Abstract
The biochemical pathway for formation of branched-chain aldehydes, which are important flavor compounds derived from proteins in fermented dairy products, consists of a protease, peptidases, a transaminase, and a branched-chain α-keto acid decarboxylase (KdcA). The activity of the latter enzyme has been found only in a limited number of Lactococcus lactis strains. By using a random mutagenesis approach, the gene encoding KdcA in L. lactis B1157 was identified. The gene for this enzyme is highly homologous to the gene annotated ipd, which encodes a putative indole pyruvate decarboxylase, in L. lactis IL1403. Strain IL1403 does not produce KdcA, which could be explained by a 270-nucleotide deletion at the 3′ terminus of the ipd gene encoding a truncated nonfunctional decarboxylase. The kdcA gene was overexpressed in L. lactis for further characterization of the decarboxylase enzyme. Of all of the potential substrates tested, the highest activity was observed with branched-chain α-keto acids. Moreover, the enzyme activity was hardly affected by high salinity, and optimal activity was found at pH 6.3, indicating that the enzyme might be active under cheese ripening conditions.
Flavor formation in cheeses is mainly the result of catabolism of milk proteins, sugar, and lipids. Degradation of proteins into amino acids followed by conversion of these amino acids by Lactococcus spp. is very important for flavor formation in semihard types of cheese, like Gouda (for reviews see references 12, 25, 38, and 43). One of the predominant amino acid conversion routes is initiated by transaminases. Transamination of amino acids yields the corresponding α-keto acids, which do not have strong flavor properties (12, 13) but are the precursors of various aldehydes, alcohols, acids, and (thio)esters. The latter compounds generally have much lower odor thresholds and strongly contribute to cheese flavor.
This is exemplified by the production of flavor compounds from leucine. Transamination of leucine results in the production of α-isocaproic acid (KICA). Interestingly, only a limited number of Lactococcus lactis strains are capable of decarboxylating this α-keto acid to 3-methylbutanal (36, 37). The decarboxylating activity is observed mainly in L. lactis strains with nondairy origins, whereas the industrial strains tested hardly showed this activity (36). Subsequently, (de)hydrogenation of 3-methylbutanal results in 3-methylbutanol or 3-methylbutyric acid, which also contributes to the flavor of cheese, but the odor thresholds for these compounds are higher than the odor threshold for 3-methylbutanal (24, 29). The rate-determining step in this route is the decarboxylation of KICA, and therefore, control of this enzyme offers good prospects for the design of starter cultures with tailored flavor production (37, 43).
The enzyme performing this reaction has not been identified. A number of thiamine pyrophosphate (TPP)-dependent α-keto acid decarboxylases have been identified in various organisms, and these enzymes include pyruvate decarboxylase (EC 4.1.1.1) (34), phenylpyruvate decarboxylase (EC 4.1.1.43) (4, 40), branched-chain 2-oxoacid decarboxylase (EC 4.1.1.72) (27), 2-oxoglutarate decarboxylase (EC 4.1.1.71) (28, 33), and indole-3-pyruvate decarboxylase (IPD) (EC 4.1.1.74) (18). In L. lactis, however, there has been only one report of partial purification of an α-keto acid decarboxylase (3), and in the genome of L. lactis IL1403 (8) a putative (partial) indole-3-pyruvate decarboxylase gene and a 2-oxoglutarate decarboxylase gene have been identified but pyruvate decarboxylase and branched-chain keto acid decarboxylase genes are absent. In the present paper we describe the identification and cloning of the gene encoding the branched-chain α-keto acid decarboxylase (kdcA) responsible for KICA conversion in L. lactis B1157. This gene was overexpressed in L. lactis by using the nisin-controlled overexpression system, and the gene product was characterized.
MATERIALS AND METHODS
Strains, plasmids, and media.
All strains and plasmids used or constructed in this study are listed in Table 1. Lactococcal strains were grown in M17 broth (Difco, Detroit, Mich.) supplemented with lactose (LM17) or glucose (GM17) as an additional carbon source at a final concentration of 0.5% (wt/vol). If appropriate, chloramphenicol or erythromycin was added to the medium to a final concentration of 5 μg/ml. Strains were grown at either 28 or 40°C. The ability of strains to produce 3-methylbutanal was determined by static headspace gas chromatography (37) analysis after 24 h of growth (optical density at 600 nm, ∼1.8) in the presence or absence of 2 mM KICA in 10-ml HS vials containing 3 ml of culture medium.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Antibiotic resistancea | Reference(s) or source |
|---|---|---|---|
| Strains | |||
| MC1061 | E. coli | 9 | |
| B1157 | Branched-chain α-keto acid decarboxylase-positive L. lactis strain | Tct, Cm | 5 |
| NZ9000 | MG1363 derivative; pepN::nisRK; standard host for NICEb | 20 | |
| MG1363 | Plasmid free, wild-type L. lactis strain, genome sequenced | 14 | |
| IL1403 | Wild-type L. lactis strain, genome sequenced | 8 | |
| B2083 | pGh9:ISSI integrated in the chromosome of B1157 at position 462 of the decarboxylase gene | Tet, Ery, Cm | This study |
| B2084 | pGh9:ISS1 integrated in the chromosome of B1157 at position 995 of the decarboxylase gene | Tet, Ery, Cm | This study |
| B2085 | NZ9000 (pNZ7500) | Ery | This study |
| Plasmids | |||
| pGh9:ISS1 | pWV01 carrying a thermosensitive replicon | Ery | 22, 23 |
| pNZ8148 | Homologous L. lactis vector for NICE system | Cm | 20 |
| pNZ7500 | pNZ8148 derivative containing a 1.8-kb fragment carrying the decarboxylase gene of B1157 | Cm | This study |
| pNZ7501 | pNZ8148 derivative containing a 1.5-kb fragment carrying the partial decarboxylase gene of B1157 | Cm | This study |
| pNZ7502 | pNZ8148 derivative containing a 1.4-kb fragment carrying the decarboxylase gene of IL1403 annotated as ipd | Cm | This study |
Tet, tetracycline, Cm, chloramphenicol; Ery, erythromycin.
NICE, nisin-controlled expression.
Escherichia coli was grown aerobically at 30°C on TY medium (Difco), which contained 200 μg of erythromycin per ml if appropriate.
DNA handling procedures.
Before the protocols used for DNA isolation were performed, L. lactis cells were pretreated for 10 min with 2 mg of lysozyme per ml in THMS buffer at 37°C. Plasmid DNA was isolated with a Micro plasmid prep kit (Amersham/Pharmacia, Uppsala, Sweden), and total DNA was isolated with an AquaPure genomic DNA isolation kit (Bio-Rad, Veenendaal, The Netherlands) used according to the manufacturer's protocols. L. lactis cells were transformed by electroporation as described previously (41). Agarose gel electrophoresis was performed as described by Sambrook et al. (30). DNA fragments were isolated from agarose gels by using a gel purification kit (Jetstar; Genomed, Bad Oeynhausen, Germany). Restriction enzymes, T4 ligase, and other DNA-modifying enzymes were purchased from Promega Corp. (Leiden, The Netherlands) or Invitrogen (Carlsbad, Calif.) and were used as recommended by the manufacturer. PCR was performed by using a Pwo polymerase PCR kit (Roche, Mannheim, Germany) and the primers listed in Table 2 with total DNA preparations as templates. Twenty-six thermal cycles with a Mastercycler (Eppendorf, Hamburg, Germany) were used with the 50-μl reaction mixtures. Each cycle started with 30 s at 95°C, which was followed by annealing for 30 s at 55°C and elongation for 3.5 min at 72°C. Sequencing was performed with a Thermo Sequenase Cy5.0 Dye terminator cycle sequencing kit (Amersham Pharmacia), and separation was performed with an Alf sequencer (Amersham Pharmacia).
TABLE 2.
PCR primers used in this study
| Primer | Sequencea |
|---|---|
| Nis_292Rev | 5′ AATTGAACGTTTCAAGCC |
| Nis_115fw | 5′ CGGCTCTGATTAAATTCTG |
| IL1403-10fw | 5′ GGAATGCCatgGATACAGTAGGAGATTACC |
| IL1403+1374Rev | 5′ AGAATCTAGATCAACTATAAGAGTTCATATGAG |
| B1157+3fw | 5′ GTATACAGTAGGACATTACC |
| B1157+623fw | 5′ CAGTAGTGATTGCAGGACAC |
| B1157+1303Rev | 5′ GTTGAAGTGAACCATCACC |
| B1157+1633Rev | 5′ GCTCAGCAAATAATTTACCC |
| B1157+1793Rev | 5′ GACTTCTCTAGATTATATCTTGCTGGTT |
| pGh9:ISS1_ER | 5′ CCGTTAAATCGACTGGCG |
| pGh9:ISS1_EF | 5′ GACTTATCAGAAAACTTTGC |
| pGh9:ISS1_HR | 5′ CGGTATCTACTGAGATTAAGG |
| pGh9:ISS1_HF | 5′ GCGCGCGTAATACGACTC |
Bases that mismatch are indicated by boldface type; the restriction sites used for cloning are underlined; and the start of a gene is indicated by lowercase letters.
Construction of a random mutant library.
A random insertion mutant library was constructed by using the thermosensitive plasmid pGh9:ISS1 as described by Maguin et al. (22, 23). The pGh9:ISS1 plasmid was isolated from E. coli(pGh9:ISS1) and transformed into L. lactis B1157. Erythromycin-resistant, plasmid-containing transformants were selected at a permissive temperature, 28°C. Twenty-five milliliters of LM17 without erythromycin was inoculated (1%) with B1157(pGh9:ISS1), and the organism was grown at 28°C for 2.5 h; this was followed by transfer of the cultures to a nonpermissive temperature, 40°C, for 2.5 h to force random plasmid integration into the genome of strain B1157. More than 8,500 integrants were selected on LM17 agar containing 2 μg of erythromycin per ml; individual colonies were organized in 384-well microtiter plates containing LM17 with erythromycin and 20% glycerol, and the plates were frozen at −80°C. It was calculated that a library consisting of 8,000 mutants was needed so that 95% of the genes were represented by an integrant, assuming random insertion into the genome, a genome size of 2.3 Mb, and an average gene size of 880 kb (8). The frequency of integration was determined to be 2.4%, and the library was analyzed for the presence of random integrants by Southern blotting. For this purpose total DNAs of 20 strains from the insertion sequence (IS) library were isolated and cut with either EcoRI or HindIII. The fragments obtained were separated on a 0.7% agarose gel, blotted, and hybridized with HindIII-cut, digoxigenin-labeled pGh9:ISS1 (Roche) as the probe. Eighty percent of the colonies gave fragments that were different sizes.
For determination of the site of integration in 3-methylbutanal-negative clones B2083 and B2084, total DNA was digested with HindIII or EcoRI and subsequently ligated by using T4 ligase. Ligation mixtures were used to transform L. Lactis MG1363 that was grown at the permissive temperature (28°C). Plasmid sequencing was performed by using the four pGh9:ISS1-derived primers (Table 2). The HR and HF primers were used for the plasmids derived from HindIII-digested total DNAs, and the ER and EF primers were used for the plasmids originating from the EcoRI-digested total DNAs.
Identification of decarboxylase knockouts.
Decarboxylase-negative clones in the mutant library were identified by fast screening of volatile compounds produced in miniaturized fermentations as described previously (36). Briefly, the method is based on the growth of individual mutants in a 96-well stainless steel block containing 2-ml wells filled with 1 ml of LM17 with 2 mM KICA. The block is closed airtight, and after fermentation for 24 h at exactly 40°C, individual headspace samples (100 μl) were analyzed by using direct inlet mass spectrometry (Trace-MS; Interscience, Breda, The Netherlands) at a throughput rate of one sample per minute (36). The amount of 3-methylbutanal present in the sample was calculated from the response at m/z = 58.
Overexpression of the decarboxylase gene.
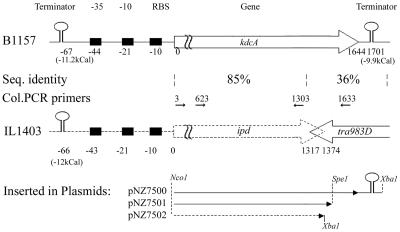
To further analyze the function of the decarboxylase gene products, plasmids were constructed for nisin-dependent overexpression of the ipd gene from IL1403 (1,374 bp), the kdcA gene from B1157 (1,644 bp plus 166 bp), and a truncated kdcA gene from B1157 (1,569 bp, missing 75 bp) (Fig. 1). The fragments were obtained by PCR by using two primer pairs, primers IL1403-10fw and IL1403+1374R and primers IL1403-10fw and B1157+1793Rev (Table 2), with total DNAs of L. lactis IL1403 and L. lactis B1157, respectively. Thirty thermal cycles with the following steps were used: denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and elongation at 72°C for 120 s. The PCR products obtained were digested with Nco1-XbaI and cloned into pNZ8148, resulting in plasmids pNZ7502 (containing IL1403 ipd) and pNZ7500 (containing the B1157 decarboxylase gene). The PCR product obtained from B1157 was also digested with SpeI and NcoI and cloned into pNZ8148, generating pNZ7501 (containing the B1157 decarboxylase gene lacking 75 bp at the 3′ terminus). The identities and integrities of the inserts cloned in pNZ8148 were confirmed by sequencing. All plasmids were introduced into strain NZ9000 (20) and used for nisin-controlled overexpression of the cloned (partial) genes by previously established procedures (10).
FIG. 1.
Schematic representation of the DNA region carrying the kdcA gene in L. lactis B1157 and the ipd gene in strain IL1403. The fragments fused with pNZ8148 for overexpression in L. lactis NZ9000 are shown below the schematic diagrams. Between the two regions the sequence homologies (Seq. identity) and the positions of the PCR primers used for screening of L. lactis strains (Col.PCR primers) are indicated.
Colony PCR.
L. lactis strains B1021, B1152, B1156, B1157, B1158, B1165, B1231, B1233, B1235, B1236, B1275, B1492, B2085, B234, B25, B26, B27, B28, B31, B4, B40, B42, B48, B643, B697, B698, B700, B76, B77, B78, B78, B79, B80, B81, B83, IL1403, NZ9000(pNZ7502), NZ9000, and SK110, obtained from the NIZO culture collection, were grown overnight in 1 ml of GM17 broth. A 0.5-μl portion of a bacterial cell suspension was treated twice for 30 s in a microwave at 750 W. Four PCRs were performed for each strain by using the four possible combinations of primers B1157+3fw, B1157+623fw, B1157+1303Rev, and B1157+1633Rev (Table 2 and Fig. 1). Using these primers should have resulted in either four fragments (complete kdcA gene present), two fragments (truncated kdcA gene present), or no fragments (no kdcA gene present). Thirty thermal cycles with the following steps were used: denaturation at 95°C for 30s, annealing at 40°C for 30 s, and elongation at 72°C for 120 s.
Overproduction of decarboxylases.
For overproduction of decarboxylases, an overnight culture of L. lactis NZ9000 harboring either pNZ7500, pNZ7501, or pNZ7502 was transferred (1%) to fresh medium (GM17) and grown until an optical density at 600 nm of 0.5 was reached. The cells were induced with 0.2 or 2 ng of nisin ml−1 and grown for another 2 h. The cells were then harvested, and the cell extracts (CFEs) were prepared as described below. These extracts were used for the enzymatic assays and protein analysis.
Preparation of CFEs.
Cells were harvested by centrifugation and washed twice in 25 mM sodium citrate buffer (pH 6.5). The cells were disrupted by the Fast Prep method (Bio 101, Savant, Holbrook, N.Y.) twice for 30 s. The supernatant after centrifugation at 20,000 × g for 15 min was used as the CFE. All steps were performed at temperatures below 6°C.
Protein analysis.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) with a 12% polyacrylamide gel in a mini Protean II system (Bio-Rad) was used to analyze the production of the protein. Protein concentrations were determined in triplicate by using a BCA protein assay reagent kit obtained from Pierce Inc. (Rockford, Ill.). A 30-μl sample was mixed with 240 μl of work reagent. After incubation at 37°C for 20 min, the optical density at 562 nm was measured with a Spectra max (Molecular Devices, Sunnydale, Calif.). Triplicate measurements were obtained independently.
KdcA assay.
The α-keto acid decarboxylase assay with KICA was performed as described previously (37). In essence, an enzyme sample was incubated for 3 h at 30°C in a closed 10-ml HS vial at pH 6.3 (50 mM sodium citrate) with 10 mM KICA as the substrate and with 1 mM MgCl2 and 0.1 mM TPP as cofactors. After 3 h the reactions were quenched by lowering the pH to between 2 and 3 with 6 N HCl. The production of 3-methylbutanal was quantified by static headspace gas chromatography as described previously (37). Gas chromatography enabled detection of the aldehyde of interest only, and a CFE of strain MG1363 was used as a negative control. The conversion rate was calculated by dividing the amount of product formed (in micromoles) by the reaction time (in minutes) and the protein content of the sample (in milligrams).
Characterization of KdcA.
For the characterization experiments CFE of overexpressing strain B2085 was used in the KdcA assay, and the pH was varied between 4 and 9 by using sodium citrate, sodium phosphate, and Tris buffers. NaCl was added at concentrations ranging from 0.05 to 1.1 M. The substrate specificity of the decarboxylase was determined by measuring the decreases in the specific substrates by reverse-phase high-performance liquid chromatography or ion-exchange high-performance liquid chromatography as described previously (35). The reaction mixture described above was used. The amount of cell extract of strain B2085 was adjusted for each substrate used; our aim was conversion of about 30% in 3 h. The following substrates were used at a final concentration of 10 mM (all substrates were obtained from Sigma, Zwijndrecht, The Netherlands, or Fisher, Landsmeer, The Netherlands) (2-oxo = α-keto): 2-oxo-4-methylpentanoic acid (α-ketoisocaproic acid or KICA), 2-oxo-3-methylpentanoic acid, 2-oxo-3-methylbutanoic acid, 2-oxohexanoic acid, 2-oxopentanoic acid, 2-oxobutanoic acid, pyruvic acid, 4-methylthio-2-oxobutanoic acid, phenylpyruvic acid, indole 3-pyruvate, 3-(4-hydroxyphenyl)-2-oxopropanoic acid, 2-oxopentanedioic acid, 4-methylpentanoic acid, 3-methylbutanoic acid, and 2-methylpropanoic acid. The conversion rate was calculated by dividing the amount of substrate converted (in micromoles) by the reaction time (in minutes) and the protein content of the sample (in milligrams).
Sequence homology analysis and homology modeling.
Homology relationships were established by using BLASTP (1) and TBLASTX (2), and multiple-sequence alignments were constructed with ClustalW (15). Homology modeling of KdcA and IPD of L. lactis IL1403 was performed with the program WHAT IF (39) by using the structure of yeast pyruvate decarboxylase (PDB code 1QPB) as a template. The 1QPB structure consists of a dimer with bound Mg2+ ions, TPP, and pyruvamide. The results of the modeling were visualized by using Protein Explorer (http://molvis.sdsc.edu/protexpl/frntdoor.htm; copyright 1998-2003 by Eric Martz).
Nucleotide sequence accession number.
The sequence of the kdcA gene has been deposited in the GenBank database under accession number AY548760.
RESULTS
Identification of the branched-chain α-keto acid decarboxylase gene.
Previous work indicated the importance of a branched-chain keto acid decarboxylase in the formation of flavor-active compounds by a number of L. lactis strains (36, 37). So far, no gene encoding a decarboxylase with activity toward branched-chain α-keto acids in L. lactis has been cloned, nor did the (preliminary) genome sequences of lactic acid bacteria reveal a clear candidate gene for this activity (8,17; http://genome.jgi-psf.org/draft_microbes/laccr/laccr.home.html). In order to identify the decarboxylase gene, an L. lactis B1157 IS mutant library was constructed by using the thermosensitive plasmid pGh9:ISS1. The library was validated by Southern blotting and restriction analysis (results not shown). Screening of 2,500 clones of the IS mutant library by using direct inlet mass spectrometry resulted in identification of two clones that had lost the capacity to produce 3-methylbutanal from KICA. These strains were designated B2083 and B2084. The integration sites were identified by recovering the chromosomal region flanking the integrated pGh9:ISS1 plasmid. This was done by ligation of either HindIII- or EcoRI-digested chromosomal DNAs of these strains. The ligation mixtures were used to transform L. lactis MG1363, and the cloned genomic regions were analyzed by restriction analysis. The plasmids that were recovered from strains B2083 and B2084 contained 6.2- and 8-kb inserts, respectively, and in the case of EcoRI digestion, the plasmid recovered from strain B2083 contained a 9.1-kb insert. The cloned flanking genomic regions were sequenced with primers annealing to pGh9:ISS1. In strain B2083 the ISS1 element integrated at position 453 of a 1,644-bp open reading frame, and in strain B2084 integration occurred at position 916 of the same open reading frame. The gene was designated the kdcA gene and was sequenced in both directions by primer walking. Upstream of this gene there are three regions that might function as a terminator (−67 bp, −11.2 kcal), a promoter (−44 to −21bp), and a ribosome binding site (−10 bp). Downstream of the kdcA gene a putative terminator (−9.9 kcal) starting at bp 1701 was identified (Fig. 1).
Homology of KdcA to other TPP-dependent decarboxylases.
A search with the BLASTP and TBLASTX programs in various protein and DNA databases showed that the gene product KdcA was homologous to indolepyruvate decarbobxylases and pyruvate decarboxylases found in various organisms. The highest sequence identity, 96%, was observed with the first 438 amino acids of the putative ipd gene product of L. lactis IL1403. The remaining C-terminal fragments of KdcA and the ipd gene product consisted of 109 and 19 residues, respectively, and did not show significant homology (Fig. 1). Moreover, in strain IL1403 the 3′ terminus of the ipd gene overlaps the IS element IS983O carrying the putative transposase gene tra983D, which is located downstream of the ipd gene (Fig. 1) (8). Typically, the ipd genes encode polypeptides consisting of approximately 550 amino acid residues (32). These data indicate that in IL1403 the ipd gene is disrupted due to the insertion of IS983O.
The amino acid sequences of the ipd gene product and KdcA were aligned with the sequences of two well-characterized and homologous keto acid decarboxylating enzymes, yeast pyruvate decarboxylase (Pdc1p_SacCe) and indolepyruvate decarboxylase of Enterobacter cloacae (IpdC_EntCl) (11, 31). KdcA, pyruvate decarboxylases, and indolepyruvate decarboxylases all depend on TPP and Mg2+ as cofactors. The levels of sequence identity observed were between 35 and 83%. Several regions were highly conserved, including the regions containing residues catalyzing the transition state intermediate during catalysis (Fig. 2) (21) and residues involved in binding of the substrate and cofactors (31). The alignment in Fig. 2 clearly demonstrates that several of the residues involved in these functions are conserved in KdcA and are absent in the ipd gene product of strain IL1403.
FIG. 2.
Amino acid sequences of the branched-chain α-keto acid decarboxylase of L. lactis strain B1157 (KdcA_LacLa; accession number gi:44921617), the putative indolepyruvate decarboxylase of L. lactis IL1403 (Ipd_LacLa; accession number gi:15673286 gi:44921617), indolepyruvate decarboxylase of E. cloacae (IpdC_EntCl; accession number gi 118333), and pyruvate decarboxylase of Saccharomyces bayanu (Pdc1p_SacCe; accession number SGDID S000004034). The ClustalW program was used to align the sequences. The numbers indicate the locations of the adjacent amino acid residues in each protein. The black shading indicates amino acids that are identical in at least three of the proteins; the gray shading indicates similar amino acids. The groups of amino acids that were considered similar were I, L, M, and V; A, G, and S; H, K, and R; D and E; N and Q; F, W, and Y; and S and T. Residues interacting with TPP in IpdC_EntCl are indicated by asterisks, and residues interacting with the substrate indolepyruvate are indicated by number signs (32). Residues essential for catalysis in Pdc1p_SacCe (21) are indicated by solid triangles.
A structural determination by X-ray crystallography of yeast pyruvate decarboxylases and IpdC_EntCl of E. cloacae has shown that the three-dimensional structures of these enzymes are very similar (32). The high levels of homology enabled us to model KdcA and the ipd gene product against the three-dimensional crystal structure of Pdc1p_SacCe (data not shown). This template was chosen since it is the only known three-dimensional structure with bound substrate and cofactor molecules, which allowed us to model these molecules as well. In the model of KdcA most of the residues likely to be involved in binding of the substrate and cofactors and in stabilization of the transition state intermediate during catalysis are identical in the template and KdcA model structures. The model suggests that larger substrates could fit into the substrate binding region, but the possible orientation could not be modeled due to a lack of template structures containing larger substrates. The model of the ipd gene product clearly showed that several of these essential residues, such as the catalytic Glu477, are absent. These results strongly imply that this gene product is not functional as an α-keto acid decarboxylase.
Overexpression of the kdcA gene.
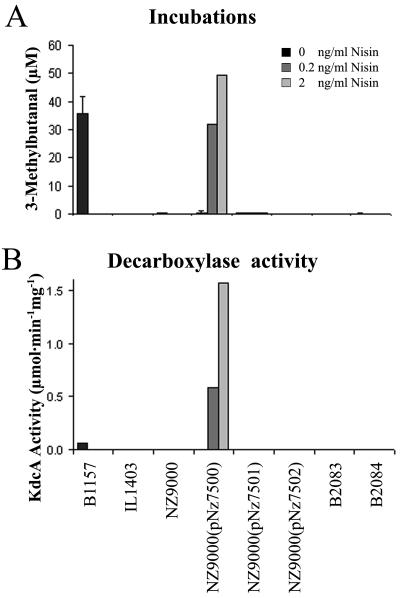
The kdcA and ipd genes were overexpressed in L. lactis for further characterization of the gene products. The kdcA gene was cloned by PCR into pNZ8148, where it was under the control of the nisA promoter, resulting in plasmid pNZ7500. Analogously, the ipd gene of strain IL1403 and the corresponding region of the kdcA gene from strain B1157 were cloned into pNZ8148, resulting in pNZ7501 and pNZ7502, in order to analyze whether the truncated gene products were inactive with KICA and could explain the decarboxylase-negative phenotype of strain IL1403. The plasmids were introduced into L. lactis strain NZ9000, which contains the nisRK signal transduction genes required for nisin-dependent inducible expression. The protein profiles of all recombinant strains and wild-type strains B2083, B2084, B1157, IL1403, and NZ9000 were analyzed by SDS-PAGE. Moreover, their abilities to produce 3-methylbutanal on GM17 were determined, and the KICA decarboxylase activity was quantified in CFEs of GM17-grown cultures. The results are shown in Fig. 3 and 4. Wild-type strain B1157 and the induced recombinant strain NZ9000(pNZ7500) produced similar levels of 3-methylbutanal. At a nisin concentration of 2 ng ml−1 strain NZ9000(pNZ7500) clearly produced an extra protein with an estimated molecular mass of 62 kDa, which is in agreement with the predicted mass deduced from the gene sequence (60.9 kDa). The decarboxylase activity in CFE of this induced strain was more than 30 times higher than the activity observed with the wild type, strain B1157 (Fig. 3). Although sequence analysis of plasmids of pNZ7501 and pNZ7502 confirmed that the cloning strategy was successful, no extra protein was detected in extracts of induced cultures of strains NZ9000(pNZ7501) and NZ9000(pNZ7501). The absence of a gene product in these cases was unexpected and may have been due to decreased stability of the mRNAs or rapid degradation of misfolded protein
FIG. 3.
(A) 3-Methylbutanal production during incubation in GM17. (B) KdcA activities of (nisin-induced) decarboxylase overexpression mutants, wild-type strain B1157, the cloning host L. lactis NZ9000, and the knockout mutants B2083 and B2084.
FIG. 4.
SDS-PAGE of knockout mutants, wild-type strains, and decarboxylase overexpression recombinants. The overexpression recombinants were induced with 0, 0.2, and 2 ng of nisin A per ml. kd, kilodaltons.
Characterization of KdcA.
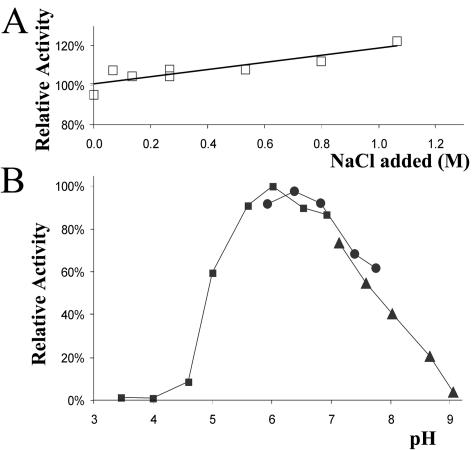
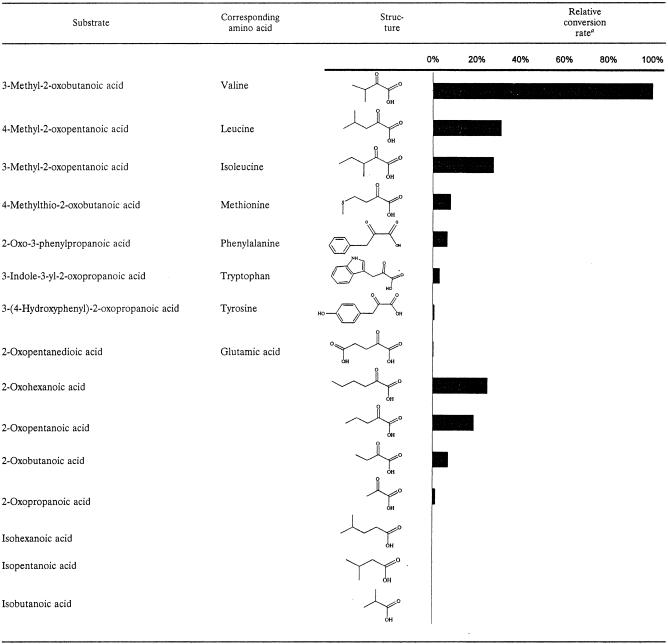
The pH optimum, salt tolerance, and substrate specificity are important parameters of flavor-forming enzymes with respect to their potential activity under cheese ripening conditions. As shown in Fig. 5B, the optimal pH was 6.3, and there was a broad pH activity profile, which corresponded well with the characteristics of known pyruvate and indole-3-pyruvate decarboxylases described in other microorganisms (16, 19, 26). The enzyme activity was not negatively affected at high salinity; in fact, it was slightly increased (Fig. 5A). The highest activity was observed with branched-chain α-keto acids derived from valine, leucine, and isoleucine, but linear α-keto acids with four and six carbon atoms and the α-keto acids derived from methionine, phenylalanine, and tryptophan (indole 3-pyruvate) could also be converted by the enzyme (Table 3). The latter substrate was of particular interest, because it is the preferred substrate for indolepyruvate decarboxylases (31) which are highly homologous to KdcA. No activity was detected with the α-keto acid α-ketoglutarate or with branched-chain aliphatic acids. No activity was detected with any of the substrates in CFE of L. lactis MG1363, indicating that KdcA was responsible for decarboxylation.
FIG. 5.
(A) Effects of salt on the activity of KdcA. (B) Effects of pH on the activity of KdcA, measured in 50 mM sodium citrate (▪), sodium phosphate (•), or Tris (▴) buffer with α-ketoisovaleric acid as the substrate.
TABLE 3.
Substrate specificity of KdcA
Individual specific conversion rates were calculated from the amount of substrate converted (as determined by high-performance liquid chromatography), the reaction time, and the protein content of the CFE of strain NZ9000(pNz7500).
Genetic organization of the kdcA gene region in decarboxylase-negative and decarboxylase-positive L. lactis strains.
A PCR strategy was used to obtain preliminary results for the genetic organization of the kdcA gene region in 30 decarboxylase-negative and 12 decarboxylase-positive L. lactis strains, because only a few lactococcal genomes were available for comparison. We used four primer sets that allowed discrimination between the presence of the complete kdcA gene and the presence of a truncated gene comparable to the ipd gene in strain IL1403 (Fig. 1 and Table 2). The absence of amplification products clearly correlated with the absence of 3-methylbutanal formation and vice versa (P > 0.95, as determined by Student's t test). The non-3-methylbutanal producers could be divided into two categories. The largest group (∼70%) did not give any PCR products, indicating absence of the kdcA gene. Strain SK11 belonged to this group, and the absence of the gene was confirmed by searching the preliminary genome sequence (www.jgi.doe.gov and www.ncbi.nlm.nih.gov, assessed 17 July 2004). Strains in the second group (∼30%) only yielded the two PCR products of the first fragment of the kdcA gene corresponding to the ipd sequence present in strain IL1403. None of the non-3-methylbutanal producers produced more than the first two PCR products; however, most (75%) of the 3-methylbutanal-producing strains produced three or four PCR products, indicating that a full-length gene was present. The majority of these strains do not have a history in dairy fermentations (wild isolates), while most of the non-3-methylbutanal-forming strains have a history of usage in the dairy industry. There was no clear correlation between the subspecies (L. lactis subsp. lactis, L. lactis subsp. lactis diacetylactis, and L. lactis subsp. cremoris) and the absence of the complete kdcA gene. However, the only L. lactis subsp. cremoris strain which was able to produce 3-methylbutanal in this experiment was B1157.
Occurrence of kdcA in lactic acid bacteria.
The BLASTN and TBLASTX search of the newly identified kdcA gene against the (preliminary) genome sequences of dairy-related bacteria (www.jgi.doe.gov and www.ncbi.nlm.nih.gov, assessed 17 July 2004) only revealed very high similarity with the ipd annotated open reading frame of L. lactis IL1403, while in the other dairy-related bacteria, including L. lactis subsp. cremoris, Lactobacillus casei, Lactobacillus brevis, Lactobacillus plantarum, Lactobacillus gasseri, Lactobacillus johnsonii NCC 533, Lactobacillus delbrueckii, Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293, and Brevibacterium linens, no orthologous gene was identified. Previously, we analyzed various strains of these species to determine their ability to produce 3-methylbutanal and the presence of the decarboxylase. The results showed that only a limited number of Lactococcus strains are able to produce 3-methylbutanal (5, 36, 37). It will be interesting to extend this analysis and elucidate whether KdcA-positive strains occur in other lactic acid bacterial species.
DISCUSSION
We cloned and sequenced the gene encoding KdcA of L. lactis strain B1157. KdcA has a broad substrate range and exhibits the highest activity with branched-chain α-keto acids. Therefore, it is best designated a branched-chain α-keto acid decarboxylase. The KdcA sequence was highly homologous to known indole pyruvate decarboxylase and pyruvate decarboxylase sequences, and characteristics like the pH optimum and molecular weight were also very similar. The structure of KdcA could easily be modeled by using yeast pyruvate decarboxylase as the template. The results indicate that KdcA is a new member of the pyruvate decarboxylase-indolepyruvate decarboxylase protein family. Moreover, they suggest that the mechanism of decarboxylation by kdcA is very similar to that in these enzymes, in which the TPP cofactor acts as an intramolecular base catalyst and Asp29, His112, His113, and Glu462 are involved in stabilization of the transition state intermediate (21, 31).
Previously, Amarita et al. described the partial purification and characterization of an α-keto acid decarboxylase from L. lactis IFPL730 active on α-keto-4-methylthiobutyrate; also, this enzyme had the highest activity with the branched-chain α-keto acids (3). Despite the fact that the molecular mass of this enzyme was estimated to be 50 kDa, various other features are very similar to features of KdcA.
Our previous activity screening analyses indicated that most lactococci do not exhibit decarboxylase activity. Our PCR-based analysis of 30 decarboxylase-negative isolates indicated that in most of these strains kdcA is completely absent, as it is in strains SK11 and MG1363. A second group has a genotype similar to strain IL1403, which is characterized by the presence of a truncated and nonfunctional homologue. Most 3-methylbutanal-producing strains seem to contain the complete gene and do not have any history in industrial dairy fermentations.
Since the decarboxylase was active at pH 5.3 and at a high salinity, which are typical conditions for Gouda cheese ripening, the enzyme has the potential to play an important role in cheese flavor formation. This was also illustrated by the malty flavor of cheese made with strain B1157 (6). Overexpressing the decarboxylase hardly resulted in increased 3-methylbutanal production in fermentations (Fig. 3). A lack of the substrate KICA was most probably the reason for the absence of high 3-methylbutanal levels. The formation of KICA by transamination has previously been shown to be suboptimal for the production of Leu-derived flavor compounds under cheese ripening conditions (7, 42). A combination of transaminase stimulation and the decarboxylase overexpression mutant might result in a major increase in 3-methylbutanal formation and also in higher levels of 2-methylpropanal and 2-methylbutanal, which are derived from valine and isoleucine, respectively. Future research should focus on the impact of the use of these kinds of strains in model systems and cheese making in order to control flavor formation better.
Acknowledgments
This work was financially supported by Stichting J. Mesdag Fonds, The Netherlands.
We thank Michiel Kleerebezem for critical reading of the manuscript. Roland Siezen and Bernadette Renckens of the Nijmegen Centre for Molecular and Biomolecular Informatics (www.cmbi.kun.nl) are thanked for the modeling of KdcA.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amarita, F., D. Fernandez Espla, T. Requena, and C. Pelaez. 2001. Conversion of methionine to methional by Lactococcus lactis. FEMS Microbiol. Lett. 204:189-195. [DOI] [PubMed] [Google Scholar]
- 4.Asakawa, T., H. Wada, and T. Yamano. 1968. Enzymatic conversion of phenylpyruvate to phenylacetate. Biochem. Biophys. Acta 170:375-391. [DOI] [PubMed] [Google Scholar]
- 5.Ayad, E. H. E., A. Verheul, C. de Jong, J. T. M. Wouters, and G. Smit. 1999. Flavour forming abilities and amino acid requirements of Lactococcus lactis strains isolated from artisanal and non-dairy origin. Int. Dairy J. 9:725-735. [Google Scholar]
- 6.Ayad, E. H. E., A. Verheul, J. T. M. Wouters, and G. Smit. 2000. Application of wild starter cultures for flavour development in pilot plant cheese making. Int. Dairy J. 10:169-179. [Google Scholar]
- 7.Banks, J. M., M. Yvon, J. C. Gripon, M. A. de la Fuente, E. Y. Brechany, A. G. Williams, and D. D. Muir. 2001. Enhancement of amino acid catabolism in Cheddar cheese using alpha-ketoglutarate: amino acid degradation in relation to volatile compounds and aroma character. Int. Dairy J. 11:235-243. [Google Scholar]
- 8.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 10.De Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyda, F., W. Furey, S. Swaminathan, M. Sax, B. Farrenkopf, and F. Jordan. 1993. Catalytic centers in the thiamin diphosphate dependent enzyme pyruvate decarboxylase at 2.4-A resolution. Biochemistry 32:6165-6170. [DOI] [PubMed] [Google Scholar]
- 12.Engels, W. J. M. 1997. Ph.D. thesis. Agricultural University, Wageningen, The Netherlands.
- 13.Gao, S., D. H. Oh, J. R. Broadbent, M. E. Johnson, B. C. Weimer, and J. L. Steele. 1997. Aromatic amino acid catabolism by lactococci. Lait 77:371-381. [Google Scholar]
- 14.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins, D., J. Thompson, T. Gibson, J. D. Thompson, D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Killenberg-Jabs, M., S. Koenig, S. Hohmann, and G. Habner. 1996. Purification and characterization of the pyruvate decarboxylase from a haploid strain of Saccharomyces cerevisiae. Biol. Chem. Hoppe-Seyler 377:313-317. [DOI] [PubMed] [Google Scholar]
- 17.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koga, J. 1995. Structure and function of indolepyruvate decarboxylase, a key enzyme in indole-3-acetic acid biosynthesis. Biochem. Biophys. Acta 1249:1-13. [DOI] [PubMed] [Google Scholar]
- 19.Koga, J., T. Adachi, and H. Hidaka. 1992. Purification and characterization of indolepyruvate decarboxylase, a novel enzyme for indole-3-acetic acid biosynthesis in Enterocloaceae. J. Biol. Chem. 267:15823-15828. [PubMed] [Google Scholar]
- 20.Kuipers, O. P., P. G. G. A. De Ruyter, M. Kleerebezem, and W. M. De Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 21.Liu, M., E. A. Sergienko, F. Guo, J. Wang, K. Tittmann, G. Hübner, W. Furey, and F. Jordan. 2001. Catalytic acid-base groups in yeast pyruvate decarboxylase. 1. Site-directed mutagenesis and steady-state kinetic studies on the enzyme with the D28A, H114F, H115F, and E477Q substitutions. Biochemistry 40:7355-7368. [DOI] [PubMed] [Google Scholar]
- 22.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maralith, P. Z. 1981. Flavour microbiology. Charles C. Thomas, Springfield, Ill.
- 25.McSweeney, P. L. H., and M. J. Sousa. 2000. Biochemical pathways for the production of flavour compounds in cheeses during ripening: a review. Lait 80:293-324. [Google Scholar]
- 26.Neale, A. D., R. K. Scopes, R. E. H. Wettenhall, and N. J. Hoogenraad. 1987. Pyruvate decarboxylase of Zymomonas mobilis: isolation, properties and genetic expression in Escherichia coli. J. Bacteriol. 169:1024-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oku, H., and T. Kaneda. 1988. Biosynthesis of branched-chain fatty acids in Bacillus subtilis. A decarboxylase is essential for branched-chain fatty acid synthetase. J. Biol. Chem. 263:18386-18396. [PubMed] [Google Scholar]
- 28.Palaniappan, C., V. Sharma, M. E. Hudspeth, and R. Meganathan. 1992. Menaquinone (vitamin K2) biosynthesis: evidence that the Escherichia coli menD gene encodes both 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylic acid synthase and alpha-ketoglutarate decarboxylase activities. J. Bacteriol. 174:8111-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sable, S., and G. Cottenceau. 1999. Current knowledge of soft cheeses flavor and related compounds. J. Agric. Food Chem. 47:4825-4836. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Schutz, A., R. Golbik, K. Tittmann, D. I. Svergun, M. H. Koch, G. Hubner, and S. Konig. 2003. Studies on structure-function relationships of indolepyruvate decarboxylase from Enterobacter cloacae, a key enzyme of the indole acetic acid pathway. Eur. J. Biochem. 270:2322-2331. [DOI] [PubMed] [Google Scholar]
- 32.Schutz, A., T. Sandalova, S. Ricagno, G. Hubner, S. Konig, and G. Schneider. 2003. Crystal structure of thiamindiphosphate-dependent indolepyruvate decarboxylase from Enterobacter cloacae, an enzyme involved in the biosynthesis of the plant hormone indole-3-acetic acid. Eur. J. Biochem. 270:2312-2321. [DOI] [PubMed] [Google Scholar]
- 33.Shigeoka, S., and Y. Nakano. 1991. Characterization and molecular properties of 2-oxoglutarate decarboxylase from Euglena gracilis. Arch. Biochem. Biophys. 288:22-28. [DOI] [PubMed] [Google Scholar]
- 34.Singer, T. P., and J. Pensky. 1952. Isolation and properties of the a-carboxylase of wheat germ. J. Biol. Chem. 196:375-388. [PubMed] [Google Scholar]
- 35.Smit, B. A., W. J. M. Engels, M. Alewijn, G. T. C. A. Lommerse, E. Kippersluijs, J. T. M. Wouters, and G. Smit. 2004. Chemical conversion of alpha-keto acids in relation to flavor formation in fermented foods. J. Agric. Food Chem. 52:1263-1268. [DOI] [PubMed] [Google Scholar]
- 36.Smit, B. A., W. J. M. Engels, J. Bruinsma, J. E. T. Van Hylckama Vlieg, J. T. M. Wouters, and G. Smit. 2004. Development of a high throughput-screening method to test flavour-forming capability of anaerobic microorganisms. J. Appl. Microbiol. 97:306-313. [DOI] [PubMed] [Google Scholar]
- 37.Smit, B. A., W. J. M. Engels, J. T. M. Wouters, and G. Smit. 2003. Diversity of l-leucine catabolism in various microorganisms involved in dairy fermentations and identification of the rate controlling step in 3-methylbutanal formation. Appl. Microbiol. Biotechnol. 64:396-402. [DOI] [PubMed] [Google Scholar]
- 38.Smit, G., A. Verheul, R. van Kranenburg, E. Ayad, R. Siezen, and W. J. M. Engels. 2000. Cheese flavour development by enzymatic conversions of peptides and amino acids. Food Res. Int. 33:153-160. [Google Scholar]
- 39.Vriend, G. 1990. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8:52-56. [DOI] [PubMed] [Google Scholar]
- 40.Vuralhan, Z., M. A. Morais, S.-L. Tai, M. D. W. Piper, and J. T. Pronk. 2003. Identification and characterization of phenylpyruvate decarboxylase genes in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 69:4534-4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wells, J. M., P. W. Wilson, and R. W. F. Le Page. 1993. Improved cloning vectors and transformation procedure for Lactococcus lactis. J. Appl. Bacteriol. 74:629-636. [DOI] [PubMed] [Google Scholar]
- 42.Yvon, M., S. Berthelot, and J. C. Gripon. 1998. Adding alpha-ketoglutarate to semi-hard cheese curd highly enhances the conversion of amino acids to aroma compounds. Int. Dairy J. 8:889-898. [Google Scholar]
- 43.Yvon, M., and L. Rijnen. 2001. Cheese flavour formation by amino acid catabolism. Int. Dairy J. 11:185-201. [Google Scholar]