Abstract
We describe the recovery of the rare species Aeromonas culicicola, so far known only in mosquitoes in India, from a drinking water supply in Spain. Typing, using enterobacterial repetitive intergenic consensus-PCR, revealed that the 27 new isolates belonged to 3 very closely related strains. These strains were genetically identified by 16S rRNA gene sequencing. Spanish strains differed from the mosquito strains in three nucleotide positions. The AHCYTOEN gene was present in these water strains, which may have a public health significance.
Aeromonas species are common inhabitants of aquatic ecosystems, although they have also been described in connection with fish and human diseases (1, 2). The most important human pathogens are Aeromonas veronii bv. sobria, Aeromonas caviae, and Aeromonas hydrophila, while Aeromonas salmonicida is the most important fish pathogen. The two major diseases associated with humans are gastroenteritis and wound infections. Gastroenteritis typically occurs after the ingestion of contaminated water or food, whereas wound infection results from exposure to contaminated water. The recently discovered species A. culicicola is a very rare and exotic taxon found only in the midgut of mosquitoes in India, from which so far only three isolates are available (14). Surprisingly, we have recently recovered a significant number of isolates of this species in a very different substrate and at a distant location from where this species was originally isolated. They were found in a drinking water supply in Spain. Here, we describe the characterization of strains of this species using molecular identification methods (16S DNA coding for RNA [rDNA]-restriction fragment length polymorphism [RFLP] and 16S rDNA sequencing) and genotyping (enterobacterial repetitive intergenic consensus [ERIC]-PCR) (3, 8, 13, 17). We have also evaluated the potential risk to human health that this finding can represent by determining the presence of the cytolytic enterotoxin gene (AHCYTOEN) in such isolates. This is a multivirulent gene that causes death in mice, hemolysis, cytotoxicity, and enterotoxicity (5, 10).
Sample collection and biochemical identification of the strains.
A total of 59 isolates of Aeromonas spp. were recovered from 494 water samples from 50 different sites in the domestic water pipelines during a routine monitoring sanitary survey. They were collected at 1- or 2-week intervals by the Instituto Municipal de Salud Publica in Zaragoza, Spain, from April to June 2002. Free chlorine residuals evaluation and recovery with ADA medium was performed as in a previous study (4). Tergitol agar, incubated for 24 h at 37°C (Difco Laboratories, Detroit, Mich.) was used in parallel in all samples. From each sample one typical colony of those grown on each medium was selected for biochemical identification, i.e., yellow ones in ADA and red ones in tergitol agar. Oxidase, glucose fermentation (Kligler agar), gas from sucrose, and esculin hydrolysis tests were used for this purpose. Strains were further confirmed by using API 20NE (BioMerieux, Marcy-l'Etoile, France). Once biochemically identified, the isolates were sent to our laboratory for molecular identification.
Aeromonas was found in 6.9% of the drinking water samples, corresponding to 20 different sampling sites within the drinking water supply system. Samples normally had concentrations of Aeromonas below 10 CFU/100 ml and an absence of fecal indicators (data not shown). Free chlorine residuals in these samples were between <0.05 and 1.5 ppm. The 59 isolates recovered were identified by using API 20NE as follows: 47 were identified as A. veronii bv. sobria, 8 were identified as A. hydrophila, and 4 were identified as A. caviae.
Molecular identification and typing.
Water isolates were reidentified using the 16S rDNA-RFLP method, which produces a unique pattern for each of the species of the genus (3, 8). In the study, we included the three isolates of A. culicicola known so far, i.e., the type strain (CECT 5761T), isolated from the midgut of the mosquito Culex quinquefasciatus, and the strains SH and SLH from Aedes aegyptii, all of them from India (14). Our results were completely different from those obtained biochemically. For instance, the 27 water isolates that had been identified biochemically as A. veronii bv. sobria showed an identical RFLP pattern that did not correspond to any previously identified Aeromonas species (3, 8). The pattern of the Spanish isolates was also different from that of the original strains of A. culicicola. The rest of the isolates were identified as follows: 21 were identified as A. veronii, 5 were identified as A. salmonicida, 4 were identified as A. hydrophila, 1 was identified as Aeromonas media, and 1 was identified as Aeromonas jandaei.
The 27 isolates and the 3 strains of A. culicicola were typed using the ERIC-PCR technique as previously described (17). The type strains of A. salmonicida (CECT 894T), A. jandaei (CECT 4228T), and A. hydrophila (CECT 839T) were used as an outgroup. PCRs and image analyses have been done as previously described (17).
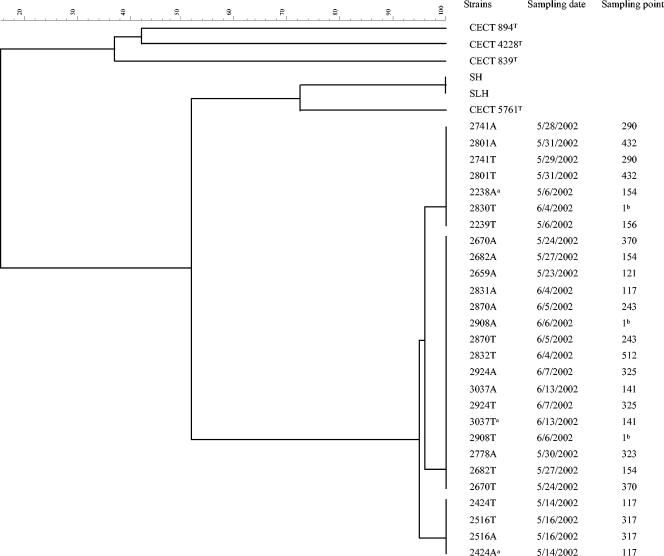
In the dendrogram obtained from the ERIC-PCR analysis (Fig. 1), the 27 isolates were split into 3 groups with a similarity of 95 to 96%. Each group contained strains with an identical ERIC pattern. At the same time, they grouped together with the three strains of A. culicicola at a similarity level of 51.78%, suggesting that they could be related to that species. Two of the isolates of A. culicicola (SH and SLH) grouped with a similarity level of 100% and grouped with a similarity of 72.22% with the third isolate (the type strain). These data revealed that the 27 isolates belong to 3 genetically very similar strains and that isolates SH and SLH are identical. All the Spanish isolates recovered from the same sample grouped together (Fig. 1). However, within each of the three subclusters, there were isolates recovered from distant sites (up to 4 km) in the water supply system.
FIG. 1.
Dendrogram based on UPGMA clustering of Dice correlation values (SD) of ERIC patterns of 27 isolates from drinking water and reference strains of A. culicicola (CECT 5761T, SH, and SLH), A. jandaei (CECT 4228T), A. hydrophila (CECT 839T), and A. salmonicida (CECT 894T). The scale represents SD values converted to percentages. A final letter A in the strain designation indicates that it was recovered from ADA medium, and T indicates tergitol. Superscript “a” indicates a strain selected for 16S rDNA sequencing. Superscript “b” indicates an isolate from water leaving the drinking water treatment plant.
16S rRNA gene sequencing.
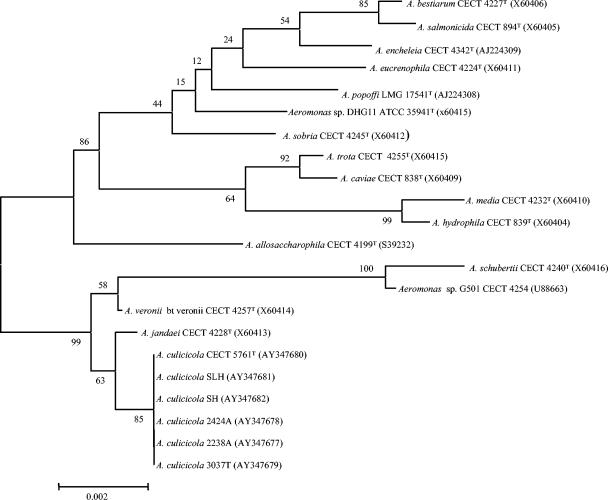
In order to confirm the identity of the Spanish isolates, we decided to sequence the 16S rRNA gene. A 1,303-bp fragment of this gene of one of the isolates (3037T) was initially sequenced in order to perform a BLAST and CLUSTAL analysis. Genomic DNA extraction, 16S rDNA amplification, and sequencing was performed as previously described (13). Sequencing was performed by using an ABI PRISM 310 genetic analyzer (Perkin-Elmer, Foster City, Calif). The higher similarity (99%) was with A. culicicola (strains SH and SLH). The sequence was completed, and two other isolates (2238A and 2424A) representative of the other two ERIC-subclusters (Fig. 1) and the three existing isolates of A. culicicola were also sequenced. The obtained sequences (1,503 bp) were compared by using CLUSTAL W with those of all the type strains of Aeromonas from GenBank (Fig. 2). A phylogenetic tree (Fig. 2) was drawn by using the neighbor-joining method with MEGA software, version 2.1 (11).
FIG. 2.
The phylogenetic tree of the 16S rRNA gene sequences of the Spanish strains (2238A, 2424A, and 3037T) and described Aeromonas species. The unrooted phylogenetic tree was drawn from 1,503 bp of the gene, using the neighbor-joining method in MEGA software. Numbers in parentheses are the GenBank sequence accession numbers.
The analysis of the sequences showed several differences (Table 1). Although Pidiyar et al. (14) indicated that the sequences of their three Indian strains were identical, we observed that the sequences of the isolates SH and SLH that they deposited in GenBank were identical to each other and differed in five nucleotides from the type strain (Table 1). The nucleotide differences found between the sequences of the three strains of A. culicicola we obtained and those of the same strain withdrawn from the GenBank (AF170914, AY130991, and AY130992) were due to double-sequencing picks at seven positions in our sequences (Table 1). The predominating nucleotides at these sites coincided with those that appeared in the sequences obtained from GenBank. Five of these variations (positions 457 to 476) were located at the hypervariable stem-loop, a region considered not phylogenetically informative (12), and the other two were located at positions 1011 and 1018. The three Spanish isolates showed less variability and presented two double-sequencing picks at positions 649 and 1308 not observed in the other strains (Table 1). Despite these differences, the three Spanish isolates were on the same branch as the three Indian isolates of A. culicicola in the phylogenetic tree, indicating that they belonged to this species (Fig. 2). A. jandaei was the species closest to A. culicicola, in agreement with the work of Pidiyar et al. (14). However, apart from the specific single-nucleotide difference reported by those authors for these two species at position 264, we have found another variation at position 1503 not described before. At this position, A. culicicola shows a G instead of the A that A. jandaei possesses.
TABLE 1.
Base differences in the 16S rDNA sequences among original and selected strains of A. culicicola
| Strain(s) | Reference | Accession no. | Base at positiona:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 457 | 458 | 469 | 474 | 475b | 649b | 1011 | 1018 | 1308b | |||
| MTCC 3249Tc | Pidiyar et al., 2002 (14) | AF170914 | C | A | T | T | G | A | C | G | C |
| This study | AY347680 | C/t | A/g | T/a | T/c | G/a | A | C/t | G/a | C | |
| SHc, SLHc | Pidiyar et al., 2002 (14) | AY130991, AY130992 | T | G | A | C | A | A | C | G | C |
| This study | AY347681, AY347682 | T/c | G/a | A/t | C/t | A/g | A | C/t | G/a | C | |
| 2238Ad | This study | AY347677 | T | G | A | C | A | A/g | C/t | G/a | C/t |
| 2424A,d 3037Td | This study | AY347678, AY347679 | T | G | A | C | A | G/a | C/t | G/a | C/t |
E. coli sequence was used as a reference.
Positions that affect restriction sites with AluI and MboI.
Sequences of A. culicicola available at GenBank.
Strains from the drinking water supply of Zaragoza.
Intraspecific variations of the 16S rRNA gene have previously been described for Aeromonas popoffii (7, 12) and in other Aeromonas spp. (18). and are not uncommon in prokaryotes (6, 9). This variability is due to the fact that nucleotide composition varies between operon copies of the ribosomal 16S multigene family and the PCR produces a consensus of the sequences (16). The two above-mentioned intraspecific variations for our strains together with the one encountered at position 475 for the Indian ones explain the mentioned differences in RFLP pattern. Contrary to what happens with the 16S rDNA intraspecific variations described for A. popoffii (7), the variations encountered in A. culicicola affected the restriction sites of the AluI and MboI enzymes. Although two RFLP patterns were observed in A. culicicola, both are specific to this species and different from those for other species of the genus.
Detection of the AHCYTOEN gene.
The presence of the AHCYTOEN gene was investigated in the Spanish A. culicicola isolates using primers (AHCF1 and AHCR1) and conditions previously described by Kingombe et al. (10). An amplicon of 232 bp, typical of the AHCYTOEN gene, was obtained for all the 27 isolates tested. The PCR product of three strains was sequenced to demonstrate that the amplified region corresponded to the gene. This cytolytic enterotoxin gene is considered a characteristic virulence trait in Aeromonas (10, 15), and its general presence in A. culicicola, indicates that this species may have public health significance.
Acknowledgments
Strains SH and SLH were provided by Yogesh S. Shouche of the National Center for Cell Science, Pune University Campus, Ganeshkhind, India.
This work was supported by grants from Fundació Ciència i Salut and from the Spanish Ministry of Health (FIS 03/1183).
REFERENCES
- 1.Altwegg, M. 1999. Aeromonas and Plesiomonas, p. 507-516. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology. American Society for Microbiology, Washington, D.C.
- 2.Austin, B., and C. Adams. 1996. Fish pathogens, p. 197-244. In B. Austin, M. Altwegg, P. J. Gosling, and S. Joseph (ed.), The genus Aeromonas. John Wiley & Sons Ltd., Chichester, England.
- 3.Borrell, N., S. G. Acinas, M. J. Figueras, and A. J. Martínez-Murcia. 1997. Identification of Aeromonas clinical isolates by restriction fragment length polymorphism of PCR-amplified 16S rRNA genes. J. Clin. Microbiol. 35:1671-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borrell, N., M. J. Figueras, and J. Guarro. 1998. Phenotypic identification of Aeromonas genomospecies from clinical and environmental sources. Can. J. Microbiol. 44:103-108. [DOI] [PubMed] [Google Scholar]
- 5.Chopra, A. K., C. W. Houston, J. W. Peterson, and G. F. Jin. 1993. Cloning, expression, and sequence analysis of a cytolytic enterotoxin gene from Aeromonas hydrophila. Can. J. Microbiol. 39:513-523. [DOI] [PubMed] [Google Scholar]
- 6.Clayton, R. A., G. Sutton, P. S. Hinkle, Jr., C. Bult, and C. Fields. 1995. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int. J. Syst. Bacteriol. 45:595-599. [DOI] [PubMed] [Google Scholar]
- 7.Demarta, A., M. Tonolla, A. P. Caminada, N. Ruggeri, and R. Peduzzi. 1999. Signature region within the 16S rDNA sequences of Aeromonas popoffi. FEMS Microbiol. Lett. 172:239-246. [DOI] [PubMed] [Google Scholar]
- 8.Figueras, M. J., L. Soler, M. R. Chacón, J. Guarro, and A. Martínez-Murcia. 2000. Extended method for discrimination of Aeromonas spp. by 16S rDNA-RFLP. Int. J. Syst. Evol. Microbiol. 50:2069-2073. [DOI] [PubMed] [Google Scholar]
- 9.Gurtler, V., and B. C. Mayall. 2001. Genomic approaches to typing, taxonomy and evolution of bacterial isolates. Int. J. Syst. Evol. Microbiol. 51:3-16. [DOI] [PubMed] [Google Scholar]
- 10.Kingombe, C. I. B., G. Huys, M. Tonolla, M. J. Albert, J. Swings, R. Peduzzi, and T. Jemmi. 1999. PCR detection, characterization and distribution of virulence genes in Aeromonas spp. Appl. Environ. Microbiol. 65:5293-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 12.Martínez-Murcia, A. J. 1999. Phylogenetic positions of Aeromonas DNA hybridization group 11, Aeromonas popoffi, and Aeromonas group 501. Int. J. Syst. Bacteriol. 49:1403-1408. [DOI] [PubMed] [Google Scholar]
- 13.Martínez-Murcia, A. J., S. Benlloch, and M. D. Collins. 1992. Phylogenetic interrelationships of members of the genera Aeromonas and Plesiomonas as determined by 16S ribosomal DNA sequencing: lack of congruence with results of DNA-DNA hybridizations. Int. J. Syst. Bacteriol. 42:412-421. [DOI] [PubMed] [Google Scholar]
- 14.Pidiyar, V., A. Kaznowski, N. B. Narayan, M. Patole, and Y. S. Shouche. 2002. Aeromonas culicicola sp nov., from the midgut of Culex quinquefasciatus. Int. J. Syst. Evol. Microbiol. 52:1723-1728. [DOI] [PubMed] [Google Scholar]
- 15.Sechi, L. A., A. Deriu, M. P. Falchi, G. Fadda, and S. Zanetti. 2002. Distribution of virulence genes in Aeromonas spp. isolated from Sardinian waters and from patients with diarrhoea. J. Appl. Microbiol. 92:221-227. [DOI] [PubMed] [Google Scholar]
- 16.Sneath, P. H. 1993. Evidence from Aeromonas for genetic crossing-over in ribosomal sequences. Int. J. Syst. Bacteriol. 43:626-629. [DOI] [PubMed] [Google Scholar]
- 17.Soler, L., M. J. Figueras, M. R. Chacón, J. Guarro, and A. J. Martínez-Murcia. 2003. Comparison of three molecular methods for typing Aeromonas popoffii isolates. Antonie Leeuwenhoek 83:341-349. [DOI] [PubMed] [Google Scholar]
- 18.Yáñez, M. A., V. Catalán, D. Apráiz, M. J. Figueras, and A. J. Martínez-Murcia. 2003. Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int. J. Syst. Evol. Microbiol. 53:875-883. [DOI] [PubMed] [Google Scholar]