Highlight
bHLH115 functions downstream of the E3 ligase BRUTUS and positively controls the expression of the iron-deficiency responsive bHLH transcription factors POPEYE and bHLH38/39/100/101 for iron homeostasis in Arabidopsis.
Keywords: Arabidopsis, Fe deficiency, bHLH115, BTS, iron, PYE.
Abstract
Iron (Fe) deficiency is a limiting factor for the normal growth and development of plants, and many species have evolved sophisticated systems for adaptation to Fe-deficient environments. It is still unclear how plants sense Fe status and coordinate the expression of genes responsive to Fe deficiency. In this study, we show that the bHLH transcription factor bHLH115 is a positive regulator of the Fe-deficiency response. Loss-of-function of bHLH115 causes strong Fe-deficiency symptoms and alleviates expression of genes responsive to Fe deficiency, whereas its overexpression causes the opposite effect. Chromatin immunoprecipitation assays confirmed that bHLH115 binds to the promoters of the Fe-deficiency-responsive genes bHLH38/39/100/101 and POPEYE (PYE), which suggests redundant molecular functions with bHLH34, bHLH104, and bHLH105. This is further supported by the fact that the bhlh115-1 mutant was complemented by overexpression of any of bHLH34, bHLH104, bHLH105, and bHLH115. Further investigations determined that bHLH115 could interact with itself and with bHLH34, bHLH104, and bHLH105. Their differential tissue-specific expression patterns and the severe Fe deficiency symptoms of multiple mutants supported their non-redundant biological functions. Genetic analysis revealed that bHLH115 is negatively regulated by BRUTUS (BTS), an E3 ligase that can interact with bHLH115. Thus, bHLH115 plays key roles in the maintenance of Fe homeostasis in Arabidopsis thaliana.
Introduction
Iron (Fe) is an important micronutrient for plant growth and development. Its role as a cofactor in the reduction–oxidation reaction makes it indispensable for many essential protein and enzymatic processes, including photosynthesis and respiration (Marschner, 1995; de Benoist et al., 2008). Although Fe is abundant, it mainly exists in the form of insoluble ferric oxides not directly available to plants; this is especially the case in aerobic or alkaline soils (Guerinot and Yi, 1994). Iron deficiency can limit plant growth, and subsequently reduces crop yield and quality (Guerinot and Yi, 1994; Marschner, 1995; Jeong and Guerinot, 2009; Kobayashi and Nishizawa, 2012). Iron overload can also be toxic; this is due to the production of hydroxyl radicals via the Fenton reaction. Therefore, the maintenance of Fe homeostasis is crucial for plant growth and development.
There are two major ways in which plants acquire Fe from the soil. To overcome iron-limiting conditions, non-graminaceous (strategy I) plants employ a reduction strategy to acquire Fe from soil (Hell and Stephan, 2003; Walker and Connolly, 2008; Hindt and Guerinot, 2012), whereas graminaceous plants utilize strategy II, in which phytosiderophores are released into the rhizosphere to chelate Fe (Walker and Connolly, 2008; Hindt and Guerinot, 2012). Strategy I involves three processes: rhizosphere acidification, reduction, and uptake. Upon iron deficiency, H+-ATPases pump protons out of the cells to reduce the rhizosphere pH, thus improving solubility of ferric Fe (Santi and Schmidt, 2009). Following acidification, the plasma membrane-bound FERRIC REDUCTION OXIDASE 2 (FRO2) reduces Fe on the root surface from ferric to ferrous Fe (Robinson et al., 1999). Finally, the high-affinity ferrous Fe transporter IRON-REGULATED TRANSPORTER 1 (IRT1) takes up ferrous Fe into the root epidermis (Henriques et al., 2002; Varotto et al., 2002; Vert et al., 2002). Fe absorbed by the roots is chelated with citrate or nicotianamine, and subsequently these complexes are translocated to other tissues and organs (Rogers and Guerinot, 2002; Haydon and Cobbett, 2007; Curie et al., 2009).
The regulation of transcription factors is essential for accurate control of Fe homeostasis. Several transcription factors have been characterized as regulating Fe homeostasis. FIT (FER-like Iron-deficiency-induced Transcription Factor) encodes a bHLH protein, and is a homologous gene of FER in tomato (Solanum lycopersicum) (Ling et al., 2002; Colangelo and Guerinot, 2004; Yuan et al., 2005). FIT is induced in the epidermis of the root upon iron deficiency (Colangelo and Guerinot, 2004). The fit mutant is chlorotic, and its seedling can not survive without an extra Fe supply. Expression of the Fe uptake-associated genes IRT1 and FRO2 is greatly reduced in the fit mutant (Colangelo and Guerinot, 2004). Overexpressing FIT alone does not increase FRO2 and IRT1 expression unless FIT is co-expressed with one of the bHLH subgroup Ib members (bHLH38/39/100/101) (Colangelo and Guerinot, 2004; Yuan et al., 2008; Wang et al., 2013). FIT and bHLH38/39/100/101 function as positive regulators of IRT1 and FRO2. Another bHLH family member, POPEYE (PYE), participates in Fe homeostasis by negatively regulating the expression of NICOTIANAMINE SYNTHASE 4 (NAS4), FRO3, and ZINC-INDUCED FACILITATOR1 (ZIF1) (Long et al., 2010). Recent studies suggest that bHLH34, bHLH104, and bHLH105 play key roles in maintaining Fe homeostasis by positively regulating bHLH38/39/100/101 and PYE (Zhang et al., 2015; Li et al., 2016).
Plants can sense internal and external Fe concentrations and they modulate gene expression to maintain intercellular Fe homeostasis. In mammals, FBXL5 (F-BOX AND LEUCINE-RICH REPEAT PROTEIN 5) was identified as a Fe sensor. FBXL5 contains a hemerythrin domain (HHE) for Fe binding, and an F-box domain for ubiquitination and degradation of IRP2 (IRON REGULATORY PROTEIN2), which participates in the regulation of Fe homeostasis (Salahudeen et al., 2009; Vashisht et al., 2009). Similar to FBXL5, BTS (BRUTUS) in Arabidopsis possesses HHE domains; these bind to the Fe ion and RING domains present in many E3 ligases (Selote et al., 2015). Moreover, BTS has a ubiquitination function and binds Fe –hence it is a potential Fe sensor. BTS interacts with PYE-like proteins (bHLH105/ILR3, bHLH104, and bHLH115) and targets bHLH105 and bHLH115 for 26S proteasome-mediated degradation (Selote et al., 2015). Consistent with these observations, BTS negatively regulates Fe homeostasis (Selote et al., 2015; Zhang et al., 2015). Despite the degradation of bHLH115 by BTS, it remains unclear how bHLH115 participates in the regulation of Fe homeostasis.
In this study, we investigated the function of bHLH115 by using a reverse genetic method. bhlh115 mutants displayed enhanced sensitivity to Fe deficiency, whereas overexpression of bHLH115 promoted Fe accumulation. Consistent with this, bHLH115 positively regulated expression of Fe-deficiency-responsive genes. Further analyses indicated similar molecular functions and different biological functions among bHLH subgroup IVc members. Genetic analysis also confirmed negative regulation of bHLH115 by BTS.
Materials and methods
Plant materials and growth conditions
The Arabidopsis thaliana ecotype Col-0 was used in this study. T-DNA insertion lines for bhlhl115-1 (WiscDsLox384C11) andbhlh115-2 (WiscDsLox384D9) were sourced from the Arabidopsis Biological Resource Center (ABRC; https://abrc.osu.edu/) and were confirmed using PCR with a T-DNA primer (p745) and gene-specific primers (see Supplementary Table S1 at JXB online). The bhlh34, bhlh104, bhlh105, and bts-1 mutants have been described previously (Long et al., 2010; Li et al., 2016). The homozygous mutants were crossed with each other to generate double- and triple-mutants. All knockout lines were confirmed by PCR analysis.
For phenotypic analyses of the Fe deprivation response, seeds were surface-sterilized and vernalized for 2–4 d at 4 °C before being plated on Murashige and Skoog (MS) medium. Fe-sufficient (+Fe) medium was half-MS medium with 1%sucrose, 0.8% agar, and 0.1 mM FeEDTA. Fe deficient (–Fe) medium was the same but without iron. For –Fe +Frz media, Fe-deficient (–Fe) medium was supplied with 0.05mM ferrozine. Plates were placed in a culture room at 22 °C under a 16-h light/8-h dark photoperiod.
Plasmid construction and plant transformation
Standard molecular biology techniques were used for the cloning procedures. Genomic DNA from Arabidopsis ecotype Col-0 was used as the template for amplification of the upstream regulatory promoter sequence for ProbHLH115:GUS. For the generation of bHLH115-OX transgenic plants, one HA tag-fused bHLH115 open reading frame was cloned into the binary vector pOCA30 in the sense orientation behind the CaMV 35S promoter. The bHLH105-OX construct was constructed by a similar method. bHLH34-OX and bHLH104-OX have been described previously (Li et al., 2016). Agrobacterium tumefaciens strain EHA105 was used for transformation into Col-0 plants through the floral dipping method (Clough and Bent, 1998). Transgenic plants were selected on half-strength MS agar plates with 50 µg ml–1 kanamycin.
Gene expression analysis
One microgram of total RNA extracted using the Trizol reagent (Invitrogen) was used for oligo(dT)18 primed cDNA synthesis according to the reverse-transcription protocol (Fermentas). The resulting cDNA was subjected to relative quantitative PCR using a SYBR Premix Ex TaqTM kit (TaKaRa) on a Roche LightCycler 480 real-time PCR machine, according to the manufacturer’s instructions. ACTIN2 (ACT2) was amplified as an internal control, and gene copy number was normalized to that of ACT2. For the quantification of each gene, at least three biological replicates were used. Each biological replicate contained three technical replicates. Analysis for statistical significance was performed using Student’s t-test. The qRT-PCR primers have been described previously (Li et al., 2016).
Histochemical GUS staining
GUS staining of ProbHLH115:GUS was performed using the following procedure. Briefly, fresh seedlings were fixed in ice-cold 90% (v/v) acetone for 20 min and then vacuum-filtrated for 10 min with staining solution containing 0.5 mg ml–1 5-bromo-4-chloro-3-indolyl-b-gl-ucuronic acid (Sigma) in 0.1M sodium phosphate buffer (pH 7.3). Then the samples were incubated in the dark at 37 °C until a blue-indigo color appeared. After the reaction, seedlings were rinsed in 0.1M sodium phosphate buffer (pH 7.3). The samples were then rinsed twice in 70% ethanol to remove chlorophylls.
Measurement of Fe content
Seeds were germinated and grown on iron-sufficient media for 10 d. Then fresh shoot samples were harvested separately and dried for 3d in an oven at 65 °C. Analysis of Fe content was performed using inductively coupled plasma spectroscopy. Means and standard deviations were calculated from three biological replications.
Ferric chelate reductase assays
Ferric chelate reductase assays were performed as previously described (Yi and Guerinot, 1996) with some modifications. Briefly, seeds were germinated, and grown on iron-sufficient media for 4 d, and then transferred to iron-sufficient or -deficient plates for 3 d. Roots of 10 seedlings were then weighed and placed in assay solution containing 0.1 mM Fe(III)-EDTA and 0.3 mM ferrozine. The roots were incubated in the dark for 20 min. An identical assay solution containing no plants was used as a blank. The purple-colored Fe(II)-ferrozine complex was measured at 562nm using a molar extinction coefficient of 28.6 mM–1 cm–1.
Chlorophyll measurement
Chlorophyll content was measured in 2-week-old plants grown on Fe-sufficient and Fe-deficient media. All leaves are collected and ground to powder in liquid nitrogen. The powder was resuspended in 80% acetone on ice and centrifuged for 5 min at 10000 g at 4 °C. Chlorophyll concentrations were calculated from spectroscopy absorbance measurements at 663.2, 646.8, and 470 nm (Lichtenthaler, 1987).
Chromatin immunoprecipitation (ChIP)assays
ChIP assays were performed essentially according to previously described protocols (Saleh et al., 2008). Wild-type and bHLH115-OX-14 transgenic plants grown on –Fe media for 2 weeks were used for ChIP assays. To quantify bHLH115-DNA binding, qPCR was performed according to the procedure described previously with the TUBIN2 as the endogenous control (Mukhopadhyay et al., 2008). All primers are listed in Supplementary Table S1. For the quantification of each DNA fragment, three biological replicates were used. Each biological replicate contained three technical replicates.
Transient expression assays
Agrobacterium tumefaciens strain EHA105 was used to infect tobacco leaves. Agrobacterial cells containing plasmids were infiltrated into leaves of Nicotiana benthamiana by an infiltration buffer (0.2 mM acetosyringone, 10 mM MgCl2, and 10 mM MES, PH 5.6). For the bimolecular fluorescent complementation (BiFC) assays, equal volumes of an Agrobacterium culture were mixed before infiltration into N. benthamiana leaves. After infiltration, yellow fluorescence protein (YFP) and DAPI fluorescence were observed under a confocal laser scanning microscope (Olympus, Tokyo, Japan). For the transcription activation assay, the final OD600 values were 0.1 (an internal control; Pro35S:Myc-GUS), 0.5 (reporter), and 0.5 (effector). After infiltration, plants were placed in the dark at 24 °C for 48 h before RNA extraction. The transcript abundance of GFP was normalized to GUS.
Yeast assays
For the yeast two-hybrid (Y2H) assay, the N-terminal truncated bHLH115N containing the bHLH domain was cloned into pGBKT7, and the full-length ORF of bHLH115was cloned into pGADT7. GAD-bHLH34, GAD-bHLH104, and GAD-bHLH105 have been described previously (Li et al., 2016). The combination of pGBKT7-53 and pGADT7-T was used as a positive control and the combination of pGBKT7-Lam and pGADT7-T as a negative control. Growth was determined as described in the Yeast Two-Hybrid System User Manual (Clontech). Primers used for the vector construction are listed in Supplementary Table S1. Experiments were repeated three times.
Accession numbers
Sequence data generated in this study have been lodged in the Arabidopsis Genome Initiative and GenBank/EMBL databases under the following accession numbers: bHLH34 (At3g23210), bHLH104 (At4g14410), bHLH105 (At5g54680), bHLH115 (At1g51070), bHLH38 (At3g56970), bHLH39 (At3g56980), bHLH100 (At2g41240), bHLH101 (At5g04150), FIT (At2g28160), MYB10 (At3g12820), MYB72 (At1g56160), FRO2 (At1g01580), IRT1 (At4g19690), PYE (At3g47640), BTS (At3g18290), and ACT2 (At3g18780).
Results
Loss-of-function of bHLH115 alters response to Fe deficiency
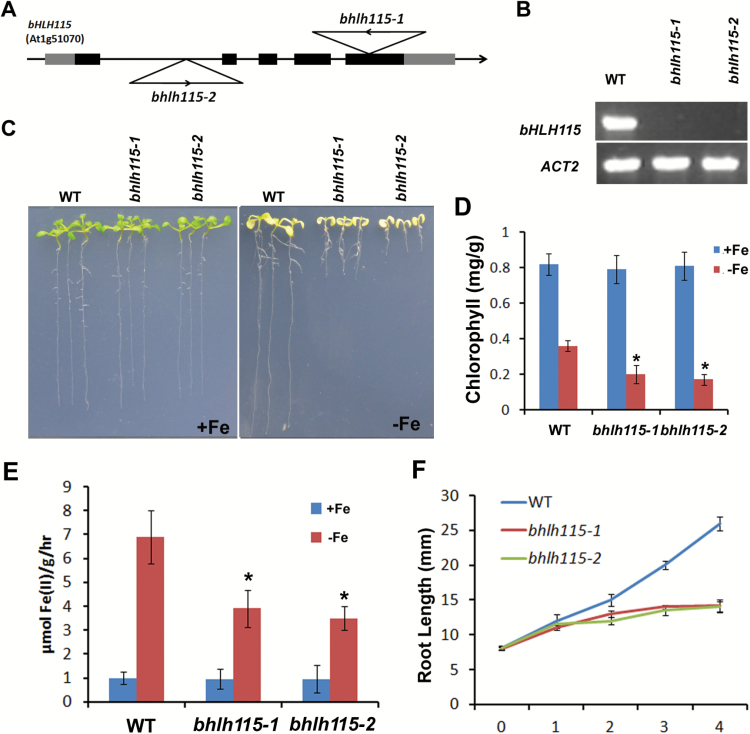
Three members of the bHLH subgroup IVc (Heim et al., 2003), bHLH34, bHLH104, and bHLH105, regulate Fe homeostasis (Zhang et al., 2015; Li et al., 2016); however, the functions of the other member, bHLH115, are not clear. We therefore screened two T-DNA insertion lines of the bHLH115 gene sourced from the Arabidopsis Biological Resource Center (Fig. 1A). Two homologous mutants, bhlh115-1 and bhlh115-2, were identified as null mutants, exhibiting loss of the full-length bHLH115 transcript (Fig. 1B). When grown vertically on Fe-sufficient medium for 10 d, no visible differences were observed between the wild-type and the two mutants. However, under Fe-deficient conditions the two mutants developed significantly shorter roots compared with the wild-type (Fig. 1C). Further investigation revealed that both the bhlh115-1 and bhlh115-2 mutants had lower chlorophyll content compared with the wild-type plants under Fe-deficient conditions, but not under Fe-sufficient conditions (Fig. 1D). Elevation of ferric chelate reductase activity in roots is a characteristic of Fe deficiency in Arabidopsis thaliana (Yi and Guerinot, 1996). A ferrozine assay was therefore used to quantify the ferric chelate reductase activity in the roots of wild-type and bhlh115 mutants. Under Fe-sufficient conditions, the ferric chelate reductase activity of the bhlh115 mutants was similar to that of the wild-type, whereas under Fe-deficient conditions activity was significantly lower in the mutants (Fig. 1E). To further investigate root responses to Fe deficiency, 4-d-old seedlings grown vertically on Fe-sufficient medium were transferred to Fe-deficient medium for 4 d. Compared with wild-type plants, primary root elongation of the mutants was strongly inhibited by Fe deficiency (Fig. 1F).
Fig. 1.
Characterization of bhlh115 mutants. (A) Location of T-DNA in the bHLH115 gene. Thick bars and lines indicate the exons and introns, respectively. Grey bars indicate the 5′ and 3′ untranslated regions. Triangles indicate the location of the T-DNA. (B) Confirmation of the bhlh115-1 and bhlh115-2 mutants. The full-length cDNA of bHLH115 was amplified by RT-PCR. (C)Images of 10-d-old seedlings that were germinated directly on +Fe or –Fe media. (D) Chlorophyll content of leaves on +Fe or –Fe media. Significant differences from the wild-type were determined by Student’s t-test, *P<0.05). (E) Iron reductase activity. Seedlings were germinated and grown on +Fe medium for 10 d and then transferred to –Fe media for 3 d. Significant differences from the wild-type were determined by Student’s t-test, *P<0.05). (F) Time-course quantification of root length of plants from 0 to 4 d after transfer to –Fe medium.
Overexpression of bHLH115 causes increased Fe accumulation
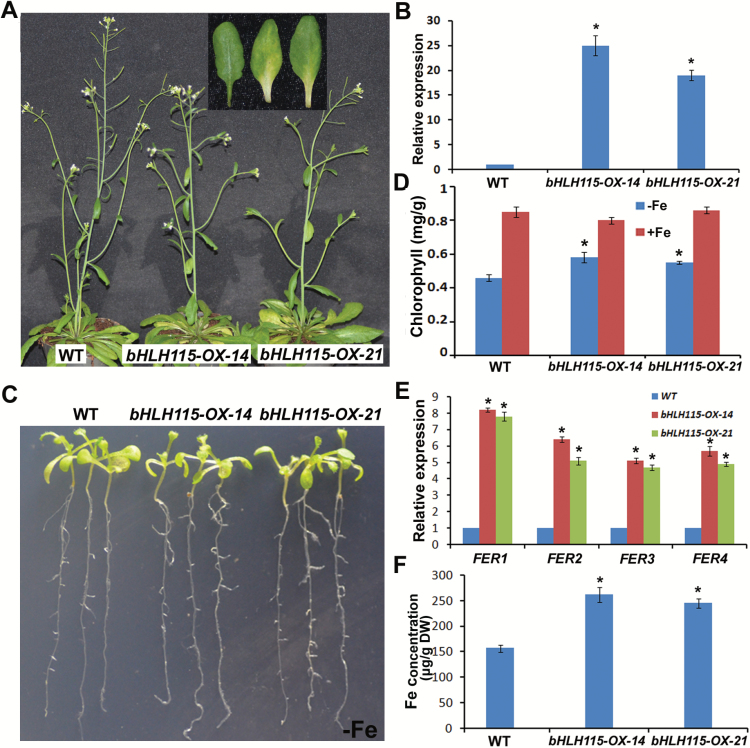
To further investigate the functions of bHLH115, we constructed bHLH115-OX transgenic plants in which the CaMV 35S promoter was used to drive expression of a HA-bHLH115 fusion gene. Forty-nine bHLH115-OX transgenic plants were obtained, 40 of which produced chlorotic rosette leaves at the reproductive stage (Fig. 2A). Two independent transgenic plant lines with elevated bHLH115 levels were used for further analysis (Fig. 2B). Considering the weak Fe-deficiency response of the bhlh115 mutants, we examined whether bHLH115-OX plants had an opposite response. When subjected to Fe-deficient medium, bHLH115-OX plant leaves were greener than those of the wild-type (Fig. 2C). Consistent with this, the chlorophyll content of bHLH115-OX leaves was higher than in the wild-type (Fig. 2D). However, they developed identical root lengths irrespective of Fe status (Fig. 2C; Supplementary Fig. S1). Having confirmed a bHLH115-regulated plant response to Fe deficiency, we speculated that bHLH115 might affect Fe accumulation in plants. Moreover, ferritins are Fe-storage proteins that sequester or release Fe upon demand (Harrison and Arosio, 1996). There are four FER genes encoding ferritins in Arabidopsis, and their expression increases in response to Fe overloading (Petit et al., 2001). Expression analysis indicated that all four FER genes were significantly up-regulated in bHLH115-OX leaves compared with the wild-type (Fig. 2E). Next, we examined the Fe concentration of 10-d-old shoots grown in Fe-sufficient conditions (Fig. 2F), and found that bHLH115-OX shoots had a higher Fe content than the wild-type.
Fig. 2.
Phenotypes of bHLH115-OX transgenic plants. (A) Images of 50-d-old plants that were grown in soil at pH 6.0–6.5. The seventh or eighth leaves are shown in the inset, from left to right: wild-type, bHLH115-OX-14, and bHLH115-OX-21. (B) Relative expression of bHLH115 in the transgenic plants. (C) Images of 10-d-old seedlings that were germinated directly on –Fe media. (D) Chlorophyll content of leaves on –Fe or +Fe media. Significant differences from the wild-type were determined by Student’s t-test, *P<0.05). (E) Relative expression of FER genes in the leaves. Shoots of 10-d-old seedlings germinated directly on +Fe media were used for RNA extraction. Significant differences from the wild-type were determined by Student’s t-test, *P<0.05). (F) Fe concentration in the leaves. Shoots of 10-d-old seedlings germinated directly on +Fe media were used for Fe measurement. Significant differences from the wild-type were determined by Student’s t-test, *P<0.05).
Expression of Fe-deficiency-responsive genes is positively correlated with bHLH115
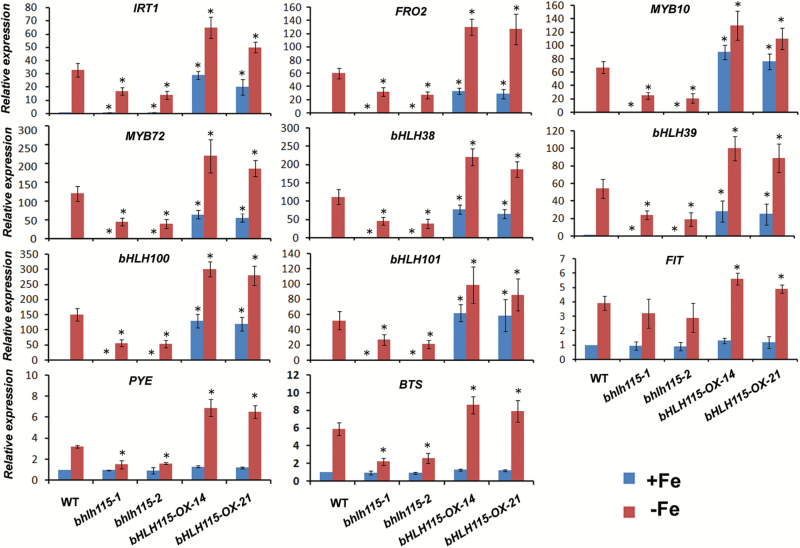
The above findings suggested that bHLH115 positively regulates the Fe-deficiency response and Fe homeostasis. As a transcription factor, bHLH115 is expected to affect the expression of many downstream genes. We therefore examined whether the susceptibility of bhlh115 mutants and the tolerance of bHLH115-OX plants to Fe deficiency were due to the altered expression of Fe-deficiency-responsive genes. The expression of the Fe uptake-associated genes IRT1 and FRO2 increases in response to Fe-deficiency conditions (Robinson et al., 1999; Connolly et al., 2002). Two MYB genes, MYB10 and MYB72, are also induced under Fe-deficient conditions (Palmer et al., 2013). Six bHLH genes, FIT, bHLH38/39/100/101, and PYE, are major regulators of Fe homeostasis and are up-regulated under Fe-deficient conditions (Colangelo and Guerinot, 2004; Wang et al., 2007; Long et al., 2010). Additionally, BTS is induced by Fe deficiency and negatively regulates Fe homeostasis (Selote et al., 2015). Under normal growth conditions, the transcript abundance of these Fe-deficiency genes was low in bhlh115 mutants and high in bHLH115-OX plants compared with the wild-type (Fig. 3). Although Fe-deficiency-responsive genes were induced by Fe deficiency in both bhlh115 mutants and bHLH115-OX plants, expression levels were lower in bhlh115 mutants and higher in bHLH115-OX plants than observed in the wild-type. This suggests that the expression of Fe-deficiency-responsive genes is positively correlated with bHLH115 expression.
Fig. 3.
Expression of Fe-deficiency-responsive genes in wild-type, mutants, and overexpression plants. Wild-type, mutants, and transgenic plants were grown on +Fe medium for 4 d, then transferred to +Fe or –Fe media for 3 d. RNA was prepared from root tissues. The data represent means (±SD) of three biological replicates. Significant differences from the wild-type were determined by Student’s t-test, *P<0.05).
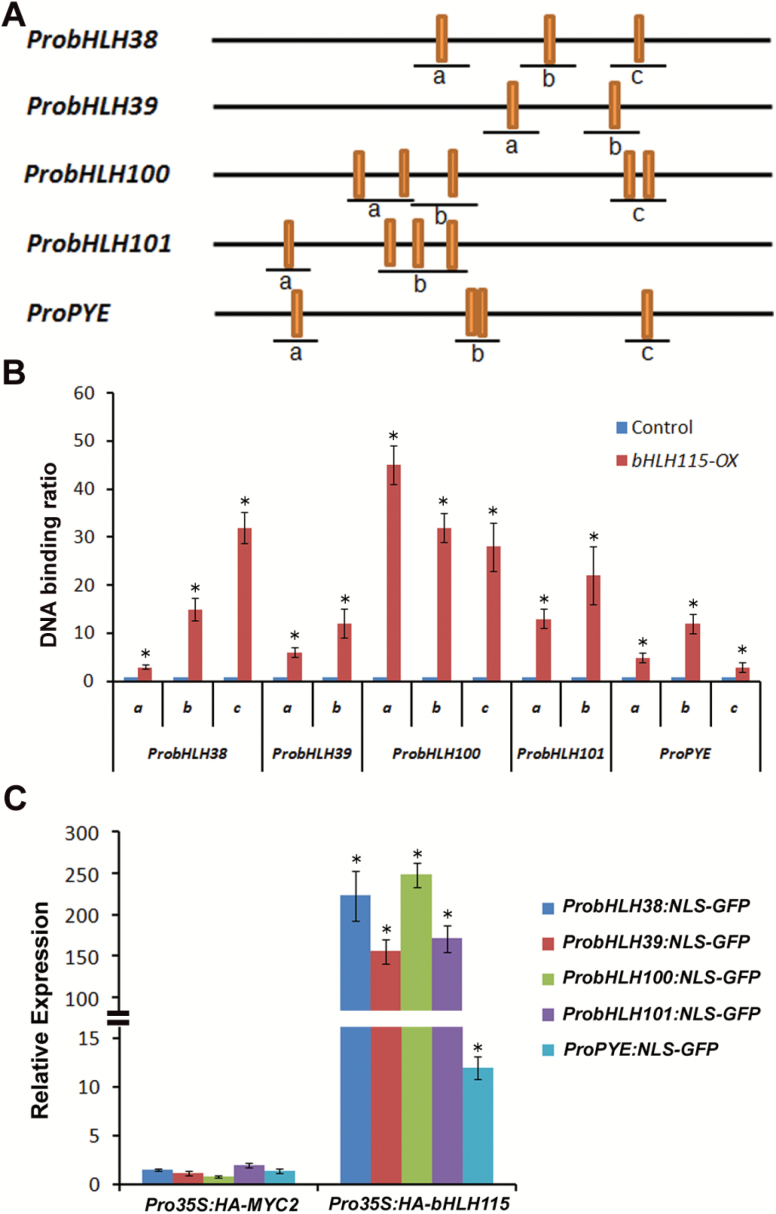
bHLH115 directly activates the transcription of bHLH38/39/100/101 and PYE
As bHLH115 is a close homologue of bHLH34, bHLH104, and bHLH105, which directly activate transcription of bHLH38/39/100/101 and PYE (Zhang et al., 2015; Li et al., 2016), we hypothesized that bHLH115 could also directly activate bHLH38/39/100/101 and PYE. To test this hypothesis, ChIP-PCR experiments were conducted using bHLH115-OX transgenic roots grown under Fe-deficient conditions. These assays revealed that promoter sequences of bHLH38/39/100/101 and PYE spanning E-box motifs (CAXXTG) (Fig. 4A) were highly enriched in anti-HA-immunoprecipitated chromatin in bHLH115-OX transgenic plants but not in wild-type plants, thus demonstrating direct binding of bHLH115 to the promoters of bHLH38/39/100/101 and PYE (Fig. 4B). Transient expression assays were performed to investigate whether bHLH115 activated the above promoters. Pro35S:HA-bHLH115 was used as an effector and ProbHLH38:NLS-GFP as a reporter. Co-expression of Pro35S:HA-MYC2 and ProbHLH38:NLS-GFP resulted in GFP transcript levels similar to that in the co-expression of the empty vector and ProbHLH38:NLS-GFP, whereas the co-expression of Pro35S:HA-bHLH115 and ProbHLH38:NLS-GFP caused high levels of GFP (Fig. 4C). These data suggest that bHLH115 activates the bHLH38 promoter. Using a similar approach, it was confirmed that bHLH115 could also activate the bHLH39, bHLH100, bHLH101, and PYE promoters.
Fig. 4.
Direct regulation of bHLH38/39/100/101 and PYE by bHLH115. (A) E-box in promoters. The bars indicate the E-box (CANNTG). The 1000-bp upstream sequence from the translation start site of the indicated gene is shown. (B) ChIP-qPCR analyses of the DNA binding ratio of bHLH115 to the promoters of bHLH38/39/100/101 and PYE genes. qPCR was used to quantify enrichment of the indicated promoters and a fragment of the TUB2 promoter containing an E-box motif was used as a negative control. The binding to the TUB2 promoter fragment in the wild-type was set to 1 and used to normalize the DNA binding ratio. Significant differences were determined by Student’s t-test, *P<0.05. (C) bHLH115 activates the promoters of bHLH38/39/100/101 and PYE. In the transient assays, Pro35S:Myc-GUS was expressed as an internal control. GFP transcript abundance was normalized to the GUS transcript. The value obtained with the empty vector as an effector was set to 1. The results are means (±SD) of three biological replicates. Significant differences were determined by Student’s t-test, *P<0.05.
bHLH34/104/105/115 share similar molecular functions
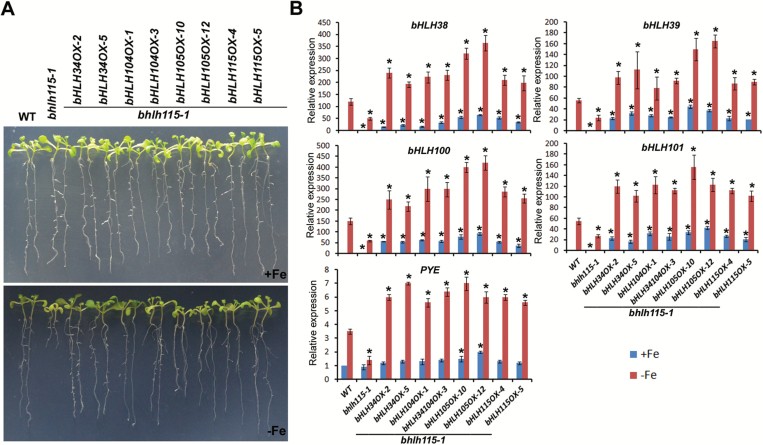
Our results and previous studies show that all four members of the bHLH subgroup IVc can activate expression of bHLH38/39/100/101 and PYE, suggesting their redundant molecular functions. Our previous work confirmed that overexpression of bHLH101 only rescued Fe-deficiency induction of IRT1 and FRO2, but not of MYB10 and MYB72, in the bhlh34 bhlh104 double-mutant (Li et al., 2016). This suggests that unknown target genes of bHLH34 and bHLH104 have yet to be identified. It remains unclear whether the four proteins share the same, as yet unidentified, target genes or have identical regulatory functions. To explore this hypothesis, each of the four genes was driven by the CaMV 35S promoter and separately introduced into the bhlh115-1 mutant plants. Fe-deficiency assays indicated that the overexpression of each gene allowed for complete rescue of Fe-deficiency sensitivity of the bhlh115-1 mutant plants under Fe-deficiency conditions (Fig. 5A). An examination of Fe-deficiency-responsive genes indicated that the expression of bHLH38/39/100/101 and PYE in bhlh115-1 was completely rescued by the overexpression of bHLH subgroup IVc members. These data suggest similar molecular functions among bHLH34/104/105/115.
Fig. 5.
Complementation of bhlh115-1 by bHLH34/104/105/115. (A) Images of 10-d-old seedlings that were germinated directly on +Fe or –Fe media. (B) Plants were grown on +Fe media for 4 d, then transferred to +Fe or –Fe media for 3 d. RNA was prepared from root tissues. The data represent means (±SD) of three biological replicates. Significant differences from the wild-type were determined by Student’s t-test, *P<0.05).
Physiological interactions between bHLH115 and bHLH34/104/105/115
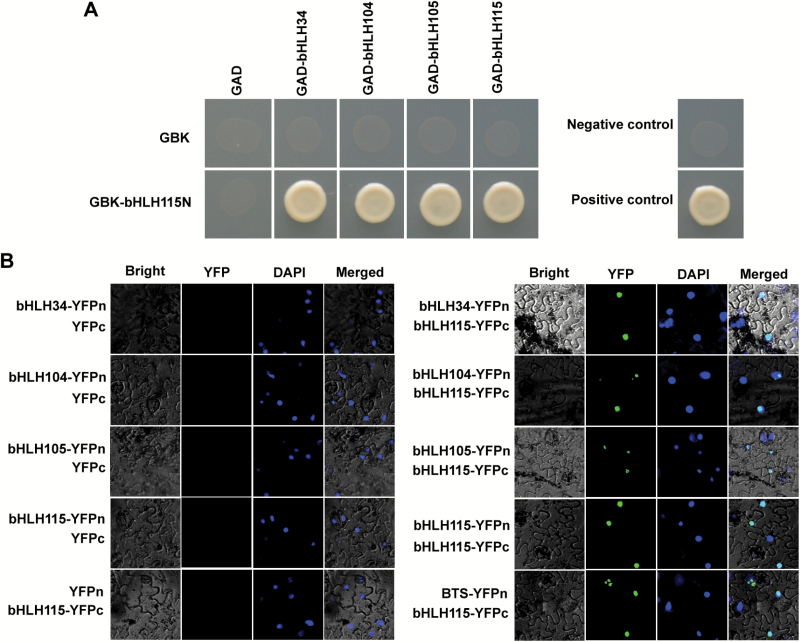
bHLH transcription factors can function as homodimers or heterodimers (Toledo-Ortiz et al., 2003). Our previous study confirmed formation of homodimers or heterodimers between bHLH34, bHLH104, and bHLH105 (Li et al., 2016). Given their similar molecular functions, we examined whether all members of bHLH subgroup IVc could interact with themselves or other members. Yeast-two-hybrid (Y2H) assays were used to test this hypothesis. Owing to the strong autoactivation caused by full-length bHLH115 fused with the BD domain (see Supplementary Fig. S2), a truncated N-terminal bHLH115 (bHLH115N) with no autoactivation was used. Full-length bHLH subgroup IVc members were fused with the AD domain. In yeasts grown on SD medium without Leu/Trp/His/Ade, bHLH115 was found to interact with each member of the bHLH subgroup IVc (Fig. 6A).
Fig. 6.
Interaction between bHLH115 and bHLH34/104/105/115. (A) Yeast two-hybrid assays. Interaction was indicated by the ability of cells to grow on synthetic dropout (SD) medium lacking Leu/Trp/His/Ade. N-terminal truncated bHLH115 was cloned into pGBKT7, and full-length cDNAs of bHLH34, bHLH104, bHLH105, and bHLH115 were cloned into pGADT7. (B) BiFC assays Of N. benthamiana leaves infiltrated with different combinations of the indicated constructs.
Bimolecular fluorescent complementation (BiFC) assays were conducted to further confirm these interactions. Full-length bHLH115 was fused with the C-terminal of YFP (bHLH115-cYFP), and full-length bHLH34/104/105/115 were fused with the N-terminal of YFP (bHLH34-nYFP, bHLH104-nYFP, bHLH105-nYFP, and bHLH115-nYFP), respectively. Transient co-expression assays were conducted in N. benthamiana leaf cells. Strong YFP signals were observed in combinations of bHLH115-cYFP with bHLH34-nYFP, bHLH104-nYFP, bHLH105-nYFP, and bHLH115-nYFP, as well as the positive control BTS-nYFP, with no GFP signals detected in the negative controls (Fig. 6B); this was in agreement with the (Y2H) assay results (Fig. 6A). The fluorescence signals co-localized with the DAPI signal from the nuclei, and this is consistent with activity of transcription factors in the nuclei.
Genetic interactions between bHLH115 and bHLH34/104/105
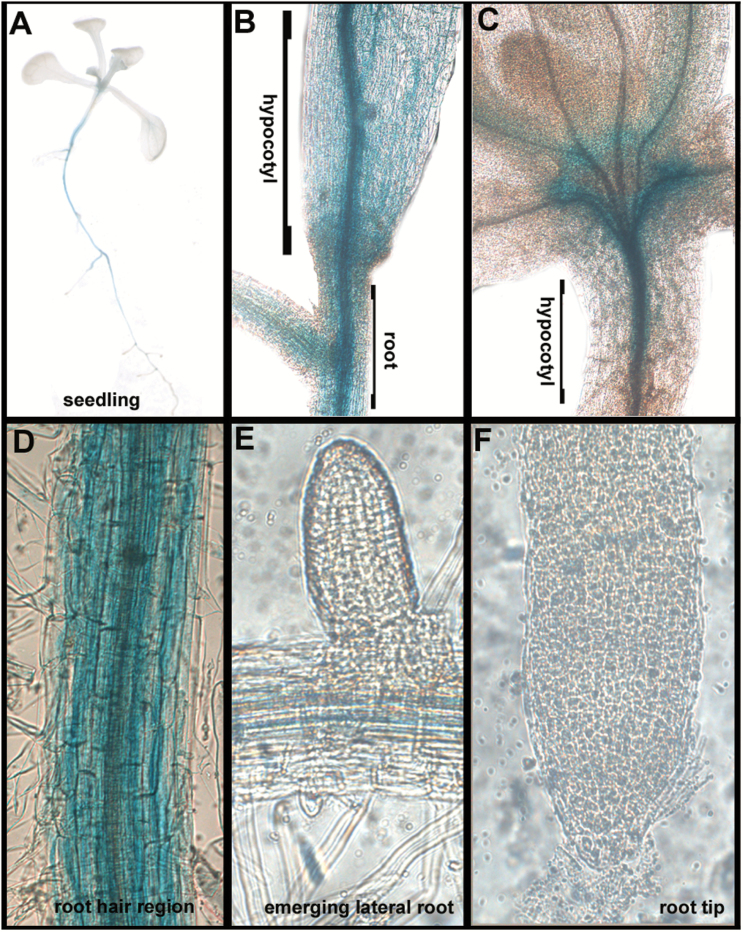
Having confirmed that bHLH34, bHLH104, bHLH105, and bHLH115 have similar regulatory functions, we wanted to examine whether bHLH115 had a specific role in specific tissues or organs. Firstly, expression of bHLH115 was determined after Fe-deficiency treatment, suggesting that its transcription was independent of Fe status (see Supplementary Fig. S3). Then, ProbHLH115:GUS transgenic plants were constructed, in which the GUS reporter gene was driven by the bHLH115 promoter. GUS staining suggested that ProbHLH115:GUS was present in roots, hypocotyls, and shoot meristems (Fig. 7). Further examination of root tissues revealed that ProbHLH115:GUS was expressed in root maturation regions, but not in initial lateral roots and root tips. The expression pattern of bHLH115 was different from that observed for bHLH34, bHLH104, and bHLH105 (Li et al., 2016), implying that bHLH115 may play non-redundant biological functions.
Fig. 7.
Expression of a ProbHLH115:GUS fusion. Images of 10-d-old seedlings that were germinated directly on +Fe medium and used for GUS staining. (A) Seedling, (B) hypocotyl and root, (C) connected region between petioles and hypocotyl, (D) root hair zone, (E) emerging lateral root, and (F) root tip.
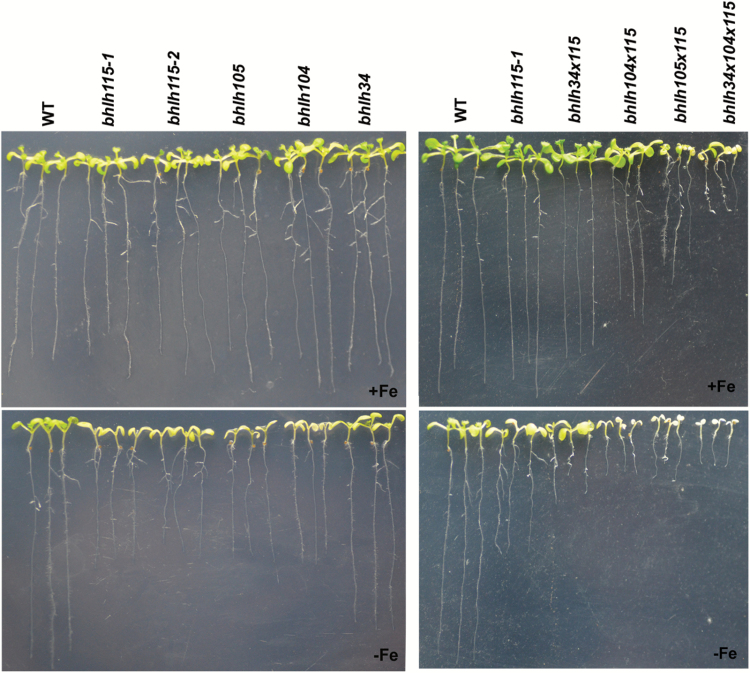
Subsequently, Fe-deficiency phenotypes of bhlh115 mutants were compared with bhlh34, bhlh104, and bhlh105 mutants. This revealed that the Fe-deficiency symptoms of the bhlh115 mutants were similar to those of bhlh104 and bhlh105, but more severe than those of bhlh34 (Fig. 8; Supplementary Fig. S4). This suggests that each of these genes may have an independent biological function. Next, three double-mutants (bhlh34 × 115, bhlh104 × 115, and bhlh105 × 115) and one triple-mutant (bhlh34 × 104 × 115) were constructed by crossing the bhlh115-1 mutant with bhlh34, bhlh104, or bhlh105 (Fig. 8). We failed to obtain other triple-mutants and quadruple-mutants. When grown on Fe-sufficient media, in comparison with the bhlh115-1 single-mutant, all multiple mutants had short roots, with triple-mutant plants having the shortest; bhlh105 × 115 and bhlh34 × 104 × 115 developed severely chlorotic leaves, and exhibited delayed growth. When grown in Fe-deficient media, bhlh104 × 115, bhlh105 × 115, and bhlh34 × 104 × 115 plants developed albino cotyledons and their shoots ceased growing before the emergence of true leaves. To further investigate root elongation in the Fe-deficient medium, 4-d-old seedlings grown in Fe-sufficient medium were transferred to Fe-deficient medium for 3d. As shown in Supplementary Fig. S5, root elongation of the various mutants was inhibited by Fe deficiency. When grown in soil, bhlh115-1, bhlh34 × 115, and bhlh104 × 115 developed normally, as did wild-type plants; in contrast, bhlh105 × 115 and bhlh34 × 104 × 115 plants could not survive unless exogenous Fe was supplied. Even with exogenous Fe application, the growth and development of bhlh105 × 115 and bhlh34 × 104 × 115 plants were severely retarded (see Supplementary Fig. S6). Taken together, bHLH115 has independent roles and also plays an additive role with bHLH34, bHLH104, and bHLH105 in Fe homeostasis.
Fig. 8.
Phenotypes of various mutants. Images of 10-d-old seedlings germinated directly on +Fe or –Fe media.
Genetic interaction between bHLH115 and BTS
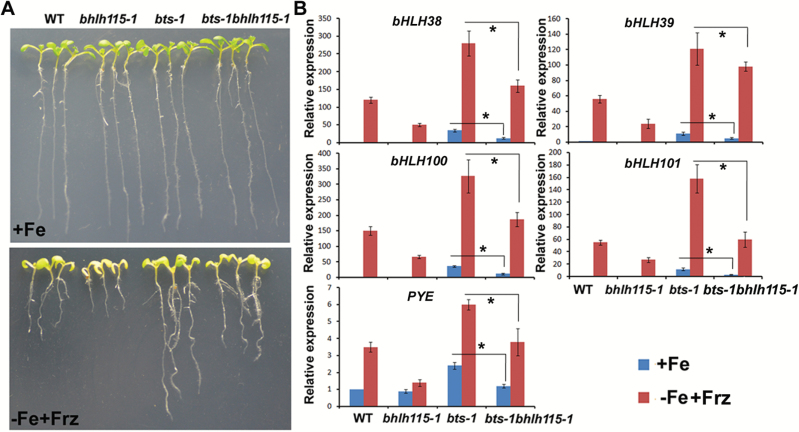
A previous study revealed that BTS interacts with bHLH115 and facilitates 26S proteasome-mediated degradation of bHLH115 proteins in the absence of Fe (Selote et al., 2015). To investigate the genetic interaction between bHLH115 and BTS, the bts-1 bhlh115-1 double-mutant was constructed by crossing bts-1 and bhlh115-1. No visible phenotype differences were observed among bhlh115-1, bts-1, and bts-1 bhlh115-1 under Fe-sufficient conditions. Another previous study confirmed that bts-1 shows enhanced Fe-deficiency tolerance under Fe-free plus ferrozine (Frz) conditions (Long et al., 2010). Consequently, we examined the Fe-deficiency tolerance of various mutants under Fe-free plus Frz conditions. The bts-1 mutant developed the longest roots, whereas bts-1 bhlh115-1 produced significantly shorter roots compared with bts-1 (Fig. 9A), suggesting that loss-of-function of bHLH115 compromised the Fe-deficiency tolerance of bts-1. We wanted to know whether the reduced Fe-deficiency tolerance of bts-1 bhlh115-1 compared with bts-1 resulted from the disrupted expression of Fe-deficiency-responsive genes, and therefore we examined the expression of these genes. Consistent with the Fe-deficiency tolerance of bts-1, the expression of bHLH38/39/100/101 and PYE was dramatically up-regulated under Fe-sufficient or Fe-free plus Frz conditions. Despite the up-regulation of bHLH38/39/100/101 and PYE in the bts-1 bhlh115-1 mutant, their expression in the bts-1 bhlh115-1 double-mutant was significantly lower than in the bts-1 mutant (Fig. 9B).
Fig. 9.
Negative regulation of bHLH115 by BTS. (A) Images of 10-day-old plants that were grown under either Fe-sufficient or Fe-free plus Frz (50 uM) conditions. (B) Expression of Fe-deficiency-responsive genes. Plants were grown on +Fe media for 4 d, then transferred to Fe-sufficient or Fe-free plus Frz media for 3 d. RNA was prepared from root tissues. Significant differences between two samples were determined by Student’s t-test, *P<0.05.
Discussion
Iron plays important roles in the regulation of many biochemical and physiological processes. When plants are exposed to Fe-deficient conditions, they can activate their Fe acquisition system for uptake. Plants have evolved a series of strategies to meet Fe demands, such as facilitating acquisition and remobilizing internal Fe. The intracellular concentration of Fe in plants is tightly controlled through the regulation of many transcription factors. For example, FIT and bHLH38/39/100/101 interact physiologically and positively regulate expression of the strategy-I associated genes IRT1 and FRO2 (Yuan et al., 2008; Wang et al., 2013). MYB10 and MYB72 directly activate NAS4 under Fe-deficient conditions (Palmer et al., 2013). PYE plays a key role in maintaining Fe homeostasis by negatively regulating ZIF1, FRO3, and NAS4 (Long et al., 2010). Previous studies showed that bHLH34/104/105 maintain Fe homeostasis by positively regulating bHLH38/39/100/101 and PYE (Zhang et al., 2015; Li et al., 2016). Here, we show that bHLH115 is a novel regulator of bHLH38/39/100/101 and PYE.
bHLH115 targets bHLH38/39/100/101 and PYE
bHLH115 is a member of the bHLH IVc subgroup. In this study, loss-of-function of bHLH115 caused a weak Fe-deficiency response, with plants displaying shorter roots under Fe-deficiency conditions (Fig. 1C). In contrast, overexpression of bHLH115 caused Fe over-accumulation (Fig. 2F). Thus, appropriate expression of bHLH115 is required for maintenance of Fe homeostasis. Several Fe-deficient mutants (frd1, man, irt1, and fit) display enhanced expression of bHLH38/39/100/101 under both Fe-sufficient and Fe-deficient conditions (Wang et al., 2007). In contrast, their expression was down-regulated in bhlh115 mutants. This suggests that bHLH38/39/100/101 function downstream of bHLH115. ChIP assays confirmed that bHLH115 binds to the promoters of bHLH38/39/100/101 and PYE (Fig. 4B). Transient expression assays indicated that bHLH115 positively regulates expression of bHLH38/39/100/101 and PYE (Fig. 4C). As target genes of bHLH115, bHLH38/39/100/101 were all up-regulated in bHLH115-OX plants (Fig. 3). Moreover, FIT expression is up-regulated in bHLH115-OX plants (Fig. 3). In agreement that FIT and bHLH38/39/100/101 synergistically regulate Fe uptake, expression of the Fe uptake-associated genes IRT1 and FRO2 was also activated in bHLH115-OX plants (Fig. 3), thus explaining the elevated Fe accumulation in these plants (Fig. 2F). Recent studies have confirmed that homologs of bHLH115 (bHLH34/104/105) positively regulate expression of bHLH38/39/100/101 and PYE (Zhang et al., 2015; Li et al., 2016). However, our results suggested that the single-mutant of bHLH115 could cause a disrupted Fe-deficiency response, indicating that bHLH115 is required for the normal Fe-deficiency response. When the bhlh115 mutant was crossed with bhlh34, bhlh104, and bhlh105 mutants, each double-mutant displayed more severe Fe-deficiency symptoms than the single-mutant (Fig. 8).
Functional similarity and differences among bHLH IVc subgroup members
The bHLH IVc subgroup contains four members, bHLH34/104/105/115, three of which, bHLH34/104/105, have been confirmed to positively regulate Fe homeostasis in Arabidopsis thaliana (Zhang et al., 2015; Li et al., 2016). Under both Fe-sufficient and Fe-deficient conditions, expression of bHLH38/39/100/101 and PYE in the bhlh115 mutants was lower than observed in wild-type plants; however, their expression was still induced by Fe limitation in bhlh115 mutants, implying that other members of the bHLH IVc subgroup may activate their expression under Fe-deficient conditions. Expression analysis confirmed that Fe-deficiency-induction of bHLH38/39/100/101 and PYE was dramatically weakened in the bhlh34 × 115, bhlh104 × 115, bhlh105 × 115, and bhlh34 × 104 × 115 mutants compared with the bhlh115-1 mutant (see Supplementary Fig. S7), indicating that bHLH34/104/105 are responsible for induction of bHLH38/39/100/101 and PYE under Fe-deficient conditions in the bhlh115-1 mutant. Thus, each member of the bHLH IVc subgroup shares similar molecular functions, namely being able to activate expression of bHLH38/39/100/101 and PYE. Genetic complementation assays demonstrated that each of the bHLH IVc subgroup members could rescue the bhlh115-1 mutant under Fe-deficient conditions, thus confirming similar regulatory functions.
Loss-of-function of anyone of bHLH34/104/105/115 can result in an abnormal Fe-deficiency response, with double- and triple-mutants displaying more severe Fe-deficiency symptoms compared with single-mutants (Fig. 8; Zhang et al., 2015; Li et al., 2016). Comparative analysis revealed their expression patterns are different (Fig. 8; Li et al., 2016). In the root maturation zone, bHLH34/104/105 were expressed in the endodermis and pericycle, whereas bHLH115 was expressed in all tissues. bHLH34/104, but not bHLH105/115, were expressed in the initial lateral roots. bHLH105was detected in the whole root tip, bHLH34/104 were expressed in the meristem of the root tip, whereas bHLH115 was not detected in the root tip. bHLH105 was not expressed in the hypocotyl, but bHLH34/104/115 were. In addition, bHLH115 was the only gene not expressed in the leaf veins, but was ubiquitously expressed in the root mature zone (Fig. 7), implying putative specific roles for bHLH115 in Fe uptake. These differential expression patterns imply specific biological functions in specific tissues. Protein interaction assays indicated potential homo- or heterodimerization among bHLH IVc subgroup members. Furthermore, overlapping expression patterns in the root, hypocotyl, and leaf were observed (Fig. 7; Li et al., 2016). This suggests they may function synergistically to maintain Fe homeostasis in specific tissues.
Recently, the chrysanthemum and apple homologs of the bHLH IVc subgroup members, CmbHLH1 and MdbHLH104, were found to positively regulate Fe accumulation by activating transcription of the gene encoding H+-ATPase (Zhao et al., 2014, 2016), suggesting conserved functions in the regulation of Fe homeostasis. We did not observed visible changes in GUS-staining in the ProbHLH115:GUS plants after Fe-deficiency treatment. Expression analysis confirmed that the bHLH115 transcript was not responsive to Fe deficiency (see Supplementary Fig. S3). Similarly, the expression of bHLH34/104/105 is not affected by Fe deficiency (Zhang et al., 2015; Li et al., 2016). Unlike Arabidopsis bHLH IVc subgroup members, transcript abundance of both CmbHLH1 and MdbHLH104 increases under Fe-deficiency conditions, indicating evolutional divergence among different plants species.
Negative regulation of bHLH IVc subgroup members by BTS
BTS is a negative regulator of Fe homeostasis, and its mutant accumulates more Fe than wild-types under Fe-sufficient conditions (Selote et al., 2015; Zhang et al., 2015). bHLH105 and bHLH115 physiologically interact with BTS in vivo and can be degraded by BTS in vitro (Selote et al., 2015). Zhang et al. (2015) demonstrated that elevated expression of bHLH105 can promote Fe accumulation in plants, and this work showed that bHLH115 had a similar function; this may explain the Fe over-accumulation observed in plants with loss-of-function of BTS. Our genetic analysis was in agreement this, revealing that the loss-of-function of bHLH115 compromised the Fe-deficiency tolerance of the bts-1 mutant. Corresponding with this, the expression of bHLH38/39/100/101 and PYE in bts-1 bhlh115-1 plants was lower than in bts-1 plants. These data suggest that bHLH115 functions downstream of BTS. Despite its interaction with BTS, the bHLH104 protein is not degraded by BTS in vitro (Selote et al., 2015). Likewise, loss-of-function of bHLH104 also alleviates Fe-deficiency tolerance of the bts-1 mutant (Zhang et al., 2015), implying a potential negative regulation of bHLH104 by BTS. However, the mechanism by which BTS affects bHLH104 functions remains unclear.
To maintain Fe concentrations within physiological limits, plants modulate processes contributing to Fe homeostasis in accordance with the external supply and internal requirements. BTS was identified as a potential Fe sensor. Both its transcripts and proteins are up-regulated under Fe-deficient conditions; however, its proteins decrease upon Fe treatment (Selote et al., 2015). It has been proposed that under Fe-deficient conditions the elevated levels of BTS facilitate degradation of bHLH105 and bHLH115, resulting in down-regulation of Fe-deficiency-responsive genes. However, this inference is not consistent with the up-regulation of Fe-deficiency-responsive genes under Fe-deficiency conditions. Therefore, other factors are likely to activate bHLH105 and bHLH115 functions by antagonizing BTS under Fe-deficiency conditions. Further investigation is required to explain this paradox.
Based on our findings and those of previous studies, we propose a working model for bHLH115 and its homologues. Under Fe-deficiency conditions, IVc subgroup bHLH transcription factors bHLH34/104/105/115 activate the expression of PYE, FIT, and bHLH38/39/100/101. FIT and bHLH38/39/100/101 synergistically activate the expression of the Fe-uptake-associated genes IRT1 and FRO2. PYE directly regulates the expression of the Fe distribution-associated genes ZIF1 and NAS4. bHLH34/104/105/115 play synergistic roles in the overlapping expression locations and each has independent roles in their specific expression locations.
Author contributions
GL and DY designed the study; GL, HZ, XL, and QA performed the research; GL, HZ, XL, and DY analysed the data. GL wrote the paper; GL, HZ, XL, and DY revised the paper. All authors read and approved the final manuscript.
Supplementary Material
Acknowledgments
We thank TAIR (The Arabidopsis Information Resource) at the Ohio State University for the T-DNA insertion mutants. This work was supported by the Youth Innovation Promotion Association of CAS, Candidates of the Young and Middle-Aged Academic Leaders of Yunnan Province [2015HB095] and the program for Innovative Research Team of Yunnan Province [2014HC017]. All authors state they have no conflicts of interest in relation to this research.
References
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML. 2004. The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. The Plant Cell 16, 3400–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EL, Fett JP, Guerinot ML. 2002. Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. The Plant Cell 14, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Cassin G, Couch D, et al. 2009. Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Annals of Botany 103, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Benoist B, McLean E, Egli I, Cogswell M. eds 2008. Worldwide prevalence of anaemia 1993–2005. Geneva, Switzerland: World Health Organization. [DOI] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y. 1994. Iron: nutritious, noxious, and not readily available. Plant Physiology 104, 815–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Arosio P. 1996. The ferritins: molecular properties, iron storage function and cellular regulation. Biochimica et Biophysica Acta 1275, 161–203. [DOI] [PubMed] [Google Scholar]
- Haydon MJ, Cobbett CS. 2007. Transporters of ligands for essential metal ions in plants. The New Phytologist 174, 499–506. [DOI] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. 2003. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Molecular Biology and Evolution 20, 735–747. [DOI] [PubMed] [Google Scholar]
- Hell R, Stephan UW. 2003. Iron uptake, trafficking and homeostasis in plants. Planta 216, 541–551. [DOI] [PubMed] [Google Scholar]
- Henriques R, Jásik J, Klein M, Martinoia E, Feller U, Schell J, Pais MS, Koncz C. 2002. Knock-out of Arabidopsis metal transporter gene IRT1 results in iron deficiency accompanied by cell differentiation defects. Plant Molecular Biology 50, 587–597. [DOI] [PubMed] [Google Scholar]
- Hindt MN, Guerinot ML. 2012. Getting a sense for signals: regulation of the plant iron deficiency response. Biochimica et Biophysica Acta 1823, 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Guerinot ML. 2009. Homing in on iron homeostasis in plants. Trends in Plant Science 14, 280–285. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK. 2012. Iron uptake, translocation, and regulation in higher plants. Annual Review of Plant Biology 63, 131–152. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang H, Ai Q, Liang G, Yu D. 2016. Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana. Plant Physiology 170, 2478–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. 1987. Chlorophylls and carotenoids, the pigments of photosynthetic biomembranes. In: Douce R, Packer L. eds. Methods in enzymology, vol. 148 New York: AcademicPress Inc., 350–382. [Google Scholar]
- Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M. 2002. The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proceedings of the National Academy of Sciences, USA 99, 13938–13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN. 2010. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. The Plant Cell 22, 2219–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H. 1995. Mineral nutrition of higher plants. London: Academic Press. [Google Scholar]
- Mukhopadhyay A, Deplancke B, Walhout AJ, Tissenbaum HA. 2008. Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nature Protocols 3, 698–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CM, Hindt MN, Schmidt H, Clemens S, Guerinot ML. 2013. MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genetics 9, e1003953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit JM, Briat JF, Lobréaux S. 2001. Structure and differential expression of the four members of the Arabidopsis thaliana ferritin gene family. The Biochemical Journal 359, 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML. 1999. A ferric-chelate reductase for iron uptake from soils. Nature 397, 694–697. [DOI] [PubMed] [Google Scholar]
- Rogers EE, Guerinot ML. 2002. FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. The Plant Cell 14, 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salahudeen AA, Thompson JW, Ruiz JC, Ma HW, Kinch LN, Li Q, Grishin NV, Bruick RK. 2009. An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 326, 722–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z. 2008. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nature Protocols 3, 1018–1025. [DOI] [PubMed] [Google Scholar]
- Santi S, Schmidt W. 2009. Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. The New Phytologist 183, 1072–1084. [DOI] [PubMed] [Google Scholar]
- Selote D, Samira R, Matthiadis A, Gillikin JW, Long TA. 2015. Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiology 167, 273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. 2003. The Arabidopsis basic/helix-loop-helix transcription factor family. The Plant Cell 15, 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto C, Maiwald D, Pesaresi P, Jahns P, Salamini F, Leister D. 2002. The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. The Plant Journal 31, 589–599. [DOI] [PubMed] [Google Scholar]
- Vashisht AA, Zumbrennen KB, Huang X, et al. 2009. Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326, 718–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. 2002. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. The Plant Cell 14, 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EL, Connolly EL. 2008. Time to pump iron: iron-deficiency-signaling mechanisms of higher plants. Current Opinion in Plant Biology 11, 530–535. [DOI] [PubMed] [Google Scholar]
- Wang HY, Klatte M, Jakoby M, Bäumlein H, Weisshaar B, Bauer P. 2007. Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226, 897–908. [DOI] [PubMed] [Google Scholar]
- Wang N, Cui Y, Liu Y, Fan H, Du J, Huang Z, Yuan Y, Wu H, Ling HQ. 2013. Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Molecular Plant 6, 503–513. [DOI] [PubMed] [Google Scholar]
- Yi Y, Guerinot ML. 1996. Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. The Plant Journal 10, 835–844. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ. 2008. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Research 18, 385–397. [DOI] [PubMed] [Google Scholar]
- Yuan YX, Zhang J, Wang DW, Ling HQ. 2005. AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Research 15, 613–621. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu B, Li M, Feng D, Jin H, Wang P, Liu J, Xiong F, Wang J, Wang HB. 2015. The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. The Plant Cell 27, 787–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Song A, Li P, Chen S, Jiang J, Chen F. 2014. A bHLH transcription factor regulates iron intake under Fe deficiency in chrysanthemum. Scientific Reports 4, 6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Ren YR, Wang QJ, Yao YX, You CX, Hao YJ. 2016. Overexpression of MdbHLH104 gene enhances the tolerance to iron deficiency in apple. Plant Biotechnology Journal 14, 1633–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.