Highlight
The roles of GmCIF1 and GmC/VIF1 as invertase inhibitors were examined in vitro in soybean (Glycine max), and post-translational elevation of extracellular invertase activity by silencing GmCIF1 expression was found to increase seed weight.
Keywords: Cell wall invertase, Glycine max, invertase inhibitor, post-translational regulation, seed weight, source and sink.
Abstract
Cell wall invertase (CWI) and vacuolar invertase (VI) play multiple functions in plant growth. As well as depending on transcriptional and post-transcriptional regulation, there is growing evidence that CWI and VI are also subject to post-translational control by small inhibitory proteins. Despite the significance of this, genes encoding inhibitors, their molecular and biochemical properties, and their potential roles in regulating seed production have not been well documented in soybean (Glycine max). In this study, two invertase inhibitor isoforms, GmCIF1 and GmC/VIF2, were characterized to possess inhibitory activities in vitro via heterologous expression. Transcript analyses showed that they were predominantly expressed in developing seeds and in response to ABA. In accordance with this, surveys of primary targets showed subcellular localizations to the apoplast in tobacco epidermis after expressing YFP-fusion constructs. Investigations using RNAi transgenic plants demonstrated marked elevations of CWI activities and improvements in seed weight in conjunction with higher accumulations of hexoses, starch, and protein in mature seeds. Further co-expression analyses of GmCIF1 with several putative CWI genes corroborated the notion that GmCIF1 modulation of CWI that affects seed weight is mainly contingent on post-translational mechanisms. Overall, the results suggest that post-translational elevation of CWI by silencing of GmCIF1 expression orchestrates the process of seed maturation through fine-tuning sucrose metabolism and sink strength.
Introduction
Invertases (EC 3.2.1.26) irreversibly catalyze the hydrolysis of sucrose into hexoses (glucose and fructose), both of which are basic nutrients and signaling molecules in plants (Koch, 2004; Rolland et al., 2006). Based on their pH optima and subcellular localizations, invertases are categorized into cytosolic neutral/alkaline invertase (CI) and acid invertase. These two types of invertases differ substantially in amino acid sequences and biochemical properties (Sturm, 1999). Acid invertases are further classified into cell wall invertase (CWI) and vacuolar invertase (VI) on the basis of their cellular targets. Unlike acid invertases, CIs are not glycosylated proteins and exhibit little sequence homology with CWIs and VIs (Ruan et al., 2010).
CWIs and VIs are recognized as essential players in sugar metabolism and sugar signaling, thereby affecting source–sink interactions and responses to environmental cues in higher plants (Ruan, 2014). The roles of VI in the major regulation of carbon metabolism, hexose distribution, cellular osmolarity, and oxidative stress defense have been extensively explored (Kohorn et al., 2006; Sergeeva et al., 2006; Nägele et al., 2010; Xiang et al., 2011; Liu et al., 2015). Suppression of VI activities alters soluble sugar conversion, thereby decreasing cold-induced sweetening in potato tubers (Bagnaresi et al., 2008; Bhaskar et al., 2010; Zhu et al., 2016) and affecting fruit size (Klann et al., 1996; Tang et al., 1999; Yu et al., 2008). In contrast, CWIs catalyze the cleavage of sucrose into hexoses, which are unloaded into sink cells of the apoplast by sucrose transporters (SUTs) from the phloem (Bihmidine et al., 2013). The hexoses released in sink tissues may go either into intracellular compartments for metabolism, polysaccharide synthesis, and gene regulation, or into extracellular compartments for fungal colonization and defense-related responses (Roitsch et al., 2003; Doidy et al., 2012). It is therefore not surprising that CWIs play multifaceted and significant roles in response to environmental stimuli and have a major influence on the development of sink organs (Rausch and Greiner, 2004; Proels and Hückelhoven, 2014). Overexpression of CWI genes delays leaf senescence and increases drought resistances in tobacco and tomato (Lara et al., 2004; Albacete et al., 2015). In Arabidopsis, CWI is considered to be an essential component of pathogen-induced plant defense (Zhao et al., 2015). AtCWI4 has been shown to be associated with floral nectar secretion (Ruhlmann et al., 2010). Ectopic expression of CWIs in Arabidopsis improves plant vegetative and reproductive growth (Heyer et al., 2004; von Schweinichen and Büttner, 2005). A null mutant of the CWI gene in maize, Mn1, led to miniature seeds as a result of the arrested endosperm development (Miller and Chourey 1992), whereas overexpression of Mn1 increased grain yield and starch content (Li et al., 2013). The critical roles of CWIs in regulation of seed development have also been well documented in rice, tomato, and cotton (Wang et al., 2008; Zanor et al., 2009; Wang and Ruan, 2012), confirming that CWIs exert multiple signaling and metabolic functions in plants.
Earlier research on the regulation of acid invertases primarily focused on transcriptional and post-transcriptional control via increases in their transcripts in response to diverse signals (Weber et al., 1996; Ehneß and Roitsch, 1997; Roitsch, 1999). However, CWI and VI are intrinsically stable proteins owing to their glycan decoration, and hence the regulation of CWI and VI may also be subject to post-translational mechanisms. An increasing body of in vitro studies have shown that CWI and VI activities are determined by small inhibitor of β-fructosidases (IFs), with sizes ranging from 15–23 kDa, with targets in the cell wall or vacuoles (Rausch and Greiner, 2004). The first putative invertase inhibitor was biochemically characterized in potato (Schwimmer et al., 1961). Thereafter in vivo inhibitory functions of cell wall invertase inhibitors (CIFs) were validated in tobacco (Greiner et al., 1998), followed by the identification of several CIFs in maize and tomato (Bate et al., 2004; Reca et al., 2008). Studies on the functional roles of vacuolar invertase inhibitors (VIFs) demonstrated a marked reduction of cold-induced sweetening in potato tubers by capping the activities of VIs (Greiner et al., 1999; Brummell et al., 2011; Liu et al., 2013; Mckenzie et al., 2013). More recently, silencing of a tomato CIF led to a delay of leaf senescence and improvements in seed filling and fruit size (Jin et al., 2009). In Arabidopsis, suppression of two invertase inhibitors resulted in an increase of seed production (Su et al., 2016), reflecting the fact that post-translational modulation of CWI is required for sugar unloading to sink tissues.
Glycine max (soybean), a legume crop, is an important source of edible proteins and oils that are widely used throughout the world (Song et al., 2013). There has been ongoing interest in improving soybean performance, although significantly enhancing seed production remains a major challenge for modern agriculture. In spite of the advances made in commonly studied plants, little is known about invertase inhibitory proteins and their physiological significance in soybean. To our knowledge, there have been no reports on the targets of invertase inhibitors, on the post-translational regulation of CWI, or on the phenotypes that arise from modification of the expression of inhibitors in soybean plants. In a bid to rectify this situation, two putative soybean invertase inhibitors, GmCIF1 and GmC/VIF2, were functionally characterized by the use of recombinant proteins. Their subcellular targets were examined by ectopic expression with fusions of fluorescent protein. The functional role of GmCIF1 was further explored in RNAi transgenic plants via the silencing of its expression. Based on transcript and metabolic analysis, our work demonstrates the physiological roles of GmCIF1 in controlling seed maturation through specifically depressing CWI activities. The substantially improved seed weight reported here provides a promising strategy for future development, and in the long term, it may facilitate increases in crop production via manipulation of the post-translational modulation of CWI.
Materials and methods
Plant materials and growth conditions
Soybean cultivar ‘Heinong 53’ (HN53) was grown in standard potting soil in a greenhouse or in growth chambers with a temperature cycling between 18 °C (night) and 24 °C (day) with 55% relative humidity under long-day (16 h light/8 h dark, 300–400 μE) conditions. In all comparative analyses of tissues, wild-type and RNAi transgenic plants were cultivated and harvested simultaneously. Tobacco (Nicotiana benthamiana) plants were maintained in a growth chamber at 25 °C under a light regime of 16 h and 300 μE.
Construction of RNA interference (RNAi) vector
A 430-bp fragment of fatty acid desaturation 2 (FAD2) intron I was amplified by using a forward primer FAD2I-F1 containing SmaI restriction sites, and a reverse primer FAD2I-R1 containing KpnI restriction sites. This PCR product was cloned into the pUC8 vector to generate pUC8-FAD2I. To form the typical RNAi structure, reverted repeat sequences of GmCIF1 (362 bp) applied in hairpin construction were amplified by using two different sets of primers: a forward primer GmCIF1-F-P1, containing the BamHI site, paired with a reverse primer GmCIF1-F-R1, containing an SmaI site, and a forward primer GmCIF1-R-P1, containing the KpnI site, paired with a reverse primer GmCIF1-R-R1, containing an SacI site. The two amplified target fragments were cloned into the pUC8-FAD2I vector to create the hairpin cassette. The hairpin cassette of GmCIF1-F/FAD2I/GmCIF1-R was further ligated into the vector pCAMBIA3300 (http://www.cambia.org/daisy/cambia/2070.html), which was used for soybean transformation (see Supplementary Fig. S3B at JXB online). The primer sequences are listed in Supplementary Table S2.
Plant transformation
Agrobacterium-mediated transformation of soybean using the bar gene as a selectable marker has been described previously (Zeng et al., 2004). Shoot initiation and elongation were primarily screened by the addition of the herbicide glufosinate to B5 basal medium (Gamborg et al., 1968) in a Petri dish. The T3 homozygous generations were used for analysis. For transient transformation, Agrobacterium tumefaciens (C58C1) cells containing the appropriate construct were grown overnight in 30 ml of YEB-medium supplemented with carbenicillin (50 μg ml–1), rifampicin (100 μg ml–1) and spectinomycin (50 μg ml–1) until the stationary phase. After centrifugation at 3000 g for 30 min at ambient temperature, the cells were re-suspended in 10–15 ml of infiltration buffer [10 mM 2-(N-morpholino) ethanesulfonic acid (MES), pH 5.9, 150 μM acetosyringone] and incubated with gentle agitation for 2 h at room temperature. The cell suspensions were adjusted to an OD600 of 1.0 by mixing with infiltration buffer. As infiltration in soybean leaves did not work properly, the lower epidermis of 4- to 5-week-old tobacco leaves were transformed by infiltrating Agrobacterium cells harboring appropriate plasmids via a needleless syringe. After 40 h, the infiltrated regions were analyzed by confocal laser scanning microscopy (CLSM).
Purification of recombinant GmCIF1 and GmC/VIF2
Coding sequences (omitting the signal peptide) of GmCIF1 and GmC/VIF2 were amplified from flowers using primers containing the GATEWAY (Invitrogen) cloning attB1 and attB2 sites and the TEV protease site (see Supplementary Table S2). PCR products were then recovered using a PCR purification kit (Thermo Scientific), sequenced, and inserted into the pDONR201 plasmid (Invitrogen) and subsequently recombined with the destination vector pETG-20A, which produced 6xHis-tagged thioredoxin A (TrxA) fusion constructs. The protein purifications were performed as described previously (Hothorn et al., 2003; Link et al., 2004). The E. coli strain Rosetta-gamiTM (DE3) (Novagen) was used as host for the protein expression. Bacteria were grown to a density of OD600 of 0.6 in lysogeny broth (LB) medium, followed by the addition of IPTG to 0.2 mM and further grown for 24 h at 18 °C. Cells were pelleted by centrifugation at 10 000 g for 15 min and extracted with 1/20 volume of lysis buffer (50 mM Na2HPO4/NaH2PO4, pH 7.0, 500 mM NaCl, 1% Triton X-100, 1 mg ml–1 lysozyme). After centrifugation at 15 000 g for 1 h, the supernatant was mixed with 0.6 g Ni-TED Protino resin (Macherey-Nagel) and stirred at 4 °C for 45 min before loading into a column. The column was washed with lysis buffer and washing buffer (50 mM Na2HPO4/NaH2PO4, pH 7.0, 500 mM NaCl, 10% glycerol), and bound fusion protein was then eluted with 10 volumes of the washing buffer containing 250 mM imidazole, and dialyzed against TEV protease cleavage buffer (50 mM Na2HPO4/NaH2PO4, pH 7.0, 200 mM NaCl). The eluted fractions were then cleaved with recombinant TEV protease for 3 h at 30 °C. The untagged inhibitor proteins were finally purified with a second elution.
Functional assays of recombinant GmCIF1 and GmC/VIF2
To test the the inhibitory activities of recombinant inhibitors against acid invertase, variable amounts of recombinant inhibitory proteins were added to a suitable invertase preparation (see below) in assay buffer in a total amount of 200 µl and incubated for 30 min at 37 °C to allow complex formation, followed by the addition of 100 µl sucrose (100 mM) and incubated for 60 min at 37 °C. The reaction was terminated by the addition of sodium phosphate buffer (1 M, pH 7.5) and heating at 95 °C for 5 min. The amount of glucose released was determined as described below. In each experiment, samples without recombinant inhibitor proteins were included to determine background absorption, which was subtracted from the other reactions.
Determination of acid invertase activity in vitro
The in vitro analysis of invertase activity was performed as described previously by Link et al. (2004). Selected tissues of soybean were ground in liquid nitrogen using a TissueLyser (Qiagen) and homogenized in 500 μl extraction buffer (30 mM MOPS, 250 mM sorbitol, 10 mM MgCl2, 10 mM KCl, and 1 mM PMSF, pH 6.0). After centrifugation (10 min, 8500 g, 4 °C), the pellets were washed once with extraction buffer plus 1% Triton X-100, and twice with extraction buffer only. The cell-wall pellets were re-suspended in 500 μl assay buffer (20 mM triethanolamine, 7 mM citric acid, and 1 mM PMSF, pH 4.6), and used for the determination of CWI activity. For VI extraction, endogenous sucrose was removed by acetone precipitation of the soluble fraction with four volumes of ice-cold acetone (20 min, −20 °C). After centrifugation (10 min, 15 000 g, 4 °C), the pellets were re-suspended in one volume of assay buffer.
For the measurement of enzyme activity, 100 μl of the obtained preparations were incubated with 100 μl sucrose (100 mM) and deionized water up to 300 μl. After incubation for 1 h at 37 °C, the reaction was stopped by the addition of 30 μl sodium phosphate (1 M, pH 7.5) and heating at 95 °C for 5 min. Assay were performed in quadruplicate, one of which was neutralized and boiled immediately after sucrose addition. The activity of this sample was subtracted from the activity of the others as background absorption. Liberated glucose was measured in a coupled enzymatic–optical assay. In this assay, 100 μl of the reaction mixture, 20 μl 30 mM ATP, 20 μl 30 mM NADP, 2 μl Hexokinase/Glucose-6-phosphate dehydrogenase suspension (340 U ml–1 HK, 170 U ml–1 G6P-DH, Roche) and up to 1 ml buffer (40 mM triethanolamine, 8 mM MgSO4, pH 7.5) were mixed and incubated for 5 min at room temperature. Formation of NADPH was measured spectrophotometrically at 340 nm and the liberated glucose was calculated according to the Lambert–Beer Law. Invertase activity was expressed in nkat g–1 fresh weight (1 nkat = 1 nmole glucose liberated per second).
Expression analysis
RNA extraction and cDNA synthesis were performed as described previously by Han et al. (2013). Selected tissues of Glycine max were ground in liquid nitrogen using a TissueLyser (Qiagen). Total RNA was extracted by using the Gene MATRIX Universal RNA purification Kit (Roboklon) according to the manufacturer’s instructions. RNA quality was determined by NanoDrop 2000 (Thermo Scientific). For a standard qRT-PCR technical application, the reactions were prepared in a 15 μl volume, containing 5 μl cDNA sample (appropriately diluted), primers, dNTP, SYBR green, and Jumpstart Taq-DNA polymerase (Sigma). The mixture was subjected to a temperature regime of 95 °C for 6 min, followed by 95 °C for 20 s, 58 °C for 20 s, and 72 °C for 20 s for 40 cycles, followed by a melt cycle from 65 °C to 95 °C. Reactions were run on a Stratagene Mx3000p QPCR Systems (Thermo Scientific). The efficiency of the primers was tested in preliminary experiments with dilutions of cDNA, producing an R2 value ≥0.99. The relative expression level of a target gene was calculated by normalizing to the geometric mean (Vandesompele et al., 2002) of multiple reference genes: EF/αb (EV279336), CYP (CF806591), Actin2/7 (BW677100), and ActinII (BW652479). The primer sequences are listed in Supplementary Table S2. Each experiment had three biological repeats, each with three technical replicates.
Transcriptome sequencing (RNA-seq) analysis of tissue-specific expressions of the soybean CWI genes was performed as described previously by Severin et al, (2010). Gene expression levels were estimated using the values of number of mapped reads per kilobase of the exon region per million mapped reads (RPKM). Data contained within the RNA-seq is publically available at the SoyBase database (http://www.soybase.org/soyseq/).
Subcellular localization of GmCIF1 and GmC/VIF2
The coding sequences (omitting the stop codon) of GmCIF1 and GmC/VIF2 were amplified by PCR, including the primers containing the Gateway (Invitrogen) attB1 and attB2 recombinant sites (see Supplementary Table S2). The PCR products were recovered by using the GeneJET PCR purification kit (Thermo Scientific) and then inserted into the pDONR201 donor plasmid, sequenced, and subsequently recombined with the binary destination vector pB7YWG2.0, yielding the pB7GmCIF1-YFP and pB7GmC/VIF2-YFP constructs. Tobacco leaves were co-infiltrated with A. tumefaciens (C58C1) harboring pB7GmCIF1-YFP (or pB7GmC/VIF2-YFP) and a cell wall-localization marker pK7BvCWI-1RFP (Rosenkranz et al., 2001). The infiltrated region of the leaves was analyzed by CLSM. Images were taken with a Zeiss LSM 510 Meta inverted microscope. The yellow fluorescent protein (YFP) was excited with a 514-nm laser line, and the emitted fluorescence was collected using a 530–600-nm band pass filter. The red fluorescent protein (RFP) was excited with 543-nm laser line and the emitted fluorescence was collected with a 560-nm long pass filter.
Quantification of sugars, protein and oil contents
Soluble sugars were extracted from 100 mg of homogenized seeds with 900 μl of 80% ethanol for 1 h at 80 °C, and then centrifuged at 12 000 g for 10 min at 4 °C. The collected supernatant was dried in a vacuum overnight. The sugar content of 350 μl of the re-dissolved solution was quantified by high-performance anion-exchange chromatography with pulsed 25 amperometric detection (HPAEC-PAD) in an ICS-3000 system (Dionex) with a CarboPac PA1 column and 15–300 mM NaOH (Fluka) in HPLC-water (VWR) as the eluent. Quantitative calculation of sugars was performed using the Chromeleon software 6.7 (Dionex). The final pellets were used for the quantification of starch content after degradation with amyloglucosidase and α-amylase, as described by Focks and Benning (1998).
Protein and oil contents in soybean seeds were determined by near-infrared spectroscopy (MATRIX-I, Bruker) with a near-infrared grain analyser as described by Delwiche (1998).
ABA treatment
Plants at 40 d after germination (DAG) were sprayed with 50 µmol ABA (dissolved in 10% ethanol) or 10% ethanol (control) once a day for 4 d. Mature leaves were harvested for RNA isolation and assessment of acid invertase activities after 5 d growing in the greenhouse.
Germination test
Seeds (60–80) of wild-type and RNAi plants were surface-sterilized with 70% ethanol, and either rinsed with water for fresh use or with 100% ethanol and air-dried. Germination tests were carried out in Petri dishes with 4 layers of autoclaved moist filter paper. After a 4-d stratification period at 4 °C in the dark, the dishes were incubated in long-day conditions of 16 h/8 h light/dark at 24/18 °C. Germination was scored by radicle emergence from the seed coat. The germination rate was documented daily for 2–5 d.
Accession numbers
Sequence data can be found in the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) under the following accession numbers: NtCIF (Y12805); NtVIF (AY145781); AtCIF1 (AT1G47960); AtC/VIF2 (AT5G64620); BvC/VIF (XP_010685378); IbC/VIF (AAM94391); CiC/VIF (KVH90191); SlCIF1 (CAA09420); SlINVINH1 (AJ010943); GmCIF1 (BT090960); GmC/VIF2 (BT091584); StINVINH1 (GU321338); StINVINH2A (GU321341); StINVINH2B (GU321342); ZmINVINNH1 (AATT24363).
Results
Cloning and characterization of GmCIF1 and GmC/VIF2
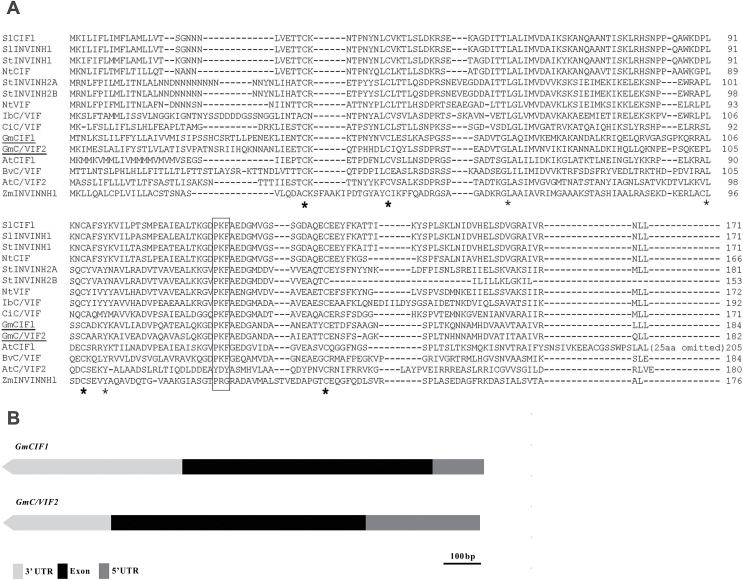
Systematic BLAST searches within the Genbank database retrieved a large number of C/VIF-like genes in the soybean genome. Two of them, GmCIF1 (BT090960) and GmC/VIF2 (BT091584), encoding putative invertase inhibitors were identified with high homologies (>40% amino acid identity) to AtCIF1 from Arabidopsis (Link et al., 2004; Su et al., 2016). A further survey of protein sequences by performing a BLAST search against the SoyBase database (http://www.soybase.org/) confirmed that GmCIF1 and GmC/VIF2 were encoded by Glyma.17G036300 and Glyma.17G036400, respectively. These two isoforms have a similar genomic structure and protein size, sharing 54% amino acid identity (Fig. 1A, B). After removal of the N-terminal signal peptides, the deduced mature proteins of GmCIF1 and GmC/VIF2 are comprised of 155 and 157 amino acid residues, respectively. The predicted molecular weight is 16.77 kDa for GmCIF1, a pI of 6.06, and 16.72 kDa for GmC/VIF2, a pI of 7.59. Protein sequence alignment with other known C/VIFs (Fig.1A) showed that GmCIF1 and GmC/VIF2 contain typical hallmarks, namely four conserved cysteine (C) residues that form two disulfide bridges to sustain protein stability, and a conserved motif (PKF) that has been demonstrated by crystallographic analysis to interact for inhibitory functions with the invertase substrate cleft (Hothorn et al., 2010). Phylogenetic analyses showed that GmCIF1 and GmC/VIF2 are clustered into a clade together with an experimentally determined AtCIF1 from Arabidopsis, whereas their locations were far away from the C/VIFs in tomato, potato, and tobacco (see Supplementary Fig. S1).
Fig. 1.
Multiple sequence alignment of functionally characterized invertase inhibitors and gene structures. (A) Protein sequence alignment was conducted using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Four cysteine residues (C) are labeled with bold asterisks, and the conserved motif is indicated by a box. Nt, Nicotiana tabacum; At, Arabidopsis thaliana; Bv, Beta vulgaris; Ib, Ipomoea batatas; Ci, Cichorium intybus; Sl, Solanum lycopersicum; Gm, Glycine max; St, Solanum tuberosum; Zm, Zea mays. (B) GmCIF1 and GmC/VIF2 showing a similar genomic structure.
Heterologous expression and inhibitory functions of GmCIF1 and GmC/VIF2
To verify their inhibitory functions in vitro, the coding sequences of GmCIF1 and GmC/VIF2 were amplified with cDNA prepared from flowers. Sequences encoding mature proteins (omitting the signal peptides) were cloned into the pETG-20A vector (see Supplementary Fig. S2A). Heterologous expression in the E. coli strain Rosetta-gami™ (DE3) yielded N-terminal TrxA fusion proteins for GmCIF1 and GmC/VIF2. Under native conditions, the fusion proteins were released by the cleavage of the TEV protease. The purified GmCIF1 and GmC/VIF2 were recovered by Ni-TED affinity chromatography to remove the His-tagged TrxA and TEV protease (Supplementary Fig. S2B). Processes during the protein purification suggested that recombinant GmCIF1 and GmC/VIF2 were soluble. The sizes of both the finally purified proteins were close to the predicted molecular weight. Further analysis of their mobility on SDS-PAGE under non-reducing conditions revealed increased shifts, reflecting the presence of active intramolecular disulfide bridges in the recombinant proteins (data not shown).
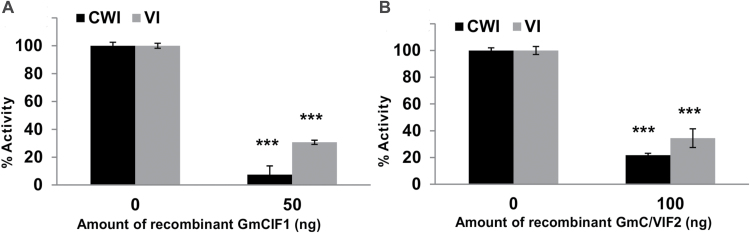
Inhibitory activities of the recombinant proteins were determined by incubating with acid invertase fractions that were extracted from leaves. Approximate 50 ng recombinant GmCIF1 was demonstrated to exert the maximum inhibitory effects and led to a significant depression of 95% in CWI activity and 70% in VI activity (Fig. 2A). Both CWI and VI were increasingly inhibited by increasing amounts of the recombinant GmCIF1 (see Supplementary Fig. S2C, D). However, incubating with even higher amounts (100 ng and 1000 ng) of the recombinant GmCIF1 did not affect the activities of CWI and VI further. In comparison with GmCIF1, the use of approximately 100 ng of the recombinant GmC/VIF2 resulted in maximum inhibition, causing an 80% decrease in CWI activity and a 75% decrease in VI activity (Fig. 2B). It is noteworthy that both recombinant GmCIF1 and GmC/VIF2 exhibited higher inhibitory affinities to CWI than VI in vivo, suggesting their putative roles as CIFs in vivo.
Fig. 2.
The in vitro inhibitory effects of the recombinant GmCIF1 and GmC/VIF2. (A) The lowest dose (50 ng) of GmCIF1 caused maxmium suppression of activities of CWI and VI. (B) The lowest dose (100 ng) of GmC/VIF2 caused maxmium suppression of activities of CWI and VI. Mature soybean leaves were harvested for the extraction of CWI and VI. The enzyme activity data represent means ±SE of at least four independent biological replicates and asterisks indicate significant differences in comparison with the control using Student’s t-test: ***P<0.001.
Expression patterns of GmCIF1 and GmC/VIF2
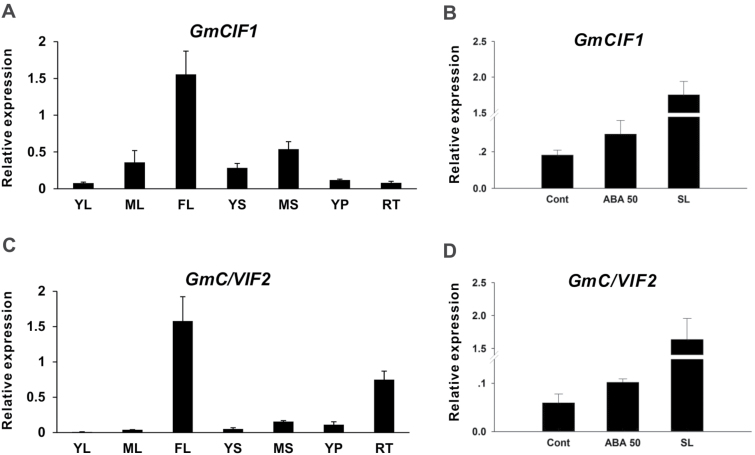
To analyze the expression characteristics of GmCIF1 and GmC/VIF2, their transcripts were investigated by qRT-PCR. The tissue-specific expression patterns were examined in seven tissues (young leaf, mature leaf, flower, young seed, mature seed, young pod, and root). The results showed that GmCIF1 and GmC/VIF2 displayed similar expression profiles; however, the transcript accumulation of GmCIF1 was more abundant than GmC/VIF2 (Fig. 3A, C). Both of them exhibited higher transcript levels in flowers and developing seeds. GmCIF1 was also observed to be expressed more in mature leaves than in young leaves (Fig. 3A), whilst GmC/VIF2 showed a relatively high expression level in roots (Fig. 3C). To test whether their transcripts are promoted in response to ABA and ABA-triggered leaf senescence, different concentrations of ABA were sprayed onto mature leaves. It was found that expression of GmCIF1 and GmC/VIF2 both increased upon 50 µM ABA treatment (Fig. 3B, D). Notably, treating with a higher concentration of ABA (200 µM) led to an accelerated process of leaf ageing, whilst the transcripts of GmCIF1 and GmC/VIF2 were significantly increased (Fig. 3B, D), suggesting that both GmCIF1 and GmC/VIF2 are ABA- and senescence-responsive genes.
Fig. 3.
Expression analyses of GmCIF1 and GmC/VIF2. (A, C) Transcript levels of GmCIF1 and GmC/VIF2 in different soybean tissues. (B, D) Expression of GmCIF1 and GmC/VIF2 in response to ABA and ABA-triggered leaf senescence. Data represent mean values ±SE of at least three independent biological replicates for qRT-PCR. GmACT2/7, GmACTII, GmEF/αb, and GmCYP were used as reference genes. YL, young leaf; ML, mature leaf; FL, flower; YS, seed 15 d after flowering (DAF); MS, seed 25 DAF; YP, young pods 10 DAF; RT, root; Cont, control; ABA 50, 50 µM ABA; SL, senescent leaves induced with 200 µM ABA.
GmCIF1 and GmC/VIF2 target to the apoplast
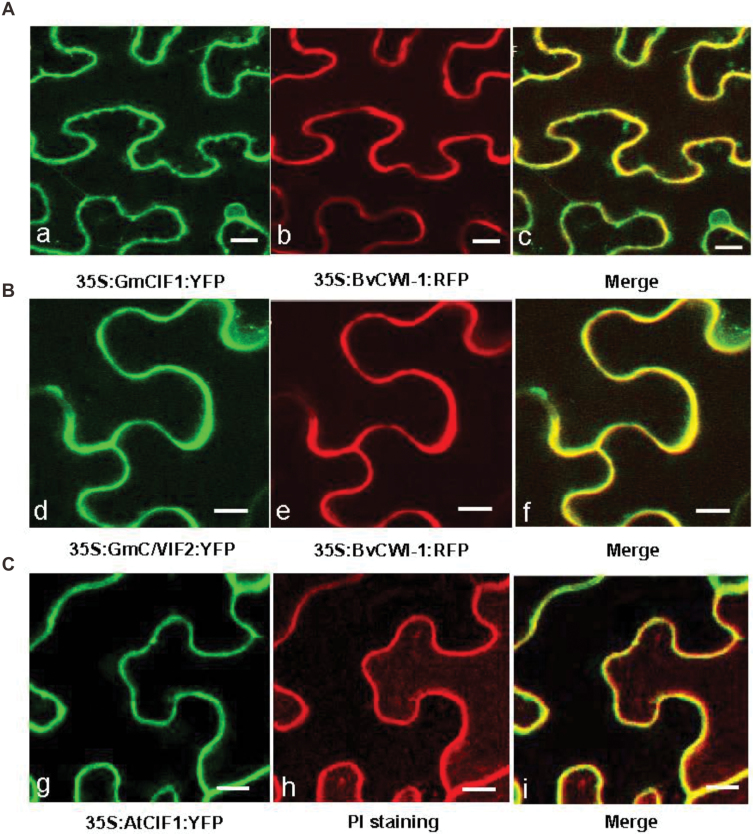
To determine the primary target of GmCIF1 and GmC/VIF2 within cells, prediction programs were applied to deduce their intracellular compartmentations. All three in silico analyses of GmCIF1 and GmC/VIF2 unanimously resulted in the highest scores for an extracellular target (see Supplementary Table S1). We then sought to verify the subcellular localizations using fluorescent-labelled GmCIF1 and GmC/VIF2. The 35S:GmCIF1:YFP and 35S:GmC/VIF2:YFP C-terminal fusion constructs were co-delivered with a RFP-fused cell wall-localization marker (35S:BvCWI-1:RFP) into tobacco leaf epidermal cells by a transient ectopic expression. By co-infiltration, a yellow fluorescent signal was visualized around the cell periphery from the overlap of the YFP signal (green) and the RFP signal (red), indicating an apoplastic localization (Fig. 4A, B). However, there was no signal captured from vacuoles. Concurrently, as a comparison, the YFP-fused Arabidopsis extracellular invertase inhibitor AtCIF1 (35S:AtCIF1:YFP) was transformed into tobacco leaves. Similarly, combined with the propidium iodide (PI) staining, overlapped fluorescent signals (yellow) between PI (red) and YFP (green) suggested an apoplastic target (Fig. 4C). Taken together, the fluorescent images of the co-localization of AtCIF1 and PI further support the view that GmCIF1 and GmC/VIF2 are primarily targeted to the apoplast, which is in agreement with the computational prediction of their subcellular localizations.
Fig. 4.
Apoplastic localization of GmCIF1 and GmC/VIF2 in tobacco leaves. Tobacco leaves were co-infiltrated with Agrobacterium culture harboring the constructs of (A) 35S:GmCIF1:YFP and 35S:BvCWI-1:RFP or (B) 35S:GmC/VIF2:YFP and 35S: BvCWI-1:RFP. (C) Apoplastic localization of AtCIF1 and PI (propidium iodide) staining. (a, d) Epidermal cells depicting YFP (green) fluorescence. (b, e) RFP (red) fluorescence in the cell wall. (c, f) Overlap of fluorescent signals (yellow) from YFP and RFP. A sugar beet CWI (BvCWI-1) was used as cell wall-localization marker.
CWI activities are increased in transgenic RNAi plants
To investigate the physiological roles of GmCIF1 and GmC/VIF2, transgenic soybeans were generated by a strategy of RNA interference (RNAi) to inhibit the expressions of GmCIF1 and GmC/VIF2. Four out of 32 independent homozygous plants with silencing of GmCIF1 were screened; however, no positive GmC/VIF2-silenced plants were identified owing to unsuccessful transformation. The selected GmCIF1-silenced plants displayed significantly decreased transcript levels of GmCIF1, ranging from 82% to 95%, in comparison with wild-type plants (Fig. 5A). To test whether these observations were due to the specific suppression of GmCIF1 expression, we seeded a BLAST search for homologs of GmCIF1 by inputting the partial coding sequence that was used for the RNAi vector construction (see Supplementary Fig. S3A, B). The search in SoyBase retrieved two genes (Glyma.07G237300 and GmC/VIF2) that exhibited the highest scores of identities (>50%). However, transcript analyses of these genes showed no apparent expression differences between the RNAi plants and the wild-type control (Supplementary Fig. S3C), suggesting that the RNA-mediated gene silencing particularly targets to GmCIF1.
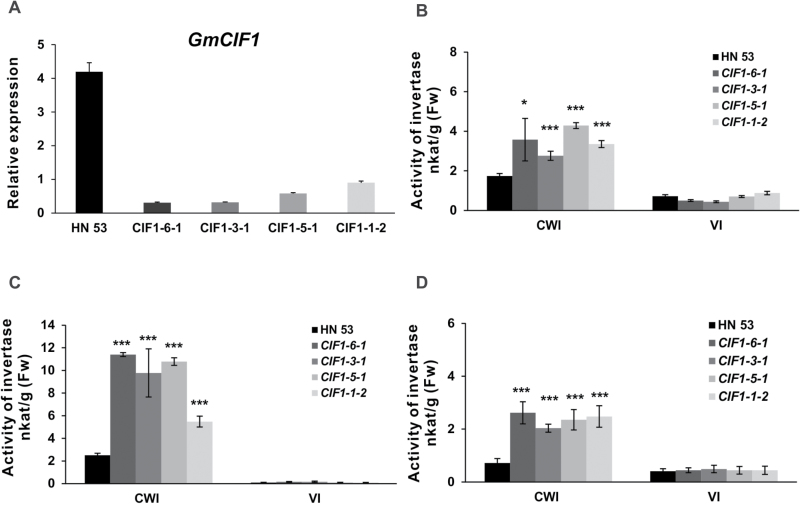
Fig. 5.
Effects on GmCIF1 expression and acid invertase activities in RNAi plants. (A) Suppression of GmCIF1 expression. (B) Increases of CWI activities in seeds at 45 d after flowering (DAF). (C) Increases of CWI activities in seeds at 65 DAF (dehydrated). (D) Increases of CWI activities in imbibed seeds (24 h). Data represent mean values ±SE of at least three independent biological replicates for qRT-PCR. GmACT2/7, GmACTII, GmEF/αb, and GmCYP were used as reference genes. The enzyme activity data represent means ±SE of at least four independent biological replicates and asterisks indicate significant differences in comparison with the control using Student’s t-test: ***P<0.001, *P<0.05.
The observed tissue-specific expression profiles of GmCIF1 prompted us to examine whether the acid invertase activities were differentially regulated in sources and sinks. In contrast to the wild-type control, analysis of enzyme activities in RNAi plants revealed that CWI activities significantly increased as seeds matured (Fig. 5B, C). In addition, significant increases of CWI activities were also detected early in seed germination (24 h) (Fig. 5D) and in mature leaves of RNAi plants (see Supplementary Fig. S4D). Particularly in dehydrated mature seeds (65 DAF), CWI activities of RNAi plants showed significantly higher levels than in wild-type plants, ranging from 1.5-fold to 4-fold higher (Fig. 5C). In contrast to CWI, VI activities appeared not to be significantly influenced as only minor changes were observed between RNAi plants and the wild-type control, except for at the early seed maturity stage (Supplementary Fig. S4A). VI activities declined to undetectable levels until the seed completed dehydration (65 DAF, Fig. 5C). Taken together, the results showed that in the RNAi transgenic soybean the silencing of GmCIF1 expression led to significant elevations of CWI activities, suggesting that CWI is the physiological target of GmCIF1.
Silencing of GmCIF1 significantly improves seed weight
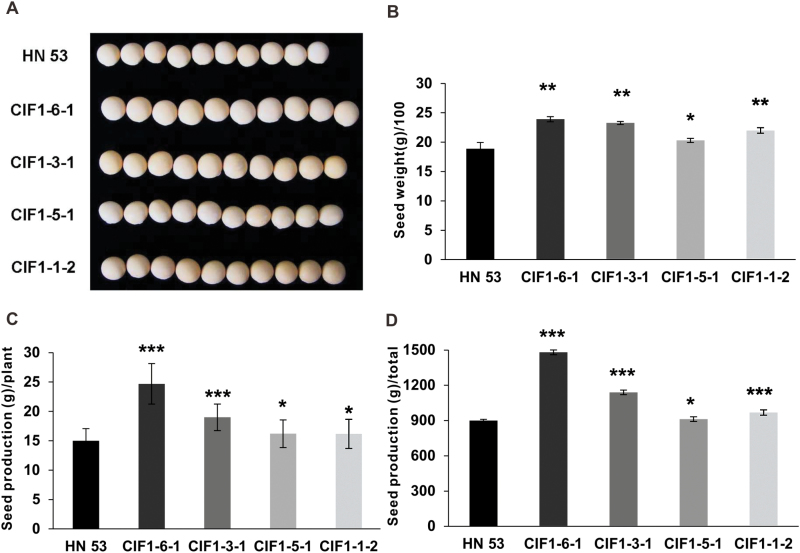
Given that the silencing of the GmCIF1 transcript increased CWI activities, we then explored whether there were any potential phenotypic changes in plant growth and development. Interestingly, the morphological analysis showed significant differences in seed weight between RNAi plants and the wild-type plants. RNAi plants produced larger seeds than the wild-type control (Fig. 6A, B; see Supplementary Fig. S5A). In the same backgrounds, analyses of the seed production per individual plant and the total across all plants showed a statistical improvement (Fig. 6C, D). However, even if very large increases in CWI activities were detected at the beginning of seed germination (24 h) of RNAi plants, the germination rate was not statistically affected (Supplementary Fig. S5B). RNAi plants appeared to grow as normally as the wild-type plants and no significant differences were recorded for the plant height in RNAi plants (data not shown). ABA perturbation caused marked depressions of activities of both CWI and VI in leaves, whereas CWI activities of RNAi plants remained at higher levels than the wild-type control plants (Supplementary Fig. S4D, E). However, RNAi plants and wild-type plants showed no differences in ABA-mediated leaf ageing (Supplementary Fig. S4B, C). Interestingly, in comparison with the wild-type control at 125 d, RNAi plants showed a prolonged plant greening period of approximately 4–6 d, which was statistically significant (Supplementary Fig. S5).
Fig. 6.
Phenotypic impacts on seed weight and production. (A) Image of seeds showing increases in size in RNAi plants (10 seeds are shown in each case). (B) Increases in the seed weight (100 seeds) in RNAi plants. (C) Seed production per individual plant increased in RNAi plants. (D) Total seed production increased in RNAi plants (40 plants). The data represent means ±SE of at least five biological replicates and asterisks indicate significant differences in comparison with the control using Student’s t-test: ***P<0.001, **P<0.01, *P<0.05.
Higher levels of hexoses and starch are accumulated in mature seeds
To test whether the increased CWI activities resulted in effects on the synthesis of primary metabolites, the contents of sugars, protein, and oil in mature seeds were quantified. Soluble sugars showed a significant increase in levels of hexoses in the RNAi backgrounds, but there were nearly no differences for maltose in comparison with the wild-type control plants (Table 1). Sucrose appeared to be more abundant than hexoses, but its content was not significantly affected in RNAi plants, except for CIF1-6-1. Starch, protein, and oil were further analyzed and it was found that more starch and protein were produced in RNAi plants. Interestingly, the percentage of oil was the same in both backgrounds (Table 1). These findings suggest that the hexoses liberated in a CWI-dependent manner during seed maturation and the elevated CWI activities are more closely associated with increases of starch and protein production in soybean mature seeds.
Table 1.
Effects on the contents of sugars, protein and oil in RNAi plants
| Genotype | Glucose | Fructose | Sucrose | Maltose | Starch | Protein | Oil |
|---|---|---|---|---|---|---|---|
| HN53 | 3.47 ± 0.31 | 0.87 ± 0.06 | 36.31 ± 4.07 | 0.80 ± 0.14 | 3.84 ± 0.04 | 40.17 ± 0.43 | 20.54 ± 0.91 |
| CIF1-6-1 | 6.18 ± 0.77*** | 2.25 ± 0.35*** | 46.78 ± 2.86*** | 0.90 ± 0.08 | 4.59 ± 0.06*** | 42.71 ± 0.36*** | 20.41 ± 0.18 |
| CIF1-3-1 | 4.28 ± 0.13*** | 1.78 ± 0.62** | 38.30 ± 1.10 | 0.90 ± 0.07 | 4.64 ± 0.18*** | 42.79 ± 0.31*** | 20.30 ± 0.17 |
| CIF1-5-1 | 4.95 ± 0.09*** | 1.88 ± 0.06*** | 38.35 ± 0.92 | 0.95 ± 0.25 | 4.38 ± 0.03*** | 42.88 ± 0.17*** | 20.63 ± 0.23 |
| CIF1-1–2 | 3.83 ± 0.06* | 1.70 ± 0.52** | 36.50 ± 1.41 | 0.82 ± 0.03 | 3.90 ± 0.05* | 41.92 ± 0.44*** | 20.12 ± 0.21 |
Data represent mean ±SE of at least six biological replicates. Valuesare mg g−1 fresh weight of samples for sugar quantifications (glucose, fructose, sucrose, and maltose) and percentage (%) for starch, protein, and oil. Asterisks indicate significant differences in comparison with the control as determined by Student’s t-test: ***P<0.001, **P<0.01, *P<0.05.
Identification of CWI genes co-expressed with GmCIF1 during seed maturation
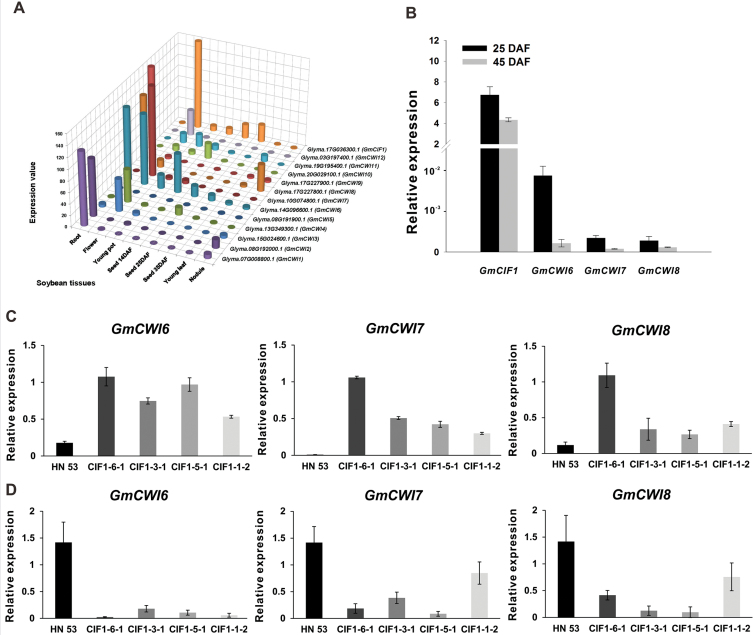
The specific modifications of CWI activities at seed maturity stages prompted us to examine whether regulation of CWI mainly depends on post-translational mechanisms. We isolated CWI genes in the soybean genome and analyzed their expressions. BLAST searches from the SoyBase database revealed the presence of 12 putative CWI genes, showing high homologies with GmCWINV1 (Dimou et al., 2005). Multiple sequence alignment and phylogenetic analyses with other known CWIs and VIs in Arabidopsis and rice suggested that they had well-conserved structures of the CWI gene family (see Supplementary Figs S6 and 7). Based on the initial RNA-seq analysis, six putative CWI genes (GmCWI3, 4, 6, 7, 8, 12) displayed high transcript levels in flowers and developing seeds (Fig. 7A). Further evaluation by qRT-PCR verified that only three isogenes (GmCWI6, 7, and 8) were potentially co-expressed with GmCIF1 at early and mid-stages of seed maturation (25 and 45 DAF) (Fig.7B). In the RNAi backgrounds, expression of CWI genes (GmCWI6, 7, and 8) increased in seeds at 25 DAF; however, it drastically decreased in seeds at 45 DAF (Fig. 7C, D), which does not relate to the increased CWI activities observed at this developmental stage. Because of low affinity in soybean, the detection of the GmCIF1 protein in the wild-type plants was not successful when using polyclonal antibodies obtained from tobacco, maize, sugar beet, and chicory (data not shown). Overall, identification of novel CWI genes in the soybean genome by examining their tissue-specific expression profiles indicated that at least three CWI isoforms appear to be co-expressed with GmCIF1 during seed maturation.
Fig. 7.
Expression patterns of CWI genes and GmCIF1 in soybean tissues. (A) RNA-seq expression of putative CWI genes in different tissues. (B) Co-expression of GmCIF1 and CWI genes in seeds at 25 and 45 d after flowering (DAF) of wild-type plants. (C) CWI gene expression in seed at 25 DAF. (D) CWI gene expression in seed at 45 DAF. The RNA-seq results are given in RPKM (reads per kilobase transcript per million reads) expression values. Data represent mean values ±SE of at least three independent biological replicates for qRT-PCR. GmACT2/7, GmACTII, GmEF/αb, and GmCYP were used as reference genes.
Discussion
GmCIF1 plays a functional role as a cell wall invertase inhibitor in soybean
In this study, our primary goal was to isolate and determine the physiological significance of cell wall invertase inhibitor (CIF) genes in post-translational regulation of cell wall invertase (CWI) in soybean plants. Sequence comparisons and phylogenetic analyses revealed that GmCIF1 and GmC/VIF2 share conserved protein structures with other known C/VIFs (Fig. 1). At the same time, functional assays in vitro confirmed that GmCIF1 and GmC/VIF2 are indeed invertase inhibitors and, in particular, they showed higher affinities towards CWI than VI (Fig. 2). Consistent with in silico predictions of extracellular compartmentation (Supplementary Table S1), ectopic expression of YFP fusion constructs showed that the subcellular localizations of GmCIF1 and GmC/VIF2 are confined to the apoplast in tobacco epidermal cells (Fig. 4), indicating that GmCIF1 and GmC/VIF2 may function as extracellular invertase inhibitors in vivo.
A tomato CIF, INVINH1, was found to show high expression in developing seeds as well as in response to ABA perturbations (Jin et al., 2009). Similarly, AtCIF1 in Arabidopsis was shown to be an ABA-responsive gene and produced transcript abundances in flowers and seeds (Su et al., 2016). Moreover, our tissue-specific expression analyses determined that GmCIF1 and GmC/VIF2 were predominantly expressed in flowers and expression appeared to increase incrementally with seed development (Fig. 3A, C). In addition, both of their transcripts were also significantly induced in response to ABA and ABA-triggered leaf senescence (Fig. 3B, D). The observed predominant expression properties of GmCIF1 and GmCIF2 in tissues as well as the response to the phytohormone cue provide evidence of their potential roles in seed development.
Transgenic soybean plants were generated by RNA-mediated silencing of GmCIF1 in order to investigate its effects on acid invertases in vivo (Fig. 5A; see Supplementary Fig. S3A, B). Notably, specific suppression of GmCIF1 expression resulted in a significant increase of CWI activities in RNAi plants, particularly at late stages of seed maturity, whereas VI activities appeared to be unaffected, suggesting a high specificity of GmCIF1 against the extracellular invertase (Fig. 5B, C). These results reinforce the view that the functional roles of GmCIF1 are compatible with the in vitro effects of the recombinant protein. Finally, expression analyses of several putative CWI genes (Fig. 7A–D) and enzyme activities suggest the significance of GmCIF1 in the control of CWI activities during seed maturation. Taken together, our data provide support for the model that GmCIF1 plays physiological roles as a CIF, which impacts on seed maturation by capping CWI activities.
Extracellular invertase activities are required for seed development
A large amount of evidence has indicated that CWIs are responsible for sugar partitioning in the apoplast of sinks and that they are involved in the regulation of seed and fruit development in a variety of species (Ruan et al., 2010; Ruan, 2014). Improved seed production has been postulated as a result of elevated extracellular invertase activities in Arabidopsis (Heyer et al. 2004). Investigation of a rice extracellular invertase gene, OsGIF, revealed that it functions as a determinant of grain filling (Wang et al., 2008). Constant expression of CWIs in maize substantially improved the grain weight and starch content (Li et al., 2013). Similarly, it is not surprising that elevation of CWI activities through silencing or knock-out of CIFs has been shown to bring about improvements in seed weight and production (Jin et al., 2009; Su et al., 2016). These reports suggest an association between functional CWI and sink strength. In agreement with this, the specific suppression of GmCIF1 expression significantly improved seed weight and production concurrent with markedly elevated CWI activities (Figs 5C and 6A). CWI activities were observed to be higher than VI activities at the mid- and late stages of seed maturity, indicating the fundamental role of CWI, but not VI, in controlling seed development in soybean.
CWI typically catalyzes hydrolysis of sucrose and releases hexoses into the apoplast; hence, the hexoses released are spatially and temporally associated with CWI activities (Roitsch, 1999; Weschke et al., 2003). Starch serves as the major storage form of carbohydrate in developing seeds (Li et al., 2013). After seed germination, the reserved starch degrades into soluble sugars, which fuel cellular respiration to produce energy for seedling growth (Zeeman et al., 2010). Soluble sugars were quantified in mature seeds of the RNAi plants; however, only glucose and fructose were positively affected, reflecting a correlation with the increased CWI activities. Starch content significantly increased in RNAi backgrounds (Table 1). These results suggested that silencing of GmCIF1 may liberate extra CWI activities, which most likely enhance sucrose hydrolysis and starch storage in sinks. Seed protein content statistically increased, whereas the oil content was barely affected in several lines of the RNAi plants (Table 1). These data are in accordance with the fact that seed protein is negatively associated with seed oil content in soybean (Proulx and Naeve, 2009; Eskandari et al., 2013). The improved seed weight and protein content result from the over-accumulation of hexoses, which have been shown to serve not only as nutrients but also as signals to induce storage-associated cell differentiation in seed development (Wobus and Weber, 1999). Moreover, sugar transport from source to sink is one of the major determinants of plant growth (Lemoine et al., 2013), and therefore we hypothesize that the modified sink strength may initially benefit from the improved efficiency of sugar production in source leaves. This notion was supported by the finding of increased CWI activities in sources leaves of RNAi plants (Supplementary Fig. S4D). The increased CWI activities correlate with the increase of photosynthetic capacity in the sources (Dai et al., 1999; Jin et al., 2009). Taken together, the elevation of CWI by the silencing of its inhibitor may facilitate sucrose unloading and release more hexoses, which are required for starch and protein synthesis during seed maturation.
GmCIF1-mediated release of CWI repression depends on post-translational mechanisms
A previous study showed that NtCIF was co-expressed with CWI in tobacco suspension-cultured cells throughout the entire culture period (Krausgrill et al., 1998). The presence of a CWI inhibitor was demonstrated with spatial and temporal expression coupled with a CWI gene, Lin5, in the phloem parenchyma of young fruit (Jin et al., 2009). More recently, co-expression of CWIs and CIFs has been shown in cotton and Arabidopsis during seed development and germination (Wang et al., 2012; Su et al., 2016), suggesting that post-translational regulation of CWI accompanied by the co-expression of CWI genes and their inhibitors is a common phenomenon in plants. To test this view, a number of putative CWI isogenes were identified in the soybean genome based on a search for homologs (see Supplementary Fig.s S6 and S7). Among the 12 identified CWI genes in the soybean genome, three isoforms were shown by the presence of transcripts during seed maturation (25 and 45 DAF) on the basis of expression analysis (Fig.7A, B). Combined with expression patterns of GmCIF1 in the wild-type plants (Figs 3A and 7A, B), these results suggest that at least three CWI genes are potentially co-expressed with GmCIF1 at the onset of seed maturity. Co-expression of CWI genes and GmCIF1 provides the potential for the direct interaction of their protein products. Moreover, such co-operation would help the efficient translocation of the hydrolyzed product to sink tissues via fine-tuning of enzyme activities (Ehneß and Roitsch, 1997). However, as concluded in a previous study on maize (Bate et al., 2004), it remains a big challenge to determine which CWI gene(s) would be inhibited by GmCIF1 as more than three possible CWIs targets for GmCIF1 can be envisioned.
Increased invertase activities in sink tissues are probably due to post-translational regulation via the alteration of levels of inhibitor proteins, as reported in a variety of species (Greiner et al., 1999; Jin et al., 2009; Leskow et al., 2016; Su et al., 2016). The increased CWI transcripts were found to be concomitant with elevated CWI activities at early stages of seed maturity (25 DAF) in the RNAi backgrounds (see Supplementary Fig. S4A). Interestingly, in the RNAi backgrounds, decreased but extremely low levels of CWI gene transcripts (GmCWI6, 7, and 8) were detected at the mid- or late stages of seed maturation (45 and 65 DAF), which were not in concert with the aforementioned significant elevations of CWI activities at these times (Fig. 5B, C). By contrast, GmCIF1 expression appeared to be more abundant at these stages of seed maturity in the wild-type background (Fig 7A, B). The inconsistency observed between the enzyme activities and gene expressions of CWI supports the idea that, at least during the seed maturation, CWIs are regulated at a post-translational level by GmCIF1. Moreover, along with the significantly elevated CWI activities in the RNAi backgrounds and concurrent with increases of GmCIF1 expression in the wild-type background, the post-translational regulation of CWI correlates more closely with the amount of inhibitor transcripts (or activities), suggesting a pivotal role for GmCIF1 later in the control of primary metabolism and biomass accumulation.
Conclusions
The search for the primary target of GmCIF1 in combination with enzyme assays and RNAi analysis revealed that GmCIF1 specifically inhibits CWI. We provide evidence that GmCIF1 plays a fundamental role in the post-translational control of extracellular invertase, which results in an impact on seed development and sucrose hydrolysis at a tissue-specific level. Silencing of GmCIF1 significantly elevated CWI activities in soybean plants. Our results are instrumental in helping to decipher how invertase inhibitors regulate CWI during seed maturation. Moreover, unravelling the mechanisms underlying the post-translational modulation of extracellular invertase in soybean has implications for plant biotechnology and may provide new cues for improving crop performance through manipulating the interaction of CWI and its inhibitory proteins. The potential roles of GmCIF1 in resistance to biotic and abiotic stresses as well as its impact on nutrient use efficiency will be the subject of future investigations.
Supplementary Material
Acknowledgments
The authors would like to thank the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), National Soybean Systems of Industry and Technology (CARS-04-PS05), National Special and Key Projects for GMO Research (2016ZX08004-002) for funding this research. Thanks to Dr Julia Wong and Dr Perrin H. Beatty for their support with the final revision of the manuscript. The authors declare that they have no conflicts of interest in relation to this work.
References
- Albacete A, Cantero-Navarro E, Großkinsky DK, et al. 2015. Ectopic overexpression of the cell wall invertase gene CIN1 leads to dehydration avoidance in tomato. Journal of Experimental Botany 66, 863–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnaresi P, Moschella A, Beretta O, Vitulli F, Ranalli P, Perata P. 2008. Heterologous microarray experiments allow the identification of the early events associated with potato tuber cold sweetening. BMC Genomics 9, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate NJ, Niu X, Wang Y, Reimann KS, Helentjaris TG. 2004. An invertase inhibitor from maize localizes to the embryo surrounding region during early kernel development. Plant Physiology 134, 246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar PB, Wu L, Busse JS, Whitty BR, Hamernik AJ, Jansky SH, Buell CR, Bethke PC, Jiang J. 2010. Suppression of the vacuolar invertase gene prevents cold-induced sweetening in potato. Plant Physiology 154, 939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihmidine S, Hunter CT, III, Johns CE, Koch KE, Braun DM. 2013. Regulation of assimilate import into sink organs: update on molecular drivers of sink strength. Frontiers in Plant Science 4, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Chen RK, Harris JC, Zhang H, Hamiaux C, Kralicek AV, McKenzie MJ. 2011. Induction of vacuolar invertase inhibitor mRNA in potato tubers contributes to cold-induced sweetening resistance and includes spliced hybrid mRNA variants. Journal of Experimental Botany 62, 3519–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai N, Schaffer A, Petreikov M, Shahak Y, Giller Y, Ratner K, Levine A, Granot D. 1999. Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. The Plant Cell 11, 1253–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwiche SR. 1998. Protein content of single kernels of wheat by near-infrared reflectance spectroscopy. Journal of Cereal Science 27, 241–254. [Google Scholar]
- Dimou M, Flemetakis E, Delis C, Aivalakis G, Spyropoulos KG, Katinakis P. 2005. Genes coding for a putative cell-wall invertase and two putative monosaccharide/H+ transporters are expressed in roots of etiolated Glycine max seedlings. Plant Science 169, 798–804. [Google Scholar]
- Doidy J, Grace E, Kühn C, Simon-Plas F, Casieri L, Wipf D. 2012. Sugar transporters in plants and in their interactions with fungi. Trends in Plant Science 17, 413–422. [DOI] [PubMed] [Google Scholar]
- Ehneß R, Roitsch T. 1997. Co-ordinated induction of mRNAs for extracellular invertase and a glucose transporter in Chenopodium rubrum by cytokinins. The Plant Journal 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Eskandari M, Cober ER, Rajcan I. 2013. Genetic control of soybean seed oil: II. QTL and genes that increase oil concentration without decreasing protein or with increased seed yield. Theoretical and Applied Genetics 126, 1677–1687. [DOI] [PubMed] [Google Scholar]
- Focks N, Benning C. 1998. wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiology 118, 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. 1968. Nutrient requirements of suspension cultures of soybean root cells. Experimental Cell Research 50, 151–158. [DOI] [PubMed] [Google Scholar]
- Greiner S, Krausgrill S, Rausch T. 1998. Cloning of a tobacco apoplasmic invertase inhibitor. Proof of function of the recombinant protein and expression analysis during plant development. Plant Physiology 116, 733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner S, Rausch T, Sonnewald U, Herbers K. 1999. Ectopic expression of a tobacco invertase inhibitor homolog prevents cold-induced sweetening of potato tubers. Nature Biotechnology 17, 708–711. [DOI] [PubMed] [Google Scholar]
- Han M, Heppel SC, Su T, Bogs J, Zu Y, An Z, Rausch T. 2013. Enzyme inhibitor studies reveal complex control of methyl-D-erythritol 4-phosphate (MEP) pathway enzyme expression in Catharanthus roseus. PLoS ONE 8, e62467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer AG, Raap M, Schroeer B, Marty B, Willmitzer L. 2004. Cell wall invertase expression at the apical meristem alters floral, architectural, and reproductive traits in Arabidopsis thaliana. The Plant Journal 39, 161–169. [DOI] [PubMed] [Google Scholar]
- Hothorn M, Bonneau F, Stier G, Greiner S, Scheffzek K. 2003. Bacterial expression, purification and preliminary X-ray crystallographic characterization of the invertase inhibitor Nt-CIF from tobacco. Acta Crystallographica. Section D, Biological Crystallography 59, 2279–2282. [DOI] [PubMed] [Google Scholar]
- Hothorn M, Van den Ende W, Lammens W, Rybin V, Scheffzek K. 2010. Structural insights into the pH-controlled targeting of plant cell-wall invertase by a specific inhibitor protein. Proceedings of the National Academy of Sciences 107, 17427–17432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Ni DA, Ruan YL. 2009. Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. The Plant Cell 21, 2072–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann EM, Hall B, Bennett AB. 1996. Antisense acid invertase (TIV1) gene alters soluble sugar composition and size in transgenic tomato fruit. Plant Physiology 112, 1321–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch K. 2004. Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Current Opinion in Plant Biology 7, 235–246. [DOI] [PubMed] [Google Scholar]
- Kohorn BD, Kobayashi M, Johansen S, Riese J, Huang LF, Koch K, Fu S, Dotson A, Byers N. 2006. An Arabidopsis cell wall-associated kinase required for invertase activity and cell growth. The Plant Journal 46, 307–316. [DOI] [PubMed] [Google Scholar]
- Krausgrill S, Greiner S, Köster U, Vogel R, Rausch T. 1998. In transformed tobacco cells the apoplasmic invertase inhibitor operates as a regulatory switch of cell wall invertase. Plant Journal 13, 275–280. [Google Scholar]
- Lara MEB, Garcia M-CG, Fatima T, Ehneß R, Lee TK, Proels R, Tanner W, Roitsch T. 2004. Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. The Plant Cell 16, 1276–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine R, La Camera S, Atanassova R, et al. 2013. Source-to-sink transport of sugar and regulation by environmental factors. Frontiers in Plant Science 4, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leskow CC, Kamenetzky L, Dominguez PG, et al. 2016. Allelic differences in a vacuolar invertase affect Arabidopsis growth at early plant development. Journal of Experimental Botany 67, 4091–4103. [DOI] [PubMed] [Google Scholar]
- Li B, Liu H, Zhang Y, Kang T, Zhang L, Tong J, Xiao L, Zhang H. 2013. Constitutive expression of cell wall invertase genes increases grain yield and starch content in maize. Plant Biotechnology Journal 11, 1080–1091. [DOI] [PubMed] [Google Scholar]
- Link M, Rausch T, Greiner S. 2004. In Arabidopsis thaliana, the invertase inhibitors AtC/VIF1 and 2 exhibit distinct target enzyme specificities and expression profiles. FEBS letters 573, 105–109. [DOI] [PubMed] [Google Scholar]
- Liu S, Lan J, Zhou B, et al. 2015. HbNIN2, a cytosolic alkaline/neutral-invertase, is responsible for sucrose catabolism in rubber-producing laticifers of Hevea brasiliensis (para rubber tree). The New Phytologist 206, 709–725. [DOI] [PubMed] [Google Scholar]
- Liu X, Lin Y, Liu J, Song B, Ou Y, Zhang H, Li M, Xie C. 2013. StInvInh2 as an inhibitor of StvacINV1 regulates the cold-induced sweetening of potato tubers by specifically capping vacuolar invertase activity. Plant Biotechnology Journal 11, 640–647. [DOI] [PubMed] [Google Scholar]
- McKenzie MJ, Chen RK, Harris JC, Ashworth MJ, Brummell DA. 2013. Post-translational regulation of acid invertase activity by vacuolar invertase inhibitor affects resistance to cold-induced sweetening of potato tubers. Plant, Cell & Environment 36, 176–185. [DOI] [PubMed] [Google Scholar]
- Miller ME, Chourey PS. 1992. The maize invertase-deficient miniature-1 seed mutation is associated with aberrant pedicel and endosperm development. The Plant Cell 4, 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nägele T, Henkel S, Hörmiller I, Sauter T, Sawodny O, Ederer M, Heyer AG. 2010. Mathematical modeling of the central carbohydrate metabolism in Arabidopsis reveals a substantial regulatory influence of vacuolar invertase on whole plant carbon metabolism. Plant Physiology 153, 260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proels RK, Hückelhoven R. 2014. Cell-wall invertases, key enzymes in the modulation of plant metabolism during defence responses. Molecular Plant Pathology 15, 858–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proulx RA, Naeve SL. 2009. Pod removal, shade, and defoliation effects on soybean yield, protein, and oil. Agronomy Journal 101, 971–978. [Google Scholar]
- Rausch T, Greiner S. 2004. Plant protein inhibitors of invertases. Biochimica et Biophysica Acta 1696, 253–261. [DOI] [PubMed] [Google Scholar]
- Reca IB, Brutus A, D’Avino R, Villard C, Bellincampi D, Giardina T. 2008. Molecular cloning, expression and characterization of a novel apoplastic invertase inhibitor from tomato (Solanum lycopersicum) and its use to purify a vacuolar invertase. Biochimie 90, 1611–1623. [DOI] [PubMed] [Google Scholar]
- Roitsch T. 1999. Source–sink regulation by sugar and stress. Current Opinion in Plant Biology 2, 198–206. [DOI] [PubMed] [Google Scholar]
- Roitsch T, Balibrea ME, Hofmann M, Proels R, Sinha AK. 2003. Extracellular invertase: key metabolic enzyme and PR protein. Journal of Experimental Botany 54, 513–524. [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. 2006. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annual Review of Plant Biology 57, 675–709. [DOI] [PubMed] [Google Scholar]
- Rosenkranz H, Vogel R, Greiner S, Rausch T. 2001. In wounded sugar beet (Beta vulgaris L.) tap-root, hexose accumulation correlates with the induction of a vacuolar invertase isoform. Journal of Experimental Botany 52, 2381–2385. [DOI] [PubMed] [Google Scholar]
- Ruan YL. 2014. Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annual Review of Plant Biology 65, 33–67. [DOI] [PubMed] [Google Scholar]
- Ruan YL, Jin Y, Yang YJ, Li GJ, Boyer JS. 2010. Sugar input, metabolism, and signaling mediated by invertase: roles in development, yield potential, and response to drought and heat. Molecular Plant 3, 942–955. [DOI] [PubMed] [Google Scholar]
- Ruhlmann JM, Kram BW, Carter CJ. 2010. Cell wall invertase 4 is required for nectar production in Arabidopsis. Journal of Experimental Botany 61, 395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwimmer S, Makower RU, Rorem ES. 1961. Invertase & invertase inhibitor in potato. Plant Physiology 36, 313–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva LI, Keurentjes JJB, Bentsink L, Vonk J, van der Plas LHW, Koornneef M, Vreugdenhil D. 2006. Vacuolar invertase regulates elongation of Arabidopsis thaliana roots as revealed by QTL and mutant analysis. Proceedings of the National Academy of Sciences, USA 103, 2994–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin AJ, Woody JL, Bolon YT, et al. 2010. RNA-seq atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biology 10, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song QX, Li QT, Liu YF, et al. 2013. Soybean GmbZIP123 gene enhances lipid content in the seeds of transgenic Arabidopsis plants. Journal of Experimental Botany 64, 4329–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A. 1999. Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiology 121, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Wolf S, Han M, Zhao H, Wei H, Greiner S, Rausch T. 2016. Reassessment of an Arabidopsis cell wall invertase inhibitor AtCIF1 reveals its role in seed germination and early seedling growth. Plant Molecular Biology 90, 137–155. [DOI] [PubMed] [Google Scholar]
- Tang GQ, Lüscher M, Sturm A. 1999. Antisense repression of vacuolar and cell wall invertase in transgenic carrot alters early plant development and sucrose partitioning. The Plant Cell 11, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology 3, RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schweinichen C, Büttner M. 2005. Expression of a plant cell wall invertase in roots of Arabidopsis leads to early flowering and an increase in whole plant biomass. Plant Biology 7, 469–475. [DOI] [PubMed] [Google Scholar]
- Wang E, Wang J, Zhu X, et al. 2008. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nature Genetics 40, 1370–1374. [DOI] [PubMed] [Google Scholar]
- Wang L, Ruan YL. 2012. New insights into roles of cell wall invertase in early seed development revealed by comprehensive spatial and temporal expression patterns of GhCWIN1 in cotton. Plant Physiology 160, 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Jiang N, Jia T, et al. 2012. Genome-wide association mapping of agronomic and morphologic traits in highly structured populations of barley cultivars. Theoretical and Applied Genetics 124, 233–246. [DOI] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U. 1996. Controlling seed development and seed size in Vicia faba: a role for seed coat-associated invertases and carbohydrate state. The Plant Journal 10, 823–834. [Google Scholar]
- Weschke W, Panitz R, Gubatz S, Wang Q, Radchuk R, Weber H, Wobus U. 2003. The role of invertases and hexose transporters in controlling sugar ratios in maternal and filial tissues of barley caryopses during early development. The Plant Journal 33, 395–411. [DOI] [PubMed] [Google Scholar]
- Wobus U, Weber H. 1999. Sugars as signal molecules in plant seed development. Biological Chemistry 380, 937–944. [DOI] [PubMed] [Google Scholar]
- Xiang L, Le Roy K, Bolouri-Moghaddam MR, Vanhaecke M, Lammens W, Rolland F, Van den Ende W. 2011. Exploring the neutral invertase-oxidative stress defence connection in Arabidopsis thaliana. Journal of Experimental Botany 62, 3849–3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Wang X, Zhang W, Qian T, Tang G, Guo Y, Zheng C. 2008. Antisense suppression of an acid invertase gene (MAI1) in muskmelon alters plant growth and fruit development. Journal of Experimental Botany 59, 2969–2977. [DOI] [PubMed] [Google Scholar]
- Zanor MI, Osorio S, Nunes-Nesi A, et al. 2009. RNA interference of LIN5 in tomato confirms its role in controlling Brix content, uncovers the influence of sugars on the levels of fruit hormones, and demonstrates the importance of sucrose cleavage for normal fruit development and fertility. Plant Physiology 150, 1204–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, Kossmann J, Smith AM. 2010. Starch: its metabolism, evolution, and biotechnological modification in plants. Annual Review of Plant Biology 61, 209–234. [DOI] [PubMed] [Google Scholar]
- Zeng P, Vadnais DA, Zhang Z, Polacco JC. 2004. Refined glufosinate selection in Agrobacterium-mediated transformation of soybean [Glycine max (L.) Merrill]. Plant Cell Reports 22, 478–482. [DOI] [PubMed] [Google Scholar]
- Zhao H, Xu L, Su T, Jiang Y, Hu L, Ma F. 2015. Melatonin regulates carbohydrate metabolism and defenses against Pseudomonas syringae pv. tomato DC3000 infection in Arabidopsis thaliana. Journal of Pineal Research 59, 109–119. [DOI] [PubMed] [Google Scholar]
- Zhu X, Gong H, He Q, Zeng Z, Busse JS, Jin W, Bethke PC, Jiang J. 2016. Silencing of vacuolar invertase and asparagine synthetase genes and its impact on acrylamide formation of fried potato products. Plant Biotechnology Journal 14, 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.