Abstract
Heterogametic species require chromosome-wide gene regulation to compensate for differences in sex chromosome gene dosage. In Drosophila melanogaster, transcriptional output from the single male X-chromosome is equalized to that of XX females by recruitment of the Male Specific Lethal (MSL) complex, which increases transcript levels of active genes two-fold. MSL complex contains several protein components and two non-coding roX (RNA on the X) RNAs that are transcriptionally activated by MSL complex. We previously discovered that targeting of MSL complex to the X-chromosome is dependent on the Chromatin-Linked Adapter for MSL Protein (CLAMP) zinc finger protein. To better understand CLAMP function, we used the CRISPR/Cas9 genome editing system to generate a frameshift mutation in the clamp gene that eliminates expression of CLAMP protein. We found that clamp null females die at the third instar larval stage, while almost all clamp null males die at earlier developmental stages. Moreover, we found that in clamp null females roX gene expression is activated whereas in clamp null males roX gene expression is reduced. Therefore, CLAMP regulates roX abundance in a sex-specific manner. Our results provide new insights into sex-specific gene regulation by an essential transcription factor.
Keywords: Drosophila, dosage compensation, gene regulation, transcription factor
INTRODUCTION
Many species employ a sex determination system that generates an inherent imbalance in sex chromosome copy number, such as the XX/XY system in most mammals and some insects. In this system, one sex has twice the number of X-chromosome encoded genes compared to the other. Therefore, a mechanism of dosage compensation is required to equalize levels of X-linked transcripts, both between the sexes and between the X-chromosome and autosomes (Lucchesi et al., 2005). Dosage compensation is an essential mechanism that corrects for this imbalance by coordinately regulating gene expression of most X-linked genes.
In Drosophila melanogaster, transcription from the single male X-chromosome is increased two-fold by recruitment of the Male Specific Lethal (MSL) complex. MSL complex is composed of two structural proteins, MSL1 and MSL2, three accessory proteins, MSL3, MOF (Males absent On the First), and MLE (Maleless), and two functionally redundant non-coding RNAs, roX1 (RNA on the X) and roX2 (Meller and Rattner, 2002; Lucchesi et al., 2005). We previously discovered that recruitment of MSL complex to the X-chromosome requires the zinc finger protein Chromatin-Linked Adapter for MSL Proteins (CLAMP) (Soruco et al., 2013).
In addition to its role in male MSL complex recruitment, we suggested that CLAMP has an additional non sex-specific essential function because targeting of clamp transcript by RNA interference results in a pupal lethal phenotype in both males and females (Soruco et al., 2013). Further understanding of CLAMP function in the context of the whole organism required a null mutant. However, due to the pericentric location of the clamp gene, no deficiencies or null mutations were available. Using the CRISPR/Cas9 system, we introduced a frameshift mutation in the clamp gene, leading to an early termination codon before the major zinc finger binding domain. This frameshift mutation generated the clamp2 allele, which eliminates detectable CLAMP protein production and is therefore a protein null allele. The majority of clamp2 mutant males die prior to the third instar stage. On the other hand, females die at the third instar stage, suggesting sex specific functions for CLAMP. Furthermore, CLAMP regulates the roX genes in a sex-specific manner, activating their accumulation in males and repressing their accumulation in females. Overall, we present a new tool for studying dosage compensation and suggest that CLAMP functions to assure that roX RNA accumulation is sex specific.
RESULTS
Two clamp alleles were generated using the CRISPR/Cas9 system
The clamp gene is located within pericentric heterochromatin on the left arm of chromosome two, one megabase from the centromere. Due to this chromosomal location, null mutants for the clamp gene were not previously available from Drosophila mutant collections. We therefore used the CRISPR/Cas9 genome editing system which can introduce missense or frameshift mutations through the resolution of double-stranded breaks by non-homologous end joining (Sander and Joung, 2014). To determine where to target the Cas9 endonuclease, we used the protein domain composition of CLAMP. There are two predicted domains in CLAMP: an amino-terminal glutamine-rich, low complexity domain and a carboxy-terminal zinc finger domain consisting of six canonical zinc fingers (Figure S1). We previously demonstrated that the zinc finger domain of CLAMP is sufficient for DNA interactions (Soruco et al., 2013). Therefore, in order to generate a clamp null allele, we used the CRISPR/Cas9 system to target specifically upstream of the zinc finger domain of the clamp gene (Figure 1A) using the best available predicted guide RNA (Gratz et al., 2014).
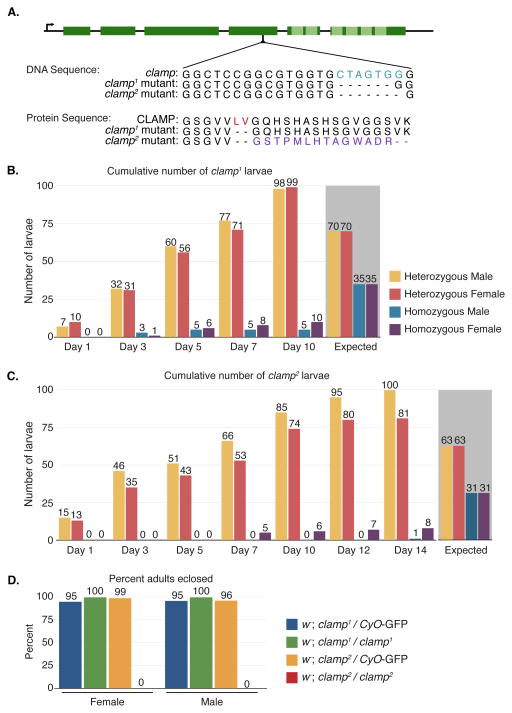
Figure 1. The clamp2 mutation is homozygous lethal and the clamp1 allele is homozygous viable.
(A) The CRISPR/Cas9-introduced frameshift is located in the fourth exon (dark green boxes) of the clamp gene, upstream of the DNA binding domain containing six zinc fingers (light green boxes). The homozygous viable clamp1 mutation is a six base pair deletion (blue), resulting in a deletion of two amino acids (red) and an in-frame shift of the protein sequence. The homozygous lethal clamp2 mutation consists of the same six base pair deletion (blue), with an additional seventh base removed. This causes a frameshift of the protein sequence (purple) resulting in an early termination codon.
(B) The cumulative number of larvae counted for both male and female clamp1 heterozygous and homozygous animals is shown. In total, 212 larvae were counted. Day 1 indicates the first day in which wandering third instar larvae began emerging. Homozygous males (blue) and females (purple) began emerging two days after their heterozygous siblings. The expected number of larvae out of 212 for each sex and genotype is indicated with the grey background.
(C) The cumulative number of larvae counted for both male and female clamp2 heterozygous and homozygous animals. In total, 190 larvae were counted. Day 1 indicates the first day in which wandering third instar larvae began emerging. Homozygous females (purple) began emerging after seven days, while we observed a single homozygous male (blue) on Day 14. The expected number of larvae out of 190 for each sex and genotype is indicated with the grey background.
(D) The percent heterozygous and homozygous clamp1 and clamp2 adults that eclosed were counted from larvae collected in (B) and (C). While almost all heterozygous and homozygous clamp1 mutants eclosed, only the heterozygous clamp2 mutants eclosed.
We generated two different mutations using the CRISPR/Cas9 system, and we balanced each with a homozygous lethal CyO second chromosome balancer carrying a larval GFP marker to allow us to track both larval and adult genotypes. Visual inspection of the wing phenotype in adult animals revealed that one mutation was homozygous viable (clamp1) while the other was not (clamp2). Sequencing of the targeted region indicated that the clamp1 homozygous viable animals carry a six base pair deletion in the clamp locus, resulting in the loss of two amino acids and an in-frame shift of the amino acid sequence (Figure 1A, Figure S1). The homozygous lethal clamp2 allele carries the same six base pair deletion with an additional seventh base deleted (Figure 1A, Figure S1). The seven base pair deletion causes a frameshift in the amino acid sequence, leading to an early stop codon occurring 14 amino acids after the mutation (Figure 1A, Figure S1).
In order to quantify the viability of homozygous clamp1 and clamp2 mutants, we first scored for the presence (in heterozygotes) or absence (in homozygotes) of GFP fluorescence in larvae and then counted the number of adult flies that eclosed from each class. Over a period of ten days, we would have expected to see 35 larvae each of homozygous clamp1 males and females out of the 212 larvae counted based on Mendelian ratios (Figure 1B). However, a total of ten homozygous clamp1 females and five homozygous clamp1 males were observed (Figure 1B, Figure S2A, Table S2). Similarly, while we would have expected to see 31 homozygous clamp2 males and females out of the 190 larvae counted over a period of 14 days, a total of eight homozygous clamp2 females and only one homozygous clamp2 male larva were observed (Figure 1C, Figure S2B, Table S2). Using a chi-squared test, we calculated that both clamp1 and clamp2 homozygous mutant larvae occur at frequencies significantly lower than expected from Mendelian ratios (Table S2, clamp1 χ2 = 66.82, p <0.00001 Table S3; clamp2 χ2 = 74.30, p <0.00001 Table S4). Furthermore, we found that despite occurring at low frequencies, clamp1 homozygous mutants are not developmentally delayed. In contrast, the clamp2 homozygous mutants are delayed by approximately seven days compared to their heterozygous siblings (Figure 1B, Figure 1C).
In order to quantify the adult viability defects caused by the clamp1 and clamp2 alleles, we compared the number of curly-winged (heterozygous) versus straight-winged (homozygous) adult flies that eclosed from previously genotyped larvae. We observed that almost all clamp1 heterozygous and homozygous mutants eclosed (Figure 1D, Table S2). In contrast, homozygous clamp2 female larvae die as third instar larvae and only the heterozygous clamp2 mutants eclosed, suggesting that the clamp2 mutation is homozygous lethal in both males and females before adulthood (Figure 1D, Table S2).
The clamp2 allele results in a homozygous lethal phenotype, suggesting that the mutation could be a recessive loss-of-function mutation. However, there are no deficiencies available to determine whether clamp2 is a genetic null. To determine if the homozygous lethality is due to the frameshift mutation in the clamp gene rather than another mutation in the genetic background, we generated a transgenic fly line containing a clamp transgene inserted on the third chromosome (Venken et al., 2006). The clamp transgene insertion stock contains a 12.5 kb region encompassing the clamp coding region and all putative upstream regulatory regions, but not any neighboring genes. We found that clamp2 homozygous lethality is rescued in both male and female flies when one copy of the clamp transgene rescue construct is present. Therefore, the lethality in the clamp2 homozygous mutants is caused by a loss of the clamp gene function and not a second site mutation.
The clamp1 allele complements the homozygous null clamp2 allele
The clamp1 allele is a homozygous viable mutation, despite producing significantly fewer animals than expected from Mendelian ratios (Figure 1B, Figure S2A, Table S2, Table S3). We asked whether the delay in development could be explained by an impact of the clamp1 allele on CLAMP expression. To determine how clamp1 affects clamp mRNA accumulation, we measured the production of clamp transcript by an established qRT-PCR assay in third instar larvae by comparing homozygous clamp1 mutants with the w−; clamp2; P{CLAMP} transgenic line (Soruco et al., 2013). We normalized transcript abundance in male and female clamp1 mutant larvae to the respective sex of the transgenic rescue line to control for genetic background. In addition, we used three normalizing control genes (gapdh, rpl32, and ras64b). We analyzed the results first by ANOVA to test for differences in means, followed by a Tukey post-hoc test to identify samples with statistically significant changes. We found no significant change in clamp transcript abundance in the clamp1 heterozygous or homozygous larvae compared to the rescue control (Figure S3A). Therefore, the clamp1 mutation does not dramatically change abundance of the clamp transcript.
To determine if the clamp1 allele affects production of CLAMP protein, we performed Western blotting of protein extracted from the salivary glands of third instar larvae, because whole larvae have large quantities of fat, making western blotting difficult. Our analysis revealed that the deletion of two amino acids in the clamp1 mutants does not detectably affect CLAMP protein production compared to controls (Figure S3B). To determine whether CLAMP protein produced from the clamp1 allele localizes to chromatin, we performed polytene chromosome immunostaining for CLAMP in homozygous and heterozygous clamp1 male and female larvae. In wild type male and female animals, CLAMP localizes to many sites throughout the genome (Figure S3C, y−w− male and female). Localization of CLAMP in the clamp1 mutant animals is not measurably different from wild type CLAMP at the resolution of polytene chromosomes (Figure S3C). We therefore concluded that the clamp1 mutants produce sufficient CLAMP protein to allow the animals to survive to adulthood.
Because our results suggest the clamp1 allele produces functional clamp protein, we hypothesized that the homozygous viable clamp1 allele could complement the clamp2 homozygous lethal allele. To test this, we crossed the heterozygous clamp1 and clamp2 stocks to generate w−; clamp1/clamp2 animals. We found that these animals are viable and survive to adulthood (data not shown) indicating that the clamp1 allele complements the homozygous lethality of the clamp2 allele.
While performing the complementation crosses, we discovered that clamp1 homozygous males are sterile. We hypothesized that if this phenotype is caused by the mutation in the clamp1 allele, we would expect the following two observations: 1) w−; clamp1/clamp2 heteroallelic males should also be sterile; 2) male sterility would be rescued by the CLAMP rescue transgene. We determined that w−; clamp1/clamp2 males are viable and fertile (data not shown), while w−; clamp1; P{CLAMP} males are sterile (data not shown). Therefore, we concluded that the clamp1 stock has a second site mutation that is linked to the clamp1 allele and results in male sterility. It is possible that this unknown second site mutation could contribute to the delay in development in the homozygous animals. Overall, we determined that the phenotypes we observed in the clamp1 mutants could not be attributed to the clamp1 allele. Therefore, we focused on characterizing the clamp2 allele because the homozygous lethal phenotype caused by this allele is rescued by the CLAMP transgene.
The clamp2 mutation is a protein null allele
Because our goal was to create a clamp protein null allele, we focused on characterizing the clamp2 allele that is homozygous lethal, a phenotype that is rescued by the CLAMP transgene (Figure 1A). First, we determined the developmental stage when the last homozygous clamp2 male larvae die compared with females. However, it is difficult to phenotypically determine the sex of larvae prior to the third instar stage. Therefore, we developed a PCR assay to measure the presence of male larvae by amplification of the Y-chromosome gene kl-5. We extracted genomic DNA from ten first or second instar larvae that were either homozygous (GFP−) or heterozygous (GFP+) for the clamp2 allele, as determined by GFP fluorescence produced from the CyO balancer chromosome. We were unable to detect the kl-5 Y-chromosome gene in GFP− larvae after the second instar stage, indicating that the last clamp2 homozygous males die between the second and third instar developmental stages (Figure S4A). In contrast, homozygous clamp2 females can survive to the third instar stage (Figure 1C, Figure S2B, Table S2, Table S4). Overall, we observed sexually dimorphic phenotypes caused by the clamp2 allele, suggesting that most homozygous males die earlier in development than females.
To determine how clamp2 affects clamp mRNA accumulation, we measured the production of clamp transcript in third instar larvae using qRT-PCR. We compared abundance of clamp mRNA in male and female clamp2 mutants to the same sex of the w−; clamp2; P{CLAMP} transgenic line. Although male homozygous clamp2 larvae are very rare, we were able to collect enough larvae to perform qRT-PCR due to the sensitive nature of the assay. We determined that there is no statistically significant change in clamp transcript abundance in clamp2 heterozygous or homozygous larvae (Figure 2A). Therefore, the clamp2 mutation has no significant effect on the abundance of the clamp transcript.
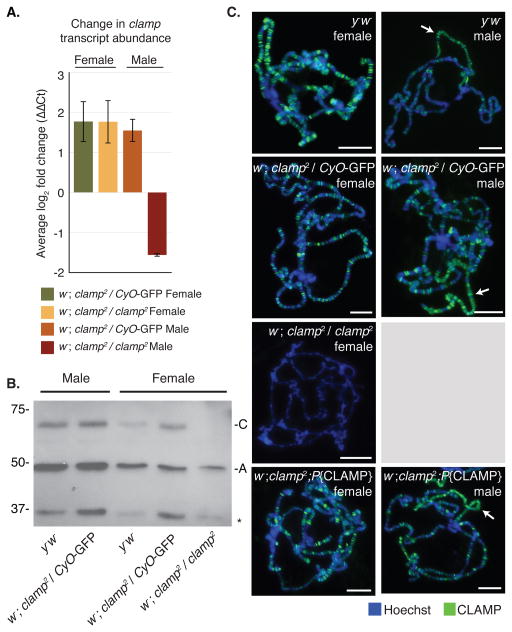
Figure 2. The clamp2 mutation is a protein null allele.
(A) Quanti tative Real-Time PCR indicates no significant change in clamp transcript abundance in male or female third instar larvae heterozygous or homozygous for the clamp2 allele. Plotted is the average log2 fold change (ΔΔCt) from three biological replicates after internal normalization to three genes (gapdh, rpl32, ras64b). Female and male samples were normalized to the respective sex of w−; clamp2; P{CLAMP} transgenic larvae. Error bars show +/− 1 standard error of the mean (S.E.M., **p<0.01, *p<0.05).
(B) Western blotting indicates that no full length CLAMP protein (“C”) is produced in homozygous clamp2 females. Loading control is Actin (“A”). Although a background band is present in all samples at 37kDa (*), a truncated form of CLAMP is not apparent as a result of the clamp2 mutation.
(C) There is no difference in CLAMP (green) localization on polytene chromosomes of heterozygous clamp2 male and female larvae compared to respective y−w− wild type controls. CLAMP does not localize to chromosomes in homozygous clamp2 females. The clamp2 homozygous mutant chromosomes are thinner than wild type and heterozygous clamp2 chromosomes. CLAMP immunostaining is rescued in clamp2 homozygotes when a 12.5 Kb genomic region encompassing the clamp gene is inserted onto the third chromosome (w−; clamp2; P{CLAMP}). We did not perform polytene chromosome spreads from homozygous clamp2 male animals due to poor gland development (grey box). White arrows indicate the male X-chromosome. Scale bars are 0.02mm.
To determine how the clamp2 mutation affects protein accumulation, we performed Western blot analysis on protein extracted from whole salivary glands of third instar larvae. We found that homozygous clamp2 female larvae do not produce full-length CLAMP (61 kDa), despite producing clamp mRNA (Figure 2A, Figure 2B, Figure S4B). We could not test protein abundance from homozygous clamp2 males because we could not collect sufficient homozygous male larvae for Western blot analysis. The clamp2 frameshift mutation generates an early termination codon, which is predicted to result in a truncated protein with a molecular weight of 37 kDa. The CLAMP antibody is specific to the amino-terminus and therefore should detect truncated protein. Although a background band is present in all samples around 37 kDa, we do not observe accumulation of truncated CLAMP specifically in clamp2 mutants (Figure 2B). We previously observed this background band and it is not ablated after clamp RNAi, suggesting that it is non-specific (Larschan et al., 2012). Therefore, it is likely that any CLAMP protein produced in the clamp2 mutant fails to accumulate. Furthermore, even if truncated CLAMP protein is produced below the level of immunoblotting detection, it would not contain the zinc finger DNA binding domain (Figure 1A).
As an alternate approach to detect any remaining CLAMP protein in the homozygous clamp2 mutant, we examined the localization of the CLAMP protein on polytene chromosomes. CLAMP localizes to many sites throughout the genomes of both male and female wild type animals (Figure 2C, y−w− male and female). Localization of CLAMP in heterozygous mutant males and females is not visibly distinct from that in wild type controls (Figure 2C, w−; clamp2/CyO-GFP). In contrast, CLAMP staining in clamp2 homozygous female larvae indicated that if CLAMP is produced from the clamp2 allele, it does not localize to polytene chromosomes (Figure 2C, w−; clamp2/clamp2). Homozygous male polytene chromosomes could not be obtained due to lack of viable animals (Figure 1C). Importantly, the CLAMP rescue transgene generates a functional CLAMP protein that localizes to polytene chromosomes in both male and female homozygous clamp2 animals (Figure 2C, w−; clamp2; P{CLAMP}). Polytene chromosome immunostaining further supports our conclusion that the clamp2 mutation is a protein null allele. Additionally, we observed that the chromosomes in clamp2 homozygous females are thinner than normal, a phenotype that has been previously observed in mutants for chromatin remodelers (Deuring et al., 2000).
To determine whether the disruption of chromosome morphology that we observed on interphase chromosomes also occurs on mitotic chromosomes, we performed mitotic chromosome spreads from third instar larval neuroblasts (Figure S4C). Dramatic changes in mitotic chromosome morphology were not observed in the mitotic chromosome spreads (Figure S4C). Therefore, it is likely CLAMP is more important to maintain the chromatin organization of interphase chromosomes than mitotic chromosomes.
CLAMP differentially regulates roX genes in males and females
We originally identified CLAMP as a transcription factor essential for directly linking the MSL complex to the X-chromosome in males (Larschan et al., 2012; Soruco et al., 2013). However, CLAMP also localizes to thousands of promoters throughout the genome (Figure 2C) and therefore has the potential to regulate additional transcripts (Soruco et al., 2013). Chromatin Immunoprecipitation followed by sequencing (ChIP-seq) demonstrates that CLAMP localizes to the known regulatory regions of both roX genes, which are among the strongest MSL complex recruitment sites (Soruco et al., 2013). In addition, we previously determined that CLAMP positively regulates the transcription of roX2 based on experiments performed in Drosophila (S2) cells and male larvae after clamp RNAi, likely because it recruits MSL complex, which is known to activate roX transcription (Bai et al., 2004; Soruco et al., 2013).
Because we previously determined that CLAMP regulates roX2 from clamp RNAi experiments, we determined the effect of the clamp2 allele on roX accumulation in vivo in third instar larvae using qRT-PCR. The roX genes are not normally expressed in wild type female larvae due to the absence of MSL complex, which activates their transcription in males (Meller et al., 1997; Meller, 2003; Bai et al., 2004). We found that there was a large increase in the amount of both roX1 and roX2 transcripts in homozygous clamp2 female larvae when normalized to female rescue controls (Figure 3A, yellow bars). Consistent with our previous findings that CLAMP promotes transcription of roX2 in male S2 cells, we found a large reduction in roX2 transcript levels in clamp2 homozygous males (Figure 3B, red bars). In contrast, we did not see a significant decrease in roX1 levels, consistent with our prior analysis of roX transcript abundance after clamp RNAi (Soruco et al., 2013). Therefore, CLAMP differentially regulates roX genes in males and females.
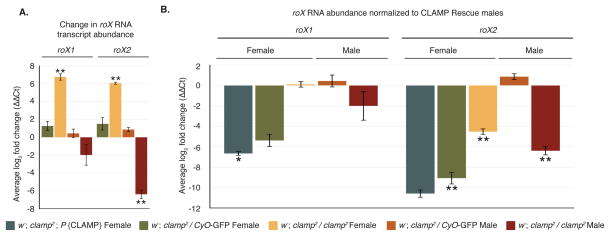
Figure 3. CLAMP regulates transcript abundance of roX differentially in males and females.
(A) The average log2 fold change for roX1 and roX2 abundance as measured by qRT-PCR in homozygous clamp2 females indicates that females have a significant increase in the abundance of both roX1 and roX2, while homozygous clamp2 males have a significant decrease in roX2 abundance. Shown is the average log2 fold change (ΔΔCt) of three biological replicates for roX1 and roX2 after normalization to three internal genes and compared to the respective sex of w−; clamp2; P{CLAMP} transgenic animals as in Figure 2A. (Error bars are +/− 1 S.E.M., **p<0.01, *p<0.05).
(B) The samples from Figure 3A were normalized to w−; clamp2; P{CLAMP} rescue males and show that roX1 transcript abundance in homozygous clamp2 females is statistically indistinguishable from males. The abundance of roX2 in homozygous clamp2 females is statistically 16-fold depleted compared to w−; clamp2; P{CLAMP} rescue males. (Error bars are +/− 1 S.E.M., **p<0.001, *p<0.005).
The large increase in abundance of the roX transcripts in clamp2 homozygous female larvae (Figure 3A) led us to ask how these levels compared to roX expression in wild type males. Therefore, we reanalyzed roX abundance by normalizing all transcript levels to clamp2 homozygous males carrying the rescue P{CLAMP} transgene (Figure 3C). We discovered that activation of roX1 in clamp2 homozygous mutant females leads to a similar abundance of roX1 as in males (Figure 3C, yellow bars). Therefore, the clamp2 mutation results in roX1 being expressed in females at similar level to that of males. The abundance of roX2 in homozygous clamp2 females is reduced compared to rescued males but is similar to that present in clamp2 males. Therefore, the absence of CLAMP leads to similar basal levels of roX2 abundance in both males and females and activates roX1 to similar transcript abundance levels as seen in males.
The increase in roX expression in clamp2 homozygous females compared to controls led us to hypothesize that homozygous clamp2 females could be dying at the third instar larval stage due to this increase in roX expression. To test whether lethality in clamp2 homozygous females is caused by the increase in roX expression (Figure 3), we generated a triple mutant fly line that is homozygous null for both roX genes and homozygous for the clamp2 allele. RoX null females are usually viable (Meller and Rattner, 2002). However, the combined loss of both roX genes does not rescue the homozygous lethality of the clamp2 allele (data not shown). Thus we conclude that the increased expression of roX RNAs is not the sole cause of the lethality seen in clamp2 homozygous females. Because CLAMP occupies thousands of promoters genome-wide (Figure 2C) (Soruco et al., 2013), the lethality of the clamp2 allele is likely caused by changes in regulation of multiple genes.
Ectopic MSL complex does not form in clamp2 homozygous females
In males, the roX genes are targeted by MSL complex for increased expression. To determine whether the increase in roX gene transcription in clamp2 homozygous females is caused by MSL complex component induction, we quantified transcript abundance of all MSL complex component genes: msl1, msl2, msl3, mle, and mof. We also compared transcriptional changes in mutant females with those in mutant males to determine if any changes are sex-specific. We found significantly increased msl1, msl3, and mof transcript abundance in clamp2 heterozygous and homozygous females compared to rescue controls (Figure 4A, green and yellow bars). We also observed increased msl2 transcript abundance in clamp2 homozygous females compared to controls (Figure 4A, yellow bars). Interestingly, changes in msl transcript abundance are not sensitive to clamp gene dosage. Unlike the roX genes, the MSL complex components encoding genes do not have clear CLAMP binding sites in their regulatory regions (Soruco et al., 2013). Therefore, it is possible that changes in msl gene expression are due to other regulatory cascades that are altered in clamp mutant larvae. In contrast, the regulatory regions of the roX genes are two of the strongest CLAMP binding sites in the genome. Thus, it is unlikely that the regulation of roX RNA expression (Figure 3A) occurs through an indirect mechanism.
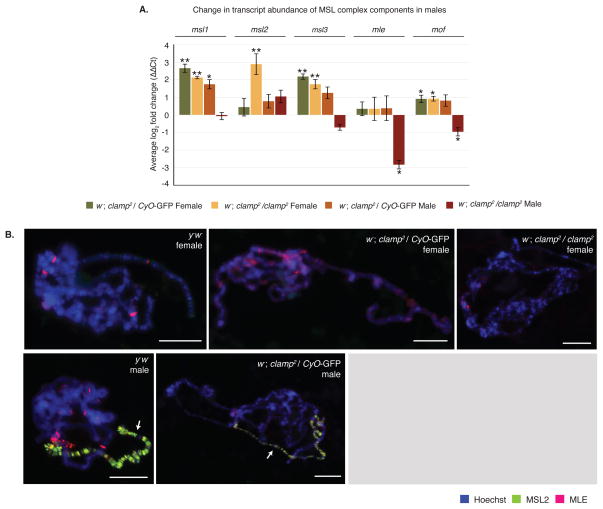
Figure 4. Ectopic MSL complex is not formed in clamp2 females.
(A) qRT-PCR shows that CLAMP regulates transcription of some MSL complex component genes in female larvae. In both heterozygous and homozygous clamp2 females, there are significant increases in abundance for the msl1, msl3, and mof transcripts. There is also a significant increase in msl2 abundance in homozygous clamp2 females. There are significant decreases in transcript abundance of mle and mof in clamp2 homozygous males. Normalization was performed on three biological replicates using three internal normalization genes. Samples were normalized to the respective sex of the w−; clamp2; P{CLAMP} rescue animal. (Error bars are +/− 1 S.E.M., **p<0.01, *p<0.05).
(B) Polytene chromosome immunostaining shows that the core MSL complex component MSL2 (green) is expressed only in males and localizes to the X-chromosome. The accessory protein, MLE (red), is expressed in both males and females and localizes throughout the genome. There is no change in localization of these proteins in heterozygous or homozygous clamp2 mutants. Reduced numbers of homozygous clamp2 male animals and their poorly developed salivary glands prevented generation of polytene chromosome spreads for this genotype (grey box). White arrows indicate the X-chromosome. Scale bars are 0.02mm.
In clamp2 homozygous males, most MSL complex transcripts did not show significant changes compared to controls (Figure 4A). The strongest perturbation we observed was an 8-fold reduction in mle transcripts in clamp2 homozygous males compared to controls. However, reduction in mle transcript levels would not explain the complete loss of MSL complex recruitment we previously reported in males after constitutive CLAMP RNAi (Soruco et al., 2013) because significant MSL complex recruitment is observed in the absence of MLE (Kelley et al., 1999).
Increased transcripts of MSL complex components and roX RNAs in clamp2 homozygous females (Figure 3, Figure 4A) could promote ectopic MSL complex formation, a situation that is known to cause female lethality (Kelley et al., 1995). Therefore, we looked for ectopic formation of MSL complex in clamp2 homozygous females by immunostaining polytene chromosomes for the MSL2 and MLE components of the MSL complex. We did not detect ectopic localization of MSL2 on either the X-chromosome or autosomes in clamp2 heterozygous or homozygous females (Figure 4B, w−; clamp2/CyO-GFP and w−; clamp2/clamp2). Furthermore, we detected MLE in similar non-X-specific patterns on clamp2 mutant and wild type female polytene chromosomes, consistent with its known localization pattern (Figure 4B, y−w− and w−; clamp2/clamp2) (Cugusi et al., 2015). Therefore, the large increases in roX transcripts observed in clamp2 homozygous females are not likely to be due to MSL complex-mediated roX activation. However, it is possible that any MSL complex formed in clamp2 homozygous females would be unable to localize to chromatin in the absence of CLAMP, leading to its destabilization and protein degradation.
DISCUSSION
We previously demonstrated that CLAMP has an essential role in MSL complex recruitment to the male X-chromosome. In addition, we suggested that CLAMP has an essential role in the viability of both males and females (Soruco et al., 2013). However, we could not perform in vivo studies to further investigate CLAMP function because there was no available null mutant line. In the current manuscript, we present a CLAMP protein null mutant and determine that this protein is essential in both sexes. This allele will provide a key tool for future in vivo studies on the role of CLAMP in dosage compensation, as well as identification of the essential function of CLAMP in both sexes.
Our initial characterization of the clamp2 protein null allele revealed sexually dimorphic roles for CLAMP in regulation of the roX genes. We observed that CLAMP promotes roX2 transcription in males but represses transcription of both roX genes in females. It is likely that recruitment of MSL complex to the roX2 locus by CLAMP promotes roX2 expression in males. In females, where MSL complex is not present, CLAMP may function to repress these loci as an additional mechanism to ensure that dosage compensation is male-specific. Additionally, we determined that most clamp2 homozygous males die earlier in development than clamp2 homozygous females. Earlier lethality in males is likely due to a misregulation of the dosage compensation process as a result of the loss of CLAMP-mediated MSL complex recruitment. However, CLAMP is enriched at the 5′ regulatory regions of thousands of genes across the genome. Therefore, it is likely that other non-sex specific regulatory pathways are disrupted resulting in female lethality.
Furthermore, CLAMP is an essential protein because our CRISPR/Cas9-generated protein null clamp allele is homozygous lethal in both males and females. These results indicate that CLAMP has a previously unstudied non-sex specific role that is essential to the viability of both males and females. An interesting observation that arose from our characterization is that polytene chromosome organization is disrupted in clamp2 mutant females, suggesting that CLAMP may play a role in regulation of genome-wide chromatin organization of interphase chromosomes. A function in regulating chromatin organization provides one possible explanation for how CLAMP performs sexually dimorphic functions. For example, CLAMP may repress of roX expression in females by promoting the recruitment of a repressive chromatin-modifying factor in the absence of MSL complex. In contrast, CLAMP may activate roX2 in males by creating a chromatin environment permissive for MSL complex recruitment in males. Although roX1 and roX2 are functionally redundant, our results suggest that CLAMP specifically activates roX2 but not roX1 in males. Interestingly, Villa et al. recently reported that roX2, but not roX1, is likely to be an early site of MSL complex recruitment (Villa et al., 2016), suggesting that CLAMP may function early in the process of dosage compensation.
Overall, our newly generated clamp2 protein null allele provides an important tool to study how the essential CLAMP protein regulates its many target genes in vivo. The generation of the clamp2 allele will facilitate future studies that will reveal a mechanistic understanding of how a single transcription factor can promote different sex-specific functions within an organism.
MATERIALS AND METHODS
Generation and validation of clamp mutant fly line using CRISPR/Cas9 technology
We used the FlyCRISPR Optimal Target Finder tool available from the University of Wisconsin to design a CRISPR target sequence for clamp (Gratz et al., 2014). We cloned target sequence oligonucleotides for clamp (sense: 5′-CTT CGG CTC CGG CGT GGT GCT AGT-3′ and antisense: 5′-AAA CAC TAG CAC CAC GCC GGA GCC-3′) into the pU6-BbsI-chiRNA plasmid (Addgene #45946), following the protocol outlined on the FlyCRISPR website. We validated correct ligation of the clamp CRISPR target sequence into the pU6-BbsI-chiRNA plasmid by Sanger sequencing using universal M13 primers.
The commercial service Genetic Services, Inc. microinjected the validated pU6-BbsI-chiRNA plasmid containing the clamp target sequence into germline-expressing Cas9 flies (y1, w1118;+; PBac{vas-Cas9, U6-tracrRNA}VK00027). Flies containing a single mutation were returned balanced over the Curly of Oster (CyO) second chromosome balancer. From these progeny, we identified the CRISPR/Cas9-generated mutations by PCR across the target region (Forward: 5′-ACA ACT GAA GGG TTT GGA CGG-3′, Reverse: 5′-CAT GCA GGC TGA ACA AAC AG-3′), followed by Sanger sequencing (Forward: 5′-TCT GCA GGA CAA ACA CCT TG-3′; Reverse: 5′-CCC AAG CAC AAC TTC AGC AAA-3′). From this validation, we isolated two independent clamp alleles: 1.) y1, w1118; clamp1/CyO; and 2.) y1, w1118; clamp2/CyO;.
Generation and validation of clamp rescue transgene and fly line
We generated a clamp rescue construct using the P(acman) system that utilizes a conditionally amplifiable bacterial artificial chromosome (BAC) clone, recombineering, and bacteriophage phiC31 mediated insertion at a genomic attB site (Venken et al., 2006). We designed primers for two homology arms to capture a 12.5 kb region spanning the entire clamp locus (3.5kb), including the presumed promoter (Left Homology Arm (1.2kb) Forward: 5′-ACC GGC GCG CCG CAG AAG GAA GAG TTT CCG A-3′, Reverse: 5′-CGC GGA TCC AAG TCC TGG CCT AAG CCC TA-3′; Right Homology Arm (800bp) Forward: 5′-CGC GGA TCC TTT TGT GCA TGG TCA ACC ACG-3′, Reverse: 5′-ACC TTA ATT AAG GGC AAA CAT ATT TCG CAC GAT AC-3′). We amplified homology arms off a conditionally amplifiable P(acman) BAC clone, Ch322 20C06 (BacPac Resources) using Copy Control (Epicenter) reagent for vector amplification. We simultaneously cloned the arms into the PacMan vector 3XP3-eGFP-attB-Amp (gift from Koen Venken) at the multicloning site (MCS) using the engineered restriction sites AscI-BamHI (left) and BamHI-PacI (right) in a three-component ligation. We identified positive colonies via Sanger sequencing across the MCS. Using BamH1, we linearized the intermediate vector and purified the product. Next, the linearized vector was transformed into E. coli that we had previously transformed with the clamp containing BAC clone Ch322 20C06 and expressing the mini-lambda vector encoding the phiC31 recombinase (SW102, NCI BRB Preclinical Repository). We identified positive colonies via sequencing across the left and right homology arm junction.
Genetic Services, Inc. microinjected the full clamp rescue construct into D. melanogaster embryos containing the attB-docking site (VK33) on Chromosome 3L band 65B2 (Venken et al., 2006). We identified clamp rescue construct transgenics using 3xP3-EGFP expression, and maintained the subsequent stock in the homozygous state (y1, w1118;+; P{3xP3-EGFP, clamp = CLAMP}).
Genetic manipulation of clamp mutant alleles and quantification of phenotypes
To generate clamp1 and clamp2 mutant lines with a larval phenotypic marker, we used standard methods to cross the original balanced stocks to a CyO-GFP stock that expresses GFP at all stages of larval development (w1118; snascl/CyO, P{ActGFP.w−}CC2). The resulting w1118; clamp1/CyO-GFP and w1118; clamp2/CyO-GFP stocks (referred to in text as clamp1 and clamp2, respectively) express both larval and adult phenotypic markers and were used for all remaining experiments. In addition, we generated a balanced clamp2 stock that expresses GFP under regulation of the twist promoter (Bloomington stock #6662: w1118; In(2LR)Gla, wgGla−1/CyO, P{w[+mC]=GAL4-twi.G}2.2, P{UAS-2xEGFP}AH2.2)). The resulting w−; clamp2/twi-GFP stock was used to generate the mitotic spreads. All other experiments utilized the CyO-GFP stock expressing GFP under the regulation of Actin.
To assess larval viability, we collected third instar larvae from either a w1118; clamp1/CyO-GFP (212 larvae collected) or w1118; clamp2/CyO-GFP (190 larvae collected) heterozygous cross. For each larva, we visually determined the sex and clamp genotype. From these larvae, we monitored eclosion of the pupae into adult flies.
To test if the clamp2 mutation can be rescued by a clamp transgene, we crossed the w1118; clamp2/CyO-GFP stock to the y1, w1118;+; P{CLAMP} rescue line and scored viability at the adult stage by wing phenotype. The resulting w1118; clamp2; P{CLAMP} line (referred to as w−; clamp2; P{CLAMP} in text) was maintained as a stock in the homozygous state and used for all quantitative analyses.
Sample collection for western blotting and PCR for kl-5 gene
We tested for the presence of the Y-chromosome gene kl-5 in first, second, and third instar larvae of the following animal genotypes: 1) y1, w1118;+; (referred to as y−w−) 2) w1118; clamp2/CyO-GFP, and 3) w1118; clamp2/clamp2. We collected and pooled 10 larvae each of the first and second instar developmental stage. For third instar larvae, we dissected 10 salivary glands of sexed males and females of each genotype in cold PBS. As an additional control, we tested 10 adult male and adult female whole flies. We flash froze all samples in liquid nitrogen and homogenized with a steel bead using on a Retsch MM300 TissueLyser Mixer Mill. Next, we suspended the homogenized samples in 30uL of lysis buffer (10mM Tris-HCl pH8.0, 1mM EDTA, 25mM NaCl, 0.2mg/ml Proteinase K, 1ng/uL RNase) and incubated at 37°C for 30 minutes, followed by a 5 minute incubation at 90°C. We purified genomic DNA by standard phenol:chloroform extraction using Phase-lock tubes (5 Prime) per the manufacturer’s instructions, followed by ethanol precipitation.
We tested purified genomic DNA for the presence of the kl-5 gene by PCR using the following primers (Forward: 5′-ATC GCA AAC GAG TGG TCT CA-3′; Reverse: 5′-TGT ATC AAG GGC AGG CAT CC-3′). As a genomic DNA loading control, we amplified the clamp locus with the PCR primers used to identify the mutation.
Quantitative Real Time PCR (qRT-PCR)
To analyze transcript abundance, we used TRIzol (Thermo Fisher Scientific) following the manufacturer’s instructions to extract total RNA from three biological replicates of five third instar larvae from each genotype. We reverse-transcribed one microgram of total RNA using the SuperScript VILO cDNA Synthesis Kit (Life technologies) by following the manufacturer’s protocol. Three technical replicates for each target transcript were amplified using SYBR Green (Life Technologies) on an Applied Biosystems StepOnePlus™ Real-Time PCR System. Primers were used at a concentration of 200nM to amplify targets from 2ng of cDNA. Primer sequences for qRT-PCR are in Table S1. We calculated transcript abundance using the standard ΔΔCt method using gapdh, rpl32, ras64b as internal controls (Livak and Schmittgen, 2001). We normalized female mutant samples to the female w1118; clamp2; P{CLAMP} transgenic rescue, except where specified. We normalized male mutant samples to the male w1118; clamp2; P{CLAMP} transgenic rescue. We tested statistical significance by performing an ANOVA multiple comparison test on the mean ΔCt values, followed by a Tukey post hoc analysis for multiple comparison correction.
Western blotting
We dissected salivary glands from third instar larvae in cold PBS and froze samples in liquid nitrogen. We extracted total protein from the samples by homogenizing in lysis buffer (50mM Tris-HCl pH 8.0, 150mM NaCl, 1% SDS, 0.5X protease inhibitor) using a small pestle. After a five-minute incubation at room temperature, we cleared the samples by centrifuging at room temperature for 10 minutes at 14,000 × g. To blot for CLAMP and Actin, we ran 5 micrograms of total protein on a Novex 10% Tris-Glycine precast gel (Life technologies). We transferred proteins to PVDF membranes using the iBlot transfer system (ThermoFisher Scientific) and probed the membranes for CLAMP (1:1000, SDIX) and Actin (1:400,000, Millipore) using the Western Breeze kit following the manufacturer’s protocol (ThermoFisher Scientific).
Relative expression of protein for CLAMP was quantified using the gel analysis tool in ImageJ software following the guidelines outlined on the website (Schneider et al., 2012). For each genotype, we first internally normalized the amount of CLAMP protein to Actin. Next, we determined relative expression of protein by comparing the Actin normalized quantities to sex of respective y1, w1118;+; (y−w−) sample.
Chromosome squashes and immunostaining
We prepared larval polytene chromosome squashes as previously described (Cai et al., 2010) and mitotic chromosome spreads from larval neuroblasts following method #3 as described (Pimpinelli et al., 2000). We stained polytene chromosomes with anti-CLAMP (rabbit, 1:1000, SDIX), anti-MLE (rabbit, 1:500, gift from M. Kuroda), or anti-MSL2 (rat, 1:500, gift from P. Becker) primary antibodies. We used DAPI to stain mitotic chromosomes. For detection, we used all Alexafluor secondary antibodies at a concentration of 1:1000. We visualized polytene chromosome slides at 40X on a Zeiss Axioimager M1 Epifluorescence upright microscope with the AxioVision version 4.8.2 software. We visualized mitotic spreads at 60X on a Zeiss LSM 800 confocal microscope with Airyscan using Zen Blue software.
Data Availability
Drosophila stocks are available upon request. Table S1 contains primer sequences for qRT-PCR.
Supplementary Material
An alignment of the clamp1 and clamp2 amino acid sequences to the wild type CLAMP amino acid sequence shows the location of the two amino acid deletion generated by the clamp1 allele (dashes in red). Red amino acids indicate the frame shift in the clamp2 allele that results in an early stop codon. Amino acids in blue specify the difference between two CLAMP isoforms, one excluding (CLAMP-PA) and one including (CLAMP-PB) five amino acids. The six known zinc fingers are annotated with lines.
(A) Over a period of ten days, a total of 212 clamp1 larvae were collected. For each larva, the sex and clamp1 genotype was determined. Plotted is the number of male and female heterozygous and homozygous clamp1 animals collected at each time point.
(B) Over a period of fourteen days, a total of 190 clamp2 larvae were collected. For each larva, the sex and clamp2 genotype was determined. Plotted is the number of male and female heterozygous and homozygous clamp2 animals collected at each time point.
(A) There are no significant changes in clamp transcript abundance in male or female clamp1 heterozygous or homozygous mutant animals as measured by qRT-PCR. Shown is the average log2 fold change of three biological replicates after internal normalization to three reference genes (GAPDH, Rpl32, and Ras64B). Each sex is normalized to the respective sex of the w−; clamp2; P{CLAMP} rescue animal. (Error bars are +/− 1 S.E.M., **p<0.01, *p<0.05).
(B) Protein was extracted from the salivary glands of clamp1 heterozygous and homozygous males (left) and females (right) and Western blotted for CLAMP and Actin. Full length CLAMP (67 kDa) is produced in clamp1 males and females (“C”). Below each graph is the quantification of each western blot. Plotted is the relative amount of CLAMP protein after internal normalization to actin and relative to y−w−.
(C) Polytene chromosome immunostaining demonstrates there is no difference in CLAMP (green) localization of heterozygous or homozygous clamp1 male and female larvae compared to respective y−w− wild type controls.
(A) DNA from first, second, and third instar larvae was tested for the Y-chromosome gene kl-5 to determine the presence or absence of male clamp2 mutant animals. For first and second instar larvae, ten larvae were pooled together. The clamp2 genotype of each sample is indicated above (“+” = wild type clamp, “−” = clamp2 allele). The sex of the animals is also indicated above for third instar animals where it was possible to anatomically determine the sex of the animals. PCR of genomic DNA determined that kl-5 is present in pooled homozygous clamp2 first and second instar larvae. Therefore, most homozygous clamp2 male larvae die between the second and third instar larval stages.
(B) Quantification of the anti-CLAMP Western blot in Figure 2B shows the relative amount of CLAMP protein after internal normalization to Actin and relative to y−w−. There is no CLAMP protein produced in homozygous clamp2 female larvae.
(C) Mitotic chromosomes from female third instar brain neuroblasts are present in all conditions evaluated including clamp2 null animals. The indicated insets below each panel show a 4X magnification of the boxed chromosomes. The top panels are optical stacks of 4 × 0.3 um each. Scale bar represents 20μm.
Table S1: Primer sequences of targets tested by qRT-PCR
The number of clamp1 and clamp2 heterozyous and homozygous male and female larvae counted are shown (“Larvae” row). Of those larvae counted, the number of adults that eclosed is indicated (“Adults eclosed” row). The number in parentheses is the percent eclosed.
The number of observed (O) larvae for homozygous and heterozygous clamp1 male or female larvae are indicated. The expected (E) number of each genotype of larvae was calculated using the total number of larvae counted (212). From these numbers, a Chi-squared statistic was calculated. To test for significance, the Chi-squared threshold was determined for 3 degrees of freedom.
The number of observed (O) larvae for homozygous and heterozygous clamp2 male or female larvae are indicated. The expected (E) number of each genotype of larvae was calculated using the total number of larvae counted (190). From these numbers, a Chi-squared statistic was calculated. To test for significance, the Chi-squared threshold was determined for 3 degrees of freedom.
Acknowledgments
We thank Dr. Koen Venken for providing us with the PacMan vector. We would also like to thank John Urban for valuable input on experimental analysis and critical review of the manuscript. Additional thanks to Dr. Mitzi Kuroda and Dr. Peter Becker for providing antibodies. J.A.U. was supported by National Institutes of Health award F31GM108423. All experiments were supported by National Institutes of Health R01GM098461-1, American Cancer Society Research Scholar 123682-RSG-13-040-01-DMC, and a Pew Biomedical Scholars program grant.
List of abbreviations
- MSL complex
Male-specific lethal complex
- CLAMP
Chromatin-linked adaptor for MSL Proteins
- roX
RNA on the X
LITERATURE CITED
- Bai X, Alekseyenko AA, Kuroda MI. Sequence-specific targeting of MSL complex regulates transcription of the roX RNA genes. EMBO J. 2004;23:2853–2861. doi: 10.1038/sj.emboj.7600299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W, Jin Y, Girton J, Johansen J, Johansen KM. Preparation of Drosophila polytene chromosome squashes for antibody labeling. J Vis Exp. 2010 doi: 10.3791/1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugusi S, Kallappagoudar S, Ling H, Lucchesi JC. The Drosophila Helicase Maleless (MLE) is Implicated in Functions Distinct From its Role in Dosage Compensation. Mol Cell Proteomics. 2015;14:1478–1488. doi: 10.1074/mcp.M114.040667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuring R, et al. The ISWI Chromatin-Remodeling Protein Is Required for Gene Expression and the Maintenance of Higher Order Chromatin Structure In Vivo. Mol Cell. 2000;5:355–365. doi: 10.1016/s1097-2765(00)80430-x. [DOI] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O’Connor-Giles KM. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196:961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- Kelley RL, Solovyeva I, Lyman LM, Richman R, Solovyev V, Kuroda MI. Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell. 1995;81:867–877. doi: 10.1016/0092-8674(95)90007-1. [DOI] [PubMed] [Google Scholar]
- Larschan E, Soruco MML, Lee OK, Peng S, Bishop E, Chery J, Goebel K, Feng J, Park PJ, Kuroda MI. Identification of chromatin-associated regulators of MSL complex targeting in Drosophila dosage compensation. PLoS Genet. 2012;8:e1002830. doi: 10.1371/journal.pgen.1002830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- Meller VH. Initiation of dosage compensation in Drosophila embryos depends on expression of the roX RNAs. Mech Dev. 2003;120:759–767. doi: 10.1016/s0925-4773(03)00157-6. [DOI] [PubMed] [Google Scholar]
- Meller VH, Rattner BP. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 2002;21:1084–1091. doi: 10.1093/emboj/21.5.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller VH, Wu KH, Roman G, Kuroda MI, Davis RL. roX1 RNA Paints the X Chromosome of Male Drosophila and Is Regulated by the Dosage Compensation System. Cell. 1997;88:445–457. doi: 10.1016/s0092-8674(00)81885-1. [DOI] [PubMed] [Google Scholar]
- Pimpinelli S, Bonaccorsi S, Fanti L, Gatti M. In: Preparation and Analysis of Drosophila Mitotic ChromosomesDrosophila Protocols. Sullivan W, Ashburner M, Hawley RS, editors. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2000. p. 728. [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soruco MML, et al. The CLAMP protein links the MSL complex to the X chromosome during Drosophila dosage compensation. Genes Dev. 2013;27:1551–1556. doi: 10.1101/gad.214585.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJT, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- Villa R, Schauer T, Smialowski P, Straub T, Becker PB. PionX sites mark the X chromosome for dosage compensation. Nature. 2016;537:244–248. doi: 10.1038/nature19338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
An alignment of the clamp1 and clamp2 amino acid sequences to the wild type CLAMP amino acid sequence shows the location of the two amino acid deletion generated by the clamp1 allele (dashes in red). Red amino acids indicate the frame shift in the clamp2 allele that results in an early stop codon. Amino acids in blue specify the difference between two CLAMP isoforms, one excluding (CLAMP-PA) and one including (CLAMP-PB) five amino acids. The six known zinc fingers are annotated with lines.
(A) Over a period of ten days, a total of 212 clamp1 larvae were collected. For each larva, the sex and clamp1 genotype was determined. Plotted is the number of male and female heterozygous and homozygous clamp1 animals collected at each time point.
(B) Over a period of fourteen days, a total of 190 clamp2 larvae were collected. For each larva, the sex and clamp2 genotype was determined. Plotted is the number of male and female heterozygous and homozygous clamp2 animals collected at each time point.
(A) There are no significant changes in clamp transcript abundance in male or female clamp1 heterozygous or homozygous mutant animals as measured by qRT-PCR. Shown is the average log2 fold change of three biological replicates after internal normalization to three reference genes (GAPDH, Rpl32, and Ras64B). Each sex is normalized to the respective sex of the w−; clamp2; P{CLAMP} rescue animal. (Error bars are +/− 1 S.E.M., **p<0.01, *p<0.05).
(B) Protein was extracted from the salivary glands of clamp1 heterozygous and homozygous males (left) and females (right) and Western blotted for CLAMP and Actin. Full length CLAMP (67 kDa) is produced in clamp1 males and females (“C”). Below each graph is the quantification of each western blot. Plotted is the relative amount of CLAMP protein after internal normalization to actin and relative to y−w−.
(C) Polytene chromosome immunostaining demonstrates there is no difference in CLAMP (green) localization of heterozygous or homozygous clamp1 male and female larvae compared to respective y−w− wild type controls.
(A) DNA from first, second, and third instar larvae was tested for the Y-chromosome gene kl-5 to determine the presence or absence of male clamp2 mutant animals. For first and second instar larvae, ten larvae were pooled together. The clamp2 genotype of each sample is indicated above (“+” = wild type clamp, “−” = clamp2 allele). The sex of the animals is also indicated above for third instar animals where it was possible to anatomically determine the sex of the animals. PCR of genomic DNA determined that kl-5 is present in pooled homozygous clamp2 first and second instar larvae. Therefore, most homozygous clamp2 male larvae die between the second and third instar larval stages.
(B) Quantification of the anti-CLAMP Western blot in Figure 2B shows the relative amount of CLAMP protein after internal normalization to Actin and relative to y−w−. There is no CLAMP protein produced in homozygous clamp2 female larvae.
(C) Mitotic chromosomes from female third instar brain neuroblasts are present in all conditions evaluated including clamp2 null animals. The indicated insets below each panel show a 4X magnification of the boxed chromosomes. The top panels are optical stacks of 4 × 0.3 um each. Scale bar represents 20μm.
Table S1: Primer sequences of targets tested by qRT-PCR
The number of clamp1 and clamp2 heterozyous and homozygous male and female larvae counted are shown (“Larvae” row). Of those larvae counted, the number of adults that eclosed is indicated (“Adults eclosed” row). The number in parentheses is the percent eclosed.
The number of observed (O) larvae for homozygous and heterozygous clamp1 male or female larvae are indicated. The expected (E) number of each genotype of larvae was calculated using the total number of larvae counted (212). From these numbers, a Chi-squared statistic was calculated. To test for significance, the Chi-squared threshold was determined for 3 degrees of freedom.
The number of observed (O) larvae for homozygous and heterozygous clamp2 male or female larvae are indicated. The expected (E) number of each genotype of larvae was calculated using the total number of larvae counted (190). From these numbers, a Chi-squared statistic was calculated. To test for significance, the Chi-squared threshold was determined for 3 degrees of freedom.
Data Availability Statement
Drosophila stocks are available upon request. Table S1 contains primer sequences for qRT-PCR.