Abstract
PCR assays for analyzing resistance-nodulation-division transporters from solvent- and drug-resistant bacteria in soil were developed. Sequence analysis of amplicons showed that the PCR successfully retrieved transporter gene fragments from soil. Most of the genes retrieved from petroleum-contaminated soils formed a cluster (cluster PCS) that was distantly related to known transporter genes. Competitive PCR showed that the abundance of PCS genes is increased in petroleum-contaminated soil.
The molecular mechanisms for the resistance of bacteria to organic solvents have been studied for gram-negative solvent-resistant bacteria (24, 29). These studies have revealed a number of mechanisms, including (i) metabolism of solvent compounds to nontoxic metabolites (24), (ii) rigidification of the cell membrane by modifying phospholipids (20), (iii) alteration in the cell surface to make it less permeable (39), (iv) formation of vesicles for removing solvents from a cell (11), and (v) efflux of toxic compounds in an energy-dependent process (10, 24). Among the most intensively studied solvent-resistant bacteria is Pseudomonas putida DOT-T1E, which can thrive in the presence of 1% toluene (22). Molecular studies have revealed that the high solvent resistance of this organism is ascribable to three resistance-nodulation-division (RND) efflux pumps (23, 25). The RND pumps of strain DOT-T1E have been grouped into the HAE1 (for hydrophobe/amphiphile efflux 1) family that also includes multidrug efflux pumps of gram-negative bacteria (32, 43).
Bioremediation has been considered as an attractive decontamination strategy due to relatively low cost and small impact to the environment. For environments heavily contaminated with petroleum fuels, the use of bioremediation is limited, since it is only applicable after the mass of petroleum is reduced by physical and/or chemical means (26). One reason for this limited use of bioremediation would be that a high concentration of petroleum is lethal to bacteria or at least suppresses bacterial activities. Nevertheless, bacteria that can grow under these conditions have been found (6, 34), suggesting that they may have the ability to resist high concentrations of petroleum. Since bacteria generally have narrow substrate ranges, they should exclude noncatabolizable substrates from cells for their survival. We can assume that molecular mechanisms found in laboratory isolates of solvent-resistant bacteria may also operate in bacteria inhabiting petroleum-contaminated environments, although no ecological evidence to support this idea has been provided.
To date, a number of studies have used PCR-mediated molecular approaches to analyze genes coding for hydrocarbon-degradative enzymes in petroleum-contaminated environments (summarized in reference 38), suggesting that catabolic enzymes homologous to those in laboratory isolates also function in petroleum-contaminated sites. Similarly, PCR assays have also been used for the detection of tetracycline efflux (tet) genes of gram-negative bacteria (1); the study analyzed the distribution of tet genes in lagoons and groundwater close to swine production facilities, suggesting that tet gene pools are present in the environment (1). The primary purpose of the present study was to develop PCR-mediated molecular approaches for analyzing RND transporter genes relevant to solvent and/or drug resistance (HAE1 transporters) in the environment. In addition, we also sought to determine whether, in addition to hydrocarbon-catabolic enzymes, solvent-efflux pumps are also important for bacteria to thrive in petroleum-contaminated soils.
Phylogeny of bacterial RND transporters in the databases.
An RND efflux pump comprises of three subunits: an RND transporter, a membrane fusion protein and an outer-membrane protein (43). RND pumps have been found in all major domains and constitute a superfamily of transporter proteins with a variety of substrates, including organic solvents, antibiotic drugs, and heavy metals (32). Phylogenetic analysis of RND transporters has shown that they could be separated into seven distinct families, including HAE1 (hydrophobe/amphiphile efflux pumps of gram-negative bacteria), HME (heavy-metal efflux pumps), SecDF (SecDF protein secretion accessory proteins), and NFE (nodulation factor exporters) families (32).
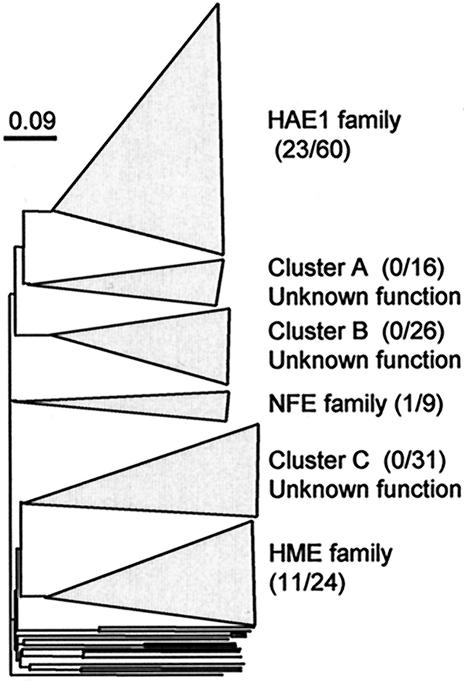
In our analysis, 217 sequences of bacterial RND transporters were found in the GenBank database; among them, functions of 35 transporters have been experimentally identified, whereas the remaining 182 were hypothetical proteins found in genome-sequenced bacteria. These sequences were aligned by the profile alignment technique of CLUSTAL W version 1.7 (31), and the alignment was refined by visual inspection. A phylogenetic tree was constructed by the neighbor-joining method (27) with the njplot program in CLUSTAL W, version 1.7. Nucleotide positions at which any sequence had a gap or an ambiguous base were not included in the phylogenetic calculation. Phylogenetic analysis based on the amino acid sequences of these bacterial transporters found six distinct clusters (Fig. 1), some of which correspond to the previously characterized families (32). A cluster corresponding to the HAE1 family (comprised of 83 transporter sequences) includes all known drug or solvent resistance RND transporters (23 sequences), together with 60 hypothetical transporters. Other clusters in Fig. 1 correspond to HME (heavy-metal efflux pumps) and NFE (nodulation factor exporters) families, whereas the remaining three clusters only included hypothetical transporters, and their functions are unknown.
FIG. 1.
Phylogenetic analysis of RND transporter genes (217 genes) retrieved from the GenBank database (the complete phylogenetic tree can be provided by request to the corresponding author). The numbers of functionally identified genes/the numbers of hypothetical genes are given in parentheses.
Development of PCR for specific amplification of HAE1 gene fragments.
Based on the phylogenetic analysis (Fig. 1), amino acid sequences of the RND transporters were rigorously compared. We found two regions that were conserved among the HAE1 transporter sequences (Fig. 2); they were used to design PCR primers (A24f2 and A577r2 in Table 1). In order to reduce biases in template/product ratios occurring due to the use of degenerate primers (21), inosine residues were incorporated into the primers at some degenerate positions. The utility of inosine residues for PCR primers has been reported previously (4, 35). A study has demonstrated the utility of inosine-containing primers for amplifying 16S rRNA gene fragments from environmental DNA samples (35). Other studies have suggested that interactions of inosine with the four natural bases exhibit different binding energies, although the differences were much less than the differences between the proper Watson-Crick base pairs and mismatch pairs (9, 15).
FIG. 2.
Comparison of sequences of representative RND transporters in nucleotide regions corresponding to the PCR primers for the specific detection of HAE1 genes. HP, hypothetical protein.
TABLE 1.
PCR primers used in this study
| Primera | Sequence (5′ to 3′)b | Specificity | Source or reference |
|---|---|---|---|
| A24f2 | CCSRTITTYGCITGGGT | HAE1 transporter genes | This study |
| A577r2 | SAICCARAIRCGCATSGC | HAE1 transporter genes | This study |
| PCS1f | GAICCIGAIITIGCITGGIC | PCS transporter genes | This study |
| PCS2r | ACTTCICCIACICCSGGIAC | PCS transporter genes | This study |
| T7W | TAATACGACTCACTATAGGGC | pGEM-T vector | 34 |
| SP6W | ATTTAGGTGACACTATAGAATACTC | pGEM-T vector | 34 |
f, forward PCR primer; r, reverse PCR primer.
According to IUC codes for bases. I, inosine.
In order to examine the utility of the designed PCR primers (A24f2 and A577r2), they were used to amplify RND transporter gene fragments from reference strains listed in Table 2. Amplification was performed with a Progene thermal cycler (Techne) by using a 50-μl mixture containing 1.25 U of Taq DNA polymerase (AmpliTaq Gold; Applied Biosystems), 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.01% (wt/vol) bovine serum albumin, each deoxynucleoside triphosphate at a concentration of 200 μM, 25 pmol of each primer, and approximately 10 ng of template DNA. The amplification conditions were as follows: 10 min of activation of the polymerase at 94°C, followed by 35 cycles of 1 min at 94°C, 1 min at 50°C, and 2 min at 72°C, and finally by 10 min of extension at 72°C. Amplified fragments were analyzed by agarose gel electrophoresis (1.5% [wt/vol]) in Tris-borate-EDTA buffer (28). The PCR conditions were primarily optimized (30 cycles of amplification) using DNA from the reference strains, whereas the PCR cycle was later increased to 35 to obtain sufficient amounts of the PCR product from soil DNA samples. Table 3 shows that these primers successfully amplified DNA fragments with the expected size from the strains bearing HAE1 genes, whereas no amplification was detected from strains whose genomic analyses found no HAE1 transporters.
TABLE 2.
PCR amplification of the HAE1 and PCS gene fragments from reference strainsa
| Strain | HAE1 geneb | PCRc
|
Reference | |
|---|---|---|---|---|
| A24f/A577r | PCS1f/PCS2r | |||
| Synechocystis sp. strain PCC 6803 | − | − | − | 17 |
| Sinorhizobium meliloti | + | + | − | 5 |
| Bacillus subtilis | − | − | − | 12 |
| Geobacter sulfurreducens | + | + | − | 16 |
| Streptomyces coelicolor | − | − | − | 2 |
| Corynebacterium glutamicum | − | − | − | 7 |
| Pseudomonas putida KT2440 | + | + | − | 18 |
| Pseudomonas putida DOT-T1E | + | + | − | 25 |
| Escherichia coli | + | + | − | 3 |
| Agrobacterium tumefaciens | + | + | − | 42 |
| Mesorhizobium loti | + | + | − | 8 |
| Clostridium acetobutylicum | − | − | − | 19 |
| Deinococcus radiodurans | − | − | − | 40 |
Bacterial strains were obtained from culture collection centers (DSMZ [German Collection of Microorganisms and Cell Cultures] and MBIC [Marine Biotechnology Institute Culture collection]). DNA was extracted from these strains as described elsewhere (14).
+, Strain known to have HAE1 gene(s); −, strain known to have no HAE1 gene.
+, Expected-size fragment was amplified; −, expected-size fragment was not amplified.
TABLE 3.
Soil samples used in this study
| Site | Description | Mean ± SD
|
||
|---|---|---|---|---|
| TPHa (mg kg−1) | DNAb (μg) | DNA recoveryc (%) | ||
| SC | Fuel oil contaminated soil obtained from an oil storage tank yard in Shizuoka | 15,000 ± 3,500 | 4.9 ± 0.8 | 24 ± 3.2 |
| SN | Uncontaminated soil obtained from a site proximate to the SC soil sampling site | BDLd | 4.1 ± 0.2 | 31 ± 5.7 |
| TH | Coal and oil sludge dumping site in Aichi | 2,900 ± 700 | 2.0 ± 0.3 | 19 ± 4.0 |
| SS | Heavy fuel oil-contaminated soil obtained from an oil storage tank yard in Fukuoka | 44,000 ± 5,200 | 0.9 ± 0.1 | 18 ± 5.2 |
| KM | Uncontaminated garden soil in Kamaishi | BDLd | 8.0 ± 1.8 | 34 ± 3.8 |
TPH, total petroleum hydrocarbon (lower detection limit, 50 mg kg−1) determined by the infrared method by using an OCMA-350 TPH analyzer (Horiba) after extraction with solvent CFC-316 (Horiba) (n = 3).
That is, the amount of DNA extracted from 0.5 g (wet weight) of soil as quantified by comparing the band intensity with the intensity of bands of the molecular size markers (n = 3).
As estimated by the recovery of the phc gene (37) introduced into soil prior to DNA extraction (n = 3).
BDL, below detection limit.
Detection and analysis of HAE1 genes in soils.
Primer set A24f2-A577r2 was used to amplify HAE1 gene fragments from DNA extracted from the soil samples listed in Table 3. Soil samples were obtained by using a core sampler in triplicate, transported at 4°C, and stored at −20°C. The SC and SN soils were obtained from the same area where there had previously been oil storage tanks; SC was contaminated with fuel oils for a long time (over 30 years), whereas spilled oil did not reach to the SN soil. The SS soil was most heavily contaminated with petroleum (mostly heavy fuel oil). The TH soil was mostly contaminated with polycyclic aromatic hydrocarbons derived from waste coals, and the TPH concentration in the TH soil was much lower than in the SC and SS soils.
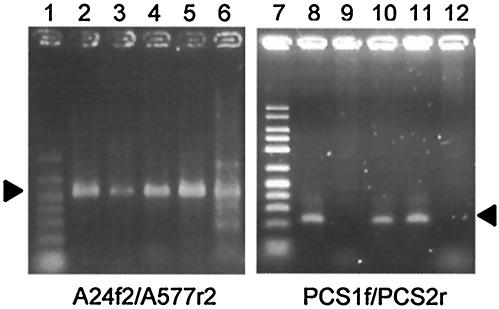
DNA was extracted from 0.5 g of soil by using a FastDNA Spin kit (Qbiogene) according to the manufacturer's instruction with following modifications. Skim milk (20 mg) was added to 0.5 g of soil before the extraction was begun in order to increase the efficiency of DNA extraction; previous studies have shown that skim milk can prevent DNA from adsorbing onto soil matrices (30, 33). The extracted DNA was dissolved in 100 μl of the DES solution supplied in the kit and quantified by measuring the band intensity in agarose gel electrophoresis (28) (Table 3), in which DNA size markers (FMC Bioproducts) were used as the quantitative standards. DNA recovery efficiency of the above-mentioned method was evaluated by quantifying the phc gene in the DNA extract solution obtained from each of the soil samples inoculated with Comamonas sp. strain rN7(R503) (harboring one copy of the phc gene per genome, inoculated at 107 cells g of soil−1) by using a method described previously (37). As presented in Table 3, the DNA recovery efficiency varied from 18 to 34%, which was incorporated into the estimation of copy numbers of RND genes in soil (see below). Figure 3 shows results of PCR using A24f2-A577r2, showing that DNA fragments with the expected sizes (ca. 550 bp) were successfully amplified from these soil DNA samples.
FIG. 3.
PCR amplification of HAE1 (lanes 2 to 6) and PCS (lanes 8 to 12) fragments from the soil DNA samples. The soil DNA samples included SC (lanes 2 and 8), SN (lanes 3 and 9), TH (lanes 4 and 10), SS (lane 5 and 11), and KM (lanes 6 and 12). Lanes 1 and 7 show molecular size markers (50 to 1,000 and 50 to 2,500 bp, respectively [FMC Bioproducts]). The positions of the expected HAE1 and PCS fragments are indicated with arrowheads.
Sequence analyses of PCR products.
In order to know if the amplified fragments actually contained HAE1 gene fragments, each amplified fragment was cloned into E. coli and sequenced. Amplified fragments were purified by electrophoresis, ligated into the pGEM-T vector (Promega), and cloned into E. coli as described previously (36). Vector-harboring clones were selected on Luria-Bertani plates (28) supplemented with ampicillin (50 μg ml−1). PCR-amplified fragments were recovered from each colony by PCR with the primers T7W and SP6W (these primers target pGEM-T vector sequences flanking the insertion; see Table 1) as described previously (36). Clones containing appropriate insert sizes were selected by an electrophoretic analysis, and their nucleotide sequences were determined as described previously (36).
As shown in Table 4, the sequence analysis of 126 clones in all produced 23 sequence types (classified as a unique clone or group of clones with sequence similarity of 0.9). These nucleotide sequences have been deposited in the DDBJ, EMBL, and National Center for Biotechnology Information nucleotide sequence databases under accession numbers AB161948 to AB161970. Most of the amplified fragments were affiliated with the HAE1 family (Table 4 and Fig. 4), demonstrating that the PCR could specifically amplify HAE1 gene fragments. The GenBank database search (Table 4) revealed that most of the HAE1 transporter fragments retrieved from the soils showed low homologies (<70%) to HAE1 transporters previously found in isolated organisms. A homologue (SN45) of the Ttg transporters of P. putida DOT-T1E (23, 25) was obtained from the SN and TH soils but not from the SC and SS soils (Table 4). We consider that this was due to the substrate specificity of Ttg-type transporters; these transporters preferentially efflux aromatic compounds, whereas aliphatic compounds share the largest portion of petroleum.
TABLE 4.
HAE1 sequence types obtained from the soil samples
| Sequence type | No. of clones
|
Highest match in the databasesa (% identity [amino acids]) | ||||
|---|---|---|---|---|---|---|
| SC | SN | TH | SS | KM | ||
| SC3 | 1 | HP Escherichia coli K-12 NP_417971 (54) | ||||
| SC30 | 8 | 5 | HP Yersinia pestis NP_406608 (56) | |||
| SC31 | 2 | 1 | 2 | HP Escherichia coli CFT073 NP_756187 (53) | ||
| SC40 | 2 | HP Escherichia coli K-12 NP_417971 (64) | ||||
| SC44 | 1 | AcrB Proteus mirabilis AAL32126 (56) | ||||
| SC50 | 7 | EnvD Escherichia coli AAA58070 (55) | ||||
| SN1 | 23 | HP Salmonella enterica NP_455072 (62) | ||||
| SN25 | 1 | HP Rhodopseudomonas palustris ZP_00011918 (65) | ||||
| SN36 | 1 | HP Escherichia coli O157 NP_290094 (51) | ||||
| SN45 | 1 | 2 | HP Pseudomonas fluorescens AAQ92181 (97) | |||
| SN49 | 1 | EefB Enterobacter aerogenes CAD48862 (59) | ||||
| TH2 | 16 | HP Azotobacter vinelandii ZP_00091148 (63) | ||||
| TH14 | 3 | HP Burkholderia fungorum ZP_00028911 (56) | ||||
| TH31 | 1 | HP Pseudomonas syringae ZP_00127062 (83) | ||||
| TH34 | 1 | EnvD Escherichia coli AAA58070 (62) | ||||
| SS4 | 18 | HP Escherichia coli K-12 NP_417971 (52) | ||||
| SS25 | 1 | HP Escherichia coli CFT073 NP_756187 (60) | ||||
| KM3 | 7 | HP Salmonella entrica NP_455072 (58) | ||||
| KM4 | 10 | HP Pseudomonas putida KT2440 NP_743544 (70) | ||||
| KM5 | 1 | HP Xylella fastidiosa 9a5c NP_299373 (65) | ||||
| KM12 | 2 | HP Xanthomonas campestris NP_637722 (51) | ||||
| KM18 | 1 | AcrB Erwinia amylovora AAQ21216 (46) | ||||
| KM20 | 1 | HP Escherichia coli K-12 NP_414995 (54) | ||||
| Non-HAE1 | 1 | 3 | 0 | 0 | 2 | |
| Total | 22 | 31 | 23 | 26 | 24 | |
HP, hypothetical protein.
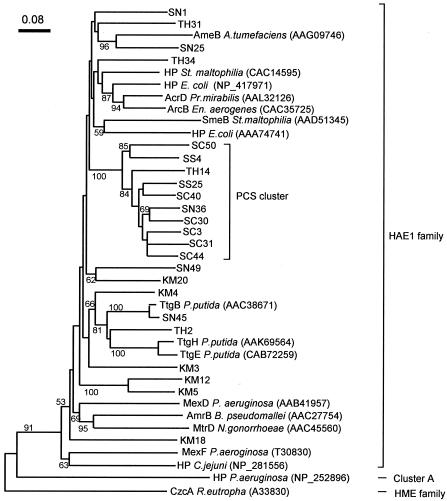
FIG. 4.
Neighbor-joining three showing phylogenetic positions of the HAE1 sequence types obtained from the soil samples (Table 4). Nucleotide positions at which any sequence had a gap or an ambiguous base were not included in the phylogenetic calculations. Numbers at the branch nodes are bootstrap values (per 100 trials); only values >50 are shown. HP, hypothetical protein.
Figure 4 also shows that the sequence types obtained from petroleum-contaminated soils formed a cluster (designated cluster PCS) that was distantly related to the known HAE1 transporters. In particular, even though the SC and SN samples were obtained from the same area (i.e., the same soil texture and climate conditions), PCS transporters were obtained mostly from the SC sample. These results allowed us to assume that petroleum contamination has resulted in the enrichment of PCS transporters in soil.
Development of PCR for specific amplification of PCS gene fragments.
In order to further analyze PCS transporters in soil samples, PCR primers (PCS1f and PCS2r in Table 1) specific for the PCS cluster were designed by comparing HAE1 sequences retrieved from the databases and those cloned from the soil samples. PCR was performed as described above except the annealing temperature was 51°C and the amount of each primer was 20 pmol. The specificity was checked by attempting PCR amplification from the reference strains (Table 3): HAE1 clones obtained from the soil samples (clones containing the sequence types listed in Table 4) and DNA samples extracted from the soils (Fig. 3). No reference strains were positive in PCR for the PCS fragments (Table 3), and PCR products with the expected size (approximately 240 bp) were obtained only from the sequence types affiliated with the PCS cluster (data not shown). Figure 3 shows PCR amplification of PCS fragments from the five soil DNA samples listed in Table 1. It can be seen that bands with the expected size were obtained only from the petroleum-contaminated soils (i.e., the SC, TH, and SS soils). These results indicate that PCR with PCS1f and PCS2r can be used to specifically detect PCS genes in both bacteria and soil samples.
Estimation of abundance of HAE1 and PCS genes in soil by cPCR.
Competitor fragments were produced using the competitive DNA construction kit (Takara Shuzo). For competitive PCR (cPCR) with the primers A24f2 and A577r2, a 334-bp competitor was produced by PCR with the primers A24f2-200F (5′-CCSRTITTYGCITGGGTGTAATAGCGATGCGTAATGA-3′) and A577r2-500R (5′-SAICCARAIRCGCATSGCAATACATCAAACGCCGCGAC-3′) and the template DNA fragments supplied in the kit. For cPCR with the primers PCSf1 and PCSr2, a 340-bp competitor was produced by PCR with the primers PCS1f-F (5′-GAICCIGAIITIGCITGGICGTACGGTCATCATCTGACAC-3′) and PCS2r-300R (5′-ACTTCICCIACICCSGGIACCGCCATCCTGGGAAGACTCC-3′) from the template DNA fragments supplied in the kit. PCR conditions in cPCR were as described above, except for the competitor fragment being added at a known copy number. A total of 2 μl of the PCR product was analyzed by electrophoresis through 1.5% (wt/vol) agarose gels with Tris-borate-EDTA buffer, and the gels were photographed after staining with SYBR Gold (FMC Bioproducts). The band intensities of the target and competitor fragments were quantified by using the Multianalyst program supplied with Gel Doc 2000 (Bio-Rad). To determine a copy number of the target fragment in a soil DNA sample, at least five PCR assays with different competitor numbers (decimal dilution series) were conducted in triplicate. Relative amplification efficiencies of the target to competitor fragments in the HAE1 and PCS PCR systems were analyzed by PCR with reaction mixtures containing equal numbers of the target (purified PCR products from PCS clones) and competitor fragments (105 to 109 copies of each fragment per PCR mixture). In the HAE1 PCR system, the relative amplification efficiency varied from 0.9 to 1.4, with the mean value of 1.2, whereas that in the PCS system varied from 0.3 to 0.6, with the mean value of 0.4; the mean values were used as the relative amplification efficiencies for estimating target copy numbers. A copy number of the target fragment was estimated by considering the band intensity, the length of the fragment, the copy number of the competitor, and the relative amplification efficiency as described by Lee et al. (13). Interpretation of the results of cPCR for estimating RND genes in soil should, however, be done carefully (as are also the cases of other PCR quantification methods), because heterogeneity in the sequence of unknown genes may cause a variation in the PCR amplification efficiency (41).
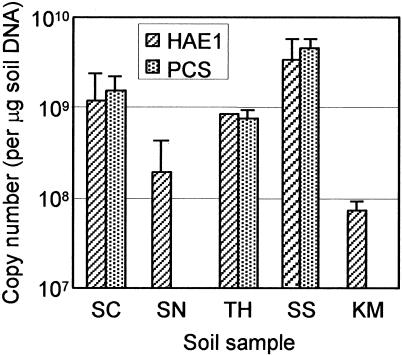
The results of cPCR (Fig. 5) show that copy numbers of the HAE1 genes in the petroleum-contaminated soils were higher than those in the uncontaminated soil samples. It is also shown that PCS genes shared significant proportions of HAE1 genes in the petroleum-contaminated samples. The highest values for copy numbers of HAE1 and PCS genes were estimated for the SS sample that was most heavily contaminated with petroleum among the soil samples analyzed. It is therefore likely that PCS transporters play important roles in soils heavily contaminated with petroleum. Unfortunately, however, since bacteria possessing PCS transporters have not yet been isolated and characterized in the laboratory, the functions of these putative efflux pumps are currently unclear. The PCR assay developed here will be useful for screening bacterial strains isolated from the petroleum-contaminated soils for identifying strains possessing PCS transporters.
FIG. 5.
Copy numbers of HAE1 and PCS genes in soil as estimated by cPCR. Values are means ± the standard deviations (n = 3).
Acknowledgments
This study was supported by the Human Frontier Science Program (RGY0021/2002).
We thank Yo Takahata for providing petroleum-contaminated soil samples.
REFERENCES
- 1.Aminov, R. I., J. C. Chee-Sanford, N. Garrigues, B. Teferedegne, I. J. Krapac, B. A. White, and R. I. Mackie. 2002. Development, validation, and application of PCR primers for detection of tetracycline efflux genes of gram-negative bacteria. Appl. Environ. Microbiol. 68:1786-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Candrian, U., B. Furrer, C. Hofelein, and J. Luthy. 1991. Use of inosine-containing oligonucleotide primers for enzymatic amplification of different alleles of the gene coding for heat-stable toxin type I of enterotoxigenic Escherichia coli. Appl. Environ. Microbiol. 57:955-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Puehler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, P. Y., and B. K. Kinkle. 2001. Mycobacterium diversity and pyrene mineralization in petroleum-contaminated soils. Appl. Environ. Microbiol. 67:2222-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalinowski, J., B. Bathe, D. Bartels, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmanns, L. Gaigalat, A. Goesmann, M. Hartmann, K. Huthmacher, R. Kramer, B. Linke, A. C. McHardy, F. Meyer, B. Mockel, W. Pfefferle, A. Puhler, D. A. Rey, C. Ruckert, O. Rupp, H. Sahm, V. F. Wendisch, I. Wiegrabe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5-25. [DOI] [PubMed] [Google Scholar]
- 8.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:381-406. [DOI] [PubMed] [Google Scholar]
- 9.Kawase, Y., S. Iwai, H. Inoue, K. Miura, and E. Ohtsuka. 1986. Studies on nucleic acid interactions: I. Stabilities of miniduplexes dG2A4XA4G2-dC2T4YT4C2 and self-complementary dGGGAAXYTTCCC containing deoxyinosine and other mismatched bases. Nucleic Acids Res. 14:7727-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kieboom, J., J. J. Dennis, J. A. de Bont, and G. J. Zylstra. 1998. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J. Biol. Chem. 273:85-91. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi, H., K. Uematsu, H. Hirayama, and K. Horikoshi. 2000. Novel toluene elimination system in a toluene-tolerant microorganism. J. Bacteriol. 182:6451-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Briginell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, N. J. Cummings, R. A. Danie, F. Denizot, K. M. Devine, A. Dusterhoft, S. D. Ehrlich, P. T. Emmerson, K. D. Entian, J. Errington, C. Fabret, E. Ferrari, D. Foulger, C. Fritz, M. Fujita, Y. Fujita, S. Fuma, A. Galizzi, N. Galleron, S. Y. Ghim, P. Glaser, A. Goffeau, E. J. Golightly, G. Grandi, G. Guiseppi, B. J. Guy, K. Haga, J. Haiech, C. R. Harwood, Y. Hasahara, A. Henaut, H. Hilbert, S. Holsappel, S. Hoson, M. F. Hullo, M. Itaya, L. Jones, B. Joris, D. Karamata, M. Klaerr-Blanchard, C. Klein, Y. Kobayashi, P. Koetter, G. Koningstein, S. Krogh, M. Kumano, K. Kurita, A. Lapidus, S. Lardinois, J. Lauer, V. Lazarevic, S. M. Lee, A. Levine, H. Liu, S. Masuda, C. Mauel, C. Medigue, N. Medina, R. P. Medallo, M. Mizuno, D. Moestl, S. Nakai, M. Noback, D. Noone, M. O'Reilly, K. Ogawa, A. Ogiwara, B. Oudega, S. H. Park, V. Parro, T. M. Pohl, D. Portetelle, S. Porwollik, A. M. Prescott, E. Presecan, P. Pujic, B. Purnelle, G. Rapoport, M. Rey, S. Reynolds, M. Rieger, C. Rivolta, E. Rocha, B. Roche, M. Rose, Y. Sadaie, T. Sato, E. Scanlan, S. Schleich, R. Schroeter, F. Scoffone, J. Sekiguchi, A. Sekowska, S. J. Seror, P. Serror, B. S. Shin, B. Soldo, A. Sorokin, E. Tacconi, T. Takagi, H. Takahashi, K. Takemaru, M. Takeuchi, A. Tamakoshi, T. Tanaka, P. Terpstra, A. Tognini, V. Tosato, S. Uchiyama, M. Vandenbol, F. Vannier, A. Vassarotti, A. Viari, R. Wambutt, E. Wedler, H. Wedler, T. Weitzenegger, P. Winters, A. Wipat, H. Yamamoto, K. Yamane, K. Yasumoto, K. Yata, K. Yoshida, H. Yoshikawa, H. F. Yoshikawa, E. Zumstein, and A. Danchin. 1998. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 13.Lee, S. Y., J. Bollinger, D. Bezdicek, and A. Ogram. 1996. Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl. Environ. Microbiol. 62:3787-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 15.Martin, F. H., M. M. Castro, F. Aboulela, and I. Tinoco, Jr. 1985. Base pairing involving deoxyinosine: implications for probe design. Nucleic Acids Res. 13:8927-8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Methe, B.A., K. E. Nelson, J. A. Eisen, I. T. Paulsen, W. Nelson, J. F. Heidelberg, D. Wu, M. Wu, N. Ward, M. J. Beanan, R. J. Dodson, R. Madupu, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, M. Gwinn, J. F. Kolonay, S. A. Sullivan, D. H. Haft, J. Selengut, T. M. Davidsen, N. Zafar, O. White, B. Tran, C. Romero, H. A. Forberger, J. Weidman, H. Khouri, T. V. Feldblyum, T. R. Utterback, S. E. van Aken, D. R. Lovley, and C. M. Fraser. 2003. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments Science 302:1967-1969. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura, Y., T. Kaneko, M. Hirosawa, N. Miyajima, and S. Tabata. 1998. CyanoBase, a www database containing the complete nucleotide sequence of the genome of Synechocystis sp. strain PCC6803. Nucleic Acids Res. 26:63-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. Cris Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. Eisen, K. N. Timmis, A. Dusterhoft, B. Tummler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 19.Nolling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinkart, H. C., and D. C. White. 1997. Phospholipid biosynthesis and solvent tolerance in Pseudomonas putida strains. J. Bacteriol. 181:3256-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in product-to-template ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramos, J. L., E. Duque, M. J. Huertas, and A. Haidour. 1995. Isolation and expansion of the catabolic potential of a Pseudomonas putida strain able to grow in the presence of high concentrations of aromatic hydrocarbons. J. Bacteriol. 177:3911-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramos, J. L., E. Duque, P. Godoy, and A. Segura. 1998. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT-T1E. J. Bacteriol. 180:3323-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramos, J. L., E. Duque, M. T. Gallegos, P. Godoy, M. I. Ramos-Gonzalez, A. Rojas, W. Teran, and A. Segura. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56:743-768. [DOI] [PubMed] [Google Scholar]
- 25.Rojas, A., E. Duque, G. Mosqueda, G. Golden, A. Hurtado, J. L. Ramos, and A. Segura. 2001. Three efflux pumps are required to provide efficient tolerance to toluene in Pseudomonas putida DOT-T1E. J. Bacteriol. 183:3967-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ron, E. Z., and E. Rosenberg. 2002. Biosurfactants and oil bioremediation. Curr. Opin. Biotechnol. 13:249-252. [DOI] [PubMed] [Google Scholar]
- 27.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Segura, A., E. Duque, G. Mosqueda, J. L. Ramos, and F. Junker. 1999. Multiple responses of gram-negative bacteria to organic solvents. Environ. Microbiol. 1:191-198. [DOI] [PubMed] [Google Scholar]
- 30.Takada-Hoshino, Y., and N. Matsumoto. 2004. An improved DNA extraction method using skim milk from soils that strongly adsorb DNA. Microbes Environ. 19:13-19. [Google Scholar]
- 31.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tseng, T. T., K. S. Gratwick, J. Kollman, D. Park, D. H. Nies, A. Goffeau, and M. H. Saier, Jr. 1999. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1:107-125. [PubMed] [Google Scholar]
- 33.Volossiouk, T., J. E. Robb, and R. N. Nazar. 1995. Direct DNA extraction for PCR-mediated assays of soil organisms. Appl. Environ. Microbiol. 61:3972-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe, K., K. Watanabe, Y. Kodama, K. Syutsubo, and S. Harayama. 2000. Molecular characterization of bacterial populations in petroleum-contaminated groundwater discharged from underground crude-oil-storage cavities. Appl. Environ. Microbiol. 66:4803-4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe, K., Y. Kodama, and S. Harayama. 2001. Design and evaluation of PCR primers to amplify bacterial 16S ribosomal DNA fragments used for community fingerprinting. J. Microbiol. Methods 44:253-262. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe, K., Y. Kodama, N. Hamamura, and N. Kaku. 2002. Diversity, abundance and activity of archaeal populations in oil-contaminated groundwater accumulated at the bottom of an underground crude oil storage cavity. Appl. Environ. Microbiol. 68:3899-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe, K., M. Teramoto, and S. Harayama. 2002. Stable augmentation of activated sludge with foreign catabolic genes harbored by an indigenous dominant bacterium. Environ. Microbiol. 4:577-583. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe, K., and N. Hamamura. 2003. Molecular and physiological approaches to understanding the ecology of pollutant degradation. Curr. Opin. Biotechnol. 14:289-295. [DOI] [PubMed] [Google Scholar]
- 39.Weber, F. J., and J. A. M. de Bont. 1996. Adaptation mechanisms of microorganisms to the toxic effects of organic solvents on membranes. Biochim. Biophys. Acta 1286:225-245. [DOI] [PubMed] [Google Scholar]
- 40.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, K. W. Minton, R. D. Fleischmann, K. A. Ketchum, K. E. Nelson, S. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wintzingerode, F. V., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 42.Wood, D. W., J. C. Setubal, R. Kaul, D. Monks, L. Chen, G. E. Wood, Y. Chen, L. Woo, J. P. Kitajima, V. K. Okura, N. F. Almeida, Jr., Y. Zhou, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, D. Guenthner, T. Kutyavin, R. Levy, M. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, D. Gordon, J. A. Eisen, I. Paulsen, P. Karp, P. Romero, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Zhao, M. Dolan, S. V. Tingey, J. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 43.Zgurskaya, H. I., and H. Nikaido. 2001. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219-225. [DOI] [PubMed] [Google Scholar]