Abstract
Muscle spindles are skeletal muscle sensory organs involved in the sensation of position and movement of the body. We have explored the capability of phase contrast computed tomography to visualise muscle spindles in murine skeletal muscle. In particular, we have validated the visualisation of nerve fibres through phase contrast computed tomography using light microscopy on stained histological sections. We further present the first three‐dimensional visualisation of muscle spindles in mouse soleus skeletal muscle in conjunction with the neurovascular bundle associated with it.
Keywords: computed tomography, muscle spindles, neurovascular bundle, phase contrast, synchrotron radiation
Introduction
Muscle spindles are mechanoreceptors located in skeletal muscles and are involved in the detection of position and movement (Banks, 2005). Muscle spindles consist of a varying number of intrafusal muscle fibres that are surrounded by a capsule of connective tissue (Craigmyle, 1986). An external capsule surrounds the internal capsule. Blood capillaries are sometimes also found within the capsular layers of the spindles. The intrafusal fibres are smaller and shorter than the regular contractile muscle fibres. The diameters of the intrafusal muscle fibres depend on their type, i.e. chain type, bag type 1 or bag type 2 (Banks et al. 1977), with the bag type fibres being considerably larger (Thornell et al. 2015). The number of muscle spindles varies between different muscles (McComas, 1996) and between different strains of the same type of mammal (Lionikas et al. 2013). The number of intrafusal fibres may also vary but is reported to be between two and eight in mouse soleus muscle (Lionikas et al. 2013).
Muscle spindles have to date been visualised using mainly light and electron microscopy, after enhancing contrast of features of interest with staining agents. However, the disadvantage of light microscopy lies in the destructive preparation necessary for imaging, i.e. the cutting of the specimen, and the specificity of staining agents for a limited number of different soft tissue features. Recently, Fratini et al. (2015) have highlighted the advantages of phase contrast imaging through synchrotron radiation‐based computed tomography (SR CT) to study soft tissue, in particular the neurovascular bundle, non‐destructively and without the addition of contrast agents.
X‐ray phase contrast imaging makes use of the difference of the phase shift that X‐rays experience when passing through a sample rather than the difference in X‐ray absorption. The X‐ray absorption of (unstained) soft tissue samples can be very low, while the induced phase shift offers an alternative approach to obtain a good image contrast to study the components of soft tissues such as kidney, liver or muscle. As the X‐rays hit the sample, the phase of the X‐ray wave is shifted and by applying an intermediate step of phase retrieval before three‐dimensional (3D) reconstruction of the CT data, this difference in phase shift can be translated into image contrast within the two‐dimensional (2D) radiographic projections. Although Fratini et al. (2015) provided an excellent proof of concept of the method, the reported histological validation was not undertaken in the same specimen that had previously been imaged using phase contrast SR CT. We have therefore performed a quantitative validation of the technique to visualise nervous tissue and have determined mean values for numbers such as cross‐sectional muscle fibre counts and numbers of muscle spindles, which can be compared with values provided in the literature by application of gold standard techniques, such as light microscopy of stained sections.
Materials and methods
Sample preparation
All animal procedures were in accordance with the regulations of the United Kingdom Animals (Scientific Procedures) Act 1986 and were conducted under Home Office Licence number 70‐6457. The study received institutional approval from the University of Southampton Biomedical Research Facility Research Ethics Committee.
Male C57BL/6 mice (n = 5) were maintained under controlled conditions. At 15 weeks of age the mice were killed by cervical dislocation. The soleus muscle of the right leg was dissected and fixed in 10% formaldehyde at 4°C overnight. It was then dehydrated in a graded series of methylated spirit and embedded in paraffin wax in coffin moulds. The wax block was trimmed for imaging using an industrial razor blade and glued onto a scanning electron microscopy stub that was clamped into a standard pin chuck. Mean (standard deviation) weight at the time of killing was determined in littermates to be 31.7 g (1.1).
Phase contrast synchrotron radiation‐based computed tomography
Scanning of the muscles using SR CT was performed at the TOMCAT beamline of the Swiss Light Source (SLS) at the Paul Scherrer Institut in Villigen, Switzerland. All scans were performed at 14 keV, at a voxel size of 0.77 μm, an exposure time of 180 ms, where 1601 projections were acquired over an angular range of 180°. For each scan we additionally recorded 32 dark field and 160 flat field images to correct the raw projections. A non‐zero propagation or sample‐to‐detector distance of 60 mm was chosen so that the free‐space propagation of the (coherent) X‐rays transforms the phase modulation into intensity variations providing image contrast, recorded as projections on the charge‐coupled device (CCD) detector. The phase of the projection images was then retrieved using an in‐house implementation of the Paganin single‐distance non‐iterative phase retrieval algorithm (Paganin et al. 2002) and subsequently reconstructed using an in‐house implementation (Marone & Stampanoni, 2012) of the Gridrec algorithm (Dowd et al. 1999) at TOMCAT.
Histological validation
For validation of the nerve fibres, histological sectioning was performed on the previously imaged muscles using 4‐μm‐thick serial sections taken from the belly of the muscle. The sections were stained with an antiPGP9.5 antibody (ab108986, Abcam, clone EPR4118, monoclonal rabbit, isotype IgG, dilution 1 : 750) in combination with a biotinylated secondary goat anti‐rabbit antibody and DAB as chromagen (with antigen retrieval by microwaving) to stain for nerve fibres. AntiPGP9.5 labels neuronal cell bodies and axons in the central and peripheral nervous system. Mayer's haematoxylin was used for counterstaining. The validation of the antibody stains was performed on three sections from all muscles. The SR CT volume was visually aligned using the ‘simple registration’ tool in vgstudio max 2.0 (Volume Graphics GmbH, Germany) to find slices corresponding to the histological sections. To account for the fact that the histological slides were thicker (4 μm) than the height of a voxel in the SR CT image (spatial resolution was 0.77 μm), a maximum intensity projection (Schindelin et al. 2012) was applied over seven SR CT slices to allow for the upper and lower boundaries of the histological slice to lie between voxels. This operation joined the consecutive images into one slice with the greyscale at each point in the resulting slice being the maximum of all greyscales in that respective point over the seven slices. The resulting slice was then compared with the slide imaged using histology. See Fig. 1A,B for a comparison of a PGP9.5 stained slice and a corresponding SR CT image. For quantitative comparison, the nerve fibres were segmented in both image types. For the nerve segmentation on histology sections, the colour base corresponding to the DAB chromagen was isolated to facilitate the image segmentation. The images of this colour channel were manually segmented using avizo fire 9.0 (FEI, Hillsboro, OR, USA); the same was done with the corresponding phase‐retrieved SR CT images. The nerve area was computed using the avizo ‘volume fraction’ tool to count the labelled voxels. Linear regression was used to compare the counts of nerve area obtained from the histological slides and the SR CT images using origin 8.1 (OriginLab Corp., Northampton, MA, USA) (Fig. 1C).
Figure 1.
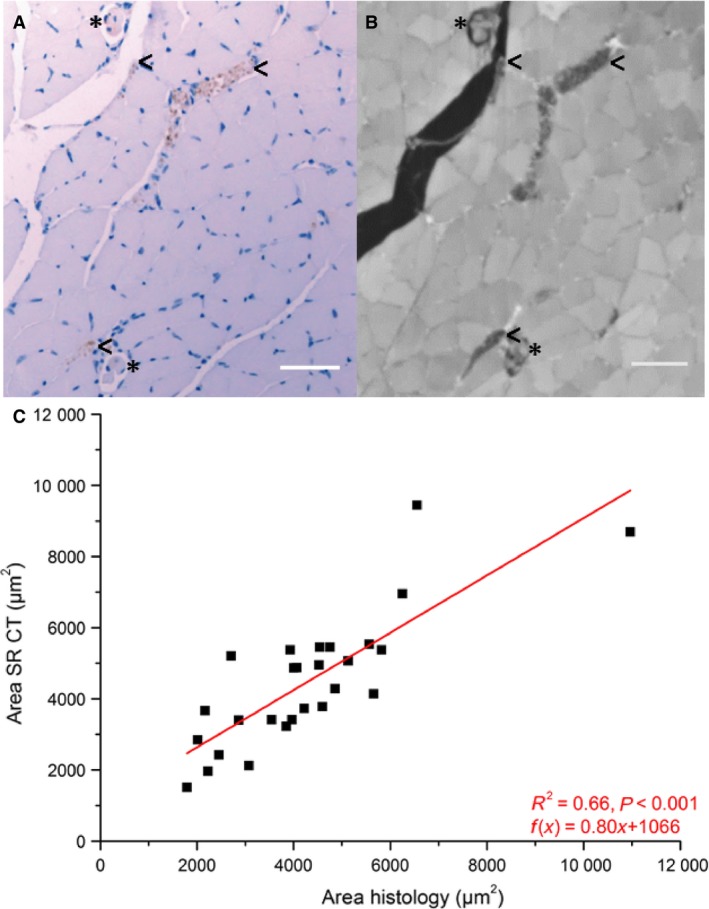
Comparison of identified and measured nerve area (C) in corresponding histology (A) and SR CT (B) images. Nerves in (A) are stained in brown by DAB chromagen and Mayer's haematoxylin was used as counterstain. The characteristic structure of both nerve tissue (<) and muscle spindles (*) is visible in the SR CT image (B). Scale bars: 50 μm. After segmentation the nerve area was measured in the corresponding images of the two imaging techniques. A linear regression line was fitted to the counts using origin 8.1. The cross‐sectional area determined by SR CT appears to be consistently lower than if it were determined by histology. This is likely due to blurring of the SR CT images during the alignment process and discussed below.
Soft tissue feature visualisation and analysis
The 3D structure of muscle spindles was segmented using semi‐automatic tools provided in avizo fire 9.0 and visualised therein. To do this, structures were segmented manually in every 10th–20th slice and the segmentation was expanded into the whole 3D volume by linear interpolation. All of the measures described in the following have been derived from SR CT data. Muscle volume was determined from the SR CT data using volume fraction counting in fiji/imagej (Schindelin et al. 2012), i.e. counting the number of voxels of muscle tissue and taking into account the image voxel size. The number of muscle fibres per cross‐section (in the belly region of the muscle), number of muscle spindles and the number of intrafusal fibres per spindle were counted manually. The distribution of muscle spindles was characterised using the BoneJ thickness plugin (Hildebrand & Rüegsegger, 1997; Doube et al. 2010) in fiji/imagej (Schindelin et al. 2012) to measure the spacing between muscle spindles within the muscle. Finally, the average area fraction of muscle spindle to muscle tissue was determined for each SR CT cross‐section through the muscle volume as a second measure of spindle distribution.
Results
Histological validation
Figure 1(A,B) shows an example of a reconstructed slice of the acquired SR CT data, together with a corresponding histological image. The nerve fibres can be clearly seen for both imaging techniques. Figure 1C shows the comparison between the nerve areas estimated from SR CT and histology images. These data suggest that although there was considerable consistency in the identification of nerves using the two imaging techniques, the cross‐sectional area of nerves fibres determined from SR CT images was lower than the area determined from histological sections.
Soft tissue feature visualisation and analysis
Figure 2(A,B) shows examples of muscle spindles observed in a reconstructed slice of the acquired SR CT data, together with a corresponding histological image. The structure of the spindles and intrafusal fibres (if) can clearly be distinguished in the SR CT images. Due to the non‐destructive and 3D nature of SR CT imaging, we were also able to follow the intrafusal fibres both within the external capsule (eca) (Fig. 2C), and also throughout the whole muscle until they merged into the muscle fascia (not shown). We were able to visualise in 3D the dedicated arterioles (art) that supply the muscle spindles and run along them. However, not every muscle spindle was accompanied by such an arteriole, which is contradictory to the angiotypic hypothesis presented by Kokkorogiannis (2004).
Figure 2.
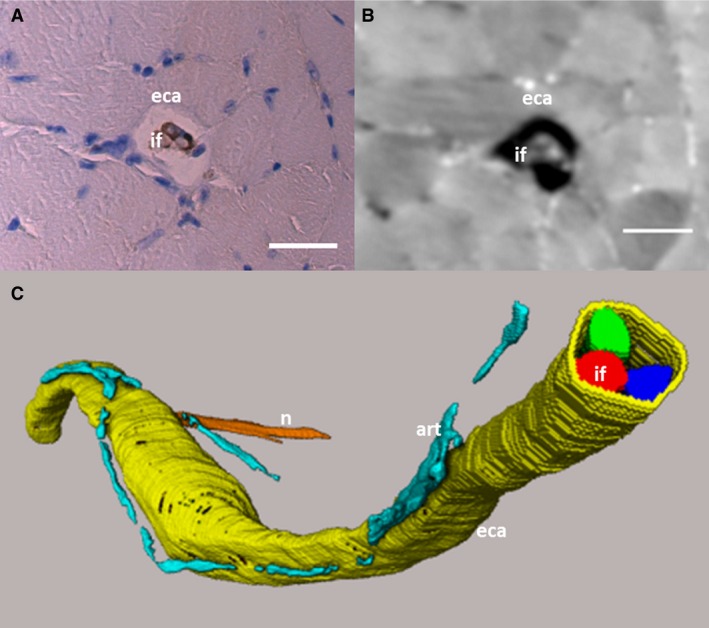
Histological and SR CT slice of muscle spindle and 3D volume rendering. (A) Histological slice corresponding to (B) the SRCT image. The external capsule (eca) and intrafusal fibres (if) can be distinguished in the SR CT image. Scale bars: (A,B) 30 μm. (C) The different intrafusal fibres (red, blue, green) and the external capsule (yellow) have been segmented from the SR CT data, along with the lumen of small arterioles (art, turqoise) running along the spindle and one of the supplying nerves (n, orange). The segmented data is visualised through volume rendering. The S‐shape of the muscle spindles is due to the muscle not being stretched during fixation, which led to a distortion of the muscle.
The mean volume of the soleus muscles was determined as 5.21 ± 0.40 mm3 (mean ± SD). The number of muscle fibres per SR CT cross‐section in the medial region of the muscle was 1013 ± 45.
We derived an average of 11.8 ± 0.7 muscle spindles per muscle with a mean of 3.7 ± 0.1 intrafusal fibres per muscle spindle. The number of spindles was greatest in the belly of the muscle (Fig. 3). The separation between muscle spindles and the muscle boundary was 0.28 ± 0.15 μm (maximum 0.51 μm).
Figure 3.
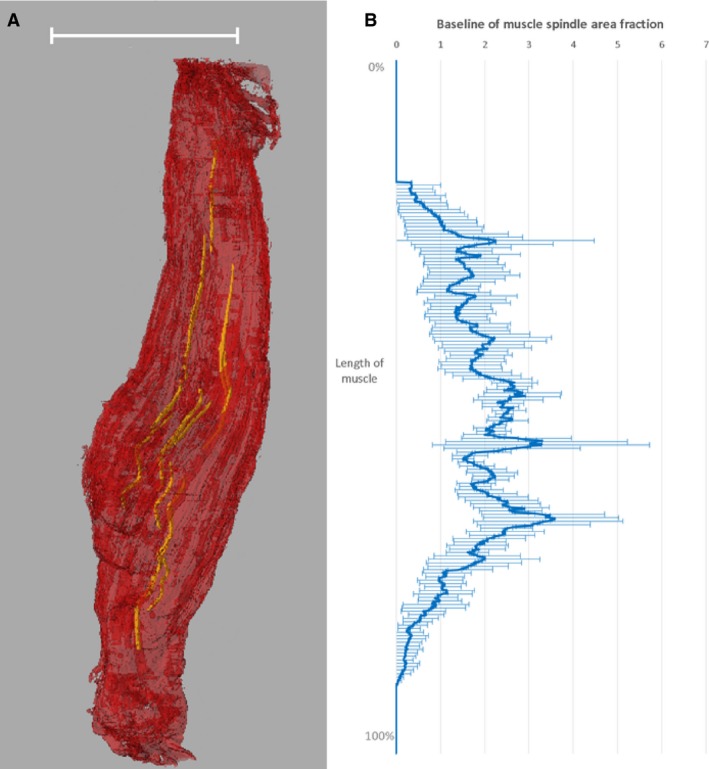
3D volume rendering of muscle spindles in mouse soleus muscle (A) and average muscle spindle area fraction per muscle tissue cross‐section over the length of the muscle (B). The representation of the muscle spindles has been derived from SR CT data. Scale bar: 3 mm. The baseline of the muscle spindle area fraction has been determined by averaging the values for n = 5 muscles.
Discussion
Phase contrast SR CT imaging is a relatively novel technique for imaging soft tissue non‐invasively in 3D at high spatial resolutions. The values presented here for mean number of muscle spindles per muscle (11.8 ± 0.7) and intrafusal fibres per spindle (3.7 ± 0.1) in the mouse soleus muscle, as derived from SR CT data, are consistent with those reported by (Lionikas et al. 2013) using light microscopy (Table 1). The number of spindles per muscle reported by Lionikas et al. (2013) for the C57BL/6 mouse strain was between 10 and 11, with a mean number of intrafusal fibres of 3.9. Using phase contrast SR CT imaging we observed 11.8 ± 0.7 neuromuscular spindles per muscle and 3.7 ± 0.1 intrafusal fibres per spindle (Table 1). Banks (2006) counted 14 neuromuscular spindles per mouse soleus muscle using serial sectioning for light microscopy, where the mouse strain remained unnamed. In our study in male C57BL/6 mice, the soleus muscle had a lower percentage of spindles with four intrafusal fibres, accounting for 68.5% of all spindles, compared with 87.5% reported by Lionikas et al. (2013). Furthermore, the mean muscle extrafusal fibre count in the medial region of the soleus muscle obtained using phase contrast SR CT did not differ significantly from that reported by Lionikas et al. (2013) using histology, i.e. 1013 ± 45 vs. 949 ± 61, respectively (Table 1). The mean cross‐sectional area per extrafusal fibre (1081 ± 103 μm2) we observed was lower than previously reported by Lionikas et al. (2013), namely 1423 ± 155 μm2 for type I and 1393 ± 160 μm2 for type IIa. This difference in area may be due to the different preparation protocols used, i.e. in this study the muscle was fixed in formalin, which will most likely have led to shrinkage. In Lionikas et al. (2013), however, the muscles were frozen in isopentane cooled in liquid nitrogen for cryosectioning.
Table 1.
Quantitative morphometry of muscle and neuromuscular spindles determined by phase contrast SR CT imaging performed on n = 5 muscles and compared with values determined by histology in the literature (Lionikas et al. 2013)
| SR CT (mean ± SD) | Histological values (Lionikas et al. 2013) | |
|---|---|---|
| Muscle volume (mm3) | 5.2 ± 0.4 | – |
| Number of neuromuscular spindles | 11.8 ± 0.7 | 10.66 |
| Number of muscle fibres per cross‐section | 1013 ± 45 | 949 ± 61 |
| Number of intrafusal fibres | 3.7 ± 0.1 | 3.9 |
| Number of spindles per unit muscle volume (1 mm−3) | 2.3 ± 0.3 | – |
| Number of muscle fibres per neuromuscular spindle | 89 ± 8 | 89a |
| Number of muscle fibres per unit muscle volume (1 mm−3) | 196 ± 18 | – |
| Mean cross‐sectional area per muscle fibre (μm2) | 1081 ± 103 |
1423 ± 155 (type I) 1393 ± 160 (type IIa) |
Computed from data in Lionikas et al. (2013).
The difference between cross‐sectional areas of nerves estimated from SR CT and histology may also be due to shearing introduced by microtoming of the muscle for histology and limited image contrast in the SR CT datasets (Supporting Information Fig. S1). The specific staining for nerve fibres in histology images facilitated the differentiation of even small nerve fibres, whereas this was not always possible in the SR CT images. However, the consistency of our results with published material suggests that phase contrast SR CT is suited to identify and to characterise quantitatively the structure of soft tissues in skeletal muscle in 3D. Furthermore, 3D SR CT has the advantage that scanning times are short, i.e. each scan was completed within 6 min, with five to six scans necessary to cover the whole length of the soleus muscle, resulting in an image acquisition time of roughly half an hour. Phase retrieval and CT reconstruction can be performed simultaneously with scanning, resulting in a maximum processing time of 45–60 min for the creation of the 3D dataset of the whole muscle. In our study, data segmentation was the bottleneck, with the segmentation of all neuromuscular spindles in one muscle taking between 5 and 6 hours. However, analysis of the segmented structures was fast, with imagej plugins running between a few seconds for volume counting and minutes for the determination of space separations. In contrast to this, in histological studies serial sectioning of a whole muscle of ~ 1 cm length at 20 μm sectioning thickness would result in 500 sections. Following staining (e.g. PGP9.5, ~ 1.5 days), image acquisition would optimally take a minimum of 6 days to assess the muscle at high‐resolution using an automated slide scanning microscope (Hannon‐Fletcher & Maxwell, 2009), and 3D image registration of individual slices and feature segmentation is not included in this estimation. Furthermore, phase contrast SR CT leaves the muscle intact, thus enabling subsequent analysis using other methods, compared with histological sectioning, which is destructive.
The main disadvantage of SR CT imaging is the availability of, and access to, the few SR sources worldwide (http://www.lightsources.org/regions). Allocation of scanning time is generally granted on a competitive basis, with proposals submitted typically half a year in advance. Lab‐based micro‐computed tomographic systems, which allow for (partially) coherent X‐ray generation at high spatial resolutions and hence for phase contrast‐based high‐resolution CT are available (Bidola et al. 2015). However, scanning times are considerably longer (several hours) due to the much lower X‐ray flux provided by lab‐based X‐ray sources compared with SR light sources, and implementation of phase contrast CT calls for some expert knowledge in terms of X‐ray imaging and image processing.
A methodological limitation of both light microscopy for histology and CT imaging is the possible shrinkage of the muscle during fixation and embedding procedures (the muscle was not pinned down during fixation), potentially leading to errors in the characterisation of the muscle spindles within the muscle body. This would distort any 2D and 3D measurement of distribution or separation and the distortion may vary between different dietary groups due to disparate fat content. This is not a matter that can currently be resolved, as the soft tissue needs to be immobilised for (high‐resolution) imaging. Finally, it needs to be noted that although providing increased soft tissue contrast, the Paganin algorithm mainly acted as a low‐pass, i.e. smoothing, filter in this application. This is because the assumptions under which the algorithm was derived were not valid (i.e. non‐homogeneous sample). Therefore, the differing greyscales of muscle fibres in the SR CT images cannot be directly linked to functionality. Thus, no further comparison using enzyme histochemistry or anti‐myosin heavy chain immunohistochemistry was performed.
Conclusion
We have presented the first 3D visualisation and quantification of muscle spindles and their intrafusal fibres and the supplying neurovascular bundle in an intact muscle. A validation of computed tomographic phase contrast imaging through cross‐correlation with standard histological techniques was provided. Phase contrast SR CT has proven to be a valuable approach for imaging of such soft tissues due to its non‐invasive 3D nature. Higher‐resolution imaging at nano‐focus beamlines, such as holotomography or ptychography (Thibault et al. 2014), may enable further insight into the 3D inner make‐up of the different soft tissue features. Using phase contrast SR CT, it is possible to study and interpret the morphology and distribution of the intramuscular soft tissue features in 3D, so further informing our understanding of the role of muscle spindles.
Data statement
All data supporting this study are openly available from the University of Southampton repository at http://doi.org/10.5258/SOTON/405591.
Funding
The authors would like to acknowledge funding by the Engineering and Physical Sciences Research Council (EPSRC) for the EPSRC doctoral training grant of author B. Zeller‐Plumhoff and the British Heart Foundation for grant PG/12/18/29453.
Conflict of interest
There are no conflicts of interest.
Supporting information
Fig. S1. Overlay of corresponding histological and SR CT slices.
Acknowledgements
The phase contrast synchrotron‐based computed tomography data was obtained at the TOMCAT beamline of the Swiss Light Source (SLS). We acknowledge the Paul Scherrer Institut, Villigen, Switzerland for provision of synchrotron radiation beamtime at the TOMCAT beamline of the SLS and would like to thank Pablo Villanueva and Alessandra Patera for their assistance. Finally, we would like to thank Prof. Ian Sinclair for valuable discussions that made our work possible and the μ‐VIS X‐ray Imaging Centre at the University of Southampton for providing the tools for image processing.
References
- Banks R (2005) The muscle spindle In: Peripheral Neuropathy, 4th edn (eds Dyck P, Thomas P.), pp. 131–150. Philadelphia, Pennsylvania: Saunders. [Google Scholar]
- Banks RW (2006) An allometric analysis of the number of muscle spindles in mammalian skeletal muscles. J Anat 208, 753–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks RW, Harker DW, Stacey MJ (1977) A study of mammalian intrafusal muscle fibres using a combined histochemical and ultrastructural technique. J Anat 123, 783–796. [PMC free article] [PubMed] [Google Scholar]
- Bidola PM, Zanette I, Achterhold K, et al. (2015) Optimization of propagation‐based phase‐contrast imaging at a laboratory setup. Opt Express 23, 30000–30013. [DOI] [PubMed] [Google Scholar]
- Craigmyle MBL (1986) A Colour Atlas of Histology. London: Wolfe Medical. [Google Scholar]
- Doube M, Klosowski MM, Arganda‐Carreras I, et al. (2010) BoneJ: free and extensible bone image analysis in ImageJ. Bone 47, 1076–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd BA, Campbell GH, Marr RB, et al. (1999) Developments in synchrotron x‐ray computed microtomography at the National Synchrotron Light Source. Dev X‐Ray Tomogr II 3772, 224–236. [Google Scholar]
- Fratini M, Bukreeva I, Campi G, et al. (2015) Simultaneous submicrometric 3D imaging of the micro‐vascular network and the neuronal system in a mouse spinal cord. Sci Rep 5, 8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon‐Fletcher M, Maxwell P (2009) Advanced techniques in diagnostic cellular pathology. Chichester: John Wiley & Sons. [Google Scholar]
- Hildebrand T, Rüegsegger P (1997) A new method for the model‐independent assessment of thickness in three‐dimensional images. J Microsc 185, 67–75. [Google Scholar]
- Kokkorogiannis T (2004) Somatic and intramuscular distribution of muscle spindles and their relation to muscular angiotypes. J Theor Biol 229, 263–280. [DOI] [PubMed] [Google Scholar]
- Lionikas A, Smith CJ, Smith TL, et al. (2013) Analyses of muscle spindles in the soleus of six inbred mouse strains. J Anat 223, 289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marone F, Stampanoni M (2012) Regridding reconstruction algorithm for real‐time tomographic imaging. J Synchrotron Radiat 19, 1029–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McComas AJ (1996) Skeletal muscle: form and function. Champaign: Human Kinetics. [Google Scholar]
- Paganin D, Mayo SC, Gureyev TE, et al. (2002) Simultaneous phase and amplitude extraction from a single defocused image of a homogeneous object. J Microsc 206, 33–40. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, et al. (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault P, Guizar‐Sicairos M, Menzel A (2014) Coherent imaging at the diffraction limit. J Synchrotron Radiat 21, 1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornell LE, Carlsson L, Eriksson PO, et al. (2015) Fibre typing of intrafusal fibres. J Anat 227, 136–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Overlay of corresponding histological and SR CT slices.


