Abstract
The presence of Ralstonia solanacearum biovar 2 in the watercourses of European countries is increasing, but little is known about its ecology in aquatic habitats. The detection of this pathogen in 2000 in one Spanish river led us to study its population density at different locations on the river over a period of 3 years. During 2000 and 2001, the pathogen was recovered at low densities (10 to 80 CFU/ml) by direct plating on modified SMSA agar from water samples at 14°C or higher, but its isolation was usually unsuccessful at temperatures below 9°C. To monitor the pathogen's abundance in winter, we used two liquid selective media for enrichment (at 29 and 35°C) and compared them by using spiked river water samples: modified Wilbrink broth (MWB) was more efficient than modified SMSA broth for double-antibody-sandwich indirect enzyme-linked immunosorbent assay (DASI-ELISA) detection of R. solanacearum. Enrichment in MWB at both temperatures allowed us to recover R. solanacearum cells that were nonculturable on solid media up to 25 days after their entry into the viable but nonculturable state. When we applied this technique to water samples during the cold months of 2001 and 2002, we obtained the best detection results by the most-probable-number method after enrichment at 35°C with MWB. The enrichment protocol was combined with DASI-ELISA and validated by Co-PCR to detect both naturally and artificially starved and cold-stressed cells in water, which were still infective. Overall, the data from this study demonstrate the effects of temperature variation on the population and culturability of R. solanacearum cells on solid media and their survival at low temperatures.
Ralstonia solanacearum (43) is the causal agent of bacterial wilt of potatoes and many other cultivated plants (18). It represents the major limitation on the production of solanaceous crops all around the world (17), and it is a destructive pathogen (27), as more plants have been severely affected by wilt disease than by any other bacterial disease (26). In Europe, many reports have demonstrated that R. solanacearum biovar 2 has the unexpected ability to survive in waterways at low temperatures in Northern countries such as the United Kingdom (12) and The Netherlands (21, 22). The first report of the detection of R. solanacearum in river ways came from Sweden (35) and was followed by reports of different outbreaks in England in 1992 and 1996 (13) and in The Netherlands (21), Belgium, and France (20, 22) in 1995. More recently, the bacterium was also isolated in Spain (36) and Scotland (41). In most of these countries, the symptoms of bacterial wilt were observed on susceptible plants after irrigation with contaminated water. Furthermore, most of these outbreaks were related to watercourses in which infected Solanum dulcamara plants were present. In aquatic ecosystems, this weed and others such as Urtica dioica (40) have been reported to be reservoirs of the bacterial wilt pathogen (10, 21).
R. solanacearum cell survival in water for variable periods depending on the temperature and the inoculum density was first reported in 1956 (25). Recent studies (39) of the effects of temperature and other environmental factors on the survival and physiology of R. solanacearum in sterilized irrigation water have confirmed these first data. van Elsas et al. (39) demonstrated that, like many other bacterial species, this organism must also be able to cope with extended periods of low nutrient and energy availability. However, these authors and others (3, 4, 39) have also shown that the apparent disappearance of R. solanacearum from water samples during cold periods can be due to the entry of this bacterium into a viable but nonculturable (VBNC) state. This state, in which cells progressively lose the ability to form colonies on solid media yet remain viable, was first described by Xu et al. (42) and has been proposed as a survival strategy of some bacteria under adverse environmental conditions (32, 38).
Thus, the isolation of R. solanacearum biovar 2 from contaminated water is generally easy during summer, since the densities of the bacterium are relatively high and the organism is actively multiplying (12, 13), but it is difficult during winter (3, 4, 12, 13). This is probably due to the entry of this bacterium into the VBNC state and/or because the low numbers of culturable free-living cells present in the waterways are at or below the detection limits of the techniques used. This may lead to underestimations of the populations of the bacterium, with important consequences for agriculture because of the waterborne transmission of the bacterial wilt disease (4, 30, 37, 39, 40). It is also important that the current European Union Directive, 98/57/EC (2), requires isolation of the pathogen and a demonstration of its pathogenicity for official confirmation of a bacterial wilt outbreak. Therefore, methodologies that facilitate isolation when the target occurs in low numbers and/or an altered physiological state are still needed.
The aim of this study was to determine the population density and survival of R. solanacearum in one Spanish river over a period of 3 years and to improve its detection at low temperatures. We have applied an enrichment procedure that can be combined with isolation and with serological and molecular methods. When the water temperature was below 10°C, the samples were also analyzed by the most-probable-number (MPN) procedure (31), which is widely used for standard analyses of water but has not been reported previously for R. solanacearum detection. The abilities of these methods to detect starved and low-temperature-stressed cells of this bacterium were also evaluated. River water isolates recovered at low temperatures as well as starved and VBNC cells were inoculated into tomato plants for evaluation of their pathogenicity.
MATERIALS AND METHODS
Routine analysis of R. solanacearum in river water samples.
From March 2000 to August 2001, 65 water samples were collected at five different sites in the Tormes River, which lies in northwest Spain, and were analyzed for R. solanacearum detection (Fig. 1). The criterion for the selection of sampling sites was proximity to a confirmed potato outbreak of R. solanacearum in the late fall of 1999 in a field that was irrigated with such water. The potato plants were eradicated according to the instructions of European Directive 98/57/EC (2). Water samples of approximately 500 ml were collected in sterile screw-cap plastic containers as part of a routine analysis of R. solanacearum. Samples were taken at a depth of 30 to 40 cm and within 2 m of the bank and were kept at 4°C until processing, which was done within 24 h. The water temperature was routinely recorded at the sampling time, whereas the pH was measured in the laboratory. The sampling of water was discontinued in the fall and/or winter, when samples were negative for the direct isolation of R. solanacearum biovar 2 for 2 to 3 months or because of the difficulties of taking samples when the surface of the river was frozen. Culturable cells of the pathogen were detected in duplicate on modified semiselective medium, South Africa (SMSA) agar (11). To this end, 100 μl of 10-fold dilutions in sterile 10 mM phosphate-buffered saline solution, pH 7.2 (PBS), were plated onto modified SMSA agar and incubated at 29°C for 48 to 72 h. Presumptive R. solanacearum colonies were purified on yeast extract-peptone-glucose agar (YPGA) (29) before biovar determinations and were confirmed by indirect immunofluorescence (IIF) (2) and/or double-antibody-sandwich indirect enzyme-linked immunosorbent assays (DASI-ELISAs) (7) as described below.
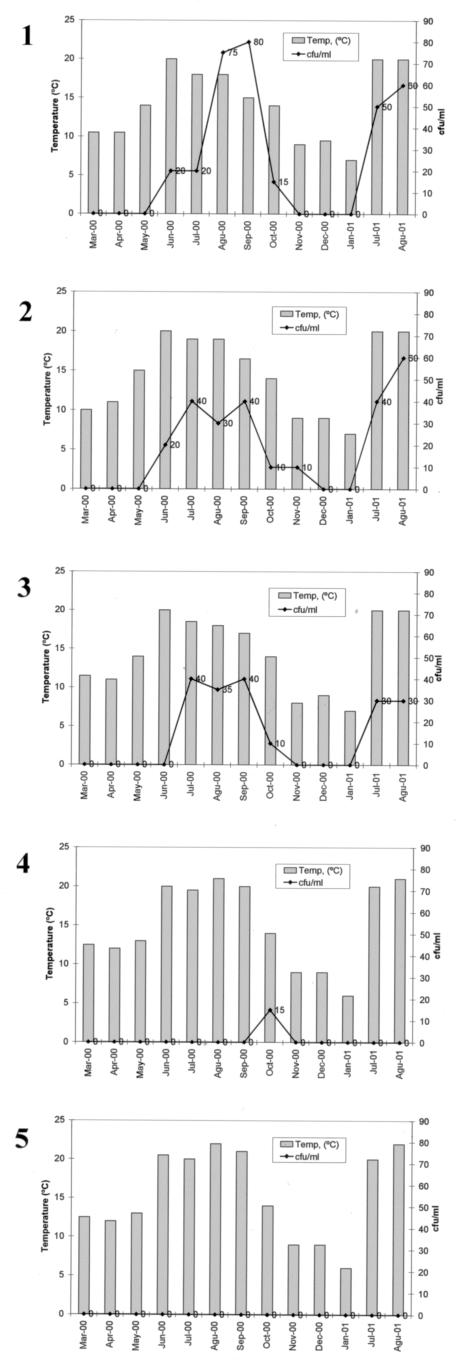
FIG.1.
Detection of R. solanacearum biovar 2 cells by direct isolation on modified SMSA medium during a survey of the Tormes river in northwest Spain from March 2000 to August 2001. Water samples were collected at five sites (1 to 5) on the river at different distances, both upstream and downstream, from a confirmed potato outbreak. Sampling sites were numbered according to their location from the first site as follows: first sample site (0 km), second site (3 km), third site (6 km), fourth site (9 km), and fifth site (12 km). The potato field was equidistantly located between sites 2 and 3. The dates indicate the sampling months during the years 2000 and 2001. The populations of R. solanacearum (from 0 to 80 CFU/ml) and the river temperatures at the time of water sampling are indicated in the figure.
Optimizing the recovery of R. solanacearum at low temperatures by enrichment. (i) Bacterial strains.
Three reference and three Spanish strains of R. solanacearum were used. In addition, 33 unidentified bacterial isolates from different Spanish freshwater sources were included (Table 1). R. solanacearum strains and non-Ralstonia water isolates were grown on YPGA at 29 and 25°C, respectively, for 48 h. The bacterial concentrations (CFU per milliliter) of R. solanacearum strains were estimated by linear regression analysis (for 109 CFU/ml, optical density at 600 nm = 0.5).
TABLE 1.
R. solanacearum strains and unknown freshwater bacterial isolates used for this study
| Species | Collectiona | Biovar | Geographic origin | Host or source |
|---|---|---|---|---|
| R. solanacearum | NCPPB 325T | 1 | United States | Lycopersicon esculentum |
| PD 2762 | 2 | Netherlands | Solanum tuberosum | |
| PD 2763 | 2 | Netherlands | Solanum tuberosum | |
| IVIA 1602.1 | 2 | Spain | Solanum tuberosum | |
| IVIA 2167 | 2 | Spain | River water | |
| IVIA 2528.55 | 2 | Spain | River water | |
| Thirty-three unidentified bacterial isolates | IVIA | Spain | Freshwater |
NCPPB, National Collection of Plant Pathogenic Bacteria, Central Science Laboratory, Sand Hutton, York, United Kingdom; PD, Collection of Plant Protection Service, Wageningen, The Netherlands; IVIA, Collection of Plant Pathogenic Bacteria, Instituto Valenciano de Investigaciones Agrarias, Moncada, Spain.
(ii) IIF.
A polyclonal 1546-H IVIA antiserum against a Spanish R. solanacearum biovar 2 strain was obtained as previously described (7). This antiserum was used for IIF according to the method of De Boer (9) to confirm the identification of pure cultures as R. solanacearum. Only staining similar to that exhibited by the homologous strain (positive control) was scored as positive. Sterile filtered PBS was used as a negative control.
(iii) DASI-ELISA.
DASI-ELISA was performed basically as described by Caruso et al. (7) by use of a detection kit supplied by Plant Print Diagnostics, Valencia, Spain, that included the specific monoclonal antibody (MAb) 8B-IVIA. Spiked and environmental water samples (see below) were assayed at least in duplicate in separate assays by the DASI-ELISA technique. The positive control was R. solanacearum biovar 2 strain PD 2762, which was used to raise MAb 8B-IVIA, and the negative control was Chryseobacterium indologenes strain P 27. Optical density values of more than twice that of the negative control were considered positive.
To exclude the possibility of cross reactions of MAb 8B-IVIA and the polyclonal 1546-H IVIA antiserum with indigenous microbiota from freshwater, we previously tested a selection of 33 bacterial isolates from different Spanish water sources (Table 1) by DASI-ELISA and IIF to determine their efficiencies in survival studies with the brown rot pathogen. All of the isolates were negative with the 8B-IVIA MAb, but a few cross reactions were observed with the 1546-H IVIA polyclonal antiserum.
(iv) PCR.
PCR detection of R. solanacearum in environmental water samples was performed according to the cooperational PCR (Co-PCR) methodology described by Olmos et al. (34) for viruses and modified by Caruso et al. (8) for R. solanacearum. The positive and negative controls were R. solanaceraum biovar 2 strain IVIA 2528.55 and sterile PBS, respectively. This technique was also used to confirm the presence of R. solanacearum in enrichment tubes and the identities of suspected R. solanacearum colonies.
Optimized recovery of R. solanacearum from spiked water samples.
To improve the recovery and detection of R. solanacearum biovar 2 from water samples at low temperatures, we performed an enrichment step at two incubation temperatures (29 and 35°C) with two selective liquid media, modified Wilbrink broth (MWB) (7) and modified SMSA broth (11). Water samples spiked with five R. solanacearum strains (Table 1) were used. R. solanacearum cultures were grown for 72 h at 29°C in YPGA and serially diluted from 107 to 1 CFU/ml in sterile PBS, and 0.5-ml aliquots were inoculated into 4.5 ml of modified SMSA broth and MWB and incubated at 29 and 35°C for 48 to 72 h with gentle shaking (125 rpm) in a Lab-line orbital shaker (model 4628). Sterile PBS was used as a negative control. All tubes that exhibited turbidity after 48 to 72 h of shaking incubation were processed by DASI-ELISA, Co-PCR, and plating on YPGA and modified SMSA agar. All assays were performed at least twice in separate experiments.
Optimized recovery of R. solanacearum from water microcosm samples.
Microcosms of 200 ml of doubly deionized water and freshwater from the river in which R. solanacearum was detected were prepared in 500-ml flasks. Water samples were filtered through 0.2-μm-pore-size membrane filters (Millipore) and autoclaved (20 min, 121°C) to remove any bacterial cells. Microcosms were inoculated with 107 CFU of R. solanacearum biovar 2 strain IVIA 1602.1 or IVIA 2528.55/ml, as previously described (4). To obtain starved cells, we maintained half of the microcosms at 25°C, and to induce the formation of nonculturable cells, we incubated the other half at 4°C (4).
Experimental microcosms were sampled at time zero and weekly, except for those that were incubated at 4°C, which were sampled every 2 days beginning on day 20. The microcosms were analyzed for culturable, viable, and total cell counts for at least 2 months. Plate counts were determined on YPGA and modified SMSA agar. When no colonies were recovered from plate counts of water microcosms that were incubated at 4°C (after plating of 1 ml), 2.5-ml aliquots were enriched in liquid medium every 2 days to determine their culturability in liquid medium. Direct viable counts were determined by a modification of the method of Kogure et al. (28), as described by van Elsas et al. (39), and total cell counts were determined by acridine orange epifluorescence as described by Oliver (33). Cells were examined at a magnification of ×1,250 with a Leitz epifluorescence microscope using blue light excitation and a 515-nm filter. All experiments were performed in duplicate in separate assays. We also evaluated whether IIF, DASI-ELISA, and Co-PCR were able to detect R. solanacearum cells in water microcosms after 2 months of starvation at 25°C and within a month in the VBNC state at 4°C. To this end, we tested aliquots of both starved and nonculturable cells directly from microcosms after 10-fold serial dilutions, using cells grown on YPGA as a positive control.
Optimized recovery of R. solanacearum biovar 2 from natural water samples at low temperatures.
Thirty-one water samples from the river Tormes from the same sites that had been sampled in 2000 were analyzed from September 2001 to March 2002 by the optimized methodology described above. Water sampling was interrupted in spring 2002 because of the dilution effect of heavy rainfalls. The numbers of river water bacteria that were culturable on YPGA were also determined. The concentration of the pathogen was also determined by filtering 10 and 100 ml of each water sample through 0.2-μm-pore-size membrane filters (Millipore). The filters were incubated in duplicate in 20 ml of MWB at 29 and 35°C. When the river water temperature was below 14°C, water samples were further analyzed by direct plating of 1 ml onto modified SMSA agar plates (15-cm-diameter) and by the three-tube MPN method (31) with MWB. For MPN testing, 10, 1, and 0.1 ml were transferred to tubes containing 10 ml of MWB (double strength for tubes inoculated with 10 ml of water samples). Samples from enrichment tubes from all experiments that exhibited turbidity after 48 to 72 h for up to 10 days of shaking incubation at 29 and 35°C were streaked onto modified SMSA agar, and Ralstonia-like colonies were processed by DASI-ELISA and Co-PCR as described above. The remaining tubes with apparently no growth for up to 10 days were also tested. The MPN of R. solanacearum cells in each water sample was estimated by determining the number of tubes in each group that yielded positive detection of R. solanacearum by direct isolation on modified SMSA agar and DASI-ELISA according to the MPN determination method described in Standard Methods for the Examination of Water and Wastewater (1). The biovars of R. solanacearum water isolates were determined as previously described (14).
Pathogenicity assays.
The pathogenicity of 13 R. solanacearum biovar 2 isolates recovered from analyzed water samples of the river Tormes at low temperatures was assayed by the use of 3-week-old tomato plants grown in pots according to a standard technique (19). In addition, starved cells from river water microcosms that were maintained for 2 months at 25°C and VBNC cells that were maintained at 4°C (in the VBNC state for about 1 month) were inoculated into tomato plants by two different methods, either stem inoculation (19) with a 10-μl aliquot of each microcosm or watering of the sterile substrate of the pots with 10 ml of the same microcosms. The population densities of culturable cells at the time of inoculation were 107 CFU/ml for microcosms at 25°C and <1 CFU/ml for those kept at 4°C. All pathogenicity assays were done in duplicate, with groups of six plants for each bacterial strain, physiological state, or way of inoculation, within a climatic room set for a photoperiod of 8 h, 26°C, and 70 to 80% humidity under quarantine conditions. The Spanish strain of R. solanacearum, IVIA 1602.1, and sterile PBS were used as positive and negative controls, respectively.
Statistical analysis.
The nonparametric Spearman coefficient was used to determine the correlation between water temperature and the numbers of R. solanacearum cells stratified by their proximity to a contaminated field. Since no R. solanacearum colonies were recovered from any of the water samples from the most distant sampling point (site 5), these null data were not included in the statistical analysis. The effect of both variables (water temperature and distance) on the cell numbers of R. solanacearum in water samples was analyzed by negative binomial regression. Data (means of two determinations) from 2 different years were analyzed separately.
RESULTS
Occurrence of R. solanacearum in a Spanish river.
The detection of R. solanacearum biovar 2 in a Spanish potato field irrigated with water from the Tormes river was the beginning of an official survey performed according to European Directive 98/57/EC (2). For this study, R. solanacearum was isolated from 22 of 65 samples analyzed (34%) from March 2000 to August 2001 (Table 2). The incidence by site as well as the temperature of water samples yielding positive isolation of the bacterial wilt pathogen at each location are shown in Fig. 1 and Table 2. The temperature range of surface water samples contaminated with R. solanacearum varied from 9 to 20°C, being quite similar for all sites analyzed (Fig. 1). Most of the water samples (95.5%) in which the pathogen was detected had temperatures above 14°C, with pathogen detection usually being unsuccessful at lower temperatures (Fig. 1 and Table 2). The populations of the pathogen during warmer months ranged from 10 to 80 CFU/ml (Table 2). The lowest temperature at which the pathogen was directly recovered on solid medium was 9°C. Figure 1 also illustrates how the densities of R. solanacearum declined as the water temperature decreased and rose as the water temperature increased. A statistical analysis of the data by the use of Spearman's correlation coefficient (rs) showed a significant positive correlation between the levels of R. solanacearum and water temperatures for sampling sites 1, 2, and 3, situated 0, 3, and 6 km, respectively, from the first sampling site (Table 3).
TABLE 2.
Incidence and populations of R. solanacearum biovar 2 in water samples by direct isolation on modified SMSA agar from March 2000 to August 2001
| Site | Distance (km)a | No. of positive samples/total no. of samples
|
Concn (CFU/ml) of bacteriumb | |
|---|---|---|---|---|
| ≥14°C | <14°C | |||
| 1 | 0 | 7/13 | 0/13 | 15-80 |
| 2 | 3 | 7/13 | 1/13 | 10-60 |
| 3 | 6 | 6/13 | 0/13 | 10-40 |
| 4 | 9 | 1/13 | 0/13 | 15 |
| 5 | 12 | 0/13 | 0/13 | 0 |
| Total | 21/65 | 1/65 | ||
Distance downstream from the first sampling site. The confirmed potato outbreak was located between sites 2 and 3 at similar distances.
Data are from analyses of water samples in duplicate and show ranges of concentrations.
TABLE 3.
Correlation between water temperature and levels of R. solanacearum biovar 2 for each sampling site from March 2000 to August 2001
| Year | Site | Distance (km)a | rsb | P value |
|---|---|---|---|---|
| 2000 | 1 | 0 | 0.827 | 0.013 |
| 2 | 3 | 0.706 | 0.034 | |
| 3 | 6 | 0.612 | 0.066 | |
| 4 | 9 | 0.400 | 0.230 | |
| 2001 | 1 | 0 | 0.814 | 0.069 |
| 2 | 3 | 0.900 | 0.044 | |
| 3 | 6 | 0.814 | 0.069 | |
| 4 | 9 | 0.043 | 0.924 |
Distance downstream from the first sampling site. The confirmed potato outbreak was located between sites 2 and 3 at similar distances.
Spearman's correlation coefficient.
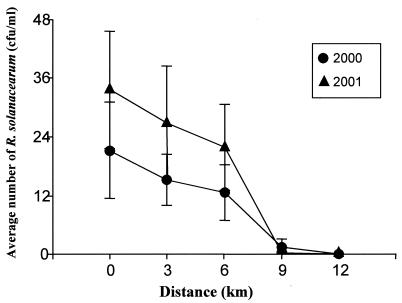
As shown in Fig. 1 and Table 2, R. solanacearum was found more frequently and at higher concentrations in water samples from sites 1 and 2, followed by site 3, and was found less frequently and at lower densities in water samples from site 4 (Fig. 2). The water samples from site 5 were always negative. Furthermore, for 2001 a regression model revealed that the levels of the pathogen in river water were significantly related to the distance from the first site (P < 0.0002), regardless of the water temperature. It also confirmed the effect of temperature on the density of R. solanacearum in water (P < 0.0001). Given that in 2000 there were few and quite variable data for each distance, the analysis did not detect differences among distances. A slight effect for temperatures was also found in this year (P < 0.01).
FIG. 2.
Relationship between mean levels of R. solanacearum biovar 2 in river water (CFU per milliliter) and proximity to the first sample site from March 2000 to August 2001, regardless of the water temperature. Water samples were collected at five sites at distances downstream of the first site of 0, 3, 6, 9, and 12 km. The detected potato outbreak was equidistant from sites 2 (3 km) and 3 (6 km). Bars on the figure represent standard errors.
Optimized recovery of R. solanacearum from spiked water samples and microcosms.
Before field studies, enrichments in modified SMSA broth and MWB at 29 and 35°C were assayed by the use of spiked river water samples to improve the recovery of low populations of R. solanacearum. Detection of the pathogen by isolation on modified SMSA agar after enrichment in these two liquid media at both temperatures was successful in all cases (data not shown), but detection by DASI-ELISA was achieved in only 74 of 120 enrichments analyzed (Table 4). However, the enriched tubes that were negative by DASI-ELISA were false negatives, since the presence of R. solanacearum was confirmed both by direct isolation on modified SMSA agar and by Co-PCR. In fact, the colonies recovered from these tubes were nonfluidal, and this type of morphology of R. solanacearum has been reported to give negative results in the DASI-ELISA protocol employed (7). When we compared the two selective media, as shown in Table 4, enrichment in modified SMSA broth gave a large number of false-negative tubes (41 of 60) by DASI-ELISA, while only 5 of 60 tubes were found to be falsely negative when MWB was used. Since MWB provided positive results for 55 of 60 DASI-ELISAs, we selected this medium for the enrichment of natural water samples. Regarding the incubation temperature, the rate of detection by DASI-ELISA was only a bit higher at 35°C (43 of 60) than at 29°C (31 of 60), and thus both temperatures were applied for analyses of environmental water samples.
TABLE 4.
Comparison of two enrichment media at two temperatures by DASI-ELISA detection of R. solanacearum biovar 2 in spiked river water samples
| Enrichment mediumb | Temp (°C) | No. of positive enrichmentsa/no. of enrichments analyzed
|
|||
|---|---|---|---|---|---|
| Assay 1 | Assay 2 | Assay 3 | Total | ||
| Modified SMSA broth | 29 | 3/10 | 0/10 | 2/10 | 5/30 |
| 35 | 6/10 | 5/10 | 3/10 | 14/30 | |
| MWB | 29 | 8/10 | 8/10 | 10/10 | 26/30 |
| 35 | 9/10 | 10/10 | 10/10 | 29/30 | |
Positive detection of R. solanacearum by DASI-ELISA. Enriched tubes that were negative for detection yielded nonfluidal colonies after plating on modified SMSA agar, which were confirmed as R. solanacearum by Co-PCR.
The numbers of culturable cells of the two Spanish R. solanacearum isolates recovered from sterile water microcosms on modified SMSA agar remained at similar levels to those at the inoculation time (approximately 107 CFU/ml) after 2 months under starvation conditions at room temperature. In contrast, in the microcosms that were maintained at 4°C, a decline in culturability on solid medium to <1 CFU/ml was observed over a period of 24 days (on average), while direct viable counts showed that approximately 90% of the cells were viable (total cell counts ranged from 7.9 × 107 to 1.1 × 107 cells/ml, while viable counts varied from 5.7 × 106 to 1.5 × 106 cells/ml). However, enrichment in MWB allowed the recovery of R. solanacearum biovar 2 cells that had been nonculturable on solid medium up to approximately 1 week after their loss of culturability on solid medium, regardless of the strain assayed, the type of water used for microcosm preparation, and the incubation temperature (Table 5). In some cases, recovery was possible even after 25 days by sampling volumes of 10 ml in triplicate (mostly in one of three enriched tubes). This was not the case for direct plating on solid SMSA medium. Recovered cells were confirmed as R. solanacearum by plating on modified SMSA agar, DASI-ELISA, and Co-PCR.
TABLE 5.
Detection of R. solanacearum biovar 2 starved and VBNC cells before and after enrichment in MWB in river water microcosms inoculated with this bacterium
| Final concn (cells/ml) | Detection resulta
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Before enrichment
|
After enrichmentb
|
|||||||
| Modified SMSA agar | IIF | DASI-ELISA | Co-PCR | Modified SMSA agar | IIF | DASI-ELISA | Co-PCR | |
| Control cells | ||||||||
| 106 | + | + | + | + | + | + | + | + |
| 105-103 | + | + | ±d | + | + | + | + | + |
| 102-10 | + | ±d | − | + | + | + | + | + |
| 1-10−2 | − | − | − | + | ND | ND | ND | + |
| Starved cells (25°C) | ||||||||
| 106 | + | + | + | + | + | + | + | + |
| 105-103 | + | + | ±d | + | + | + | + | + |
| 102-10 | + | ±d | − | + | + | + | + | + |
| 1-10−2 | − | − | − | + | ND | ND | ND | + |
| VBNC cellsc (4°C) | ||||||||
| 106 | − | + | + | + | + | + | + | + |
| 105-103 | − | + | ±d | + | ±e | + | + | + |
| 102-10 | − | ±d | − | ±f | ±e | ±e | ±e | ±f |
| 1-10−2 | − | − | − | ±f | ND | ND | ND | ±f |
The detection of stressed cells was done after 10 days to 1 month in the VBNC state. The reported detection limits of IIF, DASI-ELISA, and Co-PCR for R. solanacearum cells are 103 to 102 cells/ml, 105 cells/ml, and 10−2 cells/ml, respectively (2, 6, 7, 8).
Enrichment was done by the method of Caruso et al. (7) for 72 h at 29 and 35°C. ND, not determined.
Less than one culturable R. solanacearum cell per 0.1 ml of solid medium, but 107 cells/ml by total microscopic counts after less than 1 month in the VBNC state.
Variable results depending on the number of cells assayed.
Variable results depending on the number of cells in the VBNC state, the enriched sampling volumes, and the number of cells enriched in liquid medium.
Variable results depending on the DNA integrity of the cells.
Table 5 shows the sensitivities of detection of R. solanacearum in inoculated microcosms by serological and molecular techniques before and after the optimized enrichment step. As expected, only control and starved cells were detected by plating on modified SMSA agar before enrichment, up to 10 cells/ml. However, both starved and nonculturable cells of R. solanacearum biovar 2 were stained by IIF using the polyclonal 1546-H IVIA antibody after approximately 1 month of starvation or of being in the VBNC state. These stressed cells also gave a similar reaction by DASI-ELISA to that of nonstressed cells used as a control. Regarding PCR, both starved and VBNC cells were amplified by Co-PCR, giving a band of 408 bp. No differences were observed in the detection limits of Co-PCR for starved and control cells (10−2 cells/ml), but Co-PCR was less sensitive for nonculturable cells (1 to 103 cells/ml) (Table 5).
Optimized detection of R. solanacearum in naturally contaminated river water samples.
To determine the efficiency of recovery of R. solanacearum from environmental water samples at cold temperatures by the optimized procedure described above, we analyzed some samples at 18°C and continued the survey at the same sites at decreasing temperatures. Table 6 shows the detection results for R. solanacearum in river water at different times and temperatures, before and after enrichment at 29 and 35°C in MWB and from September 2001 to March 2002. The pHs of the water samples were between 6.5 and 7.3. Direct isolation of R. solanacearum was always possible when the water temperature was 14°C or higher. In September and October 2001, when the water temperature varied from 18 to 14°C, total culturable bacteria counts on YPGA incubated for 72 h at 25°C were about 3 × 103 CFU/ml, of which approximately 1.2% were R. solanacearum cells (from 5 to 65 CFU/ml). Starting in November, when the surface water temperature decreased to 7 to 8°C, we also analyzed the samples by plating 1 ml of water onto large petri plates. For water samples below 14°C, the lowest temperature for which the pathogen was recovered on solid medium was 8°C in November (1 CFU/ml), for only one of the five samples analyzed and by using 15-cm-diameter petri plates. Below 14°C, the numbers of mesophilic culturable bacteria also showed a tendency to decrease about 1 log unit (data not shown). All R. solanacearum-like colonies recorded were confirmed by DASI-ELISA and Co-PCR. Before enrichment, all water samples were negative for R. solanacearum detection by DASI-ELISA because the cell numbers of the pathogen were below the detection limit of this technique. Conversely, all of them were positive by Co-PCR. The detection of the pathogen was confirmed by IIF when the water temperature was above 14°C (data not shown).
TABLE 6.
Detection of R. solanacearum biovar 2 in natural river water samples at decreasing temperatures before and after up to 10 days of enrichment at 29 and 35°C in MWB. The positive enrichment of R. solanacearum was confirmed by growth on modified SMSA agar and DASI-ELISA
| Mo and yr of sampling | Water temp (°C) | Bacterial count (CFU/ml) on modified SMSA agar | No. of positive samples/no. of samples analyzed before enrichment
|
Enrichment temp (°C) | No. of positive enrichments/no. of enrichments analyzed
|
No. of MPN-positive samples/no. of samples | MPN range of R. solanacearum cells/100 ml | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Water vol sampled (ml)a
|
MPNd
|
||||||||||||
| DASI-ELISA | Co-PCR | 0.5b | 10c | 100c | 10 | 1 | 0.1 | ||||||
| September 2001 | 18 | 55-65 | 0/3 | 3/3 | 29 | 1/3 | 0/3 | 0/3 | ND | ND | ND | ND | ND |
| 35 | 3/3 | 3/3 | 3/3 | ND | ND | ND | ND | ND | |||||
| October 2001 | 14 | 5-25 | 0/3 | 3/3 | 29 | 3/3 | 0/3 | 0/3 | ND | ND | ND | ND | ND |
| 35 | 3/3 | 3/3 | 3/3 | ND | ND | ND | ND | ND | |||||
| November 2001 | 7-8 | 0-1 | 0/5 | 5/5 | 29 | 0/5 | 0/5 | 0/5 | 7/15 | 3/15 | 0/15 | 4/5 | <3-15 |
| 35 | 0/5 | 3/5 | 3/5 | 13/15 | 3/15 | 0/15 | 5/5 | 9-43 | |||||
| February 2002 | 6.4-6.9 | 0 | 0/5 | 5/5 | 29 | 0/5 | 0/5 | 0/5 | 0/15 | 0/15 | 0/15 | 0/5 | <3 |
| 35 | 0/5 | 0/5 | 0/5 | 2/15 | 0/15 | 0/15 | 1/5 | <3-9 | |||||
| March 2002 | 6.4-8.4 | 0 | 0/5 | 5/5 | 29 | 0/5 | 0/5 | 0/5 | 0/15 | 0/15 | 0/15 | 0/5 | <3 |
| 35 | 0/5 | 0/5 | 0/5 | 1/15 | 0/15 | 0/15 | 1/5 | <3-4 | |||||
Water samples were analyzed in duplicate.
Regular enrichment (1:10) in MWB.
Volume of filtered sample followed by enrichment in MWB.
Series of three tubes per sample volume. ND, not determined.
The enrichment of various water volumes in MWB at 29°C was less efficient than direct plating of water samples at temperatures of about 14°C or higher in September and October 2001 (Table 6), even after the filter-mediated concentration of 100 ml of water, probably because of the overgrowth of competitor bacteria and/or the presence of lytic phages in the river (30). In contrast, the enrichment at 35°C increased R. solanacearum populations in water samples and thus the detection sensitivity by DASI-ELISA.
When the water temperature was below 14°C (ranging from 6.4 to 8.4°C), from November 2001 to March 2002, the samples were further analyzed by the MPN method because they were negative for the detection of R. solanacearum by plate counts, except for one sample from November, and because the regular enrichment (1:10) in MWB was also unsuccessful. The enrichment of membrane filters in MWB only gave positive detection when it was performed at 35°C (for 3 of 15 water samples collected at ≤8°C), regardless of the volume of water that was filtered (10 or 100 ml) (Table 6). The best detection results were obtained by the MPN procedure performed at 35°C, since 7 of 15 water samples were positive after enrichment in MWB. In November 2001, 13 of 15 MPN sample tubes of 10 ml were positive after enrichment at 35°C compared to 7 positive tubes obtained at 29°C, while in February and March 2002 R. solanacearum was only detected by the MPN method at 35°C (Table 6). Furthermore, the most probable number of R. solanacearum cells per 100 ml of water was also higher for the enrichment performed at 35°C (<3 to 43 cells/100 ml) than for that performed at 29°C (<3 to 15 cells/100 ml). The R. solanacearum-like colonies obtained were confirmed as described above. Surprisingly, the morphology of the colonies after enrichment at 35°C was typically fluidal, according to Kelman's definition (24), while those recovered after enrichment at 29°C were mainly nonfluidal but were identified as R. solanacearum by Co-PCR. They were classified as biovar 2.
Assays for pathogenicity of the 13 R. solanacearum biovar 2 river water isolates recovered after enrichment and the MPN procedure for young tomato plants gave positive results, regardless of the river temperature at the time of water sampling. Furthermore, cells that were starved for 2 months in river water microcosms at 25°C were also able to produce wilting in 11 of 12 tomato plants that were inoculated via the stem and in 8 of 12 plants after inoculation by watering. No disease symptoms were observed in plants that were inoculated in the stem with low-temperature-stressed cells after 1 month in the VBNC state, while 1 of 12 plants that were irrigated with 10 ml of these same cells showed wilting symptoms. Ralstonia-like colonies were isolated from all affected tomato plants and were confirmed to be R. solanacearum.
DISCUSSION
An investigation of the population densities of R. solanacearum during the first 2 years of analyses at five sites on a Spanish river associated with an outbreak of bacterial wilt in potato plants has demonstrated that the isolation of this pathogen on solid media is correlated with the water temperature, while previous studies have only suggested a relationship that was not statistically confirmed (12, 21). In the analyses of 2000, R. solanacearum began to be detected in river water mostly in early July, when the temperature rose above 14°C. The abundance of this bacterium was relatively higher (from 30 to 80 CFU/ml) in August and/or September and then dramatically decreased until its apparent disappearance in November, when water temperatures were below 14°C. In 2001, R. solanacearum was detectable by plating in summer and again became nondetectable in November. These results agree with previous studies in the United Kingdom and The Netherlands, where the levels of this bacterium in waterways were usually 10 cells/ml, dropping below the detection limit when exposed to cold temperatures (12, 22). Higher densities (103 cells/ml) have been described in the summer in The Netherlands, probably due to the proximity of the sampled sites to industrial plants for potato processing (21).
Surprisingly, the populations of R. solanacearum biovar 2 in the Spanish river were not highest in the proximity of the potato field where the outbreak of brown rot was first detected. In contrast, the results indicated that the source of the infection came from water upstream of the outbreak site, since from the first sampling site the pathogen concentration decreased, regardless of the water temperature. It has been described that the persistence of this pathogen in natural water is strongly affected by other members of the aquatic microbiota (39), and we have already reported the isolation of R. solanacearum lytic phages from the same river (30).
Two techniques that were developed in our laboratory were applied to improve the detection of R. solanacearum from water at low temperatures (enrichment-ELISA and Co-PCR). Enrichment in modified SMSA broth at 29°C has been recommended for the examination of low numbers of R. solanacearum cells in asymptomatic potato tubers and water samples (2, 13, 21). We recently reported that enrichment in MWB at 29°C improved ELISA detection of the target in potato extracts (7), but its efficiency for water samples had not been investigated. A comparative evaluation of these two liquid media by the use of spiked water samples showed that MWB is superior to modified SMSA broth for DASI-ELISA detection of the brown rot pathogen. Furthermore, MWB favored the multiplication of typical fluidal cells of the pathogen. Moreover, to prevent the overgrowth of competitor aquatic bacteria observed at 29°C, we also assayed enrichment at 35°C. This temperature was selected because R. solanacearum can grow at temperatures up to 39°C (16), and temperatures of 35°C or higher rarely occur in temperate aquatic environments (38). A comparative study of these two enrichment temperatures showed a few more positive results by DASI-ELISA detection at 35°C than at 29°C, but the reason for this is still under study.
On the other hand, currently the detection of R. solanacearum in aquatic ecosystems is mainly based on cultivation methods, but this bacterium can lose its culturability on solid media (VBNC state) in response to winter temperatures (4, 39). In our study, starved cells of R. solanacearum in river water microcosms at 25°C were able to survive in a culturable state for at least 2 months, while those held at 4°C became nonculturable within 1 month according to our previous data (3) and those of van Elsas et al. (39), who used a Dutch strain. However, these VBNC cells with altered requirements are able to respond to the addition of nutrients in a liquid form (28). In fact, growth in liquid media by the MPN method has been applied by several authors as an alternative for quantifying viability (23, 38), but this approach has not been assayed before for the recovery of stressed cells of R. solanacearum. By using this procedure, we were able to isolate on modified SMSA agar R. solanacearum cells that had been nonculturable for up to 25 days in a water microcosm. However, the recovery of these VBNC cells was possible within a limited period of time and also depended on the sampled volume, suggesting that it was due to regrowth rather than resuscitation, as stated by Kell et al. (23). In addition, while direct isolation methods only allow the detection of culturable cells on solid medium, the DASI-ELISA and Co-PCR assay applied here showed their usefulness for survival and epidemiological studies of starved and VBNC cells.
When such methodologies were applied to the detection of R. solanacearum in naturally contaminated river water during the colder months in 2001 and 2002, we confirmed the laboratory data obtained with spiked water samples. Enrichment in MWB at 35°C was found to be superior to enrichment at 29°C for recovering R. solanacearum cells in low populations at cold temperatures, while direct isolation was preferred when the water temperature exceeded 14°C. Furthermore, when water samples were collected at 7 to 8°C, isolation was only successful when they were enriched in MWB after filter concentration or by the MPN method, with enrichment at 35°C also being more efficient than that at 29°C. Incubation at 35°C probably prevents the overgrowth of native river water bacteria and/or the activity of lytic phages that are present in the river (30). These results demonstrated the efficiency of the optimized enrichment in liquid medium.
Although filtration has been recommended to increase the sensitivity of detection of R. solanacearum (13, 22), during the winter season of 2001 to 2002 the recovery of the bacterium from water samples was unsuccessful after filter concentration and became progressively more difficult even after enrichment by the MPN method. This may have been due to the concentration of competitor aquatic bacteria and/or other factors when the target occurs in an altered physiological state. However, when the water temperature became favorable again, R. solanacearum also became detectable by plating on modified SMSA agar. Whether these cells came from the multiplication of a few surviving cells, the recovery of some VBNC cells, and/or alternative reservoirs must still be determined. It has been hypothesized that this bacterium can overwinter on the roots of the semiaquatic weed S. dulcamara rather than persist freely in water (13, 21, 40). This plant was present in the sampled river, and R. solanacearum was isolated from it at different times. The bacterium was found more frequently in plants close to the sites with higher levels of the pathogen (unpublished data). This supports our hypothesis that the infection source came from river water upstream of the detected potato outbreak.
Pathogenicity experiments with R. solanacearum strains isolated from river water confirmed that naturally starved cells and cells stressed by low temperatures are able to cause infections in tomato plants, including starved cells maintained in river water microcosms for 2 months at 25°C. Furthermore, starved cells were able to infect tomato plants even after being kept under these conditions for 1 year (5). VBNC cells were mostly avirulent, but in one case they were able to reproduce the symptoms of the disease after inoculation by watering. These results agree with those of Grey and Steck (15), who found wilted tomatoes after planting them in a soil with VBNC cells of R. solanacearum. Thus, the maintenance of the pathogenicity of R. solanacearum under adverse conditions supports the results of previous studies and practical observations showing the importance of freshwater as a reservoir and a vehicle for the transmission of the pathogen, even during colder months.
In conclusion, this is one of the first comprehensive studies of the seasonal variation in brown rot pathogen abundance at different sites on a contaminated river during a 3-year period, and it shows the effects of temperature on R. solanacearum populations. Since the presence of biovar 2 of R. solanacearum in waterways is a threat to crops, even at low temperatures, we have optimized an enrichment assay with MWB at 35°C by use of the MPN procedure that can be combined with DASI-ELISA and validated by Co-PCR to detect both starved and stressed cells in watercourses during colder months. Both methodologies have shed light on the hidden life of R. solanacearum biovar 2 under the oligotrophic conditions of aquatic ecosystems and thus will help us to develop new control strategies.
Acknowledgments
P. Caruso thanks the Instituto Valenciano de Investigaciones Agrarias for a Ph.D. grant, and E. G. Biosca thanks the Ministerio de Ciencia y Tecnología of Spain for a contract within the Ramón y Cajal programme. This work was funded by the following projects: FAIR 5-CT97-3632 and QLK 3-CT-2000-01598 of the European Union and FD 1997-2279 of the Ministerio de Educación y Ciencia of Spain.
We thank the Consejería de Agricultura de Castilla-León for collecting river water samples, E. Carbonell and J. Pérez Panadés for statistical analysis, and E. Marco-Noales for helping with pathogenicity assays and critical review of this article. We are also grateful to J. Janse and J. Elphinstone for sending reference strains and to J. Elphinstone, D. Caffier, and J. van Vaerenbergh for supplying unpublished data. We also thank Campbell College for English language revisions.
REFERENCES
- 1.American Public Health Association. 1992. 9221 C. Estimation of bacterial density, p. 9-45-9-52. In A. E. Greedberg, L. S. Clesceri, and A. D. Eaton (ed.), Standard methods for the examination of water and wastewater, 18th ed. American Public Health Association, Washington, D.C.
- 2.Anonymous. 1998. Council Directive 98/57/EC of 20 July 1998 on the control of Ralstonia solanacearum (Smith) Yabuuchi et al. Off. J. Eur. Communities L235/1:21.08.98. [Google Scholar]
- 3.Biosca, E. G., P. Caruso, B. Álvarez, E. Marco-Noales, and M. M. López. 2002.. Supervivencia de R. solanacearum biovar 2 en agua: inducción del estado viable no cultivable (VBNC) a bajas temperaturas, p. 38. Abstr. 11th Congr. Soc. Española Fitopatol., Almería, Spain.
- 4.Biosca, E. G., P. Caruso, E. Bertolini, B. Álvarez, J. L. Palomo, M. T. Gorris, and M. M. López. Improved detection of Ralstonia solanacearum in culturable and VBNC state from water samples at low temperatures. In C. Allen, P. Prior, and C. Hayward (ed.), Bacterial wilt disease and the Ralstonia solanacearum species complex, in press. APS Press, St. Paul, Minn.
- 5.Biosca, E. G., E. Marco-Noales, and M. M. López. Unpublished results.
- 6.Caruso, P., P. Llop, J. L. Palomo, P. Garcia, C. Morente, and M. M. López. 1998. Evaluation of methods for detection of potato seed contamination by Ralstonia solanacearum, p. 128-132. In P. Prior, C. Allen, and J. Elphinstone (ed.), Bacterial wilt disease. Molecular and ecological aspects. Springer-Verlag, Heidelberg, Germany.
- 7.Caruso, P., M. T. Gorris, M. Cambra, J. L. Palomo, J. Collar, and M. M. López. 2002. Enrichment double-antibody sandwich indirect enzyme-linked immunosorbent assay that uses a specific monoclonal antibody for sensitive detection of Ralstonia solanacearum in asymptomatic potato tubers. Appl. Environ. Microbiol. 68:3634-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caruso, P., E. Bertolini, M. Cambra, and M. M. López. 2003. A new and sensitive co-operational polymerase chain reaction (Co-PCR) for a rapid detection of Ralstonia solanacearum in water. J. Microbiol. Methods 55:257-272. [DOI] [PubMed] [Google Scholar]
- 9.De Boer, S. H. 1990. Immunofluorescence. Bacteria, p. 295-298. In R. Hampton, E. Ball, and S. De Boer (ed.), Serological methods for detection and identification of viral and bacterial plant pathogens. APS Press, St. Paul, Minn.
- 10.Elphinstone, J. G. 1996. Survival and possibility for extinction of Pseudomonas solanacearum (Smith) Smith in cool climates. Potato Res. 39:403-410. [Google Scholar]
- 11.Elphinstone, J. G., J. Hennessy, J. K. Wilson, and D. E. Stead. 1996. Sensitivity of different methods for the detection of Ralstonia solanacearum in potato tuber extracts. Bull. OEPP 26:663-678. [Google Scholar]
- 12.Elphinstone, J. G., H. M. Stanford, and D. E. Stead. 1998. Survival and transmission of Ralstonia solanacearum in aquatic plants of Solanum dulcamara and associated surface water in England. Bull. OEPP 28:93-94. [Google Scholar]
- 13.Elphinstone, J. G., H. M. Stanford, and D. E. Stead. 1998. Detection of Ralstonia solanacearum in potato tubers, Solanum dulcamara and associated irrigation water extracts, p. 133-139. In P. Prior, C. Allen, and J. Elphinstone (ed.), Bacterial wilt disease. Molecular and ecological aspects. Springer-Verlag, Heidelberg, Germany.
- 14.French, E. R., L. Gutarra, P. Alley, and J. Elphinstone. 1995. Culture media for Pseudomonas solanacearum isolation, identification and maintenance. Fitopatologia 30:126-130. [Google Scholar]
- 15.Grey, B. E., and T. R. Steck. 2001. The viable but nonculturable state of Ralstonia solanacearum may be involved in long-term survival and plant infection. Appl. Environ. Microbiol. 67:3866-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayward, A. C. 1976. Systematics and relationship of Pseudomonas solanacearum, p. 6-21. In L. Sequeira and A. Kelman (ed.), Proceedings of the 1st International Conference and Workshop on the Ecology and Control of Bacterial Wilt Caused by Pseudomonas solanacearum. North Carolina State University, Raleigh, N.C.
- 17.Hayward, A. C. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29:65-87. [DOI] [PubMed] [Google Scholar]
- 18.Hayward, A. C. 1994. The hosts of Pseudomonas solanacearum, p. 9-24. In A. C. Hayward and G. L. Hartman (ed.), Bacterial wilt: the disease and its causative agent, Pseudomonas solanacearum. CAB International, Wallingford, United Kingdom.
- 19.Janse, J. D. 1988. A detection method for Pseudomonas solanacearum in symptomless potato tubers and some data on its sensitivity and specificity. Bull. OEPP 18:343-351. [Google Scholar]
- 20.Janse, J. D. 1996. Potato brown rot in western Europe—history, present occurrence and some remarks on possible origin, epidemiology and control strategies. Bull. OEPP 26:679-695. [Google Scholar]
- 21.Janse, J. D., F. A. X. Araluppan, J. Schans, M. Wenneker, and W. Westerhuis. 1998. Experiences with bacterial brown rot Ralstonia solanacearum biovar 2, race 3, in The Netherlands, p. 147-152. In P. Prior, C. Allen, and J. Elphinstone (ed.), Bacterial wilt disease. Molecular and ecological aspects. Springer-Verlag, Heidelberg, Germany.
- 22.Janse, J. D., and J. Schans. 1998. Experiences with the diagnosis and epidemiology of bacterial brown rot (Ralstonia solanacearum) in The Netherlands. Bull. OEPP 28:65-67. [Google Scholar]
- 23.Kell, D. B., A. S. Kaprelyants, D. H. Weichart, C. R. Harwood, and M. R. Barer. 1998. Viability and activity in readily culturable bacteria: a review and discussion of the practical issues. Antonie Leeuwenhoek 73:169-187. [DOI] [PubMed] [Google Scholar]
- 24.Kelman, A. 1954. The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on a tetrazolium medium. Phytopathology 44:693-695. [Google Scholar]
- 25.Kelman, A. 1956. Factors influencing viability in cultures of Pseudomonas solanacearum. Phytopathology 46:16-17. [Google Scholar]
- 26.Kelman, A. 1998. One hundred and one years of research on bacterial wilt, p. 1-5. In P. Prior, C. Allen, and J. Elphinstone (ed.), Bacterial wilt disease. Molecular and ecological aspects. Springer-Verlag, Heidelberg, Germany.
- 27.Kelman, A., G. L. Hartman, and C. Hayward. 1994. Introduction, p. 1-7. In A. C. Hayward and G. L. Hartman (ed.), Bacterial wilt: the disease and its causative agent, Pseudomonas solanacearum. CAB International, Wallingford, United Kingdom.
- 28.Kogure, K., U. Simidu, and N. Taga. 1979. A tentative direct microscopic method for counting living marine bacteria. Can. J. Microbiol. 25:347-352. [DOI] [PubMed] [Google Scholar]
- 29.Lelliot, R. A., and D. E. Stead. 1987. Methods for the diagnosis of bacterial diseases of plants, p. 216. In T. F. Preece (ed.), Methods in plant pathology, vol. 2. Blackwell Scientific Publications, Oxford, United Kingdom. [Google Scholar]
- 30.López, M. M., and E. G. Biosca. Potato bacterial wilt management: new prospects for an old problem. In C. Allen, P. Prior, and C. Hayward (ed.), Bacterial wilt disease and the Ralstonia solanacearum species complex, in press. APS Press, St. Paul, Minn.
- 31.McCrady, M. H. 1915. The numerical interpretation of fermentation tube results. J. Infect. Dis. 17:183. [Google Scholar]
- 32.McDougald, D., S. A. Rice, D. Weichart, and S. Kjelleberg. 1998. Nonculturability: adaptation or debilitation? FEMS Microbiol. Ecol. 25:1-9. [Google Scholar]
- 33.Oliver, J. D. 1987. Heterotrophic bacterial populations of the Black Sea. Biol. Oceanogr. 4:83-97. [Google Scholar]
- 34.Olmos, A., E. Bertolini, and M. Cambra. 2002. Simultaneous and co-operational amplification (Co-PCR): a new concept for detection of plant viruses. J. Virol. Methods 106:15-23. [DOI] [PubMed] [Google Scholar]
- 35.Olson, K. 1976. Experiences of brown rot caused by Pseudomonas solanacearum (Smith) Smith in Sweden. Bull. OEPP 6:199-207. [Google Scholar]
- 36.Palomo, J. L., P. Caruso, M. T. Gorris, M. M. López, and P. García-Benavides. 2000. Comparación de métodos de detección de Ralstonia solanacearum en aguas superficiales, p. 120. Abstr. 10th Congr. Soc. Española Fitopatol., Valencia, Spain.
- 37.Person, P. 1998. Successful eradication of Ralstonia solanacearum from Sweden. Bull. OEPP 28:113-119. [Google Scholar]
- 38.Roszak, D. B., and R. R. Colwell. 1987. Survival strategies of bacteria in the natural environment. Microbiol. Rev. 51:365-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Elsas, J. D., P. Kastelein, P. M. de Vries, and L. S. van Overbeek. 2001. Effects of ecological factors on the survival physiology of Ralstonia solanacearum bv. 2 in irrigation water. Can. J. Microbiol. 47:1-13. [DOI] [PubMed] [Google Scholar]
- 40.Wenneker, M., M. S. W. Verdel, R. M. W. Groeneveld, C. Kempenaar, A. R. van Beuningen, and J. D. Janse. 1999. Ralstonia (Pseudomonas) solanacearum race 3 (biovar 2) in surface water and natural weed hosts: first report on stinging nettle (Urtica dioica). Eur. J. Plant Pathol. 105:307-315. [Google Scholar]
- 41.Wood, J. R., K. Breckenridge, and J. M. Chard. 2002. Eradicating Ralstonia solanacearum from Scottish rivers, p. 31. Abstr. 3rd Int. Bacterial Wilt Symp. (IBWS), Nelspruit, South Africa.
- 42.Xu, H., N. Roberts, F. Singleton, R. Atwell, D. Grimes, and R. Colwell. 1982. Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. FEMS Microbiol. Ecol. 8:313-323. [DOI] [PubMed] [Google Scholar]
- 43.Yabuuchi, E., Y. Kosako, I. Yano, H. Hotta, and Y. Nishiuchi. 1995. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov. Proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol. Immunol. 39:897-904. [DOI] [PubMed] [Google Scholar]