Abstract
The origin and progression of cancer is widely viewed as “somatic evolution” driven by the accumulation of random genetic changes. This theoretical model, however, neglects fundamental conditions for evolution by natural selection, which include competition for survival and a local environmental context. Recent observations that the mutational burden in different cancers can vary by 2 orders of magnitude and that multiple mutations, some of which are “oncogenic,” are observed in normal tissue suggests these neglected Darwinian dynamics may play a critical role in modifying the evolutionary consequences of molecular events. Here we discuss evolutionary principles in normal tissue focusing on the dynamical tension between different evolutionary levels of selection. Normal somatic cells within metazoans do not ordinarily evolve because their survival and proliferation is governed by tissue signals and internal controls (e.g. telomere shortening) that maintain homeostatic function. The fitness of each cell is, thus, identical to the whole organism, which is the evolutionary level of selection. For a cell to evolve, it must acquire a self-defined fitness function so that its survival and proliferation is determined entirely by its own heritable phenotypic properties. Cells can develop independence from normal tissue control through randomly accumulating mutations that disrupt its ability to recognize or respond to all host signals. A self-defined fitness function can also be gained non-genetically when tissue control signals are lost due to injury, inflammation, or infection. Accumulating mutations in cells without a self-defined fitness function will produce no evolution - consistent with reports showing mutations, including some that would ordinarily be oncogenic, are present in cells from normal tissue. Furthermore, once evolution begins, Darwinian forces will promote mutations that increase fitness and eliminate those that do not. Thus, cancer cells will typically have a mutational burden similar to adjacent normal cells and many (perhaps most) mutations observed in cancer cells occurred prior to somatic evolution and may not contribute to the cell’s malignant phenotype.
Keywords: cancer evolution somatic mutations theory, somatic evolution, fitness function
Introduction
Since proposed by Nowell [1], the conceptual model of carcinogenesis as an evolutionary process has become widely accepted [2]. Typically, the transition from normal cells to cancer, illustrated by the classic Fearon-Vogelstein model [3], is assumed to require a sequence of accumulating genetic or epigenetic changes that produce corresponding pre-malignant phenotypes. Eventually a threshold is crossed and a fully-transformed population of cancer cells emerges. Thus, “somatic evolution” in cancer biology is established as a fundamentally genetic process governed by random accumulating mutations. This gene-centric model of cancer is almost universally accepted [2, 4, 5].
Clearly, cancer is associated with accumulating genetic mutations and targeting the protein products of key (“driver”) mutations can significantly alter tumor growth and progression. On the other hand, several observations seem at odds with the standard genetic mutation model [6, 7]. For example, the number of observable mutations in cancers from different organs can vary by 2 orders of magnitude [8]. Defects in DNA repair genes such as BRCA1 and 2 increase the risk of cancer in some organs but not in others [9, 10]. Normal, non-cancerous cells may possess multiple genetic mutations, some of which would ordinarily be viewed as oncogenic [11, 12] and apparently normal cells with “oncogenic mutations” have been observed adjacent to tumors [13]. In this edition, Liggett et. al. point out additional issues and possible inconsistencies. For instance, the incidence of cancer increases late in life even though most replications and mutations occur early in life. Furthermore, there is a lack of clear correlations between cancer rate and longevity or body size across the Animal Kingdom [14]. Whether these are resolved with future investigations or are what Thomas Kuhn labelled “anomalies” [15] that usher in a new view of cancer evolution remains to be seen.
To address these issues, we will conceptually divide the key components of carcinogenesis into 1) genetic and epigenetic changes and 2) cell proliferation and cell turnover via cell mortality and replacement. The former defines the molecular mechanisms that are, of course, critical for the transition from normal to cancer. However, cell proliferation and cell turnover, which ultimately determine the clinical significance of any cancer cell, are determined by complex and often subtle evolutionary dynamics that govern the survival, reproduction and ultimately growth of cancer populations within the context of an often spatially and temporally heterogeneous environment.
Darwinian Dynamics, a review
Evolution by natural selection requires three conditions [16, 17]: 1. A population of individuals with heritable phenotypic variation. 2. A potential for population growth that exceeds ecological limits so that not all individuals can survive and proliferate (a “struggle for existence”) and 3. The heritable phenotypic properties influence survival and proliferation.
It is commonly assumed that evolution in cancer cells is simply an extension of the conventional view of cancer as “a disease of the genes.” In the sense that mutations contribute to “heritable phenotypic variation,” this connection is accurate but incomplete. It is important to recognize that a cancer cell has access to the entire human genome to deploy adaptive strategies that use the molecular technology in normal processes. Thus, increased or decreased expression of normal genes involved in, for example, wound healing, fetal development, and xenobiotic metabolism also contribute to the “heritable phenotypic variation.” These epigenetic events can produce evolution in the absence of mutations [2].
Interestingly, during the 100 years following Darwin’s seminal observation, students of evolution did not know that the “mechanism of inheritance” was molecular genetics. Nevertheless, they established a sophisticated understanding of evolution exclusively by defining the interactions between environmental properties which comprise selection forces and the organism’s phenotypic properties which serve as adaptive strategies. While arguably less quantitative than modern molecular studies, this approach had the advantage of forming logical and clearly discernible cause-effect links between an evolving organism and its competitors, predators, and [19] environment. In these fundamental studies, investigation of genetics was unnecessary and, in fact, may have caused confusion because of the complex inter-relationships of individual genes and organismal adaptive strategies. It is an interesting exercise to ask if a modern Darwin sitting in the Beagle performing microarrays on tissue samples of finches from the different Galapagos Islands could have produced “On the Origin of Species.”
Here we focus on the roles of the “struggle for existence” and “environmental context” in Darwinian dynamics and the resulting distinction between somatic evolution of cancer and the Somatic Mutation Theory. Indeed, because of their uniquely important role in the transition from normal somatic cells to cancer, these dynamics ultimately play a critical role in both somatic evolution and the conventional view of cancer as a disease of the genes.
The evolutionary unit of selection
Evolution is the change in the heritable characteristics of a population with time. In response to natural selection it is the population that evolves, not the individual. Yet, the population is composed of individuals which represent the “units of selection”. The individual’s phenotypes are subject to evolutionary triage so heritable changes that increase fitness become more common while those that decrease fitness are typically lost. Importantly, the fitness contribution of each gene is not fixed but rather is dependent on local environmental selection forces which can vary over time and space.
Consider a population of single cell protists such as Paramecium. The individual cell is the primary level of selection, and each individual Paramecium is the unit of selection and all of the Paramecium within the population compete with each other. Natural selection favors those with heritable traits that survive and proliferate better in this struggle for [20] existence. Thus, each Paramecium cell possesses a self-defined fitness function because its survival and proliferation is dependent on its own heritable phenotypic properties. The organelles, cell membrane and molecular structures that comprise the Paramecium represent heritable traits that contribute to its fitness but the whole cell remains the unit of selection.
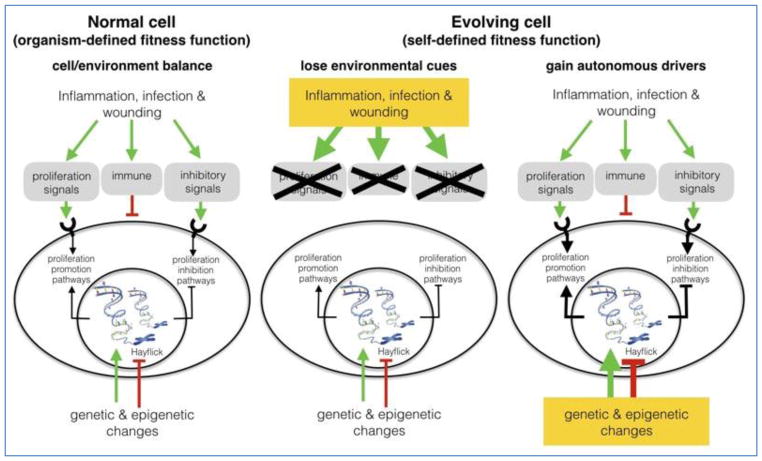
In normal human tissue, like all metazoans (multi-cellular organisms), the cells are not normally the unit of selection. That is, the whole organism is the level of biological organization shaped by natural selection. The cells of the multi-cell organism are components of the traits that contribute to the fitness of this whole organism. The organism in total possesses a fitness function but its constituent cells do not so that their proliferation and survival is governed entirely by tissue control mechanisms (Figure 1).
Figure 1.
A normal epithelial cell (left) is non-evolving because its survival and proliferation are entirely governed by tissue controls acting though intracellular pathways. Somatic evolution toward cancer requires a self-defined fitness function meaning that the cell’s survival and proliferation are determined by its own heritable phenotypic properties. One route to a self-defined fitness function (right) is the accumulation of random mutations which cumulatively turn off the cell’s reception or response to tissue messages. A second pathway (center) is loss of tissue signal due to inflammation, wounding, or infection. In this case, cells within the tissue are rendered independent and have an opportunity to evolve so that any prior mutations are suddenly relevant to its survival and proliferation. This cell now can potentially evolve toward a cancer phenotype before the tissue recovers and restores normal control signals.
We propose that a necessary condition for carcinogenesis is the evolutionary transition of somatic cells from a functioning member of a multicellular society to a single cell protist.
Evolution and the Somatic Cell
“Somatic evolution” is a term frequently applied to cellular-environmental interactions that lead to carcinogenesis. But, do somatic cells evolve? The answer is generally no – because normal cells proliferate only in response to tissue signals. As noted above, evolution by natural selection requires a direct link between the individual’s heritable properties and its survival, proliferation and success at increasing in numbers relative to other cells. Normal human cells survive and proliferate only when directed to do so by tissue signals. While some immune cells proliferate at the expense of others based on their stimulation by foreign antigens [21], normal epithelial and mesenchymal cells do not appear to compete with each other in a “struggle for existence”, and their proliferation is governed by the tissue collective. Thus, in conventional evolutionary formalism, the evolutionary unit of selection is the multicellular organism, which relies on the homeostatic functioning of multiple organs and their constituent cellular populations to survive and reproduce. In other words, the fitness of normal somatic cells is identical to that of the whole organism.
Somatic evolution during carcinogenesis appears to be a special case of Darwinian dynamics because it requires a transition from non-evolving cells to ones that are capable of evolution. That is, highly-regulated, integrated, “normal” cells that form functioning multicellular tissue cannot evolve since their proliferation is entirely governed by normal tissue signals. In contrast, cancers are chaotic populations (chaotic in the sense of not following the homeostatic directives of the whole organism) of cells that compete with each other for limited resources and evolve phenotypes that maximize fitness given these environmental circumstances. Between the states of normal and truly cancerous cells exists an intermediate series of steps in which the normal cells must develop independence to all of the probably several tissue growth constraints that control its proliferation. That is, evolution by natural selection cannot occur until a normal mammalian cell becomes capable of evolving.
Although at first glance this appears to be a tautology, we propose that, in fact, it represents an extraordinary transition in the evolutionary unit of natural selection. That is, somatic evolution will not occur until a cell’s survival and proliferation is no longer governed by tissue signals and is instead directly and solely linked to its own heritable phenotypic properties. This is a “self-defined fitness function”.
Tension between levels of selection
In most organisms, one level of biological organization becomes the primary unit of selection – the level that most completely exhibits a self-defined fitness function [22, 23] But, in all metazoans, a suppressed tension exists. Because many individual cells within the organisms have the capacity to proliferate and survive, they also have the potential to evolve but only if they acquire a self-defined fitness function. That is, an individual cell may become a unit of selection within the whole organism only if it becomes isolated from the control mechanisms of the surrounding tissue so that its survival and proliferation is governed not by tissue commands but by its own properties interacting with the environment. This somatic cell isolated from normal tissue controls can now evolve toward cancer.
Interestingly, tension between levels of selection are observed in [24] nature. Slime molds (Dictyostelium discoideum) provide a prime example [25]. Much of the time slime molds exist as free-ranging, haploid, single cell amoeba that can divide mitotically (asexual reproduction) or on occasion sexually by two cells fusing as a diploid complete with crossing over and recombination [26]. The free-ranging cells compete among themselves for resources and survival. But, for dispersal and persistence during inclement conditions, these cells gather together into mobile “slugs” that both move and later produce a stalk topped by a fruiting body. The stalk is built of individuals that forgo reproduction and differentiate into cells that provide structure. Only a small fraction of the original cells reach the top of the stalk and become spores that disperse by wind and water to become the next generation of free-ranging cells. During the free-ranging phase, the single cells are shaped by natural selection based on their competition for resources and ability to survive and reproduce -- they are a unit of selection. But, as slugs, these collectives of cells are competing to produce more successful spores than other slugs. These collectives exhibit specialized cells with division of function, and the whole slug acts as the level of biological organization subjected to natural selection. That is, the slug is now the unit of selection so that the life history of slime molds represents a balance between two levels of selection [27].
Further, multicellular organisms themselves can function as components of a higher level of selection. Consider the eusocial insects such as ants and honeybees. An individual ant is not a self-defined fitness function. It is a component of the whole colony. Natural selection operates to maximize colony fitness (the survival and proliferation of colonies) and the various castes of ants represent the heritable traits of the colony [28]. A society may be composed of individuals that are the units of selection. The individual’s fitness maximizing strategy is to contribute to and gain from the society. Examples of such tight societies include naked mole [29] rats, colonially nesting birds such as sociable weavers,[30][31] – sponges are even described as “parazoan” rather than metazoans like mammals. This mean that sponges do not have tissues and organs and some or all of the component cells can reproduce asexually and/or sexually (not unlike the slime molds in some[32] respects).
Transition to self-defined fitness function in somatic cells
How does a normal cell develop a self-defined fitness function? In general, we propose that this state requires a substantial if not total loss of communication between the cell and its local environment. Clearly, one mechanism is driven by intracellular events in which all of the cell’s receivers or processors of tissue signals become defective – consistent with the Somatic Mutation Theory.
Although it is not clear how many tissue control pathways exist (and they may vary from tissue to tissue) we might reasonably expect them to be redundant and robust to perturbations so that the accumulation of all of the necessary mutations by random events would likely be rare. Indeed, it is evident that, during a lifetime, normal cells in virtually every organ (particularly those subject to a mutagen such as skin exposed to UV light) will accumulate a significant number of diverse mutations while maintaining normal morphology and function. This is readily explained - because they lack a self-defined fitness function, they will not evolve as a result of the mutations and will remain phenotypically normal. This is, in fact, observed in normal epithelial cells [8, 33, 34]. Clearly, exposure to a mutagenic environment, such as radiation, will increase the number of mutations and, thus, the probability of achieving some threshold of inborn errors sufficient to achieve isolation from tissue controls and a self-defined fitness function. In a non-mutagenic environment, one would have to assume that this threshold is achieved only through an improbable distribution of random events that just happen to cause cellular isolation.
Is there an alternative to slowly accumulating somatic mutations as a mechanism to confer a self-defined fitness function? Yes. The equivalent dynamics can develop through a perturbation of the local tissue. Interestingly, this could be due to an extension of the Mutational Theory if genetic defects occur in surrounding cells that result in, for example, continuous over-expression of growth factors. However, it seems more likely that a cell’s environment can be globally disrupted by inflammation, injury, or infection. The local epithelial cells will become isolated not because of internal mutations but due to absent or uncoordinated signals from the surrounding host tissue. Thus, apparently normal epithelial cells will be cast adrift through disruption of the usual control signals from the extant local tissue. This also results in a self-defined fitness function and permits the cell to evolve at least until normal tissue function is restored.
It is likely that re-establishment of tissue control over each remaining epithelial and mesenchymal cell will usually occur as the tissue disruption resolves and healing progresses. However, during the period in which the cell possesses a self-defined fitness function, sufficient evolution may have occurred due to pre-existing mutations or accumulation of new heritable events so that it can adapt and overcome the control signals emerging from the healing tissue. Furthermore, because the cell is evolving, each new gene mutation becomes subject to “evolutionary triage” [35] so that favorable mutations will result in increased proliferation and unfavorable mutations will be lost and therefore not observed within the population. Because this Darwinian optimization eliminates mutations that negatively affect fitness, the number of observable genetic changes in an evolving population will be fewer than the number that would be found if the cell was not subject to evolutionary forces. Thus, many of the genetic changes found in cancer cells may have occurred prior to the onset of somatic evolution and actually have no role in the cancer formation.
Re-interpreting initiation and promotion experiments
It is interesting to re-interpret the classical initiation- promotion carcinogenesis experiments [36] in light of these evolutionary principles. Initiation requires application of a mutagen that increases local mutation burden but does not typically lead to tumor formation [37, 38]. This requires a promotion event that typically disrupts local tissue through injury or inflammation[38]. In the Somatic Mutation Theory, this wounding event is thought to promote cancer formation by increasing cellular proliferation during healing. In our model, a critical role of inflammation and wound healing is the disruption of local tissue signals which allows some cells to transiently develop a self-defined fitness function. Importantly, because this cell can now evolve, its fitness is affected by the genetic changes that occurred during the previous applications of mutagens. Furthermore, the inflammatory changes in the tissue during this time provide harsh environmental conditions that may accelerate mutation both by establishing a steep fitness gradient and increasing the mutation rate. This, along with any increased proliferation during healing, greatly increases the probability that a tumor population will emerge at the site.
Clinical relevance
Our model of carcinogenesis makes a number of predictions that should be testable. Notably, it is likely that many, perhaps most, mutations observed in cancer cells will have occurred prior to somatic evolution. That is, they accumulated randomly when the cell did not possess a self-defined fitness function. Furthermore, since somatic cells begin evolving with a legacy of mutations from their time as a normal cell, many mutations observed in cancer cells will have little if any role in their malignant phenotype. However, once the cell begins evolving, the optimization of Darwinian dynamics (evolutionary triage) will efficiently select for favorable mutations while unfavorable mutation will be lost because less fit cells are eliminated. As a result, we anticipate that the mutational burden of cancer cells will likely be very similar to the adjacent normal tissue. And, many (but not all) of the mutations in the cancer cells will also be observed in their adjacent normal counterparts. These predictions are generally consistent with clinical observations of a significant mutational burden in normal skin and breast cells that are similar to that of corresponding cancers. Furthermore, we propose that the large variations in mutational burdens in different cancers may be due primarily to molecular events that occurred prior to the events that allowed local cells to evolve.
Conclusion
Necessary and sufficient conditions for evolution by natural selection include: heritable phenotypic variability, a “struggle for existence”, and conditions such that the former governs the outcomes of the latter. In the context of normal and cancer tissues, a useful shorthand is that evolution will occur only in a somatic cell with a “self-defined fitness function.” While somatic evolution is an accurate description of carcinogenesis it is important to recognize the critical first step is transition from a cell that cannot evolve to one that can.
By self-defined fitness function, we mean that the cell’s survival and proliferation is determined solely by its own heritable properties. In contrast, normal somatic cells are controlled by tissue signals so that their survival and proliferation are governed by external signals. Until a cell acquires self-defined fitness function, any mutations that occur within it will not result in evolution so that normal cells in normal tissue may contain multiple mutations without any apparent effect.
Of course, randomly accumulating mutations can interrupt each of the control pathways resulting in the necessary transition to independence. However, we note that individual cells may become the unit of selection when tissue perturbations due, for example, to injury, infection or inflammation result in loss of normal controls. Thus, while mutations may render the cell deaf and blind to external messages, a similar state may result when tissue damage temporarily interrupts the formation of those messages. In the cellular transition from normal to malignant cells there is a likely interplay between normal and perturbed tissue dynamics with the latter being sufficient and perhaps necessary to permit the accumulation and triaging of heritable variation via periods of cell turnover. Finally, we note that inflammation is viewed as a “hallmark” of primary and metastatic cancer. Thus, it is possible that cancer cells may actively induce an inflammatory “niche” as an ongoing strategy to disrupt normal tissue control mechanisms and maintain the self-defined fitness function.
More generally, cancer has been described as a “disease of the genes” and, indeed, heritable phenotypic variation is a necessary condition for natural selection. But, a “disease of the genes” is sufficient only if these variations govern cellular survival and proliferation. When mutations occur in a cell that does not possess a self-defined fitness function, no evolution will result. Cancer has also been described as a disease of “unregulated or unlimited proliferation [39]”. The capacity to proliferate indefinitely is necessary for a cell to evolve. But, it is sufficient only in if there is heritable phenotypic variation within the population that affects their survival and proliferation. Thus, neither of the conventional paradigms is complete but the two together, heritable variation and indefinite proliferation, do provide the necessary and sufficient conditions for cancer initiation and cancer progression via evolution by natural selection.
Acknowledgments
This work is supported by the Moffitt Physical Science Oncology Center (U54CA193489) and a grant by the V Foundation. The authors also thank Dr. Jill Gallaher for gracious and skillful help in designing the figures for the manuscript
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 2.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 4.Gerlinger M, McGranahan N, Dewhurst SM, Burrell RA, Tomlinson I, Swanton C. Cancer: evolution within a lifetime. Annu Rev Genet. 2014;48:215–236. doi: 10.1146/annurev-genet-120213-092314. [DOI] [PubMed] [Google Scholar]
- 5.Vaux DL. In defense of the somatic mutation theory of cancer. Bioessays. 2011;33:341–343. doi: 10.1002/bies.201100022. [DOI] [PubMed] [Google Scholar]
- 6.Soto AM, Sonnenschein C. The somatic mutation theory of cancer: growing problems with the paradigm? Bioessays. 2004;26:1097–1107. doi: 10.1002/bies.20087. [DOI] [PubMed] [Google Scholar]
- 7.Brucher BL, Jamall IS. Somatic Mutation Theory - Why it's Wrong for Most Cancers. Cell Physiol Biochem. 2016;38:1663–1680. doi: 10.1159/000443106. [DOI] [PubMed] [Google Scholar]
- 8.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, van 't Veer L, Vincent-Salomon A, Waddell N, Yates LR, PedBrain I, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR I Australian Pancreatic Cancer Genome, IBC Consortium, IM-S Consortium. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brose MS, Rebbeck TR, Calzone KA, Stopfer JE, Nathanson KL, Weber BL. Cancer risk estimates for BRCA1 mutation carriers identified in a risk evaluation program. J Natl Cancer Inst. 2002;94:1365–1372. doi: 10.1093/jnci/94.18.1365. [DOI] [PubMed] [Google Scholar]
- 10.Levy-Lahad E, Friedman E. Cancer risks among BRCA1 and BRCA2 mutation carriers. Br J Cancer. 2007;96:11–15. doi: 10.1038/sj.bjc.6603535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yadav VK, DeGregori J, De S. The landscape of somatic mutations in protein coding genes in apparently benign human tissues carries signatures of relaxed purifying selection. Nucleic Acids Res. 2016;44:2075–2084. doi: 10.1093/nar/gkw086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM, Stebbings L, Menzies A, Widaa S, Stratton MR, Jones PH, Campbell PJ. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng G, Lu Y, Zlotnikov G, Thor AD, Smith HS. Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science. 1996;274:2057–2059. doi: 10.1126/science.274.5295.2057. [DOI] [PubMed] [Google Scholar]
- 14.Caulin AF, Maley CC. Peto's Paradox: evolution's prescription for cancer prevention. Trends Ecol Evol. 2011;26:175–182. doi: 10.1016/j.tree.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chinn CA, Brewer WF Center for the Study of Reading Urbana IL. The Role of Anomalous Data in Knowledge Acquisition A Theoretical Framework and Implications for Science Instruction. Distributed by ERIC Clearinghouse, Place Published; 1993. Technical Report No. 583. [Google Scholar]
- 16.Brown JS. Why Darwin would have loved evolutionary game theory. Proc Biol Sci. 2016;283 doi: 10.1098/rspb.2016.0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox CF. Charles Darwin and the mutation theory. Annals of the New York Academy of Sciences. 1908;18:431–451. [Google Scholar]
- 18.Darwin CR. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. James Murray, Place Published; 1859. [PMC free article] [PubMed] [Google Scholar]
- 19.Lack D. Darwin's Finches. Cambridge University Press, Place Published; 1947. [Google Scholar]
- 20.Nyberg DaBP. High Levels of Phenotypic Variability of Metal and Temperature Tolerance in Paramecium. Evolution. 1983;37:341–357. doi: 10.1111/j.1558-5646.1983.tb05544.x. [DOI] [PubMed] [Google Scholar]
- 21.Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. 2007;39:1461–1468. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
- 22.Michod RE. Evolution of the individual. Am Nat. 1997;150(Suppl 1):S5–S21. doi: 10.1086/286047. [DOI] [PubMed] [Google Scholar]
- 23.Michod RE, Roze D. Transitions in individuality. Proc Biol Sci. 1997;264:853–857. doi: 10.1098/rspb.1997.0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rinkevich B. A critical approach to the definition of Darwinian units of selection. Biol Bull. 2000;199:231–240. doi: 10.2307/1543179. [DOI] [PubMed] [Google Scholar]
- 25.Baldauf SL, Doolittle WF. Origin and evolution of the slime molds (Mycetozoa) Proc Natl Acad Sci U S A. 1997;94:12007–12012. doi: 10.1073/pnas.94.22.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robson GE, Williams KL. The mating system of the cellular slime mould Dictyostelium discoideum. Curr Genet. 1980;1:229–232. doi: 10.1007/BF00390948. [DOI] [PubMed] [Google Scholar]
- 27.Strassmann JE, Zhu Y, Queller DC. Altruism and social cheating in the social amoeba Dictyostelium discoideum. Nature. 2000;408:965–967. doi: 10.1038/35050087. [DOI] [PubMed] [Google Scholar]
- 28.Martens J. Organisms in evolution. Hist Philos Life Sci. 2010;32:373–400. [PubMed] [Google Scholar]
- 29.Burda H, Honeycutt R, Begall S, et al. Are naked and common mole-rats eusocial and if so, why? Behavioral Ecology and Sociobiology. 2000;47:293–303. [Google Scholar]
- 30.Covas R, Du Plessis M, Doutrelant C. Helpers in colonial cooperatively breeding sociable weavers Philetairus socius contribute to buffer the effects of adverse breeding conditions. 2008;63:103–112. [Google Scholar]
- 31.Buss LW. Evolution, development, and the units of selection. Proc Natl Acad Sci U S A. 1983;80:1387–1391. doi: 10.1073/pnas.80.5.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsurumi MR, HM Sexual versus asexual reproduction in an oviparous rope-form sponge, Aplysina cauliformis (Porifera; Verongida) nvertebrate Reproduction & Development. 1997;32:1–9. [Google Scholar]
- 33.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev. 2014;24:52–60. doi: 10.1016/j.gde.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gatenby RA, Cunningham JJ, Brown JS. Evolutionary triage governs fitness in driver and passenger mutations and suggests targeting never mutations. Nat Commun. 2014;5:5499. doi: 10.1038/ncomms6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Becker FF. Presidential address. Recent concepts of initiation and promotion in carcinogenesis. Am J Pathol. 1981;105:3–9. [PMC free article] [PubMed] [Google Scholar]
- 37.Shubik P, Baserga R, Ritchie AC. The life and progression of induced skin tumors in mice. Br J Cancer. 1953;7:342–351. doi: 10.1038/bjc.1953.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peraino C, Fry RJ, Staffeldt E. Effects of varying the onset and duration of exposure to phenobarbital on its enhancement of 2-acetylaminofluorene-induced hepatic tumorigenesis. Cancer Res. 1977;37:3623–3627. [PubMed] [Google Scholar]
- 39.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]