Highlight
The associations and causal relations revealed between 76 metabolites from 135 winter wheat lines and 17 372 SNP markers allow the genetic interventions on sets of metabolites to be dissected.
Keywords: Genome-wide association, metabolomics, mQTL, network analysis, Triticum aestivum, winter wheat
Abstract
We investigated associations between the metabolic phenotype, consisting of quantitative data of 76 metabolites from 135 contrasting winter wheat (Triticum aestivum) lines, and 17 372 single nucleotide polymorphism (SNP) markers. Metabolite profiles were generated from flag leaves of plants from three different environments, with average repeatabilities of 0.5–0.6. The average heritability of 0.25 was unaffected by the heading date. Correlations among metabolites reflected their functional grouping, highlighting the strict coordination of various routes of the citric acid cycle. Genome-wide association studies identified significant associations for six metabolic traits, namely oxalic acid, ornithine, L-arginine, pentose alcohol III, L-tyrosine, and a sugar oligomer (oligo II), with between one and 17 associated SNPs. Notable associations with genes regulating transcription or translation explained between 2.8% and 32.5% of the genotypic variance (pG). Further candidate genes comprised metabolite carriers (pG 32.5–38.1%), regulatory proteins (pG 0.3–11.1%), and metabolic enzymes (pG 2.5–32.5%). The combinatorial use of genomic and metabolic data to construct partially directed networks revealed causal inferences in the correlated metabolite traits and associated SNPs. The evaluated causal relationships will provide a basis for predicting the effects of genetic interferences on groups of correlated metabolic traits, and thus on specific metabolic phenotypes.
Introduction
Metabolomics is complementing the molecular breeding toolbox, facilitating the prediction of a number of important agronomic traits (Saito and Matsuda, 2010). Metabolite signatures are associated with plant growth (Meyer et al., 2007) and have been used to predict complex traits such as biomass (Riedelsheimer et al., 2012a) and grain yield (Xu et al., 2016). Furthermore, metabolites play key roles in plant defence against abiotic and biotic stresses (Buchanan et al., 2015). Hence, metabolite profiling has become a central component in obtaining a more holistic view of the effects of disease and abiotic stress on plant performance (Moco et al., 2007; Deborde et al., 2011; Kueger et al., 2012). This also holds true for fruit and seed traits in tomato (Solanum lycopersicum) (Schauer et al., 2006; Toubiana et al., 2012; Sauvage et al., 2014). The increasing role of metabolites as indirect traits in plant breeding requires a greater understand of the details of their genetic architecture.
Linkage mapping has been applied to dissect the genetic architecture of metabolite abundance in Arabidopsis thaliana (Keurentjes et al., 2006; Lisec et al., 2009) and rice (Oryza sativa) (Gong et al., 2013). Several quantitative trait loci (QTL) have been detected that control metabolite abundance; they are referred to as metabolomic QTL (mQTL). It is believed that genetic variation in metabolite abundance could be important in the adaptations to specific environmental conditions that allow particular genotypes to grow. mQTL analyses have helped to uncover co-regulated compounds and metabolic pathways, such as for glucosinolate (Keurentjes et al., 2006) and flavonoid (Gong et al., 2013) biosynthesis. Variation in the abundance of metabolites in the rice grain is largely determined by mQTL acting in concert with various regulatory factors (Matsuda et al., 2012). Several mQTL have been identified in maize (Zea mays), both in the grain (Zheng et al., 2008; Yan et al., 2011; Wen et al., 2014) and in the leaf (Riedelsheimer et al., 2012b; Wen et al., 2015). The above-outlined genome-wide mapping studies revealed several putative mQTL, suggesting that metabolite abundance has a complex genetic architecture in plants.
Wheat (Triticum aestivum L.) is one of the world’s most important crops, providing 20% of the total calories for the world’s population. Despite its economic relevance, only one study based on 233 doubled haploid lines has performed genome-wide mQTL mapping (Hill et al., 2015). This QTL study validated the utility of combining agronomic and metabolomics traits as an approach for identifying potential trait enhancement targets for breeding.
Here, an attempt has been made to associate the varied abundance of a set of 76 leaf metabolites with allelic status at a large number of genetic marker loci. The analysis was based on a panel of 135 European elite winter wheat lines, grown in three different environments. Some of the mQTL identified have been contextualized by applying a partial directed network analysis.
Materials and methods
Plant material and harvest
The genetic material consisted of 135 elite winter wheat lines adapted to Central Europe (Zhao et al., 2015). All genotypes were evaluated in replicated grain yield trials at three locations (Böhnshausen, Hadmersleben, and Hohenheim) in Germany in 2012 (Longin et al., 2013). Lines were grown in an incomplete block design with two replicates at each location. For each line, 10 flag leaf samples were randomly selected per replicate and location at the time when >60% of the lines had reached BBCH-69 (Lancashire et al., 1991). All plots were sampled from 9 to 11 am within 120 min as outlined in detail by Zhao et al. (2015). Flag leaf samples were snap-frozen in liquid nitrogen, freeze-dried, and powdered in an MM200 ball mill (Retsch, Haan, Germany).
Metabolite profiling
A mixed sample was assembled composed of equal amounts from each individual sample in order to capture systematic shifts during extraction and measurement. The mixed sample was processed in the same way as all other samples and its data were used to normalize for shifts during metabolite profile analysis across the data set for all three locations. All analyses were performed in duplicate. Polar metabolites were extracted twice with 500 μl of 70% methanol per 20 mg dry weight (DW), with 50 µM 13C6/D7 glucose as an internal standard (Sigma-Aldrich, Munich, Germany). The suspension was centrifuged (20000 g, 4°C, 20 min) and the two supernatants pooled. For phase separation, 1000 μl of water and 500 μl of chloroform were added. After centrifugation (20000 g, 4°C, 10 min), 30 µl of the upper phase was transferred to a glass vial and dried in a vacuum centrifuge.
Derivatization was performed using a MultiPurposeSampler (Gerstel, Mülheim an der Ruhr, Germany) connected to a GC-MS system by first adding 40 μl of methoxyamine hydrochloride (20 mg/ml in pyridine) for 2 h at 37°C, and then adding 40 μl of N-methyl-N-(trimethylsilyl) trifluoroacetamide containing the alkane retention time standard (C7-C30, 10 μg/μl, Sigma-Aldrich, Munich, Germany) for 30 min at 37°C (Lisec et al., 2006). The measuring device was an Agilent A7890 gas chromatograph (Böblingen, Germany) coupled to a Waters GCT Premier time-of-flight mass spectrometer (Milford, MA, USA). A 1 μL derivatized sample was injected in splitless mode at 240°C, and passed through a 30 m Rxi®-5Sil MS column (internal diameter 0.25 mm, and 0.25 μm film thickness) with a 5 m Integra-Guard® column (Restek, Bad Homburg, Germany) using the following parameters: 3 min at 80°C, followed by +5°C/min up to 300°C. Data were acquired with the MassLynx 4.1 software (Waters), capturing 10 spectra per second in the m/z range of 65–650 in centroid mode and applying the dynamic enhancement (DRE) mode. Compound annotation was performed by running authentic standards, determining retention indices and specific fragment spectra, and by comparing to information in the NIST database implemented in MassLynx 4.1 software and the Golm Metabolome Database (http://gmd.mpimp-golm.mpg.de/, last accessed 23 November 2016). Data processing and peak integration were performed using the QuanLynx tool implemented in MassLynx 4.1 software, utilizing specific masses for metabolite quantification (Supplementary Table S1 at JXB online).
Metabolic data analyses
The flexible Box–Cox power transformation was applied to achieve homoscedasticity of the residuals of metabolites (Piepho, 2009). Of the 85 metabolites assayed, 76 metabolites were retained after quality assessment (Supplementary Fig. S1 at JXB online) as outlined in detail by Anscombe and Tukey (1963). The adjusted entry means were estimated for the genome-wide association study (GWAS) (Zhao et al., 2014). A one-step model was used to estimate the genetic variance components of lines and the variance of genotype × environment interactions. The significance of variance component estimates was tested by comparing models using likelihood ratio tests in which halved P values were used as an approximation (Stram and Lee, 1994). Using the variance components we estimated the heritability on an entry-mean basis. We assumed fixed genetic effects and estimated the best linear unbiased estimates (BLUEs) of lines across environments. The metabolite abundances were analysed with the software package ASRemL-R 3.0 (Butler et al., 2009). Pairwise correlations between the abundances of the 76 metabolites were revealed by applying the average linkage clustering method, based on Pearson correlation coefficients. Visualization of abundant metabolite profiles in the various lines, and an integrated visualization of highly correlated metabolites in the context of metabolic pathways and networks, were based on the VANTED framework (http://www.vanted.org, last accessed 23 November 2016; Rohn et al., 2012).
Genotypic data generation
Fingerprinting of DNA from all genotypes was performed using a 90k SNP array using the Illumina Infinium assay (Gowda et al., 2014). We discarded loci which were monomorphic, had missing values of >5%, heterozygosity of >5% in inbred lines, or for which a minor allele frequency of <5% applied. After applying these quality controls, 17372 SNP markers remained for GWAS. Genomic positions were known for 14298 out of these 17372 SNP. Blast searches for candidate genes were performed against the IWGSCv2.2 database (http://pgsb.helmholtz-muenchen.de/plant/search.jsp, last accessed 23 November 2016; Nussbaumer et al. (2013)).
Genome-wide association mapping
Adjusted entry means of metabolic traits of each environment were used for genome-wide association mapping using a linear mixed model. Correction for population stratification was implemented using a kinship matrix (Jiang et al., 2015). Marker–trait association scans were conducted to detect main-effect mQTL using the following model:
where Y stands for the adjusted entry means of the 135 genotypes of each environment, β is a vector of environment effects, s is a vector of SNP effects, m is a vector of SNP × environment interaction effects, u is a vector of polygene background effects, and e is a vector of residual effects. X, S, , and Z are incidence matrices relating Y to β, s, m, and u. s was treated as a fixed effect and β, m, and u as random effects.
The significance of marker–trait associations was tested based on the Wald F statistic. The significance of SNP × environment interaction effects was determined by a likelihood ratio test (Stram and Lee, 1994). A procedure to control the false discovery rate (FDR) (Benjamini and Hochberg, 1995) was applied to correct for multiple testing at a significance level of P < 0.20. The proportion of the phenotypic variance explained by a single QTL (R2) and by all mQTL (adjusted R2) was estimated using a standard multiple regression approach (Utz et al., 2000). The proportion of the genotypic variance explained by a single mQTL and by all mQTL (pG) was determined as a proportion of the explained phenotypic variance standardized by broad-sense heritability. Association mapping analyses were performed using the ASReml-R 3.0 software (Butler et al., 2009).
Inferring causal phenotype networks
Causal phenotype networks among the metabolomic data were inferred using the heuristic QTL+phenotype supervised orientation (QPSO) search algorithm (Wang and van Eeuwijk, 2014). In the first step, undirected networks among all metabolites were pre-learnt using the PC-skeleton algorithm (Spirtes et al., 2000) implemented with the R package pcalg (Kalisch et al., 2012). In the second step, significant mQTL (P < 0.1, FDR correction) were added into the undirected network and the QPSO algorithm applied.
Results
Metabolite profiles are complex heritable traits with pronounced genotype × environment interaction
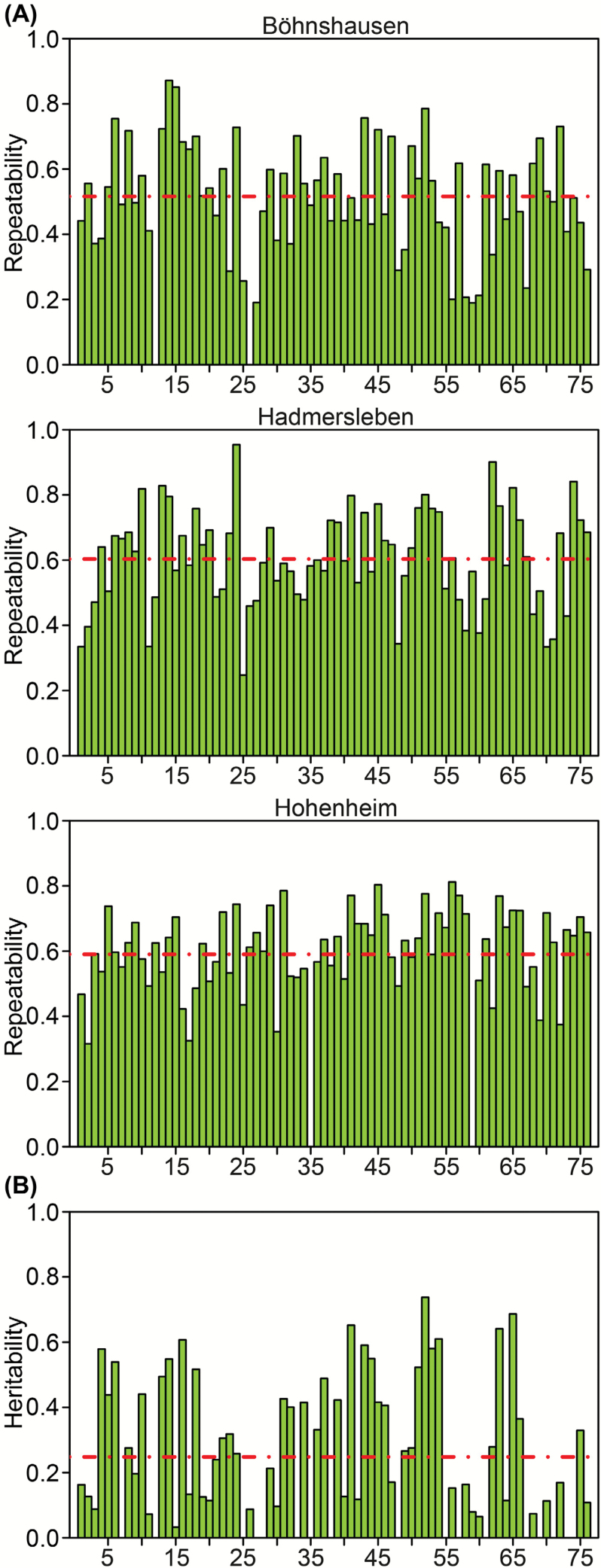
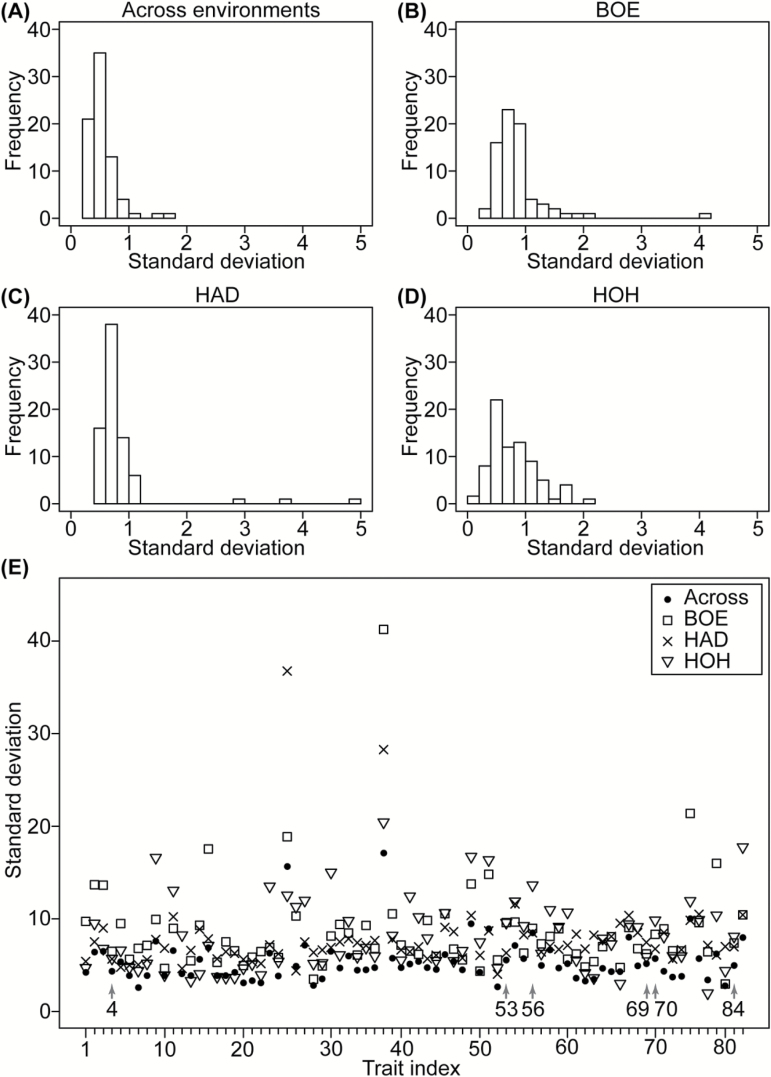
A total of 85 metabolites were recognized, but after quality evaluation, only 76 were retained for further analyses (Supplementary Tables S1 and S2; Fig. S2 at JXB online). The level of repeatability of metabolite abundance did not vary between the three sites, with a mean value of 0.52 at Böhnshausen, 0.60 at Hadmersleben, and 0.59 at Hohenheim (Fig. 1A). Furthermore, we observed no differences in the standard deviations of the genotypes across the three environments (Fig. 2). These findings indicate that the sampling and analytical strategies were appropriate and resulted in high-quality data regarding metabolite abundance.
Fig. 1.
(A) Reproducibility of metabolite abundances across the three field sites. The mean values, with 0.52 for Böhnshausen, 0.60 for Hadmersleben, and 0.59 for Hohenheim are indicated as dashed lines. (B) The heritability of abundance of the 76 metabolites across the three locations Hadmersleben, Böhnshausen, and Hohenheim. The mean value (0.25) is indicated as a dashed line.
Fig. 2.
Variation for standard deviations of metabolite profiles across and within environments. (A) Histogram for BLUEs across the three environments. Histograms for BLUEs within (B) Böhnshausen, (C) Hadmersleben, and (D) Hohenheim. (E) Individual variation of standard deviation for the investigated metabolites, red arrows indicate the six significantly associated metabolic traits.
The analyses of variance revealed a strong linear effect of the environments on the metabolite profiles (Supplementary Table S2). Individual metabolite profiles varied substantially across the three sites (Supplementary Figs S3 and S4 at JXB online), resulting in variances of genotypes that were on average only half the size of the variances of genotype × location interactions. As a result, the broad-sense heritabilities were low, with a global mean of 0.25 (Fig. 1B). In all, 24 of the metabolite abundances had a heritability of >0.4 and variances of genotypes which were significantly (P < 0.001) larger than zero (Supplementary Table S2). For 17 metabolite abundances, non-significant (P > 0.9) variation of genotypic effects was observed.
To evaluate the influence of the genetic differences in the 135 lines with respect to their developmental stage at the time the flag leaves were sampled, we estimated heritabilities for subgroups of early-, intermediate-, and late-maturing lines. The heritability estimates were similar for all three classes of lines (early 0.29, intermediate 0.23, late 0.21) (Supplementary Fig. S5 at JXB online). This implies that differences in heading date did not lead to a decrease in the heritability of flag leaf metabolite profiles in the total population of 135 lines.
Correlations among metabolites highlight the various routes of the citric acid cycle
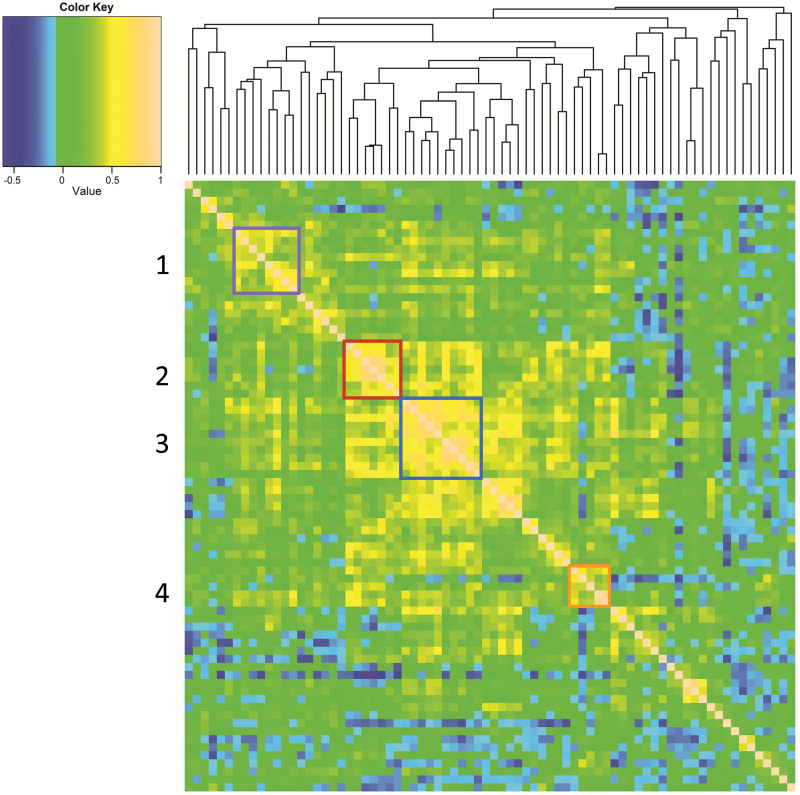
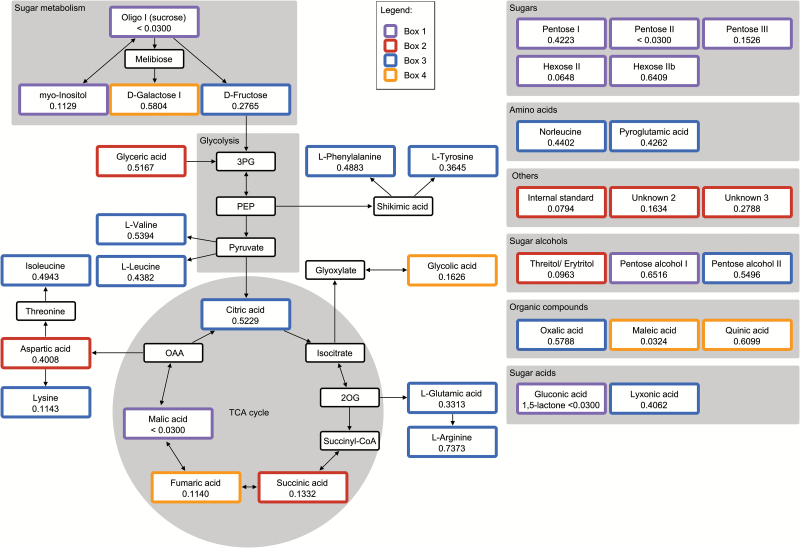
Out of the 2850 pairwise comparisons of metabolites, 794 (27.86%) were highly significantly correlated (P < 0.001), 1124 (39.44%) were moderately significantly correlated (P < 0.01), and 1462 (51.30%) were just significantly correlated (P < 0.05) (Supplementary Table S3 at JXB online). A number of regions with tightly correlated metabolites were identified, often reflecting functional relationships. Some examples are highlighted in Fig. 3, with detailed annotations given in Supplementary Fig. S6 at JXB online. One such group comprised sugars and sugar alcohols (box 1), one was a set of organic acids associated with the citric acid cycle (boxes 2 and 4), and another included various small organic acids and amino acids (box 3). Pathway analysis showed that metabolites representing the main entry points of glycolysis and the citric acid cycle clustered within box 1 (Fig. 4, shown in violet); a second group represented intermediates of amino acid and nucleic acid biosynthesis (box 2, red). The large metabolite group clustered within box 3 (blue) comprised amino acids involved in the citric acid cycle, together with key metabolites such as D-fructose and citric acid. Small organic acids related to the glycolate cycle and photorespiration clustered into box 4 (orange), implying a degree of joint regulation between photosynthesis and the citric acid/ glycolate cycles.
Fig. 3.
Correlation pattern among metabolites. Pairwise Pearson correlations are shown in a heat map representation, whereas metabolites are sorted according to correlation-based hierarchical cluster analysis. High positive correlations are represented by earth tone squares and negative ones by blue squares. The raw data are presented in Supplementary Table S4, and annotation of the cluster tree arms is shown in Supplementary Fig. S6.
Fig. 4.
Pathways involving the strongly correlated metabolite pairs indicated in Fig. 3. The most prominent groups represent elements of the citric acid cycle. Unmapped metabolites have been sorted into compound groups and are shown in the right panel. Frame colours refer to box 1 (violet), box 2 (red), box 3 (blue), and box 4 (orange). The heritability of abundance of the presented metabolites across the three locations Hadmersleben, Böhnshausen, and Hohenheim is shown within the relevant boxes.
Metabolomics and genomic diversity
We estimated distance matrices among the 135 lines based on the metabolite abundances or the genomic profiles. These matrices were only weakly associated, with a correlation coefficient of r = 0.02 (P < 0.34). A cluster analysis based on the genome-wide SNP marker data revealed the absence of a major population structure but the presence of a family structure among the 135 parental lines (Supplementary Fig. S7 at JXB online). Therefore, a linear mixed model was applied for the GWAS to correct for relatedness, modelling a kinship matrix.
GWA mapping revealed associations for six metabolic traits
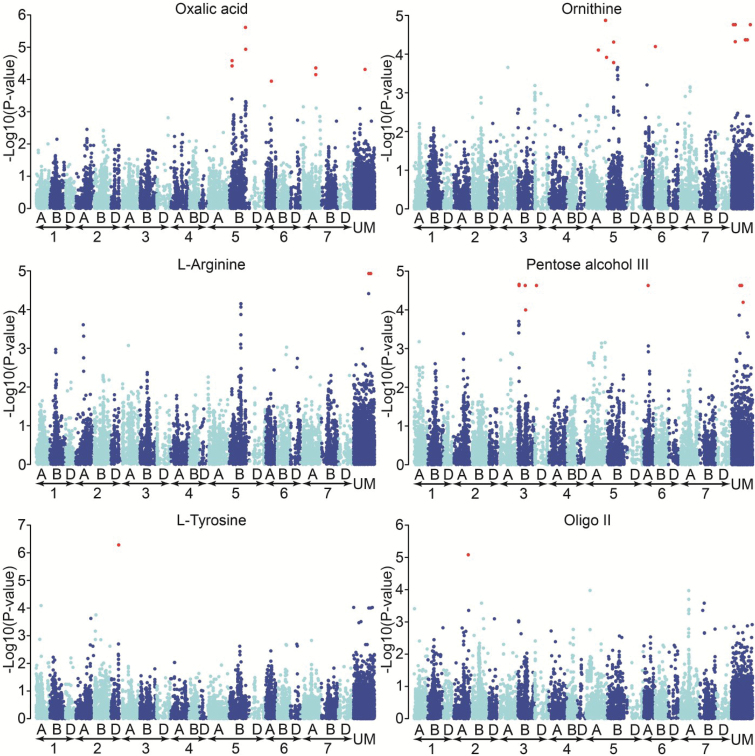
The GWA scan was applied to the 34 metabolites for which abundance was associated with a significant degree of genotypic variance. The resulting six significant SNP–metabolite abundance associations involved 40 SNPs and oxalic acid, ornithine, L-arginine, pentose alcohol III, L-tyrosine, and oligo II (Table 1 and Fig. 5). Interestingly, none of the 40 SNPs exhibited significant interaction effects with the environments (Table 1).
Table 1.
Summary of GWA mapping
| Metabolite | H 2 | Locus | Chromosome position | P SNP | P SNP×Environment | R 2 | p G | Gene ID or accession | Splice variants | Candidate gene product | Variation type |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Oxalic acid | 0.578 | IWB8786* | 5B 548.67cM | 2.43E-06 | 1 | 11.88 | 20.55 | no match | Unknown | ||
| IWB4387 | 5B 558.07cM | 1.15E-05 | 1 | 0.06 | 0.10 | no match | Unknown | ||||
| IWB14354* | 7A 532.61cM | 4.40E-05 | 1 | 6.60 | 11.42 | Traes_7DL_98245621B.2 | 1 | Transcription elongation factor (TFIIS) | N | ||
| IWB35367* | Not Mapped | 4.88E-05 | 1 | 11.11 | 19.22 | Traes_5BS_EA4A42ADB.1 | 1 | GTP-binding protein | S | ||
| IWB47615* | 5B 92.03cM | 3.78E-05 | 1 | 1.79 | 3.10 | no match | Unknown | ||||
| IWA3211 | 5B 92.03cM | 3.78E-05 | 1 | s.a. | s.a. | no match | Unknown | ||||
| IWA6946 | 5B 92.03cM | 3.78E-05 | 1 | s.a. | s.a. | no match | Unknown | ||||
| IWA6947 | 5B 92.03cM | 2.61E-05 | 1 | 0.13 | 0.22 | no match | Unknown | ||||
| IWB29877 | 7A 532.61cM | 7.10E-05 | 1 | 0.09 | 0.16 | Traes_7AL_968CDEFCB.2 | 1 | Transcription elongation factor (TFIIS) | N | ||
| IWB33544* | 6A 190.27cM | 1.14E-04 | 1 | 6.72 | 11.63 | Traes_6BS_60D771A56.1 | 1 | Galactose oxidase/kelch repeat protein | S | ||
| Ornithine | 0.27 | IWB4446* | 5A 703.91cM | 1.34E-05 | 1 | 10.28 | 38.07 | Traes_5AL_F49663738.2 | 1 | Hexose carrier protein HEX6 | N |
| IWB6885* | Not Mapped | 1.74E-05 | 1 | 12.16 | 45.04 | no match | Unknown | ||||
| IWB8628 | Not Mapped | 1.74E-05 | 1 | s.a. | s.a. | no match | Unknown | ||||
| IWB8637 | Not Mapped | 1.74E-05 | 1 | s.a. | s.a. | no match | Unknown | ||||
| IWB60850 | 5A 709.71cM | 1.74E-05 | 1 | s.a. | s.a. | Traes_4DL_FA7457EA3.1 | 1 | Hexose carrier protein HEX6 | N | ||
| IWB75174 | Not Mapped | 1.74E-05 | 1 | s.a. | s.a. | no match | Unknown | ||||
| IWB3615* | 5B 222.57cM | 4.90E-05 | 1 | 5.48 | 20.30 | no match | Unknown | ||||
| IWB8638 | Not Mapped | 4.77E-05 | 1 | 0.02 | 0.07 | no match | Unknown | ||||
| IWB46787* | Not Mapped | 4.29E-05 | 1 | 3.22 | 11.93 | Traes_5AL_A26E70EFB.1 | 1 | Unknown | S | ||
| IWB48775 | Not Mapped | 4.29E-05 | 1 | s.a. | s.a. | Traes_5AL_217A5FE69.2 | 1 | Unknown | N | ||
| IWB58986 | Not Mapped | 4.29E-05 | 1 | s.a. | s.a. | Traes_5DL_939F0D4E2.1 | 1 | Unknown | S | ||
| IWA2098* | 6B 1.28cM | 6.39E-05 | 1 | 2.89 | 10.70 | Traes_6BS_EA1DF4148.1 | 1 | Enhanced disease resistance 2-like protein | N | ||
| IWB55921* | 5A 459.12cM | 7.86E-05 | 1 | 1.61 | 5.96 | Traes_5AL_6BA7849E7.1 | 1 | Unknown | S | ||
| IWA2558 | 5A 737.3cM | 1.21E-04 | 1 | 0.00 | 0.00 | Traes_5AL_19637DE03.1 | 1 | AP-2 complex subunit alpha-2- like protein | N | ||
| IWB5567 | 5B 221.73cM | 1.66E-04 | 1 | 0.67 | 2.48 | Traes_5BL_F48317F91.1 | 1 | Probable flavin-containing monooxygenase 1 | S | ||
| IWB7364 | 5B 221.73cM | 1.66E-04 | 1 | s.a. | s.a. | no match | Unknown | ||||
| IWB43461 | 5B 221.73cM | 1.66E-04 | 1 | s.a. | s.a. | Traes_5AL_EA74144D2 | 1 | Probable flavin-containing monooxygenase 1 | S | ||
| L-Arginine | 0.737 | IWB56221* | Not Mapped | 1.17E-05 | 1 | 12.69 | 17.22 | Traes_5BL_F367A99A7.1 | 1 | Basic leucine zipper 25 | 3′ UTR |
| IWB65729 | Not Mapped | 1.17E-05 | 1 | s.a. | s.a. | X80068.1 | Basic leucin zipper 1 | 3′ UTR | |||
| Pentose alcohol III | 0.686 | IWB242* | 3B 265.41cM | 2.11E-06 | 0.44 | 22.27 | 32.46 | Traes_3B_E23F2C86F.1 | 12 | Unknown | N |
| IWB1705 | 3D 50.05cM | 2.11E-06 | 0.44 | s.a. | s.a. | Traes_3AS_0A005B552.1 | 1 | Eukaryotic translation initiation factor 3A | S | ||
| IWB25161 | Not Mapped | 2.11E-06 | 0.44 | s.a. | s.a. | Traes_3DS_6774BF63A.2 | 1 | ABC transporter G family member 7 protein | S | ||
| IWB32797 | Not Mapped | 2.11E-06 | 0.44 | s.a. | s.a. | Traes_3AS_EA87ED745.1 | 1 | Peroxisomal membrane protein PEX14 | |||
| IWB35825 | 3B 56.41cM | 2.11E-06 | 0.44 | s.a. | s.a. | no match | Unknown | ||||
| IWB56509 | 6A 156.81cM | 2.11E-06 | 0.44 | s.a. | s.a. | Traes_3DS_C4E8AD8B4.2 | 1 | Cellulose synthase catalytic subunit | S | ||
| IWB65026 | 3B 56.41cM | 2.20E-05 | 0.43 | 0.29 | 0.42 | Traes_3AS_82679750F.2 | 1 | E3 ubiquitin- protein ligase SINA-like protein 4 | S | ||
| IWB35763* | Not Mapped | 6.41E-05 | 0.46 | 0.85 | 1.24 | Traes_3AS_4E919E6081.3 | 1 | Putative histidine- rich Ca2+-binding protein | S | ||
| IWB18115* | 3B 278.07cM | 1.01E-04 | 0.39 | 1.94 | 2.83 | Traes_3AS_47780354D.2 | 1 | Transcriptional activator DEMETER | Intron | ||
| L-Tyrosine | 0.364 | IWB49741* | 2Dx 269.06cM | 5.15E-07 | 1 | 17.36 | 47.69 | no match | Unknown | ||
| Oligo II | 0.329 | IWB6807* | 2A 506.66cM | 8.33E-06 | 1 | 14.73 | 44.77 | Traes_2DL_AADC2EA0F.1 | 1 | ABC transporter E family member 2 protein | S |
Only mQTL associated with a P < 0.2 after FDR correction are listed. The chromosomal SNP positions refer to the reference annotations from the high-density 90k SNP array (Wang et al., 2014). H2: heritability; R2: proportion (%) of phenotypic variance explained by the SNP; pG: proportion (%) of genetic variance explained by the SNP; Locus: GrainGenes locus according to chromosome maps available at http://wheat.pw.usda.gov/GG3/, last accessed 23 November 2016; Gene ID of candidate gene -search against ‘TriticumAestivum_IWGSC_map_AND_unmap_CDS.fa’ at http://pgsb.helmholtz-muenchen.de/plant/search.jsp last accessed 23 November 2016; Candidate gene product -source: http://plants.ensembl.org/, last accessed 23 November 2016; s.a.: similar as above, indicating highly linked SNPs; N: variation type non-synonymous; S: variation type synonymous; 3′ UTR: 3′ untranslated region; * significant SNPs (P < 0.1, FDR correction) used for the network analysis by the QPSO algorithm; please refer to Fig. 6.
Fig. 5.
Manhattan plots displaying the GWA mapping analysis for metabolites generating a significant association signal. Significant P values (0.2 after FDR correction) are shown in red.
The sequences in which the associated SNPs were embedded (Supplementary Table S4 at JXB online) were queried against available wheat genome sequence data to define a set of candidate genes, and against available plant gene and protein sequences (http://plants.ensembl.org/, last accessed 23 November 2016) to gather any known functional information. Only 21 of the sequences could be functionally assigned. The candidate genes encoded several transporters, catalytic enzymes, proteins involved in molecular signalling processes, transcriptional and translational regulators, and one defence-related protein. Most of the annotated genes are thought to generate just a single message, but one generates 12 splicing variants (Table 1). Additionally, we analysed the type of variation for the assigned SNPs. We detected a number of synonymous (S) and non-synonymous (N) variations as well as variation in the 3′ untranslated region (UTR) for two of the assigned SNPs and one SNP with an intron. The respective sites of variation and the corresponding amino acid changes are shown in Supplementary Table S4. We observed similar variation types for functionally similar SNPs, such as for a flavin-containing monooxygenase 1 (S) and for a basic leucine zipper (bZIP) protein (3′ UTR).
Ten mQTL were mapped for oxalic acid to chromosomes 5B, 7A, and 6A (Table 1). The strongest associations involved a chromosome 5B non-annotated sequence and an unmapped sequence encoding a GTP-binding protein. Two highly associated SNPs, co-locating to chromosome 7A, marked a gene encoding a transcription elongation factor (TFIIS). Weaker associations were noted for a chromosome 5B sequence, a non-annotated sequence harbouring four SNPs on chromosome 5B, and a chromosome 6A sequence encoding a galactose oxidase/kelch repeat protein.
In total, 17 mQTL were detected for ornithine (Table 1). The strongest associations related to six SNPs with a proportion of genotypic variance >38%, two of which resided in a pair of closely linked genes, both encoding a hexose carrier protein, located on chromosome 5A. Significant associations were also noted for sequences mapping to chromosome 5B and chromosome 6B; the latter has been annotated as encoding an enhanced disease resistance 2-like protein. Among the less strongly associated SNPs, three were unmapped but likely involved in Ca2+ transport, one lay in a gene of unknown function mapping to chromosome 5A, and three were in a single site on chromosome 5B which likely corresponds to a gene encoding a flavin-containing monooxygenase 1. Additional weakly associated sequences included one unmapped locus and a second mapping to chromosome 5A encoding an AP-2 complex subunit alpha-2-like protein, probably also involved in Ca2+ transport. Our results point to the importance of Ca2+ and sugar balance for ornithine metabolism.
Nine SNPs were significantly associated with variation for pentose alcohol III abundance. The strongest association involved a gene of unknown function mapping to chromosome 3B. A second locus mapping to chromosome 6A matched the site of a gene encoding a cellulose synthase catalytic subunit. Three other significant associations involved genes encoding for an ABC transporter G family member 7 protein, a peroxisomal membrane protein PEX14 (both unmapped), and the eukaryotic translation initiation factor 3A. Among the more weakly associated SNPs were one encoding for the transcription activator DEMETER, an E3 ubiquitin-protein ligase SINA-like protein 4, an unmatched SNP at the same chromosome position, and an unmapped SNP encoding for a putative calcium-binding protein. Overall, we concluded that the associations found for pentose alcohol III involved a strong transcriptional and translational regulation.
Two L-arginine mQTL were identified, the larger one explaining 17.2% of the variance. Both informative SNPs lay within genes encoding a bZIP protein, with both yet to be chromosomally mapped.
A high proportion of the variance was explained by the sequences associated with the L-tyrosine (47.69%) and oligo II (44.77%) mQTL. The oligo II mQTL was associated with a gene encoding an ABC-transporter protein.
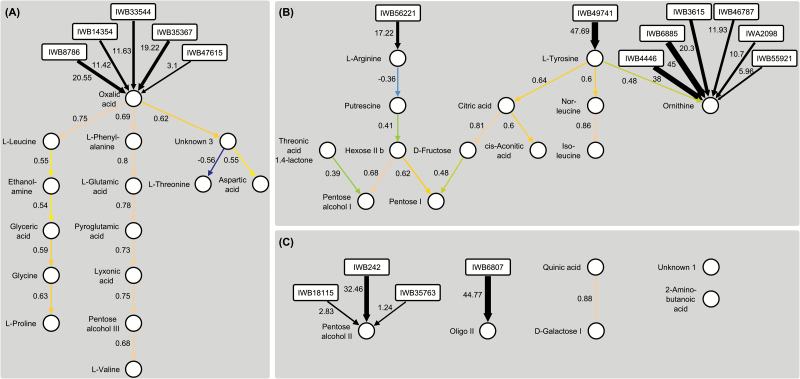
Network analysis revealed causal relationships among metabolites
Causal relationships were inferred from the reconstructed partially directed networks (Fig. 6). The five genes strongly associated with oxalic acid abundance appeared to exert a pleiotropic effect on a number of causally related metabolites, likely mediated by the concentration of oxalic acid (Fig. 6A). In the relevant metabolic network, oxalic acid lies directly upstream of L-leucine, L-phenylalanine, and an as yet unidentified metabolite (‘unknown 3’). The abundance of the latter directly affects the abundances of both L-threonine (negatively) and aspartic acid (positively). This causal relationship is of interest, because aspartic acid synthesis is thought to be catalysed by the reversible transamination of oxaloacetate, its direct precursor in the citric acid cycle; meanwhile, L-threonine is thought to be synthesized from aspartic acid via homoserine. On the other hand, oxalic acid is hydrolysed from oxaloacetate and not yet known to be a precursor or regulator of L-threonine and aspartic acid biosynthesis. In the second network branch, L-phenylalanine was shown to lie upstream of L-glutamic acid, pyroglutamic acid, lyxonic acid, pentose alcohol II, and L-valine. This constellation indicates control of various branches of amino acid biosynthesis by the concentration of L-phenylalanine, which in turn depends on that of oxalic acid. Ethanolamine, glyceric acid, glycine, and L-proline all lie downstream of L-leucine, indicating a relationship between the abundance of the branched amino acid L-leucine with that of the secondary amine L-proline, as well as with the abundance of certain glycolysis intermediates and derived small amino acids. Notably, the biosynthesis of both L-proline and L-leucine involves several enzymes acting on precursors of, respectively, L-glutamic acid and pyruvate. Because glycolysis uses precursors upstream of pyruvate, and pyruvate also lies upstream of L-glutamic acid, it seems unlikely that L-leucine exerts feedback control in both directions. Thus, the positioning of L-phenylalanine upstream of L-glutamic acid and L-valine seems to be more plausible. No functional test of these suggested causal relationships has been attempted so far, and the importance of oxalic acid in this network has yet to be confirmed.
Fig. 6.
Partially directed network graphs describing the causal relationships among the 34 metabolites showing a significant genotypic variance (P < 0.05). The QPSO algorithm inferred two directed clusters (A) and (B) as well as some unrelated metabolites (C). SNP nodes are represented by rectangles and the proportion (%) of the explained genetic variance is both given as a number and indicated by the thickness of the connecting arrows. Metabolite nodes are represented by circles and correlations are displayed both numerically and graphically by coloured arrows (the colours correspond to the heat map in Fig. 3). High positive correlations are represented in earth tones and negative ones in blue.
In the second cluster (Fig. 6B), ornithine abundance was associated with 17 SNPs but only one causal relationship with the upstream L-tyrosine could be inferred. Ornithine is derived from L-glutamic acid, a side product of the citric acid cycle, while L-tyrosine is synthesized via the shikimate pathway, which lies upstream of the citric acid cycle; a proposed causal relationship between the abundance of ornithine and that of L-tyrosine therefore seems plausible. Similarly, the positioning of the isomers norleucine and isoleucine (derived from pyruvate) downstream of L-tyrosine, as well as that of citric acid, appears to be logical. A surprising link is the negative association established between L-arginine and its breakdown product putrescine with the hexose pool (hexose II and D-fructose), pentose I, and pentose alcohol I. This causal relationship was also linked to citric acid via D-fructose, prompting the suggestion that the gene associated with L-arginine abundance could influence amino acid catabolism via the ornithine pathway, as well as influencing the size of the pools of soluble carbon-related precursors of amino acid synthesis.
Discussion
Continuing improvements in metabolomics and genomics have helped unravel the genetic basis of metabolic diversity in plants (Fiehn, 2002; Fernie and Schauer, 2009; Saito and Matsuda, 2010). While much of the effort has been focused on model species (Keurentjes et al., 2006; Lisec et al., 2009), progress has been recorded in maize (Wen et al., 2014), rice (Luo, 2015), and barley (Hordeum vulgare) (Cockram et al., 2010). Recent advances in developing the genomic toolbox in wheat (http://www.wheatgenome.org/, last accessed 23 November 2016) (Choulet et al., 2014; Marcussen et al., 2014; Pfeifer et al., 2014) also paving the way for a deeper understanding of metabolic diversity in wheat (Hill et al., 2015).
Metabolite abundance is strongly influenced by genotype × environment interactions
The reproducibility of the metabolite profiling was in line with previous findings in maize (Riedelsheimer et al., 2012a; Wen et al., 2014). Riedelsheimer et al. (2012a) acquired GC-MS-based metabolite profiles for 130 compounds from leaves of 285 maize lines. Wen et al. (2014) included 184 annotated and 799 unknown metabolites detected on LC-MS from kernels of 702 maize genotypes. Thus, comparable precision can be reached independent of the plant species, organ, and the technology applied for metabolite profiling.
We observed pronounced environment × genotype interactions, which were expected because the metabolic status of a plant is governed by the concerted action of many enzymes, each of which can respond to the growing environment (Fiehn, 2002; Tohge and Fernie, 2010; Luo, 2015). Accordingly, significant variations in metabolite levels from durum wheat were attributed to genotype × environment interactions (Beleggia et al., 2013). In tomato, the correlation with respect to metabolite abundance between the two seasons sampled was high for only about one half of the metabolites, a result revealing a substantial contribution of genotype × ripening stage interactions (Sauvage et al., 2014). In contrast, a comprehensive analysis of rice metabolome diversity based on 840 metabolites collected for 529 accessions in two environments estimated a heritability of >0.5 for 59% of the metabolites and >0.7 for 24% of them (W Chen et al., 2014). Similarly, when the performance of 175 rice lines was characterized at three environments, 68% of metabolite abundances were associated with a heritability of >0.5 (Matsuda et al., 2015). The authors suggested that these high heritability values were mainly a result of the large diversity within the investigated set of rice genotypes.
Metabolite correlations suggest a level of coordination between synthetic pathways
The strongest inter-metabolite correlations involved sugars, small organic acids, and amino acids (Fig. 3), a pattern seen also in Arabidopsis (Sulpice et al., 2010), maize (Riedelsheimer et al., 2012b; Wen et al., 2015), tomato (Sauvage et al., 2014), and rice (Matsuda et al., 2012); which also relied on GC-MS technology for metabolite profiling. Because this approach favours the profiling of primary metabolites (Fiehn, 2002; Deborde et al., 2011), the detection of the aforementioned compound classes is somehow predestined. However, besides this restriction to a sub-fraction of the metabolome, we could conclude that there exists coordinated control over sets of metabolites, representing key compounds, intermediates, or products of glycolysis and the citric acid cycle. Small organic acids related to the glycolate cycle and photorespiration behaved in a coordinated fashion, which implies a regulatory connection between photosynthesis and the citric acid/glycolate cycles. LC-MS-based metabolite profiling enables a larger structural diversity to be covered (Fernie and Schauer, 2009; Tohge and Fernie, 2010). This has made it possible to dissect the genetic components of polyamine and flavonoid biosynthesis in rice (Wen et al., 2014) and to gain insights into the genetic architecture underlying natural variation in rice primary and secondary metabolism (W Chen et al., 2014; Matsuda et al., 2015). These advancements in speed and coverage will allow for more comprehensive and large-scale elucidation of metabolite and gene–metabolite correlations in the near future.
GWAS can elucidate the genetic basis of metabolite abundance
Our GWAS revealed mQTL for six metabolites that did not exhibit significant interactions with the environment. None of them have formerly been reported in wheat (Hill et al., 2015). The mQTL accounted for around 30% of the variance for ornithine, L-tyrosine, and oligo II, and for over 58% of that for oxalic acid, L-arginine, and pentose alcohol III (Supplementary Table S2). Similar results have been reported in barley (Cockram et al., 2010). Assuming a high degree of genetic complexity for the metabolic traits in our data set, we conclude that only a small proportion of the SNPs that control polygenic traits were likely to be detected using our approach.
The 40 informative SNPs were dispersed unevenly over the genome (Table 1), and there was not a single case of an individual SNP being associated with more than one metabolite, unlike what has been observed in cognate studies on primary and secondary metabolites (Bergelson and Roux, 2010; Matsuda et al., 2012; Sauvage et al., 2014). Most of the metabolites were found to be tightly associated with only a small number of strong mQTL in our study, which might hint at mechanisms which confer tight control over these metabolite levels.
With respect to the four sequences associated with oxalic acid abundance, one putatively encodes TFIIS (a transcription elongation factor), one a GTP-binding protein, and one a galactose oxidase/kelch repeat protein. TFIIS enhances the activity of RNA polymerase II by reactivating transcription elongation complexes in which transcription has been arrested (Booth et al., 2000). In Arabidopsis TFIIS loss-of-function mutants, only around 2.3% of genes have altered transcription. This includes the upregulation of genes involved in glycoside, glucosinolate, and general metabolism and the downregulation of genes controlling secondary metabolism and the synthesis of amino acids and aromatic compounds (Grasser et al., 2009). It is unclear in which direction TFIIS affects oxalic acid levels. GTP-binding proteins are signalling proteins involved in the control of cell division, cell cycling, ribosome assembly, and protein synthesis (Miyawaki and Yang, 2014; Urano and Jones, 2014). In the filamentous fungi Botrytis cinerea, mutants lacking the expression of a small GTPase (BcCdc42) had impaired nuclear division, germination, and virulence, and a reduced ability to secrete oxalic acid (Kokkelink et al., 2011). Aluminium (Al) tolerance in wheat is highly correlated with an Al-activated release of small organic acids, such as citric acid, malic acid, and oxalic acid, and oxalic acid is also involved in intracellular Al detoxification. Genetic markers linked to Al tolerance loci are shared between rice, wheat, and barley (Kochian et al., 2004). A possible involvement of GTPase signalling in the Al ion-induced secretion of organic acids in both Arabidopsis and rye (Secale cereale) has only recently been
reported (YY Li et al., 2015). Thus the GTPase gene highlighted by our GWAS could function in Al-mediated oxalic acid level induction. Finally, galactose oxidase/kelch repeat proteins could be connected to oxalic acid abundance through their oxidation of primary alcohols, although no a priori experimental evidence for this has been found in the literature.
With respect to ornithine abundance, the candidate sequences identified encoded two hexose carrier proteins, closely linked to one another on chromosome 5A. These proteins belong to a large family of membrane proteins responsible for the binding and transport of various carbohydrates, organic alcohols, and acids (Silverman, 1991). The strong association between these loci and ornithine abundance implies that one or both gene products are intimately involved in the regulation of ornithine level. Two of the three informative SNPs lay within a chromosome 5B sequence encoding flavin-containing monooxygenase 1. Flavin monooxygenases catalyse a wide range of oxygenation reactions and have been classified into eight groups (Huijbers et al., 2014), one of which includes the YUCCAs (EC 1.14.13.168), which closely resemble ornithine monooxygenases at the primary sequence level. All plants contain these enzymes (Dai et al., 2013).
The two genes associated with L-arginine abundance both encoded bZIP proteins. Members of the bZIP superfamily of eukaryotic DNA-binding transcription factors act as master regulators of a large number of developmental and physiological processes (Alves et al., 2013). The wheat genome harbours at least 187 bZIP genes (X Li et al., 2015), one of which (TabZIB113) is identical to Traes_5BL_F367A99A7.1. This gene was differentially transcribed during wheat anther development, but its function is not known.
Genes associated with the abundance of pentose alcohol III were distributed across the genome. The putative candidate genes included one encoding a cellulose synthase catalytic subunit, a eukaryotic translation initiation factor 3A, a DEMETER transcription activator, an E3 ubiquitin-protein ligase SINA-like protein 4, and a putative calcium-binding protein. Further potentially informative associations involved two unmapped genes, one encoding a member of the G family of ABC transporters and the other the peroxisomal membrane protein PEX14. Similarly, a gene encoding a member of the E family of ABC transporters was identified as a possible candidate for an oligo II mQTL. The implied comparable regulatory mechanisms for these two sugars is reasonable, given the importance of sugars for growth (Eveland and Jackson, 2012; Kadereit et al., 2014). Nevertheless, as yet it cannot be determined whether the products of any of these genes act as direct mediators of sugar level or whether they form part of the relevant regulatory machinery.
Combining genotypic and phenotypic data uncovers novel causal relationships
While conventional correlation analysis effectively identifies interactions between metabolites or between metabolites and enzymes or genes (undirected), it often neglects the underlying metabolic and genetic networks (causalities, directed) (Sulpice et al., 2010; Tohge and Fernie, 2010; Wang and van Eeuwijk, 2014). Using recent methodology (Wang and van Eeuwijk, 2014), we revealed genetic interventions on groups of metabolites in wheat. Our analysis, based on a set of statistically significant mQTL (P < 0.1 corrected for FDR), identified oxalic acid as a key regulator or precursor of various branches of amino acid synthesis. Oxalic acid is hydrolysed via three pathways that involve glyoxylate/glycolic acid, ascorbic acid, and oxaloacetate (Kadereit et al., 2014), all of which are derived from the citric acid cycle. From the prominent position of oxalic acid in our partially directed network, we conclude that oxalic acid exerts feedback control over a number of steps in amino acid biosynthesis. Experimental evidence was provided by an in vitro study showing that oxalic acid improves protein synthesis by enhancing the ATP supply in a cell-free system derived from Escherichia coli (Kim and Swartz, 2000). Further support is supplied by the known activity of oxalic acid as a signalling molecule for a number of host–pathogen interactions (Lehner et al., 2008), as well as from its direct involvement in conferring tolerance to abiotic stress (Kochian et al., 2004). In Arabidopsis, Sulpice et al. (2010) have documented strong correlations between the synthesis of amino acids and the abundance of malic acid and some other small organic acids – but, notably, not oxalic acid.
The negative relationship between the abundance of L-arginine and its breakdown product putrescine with a side-link to upstream L-tyrosine (Fig. 6) has not been reported for any other plant species to date. Toubiana et al. (2012) observed a negative correlation between the putrescine and maltose content in tomato, while indirect evidence for the regulatory activity exerted by putrescine over several synthetic pathways comes from significant epistatic effects associated with its abundance in maize (Wen et al., 2015).
Overall, the outcome supports an extensive co-regulation of central pathways in both N and C metabolism, as previously postulated (Gutiérrez et al., 2007; Toubiana et al., 2012). However, experimental evidence on the detected SNP-metabolite relationships is needed to confirm the postulated regulatory networks. Ideally, such studies include the analysis of mutant or introgression lines, as has been done in pioneering works for Arabidopsis (Keurentjes et al., 2006; Tohge and Fernie, 2010) and rice (Gong et al., 2013; W Chen et al., 2014), as well as re-sequencing approaches as reported for maize (Wen et al., 2014). Similar prospects can be anticipated for wheat in the near future owing to the advancements in cereal transformation and sequencing technologies.
Conclusions
A number of new gene–metabolite associations and their causal relationships have been revealed in this study, confirming the potential of the metabolite GWAS approach even in a polyploid species. The limitations of this approach relate in part to the complex inheritance of metabolite abundance and in part to the limited availability of sets of genetic markers and analysable metabolites. These constraints could be overcome by additional genomic marker development and fine-mapping based on new deep-sequencing and transcriptomics technologies (Díaz et al., 2012; Kahl et al., 2012; Würschum et al., 2015). Technical improvements to metabolomic platforms (Saito and Matsuda, 2010; Deborde et al., 2011; Tohge et al., 2015) should be able to address the latter restriction, and some progress in this direction has been made in both maize (Wen et al., 2014) and rice (Matsuda et al., 2015). More sophisticated choices of germplasm can enhance the effectiveness of GWA approaches, as demonstrated in rice, for which a standard association panel has been combined with a number of conventional mapping populations (W Chen et al., 2014; Huang et al., 2010). In turn, these sophisticated functional genomics approaches rely on the establishment of novel high-throughput phenotyping platforms (D Chen et al., 2014). The recent extension of marker-assisted selection by powerful genomic selection approaches has proven applicable to metabolite markers, although with reduced accuracy when compared to genomic markers. The expectation is for further advances in knowledge-based breeding strategies, predicated on the idea of predicting plant performance from combinations of markers, which will further facilitate crop improvement in the years to come.
Author contributions
AM, H-PM, JCR, EE, VK, FL, RS, EK, and JS designed the research; AM, GL, YJ, YZ, and AH performed the research and analysed data; EE, RS, EK, and JS performed field trials and provided plant material; AM, GL, and JCR wrote the paper; HW provided support for the network analysis. The authors declare no conflict of interest.
Supplementary Material
Acknowledgements
The authors wish to gratefully acknowledge the provision of the genotypes and the conductance of field trials and phenotyping by State Plant Breeding Institute (University of Hohenheim, Germany), Nordsaat Saatzuchtgesellschaft mbH (Langenstein, Germany), Lantmännen SW Seed (Hadmersleben, Germany), Limagrain GmbH (Peine-Rosenthal, Germany), and KWS LOCHOW GmbH (Bergen, Germany). We also thank Daniel Kriegel, Martin Lietz, Petra Linow, and Annegret Wolf for their technical assistance. Financial support to AM and YZ was provided by the BMBF (FKZ0315945D).
References
- Alves MS, Dadalto SP, Gonçalves AB, De Souza GB, Barros VA, Fietto LG. 2013. Plant bZIP transcription factors responsive to pathogens: a review. International Journal of Molecular Sciences 14, 7815–7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anscombe FJ, Tukey JW. 1963. The examination and analysis of residuals. Technometrics 5, 141–160. [Google Scholar]
- Beleggia R, Platani C, Nigro F, De Vita P, Cattivelli L, Papa R. 2013. Effect of genotype, environment and genotype-by-environment interaction on metabolite profiling in durum wheat (Triticum durum Desf.) grain. Journal of Cereal Science 57, 183–192. [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery date: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300. [Google Scholar]
- Bergelson J, Roux F. 2010. Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nature Reviews. Genetics 11, 867–879. [DOI] [PubMed] [Google Scholar]
- Booth V, Koth CM, Edwards AM, Arrowsmith CH. 2000. Structure of a conserved domain common to the transcription factors TFIIS, elongin A, and CRSP70. The Journal of Biological Chemistry 275, 31266–31268. [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Gruissem W, Jones RL. (eds). 2015. Biochemistry and molecular biology of plants. Wiley Blackwell, Chichester, UK. [Google Scholar]
- Butler D, Cullis BR, Gilmour A, Gogel B. 2009. ASReml-R reference manual. The State of Queensland, Department of Primary Industries and Fisheries, Brisbane. [Google Scholar]
- Chen D, Neumann K, Friedel S, Kilian B, Chen M, Altmann T, Klukas C. 2014. Dissecting the phenotypic components of crop plant growth and drought responses based on high-throughput image analysis. The Plant Cell 26, 4636–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Gao Y, Xie W, et al. 2014. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nature Genetics 46, 714–721. [DOI] [PubMed] [Google Scholar]
- Choulet F, Alberti A, Theil S, et al. 2014. Structural and functional partitioning of bread wheat chromosome 3B. Science 345, 1249721. [DOI] [PubMed] [Google Scholar]
- Cockram J, White J, Zuluaga DL, et al. 2010. Genome-wide association mapping to candidate polymorphism resolution in the unsequenced barley genome. Proceedings of the National Academy of Sciences 107, 21611–21616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Mashiguchi K, Chen Q, Kasahara H, Kamiya Y, Ojha S, DuBois J, Ballou D, Zhao Y. 2013. The biochemical mechanism of auxin biosynthesis by an Arabidopsis YUCCA flavin-containing monooxygenase. The Journal of Biological Chemistry 288, 1448–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deborde C, Erban A, Kopka J, Goodacre R, Hall RD. 2011. Plant metabolomics and its potential for systems biology research: background concepts, technology, and methodology. Methods in Systems Biology 500, 299–336. [DOI] [PubMed] [Google Scholar]
- Díaz A, Zikhali M, Turner AS, Isaac P, Laurie DA. 2012. Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PloS One 7, e33234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveland AL, Jackson DP. 2012. Sugars, signalling, and plant development. Journal of Experimental Botany 63, 3367–3377. [DOI] [PubMed] [Google Scholar]
- Fernie AR, Schauer N. 2009. Metabolomics-assisted breeding: a viable option for crop improvement? Trends in Genetics: TIG 25, 39–48. [DOI] [PubMed] [Google Scholar]
- Fiehn O. 2002. Metabolomics—the link between genotypes and phenotypes. Plant Molecular Biology 48, 155–171. [PubMed] [Google Scholar]
- Gong L, Chen W, Gao Y, Liu X, Zhang H, Xu C, Yu S, Zhang Q, Luo J. 2013. Genetic analysis of the metabolome exemplified using a rice population. Proceedings of the National Academy of Sciences 110, 20320–20325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda M, Zhao Y, Würschum T, et al. 2014. Relatedness severely impacts accuracy of marker-assisted selection for disease resistance in hybrid wheat. Heredity 112, 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasser M, Kane CM, Merkle T, Melzer M, Emmersen J, Grasser KD. 2009. Transcript elongation factor TFIIS is involved in Arabidopsis seed dormancy. Journal of Molecular Biology 386, 598–611. [DOI] [PubMed] [Google Scholar]
- Gutiérrez RA, Lejay LV, Dean A, Chiaromonte F, Shasha DE, Coruzzi GM. 2007. Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biology 8, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CB, Taylor JD, Edwards J, Mather D, Langridge P, Bacic A, Roessner U. 2015. Detection of QTL for metabolic and agronomic traits in wheat with adjustments for variation at genetic loci that affect plant phenology. Plant Science 233, 143–154. [DOI] [PubMed] [Google Scholar]
- Huang X, Wei X, Sang T, et al. 2010. Genome-wide association studies of 14 agronomic traits in rice landraces. Nature Genetics 42, 961–967. [DOI] [PubMed] [Google Scholar]
- Huijbers MM, Montersino S, Westphal AH, Tischler D, van Berkel WJ. 2014. Flavin dependent monooxygenases. Archives of Biochemistry and Biophysics 544, 2–17. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Zhao Y, Rodemann B, et al. 2015. Potential and limits to unravel the genetic architecture and predict the variation of Fusarium head blight resistance in European winter wheat (Triticum aestivum L.). Heredity 114, 318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadereit JW, Körner C, Kost B, Sonnewald U. 2014. Strasburger—Lehrbuch der Pflanzenwissenschaften. Springer-Verlag, Berlin. [Google Scholar]
- Kahl G, Molina C, Rotter B, Jüngling R, Frank A, Krezdorn N, Hoffmeier K, Winter P. 2012. Reduced representation sequencing of plant stress transcriptomes. Journal of Plant Biochemistry and Biotechnology 21, 119–127. [Google Scholar]
- Kalisch M, Mächler M, Colombo D, Maathuis MH, Bühlmann P. 2012. Causal inference using graphical models with the R package pcalg. Journal of Statistical Software 47, 1–26. [Google Scholar]
- Keurentjes JJ, Fu J, de Vos CH, et al. 2006. The genetics of plant metabolism. Nature Genetics 38, 842–849. [DOI] [PubMed] [Google Scholar]
- Kim D-M, Swartz JR. 2000. Oxalate improves protein synthesis by enhancing ATP supply in a cell-free system derived from Escherichia coli. Biotechnology Letters 22, 1537–1542. [Google Scholar]
- Kochian LV, Hoekenga OA, Piñeros MA. 2004. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annual Review of Plant Biology 55, 459–493. [DOI] [PubMed] [Google Scholar]
- Kokkelink L, Minz A, Al-Masri M, Giesbert S, Barakat R, Sharon A, Tudzynski P. 2011. The small GTPase BcCdc42 affects nuclear division, germination and virulence of the gray mold fungus Botrytis cinerea. Fungal Genetics and Biology: FG & B 48, 1012–1019. [DOI] [PubMed] [Google Scholar]
- Kueger S, Steinhauser D, Willmitzer L, Giavalisco P. 2012. High-resolution plant metabolomics: from mass spectral features to metabolites and from whole-cell analysis to subcellular metabolite distributions. The Plant Journal 70, 39–50. [DOI] [PubMed] [Google Scholar]
- Lancashire PD, Bleiholder H, Boom T, Langelüddeke P, Stauss R, Weber E, Witzenberger A. 1991. A uniform decimal code for growth stages of crops and weeds. Annals of Applied Biology 119, 561–601. [Google Scholar]
- Lehner A, Meimoun P, Errakhi R, Madiona K, Barakate M, Bouteau F. 2008. Toxic and signalling effects of oxalic acid: oxalic acid-natural born killer or natural born protector? Plant Signaling & Behavior 3, 746–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gao S, Tang Y, Li L, Zhang F, Feng B, Fang Z, Ma L, Zhao C. 2015. Genome-wide identification and evolutionary analyses of bZIP transcription factors in wheat and its relatives and expression profiles of anther development related TabZIP genes. BMC Genomics 16, 976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Tang XL, Yang LG, Yu YX, Li XF. 2015. Possible involvement of heterotrimeric G-protein signaling in Al-induced secretion of organic acid anions in Arabidopsis and rye. Plant and Soil 388, 55–63. [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. 2006. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nature Protocols-Electronic Edition 1, 387–396. [DOI] [PubMed] [Google Scholar]
- Lisec J, Steinfath M, Meyer RC, Selbig J, Melchinger AE, Willmitzer L, Altmann T. 2009. Identification of heterotic metabolite QTL in Arabidopsis thaliana RIL and IL populations. The Plant Journal 59, 777–788. [DOI] [PubMed] [Google Scholar]
- Longin C, Gowda M, Mühleisen J, et al. 2013. Hybrid wheat: quantitative genetic parameters and consequences for the design of breeding programs. Theoretical and Applied Genetics 126, 2791–2801. [DOI] [PubMed] [Google Scholar]
- Luo J. 2015. Metabolite-based genome-wide association studies in plants. Current Opinion in Plant Biology 24, 31–38. [DOI] [PubMed] [Google Scholar]
- Marcussen T, Sandve SR, Heier L, et al. 2014. Ancient hybridizations among the ancestral genomes of bread wheat. Science 345, 1250092. [DOI] [PubMed] [Google Scholar]
- Matsuda F, Nakabayashi R, Yang Z, Okazaki Y, Yonemaru J-i, Ebana K, Yano M, Saito K. 2015. Metabolome-genome-wide association study dissects genetic architecture for generating natural variation in rice secondary metabolism. The Plant Journal 81, 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Okazaki Y, Oikawa A, et al. 2012. Dissection of genotype–phenotype associations in rice grains using metabolome quantitative trait loci analysis. The Plant Journal 70, 624–636. [DOI] [PubMed] [Google Scholar]
- Meyer RC, Steinfath M, Lisec J, et al. 2007. The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proceedings of the National Academy of Sciences 104, 4759–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki KN, Yang Z. 2014. Extracellular signals and receptor-like kinases regulating ROP GTPases in plants. Frontiers in Plant Science 5, 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moco S, Vervoort J, Bino RJ, De Vos RC, Bino R. 2007. Metabolomics technologies and metabolite identification. TrAC Trends in Analytical Chemistry 26, 855–866. [Google Scholar]
- Nussbaumer T, Martis MM, Roessner SK, Pfeifer M, Bader KC, Sharma S, Gundlach H, Spannagl M. 2013. MIPS PlantsDB: a database framework for comparative plant genome research. Nucleic Acids Research 41, D1144–D1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer M, Kugler KG, Sandve SR, Zhan B, Rudi H, Hvidsten TR, Mayer KF, Olsen OA; International Wheat Genome Sequencing Consortium 2014. Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 345, 1250091. [DOI] [PubMed] [Google Scholar]
- Piepho H-P. 2009. Ridge regression and extensions for genomewide selection in maize. Crop Science 49, 1165–1176. [Google Scholar]
- Riedelsheimer C, Czedik-Eysenberg A, Grieder C, et al. 2012. Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nature Genetics 44, 217–220. [DOI] [PubMed] [Google Scholar]
- Riedelsheimer C, Lisec J, Czedik-Eysenberg A, et al. 2012b Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proceedings of the National Academy of Sciences 109, 8872–8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohn H, Junker A, Hartmann A, Grafahrend-Belau E, Treutler H, Klapperstück M, Czauderna T, Klukas C, Schreiber F. 2012. VANTED v2: a framework for systems biology applications. BMC Systems Biology 6, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Matsuda F. 2010. Metabolomics for functional genomics, systems biology, and biotechnology. Annual Review of Plant Biology 61, 463–489. [DOI] [PubMed] [Google Scholar]
- Sauvage C, Segura V, Bauchet G, Stevens R, Do PT, Nikoloski Z, Fernie AR, Causse M. 2014. Genome-wide association in tomato reveals 44 candidate loci for fruit metabolic traits. Plant Physiology 165, 1120–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Roessner U, et al. 2006. Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nature Biotechnology 24, 447–454. [DOI] [PubMed] [Google Scholar]
- Silverman M. 1991. Structure and function of hexose transporters. Annual Review of Biochemistry 60, 757–794. [DOI] [PubMed] [Google Scholar]
- Spirtes P, Glymour CN, Scheines R. 2000. Causation, prediction, and search. MIT Press, Cambridge, MA. [Google Scholar]
- Stram DO, Lee JW. 1994. Variance components testing in the longitudinal mixed effects model. Biometrics 50, 1171–1177. [PubMed] [Google Scholar]
- Sulpice R, Trenkamp S, Steinfath M, et al. 2010. Network analysis of enzyme activities and metabolite levels and their relationship to biomass in a large panel of Arabidopsis accessions. The Plant Cell 22, 2872–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Fernie AR. 2010. Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nature Protocols 5, 1210–1227. [DOI] [PubMed] [Google Scholar]
- Tohge T, Scossa F, Fernie AR. 2015. Integrative approaches to enhance understanding of plant metabolic pathway structure and regulation. Plant Physiology 169, 1499–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubiana D, Semel Y, Tohge T, et al. 2012. Metabolic profiling of a mapping population exposes new insights in the regulation of seed metabolism and seed, fruit, and plant relations. PLoS Genetics 8, e1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Jones AM. 2014. Heterotrimeric G protein-coupled signaling in plants. Annual Review of Plant Biology 65, 365–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utz HF, Melchinger AE, Schön CC. 2000. Bias and sampling error of the estimated proportion of genotypic variance explained by quantitative trait loci determined from experimental data in maize using cross validation and validation with independent samples. Genetics 154, 1839–1849. [PMC free article] [PubMed] [Google Scholar]
- Wang H, van Eeuwijk FA. 2014. A new method to infer causal phenotype networks using QTL and phenotypic information. PloS One 9, e103997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Li D, Li X, et al. 2014. Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nature Communications 5, 3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Li K, Alseekh S, et al. 2015. Genetic determinants of the network of primary metabolism and their relationships to plant performance in a maize recombinant inbred line population. The Plant Cell 27, 1839–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würschum T, Boeven PH, Langer SM, Longin CF, Leiser WL. 2015. Multiply to conquer: copy number variations at Ppd-B1 and Vrn-A1 facilitate global adaptation in wheat. BMC Genetics 16, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Xu Y, Gong L, Zhang Q. 2016. Metabolomic prediction of yield in hybrid rice. The Plant Journal Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Yan WH, Wang P, Chen HX, et al. 2011. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Molecular Plant 4, 319–330. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Li Z, Liu G, et al. 2015. Genome-based establishment of a high-yielding heterotic pattern for hybrid wheat breeding. Proceedings of the National Academy of Sciences 112, 15624–15629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Mette MF, Gowda M, Longin CF, Reif JC. 2014. Bridging the gap between marker-assisted and genomic selection of heading time and plant height in hybrid wheat. Heredity 112, 638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Wu J, Lou X, Xu H, Shi C. 2008. The QTL analysis on maternal and endosperm genome and their environmental interactions for characters of cooking quality in rice (Oryza sativa L.). TAG. Theoretical and Applied Genetics. Theoretische und Angewandte Genetik 116, 335–342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.