Abstract
Acetohydroxy acid synthase (AHAS), which catalyzes the key reactions in the biosynthesis pathways of branched-chain amino acids (valine, isoleucine, and leucine), is regulated by the end products of these pathways. The whole Corynebacterium glutamicum ilvBNC operon, coding for acetohydroxy acid synthase (ilvBN) and aceto hydroxy acid isomeroreductase (ilvC), was cloned in the newly constructed Escherichia coli-C. glutamicum shuttle vector pECKA (5.4 kb, Kmr). By using site-directed mutagenesis, one to three amino acid alterations (mutations M8, M11, and M13) were introduced into the small (regulatory) AHAS subunit encoded by ilvN. The activity of AHAS and its inhibition by valine, isoleucine, and leucine were measured in strains carrying the ilvBNC operon with mutations on the plasmid or the ilvNM13 mutation within the chromosome. The enzyme containing the M13 mutation was feedback resistant to all three amino acids. Different combinations of branched-chain amino acids did not inhibit wild-type AHAS to a greater extent than was measured in the presence of 5 mM valine alone (about 57%). We infer from these results that there is a single binding (allosteric) site for all three amino acids in the enzyme molecule. The strains carrying the ilvNM13 mutation in the chromosome produced more valine than their wild-type counterparts. The plasmid-free C. glutamicum ΔilvA ΔpanB ilvNM13 strain formed 90 mM valine within 48 h of cultivation in minimal medium. The same strain harboring the plasmid pECKAilvBNC produced as much as 130 mM valine under the same conditions.
Corynebacterium glutamicum is used for the production of a number of amino acids, including valine. Acetohydroxy acid synthase (AHAS) (EC 4.1.3.18), also called acetolactate synthase, is the key enzyme of the pathways for biosynthesis of branched-chain amino acids (isoleucine, valine, and leucine) (Fig. 1) and therefore is a potential target for metabolic engineering. The enzyme catalyzes decarboxylation of pyruvate and its condensation either with another molecule of pyruvate to produce acetolactate (a precursor of valine and leucine) or with 2-ketobutyrate to produce acetohydroxybutyrate (a precursor of isoleucine). The reactions play a key role in determining the relative fluxes to different end products (2). Three AHAS isoenzymes, differing in their genetic determinations, biochemical properties, and regulation of both biosynthesis and activity, were described for Escherichia coli and Salmonella enterica serovar Typhimurium (26). The isoenzymes AHAS I, II, and III are encoded by the genes ilvBN, ilvGM, and ilvIH, respectively. Expression of these genes is subject to different multivalent repression by branched-chain amino acids. Expression of the ilvGMEDA operon is controlled by transcriptional attenuation mediated by all three amino acids, whereas the ilvBN operon is controlled by attenuation mediated only by valine and leucine. Expression of ilvIH is regulated by Lrp (leucine-responsive protein) (29). The large subunits (IlvB, IlvG, and IlvI) are responsible for catalytic activity of the three isoenzymes, while the small subunits mediate their inhibition (26). The activities of AHAS I and AHAS III are inhibited by valine, while AHAS II is valine resistant.
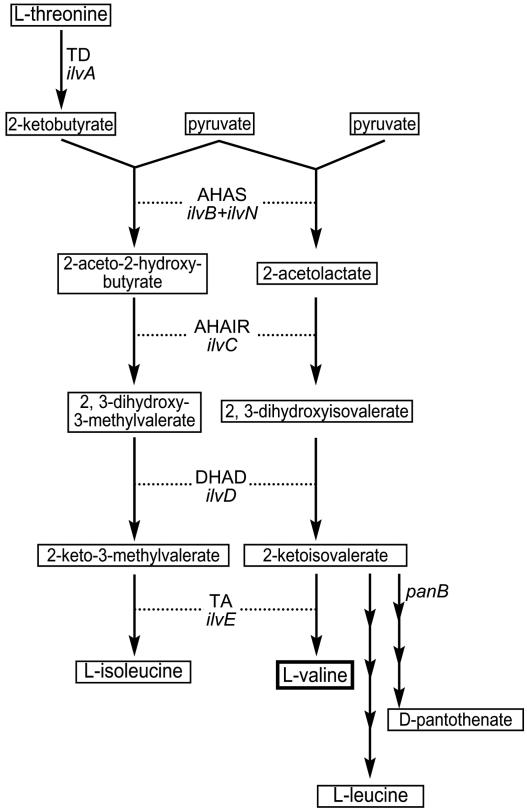
FIG. 1.
Biosynthesis of valine and isoleucine. Enzymes and the corresponding genes: AHAS (ilvB and ilvN), acetohydroxy acid synthase; TD (ilvA), threonine dehydratase; AHAIR (ilvC), acetohydroxy acid isomeroreductase; DHAD (ilvD), dihydroxyacid dehydratase; TA (ilvE), transaminase.
In C. glutamicum, only a single AHAS was found (1). The enzyme is encoded by the genes ilvB and ilvN, forming an operon with ilvC (5, 8). Expression of the operon is controlled by attenuation mediated by all three branched-chain amino acids (14). Activity of the ilvB promoter is increased about twofold under conditions of growth limitation by any of the branched-chain amino acids (valine, isoleucine, and leucine). At the same time, expression of the operon is induced in the presence of 2-ketobutyrate (8, 14). Activity of AHAS from C. glutamicum is inhibited by any of the three amino acids to approximately 50% (1, 11). However, the inhibitory effect does not exceed 50% even in the presence of all three amino acids (1). Similar to the case for AHAS II and AHAS III from E. coli and S. enterica serovar Typhimurium (26), AHAS from C. glutamicum has much higher affinity to 2-ketobutyrate than to pyruvate (1). In the presence of 2-ketobutyrate, synthesis of isoleucine is therefore preferred and synthesis of valine and leucine is reduced.
The C. glutamicum ilvBNC operon was cloned (8) and, together with ilvD, has recently been used for construction of valine- and pantothenate-producing strains (20). Further improvement of the valine-producing strain was achieved by deletion of the panBC genes, thus making more pyruvate available for valine synthesis (18). Since the inhibition of AHAS reached a maximum of only about 50% in enzyme activity assays in the presence of valine, isoleucine, and leucine, it was thought that resistance of AHAS to feedback inhibition would not provide a substantial improvement in valine production. However, chemical mutagenesis and selection with amino acid analogues led to the isolation of C. glutamicum leucine and valine producers in which AHAS activity was desensitized to valine, isoleucine, and leucine (24, 25). In Streptomyces cinnamonensis, chemical mutagenesis produced strains that were feedback resistant to valine and that overproduced the immediate valine precursor, 2-ketoisovalerate (16).
In this work, we constructed feedback-resistant mutants of AHAS by site-directed mutagenesis of ilvN, encoding the regulatory subunit of C. glutamicum AHAS. The mutated ilvN allele was cloned in the newly constructed plasmid pECKA together with other genes of the ilvBNC operon, and the most favorable mutation was also introduced into the C. glutamicum chromosome. We have shown that strains carrying the mutation ilvNM13 produced markedly higher amounts of valine.
MATERIALS AND METHODS
Bacterial strains, plasmids, oligonucleotide primers, media, and growth conditions.
E. coli DH5α (3) was used for cloning. The C. glutamicum strains and plasmids used are listed in Table 1. The primers are listed in Table 2. E. coli was cultivated in Luria-Bertani medium (21) at 37°C, and C. glutamicum was cultivated in 2× YT complete medium (21) or CGXII minimal medium (8) at 30°C. Kanamycin (20 μg/ml) was added to selective media.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| C. glutamicum strains | ||
| ATCC 13032 | Wild type | American Type Cul- ture Collection |
| ΔilvN | ilvN deletion | This work |
| ΔilvA ΔpanB | ilvA and panB deletions | This work |
| ilvNM13 | Mutation M13 in ilvN | This work |
| ΔilvA ΔpanB ilvNM13 | ilvA and panB deletions, muta- tion in ilvN | This work |
| Plasmids | ||
| pK19 | E. coli vector, Kmr | 17 |
| pK19B | E. coli vector, Kmr, no BglII site | This work |
| pKK5 | ilvBNC in pJC1 | 8 |
| pEC71 | E. coli-C. glutamicum vector, Kmr | 15 |
| pECKA | E. coli-C. glutamicum vector, Kmr | This work |
| pECKAilvBNC | ilvBNC in pECKA | This work |
| pK18mobsacB | E. coli vector for integration, Kmr | 23 |
TABLE 2.
Oligonucleotide primers
| Primer | 5′→3′ sequencea | Purpose |
|---|---|---|
| MILVNF | GCGGAGGAAGTACTGCC | ilvN mutation |
| MILVNR | CAATCAGATTAATTGCTGTTTA | ilvN mutation |
| ILVNM1 | GGACGTAGACGGTGACATTTCCCGCG | ilvN mutation |
| AT | ||
| MISBGL | GTTTAGAACTTGGCCGGAG | ilvN mutation |
| ILVNM13F | AGGACGTGACGATGACTTTTCCCGCGTAT | ilvN mutation |
| ILVNM13R | ATACGCGGGAAAAGTCATCGTCTACGTCCT | ilvN mutation |
| DELN1F | CTGAATTCCGGTGGACTGGGCACC | ilvN deletion |
| DELN1R | TTGAATGCGCTCTCTCCTTATGCCTCGGTC | ilvN deletion |
| DELN2F | TAAGGAGAGAGCGCATTCAACCTCGTGTCC | ilvN deletion |
| DELN2R | CCGAATTCAAGGAGAGGTCAGCGTC | ilvN deletion |
| DELPANBF1 | AACTGCAGCGCAAGAAGGTCT | panB deletion |
| DELPANBR1 | AATCGTCTGCATCGCGACCCAGCACAACGT | panB deletion |
| DELPANBF2 | GGGTCGCGATGCAGACGATTCGACGCATTG | panB deletion |
| DELPANBR2 | AACTGCAGGGGATTGACAA | panB deletion |
| DELILVAF1 | AGGAATTCAGAGGACTGAGCCTG | ilvA deletion |
| DELILVAR1 | TGGTTCGATAGGCCACGCCCTGGGCATGGT | ilvA deletion |
| DELILVAF2 | GGGCGTGGCCTATCGAACCAGCGGGAGCAG | ilvA deletion |
| DELILVAR2 | AAGAATTCGTACTCAGGAGTGCC | ilvA deletion |
Mutated bases are in boldface; restriction sites (ScaI, EcoRI, or PstI) are in italics.
DNA manipulations and transformation.
DNA isolation, PCR, DNA cloning, and DNA analysis were done by standard methods (21). E. coli was transformed with plasmid DNA by the method of Hanahan (3), and C. glutamicum was transformed by electrotransformation (27). DNA sequences were determined with the ABI PRISM BigDye Terminator version 3.1 cycle sequencing kit and the ABI PRISM 3100 automatic DNA sequencer (Applied Biosystems).
Construction of plasmid pECKA and cloning of the ilvBNC operon.
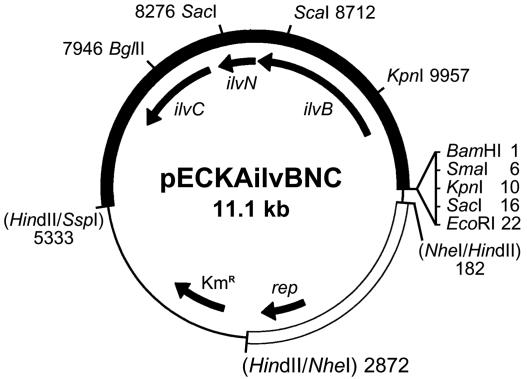
The shuttle vector pECKA (replicating in E. coli and C. glutamicum) was constructed on the basis of the E. coli vector pK19 (17) and the replicon of the C. glutamicum cryptic plasmid pBL1 (22) from pEC71 (15). First, the recognition site for restriction enzyme BglII was removed from vector pK19. Plasmid pK19 was digested with BglII, blunt ended with the Klenow enzyme, and religated, giving rise to pK19B. Next, a HindII fragment (2.7 kb) of the plasmid pEC71 was cloned into the NheI site of pK19B after blunting the ends. The resulting plasmid vector, pECKA (5.4 kb), replicates in E. coli and C. glutamicum and provides seven unique cloning sites (HindII, SalI, BamHI, SmaI, KpnI, SacI, and EcoRI), a kanamycin resistance marker, and α-complementation of β-galactosidase for cloning in E. coli. A 5.7-kb fragment of the C. glutamicum chromosome carrying the ilvBNC operon was obtained by digestion of plasmid pKK5 with SspI and BamHI. The fragment was ligated with HindII- and BamHI-digested vector pECKA, and the resulting construct was identified in the E. coli transformants. The structure of the constructed plasmid pECKAilvBNC (11.1 kb) was confirmed by restriction analysis. The restriction and genetic maps of pECKAilvBNC are shown in Fig. 2.
FIG. 2.
Map of plasmid pECKAilvBNC. The pBL1 replicon is shown as an empty box, the pK19B part is depicted as a thin line, and the cloned fragment with the ilvBNC operon is shown as a filled box.
Mutagenesis.
Site-directed mutagenesis was carried out essentially by the method of Ito et al. (6). Using oligonucleotide primers MILVNF, containing the natural ScaI site (Table 2), and MISBGL, containing a substitution removing the BglII site, a 786-bp fragment (carrying the 3′ end of ilvB and the complete ilvN gene; coordinates within the C. glutamicum genome [http://gib.genes.nig.ac.jp], 1339760 to 1340545) was amplified by PCR. Another fragment of 491 bp (3′ part of ilvN; coordinates 1340072 to 1340562) was amplified by using the degenerate mutational primer ILVNM1 and primer MILVNR. The fragments were purified and mixed, and the final PCR produced a large fragment (803 bp; coordinates 1339760 to 1340562) with primers MILVNF and MILVNR. The product was digested with ScaI and BglII and used for the substitution of the identical fragment within the wild-type ilvBNC operon cloned in vector pECKA. Only the fragments carrying the original BglII site contained the mutation. Mutations in individual clones were detected by sequencing.
Construction of chromosomal mutation and deletions.
Mutation ilvNM13 and deletions in the genes ilvN, ilvA, and panB in the chromosome were constructed by using PCR-generated fragments cloned in the vector pK18mobsacB (23). The mutation or deletion was introduced into the fragment by the method of Horton (4). The oligonucleotide primers used are listed in Table 2. DNA fragments (about 600 bp) located upstream and downstream of the internal gene region to be deleted (100 to 368 bp), extended by complementary sequences of 20 bp, were amplified. The fragments were purified and mixed, and a single large fragment (about 1.2 kb) was amplified by using the outer primers with the EcoRI or PstI sites attached. The resulting products with the deletions were digested with EcoRI or PstI and cloned into pK18mobsacB. The constructs were transferred to C. glutamicum by electrotransformation, and the clones in which the double recombination event occurred were identified by positive selection based on the conditional lethal effect of the sacB gene in C. glutamicum (7). The clones, free of the vector sequences and carrying the mutation or deletion, were confirmed by PCR and sequencing.
Determination of AHAS activity.
C. glutamicum clones were cultivated in CGXII medium to an optical density at 600 nm of 4. The cells were washed with 2% KCl at 4°C and disrupted by sonication for 30 min on ice (0.5-s pulses; amplitude, 100%) with an Ikasonic U50 sonicator (IKA Labortechnik, Staufen, Germany). The specific AHAS activity in the cell extract was determined as described previously (11). The reaction mixture (1 ml) contained 100 mM phosphate buffer (pH 7.3), 10 mM MgCl2, 100 μM thiamine pyrophosphate, 100 μM flavin adenine dinucleotide, and 50 mM pyruvate. The reaction was started by adding 100 μl of cell extract and was stopped by adding 100 μl of 50% H2SO4, which also converts acetolactate to acetoin. The concentration of acetoin was determined by the method of Westerfeld (30). The specific AHAS activity was expressed in units per milligram of protein (micromoles of acetolactate per minute per milligram of protein).
Determination of valine production.
Valine-producing strains were cultivated for 48 h in CGXII medium (60 ml in 500-ml flasks, 30°C, 120 rpm). l-Isoleucine (0.9 mM) and d-pantothenate (0.5 μM) were added to the cultures of strains with deletions ΔilvA and ΔpanB. A 1 ml aliquot of supernatant harvested from a culture was used for determination of the valine concentration. The valine concentration in the medium was determined by reversed-phase high-pressure liquid chromatography (HP1090; Hewlett-Packard, Waldbronn, Germany) with fluorimetric detection (excitation at 230 nm and emission at 450 nm) after automatic precolumn derivatization with ortho-phthaldialdehyde (12). A Hypersil octyldecyl silane column was used (particle size, 5 μm; 200 by 2.1 mm; Thermo Electron). The buffer gradient consisted of a polar phase (0.05 M sodium acetate, pH 7.2) and a nonpolar phase (methanol). Quantification was done by calculation of the valine concentration from a calibration curve. Cultivations of each clone and valine determinations were performed at least three times. Since the final optical densities at 600 nm of the cultures were similar (about 40), we could rule out an influence of the amount of biomass on valine production.
RESULTS
Inhibition of AHAS by branched-chain amino acids.
The basic biochemical characteristics of AHAS from C. glutamicum, such as affinity for the substrate (pyruvate) and cofactor (flavin adenine dinucleotide) as well as the optimal pH and temperature, have been previously described (1, 11). In addition, the 50% inhibitory concentrations for AHAS with valine, isoleucine, and leucine have been determined (0.9, 3.1, and 6 mM, respectively) (11). To identify a potential cooperative inhibiting effect of the different branched-chain amino acids on the AHAS activity, the enzyme was assayed in presence of a 5 mM concentration of each amino acid or with different combinations of two or three branched-chain amino acids (Table 3). Interestingly, whatever the test, AHAS inhibition remained at about 57%, indicating that no cumulative effect occurred.
TABLE 3.
Inhibition of activity of C. glutamicum wild-type AHAS by branched-chain amino acids
| Presence of inhibitor (5 mM)
|
Residual activity (%)a
|
|||
|---|---|---|---|---|
| Valine | Isoleucine | Leucine | Measuredb | Calculatedc |
| + | − | − | 44 | |
| − | + | − | 61 | |
| − | − | + | 62 | |
| + | + | − | 43 | 26.8 |
| + | − | + | 42 | 27.3 |
| + | + | + | 43 | 16.7 |
The activity without addition of any inhibitor is considered 100%.
The values are averages of at least three independent measurements; the standard deviations were within ± 7%.
Calculated for cumulative inhibition. This activity was calculated as follows: activity (percent) = [1−(percent inhibition with valine/100)] × [1−(percent inhibition with leucine/100)] × [1−(percent inhibition with isoleucine/100)] × 100.
Construction of AHAS mutants resistant to feedback inhibition.
Target sites for mutagenesis were designed on the basis of alignment of the amino acid sequence of the regulatory C. glutamicum AHAS subunit with the homologous amino acid sequences from E. coli and S. cinnamonensis. Within the conserved region near the N terminus of a small AHAS subunit, a mutation in the E. coli IlvH caused a complete loss of valine binding (13, 28). Similarly, amino acid alterations which were identified in S. cinnamonensis mutants resistant to inhibition by valine were found within the conserved N terminus of IlvN (9). Alignment of the N terminus of the regulatory AHAS subunit of C. glutamicum (encoded by ilvN) and the homologous regions of mutants of the E. coli and S. cinnamonensis subunits is shown in Fig. 3.
FIG. 3.
Design of mutations within C. glutamicum IlvN. The N-terminal parts of the regulatory subunits of AHAS are shown. Amino acids whose substitution decreased inhibition of AHAS by valine in E. coli (25) or S. cinnamonensis (7) are shaded, and those which are identical in two or all three sequences are in boldface. Amino acids replaced in mutants of C. glutamicum AHAS are boxed. Mutation in E. coli, 14G→D; mutations in S. cinnamonensis, 16G→D, 17V→D, 18L→F, and 30F→L.
Three amino acids (positions 20 to 22 in C. glutamicum IlvN) were chosen for substitution. The designed degenerate primer ILVNM1 allowed changes in all three amino acids. The wild-type fragment of the ilvN gene was substituted for the mutated fragment in pECKAilvBNC, and 10 clones were sequenced. Three different mutants obtained were named M8, M11, and M13. The DNA sequence and the respective amino acid compositions at positions 20 to 22 of IlvN in these mutants are shown in Table 4. To measure only the activity of AHAS with an identical mutated regulatory subunit, the deletion ΔilvN was constructed in the C. glutamicum chromosome. Since the 100-bp deletion (coordinates 1340019 to 130118) included the ilvN initiation codon, no regulatory subunit of chromosomal origin could be formed. The C. glutamicum ΔilvN strain grew extremely slowly in the minimal medium and was nearly auxotrophic for valine, isoleucine, leucine, and pantothenate. This deficiency was complemented by plasmid pECKAilvBNC. Using the C. glutamicum ΔilvN strain, the specific activity of AHAS coded by genes of the wild type or carrying the mutation M8, M11, or M13 in ilvN (on plasmid pECKAilvBNC in all cases) was measured. The values obtained in the absence of amino acids (shown as 100% in Table 5) were 0.11 U (mg of protein)−1 in the case of the wild-type ilvN and 0.04 to 0.06 U (mg of protein)−1 in the cases of the mutated ilvN. As shown in Table 5, mutations M8 and M13 caused complete resistance to inhibition by any of the three branched-chain amino acids, while in mutant M11, inhibition by isoleucine and leucine decreased only slightly and inhibition by valine even increased. The most favorable mutation (M13) was chosen for further work.
TABLE 4.
Mutations in the C. glutamicum AHAS
| Strain | DNA sequencea | Amino acid at positionb:
|
||
|---|---|---|---|---|
| 20 | 21 | 22 | ||
| Wild type | GGA ATC ATT | Gly | Ile | Ile |
| M8 | GGTGAC TTT | Gly | Asp | Phe |
| M11 | GGTGAC ATT | Gly | Asp | Ile |
| M13 | GATGAC TTT | Asp | Asp | Phe |
Mutated positions are in boldface.
Mutated amino acids at positions 20 to 22 in ilvN are in boldface.
TABLE 5.
Inhibition of AHAS activity by branched-chain amino acids in the C. glutamicum ΔilvN strain harboring pECKAilvBNC with mutations
| Mutation on plasmid | Relative activity (%) in presence of 10 mM amino acida
|
|||
|---|---|---|---|---|
| None | Valine | Isoleucine | Leucine | |
| None | 100 | 52 | 57 | 65 |
| M8 | 100 | 112 | 112 | 105 |
| M11 | 100 | 32 | 61 | 82 |
| M13 | 100 | 95 | 108 | 98 |
The values are averages of at least three independent measurements; the standard deviations were within ± 18%.
Introduction of the ilvNM13 mutation into the chromosome.
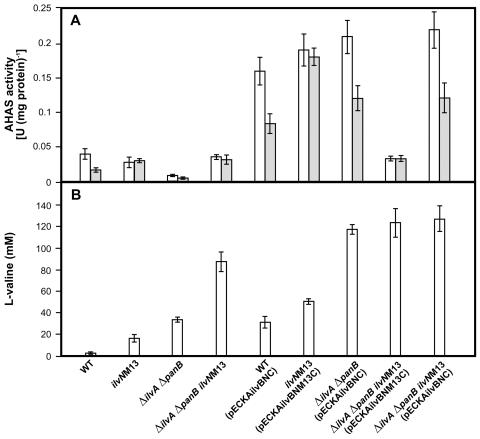
In some cases, moderate expression of a deregulated gene proved more useful for amino acid production than overexpression of the gene carried by a multicopy plasmid (19). We therefore tested expression of ilvNM13 in the chromosome. By using PCR, the 1.3-kb fragment containing the M13 mutation in ilvN was amplified and cloned in pK18mobsacB. The resulting plasmid construct was used for targeted substitution of the ΔilvN sequence for the complete ilvN sequence with the M13 mutation. The correct clone was selected according to its growth on minimal medium. The exchanged region within the chromosome was then amplified by PCR and checked by sequencing. Specific activities of AHAS of the wild type and with the mutation were determined (Table 6). As expected, the activity of the enzyme with the M13 mutation was not inhibited by any of the branched-chain amino acids at 10 mM. Moreover, in the presence of 10 mM valine, the specific activity of the mutated enzyme was 76% higher than that of the wild type [0.017 U (mg of protein)−1 for the wild type and 0.030 U (mg of protein)−1 for the mutant] (Fig. 4A). Whereas only traces of valine (1 mM) were detected in minimal medium after 48 h of cultivation of the wild-type strain of C. glutamicum, the ilvNM13 mutant formed 15.3 mM valine under the same conditions (Fig. 4B).
TABLE 6.
Inhibition of AHAS activity by branched-chain amino acids in plasmid-free strains
| Strain | Relative activity (%) in presence of 10 mM amino acida
|
|||
|---|---|---|---|---|
| None | Valine | Isoleucine | Leucine | |
| Wild type | 100 | 43 | 47 | 57 |
| ilvNM13 | 100 | 104 | 105 | 98 |
The values are averages of at least three independent measurements; the standard deviations were within ±15%.
FIG. 4.
AHAS activity and valine production in C. glutamicum strains. The means of data obtained from at least three independent cultivations are shown. Error bars show standard deviations. (A) AHAS activity without valine (open bars) and with 10 mM valine (shaded bars). (B) Valine production after 48-h cultivations in minimal medium. WT, wild type.
Construction of valine-producing strains.
To eliminate the formation of 2-ketobutyrate, the intermediate of the isoleucine synthesis pathway competing with valine synthesis, the chromosomal ilvA gene of C. glutamicum was inactivated by a 368-bp deletion (coordinates 2245496 to 2245863). Another competing pathway leading from 2-ketoisovalerate to pantothenate was cut off by constructing a 200-bp deletion (coordinates 127619 to 127818) within the panB gene. The resulting C. glutamicum ΔilvA ΔpanB strain is auxotrophic for isoleucine and pantothenate. The ilvNM13 mutation was used for further development of valine-producing strains. We used the wild-type, ilvNM13, ΔilvA ΔpanB, and ΔilvA ΔpanB ilvNM13 C. glutamicum strains as hosts for plasmids pECKAilvBNC and pECKAilvBNM13C to construct various valine producers.
Determination of AHAS activity and valine production.
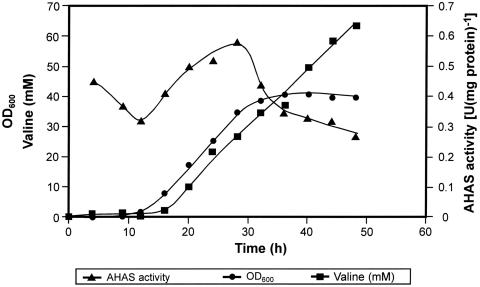
The AHAS activity, its inhibition by valine, and valine production measured in various C. glutamicum strains, as described in Materials and Methods, are shown in Fig. 4. No inhibition of AHAS activity by valine was found in all strains synthesizing only the mutated regulatory AHAS subunit (Fig. 4A). The strains carrying the ilvNM13 mutation in the chromosome produced much more valine than their wild-type counterparts, which proved a positive effect of feedback-resistant AHAS on valine production (Fig. 4B). The highest production of valine (130 mM within 48 h of cultivation) was detected in C. glutamicum ΔilvA ΔpanB ilvNM13(pECKAilvBNC) cultures. The best plasmidless strain, C. glutamicum ΔilvA ΔpanB ilvNM13, produced 90 mM valine. The profiles of valine accumulation and AHAS activity in the course of the batch cultivation of C. glutamicum ΔilvA ΔpanB ilvNM13(pECKAilvBNC) were also determined (Fig. 5). As shown in Fig. 5, valine was formed in parallel to the growth of the culture, and linear accumulation of valine continued even in stationary phase.
FIG. 5.
Growth, AHAS activity, and valine production during the batch cultivation of C. glutamicum ΔilvA ΔpanB ilvNM13(pECKAilvBNC). OD600, optical density at 600 nm.
DISCUSSION
In the strain carrying the ilvBNC operon on the vector pECKA, AHAS activity increased 4-fold while valine production increased 23-fold. A similar effect was observed after cloning of the ilvBNC operon in other plasmids (18, 20). As a result of mutations within the regulatory subunit of the enzyme, AHAS lost its regulation by inhibition. To avoid possible instability of the plasmid carrying the mutated ilvN gene (ilvNM13), we introduced the mutations into the chromosome. In the presence of 10 mM valine, the specific AHAS activity of the C. glutamicum ilvNM13 strain was higher than the activity of the wild-type enzyme, and the mutant strain produced 12-fold more valine than the wild-type strain. The mutated enzyme was also resistant to inhibition by isoleucine and leucine. This indicates that a single binding (allosteric) site for the three amino acids is present on the enzyme. As valine is the strongest inhibitor (50% inhibitory concentration, 0.9 mM) of AHAS activity (11), we assume that the binding site is occupied mainly by valine in the presence of 5 mM valine, 5 mM leucine, and/or 5 mM isoleucine. In addition, the intracellular titer of valine (in a valine-producing strain) is far higher than the concentration of leucine and isoleucine (11). Thus, in vivo, AHAS activity might be regulated mainly by valine.
The 25-fold increase of valine production in the ΔilvA strain was probably caused by blocking of the pathway to isoleucine and by growth limitation. In the ΔilvA strains, no 2-ketobutyrate is formed (Fig. 1). It has been reported that 2-ketobutyrate strongly affects the C. glutamicum AHAS activity. Eggeling et al. (1) showed that AHAS activity increased 10-fold after addition of 50 mM 2-ketobutyrate. Keilheuer et al. (8) proved that it is the higher transcription of the ilvBNC operon which is responsible for this increase. It was assumed that in the presence of a high concentration of 2-ketobutyrate, highly preferred synthesis of isoleucine resulted in a valine and leucine deficiency in the cell, which in turn derepressed the ilvBNC expression. However, under conditions of valine and leucine starvation, at most a 2.2-fold increase of AHAS activity was demonstrated (14). We have shown that the lack of 2-ketobutyrate in the cell (in ΔilvA strains) resulted in a drop in AHAS activity. When the ΔilvA strain was cultivated in the presence of 1 mM 2-ketobutyrate, AHAS activity increased by 50% (data not shown). Since we have also observed an increase of AHAS activity in the presence of 2-ketobutyrate in cells producing valine, the positive effect of 2-ketobutyrate on AHAS activity cannot be explained only by valine and leucine deficiency. Apparently, still another mechanism of induction of ilvBN transcription by 2-ketobutyrate is involved. This control may be mediated by binding of a regulatory molecule at the sequence upstream of the ilvB promoter (14).
Growth inhibition, a 2.5-fold increase of the ilvBN transcript level, and a 3.5-fold increase of specific AHAS activity were shown in ΔilvA strains in the presence of 40 mM valine (10). These consequences of a high valine concentration in the medium were attributed to the competition of valine with isoleucine for uptake by the cells and resulting isoleucine starvation (10). In the C. glutamicum ΔilvA ΔpanB ilvNM13(pECKAilvBNC) strain, we found that the specific AHAS activity was nearly twofold higher after 28 h of cultivation than after 12 h of cultivation (Fig. 5). At this point, about 30 mM valine accumulated in the medium and growth slowed, probably due to the isoleucine limitation. However, during the stationary phase, the concentration of valine in the medium increased further linearly, whereas AHAS activity dropped to the value measured after 12 h.
Since C. glutamicum AHAS can be inhibited at most to about 50% (1), it was believed that a feedback-resistant AHAS mutant could not increase valine formation in the producing strains. In this respect, our results, showing the positive effect of a feedback-resistant mutation in the regulatory AHAS subunit on valine production, are unexpected. The C. glutamicum ΔilvA ΔpanB ilvNM13 strain may be used as a basis for new generation of plasmid-free producers. The use of a producer strain which carries all modifications within the chromosome would decrease a theoretical hazard connected with plasmids carrying antibiotic resistance markers and other heterologous genes, which might be disseminated in the environment.
Acknowledgments
We thank Volker Wendisch (Jülich, Germany) and Bernd Eikmanns and Diana Fiur (Ulm, Germany) for help with the determination of valine concentrations, Stanislav Pospíšil for valuable advice, and Dana Lukavská for excellent technical assistance.
This work was supported by grants 525/01/0916 and 525/04/0548 from the Grant Agency of the Czech Republic, by a grant from the EU (VALPAN, QLK3-2000-00497), by Institutional Research Concept no. AV0Z5020903, and by the French Embassy in Prague.
REFERENCES
- 1.Eggeling, I., C. Cordes, L. Eggeling, and H. Sahm. 1987. Regulation of acetohydroxy acid synthase in Corynebacterium glutamicum during fermentation of alfa-ketobutyrate to l-isoleucine. Appl. Microbiol. Biotechnol. 25:346-351. [Google Scholar]
- 2.Epelbaum, S., R. A. LaRossa, T. K. VanDyk, T. Elkayam, D. M. Chipman, and Z. Barak. 1998. Branched-chain amino acid biosynthesis in Salmonella typhimurium: a quantitative analysis. J. Bacteriol. 180:4056-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan, D. 1985. Techniques for transformation of E. coli, p. 109-135. In D. M. Glover (ed.), DNA cloning. A practical approach, vol. 1. IRL Press, Oxford, United Kingdom. [Google Scholar]
- 4.Horton, R. M. 1995. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol. Biotechnol. 3:93-99. [DOI] [PubMed] [Google Scholar]
- 5.Inui, M., A. A. Vertes, M. Kobayashi, Y. Kurusu, and H. Yukawa. 1993. Cloning and sequence determination of the acetohydroxy acid synthase genes from Brevibacterium flavum MJ233 by using the polymerase chain reaction. DNA Sequence 3:303-310. [DOI] [PubMed] [Google Scholar]
- 6.Ito, W., H. Ishiguro, and Y. Kurosawa. 1991. A general method for introducing a series of mutations into cloned DNA using the polymerase chain reaction. Gene 102:67-70. [DOI] [PubMed] [Google Scholar]
- 7.Jäger, W., A. Schäfer, A. Pühler, G. Labes, and W. Wohlleben. 1992. Expression of the Bacillus subtilis sacB gene leads to sucrose sensitivity in the gram-positive bacterium Corynebacterium glutamicum but not in Streptomyces lividans. J. Bacteriol. 174:5462-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keilhauer, C., L. Eggeling, and H. Sahm. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 175:5595-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kopecký, J., J. Janata, S. Pospíšil, J. Felsberg, and J. Spížek. 1999. Mutations in two distinct regions of acetolactate synthase regulatory subunit from Streptomyces cinnamonensis result in the lack of sensitivity to end-product inhibition. Biochem. Biophys. Res. Commun. 266:162-166. [DOI] [PubMed] [Google Scholar]
- 10.Lange, C., D. Rittmann, V. F. Wendisch, M. Bott, and H. Sahm. 2003. Global expression profiling and physiological characterization of Corynebacterium glutamicum grown in the presence of l-valine. Appl. Environ. Microbiol. 69:2521-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leyval, D., D. Uy, S. Delaunay, J. L. Goergen, and J. M. Engasser. 2003. Characterisation of the enzyme activities involved in the valine biosynthetic pathway in a valine-producing strain of Corynebacterium glutamicum. J. Biotechnol. 104:241-252. [DOI] [PubMed] [Google Scholar]
- 12.Lindroth, P., and K. Mopper. 1979. High performance liquid chromatographic determination of subpicomole amounts of amino acids by precolumn fluorescence derivatization with o-phthalaldehyde. Anal. Chem. 51:1667-1674. [Google Scholar]
- 13.Mendel, S., T. Elkayam, C. Sella, V. Vinogradov, M. Vyazmensky, D. M. Chipman, and Z. Barak. 2001. Acetohydroxyacid synthase: a proposed structure for regulatory subunits supported by evidence from mutagenesis. J. Mol. Biol. 307:465-477. [DOI] [PubMed] [Google Scholar]
- 14.Morbach, S., C. Junger, H. Sahm, and L. Eggeling. 2000. Attenuation control of ilvBNC in Corynebacterium glutamicum: evidence of leader peptide formation without the presence of a ribosome binding site. J. Biosci. Bioeng. 90:501-507. [DOI] [PubMed] [Google Scholar]
- 15.Pátek, M., J. Hochmannová, and J. Nešvera. 1993. Production of threonine by Brevibacterium flavum containing threonine biosynthesis genes from Escherichia coli. Fol. Microbiol. 38:355-359. [DOI] [PubMed] [Google Scholar]
- 16.Pospíšil, S., J. Kopecky, V. Přikrylová, and J. Spížek. 1999. Overproduction of 2-ketoisovalerate and monensin production by regulatory mutants of Streptomyces cinnamonensis resistant to 2-ketobutyrate and amino acids. FEMS Microbiol. Lett. 172:197-204. [Google Scholar]
- 17.Pridmore, R. D. 1987. New and versatile cloning vectors with kanamycin-resistance marker. Gene 56:309-312. [DOI] [PubMed] [Google Scholar]
- 18.Radmacher, E., A. Vaitsikova, U. Burger, K. Krumbach, H. Sahm, and L. Eggeling. 2002. Linking central metabolism with increased pathway flux: l-valine accumulation by Corynebacterium glutamicum. Appl. Environ. Microbiol. 68:2246-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reinscheid, D. J., W. Kronemeyer, L. Eggeling, B. J. Eikmanns, and H. Sahm. 1994. Stable expression of hom-1-thrB in Corynebacterium glutamicum and its effect on the carbon flux to threonine and related amino acids. Appl. Environ. Microbiol. 60:126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahm, H., and L. Eggeling. 1999. d-Pantothenate synthesis in Corynebacterium glutamicum and use of panBC and genes encoding l-valine synthesis for d-pantothenate overproduction. Appl. Environ. Microbiol. 65:1973-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook, J., and D. V. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Santamaría, R., J. A. Gil, J. M. Mesas, and J. F. Martín. 1984. Characterization of endogenous plasmid and development of cloning vectors and a transformation system in Brevibacterium lactofementum. J. Gen. Microbiol. 130:2237-2246. [Google Scholar]
- 23.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 24.Tsuchida, T., and H. Momose. 1975. Genetic changes of regulatory mechanisms occurred in leucine and valine producing mutants derived from Brevibacterium lactofermentum. Agric. Biol. Chem. 39:2193-2198. [Google Scholar]
- 25.Tsuchida, T., and H. Momose. 1986. Improvement of an l-leucine-producing mutant of Brevibacterium lactofermentum 2256 by genetically desensitizing it to α-acetohydroxy acid synthase. Appl. Environ. Microbiol. 51:1024-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Umbarger, H. E. 1996. Biosynthesis of the branched-chain amino acids, p. 442-457. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 27.van der Rest, M. E., C. Lange, and D. Molenaar. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52:541-545. [DOI] [PubMed] [Google Scholar]
- 28.Vyazmensky, M., C. Sella, Z. Barak, and D. M. Chipman. 1996. Isolation and characterization of subunits of acetohydroxy acid synthase isozyme III and reconstitution of the holoenzyme. Biochemistry 35:10339-10346. [DOI] [PubMed] [Google Scholar]
- 29.Wang, Q., and J. M. Calvo. 1993. Lrp, a global regulatory protein of Escherichia coli, binds co-operatively to multiple sites and activates transcription of ilvIH. J. Mol. Biol. 229:306-318. [DOI] [PubMed] [Google Scholar]
- 30.Westerfeld, W. W. 1945. A colorimetric detection of blood acetoin. J. Biol. Chem. 161:495-502. [PubMed] [Google Scholar]