Abstract
The Gram-negative bacterium Campylobacter jejuni 81116 (Penner serotype HS:6) has a class E lipooligosaccharide (LOS) biosynthesis locus containing 19 genes, which encode for 11 putative glycosyltransferases, 1 lipid A acyltransferase and 7 enzymes thought to be involved in the biosynthesis of dideoxyhexosamine (ddHexN) moieties. Although the LOS outer core structure of C. jejuni 81116 is still unknown, recent mass spectrometry analyses suggest that it contains acetylated forms of two ddHexN residues. For this investigation, five of the genes encoding enzymes reportedly involved in the biosyntheses of these sugar residues were examined, rmlA, rmlB, wlaRA, wlaRB and wlaRG. Specifically, these genes were cloned and expressed in Escherichia coli, and the corresponding enzymes were purified and tested for biochemical activity. Here we present data demonstrating that RmlA functions as a glucose-1-phosphate thymidylyltransferase and that RmlB is a thymidine diphosphate (dTDP)-glucose 4,6-dehydratase. We also show, through nuclear magnetic resonance spectroscopy and mass spectrometry analyses, that WlaRG, when utilized in coupled assays with either WlaRA or WlaRB and dTDP-4-keto-6-deoxyglucose, results in the production of either dTDP-3-amino-3,6-dideoxy-d-galactose (dTDP-Fuc3N) or dTDP-3-amino-3,6-dideoxy-d-glucose (dTDP-Qui3N), respectively. In addition, the X-ray crystallographic structures of the 3,4-ketoisomerases, WlaRA and WlaRB, were determined to 2.14 and 2.0 Å resolutions, respectively. Taken together, the data reported herein demonstrate that C. jejuni 81116 utilizes five enzymes to synthesize dTDP-Fuc3N or dTDP-Qui3N and that WlaRG, an aminotransferase, can function on sugars with differing stereochemistry about their C-4′ carbons. Importantly, the data reveal that C. jejuni 81116 has the ability to synthesize two isomeric ddHexN forms.
Keywords: Campylobacter, fucosamine, ketoisomerase, lipooligosaccharide, quinovosamine
Introduction
Campylobacter jejuni is a leading cause of human bacterial gastroenteritis and thus represents a major public health concern worldwide. The handling and consumption of contaminated poultry products accounts for almost half of all human infections (Galanis 2007). Diarrhea and abdominal pain are typical symptoms of infection that in most cases resolve without medical intervention (Pennie et al. 1986). In more severe cases, however, infections with C. jejuni can lead to the development of Guillain-Barré syndrome, a serious and potentially fatal neurological condition (Jacobs et al. 1998). Neuropathy is thought to be induced through molecular mimicry by the lipooligosaccharide (LOS) structures of C. jejuni. These resemble human gangliosides thereby causing the development of autoimmune-induced dysfunction of the peripheral nervous system (Yuki et al. 2004).
Lipopolysaccharides (LPS) and their lower molecular weight counterparts, LOS, are located on the outer membranes of Gram-negative bacteria, and these large surface molecules are readily recognized as antigenic by the host immune systems. In an effort to increase immune evasion and survival, bacteria modify the composition of their LOSs, and indeed, they employ highly variable loci as a means of developing robust phenotypes (Moxon et al. 1994). In the case of C. jejuni, the LOS loci are located in hypervariable regions, with slip strand mismatch repair mediating phase variation (Parkhill et al. 2000).
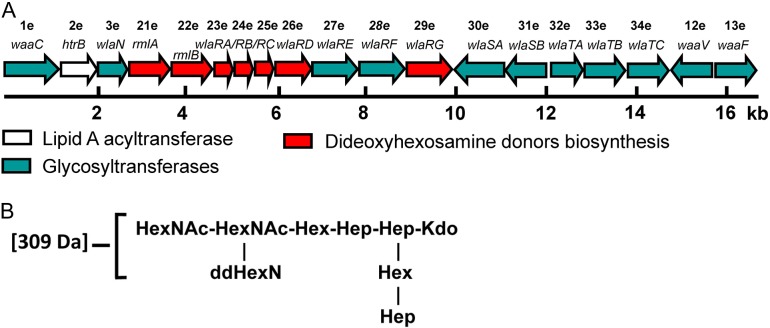
Although the LOS biosynthesis genes are found in one locus in all C. jejuni strains, variations in their order and composition do exist between classes. Currently, there are 22 extant classes of LOS found in C. jejuni strains (Richards et al. 2013). Relevant to this investigation is C. jejuni 81116, a commonly used laboratory strain, which has a class E LOS locus that does not include genes for sialic acid incorporation. The proposed functions of the class E LOS genes are provided in Table I, and as listed the locus is composed of 19 genes, encoding for putative glycosyltransferases, a lipid A acyltransferase, and a group of enzymes required for dideoxyhexosamine (ddHexN) biosynthesis (Figure 1). Although its genome has been sequenced and intensely studied, the exact chemical structure of the C. jejuni 81116 LOS is presently unknown. Mass spectrometry analysis suggests the presence of two acetylated ddHexN residues in the LOS outer core (Holden et al. 2012) (Figure 1).
Table I.
Location, name and proposed functions of the genes found in the class E LOS biosynthesis locus
| ORF #a | Gene nameb | Proposed function | Reference |
|---|---|---|---|
| 1e | waaC | Heptosyltransferase I | Klena et al. (1998), Kanipes et al. (2006) |
| 2e | htrB | Lipid A acyltransferase | Phongsisay et al. (2007) |
| 3e | wlaN | Glycosyltransferase | |
| 21e | rmlA | Glucose-1-phosphate thymidylyltransferase | This work |
| 22e | rmlB | 4,6-dehydratase | This work |
| 23e | wlaRA | 3,4-ketoisomerase | This work |
| 24e | wlaRB | 3,4-ketoisomerase | This work |
| 25e | wlaRC | N-acetyltransferase | |
| 26e | wlaRD | N-formyltransferase | Thoden et al. (2013) |
| 27e | wlaRE | Glycosyltransferase | |
| 28e | wlaRF | Glycosyltransferase | |
| 29e | wlaRG | Aminotransferase | Holden et al. (2012), this work |
| 30e | wlaSA | Glycosyltransferase | |
| 31e | wlaSB | Glycosyltransferase | |
| 32e | wlaTA | Glycosyltransferase | |
| 33e | wlaTB | Glycosyltransferase | Holden et al. (2012) |
| 34e | wlaTC | Glycosyltransferase | Holden et al. (2012) |
| 12e | waaV | Glycosyltransferase | |
| 13e | waaF | Heptosyltransferase | Oldfield et al. (2002) |
aAccording to Parker et al. (2008).
bAccording to Holden et al. (2012).
Fig. 1.
The C. jejuni LOS synthesis locus. Shown in (A) is a schematic of the LOS synthesis locus from C. jejuni strain 81116 (Pearson et al. 2007). The putative names and functions of the enzymes encoded by the open reading frame (ORF) numbers are listed in Table I. The LOS outer core structure proposed for C. jejuni 81116 is outlined in (B) (Holden et al. 2012). This figure is available in black and white in print and in color at Glycobiology online.
For this investigation, five of the genes reportedly involved in the biosynthesis of these sugar residues were examined, namely rmlA, rmlB, wlaRA, wlaRB and wlaRG. The proteins encoded by these genes were expressed and purified in Escherichia coli and their biochemical activities examined by nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry. In addition, the 3D structures of two of these proteins, WlaRA and WlaRB, were determined by single crystal X-ray diffraction, and their kinetic parameters were measured. Taken together, the data presented herein demonstrate that C. jejuni 81116 has the ability to synthesize two different ddHexN residues rather than one, and determine the identity of each ddHexN residue. Importantly, the data show that WlaRG, a sugar aminotransferase, can function on substrates that differ in their stereochemistry about the C-4′ carbon. Finally, the structural and functional analyses of WlaRA and WlaRB shed new light into our previously proposed catalytic mechanisms for the 3,4-ketoisomerases.
Results
Biochemical analyses
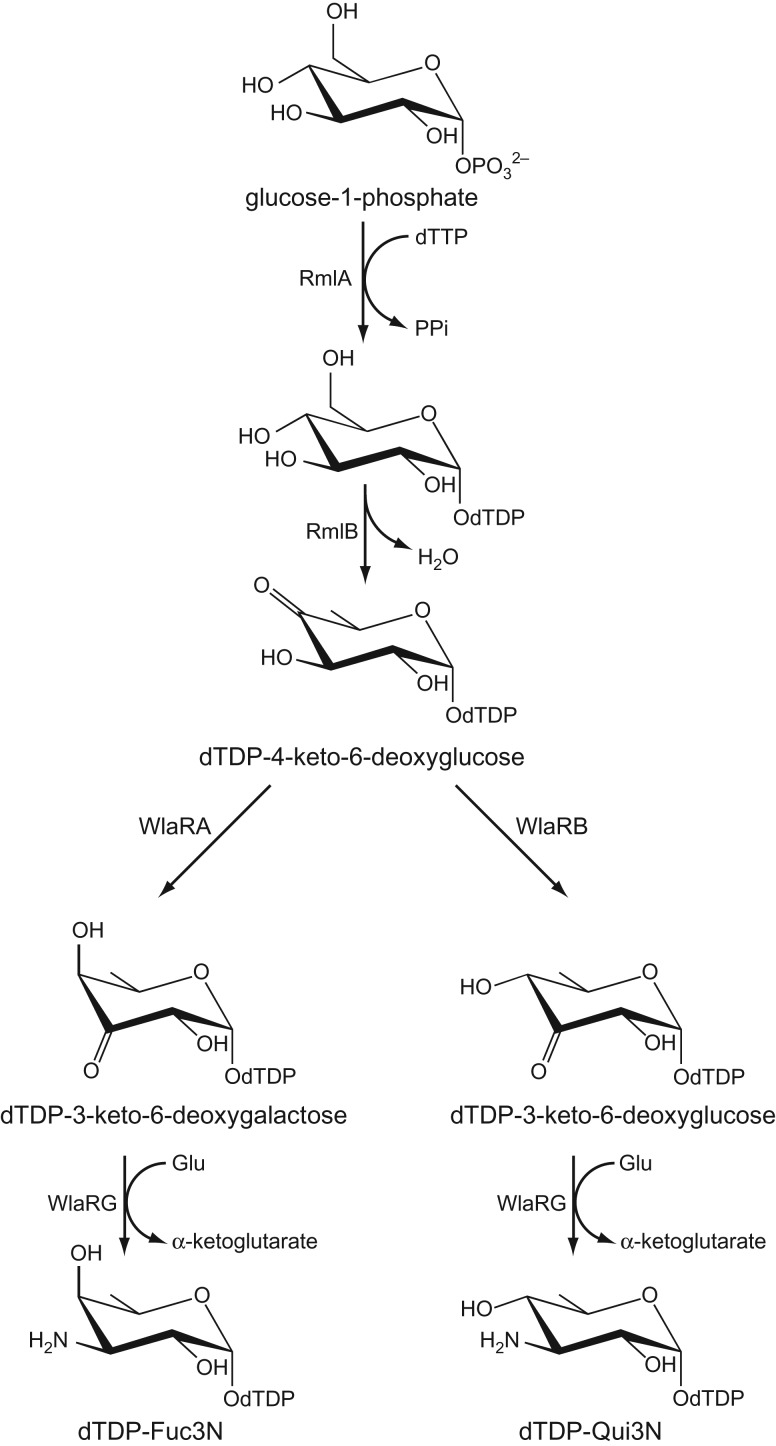
For this investigation, five genes were cloned (rmlA, rmlB, wlaRA, wlaRB and wlaRG), and the proteins encoded by them were expressed and purified to near homogeneity. To first test for the biochemical functions of the corresponding proteins, enzymatic activity assays were conducted as described in “Materials and Methods”, and product formation was monitored by capillary electrophoresis with photodiode array (CE-PDA) detection. Figure 2 outlines the putative biosynthetic pathways for the production of thymidine diphosphate (dTDP)-3-amino-3,6-dideoxy-d-galactose (dTDP-Fuc3N) or dTDP-3-amino-3,6-dideoxy-d-glucose (dTDP-Qui3N) in C. jejuni 81116. CE-PDA analysis of a reaction of RmlA with glucose-1-phosphate and thymidine triphosphate (dTTP) resulted in the appearance of a peak with a retention time of 12 min, similar to a dTDP-glucose standard (data not shown). CE-mass spectrometry (CE-MS) in negative ion mode showed a singly charged ion at m/z 563.1, which confirmed that the product was dTDP-glucose (Supplementary data, Figure S1).
Fig. 2.
Proposed biosynthetic pathway for the synthesis of dTDP-FucN and dTDPQuiN in C. jejuni 81116.
In the RmlB assay, incubating the enzyme with dTDP-glucose resulted in the appearance of a new peak by CE-PDA analysis, shifted 2 min in retention time from the substrate peak, which suggested the formation of dTDP-4-keto-6-deoxyglucose (Supplementary data, Figure S2). A similar shift in retention time was observed using both the RmlA product and commercially available dTDP-glucose. Commercially available dTDP-glucose was used for further production of dTDP-4-keto-6-deoxyglucose in order to simplify purification and analysis.
Evidence of product formation was suggested by CE-PDA analysis when the putative 3,4-ketoisomerases, WlaRA and WlaRB, were incubated with dTDP-4-keto-6-deoxyglucose (data not shown). However, the reactions did not go to completion as the area of the peak corresponding to dTDP-4-keto-6-deoxyglucose decreased only slightly. Coupling each reaction with the WlaRG aminotransferase resulted in complete conversion of dTDP-4-keto-6-deoxyglucose to products with retention times approximately 2.5 min faster than that for the substrate, indicating the formation of ddHexN products (Supplementary data, Figure S3). Interestingly, the WlaRA–WlaRG product had a retention time slightly faster than the WlaRB–WlaRG product, which suggested that distinct ddHexN residues were produced. The observation that the WlaRA and WlaRB reactions would only run to completion when coupled with the aminotransferase WlaRG suggested that the 3-keto product was possibly unstable or causing inhibition.
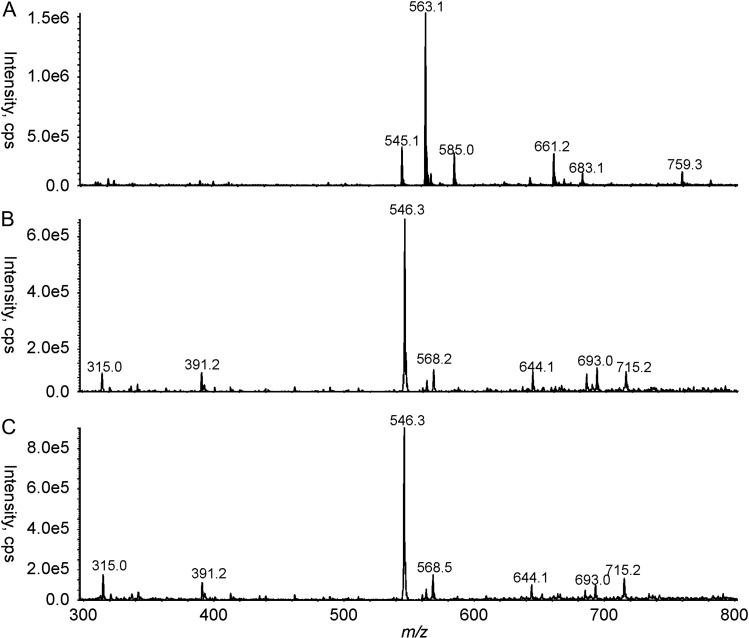
To further confirm the identities of the various reaction products, they were subsequently purified by size exclusion chromatography and analyzed by CE-MS (Figure 3). As can be seen in Figure 3A, the RmlB assay yielded a peak at m/z 563.1, corresponding to the theoretical mass of the hydrate form of dTDP-4-keto-6-deoxyglucose. The ions that corresponded to the products of the WlaRA–WlaRG and WlaRB–WlaRG coupled assays were both detected at m/z 546.3 (Figure 3B, C), which is consistent with the theoretical masses of dTDP-Fuc3N and dTDP-Qui3N. Because the final reaction mixtures of the coupled reactions did not contain the RmlB product, it can be inferred that the reactions were close to completion.
Fig. 3.
CE-MS analysis of the reaction products. Shown in (A), (B) and (C) are the extracted MS spectra for the RmlB, the WlaRA–WlaRG, and the WlaRB–WlaRG assays, respectively.
As final confirmation of the putative pathways shown in Figure 2, the products of the reactions by RmlB, RmlB–WlaRA–WlaRG or RmlB–WlaRB–WlaRG were purified and the identities of the products confirmed by both 1H NMR and 13C NMR (Supplementary data, Figures S4 and S5). The chemical shifts for dTDP-4-keto-6-deoxyglucose, dTDP-Fuc3N and dTDP-Qui3N products are found in Table II and correspond well with those previously published (Nakano et al. 2000; Pfoestl et al. 2003, 2008). These data prove that the final products of the reactions are, indeed, dTDP-Qui3N and dTDP-Fuc3N (Figure 2).
Table II.
NMR spectroscopic data of dTDP-4-keto-6-deoxyglucose (hydrate form), dTDP-α-Fuc3N and dTDP-α-Qui3N
| H/C-1 | H/C-2 | H/C-3 | H/C-4 | H/C-5 | H/C-6 | |
|---|---|---|---|---|---|---|
| 2dRiba | 6.34 | 2.38, 2.38 | 4.62 | 4.19 | 4.18, 4.18 | – |
| 85.4 | 39.9 | 72.3 | 86.7 | 66.9 | – | |
| 4-keto-6-deoxyglucoseb | 5.56 | 3.64 | 3.79 | – | 4.10 | 1.22 |
| 96.6 | 72.0 | 74.6 | NDc | 70.1 | 12.72 | |
| Fuc3Nd | 5.59 | 3.96 | 3.66 | 4.02 | 4.3 | 1.22 |
| 96.1 | 66.4 | 53.6 | 69.3 | 65.7 | 16.5 | |
| Qui3Ne | 5.59 | 3.80 | 3.44 | 3.37 | 4.02 | 1.29 |
| 95.2 | 69.7 | 56.3 | 72.5 | 70.0 | 17.3 |
aOnly one set of data is presented for 2dRib because there was no significant differences between the chemical shifts of 2dRib from the three nucleotide sugars (Nakano et al. 2000).
bThe chemical shifts for the 4-keto-6-deoxyglucose moiety correspond to the hydrate form (Pfoestl et al. 2003).
cND, not determined.
Structural analyses of WlaRA (accession No. 5TPV)
WlaRA functions as a dimer. The crystals utilized in this investigation contained three subunits in the asymmetric unit, two of which formed a dimer situated around a “local” 2-fold rotational axis and the other packing along a crystallographic dyad. The 3D structure of WlaRA was solved to a nominal resolution of 2.14 Å and was refined to an overall R-factor of 20.9%. The α-carbons for the three subunits in the asymmetric unit superimpose with root-mean-square (RMS) deviations of between 0.3 and 0.4 Å. In each subunit, several of the last C-terminal residues were disordered.
Shown in Figure 4A is a ribbon representation of the WlaRA dimer positioned around the “local” 2-fold rotational axis. The total overall buried surface area of the dimer is 3300 Å2. Each subunit of the dimer contains 10 β-strands and an α-helix defined by Tyr 123 to Lys 130. The first two β-strands from one subunit form a β-hairpin motif that extends toward the second subunit as can be seen in Figure 4A. This classical domain swapping results in each subunit containing two layers of anti-parallel β-sheet. One layer is composed of seven β-strands due to the fact that the first β-strand, defined by Tyr 3 to Ser 13, forms hydrogen bonding interactions with both its own subunit as well as with the second subunit. The second layer of the flattened barrel contains four β-strands. The fourth β-strand is punctuated by a β-bulge as a result of the dihedral angles adopted by Asp 48 (ϕ = ~63°, ψ = ~41°). The overall 3D architecture of WlaRA places it into the well-characterized cupin superfamily (Dunwell et al. 2001).
Fig. 4.
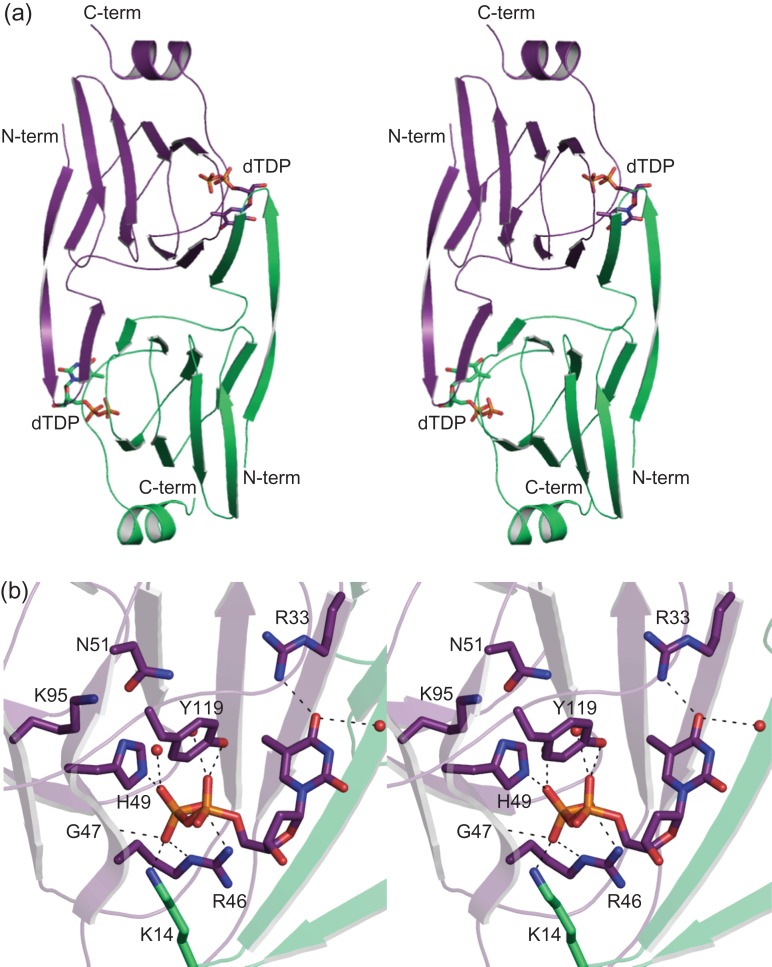
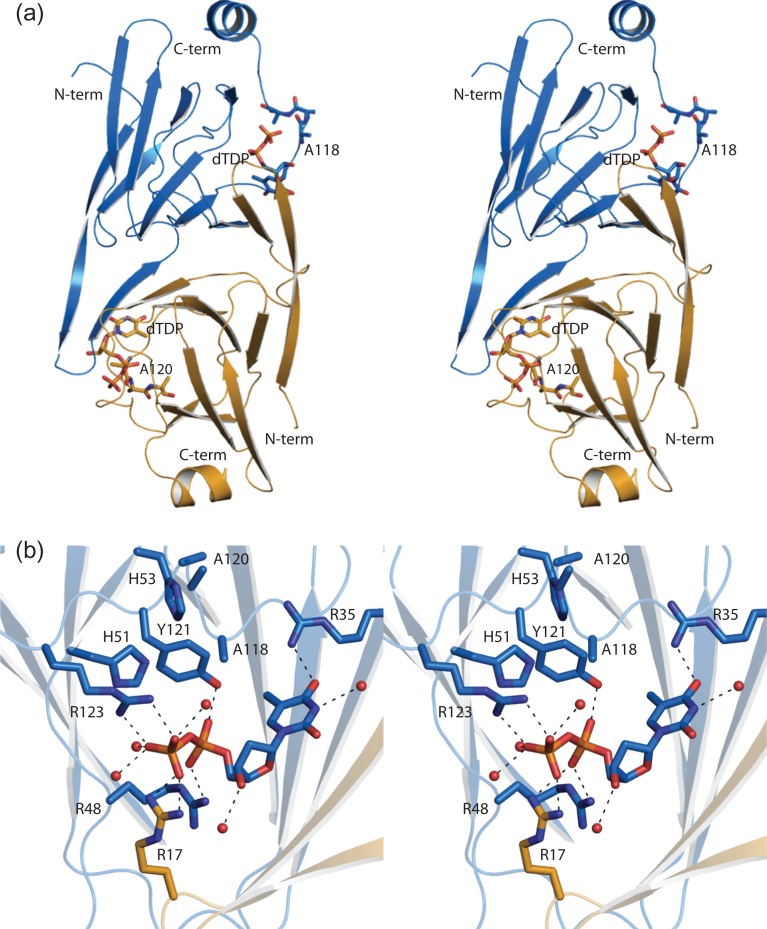
The structure of WlaRA. Shown in stereo in (A) is a ribbon representation of the WlaRA dimer. Subunits 1 and 2 are highlighted in violet and green, respectively. The bound dTDP molecules are displayed in stick representations. A close-up stereo view of the active site in subunit 1 is presented in (B). Ordered water molecules are represented by red spheres. Possible hydrogen bonding interactions, within 3.2 Å, are indicated by the dashed lines. Due to domain swapping, Lys 14 is contributed by subunit 2. This figure is available in black and white in print and in color at Glycobiology online.
WlaRA was crystallized in the presence of dTDP. A close-up view of the binding pocket is shown in Figure 4B. The ribose of the nucleotide adopts the C2′-endo pucker. A water molecule as well as the guanidinium group of Arg 33 anchors the thymine ring into the active site. The pyrophosphoryl group of the ligand lies within 3.2 Å of the side chains of Arg 46, His 49 and Tyr 119 from subunit 1 and Lys 14 from subunit 2. The backbone amide group of Gly 47 interacts with a α-phosphoryl oxygen. Two additional waters surround the pyrophosphoryl group of the ligand. Interestingly, in WlaRA, a typically conserved histidine is replaced with Asn 51 (Figure 4B). This will be discussed further below.
Structural analyses of WlaRB (accession No. 5TPU)
In order to obtain X-ray diffraction quality crystals of WlaRB, three site-directed mutations were made (E118A, K119A and E120A), and the mutant variant was crystallized. These mutations were suggested by the “surface entropy reduction” server UCLA MBI-SERp. The crystals ultimately used for the analysis contained two dimers in the asymmetric unit. The final model was refined to 2.0 Å resolution with an overall R-factor of 18.3%. Given that the α-carbons for the four subunits in the asymmetric unit superimpose with RMS deviations of between 0.2 and 0.3 Å, the following discussion refers only to the first dimer in the X-ray coordinate file. In each subunit, a few amino acid residues at the C-terminus were not visible in the electron density map.
Shown in Figure 5A is a ribbon representation of the WlaRB dimer. The buried surface area for the dimer is ~3400 Å2. The positions of the mutations required for crystallization are located in a loop near the active site and are indicated in Figure 5A. Like that observed for WlaRA, each subunit of WlaRB contains 10 β-strands and 1 α-helix defined by Tyr 125 to Ile 132. Given that the mutations were in the vicinity of the active site, the kinetic constants for the variant protein were measured and are listed in Table III. Whereas the Km for the substrate was approximately the same as that for the wild-type enzyme, the kcat was significantly reduced. Regardless, the site-directed mutations allowed for the structure of WlaRB to be determined. Still, caution must be applied when using “surface entropy reduction” as a crystallization tool given its possible effect on enzyme activity (Goldschmidt et al. 2007).
Fig. 5.
The structure of WlaRB. A stereo ribbon representation of WlaRB is presented in (A) with subunits 1 and 2 color coded in blue and yellow, respectively. The positions of the site-directed mutations made in order to produce X-ray diffraction quality crystals are shown (E118A, K119A and E120A) in stick representations. The dTDP ligands are also displayed in stick representations. A close-up stereo view of the WlaRB active site is presented in (B). Arg 17 is contributed by subunit 2 in the dimer. Possible hydrogen bonding interactions are indicated by the dashed lines. This figure is available in black and white in print and in color at Glycobiology online.
Table III.
Kinetic parameters of WlaRA and WlaRB
| Enzyme | Km (mM) | kcat (s−1) | kcat/Km (M−1s−1) |
|---|---|---|---|
| WlaRA | 0.14 ± 0.03 | 410 ± 30 | 2.9 × 106 |
| WlaRB | 0.13 ± 0.01 | 62 ± 6 | 4.8 × 105 |
| WlaRB mutant variant (E118A, K119A and E120A) | 0.19 ± 0.03 | 12 ± 1 | 6.3 × 104 |
The region surrounding the dTDP ligand in WlaRB is shown in Figure 5B. The side chain of Arg 35 and a water molecule lie within 3.2 Å of the thymine ring. The ribose, which adopts the C2′-endo pucker, interacts with an ordered water molecule. The pyrophosphoryl group of the dTDP ligand is surrounded by the side chains of Arg 48, Tyr 121 and Arg 123 from subunit 1 and Arg 17 from subunit 2.
Discussion
Studies on prokaryotic ddHexN biosynthetic pathways and the structural characterizations of their associated enzymes have been conducted on a variety of species, ranging from the Gram-positive Thermoanaerobacterium thermosaccharolyticum to the common Gram-negative E. coli and Shigella dysenteriae (Graninger et al. 2002; Davis et al. 2007; Wang et al. 2007; Thoden et al. 2009; Thoden and Holden 2014). Indeed, it has become clear that the Qui3N and Fuc3N sugars are conserved end products of these pathways, but in each case only one of the sugars has been observed in the LPS or LOS. From the results presented herein, it is now known that C. jejuni 81116 has the ability to synthesize both Qui3N and Fuc3N instead of just one, perhaps in an effort to increase fitness.
Past in vitro biochemical characterizations of the RmlA and RmlB enzymes across species have been extensive (Blankenfeldt et al. 2000; Li et al. 2006; Pfostl et al. 2008; Sha et al. 2012, Alphey et al. 2013). It is now well established that RmlA functions as a thymidylyltransferase and that RmlB catalyzes a 4,6-dehydration event. Here we confirm that both the C. jejuni RmlA and RmlB enzymes perform similar functions, respectively. We also show that WlaRA and WlaRB function as 3,4-ketoisomerases. Importantly, our data demonstrate that WlaRG has the ability to function on substrates that differ in their stereochemistry about the C-4′ carbon of the pyranose ring (Figure 2). This is in sharp contrast with most sugar aminotransferases that function on only one stereoisomer.
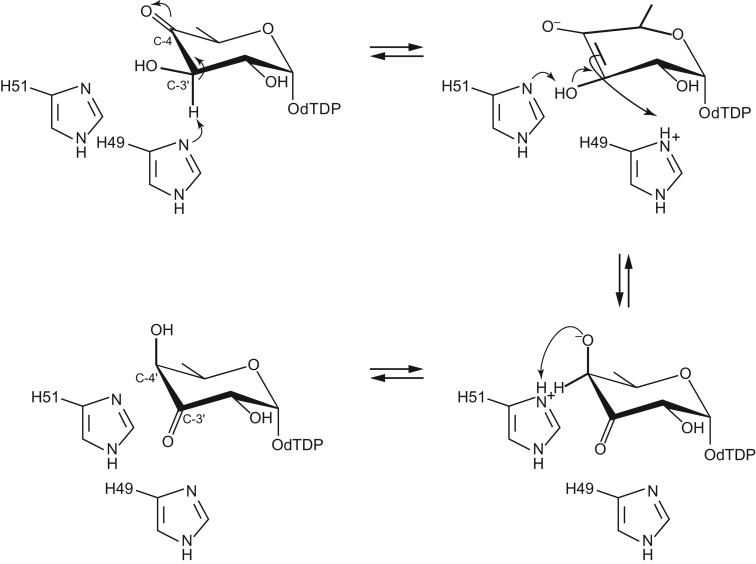
The structures of WlaRA and WlaRB solved in this investigation demonstrate that they belong to the cupin superfamily, members of which display physiological activities ranging from seed storage proteins to transcription factors. The first 3D model of a sugar 3,4-ketoisomerase to be reported was that from Aneurinibacillus thermoaerophilus (Davis et al. 2007). The enzyme, referred to as FdtA, produces dTDP-3-keto-6-deoxygalactose from dTDP-4-keto-6-deoxyglucose. At the time of the investigation, two residues, His 49 and His 51, were thought to be absolutely conserved among the 3,4-ketoisomerases. Site-directed mutagenesis experiments were subsequently conducted to probe the roles of these residues in catalysis. Specifically, the FdtA H49N and H51N mutant variants were constructed, and their activities were tested. Interestingly, whereas the H49N mutant variant displayed no measurable activity, the H51N mutant protein retained some activity, albeit significantly reduced. A catalytic mechanism was subsequently proposed as outlined in Figure 6. According to the proposal, His 49 functions as the active site base to promote removal of the proton from the C-3′ carbon and donation of it to the C-4′ carbon on the same side of the pyranose ring. His 51 was thought to shuttle a proton from the C-3′ hydroxyl group of the hexose to the C-4′ keto oxygen. The residual activity in the H51N mutant protein was speculated to be the result of another amino acid residue fulfilling the proton transfer role.
Fig. 6.
Catalytic mechanism proposed by Davis et al. (2007) for FdtA from A. thermoaerophilus, which produces dTDP-3-keto-6-deoxygalactose from dTDP-4-keto-6-deoxyglucose.
Recently, however, it has become apparent that His 51 is not as strictly conserved as was originally thought. Indeed, the amino acid sequence alignments shown in Figure 7 of the common sugar 3,4-ketoisomerases offer insight into conserved and potential active site residues. In WlaRA, His 51 is an asparagine residue. Based on the above-mentioned investigations with the FdtA H51N mutant protein, it was expected that WlaRA would display reduced enzymatic activity compared to wild-type FdtA, which has a catalytic efficiency of 2.1 × 104 M−1 s−1 (Thoden et al. 2015). A kinetic analysis was subsequently conducted on both WlaRA and WlaRB. Plots of initial velocities versus substrate concentrations for the wild-type forms of WlaRA and WlaRB are displayed in Supplementary Figure S6, and the kinetic parameters are listed in Table III. Surprisingly, the catalytic efficiency of WlaRA is 2.9 × 106 M−1 s−1, which is two orders of magnitude greater than that observed for FdtA. This would not have been predicted given our current understanding of the sugar 3,4-ketoisomerases. As a consequence, the mechanism originally proposed for FdtA must be modified given that His 51 is an asparagine residue in WlaRA. Indeed, how the proton is shuttled from the C-3′ hydroxyl group to the C-4′ keto functionality of the substrate is not obvious given the new data presented here.
Fig. 7.
Sequence alignment of QdtA, FdtA, FdtD, WlaRA and WlaRB. Stars, colons and periods indicate identity, strongly similar and weakly similar, respectively. No annotation indicates difference. This figure is available in black and white in print and in color at Glycobiology online.
The sugar 3,4-ketoisomerases are an especially intriguing class of cupins in that they function on the same substrate, dTDP-4-keto-6-deoxyglucose, but yield either dTDP-3-keto-6-deoxygalactose or dTDP-3-keto-6-deoxyglucose. All of these enzymes, regardless of whether they produce the “galacto” or “gluco” configuration, have a conserved histidine residue that serves as an active site base to initiate the reaction by abstracting the proton on the hexose C-3′ carbon (His 49 in WlaRA and His 51 in WlaRB). For those enzymes that yield the “gluco” product, there is always a conserved tyrosine residue that functions as an active site acid by protonating the C-4′ carbon on the opposite side of the hexose ring. In some instances, however, the enzymes leading to the “galacto” product, as in FdtA also have a similarly positioned tyrosine (Figure 7). Thus, the factors that determine product stereochemistry are, indeed, complicated. A recent investigation by our laboratories using FdtA and QdtA demonstrated that it was possible to convert a “galacto” enzyme into a “gluco” enzyme and vice versa by the judicious use of site-directed mutagenesis (Thoden et al. 2015). From that study, it was postulated that His 95 in FdtA was important for maintaining the stereochemical outcome of its reaction (Thoden et al. 2015).
The biochemical data presented here likewise demonstrate that WlaRA and WlaRB utilize the same substrate but yield either the “galacto” or “gluco” product, respectively (Figure 2). As would be expected, WlaRB has an appropriately positioned tyrosine to function as an active site acid (Tyr 37), whereas WlaRA has a phenylalanine in the corresponding position (Phe 35). Interestingly, however, His 95, which as discussed in the previous paragraph was thought to be important in determining the stereochemical outcome of the product, is apparently unique only to FdtA. In WlaRA, it is replaced with a lysine residue (Figure 4B). Thus, those structural features that control product stereochemistry are subtle and may extend far from the immediate vicinity of the active site.
One of the questions that arises is why C. jejuni retains both WlaRA and WlaRB? The two enzymes share only a 38% sequence conservation, yet the molecular architectures of the two proteins are, indeed, similar such that their α-carbons superimpose with an RMS deviation of 1.0 Å. Most likely C. jejuni retains both enzymes in an effort to increase variability, and not simply to ensure redundancy in its LOS locus. It would be interesting to specifically investigate several sites through deletions and substitutions to biochemically confirm their functions in the activity of the respective enzymes.
In addition to demonstrating C. jejuni 81116’s novel ability to synthesize two isomeric ddHexNs, biochemical confirmation of this LOS pathway yields additional molecular and enzymatic targets for novel treatments into campylobacteriosis and its far more serious complications. Additionally, economic and efficient biosynthesis of unusual sugars that are not commercially available or effectively synthesized through chemical means can now be considered using the enzymes characterized in this work.
Materials and methods
Plasmid construction
All genes used in this study were polymerase chain reaction (PCR) amplified from C. jejuni 81116 (Penner serotype HS:6) chromosomal DNA. The rmlA and rmlB genes were initially cloned in pCR2.1, and the sequences of the cloned amplicons were confirmed by DNA sequencing using an Applied Biosystems (Montréal, Canada) model 373 automated DNA sequencer. The inserts were transferred to the expression plasmid pF04 using the BamHI and EcoRI sites to obtain constructs that encoded a 6-His tag at the N-terminus of both RmlA (construct CJV-11) and RmlB (construct CJV-12). Constructs CJV-11 and CJV-12 were introduced into E. coli BL21-RIL cells for protein expression. The wlaRA, wlaRB and wlaRG genes were amplified with primers that included NdeI and SalI sites. The PCR products were digested with NdeI and SalI, and cloned in a version of pCWori+(-lacZ) containing the sequence of the E. coli maltose-binding protein (without the leader peptide), resulting in constructs CJV-24, CJV-26 and CJV-28 expressing wlaRG, wlaRA and wlaRB, respectively in E. coli AD202.
Protein purification for biochemical analyses
Escherichia coli BL21-RIL carrying either CJV-11 (rmlA) or CJV-12 (rmlB) were grown in 500 mL of 2 YT medium containing 70 mg/L ampicillin and 40 mg/L chloramphenicol. The cultures were incubated at 30°C until A600 = 0.7, induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and then incubated for 2 h 45 min. Escherichia coli AD202 strains carrying either CJV-24 (wlaRG), CJV-26 (wlaRA) or CJV-28 (wlaRA) were grown in 2 YT medium containing 150 mg/L ampicillin and 2 g/L glucose. The cultures were incubated at 37°C until A600 = 0.3, induced with 1 mM IPTG, and then incubated overnight at 25°C. The cells were broken using an Avestin C5 Emulsiflex cell disruptor (Avestin, Ottawa, Canada). Recombinant RmlA and RmlB included a 6-His tag at the N-terminus and were purified on Ni-NTA resin (Qiagen) using an imidazole step gradient in 50 mM sodium phosphate pH 7.7, 400 mM NaCl and 10 mM 2-mercaptoethanol. RmlA was eluted with 200 mM imidazole and RmlB was eluted with 100 mM imidazole. Recombinant WlaRA, WlaRB and WlaRG included a MalE tag at the N-terminus and were purified on amylose resin (New England Biolabs, Beverly, MA) and eluted using 10 mM maltose in 20 mM Tris, 200 mM NaCl, 5 mM β-mercaptoethanol and 1 mM ethylenediaminetetraacetic acid.
Enzyme activity assays
To test the biological role of RmlA, a reaction mixture was set up with 1.2 mM glucose-1-phosphate, 1 mM dTTP, 25 mM sodium phosphate (pH 7.5), 15 mM MgCl, yeast inorganic pyrophosphatase (Sigma-Aldrich, Oakville, Canada) and purified RmlA (CJV-11) in a final volume of 6 mL. The reaction mixture was incubated at 37°C for 6 h.
For the characterization of RlmB, the reaction mixture included 1 mM dTDP-glucose, 25 mM sodium phosphate (pH 7.5), 25 mM NaCl and purified RmlB (CJV-12) in a final volume of 18 mL. The reaction mix was incubated at 37°C for 4 h.
To confirm the enzyme activity of WlaRA, a reaction was set up with 1 mM dTDP-4-keto-6-deoxyglucose, 8 mM l-glutamic acid, 0.8 mM pyridoxal 5′-phosphate (PLP), 25 mM sodium phosphate (pH 7.5), 50 mM NaCl, 1 mL purified WlaRA (CJV-26) and 0.5 mL WlaRG (CJV-24) in a final volume of 5.75 mL. The reaction mixture was incubated at 37°C for 3 h 45 min.
Finally, to test the role of WlaRB, a reaction was set up with 1 mM dTDP-4-keto-6-deoxyglucose, 8 mM l-glutamic acid, 0.8 mM PLP, 25 mM sodium phosphate (pH 7.5), 50 mM NaCl, 0.3 mL of purified WlaRB (CJV-28) and 0.3 mL of purified WlaRG (CJV-24) in a final volume of 5.75 mL. The reaction mixture was incubated at 37°C for 2 h.
CE-mass spectrometry
The CE-MS experiments were performed using a Prince CE system (Prince Technologies, The Netherlands) that was coupled to a 4000 QTRAP mass spectrometer (AB Sciex, Canada). A sheath solution (isopropanol–methanol, 2:1) was delivered at a flow rate of 1.0 μL/min. Separations were obtained on about 90 cm length bare fused-silica capillary using 15 mM ammonium acetate in deionized water, pH 9.0. The 5 or −5 kV of electrospray ionization voltage was used for positive ion and negative ion detection modes, respectively. Tandem mass spectra were acquired in enhanced resolution scan mode (ER) with a scan rate of 250 Da/s.
NMR spectroscopy
1H NMR spectra were recorded by using a Varian (Palo Alto, CA) Inova 500 MHz spectrometer in D2O solutions at 25°C with acetone standard (2.225 ppm for 1H). For structural characterization of compounds, filtered enzymatic reactions or purified material were exchanged into 100% D2O. Structural analysis was performed using a Varian Inova 600 MHz spectrometer with a Varian 5 mm Z-gradient triple resonance cryogenically cooled probe for optimal sensitivity. All spectra (1H, gCOSY, TOCSY and 1H-13C HSQC) were referenced to an internal acetone standard (δH 2.225 ppm and δC 31.07 ppm).
Cloning of the wlaRA and wlaRB genes for X-ray analyses
The wlaRA and wlaRB genes were recloned using as templates the plasmids previously described. Primers were designed that incorporated NdeI and XhoI restriction sites. The amplified wlaRA and wlaRB genes were digested with NdeI and XhoI and ligated into pET31(b) and pET28T-3 g, respectively. The pET28T-3 g-wlaRB plasmid had been previously modified to incorporate a tobacco etch virus (TEV) protease cleavage recognition site after the N-terminal polyhistidine tag (Thoden and Holden 2005).
Protein expression and purification for X-ray analyses
The pET31b-wlaRA plasmid was used to transform Rosetta2(DE3) E. coli cells (Novagen). The cultures were grown in lysogeny broth supplemented with ampicillin (100 mg/L concentration) and chloramphenicol (50 mg/L concentration) at 37°C with shaking until A600 = 0.8. The flasks were cooled in an ice bath, and the cells were induced with 1 mM IPTG and allowed to express protein at 16°C for 24 h.
The pET28T-3 g-wlaRB plasmid was used to transform Rosetta2(DE3) E. coli cells (Novagen). The cultures were grown in lysogeny broth supplemented with kanamycin and chloramphenicol (both at 50 mg/L concentration) at 37°C with shaking until A600 = 0.8. The flasks were cooled in an ice bath, and the cells were induced with 1 mM IPTG and allowed to express protein at 16°C for 24 h.
In both cases, the cells were harvested by centrifugation and frozen as pellets in liquid nitrogen. Frozen cell pellets were subsequently disrupted by sonication on ice in a lysis buffer composed of 50 mM sodium phosphate, 20 mM imidazole, 10% glycerol and 300 mM sodium chloride (pH 8.0). The lysate was cleared by centrifugation, and both WlaRA and WlaRB were purified at 4°C utilizing Ni-NTA resin (Qiagen) according to the manufacturer's instructions. WlaRA was dialyzed against 10 mM Tris-HCl (pH 8.0) and 200 mM NaCl and concentrated to approximately 8 mg/mL based on an extinction coefficient of 0.59 (mg/mL)−1cm−1 calculated using the protein sequence and the DNAStar Lasergene software package (Madison, WI). For WlaRB, TEV protease was added in a 1:10 molar ratio to the pooled WlaRB protein solution, and dialyzed against 50 mM sodium phosphate, 300 mM sodium chloride and 20 imidazole (pH 8.0) at 4°C for 18 h. The TEV protease and any uncleaved protein were removed by passage over a Ni-NTA column, and the cleaved WlaRB protein collected and dialyzed against 10 mM Tris-HCl (pH 8.0) and 200 mM NaCl and concentrated to approximately 6 mg/mL based on an extinction coefficient of 0.89 (mg/mL)−1cm−1.
Crystallizations
Crystallization conditions for WlaRA were surveyed by the hanging drop method of vapor diffusion using a laboratory-based sparse matrix screen. The enzyme was first crystallized in 100 mM homopiperazine-1,4-bis(2-ethanesulfonic acid) (Homo-PIPES) (pH 5.0), 10% poly(ethylene) glycol 8000 and 1 M tetramethylammonium chloride in the presence of 5 mM dTDP at room temperature. These crystals often possessed a packing fault along the a/b axes. After extensive refinement experiments, the final crystallization conditions included 100 mM Homo-PIPES (pH 5.0), 8–10% poly(ethylene) glycol 8000, 1.25 tetraethylammonium chloride, 2% dimethylsulfoxide and 5 mM dTDP at room temperature. The crystals were prepared for X-ray data collection by transfer to a cryo-protectant solution composed of 100 mM Homo-PIPES (pH 5.0), 17% poly(ethylene) glycol 8000, 1.25 M tetraethylammonium chloride, 2% dimethylsulfoxide, 300 mM NaCl, 5 mM dTDP and 5% ethylene glycol. They belonged to the tetragonal space group I422 with unit cell dimensions of a = b = 177.9 Å, c = 88.6 Å and three subunits in the asymmetric unit.
Crystallization conditions for WlaRB were surveyed in a similar manner in the presence or absence of 5 mM dTDP. Crystals would appear as bundles and exhaustive screening refinements could not resolve the twinning issue. Using the surface entropy reduction prediction server, three site-directed mutations were made: E118A, K119A and E120A (Goldschmidt et al. 2007). Crystallization conditions for the “surface mutant” form of WlaRB were again surveyed. Crystals were observed growing in 100 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 7.5), 20% poly(ethylene) glycol 8000, 200 mM LiCl and 5 mM dTDP at room temperature. X-ray diffraction quality crystals were subsequently grown from 16% to 18% poly(ethylene) glycol 8000, 200 mM LiCl, 100 mM HEPES (pH 7.5) and 5 mM dTDP at room temperature. The crystals belonged to the hexagonal space group P61 with unit cell dimensions of a = b = 104.3 Å, c = 93.9 Å and two dimers in the asymmetric unit.
X-ray data collection, processing and structural analyses
The WlaRA data set was collected at Bruker AXS Inc (Madison, WI) with a Bruker AXS PHOTON II CPAD detector controlled with the Proteum software suite. The X-ray source was Ga Kα radiation (wavelength = 1.3418 Å) from a METALJET X-ray generator equipped with HELIOS MX optics and operated at 200 W. The WlaRB data set was collected in house with a Bruker AXS Platinum-135 CCD detector controlled with the PROTEUM software suite (Bruker AXS Inc.) The X-ray source was Cu Kα radiation from a Rigaku RU200 X-ray generator equipped with Montel optics and operated at 50 kV and 90 mA. Both data sets were processed with SAINT and scaled with SADABS (Bruker AXS Inc.). Relevant X-ray data collection statistics are listed in Table IV.
Table IV.
X-ray data collection statistics
| WlaRA | WlaRB | |
|---|---|---|
| Resolution limits (Å) | 50–2.14 (2.24–2.14)a | 50–2.0 (2.1–2.0)a |
| Space group | I422 | P61 |
| Unit cell (Å) | ||
| a | 177.9 | 104.6 |
| b | 177.9 | 104.6 |
| c | 88.6 | 93.9 |
| Number of independent reflections | 39,045 (4728) | 39,376 (5309) |
| Completeness (%) | 99.3 (96.2) | 99.7 (99.1) |
| Redundancy | 7.1 (5.4) | 9.5 (4.2) |
| Avg. I/avg σ(I) | 11.4 (2.8) | 12.7 (2.8) |
| Rsym (%)b | 7.1 (36.2) | 8.3 (36.0) |
aStatistics for the highest resolution bin.
bRsym = (Σ| I − Ī| / Σ I) x 100.
Structural analyses
The structure of WlaRB was solved by molecular replacement with the software package PHASER (McCoy et al. 2007) and using as the search model PDB entry 2PA7 (Davis et al. 2007). Four-fold averaging with the program DM (Cowtan 1994) resulted in an electron density map that allowed for a complete tracing of the polypeptide chain using COOT (Emsley and Cowtan 2004). The model was refined by alternate cycles of manual manipulation and refinement with REFMAC to 2.0 Å resolution (Murshudov et al. 1997).
WlaRA was also solved by molecular replacement using as a search probe the WlaRB coordinates. Three-fold averaging with DM (Cowtan 1994) yielded an electron density map that allowed the polypeptide chain to be completely traced. The model was refined to 2.14 Å resolution in a similar manner to that described above for WlaRB. Final model refinement statistics for both WlaRA and WlaRB are listed in Table V.
Table V.
Refinement statistics
| WlaRA | WlaRB | |
|---|---|---|
| Resolution limits (Å) | 50–2.14 | 50–2.0 |
| aR-factor (overall)%/no. reflections | 20.9/38,950 | 18.3/39,347 |
| R-factor (working)%/no. reflections | 20.7/37,071 | 17.9/37,330 |
| R-factor (free)%/no. reflections | 26.3/1879 | 24.5/2017 |
| Number of protein atoms | 3411 | 4308 |
| Number of heteroatoms | 296 | 389 |
| Average B values | ||
| Protein atoms (Å2) | 45.0 | 27.0 |
| Ligand (Å2) | 52.6 | 24.6 |
| Solvent (Å2) | 49.3 | 28.5 |
| Weighted RMS deviations from ideality | ||
| Bond lengths (Å) | 0.013 | 0.013 |
| Bond angles (°) | 2.1 | 2.0 |
| Planar groups (Å) | 0.009 | 0.008 |
| Ramachandran regions (%)b | ||
| Most favored | 84.8 | 89.0 |
| Additionally allowed | 15.2 | 10.5 |
| Generously allowed | – | 0.3 |
| Disallowed | – | 0.2 |
aR-factor = (Σ|Fo − Fc|/Σ|Fo|) x 100, where Fo is the observed structure-factor amplitude and Fc is the calculated structure-factor amplitude.
bDistribution of Ramachandran angles according to PROCHECK (Laskowski et al. 1993).
Kinetic analyses
The kinetic constants for WlaRA, WlaRB and the WlaRB mutant variant were determined via a coupled assay using a Beckman Coulter (Indianapolis, IN) DU-640 spectrophotometer according to the scheme displayed in Supplementary data, Figure S7. Specifically, their activities were measured by monitoring the decrease in absorbance at 340 nm as nicotinamide adenine dinucleotide phosphate (reduced) (NADPH) is oxidized to NADP+ due to the action of KijD10. Previous studies from the laboratory have verified that KijD10 from Actinomadura kijaniata functions as an NADPH-dependent C-3′ ketoreductase (Kubiak and Holden 2011) that works equally well with dTDP-3-keto-6-deoxyglucose and dTDP-3-keto-6-deoxygalactose (unpublished data). The reaction mixtures contained 50 mM HEPES (pH 7.5), 0.2 mM NADPH, 1 mg/mL KijD10 and dTDP-4-keto-6-deoxyglucose varying from 0.025 to 5.0 mM. The reactions were performed at 23°C and initiated by the addition of 0.015 μM WlaRA, 0.15 μM WlaRB or 2.4 μM WlaRB mutant variant. Plots of initial rates versus concentrations were analyzed using PRISM (GraphPad Software, Inc., San Diego, CA) and were fitted to the equation vo = (Vmax[S])/(KM + [S]). The required substrate, dTDP-4-keto-6-deoxyglucose, was prepared as previously described (Thoden and Holden 2014).
Supplementary Material
Acknowledgements
We thank Dr. Matthew M. Benning at Bruker AXS Inc for graciously collecting the X-ray data required for the structural analysis of WlaRA.
Supplementary data
Funding
Government of Canada (Genomics R&D Initiative); National Institutes of Health grant (GM115921 to H.M.H.).
Conflict of interest
None declared.
Abbreviations
CE, capillary electrophoresis; 2dRib, 2-deoxyribose; ddHexN, dideoxyhexosamine; dTDP, thymidine diphosphate; dTTP, thymidine triphosphate; Fuc3N, 3-amino-3,6-dideoxy-d-galactose; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; Homo-PIPES, homopiperazine-1,4-bis(2-ethanesulfonic acid); IPTG, isopropyl β-d-1-thiogalactopyranoside; LOS, lipooligosaccharide; LPS, lipopolysaccharide; NAD+, nicotinamide adenine dinucleotide (oxidized); MS, mass spectrometry; NADP+, nicotinamide adenine dinucleotide phosphate (oxidized); NADPH, nicotinamide adenine dinucleotide phosphate (reduced); Ni-NTA, nickel nitrilotriacetic acid; NMR, nuclear magnetic resonance; ORF, open reading frame; PCR, polymerase chain reaction; PDA, photodiode array; PLP, pyridoxal 5′-phosphate; Qui3N, 3-amino-3,6-dideoxy-d-glucose; RMS, root mean square; TEV, tobacco etch virus; Tris, tris-(hydroxymethyl)aminomethane.
References
- Alphey MS, Pirrie L, Torrie LS, Boulkeroua WA, Gardiner M, Sarkar A, Maringer M, Oehlmann W, Brenk R, Scherman MS et al. . 2013. Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa. ACS Chem Biol. 8:387–396. [DOI] [PubMed] [Google Scholar]
- Blankenfeldt W, Asuncion M, Lam JS, Naismith JH. 2000. The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA). Embo J. 19:6652–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K. 1994. ‘DM:’ An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallogr. 31:34–38. [Google Scholar]
- Davis ML, Thoden JB, Holden HM. 2007. The X-ray structure of dTDP-4-keto-6-deoxy-d-glucose-3,4-ketoisomerase. J Biol Chem. 282:19227–19236. [DOI] [PubMed] [Google Scholar]
- Dunwell JM, Culham A, Carter CE, Sosa-Aguirre CR, Goodenough PW. 2001. Evolution of functional diversity in the cupin superfamily. Trends Biochem Sci. 26:740–746. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- Galanis E. 2007. Campylobacter and bacterial gastroenteritis. CMAJ. 177:570–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt L, Cooper DR, Derewenda ZS, Eisenberg D. 2007. Toward rational protein crystallization: A web server for the design of crystallizable protein variants. Protein Sci. 16:1569–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graninger M, Kneidinger B, Bruno K, Scheberl A, Messner P. 2002. Homologs of the Rml enzymes from Salmonella enterica are responsible for dTDP-beta-l-rhamnose biosynthesis in the Gram-positive thermophile Aneurinibacillus thermoaerophilus DSM 10155. Appl Environ Microbiol. 68:3708–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden KM, Gilbert M, Coloe PJ, Li J, Fry BN. 2012. The role of WlaRG, WlaTB and WlaTC in lipooligosaccharide synthesis by Campylobacter jejuni strain 81116. Microb Pathog. 52:344–352. [DOI] [PubMed] [Google Scholar]
- Jacobs BC, Rothbarth PH, van der Meche FG, Herbrink P, Schmitz PI, de Klerk MA, van Doorn PA. 1998. The spectrum of antecedent infections in Guillain-Barré syndrome: A case-control study. Neurology. 51:1110–1115. [DOI] [PubMed] [Google Scholar]
- Kanipes MI, Papp-Szabo E, Guerry P, Monteiro MA. 2006. Mutation of waaC, encoding heptosyltransferase I in Campylobacter jejuni 81–176, affects the structure of both lipooligosaccharide and capsular carbohydrate. J Bacteriol. 188:3273–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klena JD, Gray SA, Konkel ME. 1998. Cloning, sequencing, and characterization of the lipopolysaccharide biosynthetic enzyme heptosyltransferase I gene (waaC) from Campylobacter jejuni and Campylobacter coli. Gene. 222:177–185. [DOI] [PubMed] [Google Scholar]
- Kubiak RL, Holden HM. 2011. Combined structural and functional investigation of a C-3″-ketoreductase involved in the biosynthesis of dTDP-l-digitoxose. Biochemistry. 50:5905–5917. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM. 1993. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 231:1049–1067. [DOI] [PubMed] [Google Scholar]
- Li W, Xin Y, McNeil MR, Ma Y. 2006. rmlB and rmlC genes are essential for growth of mycobacteria. Biochem Biophys Res Commun. 342:170–178. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Cryst. 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon ER, Rainey PB, Nowak MA, Lenski RE. 1994. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol. 4:24–33. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 53:240–255. [DOI] [PubMed] [Google Scholar]
- Nakano Y, Suzuki N, Yoshida Y, Nezu T, Yamashita Y, Koga T. 2000. Thymidine diphosphate-6-deoxy-l-lyxo-4-hexulose reductase synthesizing dTDP-6-deoxy-l-talose from Actinobacillus actinomycetemcomitans. J Biol Chem. 275:6806–6812. [DOI] [PubMed] [Google Scholar]
- Oldfield NJ, Moran AP, Millar LA, Prendergast MM, Ketley JM. 2002. Characterization of the Campylobacter jejuni heptosyltransferase II gene, waaF, provides genetic evidence that extracellular polysaccharide is lipid A core independent. J Bacteriol. 184:2100–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CT, Gilbert M, Yuki N, Endtz HP, Mandrell RE. 2008. Characterization of lipooligosaccharide-biosynthetic loci of Campylobacter jejuni reveals new lipooligosaccharide classes: Evidence of mosaic organizations. J Bacteriol. 190:5681–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S et al. . 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 403:665–668. [DOI] [PubMed] [Google Scholar]
- Pearson BM, Gaskin DJ, Segers RP, Wells JM, Nuijten PJ, van Vliet AH. 2007. The complete genome sequence of Campylobacter jejuni strain 81116 (NCTC11828). J Bacteriol. 189:8402–8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennie RA, Pearson RD, Barrett LJ, Lior H, Guerrant RL. 1986. Susceptibility of Campylobacter jejuni to strain-specific bactericidal activity in sera of infected patients. Infect Immun. 52:702–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfoestl A, Hofinger A, Kosma P, Messner P. 2003. Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-d-galactose in Aneurinibacillus thermoaerophilus L420-91T. J Biol Chem. 278:26410–26417. [DOI] [PubMed] [Google Scholar]
- Pfostl A, Zayni S, Hofinger A, Kosma P, Schaffer C, Messner P. 2008. Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-d-glucose. Biochem J. 410:187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phongsisay V, Perera VN, Fry BN. 2007. Expression of the htrB gene is essential for responsiveness of Salmonella typhimurium and Campylobacter jejuni to harsh environments. Microbiology. 153:254–262. [DOI] [PubMed] [Google Scholar]
- Richards VP, Lefebure T, Pavinski Bitar PD, Stanhope MJ. 2013. Comparative characterization of the virulence gene clusters (lipooligosaccharide [LOS] and capsular polysaccharide [CPS]) for Campylobacter coli, Campylobacter jejuni subsp. jejuni and related Campylobacter species. Infect Genet Evol. 14:200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha S, Zhou Y, Xin Y, Ma Y. 2012. Development of a colorimetric assay and kinetic analysis for Mycobacterium tuberculosis d-glucose-1-phosphate thymidylyltransferase. J Biomol Screen. 17:252–257. [DOI] [PubMed] [Google Scholar]
- Thoden JB, Goneau MF, Gilbert M, Holden HM. 2013. Structure of a sugar N-formyltransferase from Campylobacter jejuni. Biochemistry. 52:6114–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoden JB, Holden HM. 2005. The molecular architecture of human N-acetylgalactosamine kinase. J Biol Chem. 280:32784–32791. [DOI] [PubMed] [Google Scholar]
- Thoden JB, Holden HM. 2014. The molecular architecture of QdtA, a sugar 3,4-ketoisomerase from Thermoanaerobacterium thermosaccharolyticum. Protein Sci. 23:683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoden JB, Schaffer C, Messner P, Holden HM. 2009. Structural analysis of QdtB, an aminotransferase required for the biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-d-glucose. Biochemistry. 48:1553–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoden JB, Vinogradov E, Gilbert M, Salinger AJ, Holden HM. 2015. Bacterial sugar 3,4-ketoisomerases: Structural insight into product stereochemistry. Biochemistry. 54:4495–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xu Y, Perepelov AV, Qi Y, Knirel YA, Wang L, Feng L. 2007. Biochemical characterization of dTDP-d-Qui4N and dTDP-d-Qui4NAc biosynthetic pathways in Shigella dysenteriae type 7 and Escherichia coli O7. J Bacteriol. 189:8626–8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuki N, Susuki K, Koga M, Nishimoto Y, Odaka M, Hirata K, Taguchi K, Miyatake T, Furukawa K, Kobata T et al. . 2004. Carbohydrate mimicry between human ganglioside GM1 and Campylobacter jejuni lipooligosaccharide causes Guillain-Barré syndrome. Proc Natl Acad Sci U S A. 101:11404–11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.