Abstract
Wine yeast starters that contain a mixture of different industrial yeasts with various properties may soon be introduced to the market. The mechanisms underlying the interactions between the different strains in the starter during alcoholic fermentation have never been investigated. We identified and investigated some of these interactions in a mixed culture containing two yeast strains grown under enological conditions. The inoculum contained the same amount (each) of a strain of Saccharomyces cerevisiae and a natural hybrid strain of S. cerevisiae and Saccharomyces uvarum. We identified interactions that affected biomass, by-product formation, and fermentation kinetics, and compared the redox ratios of monocultures of each strain with that of the mixed culture. The redox status of the mixed culture differed from that of the two monocultures, showing that the interactions between the yeast strains involved the diffusion of metabolite(s) within the mixed culture. Since acetaldehyde is a potential effector of fermentation, we investigated the kinetics of acetaldehyde production by the different cultures. The S. cerevisiae-S. uvarum hybrid strain produced large amounts of acetaldehyde for which the S. cerevisiae strain acted as a receiving strain in the mixed culture. Since yeast response to acetaldehyde involves the same mechanisms that participate in the response to other forms of stress, the acetaldehyde exchange between the two strains could play an important role in inhibiting some yeast strains and allowing the growth of others. Such interactions could be of particular importance in understanding the ecology of the colonization of complex fermentation media by S. cerevisiae.
Traditionally, indigenous yeast populations were used in the alcoholic fermentation step of wine making. Due to their strong resistance to ethanol, Saccharomyces cerevisiae strains usually predominate until the later stages of fermentation. In the last 20 years, however, winemakers have begun to use pure S. cerevisiae strains in the form of active dry yeast (ADY) starters. This process allows better control of fermentation and reduces the risk of organoleptic effects resulting from the growth and metabolism of other indigenous yeasts. In some cases, wine produced with pure yeast monocultures lacks the complexity of taste and other desirable characters that originate from the indigenous yeasts (16, 35, 51). The incorporation of several wine yeast strains with different technological capabilities into the same ADY starter may help overcome these shortcomings.
Metabolic interactions in mixed strain bacterial cultures and between fungi and bacteria have been identified (12, 26, 30, 41, 43, 47), but studies of such interactions in mixed yeast strain cultures are not common. In one study of mixed strain cultures for enological purposes, an exchange of metabolites between strains was observed (20). A mathematical model also is available to simulate growth in a mixed culture of strains of S. cerevisiae with and without the KII killer toxin (28, 31), but this model is limited to the interactions due to the toxic effect of the killer toxin.
The impact of cofermentation of S. cerevisiae with other yeast species on the final organoleptic balance of the wine also has been studied. Most of these studies focused on the effects of sequential inoculation with these yeasts on the wine produced, since the growth of most indigenous non-Saccharomyces species is limited on fermentation media and is rapidly inhibited by ethanol (3, 13, 18, 29, 44, 50). In all of these studies, the analysis focused on the fermentation products and not the fermentation kinetics. Interactions between Saccharomyces strains may occur in mixed cultures (7, 20), but the underlying mechanisms have not been investigated in detail.
The objective of the present study is to understand the effect of metabolite diffusion between partners of yeast cocultures, with a special emphasis on the effect on fermentation kinetics, by-product formation, and yeast persistence in the fermentation medium. Our working hypothesis is that metabolite diffusion may occur between two yeast strains, in this case a strain of S. cerevisiae and a natural S. cerevisiae-Saccharomyces uvarum hybrid (24), with different physiological properties during alcoholic fermentations under enological conditions. The knowledge of the underlying mechanism(s) responsible for this symbiotic interaction could be of particular importance in understanding the process through which indigenous S. cerevisiae strains colonize complex fermentation media, for explaining the observed persistence of indigenous S. cerevisiae strains during fermentations inoculated with pure Saccharomyces starter cultures, and for the design of more efficient starter cultures.
MATERIALS AND METHODS
Yeast strains.
S. cerevisiae strain D254 9a2 is a spontaneous mutant of the D254 industrial wine yeast (Lallemand, Montreal, Canada) and is resistant to the mitochondrial inhibitors erythromycin and diuron. This strain is only available on demand for scientific purposes. Strain S6U is a natural hybrid of S. cerevisiae and S. uvarum selected by the Institute of Enology (Velletri, Italy) and available commercially as a dry yeast (Lallemand).
Culture media.
All media were heat sterilized (at 110°C for 20 min). The yeast strains were grown in a standard nutrient medium, YPD [10 g of yeast extract (Difco, Detroit, Mich.) per liter, 20 g of Bacto peptone (Difco) per liter, and 50 g of glucose per liter]. A synthetic fermentation medium (SM), pH 3.3, was used to simulate standard grape juice (5). This medium contained (per liter) 200 g of glucose, 6 g of citric acid, 6 g of dl-malic acid, 750 mg of KH2PO4, 500 mg of K2SO4, 250 mg of MgSO4 · 7H2O, 155 mg of CaCl2 · 2H2O, 200 mg of NaCl, 4 mg of MnSO4 · H2O, 4 mg of ZnSO4, 1 mg of CuSO4 · 5H2O, 1 mg of KI, 400 μg of CoCl2 · 6H2O, 1 mg of H3BO3, 1 mg of (NH4)6Mo7O24 · 2H2O, 20 mg of myo-inositol, 2 mg of nicotinic acid, 1.5 mg of calcium pantothenate, 250 μg of thiamine-HCl, 0.25 mg of pyridoxine-HCl, 3 μg of biotin, and 300 mg of NH4Cl. A selection medium (N-E-D) was used to differentiate between the two yeast populations in the mixed culture. This medium contained (per liter) 10 g of yeast extract (Difco), 20 g of Bacto peptone (Difco), 20 g of glycerol, and 10 g of agar in Sörensen buffer (50 mM Na2HPO4 and 50 mM NaH2PO4, pH 6.25). Following sterilization, this medium was supplemented with final concentrations of 2 g of erythromycin (in 100% ethanol) per liter and 8 mg of diuron (in 100% acetone) per liter. In the absence of diuron and erythromycin, the selection medium is referred to as N medium.
Growth conditions.
Yeast cells were precultured in 50 ml of YPD medium at 28°C without agitation for 24 h in 200-ml Erlenmeyer flasks. The fermentation medium was inoculated with 106 yeast cells ml−1. Yeast cells were cultured anaerobically in handmade glass fermentors (working volume, 1.1 liters) fitted with fermentation locks (CO2-bubbling outlets filled with water). Fermentations were carried out under isothermal conditions (24°C) with continuous magnetic stirring (500 rpm).
Cell number, viability, and culture composition.
Cells were sonicated (30 s, 10 W), and cell number and volume were determined by using an electronic particle counter (model ZB2; Beckman-Coulter, Margency, France) fitted with a 100-μm-pore-size probe. Cell viability was determined by plating approximately 250 cells (the actual number was determined by electronic particle counting) on petri dishes containing YPD medium supplemented with 20 g of agar per liter. These petri dishes were incubated at 28°C for 60 h, and the numbers of colonies were counted. The relative proportion of the two strains in the mixed cultures was determined by plating approximately 250 cells (the actual number was determined by electronic particle counting) on petri dishes containing either N or N-E-D medium. Both strains could grow on the N medium, but only D254 9a2 could grow on the N-E-D medium.
Fermentation kinetics.
The amount of CO2 released was calculated from automatic measurements (taken every 20 min) of fermenter weight (39). Loss of ethanol and water due to CO2 stripping accounted for less than 2% of the total weight loss. The rate of CO2 production was calculated by polynomial smoothing of the last 10 fermenter weight measurements. The frequency of the measurements of fermenter weight and the precision of the balance (accurate to ±0.01 g) allowed the rate of CO2 production to be calculated with a high degree of precision and reproducibility [coefficient of variation for d(CO2/dt)max = 0.8% (39)]. The fermentation progress, FP, was calculated from the amount of CO2 released from the culture medium according to the following equation:
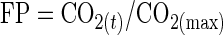 |
where CO2(t) is the cumulative amount of CO2 released at any time t, and CO2(max) is the total amount of CO2 released by the time fermentation is complete. The use of fermentation progress instead of fermentation time allows the fermentation kinetics, which are closely linked to the disappearance of substrate from the medium, to be normalized. Fermentation was considered complete when the medium contained <2 g of glucose per liter. None of the fermentations were stopped if they were <99% complete.
Identification of fermentation by-products.
Residual sugars and several organic acids were measured by high performance liquid chromatography according to the method of Reyes et al. (34). The analysis was performed on an Aminex HPX87-H column (Bio-Rad) at 45°C. The isocratic mobile phase was 0.008 N sulfuric acid in degassed, deionized water filtered through a membrane filter (pore diameter, 0.45 μm). The flow rate was 0.6 ml min−1 at a pressure of 70 × 105 Pa. Sugars and organic acids were detected simultaneously by refractometry (Agilent G1362A 1100 series; Hewlett Packard, Wilmington, Del.) and UV spectroscopy (280 nm) (Agilent G1314A 1100 series; Hewlett Packard), respectively. Measurements were calibrated with the following standard solution: 1.4% (vol/vol) ethanol, 20 g of glucose per liter, 7.2 g of malic acid per liter, 0.95 g of acetic acid per liter, 0.96 g of succinic acid per liter, and 8.4 g of glycerol per liter. The supernatants of the fermentation media were filtered and diluted at least five times with the mobile phase before analysis. Acetaldehyde dehydrogenase activity was assayed and used to estimate the acetaldehyde concentration. The reaction mixture contained 0.9 mM NAD, 50 mM KH2PO4, and 4.3 mM dithiothreitol in 100 mM triethanolamine buffer (pH 8.0). The reaction was initiated by the addition of 0.4 U of acetaldehyde dehydrogenase. The amount of NADH produced by the reaction was detected by fluorescence spectrophotometry (excitation wavelength, 340 nm; emission wavelength, 460 nm) with a Perkin Elmer LS 50B fluorescence spectrophotometer.
Rapid extraction of metabolites by quenching in cold methanol.
Metabolites were extracted as described by Gonzalez et al. (19): 10 ml of yeast culture was added to 26 ml of an ice-cold solution containing 60% (vol/vol) methanol and 70 mM HEPES (pH 7.5) and kept at −80°C until further use. The mixture was centrifuged at 5,000 × g for 30 s at −10°C. Cell pellet metabolites were extracted with 5 ml of a solution of 75% (vol/vol) boiling absolute ethanol containing 0.25 M HEPES (pH 7.5) and incubated for 5 min at 80°C. Extracts were placed on ice for 3 min and then dried for 5 min under vacuum at 70°C in a rotating evaporator (model Laborota 4000; Heidolph Instruments LLC, Cinnaminson, N.J.). The resulting residue was resuspended in a final volume of 1 to 2 ml of distilled water and stored at −80°C until use.
Measurements of intracellular NAD(H) and NADP(H) concentrations.
Metabolite concentrations were determined from NADH- or NADPH-coupled enzyme assays as described below. The amount of NADH or NADPH produced during the reaction was determined by fluorescence spectrophotometry (excitation wavelength, 340 nm; emission wavelength, 460 nm) with a Perkin Elmer LS 50B fluorescence spectrophotometer. Unless otherwise stated, enzyme assays were performed at 30°C in a reaction buffer containing 5 mM Tris-NH4Cl (pH 7.0), 0.5 mM dihydroxyacetone phosphate, and 0.5 mM α-ketoglutarate, as described by Klingenberg (25). Aliquots of 50 to 300 μl of the extracted metabolite samples were added to 1.9 ml of the reaction buffer, and a baseline reading was obtained. Twenty microliters of a 0.2 mM NADH standard solution and 20 μl of a 0.2 mM NADPH standard solution were added to give a final molarity of 4 nmol of each cofactor. NADH oxidation was initiated by the addition of 10 μl of glycerol-3-phosphate dehydrogenase (170 U ml−1; Roche catalogue no. 127 124). NADPH oxidation was initiated by the addition of 10 μl of NADPH-dependent glutamate dehydrogenase (240 U ml−1; Roche catalogue no. 127 734). NAD concentration was determined as described by Bergmeyer (6). The reaction buffer contained 0.2 M glycine (pH 9.0), 0.4 M hydrazine hydrate, and 0.2 M ethanol. The reaction was initiated by the addition of 10 μl of alcohol dehydrogenase (882 U ml−1; Roche catalogue no. 127 540). NADP concentration was determined as described by Holzer et al. (22). The reaction mixture contained 10 mM Tris-MgSO4 (pH 7.0) and 5 mM glucose-6-phosphate. The reaction was initiated by the addition of 10 μl of glucose-6-phosphate dehydrogenase (70 U ml−1; Roche catalogue no. 127 671). Cell sizes were similar in both strains studied (data not shown). Cell metabolite concentration (in millimolars) was calculated by using an estimated internal volume of 4.5 μl/108 cells for both strains (40). Metabolite extractions and measurements were performed in triplicate.
Measurement of cellular NAD(P)H fluorescence.
Cellular NADH (and NADPH) fluorescence was determined with a method adapted from that of Beauvoit et al. (4). Cells were harvested from a 2-ml culture sample and immediately suspended in a final volume of 2 ml of CMP buffer (31 mM citric acid, 45 mM dl-malic acid, 10 mM KH2PO4 [pH 3.3]). The fluorescence of NADH (bound and free forms) was monitored at 30°C with a fluorescence spectrophotometer (Perkin Elmer LS 50B). The excitation wavelength was 340 nm, and fluorescence was continuously monitored at 460 nm. Measurements were calibrated by determining the fluorescence of cells in a fully oxidized state (high NAD to NADH ratio) and of those in a fully reduced state (low NAD to NADH ratio). As previously described (4, 36), the fully oxidized state was obtained by adding an excess of acetaldehyde (3.5 mM), and the fully reduced state was obtained by adding an excess of ethanol (50 mM) and glucose (50 mM).
RESULTS
Effect of mixed culture on fermentation.
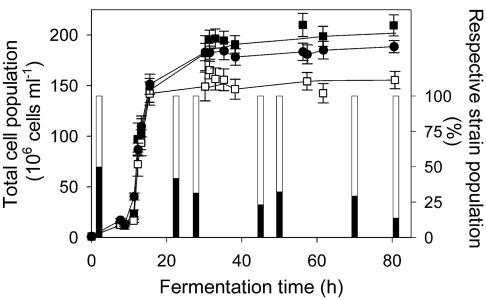
Fifty hours after inoculation with a 1:1 mixture of the two strains, D254 9a2 accounted for 70 to 75% of the total population in the mixed culture (Fig. 1). However, when the strains were cultured separately, the final S6U monoculture population was larger than the final D254 9a2 monoculture population (Fig. 1). The population of the mixed culture at the end of fermentation was larger than that predicted by the growth of the two monocultures [(184 ± 4) × 106 cells ml−1 instead of about (167 ± 7) × 106 cells ml−1]. Thus, the growth of the mixed culture is not simply the sum of the growth of the individual strains, and presumably interactions between the two strains have a substantial effect.
FIG. 1.
Cell population during fermentation of a monoculture of strain S6U (▪), a monoculture of strain D254 9a2 (□), and a mixed culture (•) inoculated with an equal amount (50:50) of each strain. Points represent the means and standard deviations of three different fermentations. Lines represent the best-fit growth curves calculated by a five-parameter sigmoid function. Stacked bars represent the respective proportion of each strain within the mixed culture as determined by differential plating (filled area, strain S6U; open area, strain D254 9a2). The strains were anaerobically cultured in SM at 24°C. All media were inoculated with 106 cells ml−1.
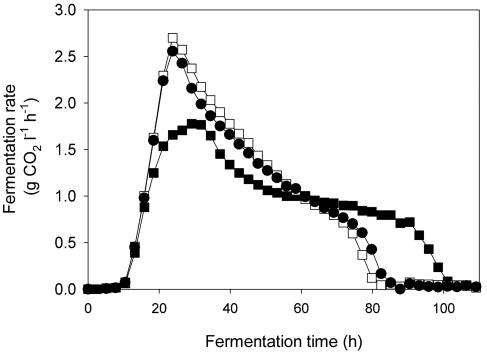
During the initial phase of the fermentation (first 20 h), there were no significant differences in the rates of fermentation between the D254 92a and S6U monocultures and the mixed culture (Fig. 2). After that, the fermentation kinetics, the duration of the fermentation, and the maximum fermentation rate of the mixed culture were not equivalent to a 70:30 average ratio of the fermentation kinetics of the two monocultures. The cells of both strains were similar in size (data not shown), so specific fermentation rates were calculated from the best-fit growth curves (Fig. 1) for each culture. The specific fermentation rate of the mixed culture also was different from that of the two monocultures (data not shown). D254 9a2 made up 70 to 75% of the total population in the mixed culture, but the overall specific fermentation kinetics of the mixed culture were more similar to those of the S6U monoculture.
FIG. 2.
Fermentation kinetics of a monoculture of strain S6U (▪), a monoculture of strain D254 9a2 (□), and a mixed culture (•). For the mixed culture, the inoculum contained equal amounts of the two strains. The strains were anaerobically cultured in SM at 24°C. All media were inoculated with 106 cells ml−1. For clarity, only 50 of the 350 data points collected are shown. Since the coefficient of variation for d(CO2/dt) with this technique was previously observed to be 0.8% (39), the results of only one experiment are given.
Effect of the mixed culture on the redox potential of the yeast strains.
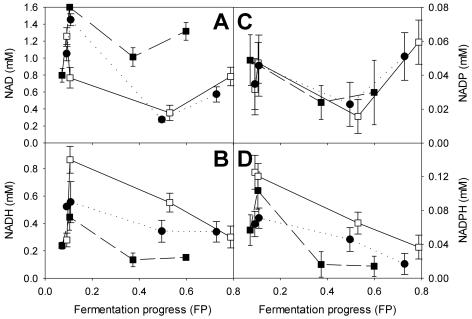
At the end of fermentation, more acetate was produced by the mixed culture than by either of the monocultures (about 410, 365, and 322 mg per liter for mixed culture, strain S6U, and strain D254 9a2, respectively). This overproduction of acetate occurred continuously throughout the fermentation and increased as the fermentation progressed (data not shown). This change in the composition of the extracellular medium could result in changes in the redox potential of one or both yeast strains in the mixed culture, possibly as a result of metabolic interactions between the two strains. The redox potential of the two monocultures differed; that is, the cellular content of NADH and NADPH was lower in strain S6U than in strain D254 9a2 (Fig. 3B and D), resulting in lower NADH to NAD and NADPH to NADP ratios in strain S6U than in strain D254 92a. The NADH and NADPH content of the mixed culture was intermediate to content values of the S6U and D254 9a2 monocultures, but the NAD content of the mixed culture was similar to that of the D254 9a2 monoculture, particularly during the second half of the fermentation (Fig. 3A). In contrast, NADP content values were very similar throughout the fermentation in all tested cultures (Fig. 3C). Such differences in the redox balance ratios were also recently observed during fermentation of three industrial wine yeast strains (8).
FIG. 3.
Intracellular content of NAD, NADH, NADP, and NADPH. ▪, monoculture of strain S6U; □, monoculture of strain D254 9a2; •, mixed culture. For the mixed culture, the inoculum contained equal amounts of the two strains. The strains were cultured anaerobically in SM at 24°C. All media were inoculated with 106 cells ml−1. The mean and standard deviation of three replicates for each point are shown.
Effect of fermentation medium renewal on the redox potential of the yeast strains.
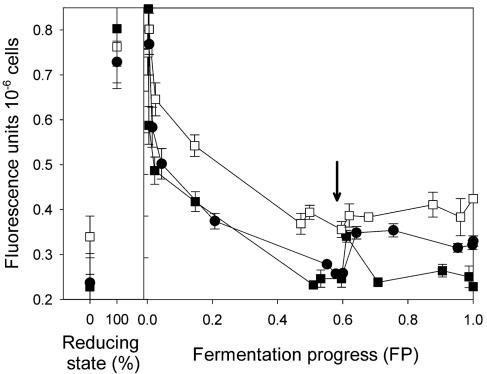
Cellular fluorescence (Fig. 4) was used to estimate cellular redox potential during fermentation (4, 23, 36). At the beginning of fermentation, the cellular fluorescence of the mixed culture was similar to that of the S6U monoculture. Removal of the fermentation products did not alter the fluorescence of strain D254 9a2, but the fluorescence of strain S6U increased sharply following medium renewal and then returned to previous levels. Fluorescence emitted by the mixed culture increased sharply following the change of medium and thereafter decreased slowly to a level similar to that of strain D254 9a2. At the end of the fermentation, the fluorescence signal of the mixed culture was higher than that in the presence of 3.5 mM acetaldehyde, which represents the fully oxidized state of intracellular reducing equivalents (Fig. 4). The NAD(P)H content of the mixed culture also decreased, but more gradually, after medium renewal.
FIG. 4.
Cellular fluorescence during fermentation. Fluorescence of harvested cells suspended in CMP buffer was monitored at 460 nm (excitation wavelength, 340 nm). Strains were cultured anaerobically in SM at 24°C. All media were inoculated with 106 cells ml−1. In preliminary experiments, spectrofluorimetric measurements were calibrated by adding 50 mM glucose and 50 mM ethanol (fully reduced state, 100%) or 3.5 mM acetaldehyde (fully oxidized state, 0%) before recording fluorescence. When the fermentation progress reached 0.6 (black arrow), cells were harvested by centrifugation (3,000 × g, 5 min, 24°C) and resuspended in a volume of fresh SM containing no nitrogen and 80 g of glucose per liter and 7% (vol/vol) ethanol. Points represent the means and standard deviations of three different fermentations. ▪, monoculture of strain S6U; □, monoculture of strain D254 9a2; •, mixed culture. For the mixed culture, the inoculum contained an equal amount of each strain.
Effect of mixed culture on the acetaldehyde levels during fermentation.
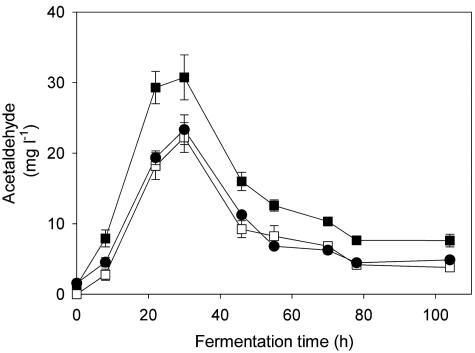
We monitored acetaldehyde levels in S6U and D254 9a2 monocultures and in the mixed culture during fermentation (Fig. 5). Strain S6U produced large amounts of acetaldehyde, particularly during the early phase of fermentation. The amount of acetaldehyde produced by strain D254 9a2 was lower than that produced by strain S6U throughout the fermentation. Even though strain S6U made up ∼30% of the total population of the mixed culture, the acetaldehyde production profile of the mixed culture was very similar to that of the D254 9a2 culture.
FIG. 5.
Acetaldehyde content of the media during fermentation. ▪, monoculture of strain S6U; □, monoculture of strain D254 9a2; •, mixed culture. For the mixed culture, the inoculum contained an equal amount of each strain. Values are the means and standard deviations of three different fermentations. The strains were cultured anaerobically in SM at 24°C. All media were inoculated with 106 cells ml−1.
DISCUSSION
Several previous studies have shown that growth or metabolism increases in S. cerevisiae and bacterial cocultures (11, 12, 26, 30, 47). This increase could result from competition between the yeast and bacteria for nutrients, particularly sugars and vitamins (3, 11, 43, 47). However, these effects also could result from the diffusion of metabolites between the two species (11, 20, 30, 47). To our knowledge, no work was initiated to identify the effect of metabolite diffusion between the partners of yeast cocultures. In order to define an experimental preliminary model allowing such a study, we examined the interactions between two yeast strains (one a strain of S. cerevisiae and the other a natural S. cerevisiae-S. uvarum hybrid) known to exhibit different physiological properties during alcoholic fermentations under enological conditions (24). Fermentation with S. uvarum is known to produce much more acetaldehyde in the resulting wines than fermentation with S. cerevisiae (9, 14). We found that the S. cerevisiae-S. uvarum hybrid (S6U) produces during fermentation large quantities of acetaldehyde that S. cerevisiae strain D254 9a2 can utilize in mixed cultures. This process results in a shift toward lower cellular NAD(P)H levels by the D254 9a2 cells in response to the acetaldehyde produced by strain S6U. This change in redox potential is linked to increases in biomass and specific fermentation rate. Acetate overproduction observed in the mixed culture could result from the activity of aldehyde dehydrogenase from strain D254 9a2 (33) on the acetaldehyde produced by strain S6U.
The present report demonstrates a clear exchange of an electron acceptor between two different yeast strains during alcoholic fermentation, as has been suggested by others (20). This exchange could alter the behavior of the mixed culture. Previous studies have shown that a continuous perfusion of monoculture fermentation medium with acetaldehyde decreases glycerol production and increases acetate and butanediol production (37, 38). This phenomenon was observed on three different yeast strains. In the present study, we did not observe any changes in glycerol or butanediol production (data not shown). Therefore, although both acetaldehyde exchange between two yeast strains and continuous perfusion with acetaldehyde increase the fermentation rate, they lead to the formation of different end products. Acetaldehyde from a natural source produced in situ might affect yeast metabolism differently than does its exogenous addition, but further research is needed to test this hypothesis.
Acetaldehyde is considered to be a leakage product of alcoholic fermentation by yeasts. Acetaldehyde is unique in that it is highly reactive and biologically toxic. Acetaldehyde also is very polar and may cause water stress in yeasts (21). There are large species and strain differences in acetaldehyde production by yeasts from 0.5 to 700 mg of acetaldehyde per liter (27). Exogenously added acetaldehyde at a concentration of ≥400 mg/liter lengthened the lag phase and decreased the exponential specific growth rate of S. cerevisiae in a medium lacking ethanol (45). In contrast to this inhibitory effect, low levels of acetaldehyde may stimulate yeast growth. For example, ≤580 mg of acetaldehyde per liter shortens the lag phase and increases the specific growth rate of S. cerevisiae in the presence of 3 to 6% (vol/vol) ethanol, although not in its absence (45, 49). In addition, very low levels of acetaldehyde (<100 mg/liter) were found to greatly reduce the lag phase of ethanol- or temperature-stressed S. cerevisiae (46). Thus, acetaldehyde seems to alleviate ethanol-induced growth inhibition. An early study showed that exogenously added acetaldehyde at a concentration of 1,000 mg/liter reduced the fermentation rate of glucose by S. cerevisiae by 30% (17). From a more fundamental point of view, such an addition of acetaldehyde initiates a transcriptional response in yeast cells that changes the expression of HSP genes (1), the genes encoding aldehyde dehydrogenases (ALD genes [2]) in order to allow the cells to use ethanol and acetaldehyde as carbon and energy sources under several circumstances.
Collectively, these data suggest that the yeast response to acetaldehyde utilizes the same mechanisms that participate in the response to other forms of stress. Thus, acetaldehyde exchange between strains could inhibit the growth of some yeast strains while encouraging the growth of others. This phenomenon could be particularly important for understanding the ecology of the colonization of complex fermentation media by S. cerevisiae after the elimination of non-Saccharomyces yeasts. During spontaneous fermentations, a succession of different indigenous S. cerevisiae yeasts could be observed throughout the fermentation stationary phase, although no arguments were found to explain such behavior (16, 48). When fermentations are inoculated with pure Saccharomyces starter cultures, the persistence of several indigenous S. cerevisiae strains during fermentation may also be observed (16). These two examples could represent interesting models for determining the role of acetaldehyde during fermentation.
There is great variation in metabolic capability among isolates of naturally occurring S. cerevisiae. The variation includes significant heterogeneity among strains in the production of ethanol, acetic acid, sulfite, and other products of metabolism (10). To our knowledge, there are very few data on the production of acetaldehyde during the colonization of complex fermentation media. The initial acetaldehyde level in a fermentation medium could encourage the growth of S. cerevisiae yeasts, e.g., D254 9a2, that can function as receptor strains and utilize the acetaldehyde produced. To test this hypothesis, effort should be made to develop a bioreactor in which differential growth of a yeast strain in a homogenous fermentation medium can be established. We are now testing a two-reservoir, hollow-fiber bioreactor (28) for use in the study of the dynamics of such mixed populations.
Over 200 different wine yeast strains are currently available for use in ADY starters; however, their physiological differences have not been systematically evaluated (9, 14, 15, 24, 32, 42). Empirical experiments have been conducted on both the laboratory and commercial scale on the effect of changes in the yeast population on the reliability of mixed fermentations and on the quality of the wines produced (13, 29, 44, 50). From a technological point of view, more ADY starters containing different mixtures of wine yeast strains are likely to be introduced to the market. Thus, the potential interactions between the yeast strains used and the potential effects (detrimental or otherwise) on the final product need to be evaluated. Including strains with opposite acetaldehyde-producing properties in the same starter could bloom the wine yeast, resulting in a higher fermentation rate or improved organoleptic properties. A general survey of the acetaldehyde-producing properties of and the physiological response to acetaldehyde of all of the commercially available wine yeast strains would therefore be valuable.
REFERENCES
- 1.Aranda, A., A. Querol, and M. del Olmo. 2002. Correlation between acetaldehyde and ethanol resistance and expression of HSP genes in yeast strains isolated during the biological aging of sherry wines. Arch. Microbiol. 177:304-312. [DOI] [PubMed] [Google Scholar]
- 2.Aranda, A., and M. del Olmo. 2003. Response to acetaldehyde stress in the yeast Saccharomyces cerevisiae involves a strain-dependent regulation of several ALD genes and is mediated by the general stress response pathway. Yeast 20:747-759. [DOI] [PubMed] [Google Scholar]
- 3.Bataillon, M., A. Rico, J. M. Sablayrolles, J. M. Salmon, and P. Barre. 1996. Early thiamin assimilation by yeasts in enological conditions: impact on alcoholic fermentation kinetics. J. Ferment. Bioeng. 82:145-150. [Google Scholar]
- 4.Beauvoit, B., M. Rigoulet, O. Bunoust, G. Raffard, P. Canioni, and B. Guérin. 1993. Interactions between glucose metabolism and oxidative phosphorylations on respiratory-competent Saccharomyces cerevisiae cells. Eur. J. Biochem. 214:163-172. [DOI] [PubMed] [Google Scholar]
- 5.Bely, M., J. M. Sablayrolles, and P. Barre. 1990. Description of alcoholic fermentation kinetics: its variability and significance. Am. J. Enol. Vitic. 40:319-324. [Google Scholar]
- 6.Bergmeyer, H. U. 1955. Zur messung von katalase aktivitäten. Biochem. Z. 327:255-258. [PubMed] [Google Scholar]
- 7.Bisson, L. F., and R. E. Kunkee. 1991. Microbial interactions during wine production, p. 37-59. In K. Zeilaus and M. K. Johnson (ed.), Mixed cultures in biotechnology. MacGraw-Hill, New York, N.Y.
- 8.Camarasa, C., A. Ortiz-Julien, S. Dequin, and J. M. Sablayrolles. 2003. Rôle de l'équilibre d'oxydoréduction dans le comportement fermentaire de trois levures oenologiques, p. 308-311. In A. Lonvaud-Funel, G. de Revel, and P. Darriet (ed.), Oenologie 2003, 7ème Symposium International d'Oenologie. Editions Tec et Doc, Paris, France.
- 9.Castellari, L., C. Zambonelli, P. Passarelli, V. Tini, and F. Coloretti. 2002. Study of the main characteristics of oenological yeast strains from the CATEV-DIPROVAL collection. Vignevini 29:91-95. [Google Scholar]
- 10.Cavalieri, D., C. Barberio, E. Casalone, F. Pinzauti, F. Sebastiani, R. K. Mortimer, and M. Polsinelli. 1998. Genetic and molecular diversity in S. cerevisiae natural populations. Food Technol. Biotechnol. 36:45-50. [Google Scholar]
- 11.Chaucheyras, F., G. Fonty, G. Bertin, J. M. Salmon, and P. Gouet. 1996. Effects of a strain of Saccharomyces cerevisiae (Levucell SC1), a microbial additive for ruminants, on lactate metabolism in vitro. Can. J. Microbiol. 42:927-933. [DOI] [PubMed] [Google Scholar]
- 12.Cheirsilp, B., H. Shimizu, and S. Shioya. 2003. Enhanced kefiran production by mixed culture of Lactobacillus kefiranofaciens and Saccharomyces cerevisiae. J. Biotechnol. 100:43-53. [DOI] [PubMed] [Google Scholar]
- 13.Ciani, M., and L. Ferraro. 1998. Combined use of immobilized Candida stellata cells and Saccharomyces cerevisiae to improve the quality of wines. J. Appl. Microbiol. 85:247-254. [DOI] [PubMed] [Google Scholar]
- 14.Ciani, M., G. Picciotti, and L. Ferraro. 1994. Evaluation of the enological aptitude of some selected wine yeasts. Ann. Fac. Agrar. Univ. Stud. Perugia 48:49-58. [Google Scholar]
- 15.Eder, R., and L. Alzinger. 2003. Influence of different yeast preparations on the acid composition of “Gruener Veltliner” wines. Mitt. Klosterneuburg 53:52-60. [Google Scholar]
- 16.Egli, C. M., W. D. Edinger, C. M. Mitrakul, and T. Henick-Kling. 1998. Dynamics of indigenous and inoculated yeast populations and their effect on the sensory character of Riesling and Chardonnay wines. J. Appl. Microbiol. 85:779-789. [DOI] [PubMed] [Google Scholar]
- 17.Freeman, G. G., and G. M. Donald. 1957. Fermentation processes leading to glycerol. II. Studies on the effects of sulfite on viability, growth, and fermentation of Saccharomyces cerevisiae. Appl. Microbiol. 5:211-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia, A., C. Carcel, L. Dulau, A. Samson, E. Aguera, E. Agosin, and Z. Günata. 2002. Influence of a mixed culture with Debaryomyces vanriji and Saccharomyces cerevisiae on the volatiles of a Muscat wine. J. Food Sci. 67:1138-1143. [Google Scholar]
- 19.Gonzalez, B., J. François, and M. Renaud. 1997. A rapid and reliable method for metabolite extraction using boiling buffered ethanol. Yeast 13:1347-1356. [DOI] [PubMed] [Google Scholar]
- 20.Grossmann, M., H. Linsenmeyer, H. Muno, and A. Rapp. 1996. Use of oligo-strain yeast cultures to increase complexity of wine aroma. Vitic. Enol. Sci. 51:175-179. [Google Scholar]
- 21.Hallsworth, J. E. 1998. Ethanol-induced water stress in yeast. J. Ferment. Bioeng. 85:125-137. [Google Scholar]
- 22.Holzer, H., D. Busch, and H. Kröger. 1958. Enzymicoptic determination of reduced and oxidized triphosphopyridine nucleotides in presence of reduced and oxidized diphosphopyridine nucleotides. Hoppe-Seyler's Z. Physiol. Chem. 313:184-193. [DOI] [PubMed] [Google Scholar]
- 23.Iwami, Y., and T. Yamada. 1999. Intracellular flux of glucose metabolism in streptococcal cells by simultaneous monitoring of fluorescence dependent on reduced nicotinamide adenine nucleotide and acid excretion under strictly anaerobic conditions. Oral Microbiol. Immunol. 14:220-224. [DOI] [PubMed] [Google Scholar]
- 24.Julien, A., J. L. Roustan, L. Dulau, and J. M. Sablayrolles. 2000. Comparison of nitrogen and oxygen demands of enological yeasts: technological consequences. Am. J. Enol. Vitic. 51:215-222. [Google Scholar]
- 25.Klingenberg, M. 1974. Nicotinamide-adenine dinucleotides (NAD, NADP, NADH, NADPH): spectrophotometric and fluorimetric methods, p. 2045-2059. In H. U. Bergmeyer (ed.), Methods of enzymatic analysis, 2nd ed., vol. 4. Academic Press, New York, N.Y. [Google Scholar]
- 26.Lee, S. S., J. K. Ha, and K. Cheng. 2000. Relative contributions of bacteria, protozoa, and fungi to in vitro degradation of orchard grass cell walls and their interactions. Appl. Environ. Microbiol. 66:3807-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, S. Q., and G. J. Pilone. 2000. An overview of formation and roles of acetaldehyde in wine making with emphasis on microbiological implications. Int. J. Food Sci. Technol. 35:49-61. [Google Scholar]
- 28.Manjarrez, E. S., C. Albasi, and J. P. Riba. 2000. A two-reservoir, hollow-fiber bioreactor for the study of mixed-population dynamics: design aspects and validation of the approach. Biotech. Bioeng. 69:401-408. [DOI] [PubMed] [Google Scholar]
- 29.Mora, J., J. I. Barbas, and A. Mulet. 1990. Growth of yeast species during the fermentation of musts inoculated with Kluyveromyces thermotolerans and Saccharomyces cerevisiae. Am. J. Enol. Vitic. 41:156-159. [Google Scholar]
- 30.Odame-Darkwah, J. K., and D. L. Marshall. 1993. Interactive behavior of Saccharomyces cerevisiae, Bacillus pumilus and Propionibacterium freudenreichii subsp. shermanii. Int. J. Food Microbiol. 19:259-269. [DOI] [PubMed] [Google Scholar]
- 31.Ramon-Portugal, F. 1997. Kinetic study and mathematical modeling of killer and sensitive Saccharomyces cerevisiae strains growing in mixed culture. Bioproc. Eng. 82:375-381. [Google Scholar]
- 32.Rauhut, D., H. Kürbel, H. H. Dittrich, and M. Grossmann. 1996. Properties and differences of commercial yeast strains with respect to their formation of sulfur compounds. Wein-Wiss. 51:187-192. [Google Scholar]
- 33.Remize, F., E. Andrieu, and S. Dequin. 2000. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae: role of the cytosolic Mg2+ and mitochondrial K+ acetaldehyde dehydrogenases Ald6p and Ald4p in acetate formation during alcoholic fermentation. Appl. Environ. Microbiol. 66:3151-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reyes, F. G. R., R. E. Wrolstad, and C. J. Cornwell. 1982. Comparison of enzymic, gas-liquid chromatographic, and high performance liquid chromatographic methods for determining sugars and organic acids in strawberries at three stages of maturity. J. Assoc. Off. Anal. Chem. 65:126-131. [Google Scholar]
- 35.Romano, P., C. Fiore, M. Paraggio, M. Caruso, and A. Capece. 2003. Function of yeast species and strains in wine flavour. Int. J. Food Microbiol. 86:169-180. [DOI] [PubMed] [Google Scholar]
- 36.Rosenfeld, E., B. Beauvoit, M. Rigoulet, and J. M. Salmon. 2002. Non-respiratory oxygen consumption pathways in anaerobically grown Saccharomyces cerevisiae: evidence and partial characterization. Yeast 19:1299-1322. [DOI] [PubMed] [Google Scholar]
- 37.Roustan, J. L., and J. M. Sablayrolles. 2002. Impact of the addition of electron acceptors on the by-products of alcoholic fermentation. Enzyme Microb. Technol. 34:142-152. [Google Scholar]
- 38.Roustan, J. L., and J. M. Sablayrolles. 2002. Modification of the acetaldehyde concentration during alcoholic fermentation and effects on fermentation kinetics. J. Biosci. Bioeng. 93:367-375. [PubMed] [Google Scholar]
- 39.Sablayrolles, J. M., P. Barre, and P. Grenier. 1987. Design of a laboratory automatic system for studying alcoholic fermentations in anisothermal enological conditions. Biotechnol. Technol. 1:181-184. [Google Scholar]
- 40.Salmon, J. M. 1997. Enological fermentation kinetics of an isogenic ploidy series derived from an industrial Saccharomyces cerevisiae strain. J. Ferment. Bioeng. 83:253-260. [Google Scholar]
- 41.Schrink, B. 2002. Synergistic interactions in the microbial world. Antonie Leeuwenhoek 66:257-261. [DOI] [PubMed] [Google Scholar]
- 42.Shimazu, Y., and M. Watanabe. 1981. Effects of yeast strains and environmental conditions on formation of organic acids in must during fermentation. J. Ferment. Technol. 59:27-32. [Google Scholar]
- 43.Shimizu, H., T. Mizuguchi, E. Tanaka, and S. Shioya. 1999. Nisin production by a mixed-culture system consisting of Lactococcus lactis and Kluyveromyces marxianus. Appl. Environ. Microbiol. 65:3134-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soden, A., I. L. Francis, H. Oakey, and P. Henschke. 2000. Effects of co-fermentation with Candida stellata and Saccharomyces cerevisiae on the aroma and composition of chardonnay wine. Austr. J. Grape Wine Res. 6:21-30. [Google Scholar]
- 45.Stanley, G. A., N. G. Douglas, E. J. Every, T. Tzanatos, and N. B. Pamment. 1993. Inhibition and stimulation of yeast growth by acetaldehyde. Biotechnol. Lett. 15:1199-1204. [Google Scholar]
- 46.Stanley, G. A., T. J. Hobley, and N. B. Pamment. 1997. Effect of acetaldehyde on Saccharomyces cerevisiae and Zymomonas mobilis subjected to environmental shocks. Biotechnol. Bioeng. 53:71-78. [DOI] [PubMed] [Google Scholar]
- 47.Sullivan, H. M., and S. A. Martin. 1999. Effects of a Saccharomyces cerevisiae culture on in vitro mixed ruminal microorganism fermentation. J. Dairy Sci. 82:2011-2016. [DOI] [PubMed] [Google Scholar]
- 48.Villanova, M., C. Martinez, C. Siero, I. Masneuf, and D. Dubourdieu. 2003. Ecology of Saccharomyces cerevisiae fermentations at a Rias Baixas appellation contrôlée winery. J. Inst. Brew. 109:305-308. [Google Scholar]
- 49.Walker-Caprioglio, H. M., and L. W. Parks. 1987. Autoconditioning factor relieves ethanol-induced growth inhibition of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 53:33-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zironi, R., P. Romano, G. Suzzi, F. Battistutta, and G. Comi. 1993. Volatile metabolites produced in wine by mixed and sequential cultures of Hanseniaspora guilliermondii or Kloeckera apiculata and Saccharomyces cerevisiae. Biotechnol. Lett. 15:235-238. [Google Scholar]
- 51.Zoecklein, B. W., J. E. Marcy, J. M. Williams, and Y. Jasinski. 1997. Effect of native yeasts and selected strains of Saccharomyces cerevisiae on glycosyl glucose, potential volatile terpenes and selected aglycones of White Riesling (Vitis vinifera L.) wines. J. Food Compost. Anal. 10:55-65. [Google Scholar]