Abstract
Microorganisms are key components for plant biomass breakdown within rumen environments. Fibrobacter succinogenes have been identified as being active and dominant cellulolytic members of the rumen. In this study, F. succinogenes type strain S85 was adapted for steady state growth in continuous culture at pH 5.75 and confirmed to grow in the range of pH 5.60–5.65, which is lower than has been reported previously. Wild type and acid tolerant strains digested corn stover with equal efficiency in batch culture at low pH. RNA-seq analysis revealed 268 and 829 genes were differentially expressed at pH 6.10 and 5.65 compared to pH 6.70, respectively. Resequencing analysis identified seven single nucleotide polymorphisms (SNPs) in the sufD, yidE, xylE, rlmM, mscL and dosC genes of acid tolerant strains. Due to the absence of a F. succinogenes genetic system, homologues in Escherichia coli were mutated and complemented and the resulting strains were assayed for acid survival. Complementation with wild-type or acid tolerant F. succinogenes sufD restored E. coli wild-type levels of acid tolerance, suggesting a possible role in acid homeostasis. Recent genetic engineering developments need to be adapted and applied in F. succinogenes to further our understanding of this bacterium.
Introduction
Understanding and overcoming plant biomass recalcitrance is a crucial step to enable industrial biofuels and biomaterials production1, 2. There is interest in examining and learning from environments where nature’s best lignocellulosic deconstruction occurs. Industrial ethanologenic bacteria and yeast are capable of ethanol production in pH ranges of pH 3.5–7.5 and pH 2–6.5, respectively3, 4. In general, anaerobic cellulolytic bacteria are not known to grow (i.e., increase microbial cell mass) below pH 6.0 and the majority of fermentative microorganisms grow within a relatively narrow pH range near circumneutral pH5. There is a biotechnological interest in fermentation processes and enzymes that can operate at lower pH values to mitigate contamination and to enhance productivity6, 7.
Herbivorous mammals have a variety of specialized compartments to breakdown and to access nutrients derived from dietary plant materials. Efficient biomass breakdown in the rumen is facilitated by a complex consortium of anaerobic microorganisms and host actions8–10, and building an expanded gene catalogue for biomass deconstruction is one area of applied interest11. Rumen microorganisms produce an abundance of different enzymes necessary for biomass deconstruction, as well as producing short chain (C2 to C6) volatile fatty acids (VFAs) and other compounds that support animal energy requirements11. Other microbial metabolic products include lactic acid, methane, hydrogen and carbon dioxide, as well as numerous cross feeding interactions and opportunities among microbiome members.
Several studies have observed large differences in rumen bacterial community composition for animals feeding on the same diet12, 13, while other studies indicate steady microbiome core taxa are maintained even as microbial communities shift following diet changes14, 15. Modern cattle diets contain rapidly fermentable feedstuffs, which results in low rumen pH for much of the day, reducing the efficiency of forage utilization and can lead to a condition known as sub-acute ruminal acidosis or chronic acidosis16. Inefficient and slow anaerobic degradation of cellulose by rumen microbial consortia has been reported as low as pH 5.3 in batch experiments for feedlot-type diets. Major rumen pH decline can reduce populations of cellulolytic bacteria and increase bacterial endotoxins leading to inflammatory responses and a negative impact on animal health16. Approaches to attenuate rumen pH decreases have included application of various feed supplements, such as probiotic Saccharomyces cerevisiae strain CNCM I-107717, Megasphaera elsdenii, strain H6F3218, flavonoids19, dicarboxylic acids malate and fumarate20, and essential oils21. Hence, there is interest in better understanding and preventing adverse outcomes associated with sub-acute ruminal acidosis.
In early rumen studies, the Gram positive Ruminococcus flavefaciens and Ruminococcus albus, and Gram negative Fibrobacter succinogenes were identified as being key active cellulolytic members in the rumen8. More recently, molecular biology tools have provided new insights into the complex community that resists cultivation and greatly expanded the cow rumen carbohydrate-active gene catalogue22. Metagenomic studies have characterized rumen microbial community structure and functions under a variety of conditions23–25. Amongst other challenges, obtaining ruminal pH samples and analyses can be complicated as ruminal pH is not homogenous and different analytical techniques can produce different results26.
In order to better understand the limitations of a key rumen fermentative microorganism at lower pH, we studied F. succinogenes type strain S85 as a model organism using chemostat cultivation and systems biology tools. Fibrobacter spp. are able to ferment xylan, glucose, cellobiose and cellulose to form succinic, acetic and formic acids27. Members of the phylum are found in termite guts and Fibrobacter spp. have been detected in diverse terrestrial and aquatic environments. A 3.84 Mb complete genome sequence for F. succinogenes S85 has been reported28. A study with 22 combinations of cellulose-fed dilution rates (D, 0.014–0.076 h−1) and pH (6.11–6.84) revealed F. succinogenes S85 could not maintain steady state growth below pH 6.1 and was washed out of continuous cultivation29, consistent with an earlier study where the same strain was washed out of cellobiose grown continuous cultures at pH 6.0 (D, 0.165 h−1)30. We hypothesized that strain S85 could grow in continuous cultivation below pH 6.0 following strain adaption and might have enhanced properties for biomass deconstruction and fermentation.
Materials and Methods
Bacterial strains and culture media
Fibrobacter succinogenes S85 (ATCC 19169T) was purchased from ATCC although the bacteria used in the experiment were a kind gift from Dr. Garret Suen at University of Wisconsin-Madison. Growth medium for F. succinogenes S85 was prepared as described previously28. Briefly, boiled modified Dehority medium (MDM) was saturated with high purity CO2 during cooling to room temperature, anaerobically dispensed and sealed in serum bottles or Hungate tubes, and sterilized by autoclave. Before inoculation, medium amendments were added, including cellobiose at a concentration of 4 g/L unless otherwise specified. The complete medium also contained 0.1% cysteine-HCl as a reducing agent and 0.001% resazurin as an oxygen indicator. Cultures were incubated at 37 °C without shaking.
Escherichia coli knockout mutant strains were purchased from the E. coli Genetic Stock Center (CGSC) at Yale University. The mutant library was originally constructed as the Keio collection31. The identity of each mutant strain was confirmed by PCR amplification of the junction region spanning the kanamycin resistance cassette and its upstream region, with the k1 and H1 PCR primers, as described previously31.
Bioreactor setup
F. succinogenes S85 was grown at 37 °C in a 500-ml (vessel capacity 1.3 L) culture using a water-jacketed BioFlo110 bioreactor (New Brunswick Scientific, Edison, NJ) for continuous growth at different dilution rates over hundreds of hours. Temperature, pH, and agitation were monitored and controlled in the bioreactor via the BioCommand 2.62 software. Continuous culture conditions were similar to those described previously29, 32. Briefly, ultrapure N2 and CO2 gases were connected to a bioreactor gas mixing unit and delivered to the bioreactor’s sparger through a sterile filter and tube. The medium feed line was connected to an anaerobic 19-L glass medium reservoir through a glass 3-way medium break tube. The gas inlet on the medium break tube was slightly pressurized with N2 gas to prevent medium from flowing backward. The medium flow rate was controlled with a MINIPULS 3 peristaltic pump (Gilson, Middleton, WI) and was measured by weighing collected effluent over a period of time. Dilution rates were varied throughout the course of the experiment and are described below. The bioreactor and medium feed carboy were kept anaerobic via gas sparging with either N2 or CO2 gases. Culture pH was maintained and controlled through a combination of gas mixing and acid addition. The controller was set to constantly sparge N2 into the culture, which maintained an anaerobic environment and gradually increased the pH due to the buffering property of carbonate in the medium. Once culture pH raised above the set point, the CO2 valve was opened by the pH control system and equilibrated the culture pH around the set point within a ±0.01 range. For conditions where pH set points were below carbonate buffering capacity (~pH 6.1), 0.5 N HCl was pumped into the bioreactor by the pH control system. Gel-reference pH electrodes were purchased from Mettler-Toledo (Woburn, MA). After autoclaving of the bioreactor and throughout the course of culturing, the electrodes were regularly checked and calibrated for accuracy, by measuring the pH of a culture sample taken from the bioreactor with a calibrated external pH meter (Fisher Scientific Accumet AB150, Pittsburg, PA). To generate acid tolerant F. succinogenes, N-methyl-N-nitro-N-nitrosoguanidine (NTG, obtained from TCI America and manufactured by Tokyo Chemical Industry Co. Ltd, Tokyo, Japan)33–35 was added to chemostat cultures at 15 µg/ml, and tolerant strains were selected by growth at lower pH based upon previous literature values29, 30. The dosage of NTG at 15 µg/ml was chosen based on previous studies for different bacteria, e.g. ref. 36, and also on preliminary experiments where a range of NTG concentrations were tested for its effects on log phase culture in Hungate tubes. Wild-type F. succinogenes treated at this level had static growth and no significant autolysis measured by optical density readings.
Biomass pre-treatment, culturing and compositional analysis
Milling of dry corn stover was conducted firstly with a Wiley Mill Model 4 (Thomas Scientific, Swedesboro, NJ) through a 2-mm mesh, and further milled through a number 60 mesh with a Wiley Mini-Mill (Thomas Scientific). The 60-mesh milled corn stover was sieved through a 12-in stainless-steel number 170 mesh (Fisher Scientific, Pittsburgh, PA) on a shaking platform at 300 rpm. The sieved corn stover was added into MDM at 6 g/L in serum bottles, de-gassed, sealed, and autoclaved for 15 min. Before inoculation, amendments, except cellobiose, were added as described above. Inocula were grown in cellobiose batch culture until OD680 reached around 1.0, at which time the cultures were washed with oxygen-depleted MDM, resuspended and injected into serum bottles at a density equivalent to OD680 0.1. Carbohydrate content in corn stover was analyzed with a quantitative saccharification assay as described before37, 38. Briefly, biomass before or after fermentation was washed to remove bacteria and then hydrolyzed in 72% w/w H2SO4 for 1 h at 30 °C and was further broken down into oligomers in 4% w/w H2SO4 at 121 °C for 1 h. Samples were then neutralized to pH ~7.0 with calcium carbonate, filtered through 0.2 µm and carbohydrate content (glucose, xylose, galactose, mannose, and arabinose) was quantified by high-performance liquid chromatography (HPLC) against known standards.
High-performance liquid chromatography (HPLC) analysis
Concentrations of cellobiose, succinate, formate and acetate in culture samples were analyzed with HPLC. Samples taken at different time points were stored at −20 °C. Before HPLC analysis, the samples were thawed, filtered through a 0.2-μm nylon filter and acidified with 2 M sulfuric acid. Twenty microliters each of the acidified samples were then applied to an Aminex HPX-87H, 300 × 7.8 mm column (Bio-Rad, Hercules, CA) on a Hitachi LaChrom Elite System (Hitachi High Technologies America, Inc., San Jose, CA) or a Waters Breeze system (Waters Corp., Milford, MA). Analysis was performed at a flow rate of 0.5 ml/min in 5 mM H2SO4 for 35 min as previously described39. Soluble fermentation products were identified by comparison with retention times and peak areas of corresponding standards.
Genomic DNA isolation and preparation for resequencing or PCR
Cells were pelleted from a 50 ml chemostat sample by centrifugation (7,600 × g, 4 °C for 4 min), flash frozen in liquid nitrogen, and stored at −80 °C. The genomic DNA (gDNA) extraction protocol was adapted from Current Protocols in Molecular Biology40. Briefly, a solution of 565 µl of TE buffer, 2 µl of 100 mg/ml RNase A, 30 µl of 10% SDS and 3 µl of 20 mg/ml proteinase K was added to a bacterial pellet and the well-mixed suspension was incubated at 65 °C for 1 h. A 100 µl aliquot of 5 M NaCl and 80 µl of 4.2% NaCl/10% cetyltrimethylammonium bromide (CTAB) were sequentially added and mixed into the bacterial lysate, followed by a 10-min incubation at 65 °C and two rounds of phenol/chloroform extractions. Genomic DNA was precipitated with 0.7 × volume of isopropanol, washed with 70% ethanol, dried in a DNA mini speed vacuum (ATR, Laurel, MD), and finally resuspended in deionized H2O. DNA quality was assessed by NanoDrop (Thermo Fisher Scientific Inc.) and quantity was determined by the Qubit broad range double stranded DNA assay (Life Technologies, Grand Island, NY). Illumina TruSeq libraries for resequencing were prepared from gDNA as described in the manufacturer’s protocols following the low throughput protocol. Final libraries were validated by Qubit and visualized with Agilent 2100 Bioanalyzer (Agilent Technologies Santa Clara, CA) for appearance and size determination. Samples were normalized using the Illumina’s Library dilution calculator and pooled into a 10 nM stock.
Illumina sequence generation and analysis
Samples were sequenced on an Ilumina MiSeq Instrument (Illumina, San Diego, CA). Paired end sequencing (2 × 251) was completed on the TruSeq genomic DNA libraries with a MiSeq V2 cartridge. Sequencing reads raw data were imported to CLC Genomics Workbench 6.5 (CLC bio, Boston, MA), trimmed, and mapped to F. succinogenes S85 genome sequence (CP001792). The trim settings for the raw data inputs used for removal of low quality sequences were: limit = 0.02, removal of ambiguous nucleotides: maximal 1 nucleotide allowed, and removal of sequences on length: minimal 20 nucleotides. Genome sequence variations were detected with the quality-based variation detection tool, requiring presence in both forward and reverse reads and at least 10 times reads coverage. Minimum neighborhood and central quality were set at 15 and 20, respectively.
RNA extraction, RNA-seq library construction and analysis
Cells were pelleted from a 50 ml chemostat sample by centrifugation (7,600 × g, 4 °C for 4 min), flash frozen in liquid nitrogen, and stored at −80 °C. Ten milliliters of TRIzol reagent (Life Technologies) were added to the frozen cell pellet and mixed by vortexing until the pellet was fully dissolved. After treating with chloroform, the aqueous phase containing RNA was further processed with a Qiagen RNeasy mini kit (Qiagen, Valencia, CA), following the manufacturer’s instructions, including the on-column DNase I (Qiagen) treatment. Quantity of RNA was measured with a NanoDrop spectrophotometer and quality was assessed with an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA) on a RNA 6000 Nano Chip. Ribosomal RNA was depleted using the Ribo-Zero rRNA Removal Kit for Gram-negative Bacteria (Epicentre, Madison, WI) following the manufacturer’s protocol. RNA-seq libraries were constructed using an Epicentre Scriptseq mRNA-Seq library preparation kit (Illumina compatible). Briefly, as previously described (1), depleted RNA samples were used to synthesize barcode-tagged cDNA. The final RNA-seq libraries were quantified with a Qubit Fluorometer and library quality was assessed with Agilent Bioanalyzer. Final libraries were normalized using the Illumina’s library dilution calculator and pooled into a 10 nM stock. Trimmed raw count data for uniquely mapping reads underwent negative binomial distribution normalization and statistical analysis in the DESeq2 software41. Differential expression relative to the reference condition, pH 6.70, was defined as genes with a fold of change ≥2 for up-regulation, or ≤−2 for down-regulation, and with an adjusted P-value ≤ 0.05 based on DESeq2 analysis.
A gene ontology enrichment analysis was performed on differentially expressed genes using the 2014 version of Clusters of Orthologous Group (COG) database42 (available at ftp://ftp.ncbi.nih.gov/pub/COG/COG2014/data). Genes that showed differential expression were grouped into their COG categories. To examine which COG categories were over-represented in differentially expressed genes, a Fisher’s exact test was used. The significance level α was adjusted by Bonferroni correction.
Plasmid construction
Expression vector pBAD24 was used in the current study. It is inducible with a wide range of arabinose concentrations and expression levels are extremely low in the absence of arabinose43. Purified plasmid DNA were digested with XbaI and SdaI within the multiple cloning site, treated with calf intestinal alkaline phosphatase (Promega) and recovered after agarose gel electrophoresis with Zymoclean Gel DNA Recovery Kit (Zymo Research, Irvine, CA). Insert fragments were amplified with PCR reactions from gDNA of E. coli BW25113, wild-type F. succinogenes S85 or acid-tolerant F. succinogenes. Primers used for PCR amplifications were listed as follows: E. coli sigS sequence, Fwd, 5′-ACTCTAGAGAGTCAGAATACGCTGAAAGTTCA, Rev, 5′-ACCCTGCAGGTGAGACTGGCCTTTCTGACA; E. coli sufD, Fwd, 5′-ACTCTAGAGGCTGGCTTACCGAACAGC, Rev, 5′-ACCCTGCAGGCCCGCACTTTGTCG; E. coli sufDSE, Fwd, 5′-ACTCTAGAGGCTGGCTTACCGAACAGC, Rev, 5′-ACCCTGCAGGAGCCAACCGGATGAAAGC. The thermocycler program was set for an initial cycle at 94 °C for 3 min, 25 cycles of 94 °C for 30 sec, 56 °C for 30 sec and 72 °C for 60–150 sec, depending on insert size. DNA polymerase for the reactions was Platinum® Taq DNA Polymerase High Fidelity (Life Technologies, Carlsbad, CA). Amplified insert fragments were purified with DNA Clean & Concentrator columns (Zymo Research), digested with XbaI and SdaI and purified again with DNA Clean & Concentrator columns. The purified inserts were ligated (T4 DNA ligase, New England BioLabs, Ipswich, MA) into the processed pBAD24, then the ligation mixture was transformed by heat shock into appropriate host competent cells. For constructs that contain coding sequences from both F. succinogenes and E. coli, the plasmid DNA of pBAD24-sufDSE, constructed as described above, was digested with XbaI and SalI to remove the whole sufD coding sequence and the first 13 bp of the downstream gene, sufS. An insert fragment amplified from either the wild-type or acid-tolerant F. succinogenes gDNA with primer pair Fwd, 5′-ACTCTAGAGAACGCTGAATTTATACAGAACTTGC and Rev, 5′-ACGTCGACGGAAAAAATCATCTTGCACCTCCTGGAATTTCCGCCATCAATCGC was digested with XbaI and SalI, purified and ligated into the processed pBAD24-sufSE to create a fusion operon containing F. succinogenes sufD and E. coli sufSE genes. The reverse primer for these constructs included the 3′ end sequences of F. succinogenes sufD, the endogenous Shine-Dalgarno sequence and the first 13 bp of sufS. As a result, this fusion operon had full-length F. succinogenes sufD fused with the last 18 bp of E. coli sufD, which contained the Shine-Dalgarno sequence, and in-frame E. coli sufS and sufE.
Results
Culturing and generating acid tolerant F. succinogenes strains
To culture F. succinogenes at a 500-ml scale in a bioreactor, wild-type F. succinogenes S85 was first grown overnight in a serum bottle with 50 ml of medium supplemented with 3 g/L cellobiose, which was then inoculated into a bioreactor that contained 500 ml of fully reduced complete medium at pH 6.70. The bioreactor was initially run in batch mode and until the culture reached early stationary phase (OD680 around 1.3), around which point chemostat growth was initiated by continuous input of medium containing 3 g/L cellobiose, at 970 ml/day (D = 0.08 h−1). Initially, the chemostat was set at pH 6.70 and maintained using dynamic mixing of N2 and CO2 gases. Under such conditions the culture maintained a stable cell density with OD680 readings around 1.3 for at least 5 volume changes and was considered growing in steady state. To monitor cellobiose consumption and succinate accumulation in the bioreactor, samples were analyzed by HPLC. Succinate concentration during steady state was around 11.5 mM. The pH set point was then lowered to 6.25.
The pH 6.25 steady-state culture was treated with NTG at 15 µg/ml to begin the selection of acid tolerant strains33–35 (Fig. 1). At the time NTG was added into the bioreactor, medium feed was stopped for the following hour, and then restarted at half the dilution rate (D = 0.04 h−1) as prior to the treatment. To create an acidic environment and select for acid tolerant strains, the chemostat culture was adjusted with HCl to pH 6.05 at 21 h post-NTG treatment, near the pH value (pH 6.1) that is thought to be the lower limit of ruminal bacteria growth in continuous culturing29, 30, 44. Cell density was observed to decline steadily from OD680 1.48 at the time of treatment to OD680 0.27 at 30 h post-treatment, which suggested cells had undergone static growth and were being washed out, and some proportion of cells also likely underwent lysis. The medium feed was then stopped, and 250 ml of fresh medium was added to dilute down any possible detrimental metabolites or residual NTG. A continued decline in culture OD680 readings was observed until below OD680 0.05 at 53 h post-treatment, while at pH 6.05. Significant growth resumed at 148 h post-treatment with the culture density reaching OD680 0.66, at which time the medium feed was reestablished at a dilution rate of around 0.04 h−1. Another drop in cell density occurred around 169 h post-treatment (Fig. 1) and growth recovered within 40 hours by using the same approaches described above. After culture recovery, culture pH was gradually lowered below 6.0 in steps of pH values of 0.05 or 0.10 over a 30-hour period. The culture density was maintained around OD680 1.3 until the pH was lowered to 5.75, when it dropped below OD680 0.05 and soon recovered again (Fig. 1) remaining stable at OD680 ~ 1.3 (Fig. 1).
Figure 1.
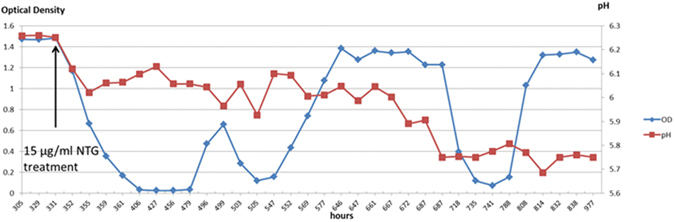
F. succinogenes growth and pH control after random mutagenesis. When the chemostat culture of wild-type F. succinogenes was grown in steady state in cellobiose medium at pH 6.25, mutagen NTG was added to induce random mutations. The pH and medium feed control units were carefully controlled to maintain viability of the culture and to screen for acid tolerant populations. As pH (red) decreased over time, the OD readings in blue shows culture adaptation to low pH stress accordingly. The mutant populations went through three major adjustments and finally reached steady state at pH 5.75. The selected culture was later confirmed to have the ability to tolerate a pH between 5.60 and 5.65 in batch culture.
Overall, after NTG treatment, the culture went through three major growth fluctuations along a series of adjustments of decreasing pH set points during a course of ~500 h. After the stable acid tolerant population had been obtained, aliquots were harvested, stored at −80 °C and were later able to be recovered in batch culture in medium pre-adjusted at pH 6.70 and pH 5.70, while the wild-type strain could only grow in pH 6.70 medium. In batch culture at pH 6.70, wild-type F. succinogenes S85 appeared to have a higher stationary phase cell density (OD680 around 1.2), while the acid tolerant strain had density readings around 0.68. Nevertheless, the doubling times for both strains were not different (Fig. 2). The selected F. succinogenes acid tolerant mutant population was later grown in a chemostat and tested for their pH lower limit. In the same chemostat settings (3 g/L cellobiose, D = 0.08 h−1), the culture successfully reached steady state at pH 5.65 and cell density readings remained stable around 1.0 for 148 h until the setpoint was further lowered to pH 5.60. The cell density then dropped below OD680 0.1 and thus the culture was unable to tolerate the new levels of acidity. Therefore, the acid tolerant F. succinogenes had a pH lower limit between pH 5.60 and pH 5.65. Metabolite profiles for cellobiose, succinate, formate and acetate in steady state at three pH set points were analyzed with HPLC (Fig. 3). The cellobiose concentration in samples from the pH 5.65 chemostat was higher (0.27 mM) than those at pH 6.70 and pH 6.10 (0.17 and 0.13 mM, respectively). Succinate concentrations increased from 11.4 mM, 12.0 mM to 12.7 mM as pH decreased from 6.70 to 6.10, and 5.65, respectively. Acetate had similar increases to succinate while formate dropped from 0.6 mM to 0.29 mM and rose to 0.8 mM at pH 6.70, 6.10 and 5.65, respectively.
Figure 2.
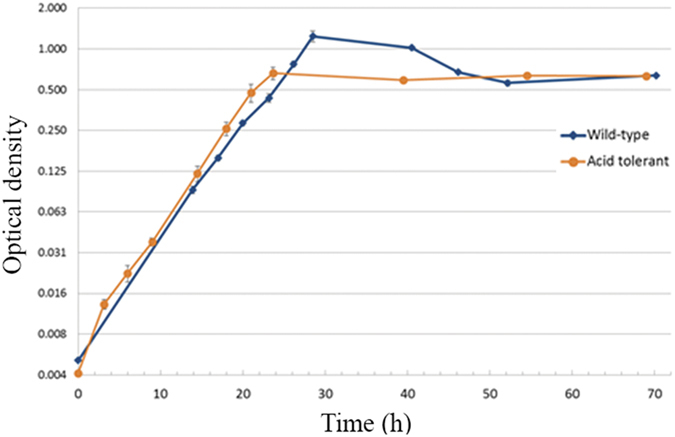
Growth curves of wild-type and acid tolerant strains of F. succinogenes in cellobiose medium. A growth curve of the acid tolerant F. succinogenes grown in 3 g/L cellobiose medium (initial pH 6.90 was measured by cell density readings and plotted against the wild type. The two had the same doubling time but the wild type reached a higher density (OD > 1.2) in late log phase while the acid-tolerant culture grew to OD around 0.7.
Figure 3.
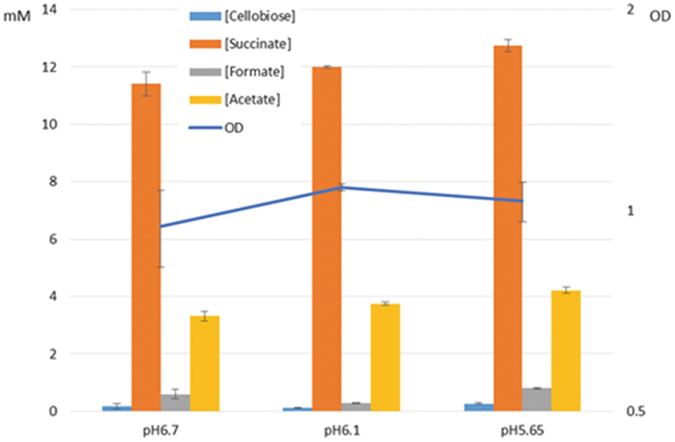
Metabolic profiles of chemostat culture in steady state at different pH values. After the chemostat culture reached steady state at the indicated pH, samples were taken for HPLC analysis. At least three samples per pH setting were analyzed. Concentrations of succinate and acetate in the culture increased as culture pH decreased, while pH 6.10 samples had the least formate concentration among the three. Concentration of cellobiose was also the highest at pH 5.65, suggesting a slight decrease of cellobiose intake or utilization despite the relatively constant cell density across conditions.
Biomass fermentation of the acid tolerant F. succinogenes strains
We compared the ability of F. succinogenes to digest fiber between wild type and acid tolerant strains in batch culture containing corn stover as the carbon source. Batch culture succinate concentrations in medium were measured as an indication of bacterial growth. In medium at pH 6.70, succinate concentration up to 72 h post-inoculation did not significantly differ between wild type and acid tolerant strains (P = 0.4). Similarly, wild type and acid tolerant strains inoculated into medium at pH 5.75 produced comparable amount of succinate (P = 0.4). However, the succinate concentration at 72 h in the high pH environment was about 2.8 times higher than that in the low pH regardless of the strains inoculated. We further examined sugar composition of the biomass before and after fermentation with a quantitative saccharification assay, a method to hydrolyze polysaccharides and analyze oligomer components from the samples. This analysis showed that there was no significant difference of glucose, xylose and mannose content in the remaining biomass after fermentation (7 days post inoculation) among samples from wild type and acid tolerant strains regardless of the pH of the medium (Supplementary Fig. 1). On the other hand, there was less galactose in the remaining biomass at pH 6.70 than at pH 5.75 when compared between the same strain (P = 0.008 between wild type and 0.02 between acid tolerant strain). Similarly, arabinose content was found to be less in samples at pH 6.70 than pH 5.75 (P = 3.6E-05 between wild type and 7.6E-04 between acid tolerant strain). However, there was no significant difference of sugar contents in corn stover after fermentation between wild type and acid tolerant strains at pH 5.75 (Supplementary Fig. 1), suggesting the acid tolerant strains digested corn stover in a similar manner to the wild type at low pH environments.
Genome sequence analysis
To identify variations in the acid tolerant strains, gDNA was prepared from a chemostat culture sample taken at pH 5.65 (time mark EFT723). Genomic DNA from wild-type F. succinogenes S85 was also prepared in parallel. This wild-type culture had been grown for two generations after we acquired it. Resequencing of the wild-type and acid tolerant populations generated overall genome sequence coverages of 398× and 309×, respectively. Raw data are available via the NCBI Sequence Read Archive (SRA) under accession SRP097623. A quality-based variant analysis was used to identify sequence variations in the wild-type and acid-tolerant strains using the published F. succinogenes S85 (CP001792) reference sequence. Table 1 lists all the detected variations (N = 8) between our wild-type strain sequence and the reference strain sequence, six of which are within coding regions. Two of the six variations are consecutive substitutions within the coding region of Fisuc_0822, an FG-GAP repeat protein gene45, 46 based on sequence homology. This results in a synonymous substitution at residue 1223 and a valine-to-isoleucine substitution at residue 1224. Another sequence variation was found within the coding region of Fisuc_1012, a gene encoding a branched-chain amino acid transport protein, AzlC, where the threonine at the 48th residue is substituted with an alanine. The remaining three variations within the coding regions of Fisuc_2022 and Fisuc_3045 are synonymous (Table 1). In addition, two single nucleotide insertions were identified in intergenic regions between Fisuc_2307 and Fisuc_2308, as well as between Fisuc_2832 and Fisuc_2833. The former insertion is an A, 45 bp and 141 bp away from the initiation codons of Fisuc_2307 and Fisuc_2308, respectively,as Fisuc_2307 is on the complement strand and Fisuc_2308 is on the forward strand. The other insertion is a T, located in the downstream region of both genes, 151 bp and 117 bp away from the stop codons of Fisuc_2832 and Fisuc_2833, respectively.
Table 1.
Variations in sequences between published F. succinogenes S85 and re-sequencing of F. succinogenes S85 in this study.
| Number | Coordinate on CP001792 | Locus Tag | Annotation | ORF direction | Position at ORF sequence | Sequence on CP001792 | Sequence determined in this study | Count | Coverage | Frequency | Average quality | Distance to initiation codon | residue change |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coding region | |||||||||||||
| 1 | 1007759 | Fisuc_0822 | FG-GAP repeat protein | — | 3670 | C | T | 115 | 116 | 99.1 | 36.87 | Val1224Ile | |
| 2 | 1007760 | Fisuc_0822 | FG-GAP repeat protein | — | 3669 | C | G | 115 | 116 | 99.1 | 36.87 | Thr1223, synonymous | |
| 3 | 1249999 | Fisuc_1012 | AzlC family protein | — | 142 | T | C | 179 | 237 | 75.5 | 37.483 | Thr48Ala | |
| 4 | 2498380 | Fisuc_2022 | Hypothetical | + | 2847 | C | T | 102 | 282 | 36.2 | 38.126 | Asp949, synonymous | |
| 5 | 2498437 | Fisuc_2022 | Hypothetical | + | 2904 | T | G | 123 | 265 | 46.4 | 35.132 | Thr968, synonymous | |
| 6 | 3746851 | Fisuc_3045 | Tryptophan synthase subunit | + | 1029 | G | T | 155 | 207 | 74.9 | 36.766 | Ala343, synonymous | |
| Noncoding region | |||||||||||||
| 1 | 2842868 | Intergenic (Fisuc_2307-Fisuc_2308) | — | A | 200 | 217 | 92.2 | 37.715 | 45, 141 | ||||
| 2 | 3495925 | Intergenic (Fisuc_2832, Fisuc_2833) | — | T | 144 | 160 | 90.0 | 37.521 | N/A | ||||
Seven single nucleotide variations were identified in the acid tolerant culture compared to the published F. succinogenes S85 sequence (Table 2). One variation is a deletion of G at nucleotide position 462 in the Fisuc_1133 coding sequence (a putative yidE permease gene). This deletion results in a frameshift mutation, generating a stop codon at the 18th codon position after the deletion. The mutant gene therefore has only 172 codons instead of 555 codons. The remaining six variations are single nucleotide substitutions, resulting in six residue changes in five genes, Fisuc_0527, Fisuc_1804, Fisuc_1945, Fisuc_2074 and Fisuc_2957, encoding SufD, XylE, RlmM, MscL and DosC, respectively.
Table 2.
Sequence variations between the acid tolerant strain and wild type of F. succinogenes S85.
| Number | Coordinate on CP001792 | Locus Tag | Gene name | Annotation | ORF direction | Position at ORF sequence | Sequence on CP001792 | Sequence determined in this study | Count | Coverage | Frequency | residue change | Gene homolog in E. coli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 612124 | Fisuc_0527 | sufD | ABC-type transport system involved in Fe-S cluster assembly, permease component | + | 514 | C | T | 61 | 151 | 63.04 | Arg172Cys | JW1671 (b1681) |
| 2 | 1396377 | Fisuc_1133 | yidE | Predicted permease, may bind an unidentified ligand | − | 462 | G | − | 200 | 200 | 100 | Frameshift. Stop codon at the 18th codon after the shift. | JW3662 (b3685) |
| 3 | 2230314 | Fisuc_1804 | xylE | sugar transporter | − | 205 | C | T | 217 | 220 | 96.14 | Ala69Thr | JW3991 (b4031) |
| 4 | 2399135 | Fisuc_1945 | rlmM (ygdE) | DNA alkylation repair | + | 331 | A | G | 199 | 199 | 94.12 | Thr111Ala | JW2777 (b2806) |
| 5 | 2561445 | Fisuc_2074 | mscL | Large conductance mechanosensitive channel protein. Critical roles in transducing physical stresses at cell membrane into an electrochemical response | − | 310 | T | C | 156 | 182 | 93 | Met104Val | JW3252 (b3291) |
| 6 | 2561669 | Fisuc_2074 | mscL | See above | − | 86 | C | A | 206 | 206 | 100 | Gly29Val | JW3252 (b3291) |
| 7 | 3649525 | Fisuc_2957 | dosC | Signal transducer; phosphorylation receiver or oxygen sensing | + | 803 | G | T | 230 | 232 | 100 | Gly268Val | JW5241 (b1490) |
RNA-seq analysis
To gain further insights into F. succinogenes physiology and gene regulation in acidic environments, global transcript profiles were generated with RNA-seq from chemostat steady state cultures at various pH set points. Transcript profiles of three samples each at pH 6.70 and 6.10 from wild-type F. succinogenes S85 chemostat were analyzed, as well as for four samples at pH 5.65 from the acid tolerant F. succinogenes S85 chemostat. RNA-seq data have been deposited in NCBI Gene Expression Omnibus (GEO) database under accession number GSE93907 and raw sequence data deposited at the NCBI SRA under accession number SRP097575. The number of reads, percentage trimmed and uniquely mapped reads, as well as normalized data are provided (Supplementary Dataset Number 1). The range of reads numbers were from 1,763,076 to 4,524,604 among all samples. Trimmed raw count data for uniquely mapping reads underwent negative binomial distribution normalization and statistical analyses in the DESeq2 software41. The overall correlation among biological replicates within each pH value and across all three pH conditions is shown in a principal component analysis (PCA) plot (Fig. 4A). The replicates for each condition clustered close to one another for the most part. The first principal component (55.5%) indicates that the mutant S85 chemostat culture at pH 5.65 was the largest variance factor. A hierarchical clustering analysis of the ten samples also demonstrates that samples within each condition grouped closely (Fig. 4B).
Figure 4.
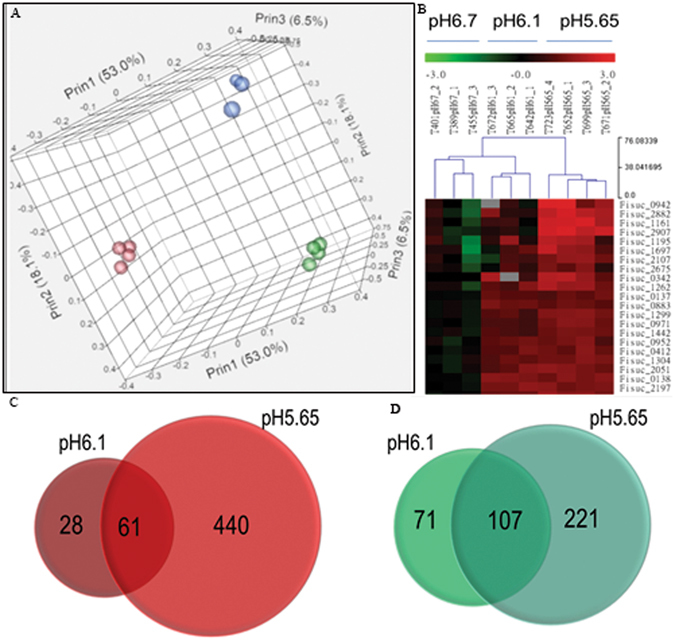
Overview of RNA-seq analysis of chemostat culture under low pH environment. (A) A principal component analysis (PCA) plot of the RNA-seq samples at three different pH levels. The close spatial arrangements of samples within each condition indicate high correlation among replicates and the distances among conditions suggest distinct gene expression profiles among conditions. (B) A partial hierarchical clustering analysis heat map of all 10 samples. Gene clustering was determined by expression level of each gene but the detailed tree structure was omitted in this graph due to complexity. (C) Numbers of genes that were significantly up-regulated (red) or (D) down-regulated (green) in pH 6.10 and pH 5.65. Numbers in the overlapped area between two circles indicate numbers of genes that were commonly regulated in both pH.
Differential expression relative to the reference condition at pH 6.70 identified 89 and 501 genes as being upregulated in pH 6.10 and 5.65 conditions, respectively, among which 61 are common between the two conditions (Fig. 4C). On the other hand, 178 and 328 genes were down-regulated, respectively in pH 6.10 and 5.65 conditions, with 107 common genes (Fig. 4D). All differential expression results are provided (Supplementary Dataset Number 1). The common up-regulated genes between pH 6.10 and 5.65 conditions include 31 hypothetical genes, a heat shock protein Hsp20 (Fisuc_1188), putative transcription regulators Fisuc_0335, Fisuc_0933 and Fisuc_1186, a diguanylate cyclase Fisuc_2957, and genes Fisuc_0137, Fisuc_0138 and Fisuc_2558, which are involved in tryptophan metabolism.
A gene ontology enrichment analysis was performed on differentially expressed genes. Among the up-regulated genes in the pH 5.65 samples, category V: defense mechanisms and category X: Mobilome: prophages, transposons were over-represented (P = 3.27E-06 and 8.00E-04, respectively) (Table 3). Two COG categories (G: Carbohydrate transport and metabolism and Q: Secondary metabolites biosynthesis, transport, and catabolism) were significantly down-regulated in the pH 5.65 samples. Four categories were over-represented from samples collected at pH 5.65 (C: Energy production and conversion, E: Amino acid transport and metabolism, G: Carbohydrate transport and metabolism, and T: Signal transduction mechanisms; Table 4). A list of all 162F. succinogenes genes annotated in category G was loaded into the KEGG database and 46 genes in the list were returned in the KEGG pathway analysis results. Among these 46 genes, the most up-regulated genes were Fisuc_1985 (1.4 fold at pH 5.65) and Fisuc_1572 (1.3 fold at pH 5.65). Fisuc_1985 is an annotated glycosidase that is involved in glycosaminoglycan degradation and beta-Lactam resistance. Conversely, an orthologous gene (Fisuc_1751) in the same category that was also annotated as K01207, was otherwise found down-regulated (1.8 fold) at pH 5.65. On the other hand, the majority of KEGG-annotated category G genes were down-regulated. Several genes down-regulated by more than 2 fold at pH 5.65 were found involved in glycolysis, such as Fisuc_1204 (glucose-6-phosphate isomerase), Fisuc_2891 (fructose-bisphosphate aldolase), Fisuc_0381 (phosphoglycerate kinase) and Fisuc_1261 (enolase). In the same carbon metabolism KEGG pathway: fsu01200, other genes that were down-regulated between 1.5 and 2 fold included Fisuc_1107 (glucokinase), Fisuc_1322 (6-phosphofructokinase 1), Fisuc_0400 (transketolase), Fisuc_0768 (2,3-bisphosphoglycerate-independent phosphoglycerate mutase) and Fisuc_0024 (ribose 5-phosphate isomerase A). These results suggest that F. succinogenes acid tolerant strains reduce the activity of the glycolysis pathway for producing pyruvate at low pH environments. We did not observe a known alternative pathway that was activated to compensate the energy production reduction through glycolysis.
Table 3.
Enrichment analysis of up-regulated genes in low pH environment.
| Category | Number of genes | p-valuea | |
|---|---|---|---|
| pH 5.65 | V, Defense mechanisms | 24 out of 79 | 3.27E-06 |
| X, Mobilome: prophages, transposons | 5 out of 8 | 8.00E-04 |
aTwo-tail fisher’s Exact test.
Table 4.
Enrichment analysis of down-regulated genes in low pH environment.
| Category | Number of genes | p-valuea | |
|---|---|---|---|
| pH 6.10 | G, Carbohydrate transport and metabolism | 26 out of 162 | 6.80E-07 |
| Q, Secondary metabolites biosynthesis, transport and catabolism | 8 out of 34 | 4.99E-04 | |
| C, Energy production and conversion | 22 out of 100 | 6.26E-04 | |
| pH 5.65 | E, Amino acid transport and metabolism | 35 out of 188 | 4.63E-04 |
| G, Carbohydrate transport and metabolism | 45 out of 162 | 1.28E-10 | |
| T, Signal transduction mechanisms | 4 out of 165 | 1.02E-04 |
Other genes possibly involved in carbon metabolism include a group of cellulases that were identified earlier28. Expression levels of those 31 cellulases in low pH conditions were compared to those at pH 6.70. At pH 5.65, only two cellulases, Fisuc_1661 (GH5 family endoglucanase28) and Fisuc_1426 (GH45 family xylanase47), were expressed differently relative to pH 6.70 (2.2 and 1.9 fold, respectively). Eleven cellulase genes were expressed within ±1.5 fold compared to pH 6.70. The other 18 cellulase genes were down-regulated more than 1.5 fold. The expression profiles for these genes at pH 6.10 were similar to pH 5.65, with only one gene up-regulated close to 2 fold (Fisuc_2033, 1.7 fold) and 13 genes down-regulated more than 1.5 fold (Supplementary Dataset Number 1). Before F. succinogenes digests cellulose, it is thought that it needs to adhere to the surface of cellulose28, 48, 49. A group of 10 proteins (Fisuc_0377, Fisuc_1474, Fisuc_1475, Fisuc_1326, Fisuc_1327, Fisuc_1979, Fisuc_2031, Fisuc_2293, Fisuc_2471 and Fisuc_2484) were identified and annotated to have fibro-slime domains and thought to be involved in adherence28, 50. In our analysis, only Fisuc_2471 was up-regulated by almost 2 fold at pH 6.10, five genes were moderately up-regulated (1.2–1.4 fold) and four genes were down-regulated (1.02–4.56 fold). At pH 5.65, Fisuc_2471 was the only up-regulated gene (1.4 fold) and the remaining nine genes were all down-regulated (1.1–9.5 fold).
A group of highly differentially expressed genes that drew our attention were genes encoding proteins with Fe-S clusters, specifically radical SAM family proteins. One of the mutations we identified in the acid tolerant F. succinogenes genome sequence is in the sufD gene, encoding a structural protein that may be involved in transfer of sulfur groups to form Fe-S clusters in the SufBCD complex51, 52. Fe-S cluster-containing proteins are involved in a pleiotropic range of cellular functions, such as gene regulation, metabolism, and electron transfer. Radical SAM proteins may be one of the groups of proteins under control of Suf and they perform functions such as methylation, isomerization, sulfur insertion, ring formation, anaerobic oxidation and protein radical formation53. Of the six significantly expressed radical SAM proteins at pH 5.65, four were up-regulated between 2.0 and 2.4 fold (Fisuc_1354, Fisuc_2052, Fisuc_2935 and Fisuc_2951) and the other two, Fisuc_0219 and Fisuc_2711, were down-regulated (2.8 and 2.4 fold). At pH 6.10, only one of the six genes was significantly up-regulated (Fisuc_2052, 2.1 fold). Analysis of differential gene results for cells collected pH 5.65 compared to pH 6.97 indicate a range of metabolic systems are involved in responding and adapting to growth in lower pH environments (Table 5). Additional RNA-seq analysis is provided as Supplementary Information.
Table 5.
Selected genes with significantly higher expression levels in pH 5.65 compared to pH 6.70.
| Locus Tag | log2 Fold Change | Adjusted P-value | Product Description (with TIGR or COG descriptions) |
|---|---|---|---|
| Fisuc_0029 | 1.3 | 2.94E-09 | lipoprotein (TIGR02167 bacterial surface protein 26-residue repeat) |
| Fisuc_0051 | 2.1 | 7.37E-13 | amidophosphoribosyltransferase (Glutamine phosphoribosylpyrophosphate amidotransferase) |
| Fisuc_0060 | 1.4 | 2.34E-05 | chaperonin Cpn10 |
| Fisuc_0159 | 3.2 | 2.93E-28 | Biotin synthase |
| Fisuc_0160 | 3.3 | 8.63E-13 | hypothetical protein (Putative threonine efflux protein) |
| Fisuc_0163 | 2.4 | 3.21E-07 | SirA family protein (Predicted redox protein, regulator of disulfide bond formation) |
| Fisuc_0539 | 2.4 | 0.000305 | type II and III secretion system protein (component PulD) |
| Fisuc_0582 | 2.1 | 2.47E-10 | Sporulation protein YtfJ |
| Fisuc_1002 | 2.8 | 7.52E-16 | Phosphotransferase system, phosphocarrier protein HPr |
| Fisuc_1095 | 1.8 | 0.02141 | Pyridoxal-5′-phosphate-dependent protein beta subunit (Cysteine synthase) |
| Fisuc_1133 | 1.4 | 8.92E-08 | YidE/YbjL duplication |
| Fisuc_1215 | 1.6 | 0.0242 | small multidrug resistance protein (Membrane transporters of cations and cationic drugs) |
| Fisuc_1277 | 3.1 | 7.56E-36 | preprotein translocase, SecE subunit |
| Fisuc_1362 | 1.4 | 1.18E-05 | lipoprotein (TIGR02167 bacterial surface protein 26-residue repeat) |
| Fisuc_1369 | 1.9 | 4.00E-11 | Rubredoxin-type Fe(Cys)4 protein |
| Fisuc_1491 | 2.4 | 1.41E-12 | NADH-ubiquinone/plastoquinone oxidoreductase chain 3 |
| Fisuc_1532 | 2.1 | 5.22E-09 | Chorismate binding-like protein (Anthranilate/para-aminobenzoate synthases component I) |
| Fisuc_1564 | 2.8 | 4.71E-07 | hypothetical protein (Predicted Na+-dependent transporter) |
| Fisuc_1593 | 2.6 | 1.4E-07 | GtrA family protein (Predicted membrane protein) |
| Fisuc_1665 | 5.1 | 1.70E-96 | phosphoadenosine phosphosulfate reductase |
| Fisuc_1668 | 4.6 | 1.53E-40 | cysteine desulfurase family protein |
| Fisuc_1673 | 3.7 | 4.34E-52 | putative RNA polymerase, sigma 70 family subunit |
| Fisuc_1758 | 1.3 | 1.57E-14 | hypothetical protein (Cell envelope:Other) |
| Fisuc_1818 | 1.9 | 0.003094 | glutamate synthase, NADH/NADPH, small subunit |
| Fisuc_1820 | 1.5 | 0.02261 | ammonium transporter |
| Fisuc_1821 | 3.0 | 1.36E-09 | nitrogen regulatory protein P-II |
| Fisuc_2091 | 2.5 | 0.001303 | Rubredoxin-type Fe(Cys)4 protein |
| Fisuc_2123 | 2.3 | 0.002766 | 4Fe-4S ferredoxin iron-sulfur binding domain protein |
| Fisuc_2558 | 1.9 | 2.99E-15 | Chorismate mutase |
| Fisuc_2559 | 2.0 | 1.03E-16 | Prephenate dehydrogenase |
| Fisuc_2908 | 2.1 | 0.000191 | (Sulfur transfer protein involved in) thiamine biosynthesis protein ThiS |
Acid survival for E. coli knockout mutants
To date, genetic manipulation tools are not available to construct targeted Fibrobacter gene knockout mutants. To investigate the function of individual genes, we used E. coli deletion strains for genes with homology to F. succinogenes genes. The six mutant genes we identified in the acid tolerant F. succinogenes all have annotated functions and homologs in wild-type E. coli BW25113 strain (Table 1). Initially acid survival percentage for each E. coli gene knockout mutant was assayed in minimal medium M9, and an acid sensitive ΔsigS mutant was included as a control54. As shown in Fig. 5(A), all the mutant strains except ΔJW1671 (ΔsufD) showed comparable acid survival percentages to the wild type, while the ΔsufD mutant survival colony count was only 2.7% of the wild type. We constructed a strain where the E. coli sufD in the sufDSE operon was replaced with the F. succinogenes sufD coding sequence and the coding frames and ribosome binding sites were retained (Fig. 5B). A ΔsufD mutant complemented with E. coli sufD alone did not restore its acid survival (Fig. 5C), likely due to polar effects on the sufSE genes in the sufABCDSE operon. The ΔsufD mutant complemented with the sufDSE three-gene operon partially restored the acid survival percentage. Although the baseline of acid survival of the ΔsufD mutant strain (without arabinose induction) seemed to be higher than the ΔsigS mutant, when induced, the complemented strains of the sufDSE fusion operon also partially restored acid survival percentage, and the sufD sequence from the acid tolerant F. succinogenes had a slightly better restoration than from the wild type.
Figure 5.
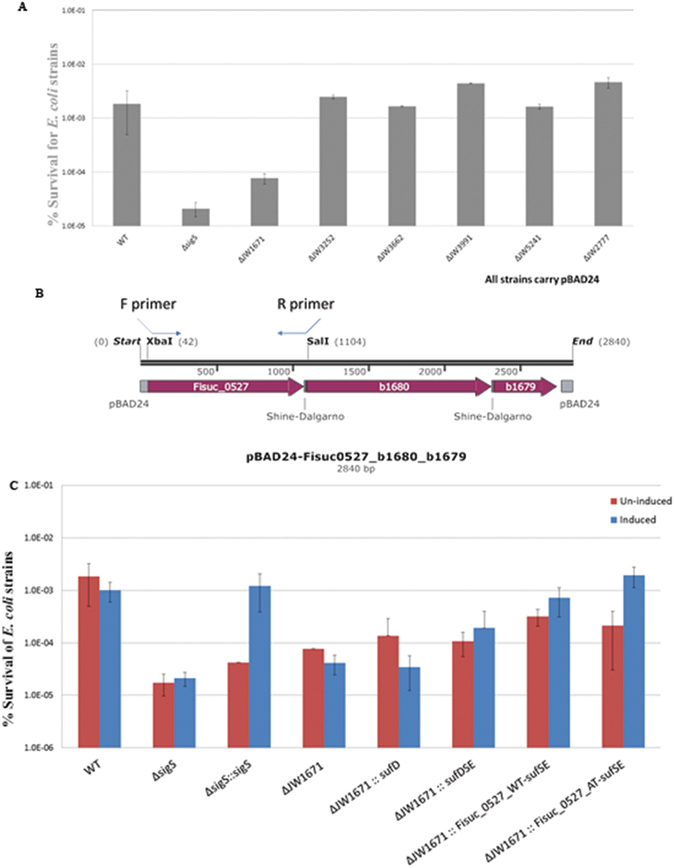
Acid survival assay of ΔJW1671 and complemented strains in E. coli. To investigate the roles of selected genes in the acid tolerant phenotype, E. coli strains deleted in individual genes homologous to acid tolerant F. succinogenes mutant genes were assayed using an acid survival CFU count method. A ΔsigS mutant was included as the acid sensitive control. (A) Among the six mutants, only the ΔJW1671 (sufD) mutant showed reduced ability to survival pH 2.5 treatment in minimal medium, compared to the wild type. (B) A linear map of the inset fragment of the fusion operon construct that was composed of F. succinogenes sufD fused with E. coli sufSE. A construct of sufDSE operon was first generated, and the sufD sequence was then replaced with the F. succinogenes counterpart. The reverse primer was designed to maintain the ribosome binding site of the immediate downstream gene. (C) Complementation of E. coli sufDSE but not sufD partially restored acid survival percentage, probably because the disruption of sufD caused a polar effect. Complementation with wild-type or acid tolerant F. succinogenes sufD restored the function of E. coli SufDSE, suggesting a possible role F. succinogenes sufD was involved in acid homeostasis.
Discussion
We demonstrated that adapted F. succinogenes strains are capable of steady state growth in continuous culture conditions at pH 5.75 and confirmed they were able to grow in the range of pH 5.60–5.65 (Fig. 1), which is lower than that which has been previously reported30. It is widely thought that the lower external pH limits for continuous growth of most ruminal bacteria are between 5.9 and 6.029, 30, 44. While industrial fermentative microorganisms, such as Zymomonas mobilis and yeast produce ethanol in the ranges pf pH 3.5–7.5 and pH 2–6.5, respectively3, anaerobic cellulolytic bacteria are not generally thought to grow below the pH 6.0 range5 and the results of this study may have broader implications for developing new biocatalysts.
Bacteria maintain their internal pH in face of changing external environments by mechanisms and responses generally called pH homeostasis. Using RNA-seq and resequencing techniques, this study has provided for the first time global insights into how F. succinogenes responds and adapts to growth in lower pH environments. When the external pH is lowered from 7.0 to 6.0, the internal pH of F. succinogenes is kept about 6.555. Additionally, it is slightly lower than 6.5 when the external pH is between 5.7–6.0, whereas it drops linearly as the external pH drops to a range lower than 5.755. Maintaining a pH gradient (ΔpH) across the membrane is of physiological importance because many cellular processes such as nutrient or ion transportation require the proton motive force (PMF) generated by transmembrane ΔpH and transmembrane electrical potential (Δψ)56. In low pH growth conditions, most bacteria take general strategies to counter the increased pH gradient across the membrane. These strategies for anaerobes include efflux of protons by proton pumps such as proton-coupled ATPases and cation-proton antiporters such as Na+/H+ and K+/H+ antiporters. In F. succinogenes, a V-type ATPase complex was identified (Fisuc_2835-Fisuc_2842), but only Fisuc_2839 (atpD) was significantly up-regulated during our acid treatment in the chemostat. AtpD is likely to contribute to the ATPase activity and Fisuc_2840 (atpI) may confer the proton transmembrane transporter activity, according to sequence analysis. However, given the observation that only one gene in the complex was up-regulated, it is not clear whether the activity of the putative proton pump was impacted during acid treatments or if this subunit was affected at the protein level. We also examined the expression of all annotated Na+/H+ and H+/Cl− antiporters. Although not statistically significant (P > 0.50 for both pHs), only one out of five genes was up-regulated in both pH conditions, while others were down-regulated (between 1.3 and 2.5 fold). A role for antiporters for acid resistance in this case is unclear.
Another pH homeostasis strategy is to alter metabolic networks to consume or reduce production of protons. For example, some enteric bacteria up-regulate glutamate and arginine decarboxylases to consume protons, and some anaerobic bacteria such as the facultative Streptococcus mutans increases the activity of malolactic fermentation to incorporate protons into lactate57. F. succinogenes lacks glutamate decarboxylases and the malolactic fermentation pathway, and the expression levels of arginine decarboxylases, Fisuc_2975 and Fisuc_2976, were not significantly altered at pH 6.10 and 5.65. Alternatively, we identified some significantly up-regulated genes that are involved in amino acid metabolism and those pathways possibly contribute to proton consumption. A glutamate synthase NADH/NADPH subunit, Fisuc_1818, which is involved in converting a glutamine and a 2-oxoglutarate into a glutamate with participation of an NADH and a proton, was up-regulated by 3.6 fold. We also found that the two NADH-quinone oxidoreductases in F. succinogenes, Fisuc_1491 and Fisuc_2345, were up-regulated by 5.3 and 2.2 fold, respectively, only at pH 5.65. In addition, a D-cysteine disulfhydrase (Fisuc_2650) that converts a cysteine to a hydrogen sulfide, ammonia and pyruvate was up-regulated only at pH 5.65 by 2.2 fold. This reaction may compensate for reduced pyruvate production via glycolysis and generate ammonia molecules, which in turn combines with a proton. Ammonia may also be generated by the reaction catalyzed by a glutamine synthetase, Fisuc_1816 (up-regulated only at pH 5.65 by 2.5 fold), which converts a glutamine to an ammonia molecule, a glutamate and generates an ATP. It is worth noting that of all 501 significantly up-regulated genes at 5.65, only 43 were mapped in the KEGG database. There may be novel metabolic pathways that are responsible for intracellular pH homeostasis in F. succinogenes and certainly many genes respond to lower pH environments at the level of transcription. This study did not account for post-translational regulation of enzyme activity.
Resequencing analysis identified only eight sequence differences between the reference genome and our S85 wild-type strain (Table 1), indicating the reference genome sequence is an appropriate reference for our study and it is of high-quality. Genome sequencing analysis for the pH 5.65 tolerant strains identified seven single-nucleotide mutations in six genes (Table 2). Those genes were xylE, encoding a sugar transporter protein with unknown sugar substrate, rlmM, encoding a DNA alkylation repair enzyme, dosC, a diguanylate cyclase gene, sufD, an Fe-S cluster assembly complex component gene, yidE, a permease gene and mscL, encoding a membrane mechanosensitive channel protein. We initially suspected that dosC, yidE or mscL was responsible for the acid tolerance phenotype, since dosC is a possible signal transducer that may regulate several target genes58, yidE belongs to a permease family that may control transport of substances, altering PMF across the membrane59, and mscL may be important for membrane integrity in stress conditions60. Given the phylogenetic uniqueness and lack of genetic manipulating tools of F. succinogenes, we chose to perform a functional assay on a well-studied model organism to test the phenotype of single knockout mutants of each gene. Our acid survival CFU counting assay of the E. coli knockout mutants indicated that all single gene knockout mutants but the sufD (JW1671) mutant demonstrated comparable survival rates to the wild type in low pH minimal medium (Fig. 5A). In E. coli, sufD is located in the sufABCDSE operon and based on the genomic context, replacing the sufD coding region with a kanamycin cassette removed the ribosomal binding sites, altering the coding frames of the two downstream genes, and thus causing a polar effect on the operon. Therefore, the single gene knockout mutant of JW1671 should be functionally a sufDSE mutant, and a single gene complementation of sufD might not be sufficient for restoring the phenotype. This phenotype prediction was supported by the acid survival plating results of the sufD gene-complemented strains, where acid survival was not restored, and the complementation of the three genes, sufDSE, showed partial restoration of the acid survival phenotype. In E. coli, the model organism for studying the SUF system, the genes sufS and sufE are important for the complete functionality of the SUF system as the biogenesis of Fe-S-proteins starts with transferring the sulfur from a free cysteine molecule catalyzed by SufS, a cysteine desulphurase, which is associated to and enhanced by the helper protein, SufE61. The sulfur is subsequently clustered with Fe2+ or Fe3+ ions and transferred to the SufBCD complex by a still unclear mechanism. SufB and SufD share 45% sequence similarity in their C-terminal 150 residues62. Initially thought to function as scaffold proteins, they form a stable complex with SufC, an ATPase that provides energy to transfer Fe-S clusters to apo-receptors52. The ATPase activity of SufC is higher when complexed with SufB and SufD than when assayed alone63. In in vivo studies, deletion of sufB, sufC or sufD impairs the function of the Suf complex64–66. These findings suggest SufB and SufD are crucial for bacterial Fe-S homeostasis, which is in turn critical to many cellular functions as the Fe-S clusters are essential cofactors in proteins that play pivotal roles in gene regulation, central metabolism, electron transfer, and DNA repair, among others51. Moreover, SufD is also reported to be indispensable for the iron acquisition process in the Fe-S cluster assembly62. In our E. coli acid survival plating assay, when the E. coli sufD coding region in the sufDSE operon on the complementing plasmid was replaced with wild-type Fisuc_0527 sequence, the survival percentage was improved as compared to the ΔsufD mutant. This indicates F. succinogenes sufD gene confers equivalent functions in the E. coli Suf machinery. Although replacing E. coli sufD with the mutant Fisuc_0527 sequence did not further increase the survival percentage significantly, further investigation into the arginine to cysteine mutation at the 172th residue in Fisuc_0527 may be an avenue for future amino acid substitution, structural and functional studies since other mutant proteins alone did not seem to have an effect on E. coli acid survival (Fig. 5C). The mutation in Fisuc_0527 may stabilize the protein structure or also the entire SufBCD complex structure, to maintain function at lower pH. Alternatively, but not exclusively, the mutation could improve iron acquisition during the Fe-S assembly process in low pH conditions and maintain regular cellular activities. It is also worth noting that in F. succinogenes, sufD (Fisuc_0527), sufS (Fisuc_0172, Fisuc_1055) and sufE (Fisuc_0329) are not in one operon as seen in E. coli. Those counterparts may execute similar functions deduced by their sequence homology, but the gene regulation mechanism is expected to be distinct. The possibility that the F. succinogenes acid tolerant phenotype results from the synergistic effect of multiple mutations cannot be ruled out and combinations of multiple-gene knockout mutants in E. coli could be assayed in the future.
When monitoring metabolite concentrations in the chemostat including cellobiose, we observed a slight increase in succinate, from 11.4 mM to 12.7 mM when pH was dropped to 5.65, as well as an acetate concentration increase and formate concentration decrease, which we interpret to be the cells rebalancing the energetics and metabolism of the system. The increased yield of succinate and acetate may be of interest for industrial applications. This result, combined with our observation that batch culture at a lower pH tended to have a slight positive pressure built up in the head space, suggests that the bacteria have an altered metabolic strategy and output. This is supported by our transcriptomic analysis that the glycolysis pathway was depressed in samples from low pH chemostat, although an alternative metabolic pathway remains to be fully elucidated. We suspect that the produced gas, possibly CO2, partially accounts for the carbon output in the low pH chemostat culture.
In the batch culture setting with biomass as carbon source, the wild type and acid-tolerant strains produced comparable amount of succinate, regardless of the medium pH. Although the acid-tolerant population was shown to be able to survive much more acidic environments, the limiting factors for biomass utilization appear to be, instead of long-term survival, the effects of pH on bacterial attachment to substrates, biofilm formation and cellulose activities, given the proposed model that F. succinogenes requires close contact to perform cellulose deconstruction by its array of secreted cellulases28. In addition, analysis of the leftover biomass sugar component after fermentation in batch culture indicated that utilizations of galactose and arabinose, although only account for a minor part of the carbon energetics, were higher in pH 6.70 than in pH 5.75. It is thought that F. succinogenes has incomplete pathways to utilize galactose, mannose, fructose and pentose, and cannot survival solely on those sugars28; however, when incubating with glucomannan or type II arabinogalactan (which contain various linkages such as β-1,4-Man and β-1,3-Gal), F. succinogenes was found to produce 0.4 mM and 0.2 mM succinate, respectively28. This may explain that although they are not the major carbon sources, galactose and arabinose could be utilized to a limited extent. The utilization of those sugars, including xylan digestion, is likely also affected by pH, therefore, differences of these sugar content were observed at two different pH values. Among the F. succinogenes cellulases that have been assayed before, optimal pH for these enzymes ranged between 5.0 and 6.567–74. Some of those enzymes retained 80% activities between pH 5.5 and 8.073, 74. On the other hand, a fewer number of characterized xylanases showed an optimal pH at 6.0 or 6.575, 76, and also retained most activities at a wide range of pH. It is thus difficult to clarify how pH affects utilization of individual sugar since the pH kinetics of most of those enzymes remain unclear, and the rest of the predicted enzymes have not been characterized.
This study has provided global insights into F. succinogenes under carbon limited continuous growth conditions at different pH levels and has shown it is possible to generate acid tolerant strains of cellulolytic anaerobic bacteria. Recent genetic engineering approaches (e.g. refs 77–79) should be adapted to enable further progress in our understanding of F. succinogenes.
Electronic supplementary material
Acknowledgements
Elanco Animal Health (Greenfield, Indiana, USA) provided funding support for these studies. We thank Dr. Jane Leedle (JL Microbiology, Inc.) for useful discussions, and Dr. Dale Pelletier/Ms. Meghan Drake (ORNL) for reviewing the manuscript. This research was performed in laboratories supported by the Bioenergy Science Center (BESC), which is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. ORNL is managed by UT-Battelle, LLC, Oak Ridge, TN, USA, for the DOE under contract DE-AC05–00OR22725.
Author Contributions
C.W., T.S., V.B. and S.B. designed the study. C.W. and S.B. conducted chemostat experiments. C.W. and M.R. conducted bioconversion assays and HPLC analyses. C.W. conducted growth and genetic experiments. D.K. generated RNA-seq and resequencing data. C.W., T.S., V.B. and S.B. analyzed growth data. C.W. and S.B. analyzed RNA-seq and resequencing data. All authors reviewed and contributed to the manuscript.
Competing Interests
V.B. is a former Elanco employee. T.S. consulted for Elanco and received compensation. C.W., D.K., M.R. and S.B. were funded by Elanco for the current study and declare no potential conflict of interest.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02628-w
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ragauskas AJ, et al. The path forward for biofuels and biomaterials. Science. 2006;311:484–489. doi: 10.1126/science.1114736. [DOI] [PubMed] [Google Scholar]
- 2.Himmel, M. E. & Picataggio, S. K. In Biomass Recalcitrance (ed. Michael E. Himmel) 1–6 (Blackwell Publishing Ltd., 2009).
- 3.Panesar PS, Marwaha SS, Kennedy JF. Zymomonas mobilis: an alternative ethanol producer. J. Chem. Technol. Biotechnol. 2006;81:623–635. doi: 10.1002/jctb.1448. [DOI] [Google Scholar]
- 4.Yang S, et al. Zymomonas mobilis as a model system for production of biofuels and biochemicals. Microbial Biotechnol. 2016;9:699–717. doi: 10.1111/1751-7915.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rubin EM. Genomics of cellulosic biofuels. Nature. 2008;454:841–845. doi: 10.1038/nature07190. [DOI] [PubMed] [Google Scholar]
- 7.Fischer CR, Klein-Marcuschamer D, Stephanopoulos G. Selection and optimization of microbial hosts for biofuels production. Metab. Eng. 2008;10:295–304. doi: 10.1016/j.ymben.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Flint, H. J. In Biomass Recalcitrance (ed. M. E. Himmel) 393–406 (Blackwell Publishing Ltd., 2009).
- 9.van Houtert MFJ. The production and metabolism of volatile fatty acids by ruminants fed roughages: A review. Anim. Feed Sci. Technol. 1993;43:189–225. doi: 10.1016/0377-8401(93)90078-X. [DOI] [Google Scholar]
- 10.Janssen, P. H. Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim. Feed Sci. Technol. 160, 1–22, doi:10.1016/j.anifeedsci.2010.07.002.
- 11.Russell JB, Muck RE, Weimer PJ. Quantitative analysis of cellulose degradation and growth of cellulolytic bacteria in the rumen. FEMS Microbiol. Ecol. 2009;67:183–197. doi: 10.1111/j.1574-6941.2008.00633.x. [DOI] [PubMed] [Google Scholar]
- 12.Golder, H. M. et al. Effects of partial mixed rations and supplement amounts on milk production and composition, ruminal fermentation, bacterial communities, and ruminal acidosis. J. Dairy Sci. 97, 5763–5785, doi:10.3168/jds.2014-8049. [DOI] [PubMed]
- 13.Jami E, Mizrahi I. Composition and similarity of bovine rumen microbiota across individual animals. Plos One. 2012;7:e33306. doi: 10.1371/journal.pone.0033306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernando SC, et al. Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microbiol. 2010;76:7482–7490. doi: 10.1128/AEM.00388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hook SE, et al. Impact of subacute ruminal acidosis (SARA) adaptation and recovery on the density and diversity of bacteria in the rumen of dairy cows. FEMS Microbiol. Ecol. 2011;78:275–284. doi: 10.1111/j.1574-6941.2011.01154.x. [DOI] [PubMed] [Google Scholar]
- 16.Abdela N. Sub-acute ruminal acidosis (SARA) and its consequence in dairy cattle: A review of past and recent research at global prospective. Achiev. Life Sci. 2016;10:187–196. doi: 10.1016/j.als.2016.11.006. [DOI] [Google Scholar]
- 17.Bach, A., Iglesias, C. & Devant, M. Daily rumen pH pattern of loose-housed dairy cattle as affected by feeding pattern and live yeast supplementation. Anim. Feed Sci. Technol. 136, doi:10.1016/j.anifeedsci.2006.09.011 (2007).
- 18.Long, M. et al. Effects of the acid-tolerant engineered bacterial strain Megasphaera elsdenii H6F32 on ruminal pH and the lactic acid concentration of simulated rumen acidosis in vitro. Res. Vet. Sci. 96, doi:10.1016/j.rvsc.2013.11.013 (2014). [DOI] [PubMed]
- 19.Balcells, J. et al. Effects of an extract of plant flavonoids (Bioflavex) on rumen fermentation and performance in heifers fed high-concentrate diets. J. Anim. Sci. 90, doi:10.2527/jas.2011-4955 (2012). [DOI] [PubMed]
- 20.Nisbet, D. J., Callaway, T. R., Edrington, T. S., Anderson, R. C. & Krueger, N. Effects of the dicarboxylic acids malate and fumarate on E. coli O157:H7 and Salmonella enterica typhimurium populations in pure culture and in mixed ruminal microorganism fermentations. Curr. Microbiol. 58, doi:10.1007/s00284-008-9351-1 (2009). [DOI] [PubMed]
- 21.Calsamiglia, S., Busquet, M., Cardozo, P. W., Castillejos, L. & Ferret, A. Invited review: Essential oils as modifiers of rumen microbial fermentation. J. Dairy Sci. 90, doi:10.3168/jds.2006-644 (2007). [DOI] [PubMed]
- 22.Hess M, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 23.De Nardi R, et al. Metagenomic analysis of rumen microbial population in dairy heifers fed a high grain diet supplemented with dicarboxylic acids or polyphenols. BMC Vet. Res. 2016;12:29. doi: 10.1186/s12917-016-0653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nathani NM, et al. Comparative evaluation of rumen metagenome community using qPCR and MG-RAST. AMB Express. 2013;3:55. doi: 10.1186/2191-0855-3-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denman SE, Martinez Fernandez G, Shinkai T, Mitsumori M, McSweeney CS. Metagenomic analysis of the rumen microbial community following inhibition of methane formation by a halogenated methane analog. Front. Microbiol. 2015;6:1087. doi: 10.3389/fmicb.2015.01087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aschenbach, J. R., Penner, G. B., Stumpff, F. & Gäbel, G. Ruminant Nutrition Symposium: Role of fermentation acid absorption in the regulation of ruminal pH12. J. Anim. Sci. 89, doi:10.2527/jas.2010-3301 (2011). [DOI] [PubMed]
- 27.Ransom-Jones E, Jones DL, McCarthy AJ, McDonald JE. The Fibrobacteres: an Important Phylum of Cellulose-Degrading Bacteria. Microbial Ecol. 2012;63:267–281. doi: 10.1007/s00248-011-9998-1. [DOI] [PubMed] [Google Scholar]
- 28.Suen, G. et al. The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist. PloS One6, doi:10.1371/journal.pone.0018814 (2011). [DOI] [PMC free article] [PubMed]
- 29.Weimer PJ. Effects of dilution rate and pH on the ruminal cellulolytic bacterium Fibrobacter succinogenes S85 in cellulose-fed continuous-culture. Arch. Microbiol. 1993;160:288–294. doi: 10.1007/BF00292079. [DOI] [PubMed] [Google Scholar]
- 30.Russell JB, Dombrowski DB. Effect of pH on the efficiency of growth by pure cultures of rumen bacteria in continuous culture. Appl. Environ. Microbiol. 1980;39:604–610. doi: 10.1128/aem.39.3.604-610.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, doi:10.1038/msb4100050 (2006). [DOI] [PMC free article] [PubMed]
- 32.Weimer, P. In US Dairy Forage Research Center. Informational Conference with Dairy and Forage Industries 53–60 (1996).
- 33.Ribbons, D. W. & Norris, J. R. Methods Microbiol. Vol. 2 (Academic Press, 1970).
- 34.Bowring SN, Morris JG. Mutagenesis of Clostridium acetobutylicum. J. Appl. Bacteriol. 1985;58:577–584. doi: 10.1111/j.1365-2672.1985.tb01714.x. [DOI] [PubMed] [Google Scholar]
- 35.Gong J, Forsberg CW. Factors affecting adhesion of Fibrobacter succinogenes subsp. succinogenes S85 and adherence-defective mutants to cellulose. Appl. Environ. Microbiol. 1989;55:3039–3044. doi: 10.1128/aem.55.12.3039-3044.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joachimsthal E, Haggett KD, Jang J-H, Rogers PL. A mutant of Zymomonas mobilis ZM4 capable of ethanol production from glucose in the presence of high acetate concentrations. Biotechnol. Lett. 1998;20:137–142. doi: 10.1023/A:1005320306410. [DOI] [Google Scholar]
- 37.Dumitrache A, et al. Consolidated bioprocessing of Populus using Clostridium (Ruminiclostridium) thermocellum: a case study on the impact of lignin composition and structure. Biotechnol Biofuels. 2016;9:31. doi: 10.1186/s13068-016-0445-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson CM, et al. Global transcriptome analysis of Clostridium thermocellum ATCC 27405 during growth on dilute acid pretreated Populus and switchgrass. Biotechnol. Biofuels. 2013;6:179. doi: 10.1186/1754-6834-6-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang S, et al. Transcriptomic and metabolomic profiling of Zymomonas mobilis during aerobic and anaerobic fermentations. Bmc Genomics. 2009;10:34. doi: 10.1186/1471-2164-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ausubel, F. M. Current protocols in molecular biology. (John Wiley & Sons, 2010).
- 41.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucl. Acids Res. 2015;43:D261–269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russell JB, Wilson DB. Why are ruminal cellulolytic bacteria unable to digest cellulose at low pH? J. Dairy Sci. 1996;79:1503–1509. doi: 10.3168/jds.S0022-0302(96)76510-4. [DOI] [PubMed] [Google Scholar]
- 45.Springer TA. Folding of the N-terminal, ligand-binding region of integrin alpha-subunits into a beta-propeller domain. Proc. Natl Acad. Sci. USA. 1997;94:65–72. doi: 10.1073/pnas.94.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loftus JC, Smith JW, Ginsberg MH. Integrin-mediated cell adhesion: the extracellular face. J. Biol. Chem. 1994;269:25235–25238. [PubMed] [Google Scholar]
- 47.Brumm P, et al. Functional annotation of Fibrobacter succinogenes S85 carbohydrate active enzymes. Appl. Biochem. Biotechnol. 2011;163:649–657. doi: 10.1007/s12010-010-9070-5. [DOI] [PubMed] [Google Scholar]
- 48.Weimer, P. J. & Odt, C. L. In Enzymatic Degradation of Insoluble Carbohydrates ACS Symposium Series (eds J. Saddler & M. Penner) 291–304 (American Chemical Society, 1996).
- 49.Kudo H, Cheng KJ, Costerton JW. Electron microscopic study of the methylcellulose-mediated detachment of cellulolytic rumen bacteria from cellulose fibers. Can. J. Microbiol. 1987;33:267–272. doi: 10.1139/m87-045. [DOI] [PubMed] [Google Scholar]
- 50.Jun HS, Qi M, Gong J, Egbosimba EE, Forsberg CW. Outer membrane proteins of Fibrobacter succinogenes with potential roles in adhesion to cellulose and in cellulose digestion. J. Bacteriol. 2007;189:6806–6815. doi: 10.1128/JB.00560-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roche B, et al. Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim. Biophys. Acta. 2013;1827:923–937. doi: 10.1016/j.bbabio.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 52.Wayne Outten F. Recent advances in the Suf Fe-S cluster biogenesis pathway: Beyond the Proteobacteria. Biochim. Biophys. Acta. 2015;1853:1464–1469. doi: 10.1016/j.bbamcr.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucl. Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski JL. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russell JB. Effect of extracellular pH on growth and proton motive force of Bacteroides succinogenes, a cellulolytic ruminal bacterium. Appl. Environ. Microbiol. 1987;53:2379–2383. doi: 10.1128/aem.53.10.2379-2383.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krulwich TA, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 2011;9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheng J, Marquis RE. Malolactic fermentation by Streptococcus mutans. FEMS Microbiol. Lett. 2007;272:196–201. doi: 10.1111/j.1574-6968.2007.00744.x. [DOI] [PubMed] [Google Scholar]
- 58.Tuckerman JR, et al. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochem. 2009;48:9764–9774. doi: 10.1021/bi901409g. [DOI] [PubMed] [Google Scholar]
- 59.Abe K, et al. Plasmid-encoded asp operon confers a proton motive metabolic cycle catalyzed by an aspartate-alanine exchange reaction. J. Bacteriol. 2002;184:2906–2913. doi: 10.1128/JB.184.11.2906-2913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levina N, et al. Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: identification of genes required for MscS activity. EMBO J. 1999;18:1730–1737. doi: 10.1093/emboj/18.7.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 62.Saini A, Mapolelo DT, Chahal HK, Johnson MK, Outten FW. SufD and SufC ATPase activity are required for iron acquisition during in vivo Fe-S cluster formation on SufB. Biochem. 2010;49:9402–9412. doi: 10.1021/bi1011546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petrovic A, et al. Hydrodynamic characterization of the SufBC and SufCD complexes and their interaction with fluorescent adenosine nucleotides. Protein Sci. 2008;17:1264–1274. doi: 10.1110/ps.034652.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Outten FW, Djaman O, Storz G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 2004;52:861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 65.Takahashi Y, Tokumoto U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 2002;277:28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 66.Nachin L, El Hassouni M, Loiseau L, Expert D, Barras F. SoxR-dependent response to oxidative stress and virulence of Erwinia chrysanthemi: the key role of SufC, an orphan ABC ATPase. Mol. Microbiol. 2001;39:960–972. doi: 10.1046/j.1365-2958.2001.02288.x. [DOI] [PubMed] [Google Scholar]
- 67.McGavin MJ, et al. Structure of the cel-3 gene from Fibrobacter succinogenes S85 and characteristics of the encoded gene product, endoglucanase 3. J. Bacteriol. 1989;171:5587–5595. doi: 10.1128/jb.171.10.5587-5595.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cavicchioli R, Watson K. Molecular cloning, expression, and characterization of endoglucanase genes from Fibrobacter succinogenes AR1. Appl. Environ. Microbiol. 1991;57:359–365. doi: 10.1128/aem.57.2.359-365.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iyo AH, Forsberg CW. Features of the cellodextrinase gene from Fibrobacter succinogenes S85. Can. J. Microbiol. 1994;40:592–596. doi: 10.1139/m94-094. [DOI] [PubMed] [Google Scholar]
- 70.Iyo AH, Forsberg CW. Endoglucanase G from Fibrobacter succinogenes S85 belongs to a class of enzymes characterized by a basic C-terminal domain. Can. J. Microbiol. 1996;42:934–943. doi: 10.1139/m96-120. [DOI] [PubMed] [Google Scholar]
- 71.Malburg LM, Jr., Iyo AH, Forsberg CW. A novel family 9 endoglucanase gene (celD), whose product cleaves substrates mainly to glucose, and its adjacent upstream homolog (celE) from Fibrobacter succinogenes S85. Appl. Environ. Microbiol. 1996;62:898–906. doi: 10.1128/aem.62.3.898-906.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Malburg SR, Malburg LM, Jr., Liu T, Iyo AH, Forsberg CW. Catalytic properties of the cellulose-binding endoglucanase F from Fibrobacter succinogenes S85. Appl. Environ. Microbiol. 1997;63:2449–2453. doi: 10.1128/aem.63.6.2449-2453.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qi M, Jun HS, Forsberg CW. Characterization and synergistic interactions of Fibrobacter succinogenes glycoside hydrolases. Appl. Environ. Microbiol. 2007;73:6098–6105. doi: 10.1128/AEM.01037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qi M, Jun HS, Forsberg CW. Cel9D, an atypical 1,4-beta-D-glucan glucohydrolase from Fibrobacter succinogenes: characteristics, catalytic residues, and synergistic interactions with other cellulases. J. Bacteriol. 2008;190:1976–1984. doi: 10.1128/JB.01667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu H, Paradis FW, Krell PJ, Phillips JP, Forsberg CW. Enzymatic specificities and modes of action of the two catalytic domains of the XynC xylanase from Fibrobacter succinogenes S85. J. Bacteriol. 1994;176:3885–3894. doi: 10.1128/jb.176.13.3885-3894.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jun HS, Ha JK, Malburg LM, Jr., Verrinder GA, Forsberg CW. Characteristics of a cluster of xylanase genes in Fibrobacter succinogenes S85. Can. J. Microbiol. 2003;49:171–180. doi: 10.1139/w03-024. [DOI] [PubMed] [Google Scholar]
- 77.Croux C, et al. Construction of a restriction-less, marker-less mutant useful for functional genomic and metabolic engineering of the biofuel producer. Clostridium acetobutylicum. Biotechnol. Biofuels. 2016;9:1–13. doi: 10.1186/s13068-015-0423-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang X, Xu M, Yang S-T. Restriction modification system analysis and development of in vivo methylation for the transformation of Clostridium cellulovorans. Appl. Microbiol. Biotechnol. 2015;100:2289–2299. doi: 10.1007/s00253-015-7141-9. [DOI] [PubMed] [Google Scholar]
- 79.Chung D, Farkas J, Huddleston JR, Olivar E, Westpheling J. Methylation by a unique a-class N4-cytosine methyltransferase is required for DNA transformation of Caldicellulosiruptor bescii DSM6725. Plos One. 2012;7:e43844. doi: 10.1371/journal.pone.0043844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


