Abstract
The use of live, genetically modified bacteria as delivery vehicles for biologics is of considerable interest scientifically and has attracted significant commercial investment. We have pioneered the use of the commensal gut bacterium Bacteroides ovatus for the oral delivery of therapeutics to the gastrointestinal tract. Here we report on our investigations of the biological safety of engineered B. ovatus bacteria that includes the use of thymineless death as a containment strategy and the potential for the spread of transgenes in vivo in the mammalian gastrointestinal tract. We demonstrate the ability of GM-strains of Bacteroides to survive thymine starvation and overcome it through the exchange of genetic material. We also provide evidence for horizontal gene transfer in the mammalian gastrointestinal tract resulting in transgene-carrying wild type bacteria. These findings sound a strong note of caution on the employment of live genetically modified bacteria for the delivery of biologics.
Introduction
The use of live genetically modified bacteria as delivery vehicles for the in situ delivery of therapeutics has to date relied on the use of specific commensal or food-grade bacteria, typically lactic acid bacteria, for heterologous gene expression. Efficacy has been demonstrated in proof of principle animal models of infectious and autoimmune diseases1–3 leading to the development of a number of sophisticated heterologous gene expression and genetic engineering systems4–6. This has culminated in clinical trials of GM-Lactococcus lactis ssp. cremoris, engineered to secrete the anti-inflammatory cytokine interleukin-10 in inflammatory bowel disease patients7. We have focused on the development of a novel protein delivery technology using the anaerobic Gram-negative human gut commensal bacterium Bacteroides ovatus for the diet-controlled delivery of therapeutic proteins to the gastrointestinal tract8–10. Following successful proof of principle studies using GM-B. ovatus engineered to secrete human cytokines for the treatment of experimental intestinal colitis8, the GM-strains were re-engineered to make them suitable for first-in-man studies. Apart from technical feasibility and clinical safety a major issue that all technologies of this kind must fully address is the biological safety of the genetically modified bacteria. Specifically, ensuring environmental containment and preventing the spread of the transgene by horizontal gene transfer as well as the survival of the genetically modified bacteria in the environment.
Results and Discussion
In vitro and in vivo horizontal gene transfer in Bacteroides ovatus
Thymineless death (TLD), the fatal process that thymine auxotrophic microorganisms undergo in response to thymine starvation, has previously been used as a biosafety strategy in for example, GM-L. lactis strains11. It was first discovered in E. coli 12 and exists in many other organisms including eukaryotes13. We reasoned that incorporating thymine auxotrophy, as employed for GM-L. lactis in a phase I clinical trial7, in addition to the anaerobic nature of B. ovatus, seemed an appropriate environmental containment strategy to adopt. A further advantage of using thymidine auxothrophy is that it facilitates a thyA based counter-selectable gene deletion and integration system for B. ovatus, similar to that described for B. fragilis 14.
Horizontal gene transfer in prokaryotes occurs via transformation, conjugation, or transduction15. As we were not aware of any reports of transduction or the occurrence of natural competence in Bacteroides, and since conjugation involving E. coli helper strains is routinely employed to genetically manipulate Bacteroides species we focused on conjugation as a potential mechanism of horizontal gene transfer. Two B. ovatus thyA deletion strains (see methods) tagged with the tetQ marker gene, GH439 (ΔthyA, cblA::tetQ) and GH440 (ΔthyA, cblA::tetQ, ΔoxyR, BACOV975_00918::tgfβ_ex1) were created (Table 1). To our knowledge a high-frequency of recombination (Hfr) phenotype involving the conjugational transfer of the whole chromosome from one cell to another has not been observed in Bacteroides species and as the only Hfr-type transfer reported to date relates to the partial transfer of the conjugative transposon CTnERL by B. thetaiotaomicron 16, we did not expect to detect chromosomal gene transfer events. In contrast, Hfr-type conjugation including the transfer of the thyA gene is well documented in E. coli 17 and, owing to the lactococcal sex factor, can occur in L. lactis 18, 19. Initially horizontal gene transfer was examined in vitro using B. ovatus V975 and GH439 or GH440 (Table 1) as potential B. ovatus donor and recipient cells during filter mating experiments in the absence of E. coli helper cells. Transconjugant colonies displaying a thyA +/tet R phenotype were readily detected at a frequency of 0.9–4.2 × 10−7 transconjugants per recipient with the frequency independent of tetracycline use. PCR confirmed the genotype of donor and recipient cells as thyA +/cblA (wt) and, ΔthyA/cblA::tetQ (GH439) or ΔthyA/cblA::tetQ/ΔoxyR/BACOV975_00918::tgfβ (GH440), respectively (Fig. 1a; GH439 and data not shown; Table 2). Horizontal gene transfer in the corresponding transconjugants was confirmed by PCR and their genotypes were determined as thyA +/cblA::tetQ (conjugation with GH439, data not shown; Table 2) and thyA +/cblA::tetQ/ΔoxyR/BACOV975_00918::tgfβ (type1), thyA +/cblA::tetQ/ΔoxyR/BACOV975_00918 (type 2) or thyA +/cblA::tetQ/ΔoxyR/BACOV975_00918/BACOV975_00918::tgfβ (type 3) (conjugation with GH440, Fig. 1b, Table 2). The latter genotype, as indicated by fragments of 2250 bp and 3019 bp corresponding to the BACOV975_00918 wt gene as well as the BACOV975_00918::tgfβ locus (Fig. 1b), suggested a possible BACOV975_00918 gene duplication event. Since strains GH439 and GH440 are derivatives of B. ovatus V975 and virtually genetically identical we were unable to ascertain which strains acted as donor. However, it is reasonable to assume that all three strains share this ability and as a result act simultaneously as recipient and donor. To determine whether horizontal gene transfer occurred in vivo in the mammalian gastrointestinal tract mice were orally gavaged with wt and GH439 bacteria sequentially with a one hour interval and feces subsequently sampled, homogenized and plated on selective media with potential transconjugants identified by PCR (see methods). Horizontal gene transfer events were identified in 5/10 colonized mice from which 48 thyA +/tet R colonies were obtained from fresh feces with 21 assessed by PCR. In all cases horizontal gene transfer was confirmed on the basis of amplicon patterns (Fig. 1) indicative of a thyA +/tet R genotype, and phenotypically by the growth of clones in liquid cultures in the presence of tetracycline without thymidine. As seen in the in vitro experiment for BACOV975_00918, some clones isolated from mouse feces produced a cblA amplicon pattern (see Fig. 1c, Table 2) indicative of a duplication of the β-lactamase gene with a 2040 bp amplicon representing the wt cblA allele as well as a 4206 bp amplicon expected for the cbl::tetQ allele and the tetQ specific 675 bp amplification product. These isolates could be grown in liquid culture without thymidine in the presence of ampicillin (200 μg/ml) and tetracycline (5 μg/ml). As these transconjugants had arisen from a horizontal gene transfer event that had taken place in the murine GI-tract, one of the isolates, number 4, was investigated further. Southern blot analysis of AvaI-restricted chromosomal transconjugant 4 DNA using a cblA and a tetQ specific probe revealed a banding pattern consistent with the presence of two cblA loci, one representing the wt and the other the cblA::tetQ topography (data not shown). Several scenarios could explain this result, the most likely of which is the random insertion of one of the alleles into the chromosome (Fig. 2a) creating a similar sized fragment in the Southern blot. Alternatively, two chromosomes may exist, each carrying one of the two alleles. In this scenario the chromosomes could either be identical bar their respective cblA loci (Fig. 2b) or, one could be smaller (Fig. 2c) due to a deletion, representing only a part of the chromosome. However, bacteria unlike higher eukaryotes, are generally haploid20 and contain a single chromosome, which is replicated during the cell cycle. Some exceptions have been reported21, 22 and mini-chromosomes have been genetically engineered for E. coli 23, although these rely on antibiotic resistance markers to be stably maintained. To distinguish these possibilities, the entire genome of transconjugant 4 was sequenced. We predicted that in the case where the cblA gene would have been randomly integrated into the chromosome two additional contigs would be found one representing the cblA::tetQ locus and the other the cblA gene in its new genetic location. In the case of an additional smaller chromosome, a contig joining genetic locations that are not adjacent on the wt chromosome would be present in addition to a contig representing the cblA::tetQ locus. In the case of two virtually identical chromosomes, we would expect all contigs to align to the wt sequence bar that representing the cblA::tetQ locus. After assembling 5.2 million paired end reads equivalent to >200-fold coverage of the wt genome into 100 contigs of ≥150 bp using the SPAdes24 software, contigs of ≥150 bp were aligned to the wt sequence25 either manually in a STADEN26 assembly or computationally using QUAST27; QUAST revealed that the contigs in the assembly represented 98.3% of the original sequence, with NG75 and LG75 values of 162375 bp and 16, respectively. Three small insertions of 63, 204 and 313 bp and two small local inversions of 1.2 and 7.2 kb, respectively, were identified in the transconjugant 4 assembly. Only two contigs could not be aligned to the wt genome. The first (151 bp) represented part of an insertion sequence (IS) element with its existence in B. ovatus V975, GH439 and transconjugant 4 confirmed by PCR, excluding it arising from the formation of a smaller chromosome. The second contained the cblA::tetQ locus, with sequences of this contig either side of the tetQ insertion being aligned perfectly to the wt sequence, confirming the Southern blot finding that transconjugant 4 harbored two chromosomes that were identical apart from their cblA locus and hence are at least diploid. We do not know whether B. ovatus is generally diploid, but it is clear it can use this ability to acquire new genes, which in turn aids in circumventing genetically engineered safety measures, for example, by maintaining a modified thyA locus whilst acquiring a functional wt gene. In this context it is noteworthy that several L. lactis subsp. cremoris and lactis strains, including MG1363, a derivative of which was used in a phase I clinical trial involving live bacteria, have been described as diploid28.
Table 1.
Bacterial Strains.
| Strain | Description | Reference |
|---|---|---|
| Bacteroides ovatus V975 | Wild type | B. ovatus strain V975 was obtained from T.R. Whitehead (USDA ARS NCAUR, Peoria, IL) |
| Bacteroides ovatus GH439 | ΔthyA, cblA::tetQ | This work |
| Bacteroides ovatus GH440 | ΔthyA, cblA::tetQ, ΔoxyR, BACOV975_00918::tgfβ_ex1 | This work |
| E. coli strain HB101 pRK201329 | F–, thi-1, hsdS20 (rB −, mB −), supE44, recA13, ara14, leuB6, proA2, lacY1, galK2, rpsL20 (strr), xyl-5, mtl-1. pRK2013 (KmR, oriColE1, RK2-Mob+, RK2-Tra+) | Clontech |
| E. coli GC10 | F−, mcrA, ∆(mrr-hsdRMS-mcrBC), Φ80d, lacZ∆M15, ∆lacX74, endA1, recA1, ∆(ara, leu) 7697araD139 galU, galK, nupG, rpsL, λ−, T1R. | GeneChoice, Inc. |
Figure 1.
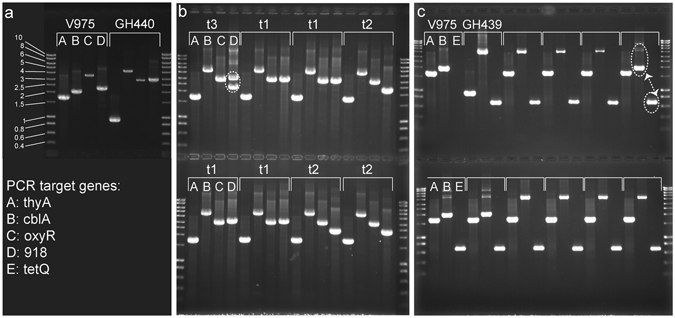
(a,b) Amplicon patterns of donor and recipient stains (a) and isolates after filter mating experiments (b). (c) Amplicon patterns of donor and recipient stains and isolates from the murine GI tract. A, B, C, D and E indicate the target (primer pairs) used in PCR, t denominates the different types (1–3) observed. The brackets above indicate tracks that belong to a specific isolate. Patterns indicating potential gene duplication events are circled. The arrow highlights the simultaneous presence of the wt cblA gene and the tetR gene in this isolate.
Table 2.
PCR amplicon sizes.
| B. ovatus strains | |||
|---|---|---|---|
| Primers | V975 | GH439 | GH440 |
| Expected fragment sizes | |||
| ΔthyA_folA_R/thyA_F (A) | 1699 | 909 | 909 |
| cblA_F/cblA_R (B) | 2040 | 4206 | 4206 |
| oxyR left_flank/oxyR_r_flank3 (C) | 3484 | ND | 2946 |
| 918_left_flank/918_right_flank (D) | 2250 | NA | 3019 |
| RT_tetQ_3′/RT-tetQ_5′ (E) | NA | 673 | 673 |
Figure 2.
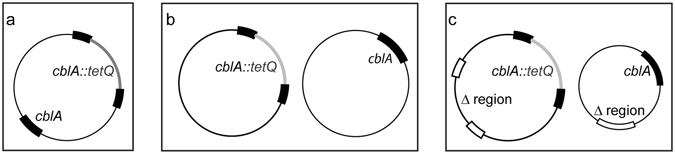
Schematic representation of 3 possible cblA gene duplication scenarios: (a) insertion of duplicated cblA gene into random position of chromosome, (b) diploid bacteria carrying 2 identical chromosomes differing only in their respective cblA allele, (c) partially diploid bacteria due to partial deletion from the 2nd chromosome with the chromosomes differing in their respective cblA allele, the gene content and synteny.
In vitro and in vivo viability of Bacteroides ovatus thyA mutants
As expected the viability of GH439 was similar to that of the wt when grown in the presence of thymidine (Fig. 3a, condition B and D). The same growth profile was observed for the wt strain in the absence of thymidine (Fig. 3a, condition C) whereas the viability of GH439 in the absence of thymidine declined immediately after inoculation (Fig. 3a, condition A) with 1% viability after 7 h and 0.001% after 24 h. This contrasted with the wt strain which, under the same conditions, showed no decline in viability over 24 h (Fig. 3a which shows that viability of cells in condition A is significantly lower than that of conditions B, C and D, at 7 and 24 h; 2-way ANOVA, p < 0.01). The rapid decline in viability seen after inoculation of the B. ovatus thyA deletion strain was not observed when bacteria were cultured under non-growing conditions and when thymidine was absent (Fig. 3b, which shows that viability of cells in condition A is significantly lower than that of conditions E, F, G, H, after 24 h; 2-way ANOVA, p < 0.01). Indeed, when GH439 bacteria lacked glucose as a carbon source (Fig. 3b, conditions E and F) the cells remained almost 100% viable after 24 h and by 72 h had retained more than 1% viability, in contrast to GH439 cells under growing conditions but lacking thymidine, which were no longer viable after 72 h (Fig. 3b, conditions A). The viability of GH439 cells inoculated under optimal environmental conditions (with thymidine but without glucose) increased slightly during the first 24 h, unlike the cells in the same conditions but without thymidine (Fig. 3b, conditions E and F). This is because YCFA is not a minimal medium and B. ovatus does not grow in minimal medium, with the yeast extract present in the medium (0.05%) most likely supporting some limited growth when thymidine is available. When GH439 cells were inoculated in medium with glucose but under non-optimal temperature and oxygen conditions, the profile was similar, independent of the presence of thymidine. Approximately 100% of cells retained their viability during the first 24 h, which then decreased slowly, probably due to oxygen stress, and after 96 h (4 days) approximately 1% of cells retained viability (Fig. 3b, conditions G and H). Thus, culture conditions that prevent bacterial growth protect bacteria from TLD.
Figure 3.
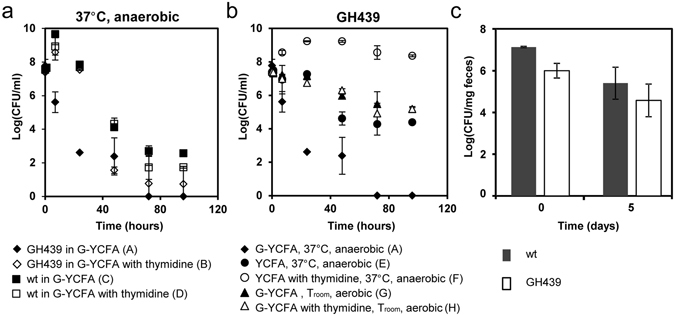
Growth and survival of B. ovatus wt and GH439 strains. (a) Growth and survival of wt and GH439 under optimal growing conditions with and without thymidine. (b) Growth and survival of GH439 under different environmental condition with and without thymidine. (c) Survival of wt and GH439 in mouse fecal samples prior to (0) and at 5 days post inoculation. In (a,b) data shown are representative of at least two independent experiments. In (c) values represent the mean of data from three independent experiments. Error bars indicate standard deviation.
To determine whether B. ovatus TLD occurs in vivo, mice were colonized with wt or GH439 bacteria their feces collected and bacterial cfu determined. The number of viable wt and GH439 bacteria after 5 days of storage of the fecal samples at ambient temperature and in the presence of oxygen were 3.9% and 6.6% of the initial counts, respectively (Fig. 3c), indicating that B. ovatus thyA deletion strains survive and can be readily acquired by a new host. Furthermore, TLD is entirely overcome through the exchange of genetic material between wt and GM bacteria, an occurrence that could also result in the escape of the transgene into an unintended population of bacteria.
Based on these results we conclude that TLD is unsuitable as a biosafety control measure in Bacteroides species and we therefore caution against its use in other bacterial species. Indeed, our work brings into sharp focus the uncertainty all biosafety control measures suffer from, in that whilst it is possible to define the genetic status of the genetically modified bacteria prior to delivery into a host, it is not possible to completely predict or control what type of bacteria are already present or enter the system at a later stage nor how the GM bacteria interact with other bacteria in their environment and how this impacts on their genetic stability and ability to escape any containment measures.
Material and Methods
Bacterial growth conditions and transformations
Escherichia coli strains were grown in Luria-Bertani medium at 37 °C. B. ovatus V975 and derivative strains (Table 1) were grown under anaerobic conditions at 37 °C in BHI medium (Oxoid, UK) supplemented with 0.001% haemin (BHIH) and 50 μg/ml thymidine when necessary. Antibiotics were added as selective agents when appropriate; erythromycin 5 μg/ml, kanamycin 50 μg/ml, rifampicin 35 μg/ml, spectinomycin 100 μg/ml, tetracycline 5 μg/ml and trimethoprim 100 μg/ml. E. coli was transformed by electroporation using a Gene Pulser II (Bio-Rad, UK). The construction of plasmids described below was performed using E. coli GC10 as the host strain. Plasmids were mobilized into B. ovatus V975 using the E. coli strain HB101 pRK201329 as a helper strain for triparental filter matings30.
Construction of B. ovatus mutants
To establish a counter-selectable gene deletion and integration system in B. ovatus first, the thyA gene was deleted from the chromosome of this strain, to enable counter-selection with trimethoprim. For this purpose a DNA fragment representing the thyA containing region of the B. ovatus chromosome void of thyA was created through splice overlap extension PCR. Firstly, the primer pairs ΔthyA_cls_F/ΔthyA_cls_R and ΔthyA_folA_F/ΔthyA_folA_R (see supplementary Table 1 for all PCR primers in study)were used with B. ovatus V975 genomic DNA as template to generate the respective PCR fragments. Second, the PCR products were used as templates for the splice PCR involving the primer pair ΔthyA_cls_F/ΔthyA_folA_R. The resulting 1595 bp product was cloned into the SmaI site of the suicide vector pFD51631 creating plasmid pGH138, which was conjugated into B. ovatus. Single-crossover transconjugants were selected on erythromycin (1 μg/ml) and subsequently plated onto BHIH agar containing thymidine and trimethoprim to select for the thyA gene deletion. The resulting colonies were screened by PCR using primer pair ΔthyA_left_flank/ΔthyA_right_flank and the resulting PCR product from a positive clone was sequenced in its entirety to confirm the integrity of the resulting thyA deletion strain denominated GH369. To allow subsequent cycles of gene deletions or insertions the suicide vector pGH139 was constructed by inserting a 905 bp PCR fragment carrying the thyA gene amplified with primer pair thyA_F/thyA_R into PshAI/ScaI restricted pFD516. Plasmid pGH139 allows for the selection of single-crossover transconjugants on BHIH agar upon conjugation into B. ovatus GH369 and the subsequent counter-selection on BHIH plates containing thymidine and trimethoprim to identify clones in which a double-crossover event has taken place. Using this system the tetracycline resistant B. ovatus GH439 was created as follows. The cblA integration target site was amplified using primer pair cblA_F/cblA_R and the resulting 2040 bp product was cloned into PvuII restricted pGH139. Next, the resulting plasmid was restricted with EcoRI and a tetQ carrying fragment, generated through PCR using primer pair tetQ_MfeI_F/tetQ_MfeI_R and pFI271632 as template, was restricted with MfeI and ligated into the EcoRI site generating pGH139_cblA_tetQ. This plasmid was then conjugated into B. ovatus GH369 and tetQ-tagged double-crossover transconjugants were isolated in a two-step process as described above. A second integration plasmid was created in a similar fashion using the primer pair 918_F/918_R and the resulting 2075 bp product was cloned into the SmaI site of pGH139 and the intrinsic EcoRV site was used as the cloning site for the tgfβ_ex1 gene acquired through gene synthesis, coding for the active form of the human transforming growth factor beta-1 (position 279–390) fused to the Bacteroides fragilis enterotoxin signal peptide. The resulting plasmid was employed to generate B. ovatus GH378 from GH369. To delete the oxyR gene from the chromosome of B. ovatus a 2661 bp PCR fragment generated with primer pair ΔoxyR_F/ΔoxyR_R was cloned into PvuII restricted pGH139 and subsequently the internal PvuII/NruI fragment was deleted from the oxyR gene resulting in pGH148, which in turn was used to generate GH397 from GH378. Finally pGH139_cblA_tetQ was used to generate B. ovatus GH440 from GH397.
Filter mating experiments
Overnight cultures of strains were grown anaerobically in BHIH containing thymidine and in the case of B. ovatus GH439 and GH440 with and without tetracycline. Cultures were then inoculated 1/20 into 20 ml BHIH containing thymidine and incubated anaerobically for 3.5 h. Subsequently donor and recipient cultures were united in 50 ml tubes, mixed and spun down at ambient temperature and 5000 g for 30 min. Culture supernatants were removed, cells were resuspended in the same media, transferred onto a 0.45 μm filter disc located on a BHIS agar plate containing thymidine and incubated anaerobically overnight. To enumerate the conjugation efficiency, cells were washed off the filter and serial dilutions plated or spotted onto either BHIS agar (V975), BHIS agar containing thymidine and tetracycline (GH439, GH440) or, BHIS agar containing tetracycline and rifampicin (transconjugants).
Animal studies
Seven-week-old male C57BL/6J mice were housed in a conventional animal facility and fed standard laboratory chow. Prior to administering B. ovatus strains, mice (two groups of 3) were treated for 7 days with 1 mg/ml ampicillin and 1 mg/ml neomycin via drinking water. Bacteria suspensions of B. ovatus GH439 (ΔthyA, cblA::tetQ) and B. ovatus V975 (wt) containing 5 × 108 CFU were administrated sequentially by orogastric gavage with a 1 h interval between gavages. Animal experiments were conducted in full accordance with the Animal Scientific Procedures Act 1986 under Home Office approval. Fresh feces were collected into sterile containers, weighed and homogenized in PBS. Serial dilutions of the suspensions were plated onto BHIH-RNAG (BHIH plates supplemented with 35 µg/ml rifampicin, 10 µg/ml neomycin, 75 µg/ml amikacin and 100 µg/ml gentamycin) to select for the B. ovatus V975, BHIH-ThymidTRNAG (plates supplemented with 50 µg/ml thymidine, 5 µg/ml tetracycline, 35 µg/ml rifampicin, 10 µg/ml neomycin, 75 µg/ml amikacin and 100 µg/ml gentamycin) was used to select for B. ovatus GH439, and BHIH-TRNAG (plates supplemented with 1.5 µg/ml tetracycline, 35 µg/ml rifampicin, 10 µg/ml neomycin, 75 µg/ml amikacin and 100 µg/ml gentamycin) to select for cells in cases of horizontal gene transfer. The plates were incubated anaerobically at 37 °C and CFU were counted 48 h later.
PCR screening for horizontal gene transfer of thyA, tetQ, BACOV975_00918 and oxyR after filter mating
Colonies obtained on BHIS agar containing tetracycline and rifampicin were assessed by colony PCR by suspending single colonies in 30 µl of sterile water and using 1 µl of this cell suspension in a 25 µl PCR reaction using GoTaq Green Master Mix (Promega) and primer pairs ΔthyA_folA_R/ΔthyA_F (a), cblA_F/cblA_R (b), oxyR left_flank/oxyR_r_flank3 (c) or 918_left_flank/918_right_flank (d) according to the manufacturer’s instruction. The resulting amplicons (Table 2) were analyzed and compared by gel electrophoresis to amplicon patterns obtained with DNA isolated from donor and recipient cultures grown for the filter mating experiment.
Screening for horizontal gene transfer of tetQ and thyA in the murine colon by PCR
Colonies obtained on BHIH-TRNAG plates were streaked onto BHIH-TRNAG plates containing 5 µg/ml tetracycline and incubated anaerobically at 37 °C. Single colonies were either analyzed by colony PCR as described above or transferred into BHIH medium supplemented with tetracycline and total DNA was extracted from the cultures 24 h later using the GeneJET Genomic DNA Purification Kit (ThermoFisher) according to the manufacturer’s instructions. To determine the genotype of the bacterial isolates with regards to thyA, cblA and tetQ, PCRs using cell suspensions or genomic DNA as template were carried out using primer pairs ΔthyA_folA_R/thyA_F (a), cblA_F/cblA_R (b) and RT_tetQ_3′/RT-tetQ_5′ (e) and the resulting amplicons (Table 2) analyzed by gel electrophoresis and compared to amplicon patterns obtained with DNA isolated from B. ovatus V975 and GH439 cultures grown in parallel.
Viability tests
B. ovatus wt and GH439 strains were grown over-night at 37 °C under anaerobic conditions, in YCFA medium supplemented with 0.5% glucose (G-YCFA) and with 50 µg/ml thymidine in the case of GH439, washed once with PBS and diluted (1/100) in YCFA or G-YCFA. YCFA and G-YCFA cultures of wt and GH439 were kept at 37 °C under anaerobic conditions or at ambient temperature under aerobic conditions. When indicated, wt and GH439 cultures were supplemented with 50 µg/ml thymidine. At different time points samples were taken diluted in PBS and plated in BHI (wt) or BHI supplemented with 50 µg/ml thymidine (GH439). The plates were incubated anaerobically at 37 °C and CFU counted 48 h later.
Electronic supplementary material
Acknowledgements
This work was supported by grants from Biotechnology and Biological Sciences Research Council (BBSRC) (BB/J004529/1 and BB/L004291/1, S.R.C.).
Author Contributions
U.W. and A.L.C. designed and performed the experiments and analyzed the data. S.R.C. and M.S. conceived the study and S.R.C. obtained the funding. U.W., A.L.C. and S.R.C. wrote the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Udo Wegmann and Ana Lucia Carvalho contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02591-6
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cano-Garrido, O., Seras-Franzoso, J. & Garcia-Fruitos, E. Lactic acid bacteria: reviewing the potential of a promising delivery live vector for biomedical purposes. Microb Cell Fact14, 137-149, doi:10.1186/s12934-015-0313-6 (2015). [DOI] [PMC free article] [PubMed]
- 2.Daniel C, Roussel Y, Kleerebezem M, Pot B. Recombinant lactic acid bacteria as mucosal biotherapeutic agents. Trends Biotechnol. 2011;29:499–508. doi: 10.1016/j.tibtech.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Robert, S. & Steidler, L. Recombinant Lactococcus lactis can make the difference in antigen-specific immune tolerance induction, the type 1 diabetes case. Microb Cell Fact13, Suppl 1:S11, doi:10.1186/1475-2859-13-S1-S11 (2014). [DOI] [PMC free article] [PubMed]
- 4.Mimee, M., Tucker, A. C., Voigt, Christopher, A. & Lu, T. K. Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell Syst1, 62–71, doi.org/10.1016/j.cels.2015.06.001 (2015). [DOI] [PMC free article] [PubMed]
- 5.Kong, W., Kapuganti, V. S. & Lu, T. A gene network engineering platform for lactic acid bacteria. Nucleic Acids Res44, e37, doi.org/10.1093/nar/gkv1093 (2015). [DOI] [PMC free article] [PubMed]
- 6.de Vos WM. Gene expression systems for lactic acid bacteria. Curr Opin Microbiol. 1999;2:289–295. doi: 10.1016/S1369-5274(99)80050-2. [DOI] [PubMed] [Google Scholar]
- 7.Braat H, et al. A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn’s disease. Clin Gastroenterol Hepatol. 2006;4:754–759. doi: 10.1016/j.cgh.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 8.Farrar MD, et al. Engineering of the gut commensal bacterium Bacteroides ovatus to produce and secrete biologically active murine interleukin-2 in response to xylan. J Appl Microbiol. 2005;98:1191–1197. doi: 10.1111/j.1365-2672.2005.02565.x. [DOI] [PubMed] [Google Scholar]
- 9.Hamady ZZR, et al. Identification and use of the putative Bacteroides ovatus xylanase promoter for the inducible production of recombinant human proteins. Microbiology. 2008;154:3165–3174. doi: 10.1099/mic.0.2008/019109-0. [DOI] [PubMed] [Google Scholar]
- 10.Hamady ZZR, et al. Xylan-regulated delivery of human keratinocyte growth factor-2 to the inflamed colon by the human anaerobic commensal bacterium Bacteroides ovatus. Gut. 2010;59:461–469. doi: 10.1136/gut.2008.176131. [DOI] [PubMed] [Google Scholar]
- 11.Steidler L, et al. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat Biotechnol. 2003;21:785–789. doi: 10.1038/nbt840. [DOI] [PubMed] [Google Scholar]
- 12.Barner, H. D. & Cohen, S. S. The induction of thymine synthesis by T2 infection of a thymine requiring mutant of Escherichia coli. J Bacteriol. 68, 80–88 (1954). [DOI] [PMC free article] [PubMed]
- 13.Ahmad SI, Kirk SH, Eisenstark A. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu Rev Microbiol. 1998;52:591–625. doi: 10.1146/annurev.micro.52.1.591. [DOI] [PubMed] [Google Scholar]
- 14.Tang, Y. P. Identification and Characterization of Genes in Bacteroides fragilis Involved in Aerotolerance. Department of Molecular Biology & Microbiology (Tufts University, 2000).
- 15.Syvanen, M. & Kado, C. I. Horizontal Gene Transfer, Edn. 2nd (Academic Press, San Diego, 2002).
- 16.Whittle G, Hamburger N, Shoemaker NB, Salyers AA. A Bacteroides conjugative transposon, CTnERL, can transfer a portion of itself by conjugation without excising from the chromosome. J. Bacteriol. 2006;188:1169–1174. doi: 10.1128/JB.188.3.1169-1174.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishibashi M, Sugino Y, Hirota Y. Chromosomal location of thymine and arginine genes in Escherichia coli and F′ incorporating them. J. Bacteriol. 1964;87:554–561. doi: 10.1128/jb.87.3.554-561.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasson MJ, et al. Characterization and exploitation of conjugation in Lactococcus lactis. Int Dairy J. 1995;5:757–762. doi: 10.1016/0958-6946(95)00030-5. [DOI] [Google Scholar]
- 19.Godon JJ, Pillidge CJ, Jury K, Gasson MJ. Characterization of an original conjugative element: The sex factor of Lactococcus lactis 712. Lait. 1996;76:41–49. doi: 10.1051/lait:19961-25. [DOI] [Google Scholar]
- 20.Levin BR, Bergstrom CT. Bacteria are different: observations, interpretations, speculations, and opinions about the mechanisms of adaptive evolution in prokaryotes. Proc Natl Acad Sci USA. 2000;97:6981–6985. doi: 10.1073/pnas.97.13.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen MT. Multiplicity of genome equivalents in the radiation-resistant bacterium Micrococcus radiodurans. J Bacteriol. 1978;134:71–75. doi: 10.1128/jb.134.1.71-75.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tobiason DM, Seifert HS. The obligate human pathogen, Neisseria gonorrhoeae, is polyploid. PLoS Biol. 2006;4:1069–1078. doi: 10.1371/journal.pbio.0040185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lobnerolesen A, Atlung T, Rasmussen KV. Stability and replication control of Escherichia coli minichromosomes. J. Bacteriol. 1987;169:2835–2842. doi: 10.1128/jb.169.6.2835-2842.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wegmann, U., Goesmann, A. & Carding, S. R. Complete genome sequence of Bacteroides ovatus V975. Genome Announc 4, e01335-16, doi:10.1128/genomeA.01335-16 (2016). [DOI] [PMC free article] [PubMed]
- 26.Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 27.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michelsen O, Hansen FG, Albrechtsen B, Jensen PR. The MG1363 and IL1403 laboratory strains of Lactococcus lactis and several dairy strains are diploid. J. Bacteriol. 2010;192:1058–1065. doi: 10.1128/JB.00900-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shoemaker NB, Getty C, Gardner JF, Salyers AA. Tn4351 transposes in Bacteroides spp and mediates the integration of plasmid R751 into the Bacteroides chromosome. J. Bacteriol. 1986;165:929–936. doi: 10.1128/jb.165.3.929-936.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith CJ, Rollins LA, Parker AC. Nucleotide sequence determination and genetic analysis of the Bacteroides plasmid, pBI143. Plasmid. 1995;34:211–222. doi: 10.1006/plas.1995.0007. [DOI] [PubMed] [Google Scholar]
- 32.Wegmann U, Horn N, Carding SR. Defining the bacteroides ribosomal binding site. Appl Environ Microbiol. 2013;79:1980–1989. doi: 10.1128/AEM.03086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


