100 mM NaCl. To gain insight into the transcriptional regulation of the AtTrxo1 gene, a phylogenomic analysis was coupled with the screening of an arrayed library of Arabidopsis transcription factors in yeast. The basic leucine zipper AtbZIP9 and the zinc finger protein AZF2 were identified as putative transcriptional regulators. Transcript regulation of AtbZIP9 and AtAFZ2 during germination was compatible with the proposed role in transcriptional regulation of AtTrxo1. Transient over-expression of AtbZIP9 and AtAZF2 in Nicotiana benthamiana leaves demonstrated an activation effect of AtbZIP9 and a repressor effect of AtAZF2 on AtTrxo1 promoter-driven reporter expression. Although moderate concentrations of salt delayed germination in Arabidopsis wild-type seeds, those of two different AtTrxo1 knock-out mutants germinated faster and accumulated higher H2O2 levels than the wild-type. All these data indicate that AtTrxo1 has a role in redox homeostasis during seed germination under salt conditions.
Introduction
Plant cells generate reactive oxygen and nitrogen species (ROS and RNS, respectively) during plant development, including maturation and germination of seeds, and they can act as signaling molecules (El-Maarouf-Bouteau and Bailly, 2008; Sanz et al., 2015). The germination of seeds proceeds in two different steps: (i) germination sensu stricto, spanning from the start of water uptake to radicle emergence; and (ii) reserve mobilization, which is considered a post-germination process (Bewley, 1997; Nonogaki, 2014). In Arabidopsis thaliana, germination sensu stricto involves rupturing of the testa and breakage of the micropylar endosperm, which occurs mainly due to the weakening of the endosperm cell walls by mannanases and other hydrolytic enzymes (Iglesias-Fernández et al., 2011, 2013; González-Calle et al., 2015). The emergence of the radicle marks the onset of post-germination events, and growth is supported by the hydrolysis of reserve compounds (proteins, lipids, carbohydrates) until the seedling becomes fully photosynthetic (Vicente-Carbajosa and Carbonero, 2005; González-Calle et al., 2014; Iglesias-Fernández et al., 2014).
To avoid the harmful effects of ROS/RNS, plants have evolved a range of redundant and elaborate mechanisms that involve metabolites and enzymes under oxidative and nitrosative stress responding to developmental and environmental cues (Mittler et al., 2011; Noctor and Mhamdi, 2014; Sevilla et al., 2015). Thioredoxins (Trxs) are ubiquitous small proteins (around 12 kDa) with oxidoreductase activity, containing two cysteines in the redox-active center that regulate the function of target proteins. Trxs are a key factor in maintaining protein dithiol/disulphide homeostasis, which modulates redox signaling during development and stress adaptation (Meyer et al., 2012; Lázaro et al., 2013). In plants, there are at least 10 families of Trxs, with more than 40 members (Meyer et al., 2012; Traverso et al., 2013), and the presence of at least 29 Trx genes has been reported in the Arabidopsis genome (Meyer et al., 2009; Belin et al., 2015). Trxs are present in almost all cellular compartments, including chloroplasts, mitochondria, apoplast, cytosol and nuclei (Meyer et al., 2012; Traverso et al., 2013). To date, the best-known mitochondrial Trxs are of the o-type, but the pea Trxo1 has been also found in the nucleus (Martí et al., 2009). Pea mitochondrial PsTrxo1 has been reported to be involved in the response mechanism against salt stress in pea leaves, in addition to mitochondrial PrxIIF and other antioxidant enzymes (Martí et al., 2011). Mitochondrial Trxo1 may function in the reductive activation of citrate synthase, alternative oxidase (AOX), and PrxIIF (Umbach and Siedow, 1993; Martí et al., 2011) thereby favouring flux through the Krebs cycle and the respiratory chain over fermentative processes (Rhoads et al., 1998; Florez-Sarasa et al., 2014). Interestingly, a recent study by Daloso et al. (2015) has revealed that mitochondrial enzymes of the TCA cycle are redox regulated, since in the mutant AtTrxo1 the enzymatic activities of ATP-cytrate lyase (ACL) and succinyl-CoA ligase (SCoAL) are drastically decreased, suggesting that Trxo1 promotes differential redox regulation of these enzymes.
In plant mitochondria, the presence of thioredoxin-dependent peroxirredoxins (Prxs) and sulfiredoxins (Srxs) has also been described (Dietz et al., 2006; Barranco-Medina et al., 2007, 2008). In this organelle, Trxs coupled with Prxs scavenge H2O2 (Konig et al., 2002; Barranco-Medina et al., 2007), and Srx (which is a small thiol reductase) catalyzes the retro-reduction of hyperoxidized (sulfinic) Prx in an ATP-dependent manner, similar to that proposed for other Srxs (Rey et al., 2007; Iglesias-Baena et al., 2010, 2011). In the past decade, our understanding of the chloroplast and cytosol Trx systems has grown significantly. Trxh isoforms constitute the largest group in the Trx family in Arabidopsis and extensive work on its function has implicated it in the seed germination process (Montrichard et al., 2003; Serrato and Cejudo, 2003; Pulido et al., 2009). In contrast, relatively few studies have addressed the possible involvement of the plant Trxo system in germination and no data are available about its transcriptional regulation. Transcriptional regulation of gene expression is driven by short DNA sequences (cis-elements) in gene promoters and by transcription factors (TFs), proteins that interact with them. Comparison of the promoter sequences of orthologous genes facilitates the finding of these cis-elements, which are conserved through evolution (‘phylogenomics’), and these are used as baits for the screening of an arrayed library of Arabidopsis TFs (Y1H, yeast one-hybrid assays; Castrillo et al., 2011).
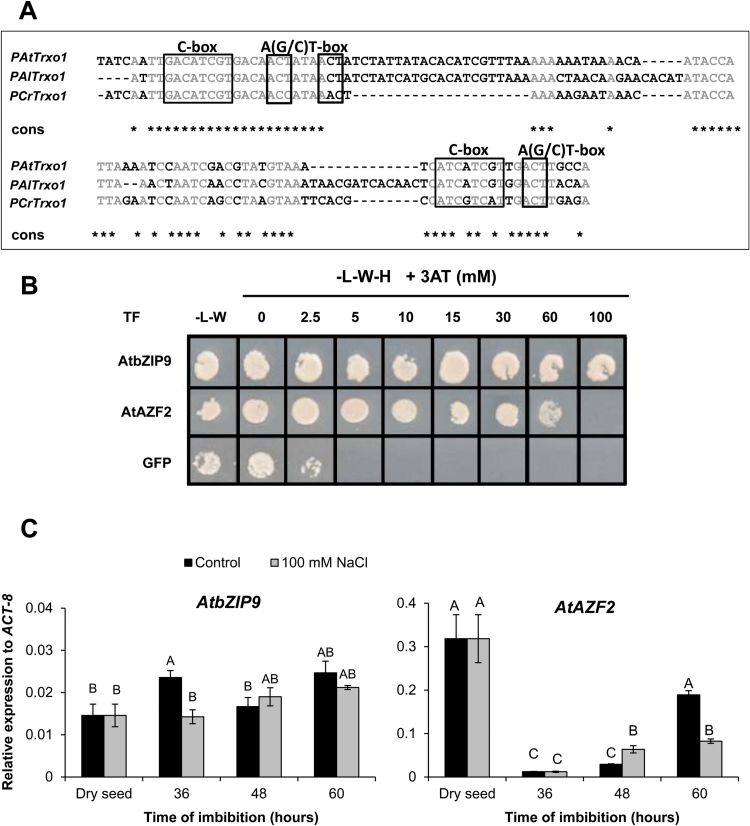
In this study, the molecular characterization of the gene AtTrxo1 from Arabidopsis has been carried out, with special emphasis on its transcriptional regulation and in its role in seed germination. For that purpose, a bioinformatic search for its putative orthologous genes and corresponding promoters within the Brassicaceae family has been done and a conserved promoter cis-element has been used as the bait to look for interacting TFs in a yeast library of circa 1200 ORF TFs from Arabidopsis (Castrillo et al., 2011). Among the different interacting TF proteins, a basic-leucine zipper AtbZIP9 and a zinc finger protein AZF2 have been identified as possible transcriptional regulators of the AtTrxo1 gene. To explore the potential physiological role of this Trxo1, we have carried out a comparison of the wild-type and two AtTrxo1 knock-out mutants, and also examined the effect of lacking Trxo1 on plant development under saline (100 mM NaCl) conditions, including specifically a deeper study of the germination process. A comparison of the germination kinetics of the wild-type and the knock-out mutants in 100 mM NaCl demonstrated that the mutants germinate faster than the wild-type seeds under these stress conditions.
Materials and methods
Plant material, growth conditions, and germination assays
Seeds of Arabidopsis thaliana ecotype Columbia (Col-0; the wild-type, WT) and two different T-DNA insertion mutants [knock-out (KO) AtTrxo1: SALK_143294C and SALK_042792] were obtained from the European Arabidopsis Stock Centre (NASC, http://Arabidopsis.info/). The homozygous plants for these T-DNA insertions were selected by PCR using gene-specific primers and a primer derived from the left border (LBb1.3) of the T-DNA (http://signal.salk.edu/tdnaprimers.2.html; see Supplementary Table S1 available at JXB online).
Plants (one per pot) were grown in substrate containing perlite:peat soil (1:3, v:v) under controlled conditions of light (150 µmol m−2 s−1 PAR), photopheriod (16/8 h light/dark), relative humidity (60%), and temperature (23/18 °C light/dark). Study of the response of the WT and KO mutant plants to salt stress was carried on 7-d-old plants exposed to water (control) and 100 mM NaCl twice a week during plant development. Plants were harvested after 28 and 42 d of growth.
Arabidopsis seeds were surface-sterilized and germinated in 0.5× Murashige and Skoog solidified medium including vitamins (Duchefa-Biochemie, Haarlem, The Netherlands) essentially as described in Iglesias-Fernández et al. (2014). Seeds (WT and KO mutants) were after-ripened at 21 °C and 30% relative humidity for 1 month before germination assays were performed.
Three replicates of 100 after-ripened seeds were imbibed in 90-mm Petri dishes on Whatman No.1 filter paper moistened with 3 ml of 100 mM NaCl (controls in 3 ml H2O). This NaCl concentration was chosen from a range of 10, 50, 75, 100, and 150 mM because it allowed the complete germination of seeds of WT and KO mutants while presenting different germination kinetics (Supplementary Fig. S2). Germination was under long-day conditions (16/8 h light/dark; light intensity of 150 µmol m−2 s−1 PAR). Seeds were not surface-sterilized in order to avoid influencing their dormancy status, and were considered germinated when radicle protrusion was visible under a magnifying lens. Germination tests were performed four times.
Generation of transgenic lines and histochemical β-glucuronidase (GUS) assays
The transgenic reporter lines PAtTrxo1::uidA were produced by fusing 1011 bp of the AtTrxo1 promoter (amplified from Arabidopsis genomic DNA by nested PCR using oligonucleotide pairs as shown in Supplementary Table S1) to the uidA reporter gene. The promoter fragment was cloned into the pDONR221 vector by the Gateway® BP recombination and then transferred by Gateway LR® recombination (Invitrogen, http://www.invitrogen.com) into the destination vector pMDC163. This construct was introduced into Agrobacterium tumefaciens strain C58C1 GV3101 by electroporation and then used to transform Arabidopsis (Col-0) by the floral dip method (Clough and Bent, 1998).
Qualitative GUS staining assays were performed as described by Jefferson et al. (1987) and Stangeland and Salehian (2002), and were visualized under a magnifying lens (Leica, Wetzlar, Germany).
Bioinformatic tools
The sequences from five different Brassicaceae (A. thaliana, A. lyrata, Brassica rapa, Capsella rubella, and Eutrema salsugineum) and two Leguminosae (Phaseolus vulgaris and Glycine max) species were obtained from the Phytozome v8.0 Database (http://www.phytozome.net;Goodstein et al., 2012). The sequence of the pea PsTrxo1 has been described in Martí et al. (2009). The deduced amino acid sequences of the 13 Trxo genes were used to construct a phylogenetic dendrogram. The alignment of these sequences was carried out with the CLUSTALW programme (Thompson et al., 1994) prior to the phylogenetic analysis, which was done by the neighbour-joining method with the MEGA 4.0 software (Tamura et al., 2007), using a bootstrap analysis with 1000 replicates, complete deletion, and the Jones–Taylor–Thornton matrix as settings. The conserved motifs within the deduced protein sequences of the 13 Trxo were identified with the MEME program (http://meme-suite.org/tools/meme;Bailey et al., 2009). Mitochondrial signal peptides and their cleavage sites were predicted using MitoProt (ExPASy tools, http://www.expasy.org/tools). The promoter sequences of Trxo1 from A. thaliana, A. lyrata, and C. Rubella were used to create pair-wise alignments (phylogenomics) using the software mVISTA Shuffle-LAGAN (http://genome.lbl.gov/vista/mvista/submit.shtml;Frazer et al., 2004) and T-Coffee (http://www.ebi.ac.uk/Tools/msa/tcoffee;Notredame et al., 2000).
Yeast one-hybrid (Y1H) assays
Yeast one-hybrid screenings were performed essentially as described by Castrillo et al. (2011). The AtTrxo1-B2-element was amplified by PCR using specific primers that contained XmaI and XbaI restriction sites (Supplementary Table S1), and this PCR product was cloned into the pTUY1H plasmid upstream of a HIS3 reporter gene to be used to transform Saccharomyces cerevisiae Y187α (MAT-α) cells. Positive colonies were visible after 2–5 d of incubation at 28 °C in a selection medium lacking leucine (L), tryptophan (W), and histidine (H) under increasing concentrations of the inhibitor 3-AT (3-amino-1,2,4- triazole; Sigma, St Louis, MO, USA).
Transient trans-activation assays
A set of three different constructs, derived from the promoter of AtTrxo1, were fused to the reporter uidA gene (GUS) and used to transform A. tumefaciens (strain C58C1 GV3101) for transient expression assays with or without the effector construct P35S::AtbZIP9, P35S::AtAZF2 and these were used to infiltrate Nicotiana benthamiana leaves. In addition, the leaves were also infiltrated with P35S::LUC (luciferase) for normalization and with P35S::P19 to avoid silencing (Jefferson et al., 1987; Voinnet et al., 2003). Relative GUS/LUC activities were determined by fluorescence and luminescence using a Genios Pro 96/384 multifunction microreader (TECAN®; Tecan Group, Männedorf, Switzerland). Three independent transformation experiments were done for each construct.
Real-time quantitative PCR assays
Total RNA was purified from different organs of Arabidopsis, including seeds at several time-points during germination (Oñate-Sánchez and Vicente-Carbajosa, 2008), and used to synthesized cDNA from 1-μg RNA samples (RT-PCR Kit from Roche Applied Science, Mannheim, Germany). The specific primers for the RT-qPCR analyses are shown in Supplementary Table S1 and the expression of the Actin 8 (ACT-8, At1g49240) gene was used to normalize the data (Graeber et al., 2011). Eco-Real-Time PCR System (Illumina, San Diego, CA, USA) was used and for each 10-μl reaction: 1 μl cDNA sample was mixed with 5 μl of FastStart Universal SYBR Green Master (Roche Applied Sciences), 0.25 μl of each primer (final concentration 500 nM), plus sterile water up to the final volume. The thermal-cycling conditions were 95 °C for 10 min, 40 cycles for 10 s at 95 °C, and 30 s at 60 °C. The melting curve was designed to increase from 55 to 95 °C and primer efficiencies were estimated from a calibration dilution curve and slope calculation (Supplementary Table S1). This analysis was performed with three different biological samples for each time point. Expression levels were determined as the number of cycles needed for the amplification to reach a threshold fixed in the exponential phase of the PCR reaction (Ct; Pfaffl, 2001).
Determination of ROS parameters: H2O2, lipid peroxidation and protein oxidation
Hydrogen peroxide content was measured in seeds using the eFox method as described by Camejo et al. (2011). Samples of 50–100 mg of seeds at different times of germination were homogenized in liquid nitrogen, re-suspended in 1 ml of acid acetone (0.13% sulphuric acid in acetone) and frozen again in liquid nitrogen. After defrosting, the cellular mixture was centrifuged at 10 000 g for 10 min at 4 °C, and the supernatant was mixed (1:5, v/v) with the assay solution (250 μM ferrous ammonium sulphate, 25 mM H2SO4, 100 μM xylenol orange, 100 mM sorbitol). After 45 min incubation at room temperature, the peroxide-mediated oxidation of Fe2+ to Fe3+ was determined by measuring the absorbance at 560 nm of the Fe3+ xylenol orange complex formed.
The level of lipid peroxidation in 50–100 mg samples of seeds was estimated by determining the concentration of ThioBarbituric Acid-Reactive Substances (TBARS) as described by Cakmak and Horst (1991).
Protein oxidation (carbonyl protein content) from 50 mg samples of seeds was measured as described by Vanacker et al. (2006) by reaction with 2,4 dinitrophenylhydrazine (Levine et al., 1990). Total soluble proteins were measured by the Bradford method, using Bovine Serum Albumin (BSA) as the standard (Vanacker et al., 2006).
Statistical analyses
Experiments were conducted in a completely randomized design. The results presented are the mean of at least three biological replicates from each experiment, and all the experiments were repeated at least three times. Data were subjected to ANOVA (one factor) using Tukey’s test (P<0.05), using the IBM SPSS Statistics 20 programme.
Results
The thioredoxin-o (Trxo) gene subfamily
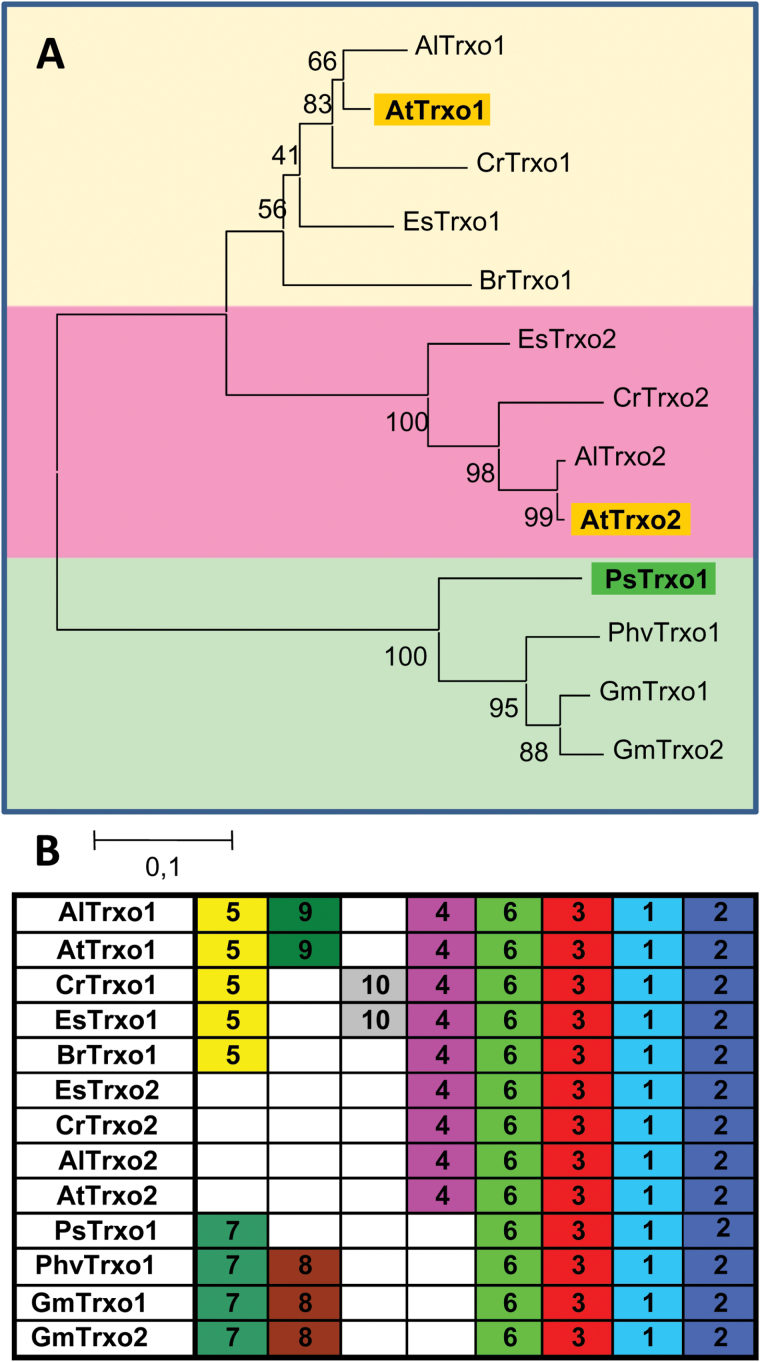
Previous studies on the thioredoxins of Pisum sativum (Leguminosae) reported for the first time the presence of a gene encoding a mitochondrial and nuclear Trxo isoform (Martí et al., 2009). In a further investigation, we searched for its putative Arabidopsis orthologous gene(s) using bioinformatic tools. Two thioredoxin-o genes, AtTrxo1 and AtTrxo2 (loci At2g35010 and At1g31020, respectively) are present in the Arabidopsis genome (Laloi et al., 2001), and a non-redundant compilation of deduced Trxo orthologs in other Brassicaceae (A. lyrata, C. rubella, E. salsugineum, B. rapa) and other Leguminosae (P. vulgaris and G. max) genomes have been annotated and used to construct a phylogenetic un-rooted tree using the neighbour-joining algorithm (Fig. 1A). The pair-wise amino acid similarities (>50%) clearly delimit three clades: two containing the Brassicaceae sequences, and one containg the Leguminosae Trxo proteins. The occurrence of common motifs analysed by the MEME software further support the three clades (Fig. 1B and Table 1). Proteins in the first clade, sharing motifs 1, 2, 3, and 6 with the other Trxo sequences, also share motifs 5 and 4. Members of the Leguminosae group have motif 7 in common instead of motif 5 and lack motif 4. Moreover, the position of the predicted intron–exon gene structures in all Trxo orthologous sequences is conserved among members of the same clade (Supplementary Fig. S1).
Fig. 1.
(A) Phylogenetic dendrogram with the deduced protein sequences of AtTrxo1, AtTrxo2, and PsTrxo1 and their orthologs from other Brassicaceae and Leguminosae as determined from databanks (see text for details); bootstrapping values are indicated on the branches. Abbreviations: Al, Arabidopsis lyrata; At, A. thaliana; Cr, Capsella rubela; Br, Brassica rapa; Es, Eutrema salsugineum; Ps, Pisum sativum; Phv, Phaseolus vulgaris; Gm, Glycine max. Accession numbers: AlTrxo1 (482466), AtTrxo1 (AT2G35010), CrTrxo1 (Carubv 10024096m.g), EsTrxo1 (Thahalv10017264m), BrTrxo1 (021942), EsTrxo2 (Thahalv10008976m), CrTrxo2 (Carubv 10010516m), AlTrxo2 (473345), AtTrxo2 (AT1G31020), PsTrxo1 (EMBL: Q257C6), PhvTrxo1 (091018134m), GmTrxo1 (05g26300.1), GmTrxo2 (08g09210.1). (B) Schematic distribution of conserved motifs among the deduced protein sequences in the phylogenetic tree in (A), identified by means of the MEME analysis. (This figure is available in colour at JXB online.)
Table 1.
Sequences of conserved amino-acid motifs (MEME analysis; Bailey et al., 2009) of the 13 deduced Trxo1 and Trxo2 protein sequences shown in Fig. 1. The conserved amino acids of the active site (WCGPC) are in bold. Motifs containing predicted mitochondrial localization signals according to the MitoProt software (Claros and Vicens, 1996) are in italics
| Motif | E-value | Consensus sequences |
|---|---|---|
| 1 | 7.3e-472 | K[AV][QR]D[GD]SL[PH][SA][VI]FYFTA[AV]WCGPCR[FL]I[SA]P[VI][IV][GLV]ELSK[KQ]YPDVTTYK[VI]DID[EQ][GE][GA][LI] |
| 2 | 6.8e-266 | [IL][GS]KL[NQ][IV][ST][AS]VPTL[HQ]FF[KQ][GN]G[SVK]K[KA][AGD]E[ILV]VG[AV]DV[ATV][KR]LK[NS][LIV][MT]E[QK]L[YF]K |
| 3 | 1.0e-091 | AG[GDA][EPR][SN][GDS][VF]V[LIV][VL][KN]SE[EA]EF[NI][NS][AI][LM][ST] |
| 4 | 4.6e-082 | [PQ][NW]S[INM][FSP][SH][QL]I[AG]RNS[LF][FL][AT]AST[IVF][GY][APV]S[IT][ED]FNF[SL]NTS |
| 5 | 1.0e-040 | MKG[NS][WFL]SI[VI]R[QK][VFI][LF][HQ]R[RQ]FSTLRSS[TRS][PT] |
| 6 | 3.4e-026 | [LF][PHL]H[RS]RS[LF][CS] |
| 7 | 6.2e-022 | [MG][AT]R[NL][LW][VIL][VF]RSLALRH[AV][IM]KN[RT][VG][RL][PT][LI][FLS][FT][NH][RAT][HILN][LH] |
| 8 | 4.1e+002 | [TF]A[TA]A[TI]ASS[QR]LSLLP |
| 9 | 1.7e+004 | RLSTS[IV]RPLV |
| 10 | 9.9e+004 | [AS]RP[LS][AM]L |
AtTrxo1 expression in vegetative and reproductive organs
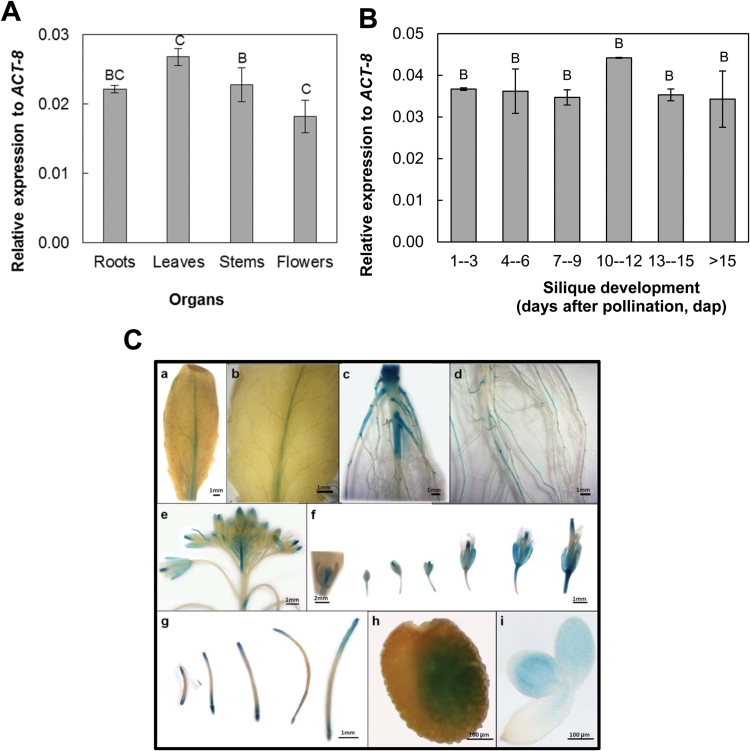
AtTrxo1 transcripts were found ubiquitously in roots, rosette leaves, stems, and flowers of Arabidopsis (Fig. 2A) and its expression was practically constant during silique development (1 to >15 d after pollination, dap; Fig. 2B). To further study the expression of the AtTrxo1 gene, stable transgenic lines of Arabidopsis (Col-0) with its promoter (–1047 bp) were produced that were transcriptionally fused to the reporter uidA gene, which encodes a β-glucuronidase (GUS) enzyme (PAtTrxo1::uidA). The length of the promoter used was selected taking into account the intergenic distance between the ATG translation initiation codon of AtTrxo1 (At2g35010) and its preceding gene At2g35000 in the Arabidopsis genome. GUS histochemical staining of these lines indicated that AtTrxo1 was expressed in adult leaves preferentially in the vascular elements (Fig. 2Ca, Cb). In roots, GUS activity was detected at the initiation of the secondary roots and at the young internodes, as well as being expressed in the crown (Fig. 2Cc, Cd). GUS was also expressed at the floral primordia, the vascular bundle of the sepals and anthers, and at the stigma of flowers (Fig. 2Ce, Cf). In fruits, GUS was expressed at the base and at the upper part of the silique throughout all the stages analysed (Fig. 2Cg). Upon seed imbibition, GUS activity in the PAtTrxo1::uidA reporter lines was detected in the cotyledons, before and after radicle protrusion (Fig. 2Ch, Ci). Due to the high expression found in seeds and considering previous results showing pea Trxo1 as a part of the mitochondrial system responding to saline stress (Martí et al., 2011), we focused on AtTrxo1 expression during germination in both control and saline conditions as well as on its transcriptional regulation during this process. Firstly, we selected 100 mM NaCl as an appropriate treatment after an analysis of the germination kinetics at 10, 50, 75, 100, and 150 mM NaCl for the WT, but also of two KO AtTrxo1 lines, as we describe below. The results for the two KO lines (KO1 and KO2) were found to be similar. A salt concentration of 100 mM NaCl was chosen because it showed the highest difference in germination rate between the WT and the KO lines whilst still allowing 100% germination (see Supplementary Fig. S2).
Fig. 2.
Expression analysis of the AtTrxo1 gene (relative to ACT-8) as determined by RT-qPCR (A) in different organs of the Arabidopsis plant, and (B) during silique development. (C) Expression of β-glucuronidase (GUS) activity driven by the AtTrxo1 whole promoter (–1047 bp) fused to the reporter uidA (GUS) gene in transgenic lines of Arabidopsis: (a, b) rosette leaf; (c, d) root system; (e) inflorescence; (f) floral to fruit development; (g) silique development; (h) seed after 24 h of imbibition; (i) same as (h) without seed coat.
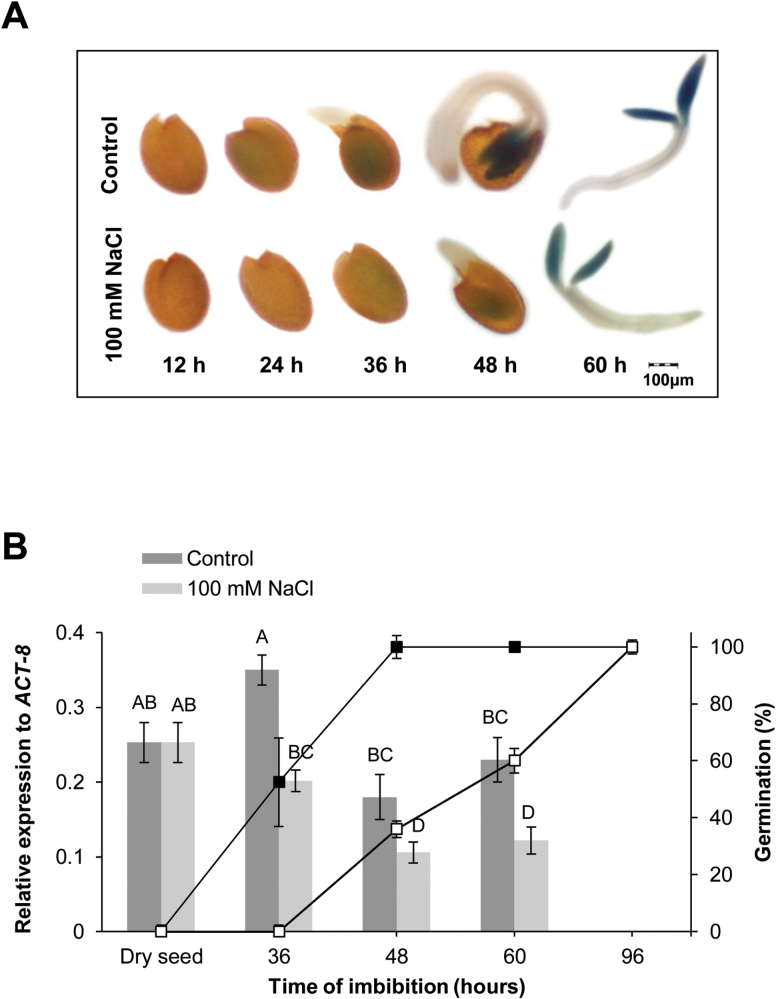
Analysis of GUS activity revealed an increase of expression of AtTrxo1 in water-imbibed seeds during germination, mainly localized in cotyledons at 48 h, and a decrease upon imbibition in the presence of 100 mM NaCl (Fig. 3A). AtTrxo1 expression was also analysed by RT-qPCR in imbibed seeds in 100 mM NaCl at 36, 48, and 60 h, using as controls seeds imbibed in water for the same period of time (Fig. 3B). AtTrxo1 transcripts were especially abundant in dry seeds after 1 month of dry storage at 21 °C (after-ripened seeds), being one order of magnitude higher than in the other organs analysed (see Fig. 2A). In germinating control seeds, at 36 h of imbibition in water (t50 value when 50% of the seeds had germinated), the AtTrxo1 transcript accumulation was similar to that found in dry seeds (Fig. 3B), decreasing thereafter until germination sensu stricto was completed (100% radicle protrusion at 48 h). In the presence of salt, a significant reduction of AtTrxo1 transcript accumulation was found at each time-point analysed and a decrease was observed during germination, which was significantly delayed (Control t100=48 h; Salt t100=96 h).
Fig. 3.
Expression of AtTrxo1 during germination. (A) Histochemical localization of PAtTrxo1::uidA (–1047 bp) during germination in water (Control) and in the presence of salt (100 mM NaCl). (B) Expression analysis of the AtTrxo1 gene by RT-qPCR upon seed germination in the absence (Control) or the presence of 100 mM NaCl. Data are means ±SE of three technical replicates of three biological samples. Different letters indicate that data are significantly different according to Tukey’s test (P<0.05). Percentage of germination is also indicated (Control, closed squares; 100 mM NaCl, open squares).
Identification of evolutionary conserved cis-motifs in the AtTrxo1 gene promoter and of the TFs recognizing the conserved B2 domain
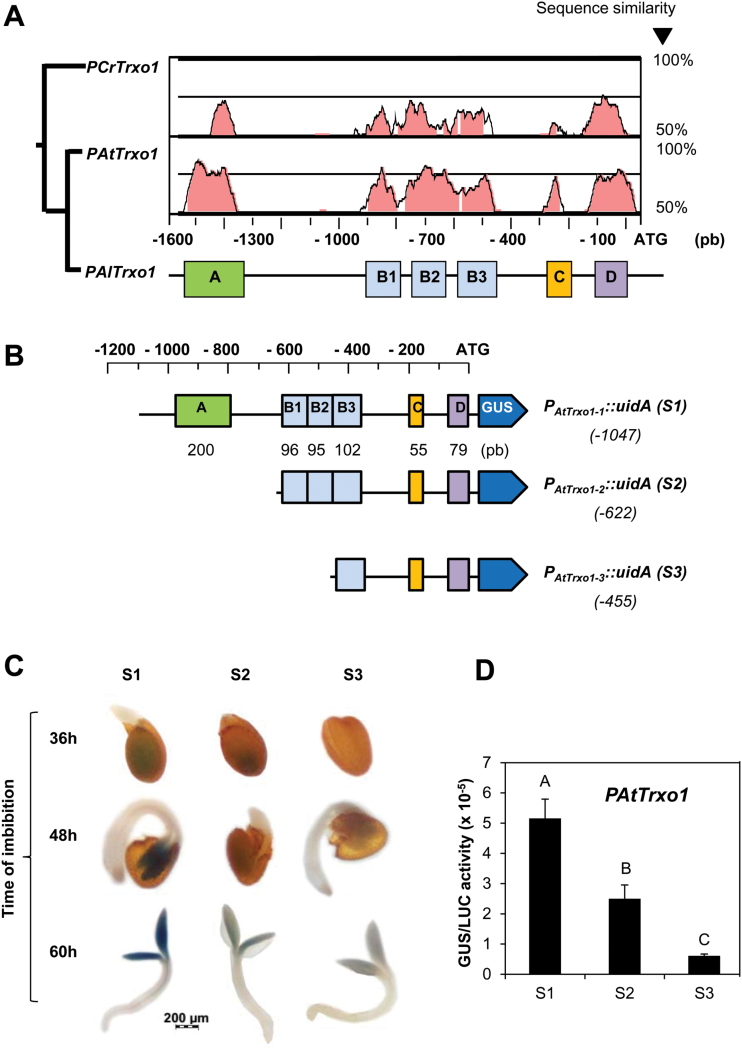
The promoter regions of the Trxo1 orthologous genes within the Brassicaceae species were searched in silico for conserved sequences (phylogenetic shadowing, Iglesias-Fernández et al., 2013). The pair-wise alignment among three promoters analysed (A. lyrata, C. rubella and A. thaliana) showed several conserved blocks upstream of the ATG named motifs A, B, C, and D (see Fig. 4A, B).
Fig. 4.
Influence of conserved domains A and B1+B2 of the AtTrxo1 promoter on gene expression upon germination. (A) Identification of conserved cis-elements in orthologus Trxo1 promoters in the Brassicaceae species Arabidopsis thaliana (At), A. lyrata (Al), and Capsella rubella (Cr). Shaded areas show conserved blocks. (B) Schematic representation of the three different promoter constructs of the AtTrxo1 gene: S1 (–1047 bp), S2 (–622 bp), and S3 (–455 bp) fused to the reporter uidA gene (GUS) for use in transient expression assays. (C) Histochemical localization of GUS expression in transformed Arabidopsis plants with serial deletions of the PAtTrxo1::uidA construct (S1, S2, S3) during seed germination. (D) Quantification of GUS activity in tobacco leaves co-infiltrated with Agrobacterium tumefaciens containing the three different promoter constructs (S1, S2, S3) described in (B). Data are means ±SE of three independent experiments. Different letters indicate significant differences according to Tukey’s test (P<0.05).
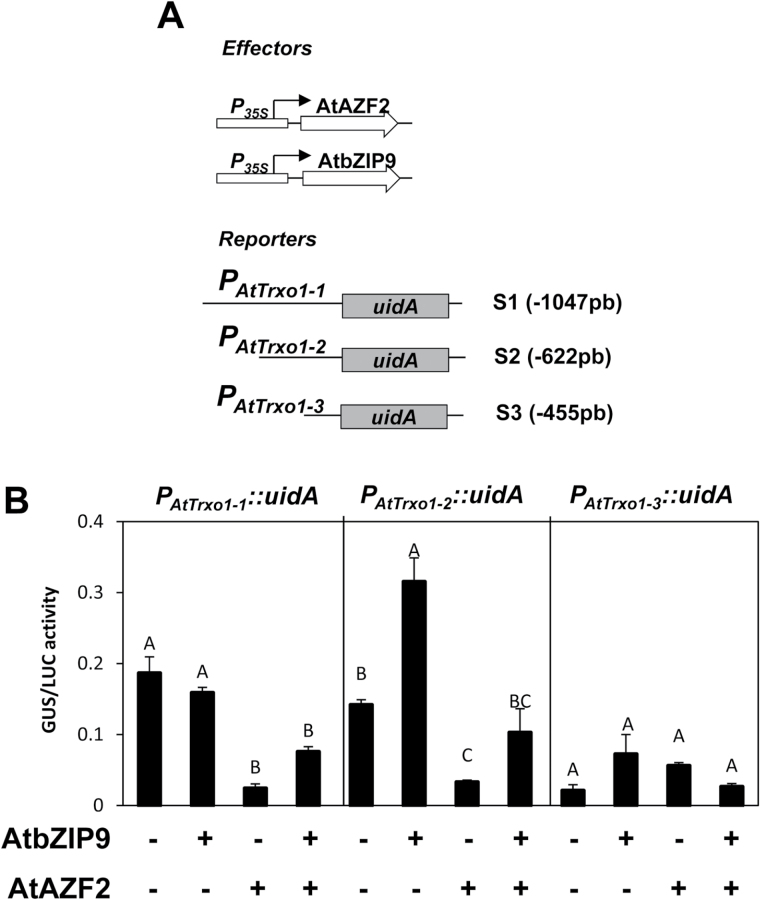
In order to explore the functional relevance of these conserved motifs, a set of deletion constructs of the AtTrxo1 promoter (PAtTrxo1) fused to the β-glucuronidase (GUS) reporter gene (uidA) were generated. These constructs are: PAtTrxo1-1::uidA (–1047 bp, S1) containing the whole promoter; PAtTrxo1-2::uidA (–622 bp, S2) lacking motif A; and PAtTrxo1-3::uidA (–455 bp, S3) deprived of motifs A, B1, and B2 (Fig. 4B). Transient expression assays by agro-infiltration of these constructs in N. benthamiana leaves showed a decrease in GUS expression with consecutive deletions (Fig. 4B, C). Homozygous transgenic Arabidopsis plants were generated with these constructs and GUS activity was evaluated during germination (Fig. 4D). The transgenic line with the PAtTrxo1-1::uidA construct showed that GUS activity was high in the cotyledons at 36 and 48 h of imbibition. Although the removal of the A motif decreased this activity, the S2 construct (PAtTrxo1-2::uidA) still retained 50% of the original GUS expression, and in the S3 construct (devoid of B1 and B2 elements) this expression was faint (Fig. 4B–D).
The conserved cis-elements identified in silico as blocks B1 (96 bp) and B2 (95 bp; Figs 4B and 5A) were selected as baits for the screening of the arrayed library of Arabidopsis TFs in yeast (Castrillo et al., 2011), and the AtbZIP9 (At5g24800) and AtAZF2 (At3g19580) that interacted with the B2 element were selected. Diploid yeast containing the plasmids B2-element–pTUY1H and AtbZIP9-pDEST22 or AtAZF2-pDEST22 were, respectively, able to grow in an auxothophic medium lacking histidine in the presence of up to 100 mM and 30 mM 3-AT a competitive inhibitor of the product of the HIS3 gene (Fig. 5B). Moreover, in the web database PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), in the block B2, a G-box (binding site for bZIP proteins) and an A(G/C)T box (binding site for AZF2 proteins) were predicted (Sakamoto et al., 2004). To determine whether AtbZIP9 and AtAZF2 could be transcriptional regulators of the AtTrxo1 gene during germination, the expression of the AtbZIP9 and AtAZF2 genes were analysed. Accumulation of AtbZIP9 transcripts was high (Fig. 5C) at 36 h when 50% of the seeds had germinated (t50) and presented a similar pattern to AtTrxo1 expression (Fig. 3B), in a manner compatible with AtbZIP9 being a transcriptional regulator of the AtTrxo1 gene. In the presence of salt, expression of the AtbZIP9 gene was lower than in control conditions only at 36 h (Fig. 5C) when no germinated seeds were observed (Fig. 3B). In contrast, the AZF2 transcript showed a significant increase in control conditions at 60 h but under salinity the increase was significant at 48 h (Fig. 5C), when 40% of the seeds were germinated (Fig. 3B), which is compatible with a role as a negative regulator of the AtTrxo1 gene.
Fig. 5.
(A) Identification of the conserved cis-element, B2, in orthologous AtTrxo1 promoters in the Brassicaceae species Arabidopsis thaliana (At), A. lyrata (Al), and Capsella rubella (Cr) by pair-wise alignment. The boxes indicate predictable cis-binding domains to AZF2 [A(G/C)T box] and bZIP9 (C-box). (B) AtAZF2 and AtbZIP9 bind to the AtTrxo1-B2-element in a yeast one-hybrid screening. Growth of diploid cells after mating of two yeast strains containing the AtTrxo1-element–pTUY1H construct and pDEST22-AD:AtAZF2, pDEST22-AD:bZIP9, or pDEST22-AD:GFP (negative control) on non-selective (–L–W) or selective medium (–L–W–H) with increasing concentrations of 3-AT. (C) Expression analysis of the AtbZIP9 and AtAZF2 genes (relative to ACT-8) as determined by RT-qPCR in dry seeds and upon germination in water or in 100 mM NaCl. Data are means ±SE of three technical replicates of three biological samples. Different letters indicate significant differences according to Tukey’s test (P<0.05).
The interaction between AtbZIP9 and AtAZF2 with the B2 element was further investigated in planta (Fig. 6) using a tobacco transient expression system (N. benthamiana leaves). As reporter constructs, the promoter fragments S1, S2, and S3 fused to the uidA (GUS) gene were used (PAtTrxo1-1::uidA, PAtTrxo1-2::uidA, and PAtTrxo1-3::uidA, Figs 4B and 6A); as effectors, plasmid constructs were used where the AtbZIP9 and /or the AtAZF2 coding sequences were under the control of the 35S CaMV promoter (P35S::AtbZIP9, P35S::AtAZF2) (Fig. 6A). A construct containing the LUC-encoding ORF (P35S::LUC) was used as an internal control for the transformation experiments. Co-transformation of P35S::AtbZIP9 and reporters enhanced the GUS activity driven by the S2 construct, but not by the S1 construct (Fig. 6B). Although the two constructs contain the B2 motif, only the S1 contains the A element. Moreover, GUS activity was not enhanced in the S3 construct, which is devoid of the A, B1, and B2 motifs. Co-transformation of P35S::AtAZF2 and the S1 and/or S2 constructs, both containing the B2 element, diminished the GUS activity driven by the S1 and S2 promoters; when both effectors were transfected, the repressor effect of AtAZF2 counteracted the transcriptional activator effect of AtbZIP9 (Fig. 6B).
Fig. 6.
Trans-activation assays using as effectors the TFs AtbZIP9 and AtAZF2, and as reporters the AtTrxo1 gene promoter driven by the expression of the uidA gene (GUS activity). (A) Schematic representation of the effector and reporter contructs used in the analysis. (B) Agro-infiltration of the effector and the reporter combinations indicated in tobacco leaves. The relative amounts of reporter and effector plasmids used in these assays correspond to the 1:1 ratio. GUS activity was relative to luciferase (LUC) activity, which was used as internal control. Data are means ±SE of three biological replicates. Different letters indicate significant differences according to Tukey’s test (P<0.05).
Growth and germination of Arabidopsis lacking Trxo1 in saline conditions
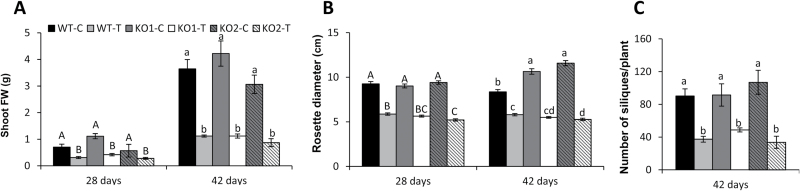
In order to gain further information on the functions of Trxo1 and its potential role during plant development and seed germination, we analysed the growth of wild-type Arabidopsis plants and two T-DNA insertion mutant lines in the thioredoxin-o1 gene (At2g35010) from the Salk collection (KO1, SALK_143294C; KO2, SALK_042792). The insertion in the Trxo1 gene was mapped to the first intron and the lack of AtTrxo1 expression in germinating seeds of these mutants (at 36 h) was confirmed by RT-qPCR analysis (Supplementary Fig. S3A, B). We then studied the growth of plants under saline conditions and we also examined germination in the presence of NaCl. After 28 and 42 d of growth in control conditions, wild-type and both KO mutant plants showed similar fresh weights (Fig. 7A), and they also showed similar rosette diameter at 28 d, although this parameter was significantly higher in the two KO lines at 42 d of growth (Fig. 7B). At 42 d of growth, the number of siliques per plant was not significantly different between the mutants and the WT plants (Fig. 7C). Analysing the response of plants grown under salt conditions, we found no apparent differences between WT and KO mutant plants, although a strong effect at 42 d was observed in all the plants with loss of chlorophyll and senescence symptoms (Supplementary Fig. S4). Treatment with 100 mM NaCl caused a similar decrease in all the physiological parameters measured in the three genotypes (Fig. 7). With this treatment, no differences were found in the fresh weight between WT and KO mutants at 28 and 42 d of development and the number of siliques was similar, while the rosette diameter was found to be smaller in the KO2 plants.
Fig. 7.
Physiological characterization of the wild-type (WT) and two knock-out (KO) AtTrxo1 mutants after 28 and 42 d growing in the absence (control, C) or presence (treated, T) of 100 mM NaCl. The shoot fresh weight (A), rosette diameter (B), and number of siliques per plant (C) were determined in plants grown in a growth chamber as described in the Material and Methods. Data are means ±SE of at least three independent experiments. Different letters (capital letters at 28 d and lower-case letters at 42 d) indicate that data at each time point are significantly different according to Tukey’s test (P<0.05).
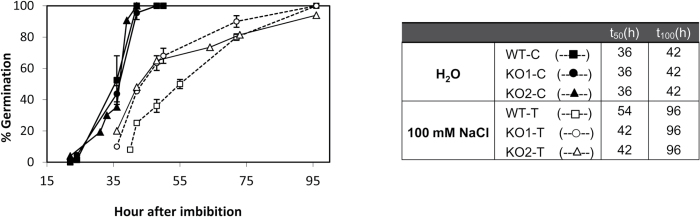
Analysing germination in control conditions, and as previously described for the KO2 mutant by Daloso et al. (2015), we found that seeds of both KO AtTrxo1 lines failed to show any differences in germination rate compared to WT water-imbibed seeds (100% of germination at 42 h; t50=36 h; Fig. 8). When 100 mM NaCl was used as the imbibition medium, the germination in the KO AtTrxo1 mutants was faster (t50=42 h) than that of the WT seeds (t50=54 h), although all of them reached 100% germination at 96 h (Fig. 8).
Fig. 8.
Germination time course of seeds imbibed in water (control C, closed symbols) or in 100 mM NaCl (treated T, open symbols) in the wild-type (WT, squares), KO1 mutant (circles), and KO2 mutant (triangles). The t50 value (time required for radicle emergence in 50% of seeds) is indicated in the table. Data are means ±SE of three technical replicates of six biological samples.
Oxidative parameters and Prx and Srx gene expression during germination in saline conditions
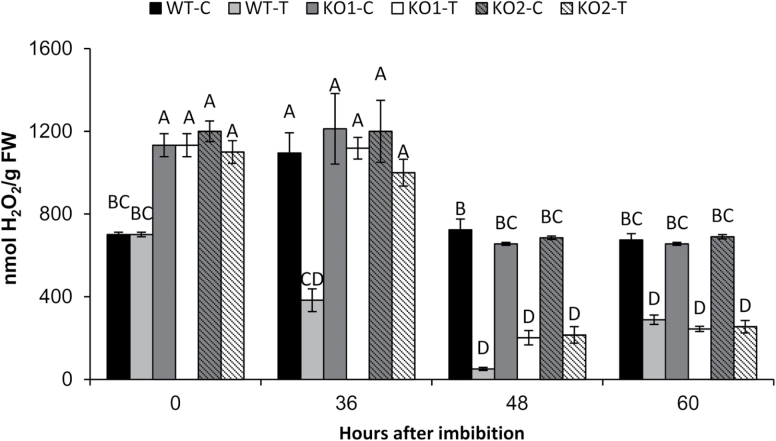
Since H2O2 plays a central role in redox homeostasis, we analysed the H2O2 content in seeds at different times of germination in WT and KO AtTrxo1 lines (KO1 and KO2), in water and in 100 mM NaCl. As shown in Fig. 9, dry seeds of the KO lines had a 1.5-fold higher content than the WT seeds, while upon germination in water (36, 48, 60 h) no significant differences were observed. In water, a high H2O2 content was observed at 36 h, it decreased at 48 h and was maintained at 60 h in all the genotypes. In saline conditions, the H2O2 content at 36 h in the WT plants decreased to approximately half of that found in control conditions, while it remained at the same value as the dry seeds in the KO mutants, in which the content represented a three-fold increase as compared with the WT seeds under these stress conditions. At 48 h, the H2O2 diminished drastically in both the WT and KO mutants, and this content remained without significant changes in both types of seeds at 60 h.
Fig. 9.
Hydrogen peroxide content (g–1 FW) in dry seeds (0 h) and germinating seeds of the wild-type (WT) and KO AtTrxo1 (KO1 and KO2) in control (C) water conditions or in the presence of 100 mM NaCl (treated, T). Data are means ±SE of three technical replicates of three biological samples. Different letters indicate significant differences according to Tukey’s test (P<0.05).
Since the patterns of germination rate and H2O2 content were similar in both KO1 and KO2 seeds, we chose the KO1 mutant to measure other oxidative parameters. Measurement of protein oxidation (carbonyl protein content) and lipid peroxidation (thiobarbituric acid reactive substances content, TBARS), as markers of oxidative stress, in the seeds of both lines under control and saline conditions revealed no significant changes between WT and mutant seeds over time under both conditions, although the presence of 100 mM NaCl decreased the lipid peroxidation level in dry seeds of both lines (Supplementary Fig. S5).
Since the Trxo1/PrxIIF/Srx system has been described as important for ROS homeostasis in mitochondria (Iglesias-Baena et al., 2011), the expression of AtPrxIIF and AtSrx genes during germination was examined both in water and in 100 mM NaCl. In dry seeds, the AtPrxIIF and AtSrx transcript content was higher in the WT than in the KO1 AtTrxo1 mutant, and both genes decreased their expression upon imbibition, but no significant differences between the lines were observed in control or even in saline conditions (Supplementary Fig. S6).
Discussion
The Arabidopsis genome has several thioredoxin genes. Laloi et al. (2001) first identified an AtTrxo1 gene encoding a thioredoxin located in mitochondria. Martí et al. (2009, 2011) found in pea leaves a double location for the PsTrxo1 encoding protein in mitochondria and in the nucleus, and demonstrated its participation in saline stress. The gene AtTrxo1 is expressed ubiquitously in the vascular elements of leaves and roots, and this expression is particularly important in dry and germinating seeds, with a high transcript accumulation at 36 h of imbibition, as was shown by RT-qPCR and GUS expression assays. This localization in the seed is similar to that of other thioredoxins, such as Trxf and Trxm from pea seedlings and Trxh from barley seeds (Shahpiri et al., 2008; Fernández-Trijueque et al., 2012). The implication of these cytoplasmic and chloroplastic Trxs in germination has been described previously (Montrichard et al., 2009; Pulido et al., 2009), but there is no information concerning the participation of mitochondrial AtTrxo1 in germination. The reactivation of metabolism that occurs during seed germination generates an important quantity of ROS and produces an increase in the content of diverse antioxidant compounds, such as flavonoids, phenols, ascorbate (ASC), and reduced glutathione (GSH), as well as increases in the expression of Trx, Prx, and CAT genes (Simontacchi et al., 1993; De Gara et al., 1997; Yang et al., 2001; De Tullio and Arrigoni, 2003). Our data indicate a high AtTrxo1 expression in cotyledons, similar to that reported for other Trxs, such as Trxh6 in Medicago truncatula (Renard et al., 2011), with a role related to the regulation of specific targets. In addition, Trxh1 and, to a lesser extent, Trxh2 are abundant in both embryonic axes and cotyledons, so the different spatial distribution of the isoforms of Trxh in Medicago suggests that they play different roles during germination, which might be related to the maintenance of the redox homeostasis during this process. In previous work, both Trxh3 and h4 were found in dry seeds of pea only in embryo axes (h4) or in both axes and cotyledons (h3), indicating that they were synthesized before germination. In contrast, they showed similar expression profiles upon imbibition, with a strong induction of expression in axes after radicle protrusion (46 h) and in cotyledons just before and after radicle protrusion (22 and 46 h; Montrichard et al., 2003). The authors suggested different roles for these Trxs during germination, such as reserve mobilization or protection against ROS. In fact, several proteases, α-amylases, and their inhibitors in the endosperm and embryo are mainly oxidized in dry seeds and are reduced to the sulfydril state after imbibition, increasing their solubilization and susceptibility to proteolysis (Lozano et al., 1996; De Gara et al., 2003; Serrato and Cejudo, 2003). In addition, in the dicotyledonous Medicago truncatula, and in the monocotyledonous Triticum sp. and Hordeum vulgare, the majority of the proteins susceptible to redox modification at the beginning of germination are targets of Trx (Wong et al., 2004; Alkhalfioui et al., 2007; Montrichard et al., 2009).
Although available information about transcriptional regulation of plant thioredoxin genes is scarce, CCA1 and DOF7 TFs have been identified as regulators of Pisum sativum chloroplast Trxf and Trxm1, encoding genes that respond to the circadian cycle and to glucose levels (Blazquez et al., 2011; Barajas-López et al., 2012). In addition, the WRKY6 TF is a positive regulator of cytosolic AtTrxh5 expression, which is also mediated by ROS under oxidative stress conditions. Moreover, AtTRXh5 was up-regulated in plants overexpressing WRKY6. This regulation is specific to the thioredoxin h family (Laloi et al., 2004). However, no information is available regarding the transcriptional regulation of Trxo1. To rectify this, the cis–trans transcriptional regulatory code of AtTrxo1 was established by phylogenomic analyses of orthologous Trxo1 gene promoters within the Brassicaceae family coupled with screenings of an arrayed library of Arabidopsis TFs (Castrillo et al., 2011). By this method, the conserved B2-element, the AtbZIP9 and the AtAZF2 TFs, were selected.
AtbZIP9 belongs to the basic leucine zipper C-subfamily of TF proteins that includes AtbZIP10, AtbZIP25, and AtbZIP63 (Jakoby et al., 2002; Lara et al., 2003). The importance of the conserved motifs in the AtTrxo1 promoter has been demonstrated in planta by transient expression in agro-infiltrated N. benthamiana leaves and by stable GUS expression (uidA gene) driven by sequential deletions of AtTrxo1 promoter constructs. These experiments indicated the relevance of block B2 as an interactor with the AtbZIP9 TF, a transcriptional activator of the AtTrxo1 gene. The co-expression of the P35S::AtbZIP9 and the PAtTrxo1-2::uidA (containing the B2 element) constructs in N. benthamiana leaves revealed the activator effect of AtbZIP9 over the AtTrxo1 promoter, and this effect disappeared when the B2 motif was deleted. Moreover, when the A motif was present (PAtTrxo1-1::uidA; Fig. 6B), the activation effect of AtbZIP9 was not observed, perhaps indicating an accession difficulty of this TF to its interacting B2 cis-motif. In addition, during germination the AtbZIP9 gene displayed a pattern of expression compatible with being a transcriptional activator of the AtTrxo1 gene. Other TF genes from the bZIP C-subfamily, namely AtbZIP10 and AtbZIP25, have been found to have expression patterns during seed maturation that temporally and spatially match with those of the seed storage protein genes (Lara et al., 2003). Nevertheless, no effect in the AtbZIP9 expression profile was observed in germination under salinity, although the decrease of Trxo1 expression with time of seed imbibition indicated that under salinity another TF with transcriptional repressor activity must be involved.
AtAZF2 belongs to the Cys-2/His-2-type zinc finger proteins that they are induced in plants by dehydration, salinity, cold stress, and ABA treatment (Sakamoto et al., 2004). Transient assays in planta with co-expression of the P35S::AZF2 and PAtTrxo1-1::uidA and PAtTrxo1-2::uidA constructs revealed the repressor effect of AtAZF2 over the AtTrxo1 gene. In addition, expression analysis of AtAZF2 during germination confirms the behaviour of this gene as a repressor of AtTrxo1, mainly under salinity. This behaviour has been already reported in seeds, where AZF2 acts as a repressor of several genes under salt stress (Sakamoto et al., 2004; Drechsel et al., 2010; Kodaira et al., 2011).
Trxo1 mutants have been used to corroborate a role for the Trx system in regulating different metabolic processes in mitochondria, although no extreme phenotype has been described, possibly due to the redundancy or overlapping functions with mitochondrial or cytosolic proteins as glutaredoxins (Daloso et al., 2015). Among abiotic stresses, salinity is one of the most important unfavorable conditions for plant yield and growth, and redox systems are considered as key players for stress sensing and signal transduction pathways (Lázaro et al., 2013). In our study on the behavior of KO AtTrxo1 plants growing in the presence of 100 mM NaCl, the lack of Trxo1 seems to be compensated for, as evidenced by the plant growth of Trxo1 insertion lines not being significantly affected under this stress situation compared to the WT plants, although this behaviour was more evident in the KO1 than in the KO2 plants, as shown by the rosette diameter. Production of ROS, particularly H2O2, increases during seed maturation in sunflower seeds, and the subsequent ability to germinate depends on a critical accumulation of this compound (Bailly et al., 2008). An increase in H2O2 has been also reported in other species under salinity conditions and it has been described as an inducer of earlier germination (Puntarulo et al., 1991; Hernandez et al., 2001; Lin et al., 2013). In our analyses, the H2O2 levels were higher in dry seeds of the KO AtTrxo1 than in the WT, although there was no detectable difference either in the germination kinetics in water or in protein oxidation or lipid peroxidation between the WT and the KO mutant. Additionally, in both of these seeds, the expression of AtPrxIIF and AtSrx did not differ significantly during germination in either water or NaCl. This behaviour was different to that found in pea plants, where Trxo1 and PrxIIF expression is increased in response to short-term salt stress (Barranco-Medina et al., 2008; Martí et al., 2011), pointing to the heterogeneity of response of the antioxidant system depending on the salt sensitivity of the cultivars, the NaCl concentration, and the duration of the stress (Lázaro et al., 2013). Salinity in imbibed seeds usually produces a delay in germination, as occurred in our experiments, although we found that KO AtTrxo1 seeds had higher H2O2 content at the beginning of germination and a faster germination rate than those of the WT (see t50 values in Fig. 8). Figure 10 is presented as a summary illustration of our main results in saline conditions, including the participation of AtbZIP9 and AtAZF2 TFs in the regulation of the AtTrxo1 gene. Among other ROS, H2O2 is known to accumulate during imbibition and early stages of germination, with mitochondria being essential producers (Zhang et al., 2014). The differential H2O2 peak in the mutant line may be one factor in the early germination shown under salt stress, and the lack of lipid or protein oxidation could be related with the different roles of H2O2 in cell wall growth and cross-talk with NO and hormones such as ABA and GA (Wojtyla et al., 2016). Similar changes as regards bringing forward or delaying germination have been described for other redox proteins, including Trxs. The fact that this divergence in the germination pattern between WT and KO seeds basically occurs in the face of saline stress may reflect a specific role for Trxo1 in the germination of seeds exposed to salt stress, which may in turn be related to specific Trxo1 targets. This behaviour is similar to some antioxidant enzymes, such as symplastic ascorbate oxidase from tobacco and Arabidopsis (Yamamoto et al., 2005), and the behaviour found in Arabidopsis RNA interference lines of the barley ortholog of AtPER1 (encoding a 1 Cys-Prx), which germinated earlier than WT seeds under salinity, whereas the over-expression of AtPER1 caused germination to be delayed (Haslekås, 2003).
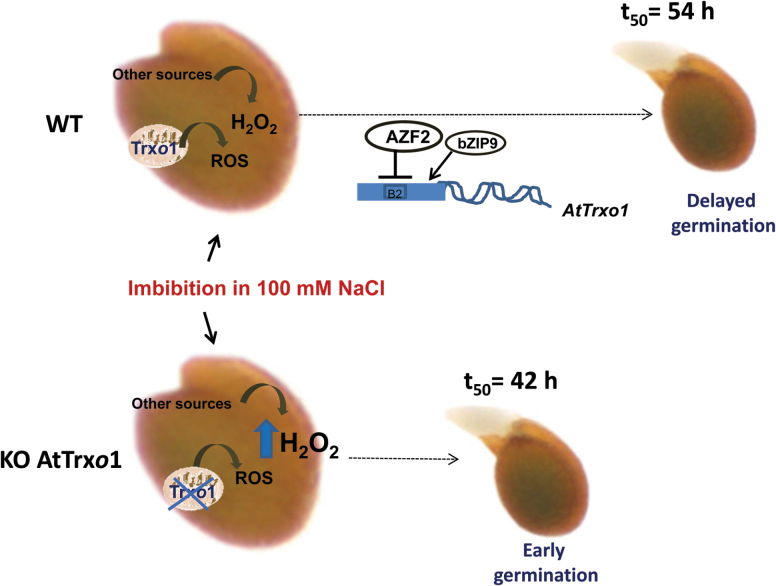
Fig. 10.
Proposed model of the germination of Arabidopsis wild-type (WT) and KO AtTrxo1 mutant seeds in the presence of 100 mM NaCl. The production of H2O2 by the mitochondria and other sources in the presence of NaCl accelerates germination of imbibed seeds and is greater (2–3 times) in KO Trxo1 mutants than in the WT seeds. t50, time when 50% of the seeds have germinated. AZF2 is a transcriptional repressor (line ending in bar) and bZIP9 is an activator (arrow).
In summary, these results indicate a role for the AtTrxo1 gene in seed germination that is more evident in 100 mM NaCl, where Trxo1 could act as a possible sensor of saline stress and an inducer of H2O2 accumulation, independently of other ROS parameters (PrxIIF and AtSrx gene expression, protein oxidation, or lipid peroxidation). In addition, for the first time, the transcriptional regulation of this gene has been investigated. Two transcription factors have been identified, bZIP9 and AZF2, that show a positive and negative role, respectively, over Trxo1 expression during germination. Further studies that are focused on determining the specific Trxo1 target proteins and/or its involvement in redox signaling pathways will help to establish the mechanism by which Trxo1 is acting during germination.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Jose L. Micol and Dr Rosa M. Ponce from the University of Miguel Hernández, Spain, for their valuable help in the selection of the KO AtTrxo1 mutants. Financial support from Spanish grants MINECO-BFU/FEDER 2014-52452-P and the Seneca Excellence Project 19876/GERM/15 (IP: F. Sevilla) and MICINN-CONSOLIDER CSD2007-00057 (IP: P. Carbonero) are gratefully acknowledged.
References
- Alkhalfioui F, Renard M, Vensel WH, Wong J, Tanaka CK, Hurkman WJ, Buchanan BB, Montrichard F. 2007. Thioredoxin-linked proteins are reduced during germination of Medicago truncatula seeds. Plant Physiology 144, 1559–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly C, El-Maarouf-Bouteau H, Corbineau F. 2008. From intracellular signaling networks to cell death: the dual role of reactive oxygen species in seed physiology. Comptes Rendus Biologies 331, 806–814. [DOI] [PubMed] [Google Scholar]
- Barajas-López J, de D, Tezycka J, Travaglia CN, Serrato AJ, Chueca A, Thormählen I, Geigenberger P, Sahrawy M. 2012. Expression of the chloroplast thioredoxins f and m is linked to short-term changes in the sugar and thiol status in leaves of Pisum sativum. Journal of Experimental Botany 63, 4887–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barranco-Medina S, Krell T, Bernier-Villamor L, Sevilla F, Lázaro JJ, Dietz KJ. 2008. Hexameric oligomerization of mitochondrial peroxiredoxin PrxIIF and formation of an ultrahigh affinity complex with its electron donor thioredoxin Trx-o. Journal of Experimental Botany 59, 3259–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barranco-Medina S, Krell T, Finkemeier I, Sevilla F, Lázaro JJ, Dietz KJ. 2007. Biochemical and molecular characterization of the mitochondrial peroxiredoxin PsPrxII F from Pisum sativum. Plant Physiology and Biochemistry 45, 729–739. [DOI] [PubMed] [Google Scholar]
- Belin C, Bashandy T, Cela J, Delorme-Hinoux V, Riondet C, Reichheld JP. 2015. A comprehensive study of thiol reduction gene expression under stress conditions in Arabidopsis thaliana. Plant, Cell & Environment 38, 299–314. [DOI] [PubMed] [Google Scholar]
- Bewley JD. 1997. Seed germination and dormancy. The Plant Cell 9, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez MA, Filichkin SA, Breton G, et al. 2011. Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PLoS ONE 6, e16907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmak I, Horst WJ. 1991. Effect of aluminium on lipid peroxidation, superoxide dismutase, catalase, and peroxidase activities in root tips of soybean (Glycine max). Physiologia Plantarum 83, 463–468. [Google Scholar]
- Camejo D, Martí MC, Jiménez A, Cabrera JC, Olmos E, Sevilla F. 2011. Effect of oligogalacturonides on root length, extracellular alkalinization and O2–-accumulation in alfalfa. Journal of Plant Physiology 168, 566–575. [DOI] [PubMed] [Google Scholar]
- Castrillo G, Turck F, Leveugle M, Lecharny A, Carbonero P, Coupland G, Paz-Ares J, Oñate-Sánchez L. 2011. Speeding cis-trans regulation discovery by phylogenomic analyses coupled with screenings of an arrayed library of Arabidopsis transcription factors. PLoS ONE 6, e21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P. 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. European Journal of Biochemistry 241, 779–786. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Daloso DM, Müller K, Obata T, et al. 2015. Thioredoxin, a master regulator of the tricarboxylic acid cycle in plant mitochondria. Proceedings of the National Academy of Sciences, USA 112, E1392–E1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gara L, de Pinto MC, Arrigoni O. 1997. Ascorbate synthesis and ascorbate peroxidase activity during the early stage of wheat germination. Physiologia Plantarum 100, 894–900. [Google Scholar]
- De Gara L, de Pinto MC, Moliterni VM, D’Egidio MG. 2003. Redox regulation and storage processes during maturation in kernels of Triticum durum. Journal of Experimental Botany 54, 249–258. [DOI] [PubMed] [Google Scholar]
- De Tullio MC, Arrigoni O. 2003. The ascorbic acid system in seeds: to protect and to serve. Seed Science Research 13, 249–260. [Google Scholar]
- Dietz KJ, Jacob S, Oelze ML, Laxa M, Tognetti V, de Miranda SM, Baier M, Finkemeier I. 2006. The function of peroxiredoxins in plant organelle redox metabolism. Journal of Experimental Botany 57, 1697–1709. [DOI] [PubMed] [Google Scholar]
- Drechsel G, Raab S, Hoth S. 2010. Arabidopsis zinc-finger protein 2 is a negative regulator of ABA signaling during seed germination. Journal of Plant Physiology 167, 1418–1421. [DOI] [PubMed] [Google Scholar]
- El-Maarouf-Bouteau H, Bailly C. 2008. Oxidative signaling in seed germination and dormancy. Plant Signaling & Behavior 3, 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Trijueque J, Barajas-López J de D, Chueca A, Cazalis R, Sahrawy M, Serrato AJ. 2012. Plastid thioredoxins f and m are related to the developing and salinity response of post-germinating seeds of Pisum sativum. Plant Science 188–189, 82–88. [DOI] [PubMed] [Google Scholar]
- Florez-Sarasa I, Lambers H, Wang X, Finnegan PM, Ribas-Carbo M. 2014. The alternative respiratory pathway mediates carboxylate synthesis in white lupin cluster roots under phosphorus deprivation. Plant, Cell & Environment 37, 922–928. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Research 32, W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Calle V, Barrero-Sicilia C, Carbonero P, Iglesias-Fernández R. 2015. Mannans and endo-β-mannanases (MAN) in Brachypodium distachyon: expression profiling and possible role of the BdMAN genes during coleorhiza-limited seed germination. Journal of Experimental Botany 66, 3753–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Calle V, Iglesias-Fernández R, Carbonero P, Barrero-Sicilia C. 2014. The BdGAMYB protein from Brachypodium distachyon interacts with BdDOF24 and regulates transcription of the BdCathB gene upon seed germination. Planta 240, 539–552. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40, D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber K, Linkies A, Wood AT, Leubner-Metzger G. 2011. A guideline to family-wide comparative state-of-the-art quantitative RT-PCR analysis exemplified with a Brassicaceae cross-species seed germination case study. The Plant Cell 23, 2045–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslekås C, Viken MK, Grini PE, Nygaard V, Nordgard SH, Meza TJ, Aalen RB. 2003. Seed 1-cysteine peroxiredoxin antioxidants are not involved in dormancy, but contribute to inhibition of germination during stress. Plant Physiology 133, 1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández JA, Ferrer MA, Jiménez A, Barceló AR, Sevilla F. 2001. Antioxidant systems and O2·–/H2O2 production in the apoplast of pea leaves. Its relation with salt-induced necrotic lesions in minor veins. Plant Physiology 127, 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Baena I, Barranco-Medina S, Lázaro-Payo A, López-Jaramillo FJ, Sevilla F, Lázaro JJ. 2010. Characterization of plant sulfiredoxin and role of sulphinic form of 2-Cys peroxiredoxin. Journal of Experimental Botany 6, 1509–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Baena I, Barranco-Medina S, Sevilla F, Lázaro JJ. 2011. The dual-targeted plant sulfiredoxin retroreduces the sulfinic form of atypical mitochondrial peroxiredoxin. Plant Physiology 155, 944–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Fernández R, Barrero-Sicilia C, Carrillo-Barral N, Oñate-Sánchez L, Carbonero P. 2013. Arabidopsis thaliana bZIP44: a transcription factor affecting seed germination and expression of the mannanase-encoding gene AtMAN7. The Plant Journal 74, 767–780. [DOI] [PubMed] [Google Scholar]
- Iglesias-Fernández R, Rodríguez-Gacio MC, Barrero-Sicilia C, Carbonero P, Matilla A. 2011. Three endo-β-mannanase genes expressed in the micropylar endosperm and in the radicle influence germination of Arabidopsis thaliana seeds. Planta 233, 25–36. [DOI] [PubMed] [Google Scholar]
- Iglesias-Fernández R, Wozny D, Iriondo-de Hond M, Oñate-Sánchez L, Carbonero P, Barrero-Sicilia C. 2014. The AtCathB3 gene, encoding a cathepsin B-like protease, is expressed during germination of Arabidopsis thaliana and transcriptionally repressed by the basic leucine zipper protein GBF1. Journal of Experimental Botany 65, 2009–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. 2002. bZIP transcription factors in Arabidopsis. Trends in Plant Science 7, 106–111. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodaira KS, Qin F, Tran LS, Maruyama K, Kidokoro S, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K. 2011. Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiology 157, 742–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Baier M, Horling F, Kahmann U, Harris G, Schurmann P, Dietz KJ. 2002. The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of photosynthetic electron flux. Proceedings of the National Academy of Sciences, USA 99, 5738–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi C, Mestres-Ortega D, Marco Y, Meyer Y, Reichheld JP. 2004. The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor. Plant Physiology 134, 1006–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi C, Rayapuram N, Chartier Y, Grienenberger JM, Bonnard G, Meyer Y. 2001. Identification and characterization of a mitochondrial thioredoxin system in plants. Proceedings of the National Academy of Sciences, USA 98, 14144–14149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara P, Oñate-Sánchez L, Abraham Z, Ferrándiz C, Díaz I, Carbonero P, Vicente-Carbajosa J. 2003. Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. The Journal of Biological Chemistry 278, 21003–21011. [DOI] [PubMed] [Google Scholar]
- Lázaro JJ, Jiménez A, Camejo D, Iglesias-Baena I, Martí Mdel C, Lázaro-Payo A, Barranco-Medina S, Sevilla F. 2013. Dissecting the integrative antioxidant and redox systems in plant mitochondria. Effect of stress and S-nitrosylation. Frontiers in Plant Science 4, 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RL, Garland D, Oliver CN, Amici A, Climent I, Lenz AG, Ahn BW, Shaltiel S, Stadtman ER. 1990. Determination of carbonyl content in oxidatively modified proteins. Methods in Enzymology 186, 464–478. [DOI] [PubMed] [Google Scholar]
- Lin Y, Yang L, Paul M, Zu Y, Tang Z. 2013. Ethylene promotes germination of Arabidopsis seed under salinity by decreasing reactive oxygen species: evidence for the involvement of nitric oxide simulated by sodium nitroprusside. Plant Physiology and Biochemistry 73, 211–218. [DOI] [PubMed] [Google Scholar]
- Lozano R, Wong J, Yee B, Peters A, Kobrehel K, Buchanan B. 1996. New evidence for a role for thioredoxin h in germination and seedling development. Planta 200, 100–106. [Google Scholar]
- Martí MC, Florez-Sarasa I, Camejo D, Ribas-Carbó M, Lázaro JJ, Sevilla F, Jiménez A. 2011. Response of mitochondrial thioredoxin PsTrxo1, antioxidant enzymes, and respiration to salinity in pea (Pisum sativum L.) leaves. Journal of Experimental Botany 62, 3863–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí MC, Olmos E, Calvete JJ, Díaz I, Barranco-Medina S, Whelan J, Lázaro JJ, Sevilla F, Jiménez A. 2009. Mitochondrial and nuclear localization of a novel pea thioredoxin: identification of its mitochondrial target proteins. Plant Physiology 150, 646–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Y, Belin C, Delorme-Hinoux V, Reichheld JP, Riondet C. 2012. Thioredoxin and glutaredoxin systems in plants: molecular mechanisms, crosstalks, and functional significance. Antioxidants & Redox Signaling 17, 1124–1160. [DOI] [PubMed] [Google Scholar]
- Meyer Y, Buchanan BB, Vignols F, Reichheld JP. 2009. Thioredoxins and glutaredoxins: unifying elements in redox biology. Annual Review of Genetics 43, 335–367. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. 2011. ROS signaling: the new wave? Trends in Plant Science 16, 300–309. [DOI] [PubMed] [Google Scholar]
- Montrichard F, Alkhalfioui F, Yano H, Vensel WH, Hurkman WJ, Buchanan BB. 2009. Thioredoxin targets in plants: the first 30 years. Journal of Proteomics 72, 452–474. [DOI] [PubMed] [Google Scholar]
- Montrichard F, Renard M, Alkhalfioui F, Duval FD, Macherel D. 2003. Identification and differential expression of two thioredoxin h isoforms in germinating seeds from pea. Plant Physiology 132, 1707–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A. 2014. Glutathione and NADPH in plant responses to H2O2. Free Radical Biology & Medicine 75, S3–S4. [DOI] [PubMed] [Google Scholar]
- Nonogaki H. 2014. Seed dormancy and germination-emerging mechanisms and new hypotheses. Frontiers in Plant Science 5, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. Journal of Molecular Biology 302, 205–217. [DOI] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Vicente-Carbajosa J. 2008. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Research Notes 1, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido P, Cazalis R, Cejudo FJ. 2009. An antioxidant redox system in the nucleus of wheat seed cells suffering oxidative stress. The Plant Journal 57, 132–145. [DOI] [PubMed] [Google Scholar]
- Puntarulo S, Galleano M, Sanchez RA, Boveris A. 1991. Superoxide anion and hydrogen peroxide metabolism in soybean embryonic axes during germination. Biochimica et Biophysica Acta 1074, 277–283. [DOI] [PubMed] [Google Scholar]
- Renard M, Alkhalfioui F, Schmitt-Keichinger C, Ritzenthaler C, Montrichard F. 2011. Identification and characterization of thioredoxin h isoforms differentially expressed in germinating seeds of the model legume Medicago truncatula. Plant Physiology 155, 1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey P, Bécuwe N, Barrault MB, Rumeau D, Havaux M, Biteau B, Toledano MB. 2007. The Arabidopsis thaliana sulfiredoxin is a plastidic cysteine-sulfinic acid reductase involved in the photooxidative stress response. The Plant Journal 49, 505–514. [DOI] [PubMed] [Google Scholar]
- Rhoads DM, Umbach AL, Sweet CR, Lennon AM, Rauch GS, Siedow JN. 1998. Regulation of the cyanide-resistant alternative oxidase of plant mitochondria. Identification of the cysteine residue involved in alpha-keto acid stimulation and intersubunit disulfide bond formation. The Journal of Biological Chemistry 273, 30750–30756. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K. 2004. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiology 136, 2734–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L, Albertos P, Mateos I, Sánchez-Vicente I, Lechón T, Fernández-Marcos M, Lorenzo O. 2015. Nitric oxide (NO) and phytohormones crosstalk during early plant development. Journal of Experimental Botany 66, 2857–2868. [DOI] [PubMed] [Google Scholar]
- Serrato AJ, Cejudo FJ. 2003. Type-h thioredoxins accumulate in the nucleus of developing wheat seed tissues suffering oxidative stress. Planta 217, 392–399. [DOI] [PubMed] [Google Scholar]
- Sevilla F, Camejo D, Ortiz-Espín A, Calderón A, Lázaro JJ, Jiménez A. 2015. The thioredoxin/peroxiredoxin/sulfiredoxin system: current overview on its redox function in plants and regulation by reactive oxygen and nitrogen species. Journal of Experimental Botany 66, 2945–2955. [DOI] [PubMed] [Google Scholar]
- Shahpiri A, Svensson B, Finnie C. 2008. The NADPH-dependent thioredoxin reductase/thioredoxin system in germinating barley seeds: gene expression, protein profiles, and interactions between isoforms of thioredoxin h and thioredoxin reductase. Plant Physiology 146, 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simontacchi M, Caro A, Fraga CG, Puntarulo S. 1993. Oxidative stress affects [alpha]-tocopherol content in soybean embryonic axes upon imbibition and following germination. Plant Physiology 103, 949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangeland B, Salehian Z. 2002. An improved clearing method for GUS assay in Arabidopsis endosperm and seeds. Plant Molecular Biology Reporter 20, 107–114. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverso JA, Micalella C, Martinez A, Brown SC, Satiat-Jeunemaître B, Meinnel T, Giglione C. 2013. Roles of N-terminal fatty acid acylations in membrane compartment partitioning: Arabidopsis h-type thioredoxins as a case study. The Plant Cell 25, 1056–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach AL, Siedow JN. 1993. Covalent and noncovalent dimers of the cyanide-resistant alternative oxidase protein in higher plant mitochondria and their relationship to enzyme activity. Plant Physiology 103, 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacker H, Sandalio L, Jiménez A, et al. 2006. Roles for redox regulation in leaf senescence of pea plants grown on different sources of nitrogen nutrition. Journal of Experimental Botany 57, 1735–1745. [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Carbonero P. 2005. Seed maturation: developing an intrusive phase to accomplish a quiescent state. The International Journal of Developmental Biology 49, 645–651. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. The Plant Journal 33, 949–956. [DOI] [PubMed] [Google Scholar]
- Wojtyla Ł, Lechowska K, Kubala S, Garnczarska M. 2016. Different modes of hydrogen peroxide action during seed germination. Frontiers in Plant Science 7, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JH, Cai N, Balmer Y, Tanaka CK, Vensel WH, Hurkman WJ, Buchanan BB. 2004. Thioredoxin targets of developing wheat seeds identified by complementary proteomic approaches. Phytochemistry 65, 1629–1640. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Bhuiyan MN, Waditee R, Tanaka Y, Esaka M, Oba K, Jagendorf AT, Takabe T. 2005. Suppressed expression of the apoplastic ascorbate oxidase gene increases salt tolerance in tobacco and Arabidopsis plants. Journal of Experimental Botany 56, 1785–1796. [DOI] [PubMed] [Google Scholar]
- Yang B, Kotani A, Arai K, Kusu F. 2001. Estimation of the antioxidant activities of flavonoids from their oxidation potentials. Analytical Sciences 17, 599–604. [DOI] [PubMed] [Google Scholar]
- Zhang H, Hu H, Greeley N, et al. 2014. STAT3 restrains RANK- and TLR4-mediated signalling by suppressing expression of the E2 ubiquitin-conjugating enzyme Ubc13. Nature Communications 5, 5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.