Abstract
Auxotrophic mutants of the yeast Saccharomyces cerevisiae are usually isolated in haploid strains because the isolation of recessive mutations in diploids is thought to be difficult due to the presence of two sets of genes. We show here that auxotrophic mutants of diploid industrial sake yeast strains were routinely obtained by a standard mutant selection procedure following UV mutagenesis. We isolated His−, Met−, Lys−, Trp−, Leu−, Arg−, and Ura− auxotrophic mutants of five sake strains, Kyokai no. 7, no. 9, no. 10, no. 701, and no. 901, by screening only 1,700 to 3,400 colonies from each treated strain. Wild-type alleles were cloned and used as markers for transformation. With HIS3 as a selectable marker, the yeast TDH3 overexpression promoter was inserted upstream of ATF1, encoding alcohol acetyltransferase, by one-step gene replacement in a his3 mutant of Kyokai no. 7. The resulting strain contained exclusively yeast DNA, making it acceptable for commercial use, and produced a larger amount of isoamyl acetate, a banana-like flavor. We argue that the generally recognized difficulty of isolating auxotrophic mutants of diploid industrial yeast strains is misleading and that genetic techniques used for haploid laboratory strains are applicable for this purpose.
Auxotrophic mutants of the yeast Saccharomyces cerevisiae have played an important role in the development of classical genetic techniques, molecular biology, and genetic engineering (48, 49). In classical genetic techniques, complementing auxotrophic mutations have often been used to select diploids in matings between MATa and MATα haploid strains (53). They have also been used as genetic markers for gene mapping by calculating meiotic recombination rates (50). In genetic engineering and molecular biology, auxotrophic mutations and the corresponding wild-type genes have been used to select yeast transformants (45). The one-step gene disruption procedure and other targeted gene manipulations involving mitotic recombination have also relied on the routine use of auxotrophs of laboratory yeast strains (46, 51). These gene manipulation systems have led to the construction of a set of 6,000 gene knockout strains (57). Thus far, progress in studies of yeast strains as model organisms has depended, in large part, on advanced genetic methods based on auxotrophic mutations and their respective marker genes. Unfortunately, the application of these methods is restricted to laboratory yeast strains and is not used for industrial yeast strains.
Industrial yeast strains are usually diploid or polyploid and carry no auxotrophic mutations (12, 17, 22). For example, yeast strains commonly used for Japanese sake are diploid and prototorophic (31, 43). Lager beer strains seem to be natural hybrids of S. cerevisiae and non-S. cerevisiae species and possess complex genomes (32a). Many wine and baking yeast strains are also polyploid and prototrophic (21, 44, 54). In addition to the polyploidy, many industrial yeast strains sporulate poorly or not at all (17, 22, 39, 56). Therefore, breeding of industrial yeasts by crossing haploid strains is not common. Instead, molecular breeding by the introduction of useful genes through transformation has been developed. For this purpose, many dominant drug resistance markers have been used due to the absence of auxotrophic mutations (2, 5). However, transformation efficiencies obtained with drug resistance markers are usually lower than those obtained with auxotrophic markers. Moreover, background colonies that are resistant to the drugs often arise in addition to the true transformants. Because many drug resistance markers are derived from bacterial antibiotic resistance genes, they cannot be used freely in industrial yeasts due to concerns of consumers about this aspect of genetically modified organisms (18, 29, 47).
If auxotrophic mutants could be derived from industrial yeasts, then recombinant DNA technology could be applied to them more easily; in addition, because auxotrophic markers are derived from the yeasts themselves, strain construction by “self-cloning,” acceptable for commercial use in Japan, would become possible. Self-cloning refers to the introduction of DNA from one strain into another strain of the same species (2, 10, 26). To our knowledge, however, auxotrophic mutants of industrial yeasts have not been isolated routinely, possibly because the yeasts are diploid or polyploid. Previous efforts to obtain auxotrophic mutants of industrial strains used positive selection, such as 5-fluoroorotic acid (5-FOA) (40) or α-aminoadipic acid (41) selection, or enrichment with nystatin (43) or benomyl (13). Thus far, the isolation of auxotrophic mutants of industrial yeasts has been thought to be difficult without the use of special selection procedures.
In this study, we compared the rates of survival of isogenic laboratory haploid and diploid strains following UV irradition or treatment with the chemical mutagen ethyl methanesulfonate (EMS). While the diploid exhibited greater survival than the haploid at the same doses of mutagen or irradiation, the lethality seemed high enough for the screening of mutants by conventional replica plating. Based on this result, we screened and obtained auxotrophic mutants of all sake strains used. Genes that complemented the auxotrophic mutations were isolated and used for constructing self-cloning yeast strains. The approach presented here is likely to change the way in which gene manipulations of industrial yeasts are performed.
MATERIALS AND METHODS
Strains and media.
The strains used in this study are listed in Table 1. Kyokai strains were obtained from the Brewing Society of Japan. Yeast strains were grown on YPD medium (1% yeast extract, 2% polypeptone, 2% glucose and, if necessary, 2% agar) at 28°C. For screening, minimal medium (MM; 0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, 2% glucose, and 2% agar) was used. Stock nutrient solutions and pools of amino acids, purines, and pyrimidines were prepared according to published procedures (49). 5-FOA medium was prepared as described previously (14). Escherichia coli strain DH5α and standard bacterial media were used.
TABLE 1.
Strains
| Strain | Description | Source or reference |
|---|---|---|
| Kyokai no. 7 | Sake yeast, MATa/MATα | BSJa |
| Kyokai no. 9 | Sake yeast, MATa/MATα | BSJ |
| Kyokai no. 10 | Sake yeast, MATa/MATα | BSJ |
| Kyokai no. 701 | Sake yeast, MATa/MATα | BSJ |
| Kyokai no. 901 | Sake yeast, MATa/MATα | BSJ |
| W303 | MATa/MATα ura3/ura3 his3/his3 leu2/ leu2 trp1/trp1 ade2/ade2 can1/can1 | 55 |
| W303-1A | MATaura3 his3 leu2 trp1 ade2 can1 | 55 |
| RAK1536 | MATa/MATα his3/his3 | Kyokai no. 7 |
| RAK1537 | MATa/MATα Met− | Kyokai no. 7 |
| RAK1546 | MATa/MATα his3/his3 lys4/lys4 | RAK1536 |
| RAK1763 | MATa/MATα leu4/leu4 | Kyokai no. 701 |
| RAK1764 | MATa/MATα Arg− | Kyokai no. 701 |
| RAK1785 | MATa/MATα Ura− | Kyokai no. 701 |
| RAK1786 | MATa/MATα Met− | Kyokai no. 10 |
| RAK1787 | MATa/MATα trp3/trp3 | Kyokai no. 10 |
| RAK1788 | MATa/MATα Trp− | Kyokai no. 9 |
| RAK1922 | MATa/MATα trp3/trp3 | Kyokai no. 901 |
| RAK1923 | MATa/MATα trp3/trp3 | Kyokai no. 901 |
| RAK1924 | MATa/MATα trp3/trp3 | Kyokai no. 901 |
| RAK1925 | MATa/MATα trp3/trp3 | Kyokai no. 901 |
| RAK1926 | MATa/MATα Trp− | Kyokai no. 901 |
| RAK1927 | MATa/MATα trp3/trp3 | Kyokai no. 901 |
| RAK1883 | MATa/MATα his3/his3 HIS3-TDH3p-ATF1 | RAK1536 |
| RAK1884 | MATa/MATα his3/his3 HIS3-TDH3p-ATF1 | RAK1536 |
| RAK2196 | MATa/MATα his3/his3 HIS3-TDH3p-ATF1 | RAK1536 |
BSJ, Brewing Society of Japan.
Mutagenesis and mutant isolation.
For the analysis of survival rates by UV mutagenesis, cells grown for 24 h at 28°C on a YPD medium plate were collected and suspended in sterile water. After the cell concentration was determined by counting, cells were spread on YPD medium plates. The plates were placed under a UV lamp (Toshiba GL15) at a distance of 35 cm and were irradiated for various periods of time (Fig. 1). Following irradiation, the plates were incubated at 28°C, and the numbers of colonies were counted to determine survival rates.
FIG. 1.
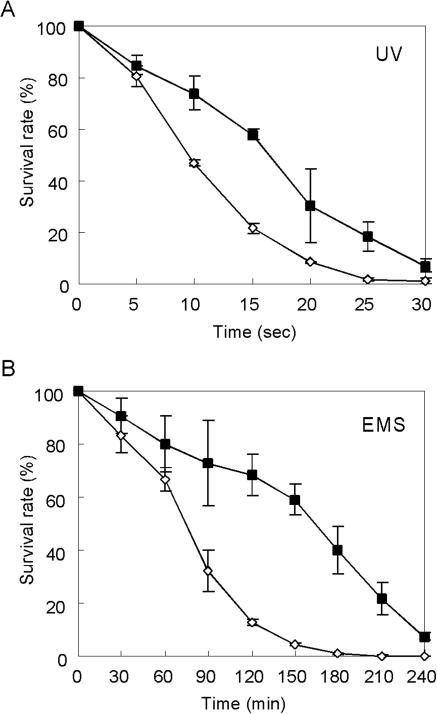
Survival rates following UV mutagenesis and EMS mutagenesis. Diploid W303 (▪) and haploid W303-1A (⋄) strains were mutagenized by UV or EMS, and survival rates were determined. Means and standard deviations were obtained from three experiments.
For the analysis of survival rates by EMS mutagenesis, cells were inoculated into liquid YPD medium and grown for 24 h at 28°C. The cells were washed once with sterile water, diluted, and suspended in 1 ml of 0.1 M sodium phosphate buffer (pH 7.0). To the suspension, 30 μl of EMS was added. After various incubation times, the cells were washed once with 1 ml of 5% sodium thiosulfate, suspended in water, and spread on YPD medium plates to determine survival rates.
In order to generate auxotrophic mutants, cells were diluted, spread on YPD medium, and exposed to UV for 20 s. Colonies grown on YPD medium plates (200 to 500 per plate) were replica plated on both MM and YPD medium plates. For strain RAK1546, MM plates supplemented with histidine were used. Colonies that failed to grow on MM plates were picked from YPD medium plates, streaked as single colonies, and tested for nutrient requirements on MM plates supplemented initially with various pools of nutrients and then with individual nutrients in order to identify the specific requirements of the mutants (49).
Cloning of complementing genes.
In order to clone the genes that complemented the auxotrophic mutations, auxotrophs were transformed with genomic libraries made from the laboratory strains by using a simple lithium acetate method (16). The transformation mixture was spread directly on MM plates. For cloning of genes complementing the Lys− mutant of strain RAK1546, MM plates supplemented with histidine were used. The genomic DNA libraries used were constructed with the vectors YEp51B (7), pRS318 (51), and pSMA112 (30). The genomic libraries in pRS318 and pSMA112 were constructed by inserting partial Sau3AI digests of chromosomal DNA from strain RAK711 (MATa ade2 ura3 trp1 his3 leu2 ste4::HIS3 sst2::lacZ-URA3 STE7-101; isogenic to W303-1A) into the vector BamHI site. Plasmids were extracted by using a Zymoprep yeast plasmid minipreparation kit (Zymo Research, Orange, Calif.) according to the manufacturer's instructions. E. coli was transformed with the extracts, and inserts from the isolated plasmids were partially sequenced by using a BigDye Terminator cycle sequencing kit and an ABI Prism 310 genetic analyzer (Perkin-Elmer Applied Biosystems).
Plasmid constructs.
The primers used and the plasmids constructed in this study are listed in Tables 2 and 3, respectively. The marker genes for plasmid construction, except for HIS3, were amplified by PCR with Kyokai no. 7 chromosomal DNA as a template. PCRs were performed with KOD Dash DNA polymerase (Toyobo, Osaka, Japan) according to the manufacturer's instructions. The PCR program was initiated at 94°C for 1 min, followed by 25 or 30 cycles at 94°C for 20 s, 54°C for 2 s, and 74°C for 1 to 3 min. TRP3 was amplified with primers TRP3-1 and TRP3-2, and LYS4 was amplified with primers LYS4-1 and LYS4-2. pBlueKS+AscI was constructed by inserting a phosphorylated AscI linker (5′-pGGCGCGCC -3′) into the SmaI site of pBluscript KS(+) (Stratagene). TRP3 and LYS4 PCR fragments were digested with AscI and BamHI and inserted into the AscI-BamHI sites of pBlueKS+AscI to form pBlueTRP3 and pBlueLYS4, respectively.
TABLE 2.
Primers
| Primer | Sequencea (5′→3′) |
|---|---|
| TRP3-1 | GGCGCGCCGAGCTCTAATATTATTTACAA |
| TRP3-2 | TTCCAAGCCGGATCCTTTAA |
| LYS4-1 | GGGGATCCCGGGATTTTGCTCGAGAT |
| LYS4-2 | GGCGCGCCTCGAGACATTCAATGTAAGA |
| ScHIS3+12 | GGCGCGCCCGGGTTTTAAGAGCTTGG |
| SacIScHIS3 | AAAGAGCTCCGTCGAGTTCAAG |
| LYS4+12 | GGCGCGCCCGGGGCTAACAGAGACCCACCG |
| LYS4SacI | AAAGAGCTCGGGTATCTCGAGACATTC |
| TRP3+12N | GGCGCGCCCGGGCAGCGGCAATGGATCC |
| TRP3SacI | AAAGAGCTCAACGCCGGCCCCAGAGGG |
| LEU4+12 | GGCGCGCCCGGGGTGCTTATCAGAAGTGCG |
| LEU4SacI | AAAGAGCTCTGCTGCCTTTGCGTGGCC |
| CEN/ARS121 | GGCGCGCCCATCACGTGCTATAA |
| CEN/ARS122 | GGCGCGCCGACGGATCGCTTGCC |
| 352ori-121 | GGCGCGCCCTTGTTACCCATCAT |
| 352ori-122 | GGCGCGCCGTATGGCTTCATTCA |
| LEU1-1 | GCCGCCTCCGCGAAATAC |
| LEU1-2 | GCATTAACAAGGGCTAGC |
| ATF1-401 | CTTCATCAGTATCACAAATACCATCAATTTATCAGCTCTCGGCGCGCCCG |
| ATF1-402 | GTTGCACGGGGGCCTGATTTTTCTCATCGATTTCATTCATTTTATGTGAT |
| TDH3-160 | CCCCTACTTGACTAATAA |
| NotI-ATF1 | GGGGCGGCCGCTCCTTACATCGAGAAGAT |
| ATF1-UPXBA | CCAATCATTAGCCGTTCCTCTAGA |
| ATF1-APA | TTTGGGCCCCTAAGGGCCTAAAAGGAGAGC |
Restriction sites are shown in italic type.
TABLE 3.
Plasmids
| Plasmid | Feature(s)a |
|---|---|
| pBlueTRP3 | TRP3 |
| pBlueLYS4 | LYS4 |
| pGG121 | TRP3 GALp-GIN11M86 |
| pGG122 | LYS4 GALp-GIN11M86 |
| pScHIS3TDH3p | HIS3 TDH3p |
| pScLYS4TDH3p | LYS4 TDH3p |
| pScTRP3TDH3p | TRP3 TDH3p |
| pScLEU4TDH3p | LEU4 TDH3p |
| pScHIS3cat | HIS3 TDH3p CEN/ARS |
| pScLYS4cat | LYS4 TDH3p CEN/ARS |
| pScTRP3cat | TRP3 TDH3p CEN/ARS |
| pScLEU4cat | LEU4 TDH3p CEN/ARS |
| pScHIS3ori | HIS3 TDH3p, 2μm origin |
| pScLYS4ori | LYS4 TDH3p, 2μm origin |
| pScTRP3ori | TRP3 TDH3p, 2μm origin |
| pScLEU4ori | LEU4 TDH3p, 2μm origin |
GALp-GIN11M86 is a counterselection marker (4).
A 1.8-kb DraIII-NotI fragment of pBlueKS+M86 (4) containing GALp-GIN11M86 was inserted into the DraIII-NotI sites of pBlueTRP3 to form pGG121. A 2.5-kb BamHI-ScaI fragment of pBlueKS+M86 was inserted into the BamHI-ScaI sites of pBlueLYS4 to form pGG122.
pPpHIS3TDH3p and pKlURA3TDH3p contained Pichia pastoris HIS3 and Kluyveromyces lactis URA3, respectively, together with the TDH3 promoter (TDH3p), and were constructed (unpublished data) on the basis of vector pT7Blue (Novagen). A 1.2-kb HIS3 fragment was amplified with primers ScHIS3+12 and SacIScHIS3 and with pGG113 (4) as a template. The AscI-SacI-digested HIS3 fragment was inserted into the AscI-SacI sites of pKlURA3TDH3p to form pScHIS3TDH3p. A 3.3-kb LYS4 fragment was amplified with primers LYS4+12 and LYS4SacI. The AscI-SacI-digested LYS4 fragment was inserted into the AscI-SacI sites of pPpHIS3TDH3p to form pScLYS4TDH3p. Similarly, 2.3-kb TRP3 and 3.3-kb LEU4 fragments were amplified with primers TRP3+12N and TRP3SacI and primers LEU4+12 and LEU4SacI, respectively, and inserted into the AscI-SacI sites of pPpHIS3TDH3p to form pScTRP3TDH3p and pScLEU4TDH3p, respectively.
A 0.5-kb CEN/ARS sequence was amplified from pRS316 (52) with primers CEN/ARS121 and CEN/ARS122. The fragment was digested with AscI and inserted into the AscI sites of pScHIS3TDH3p, pScLYS4TDH3p, pScTRP3TDH3p, and pScLEU4TDH3p to form pScHIS3cat, pScLYS4cat, pScTRP3cat, and pScLEU4cat, respectively.
A 1.4-kb fragment containing the 2μm origin was amplified from YEp352 (24) with primers 352ori-121 and 352ori-122. The fragment was digested with AscI and inserted into the AscI sites of pScHIS3TDH3p, pScLYS4TDH3p, pScTRP3TDH3p, and pScLEU4TDH3p to form pScHIS3ori, pScLYS4ori, pScTRP3ori, and pScLEU4ori, respectively.
A 1.4-kb SalI-XhoI fragment containing the HIS3 gene from pSK-HIS3 (4) was inserted into the SalI site of p305ATTDH3pATF1 (26) to form pSKHIS3-ATF1.
Strain construction.
Yeast transformation was performed by using a simple lithium acetate method (16). A 1.9-kb PCR fragment was amplified with primers ATF1-401 and ATF1-402 and with pScHIS3TDH3p as a template. The fragment contained HIS3, TDH3p, and 40 bp of sequences homologous to ATF1 at both ends. Strain RAK1536 was transformed with the fragment in order to insert HIS3-TDH3p upstream of ATF1 as described previously (35). Correct insertion of the fragment was determined by colony PCR (6) with primers TDH3p-160 and NotI-ATF1. Strain RAK1536 was also transformed with XhoI-XbaI-digested pSKHIS3-ATF1 containing the ATF1 5′-noncoding region and the HIS3-TDH3p-ATF1 coding region. Correct insertion was determined by colony PCR with primers ATF1-UPXBA and ATF1-APA.
For the analysis of the alcohol fermentation ability of the mutants, a small-scale fermentation experiment was performed. For this, 9.5 g of rice and 3.0 g of Koji (G-50; Tokushima Seiko Co., Ltd.) were suspended in 17 ml of sterile water. To this tube, 1.2 ml of a 1-day-old yeast culture in YPD medium was added. Fermentation was performed at 8°C for 8 days. The ethanol concentration was measured with the alcohol detector Alcomate (Riken Keiki Co., Ltd.).
For the analysis of flavor components, strains Kyokai no. 7, RAK1536, and RAK1884 were inoculated into 7 ml of Koji extract prepared by incubating Koji powder suspension in 2 volumes of water at 50°C for 5 h, mixed with 0.5 g of a dry-powder preparation of Aspergillus oryzae, and incubated at 15°C for 7 day. The amount of isoamyl acetate was measured as described previously (10).
RESULTS
Rates of survival of haploid and diploid strains following mutagenesis.
Rates of survival of isogenic haploid and diploid strains were determined following UV mutagenesis and EMS mutagenesis. If one assumes that the induced rate of mutation for a single gene is about 10−3 (49) in haploid cells, then the simultaneous rate of mutation for two homozygous genes in a diploid would be 10−6, which is much too low to allow detection by replica plating. If this simple calculation is applied to survival rates for diploid cells, based on induced mutations occurring in essential genes, few diploids would be expected to be killed even at doses found to be lethal for haploids. However, after exposure to UV for 30 s or incubation with EMS for 240 min, the survival rate for the diploid strain was less than 10% (Fig. 1). This result indicates that the model of simultaneous double mutagenesis for homozygous alleles in a diploid is unlikely to apply in the yeast S. cerevisiae (28). This result also suggests that even in a diploid, recessive mutations might be obtained by a conventional screening procedure. We decided to use UV mutagenesis because the survival curve for the diploid exposed to UV was closer to that of the haploid than the survival curve after EMS mutagenesis.
Auxotrophic mutants selected from industrial yeast strains.
When Kyokai strains were exposed to UV for 20 s, survival rates ranged from 8.1 to 22.0% (Table 4). Auxotrophic mutants were screened by a replica plating method. After exposure to UV, colonies grown on YPD medium plates (200 to 500 per plate) were replica plated on MM and YPD medium plates, and cells that failed to grow on MM were selected. Among 1,700 to 3,400 selected colonies, one to six auxotrophic mutants of each parental strain were obtained (Table 4). Mutation frequencies ranged from 5.3 × 10−4 to 2.0 × 10−3.
TABLE 4.
Generation of auxotrophic mutants by UV mutagenesisa
| Strain | % Survival rate | No. of:
|
Frequency of mutants | |
|---|---|---|---|---|
| Colonies selected | Mutants | |||
| Kyokai no. 7 | 22.0 | 3,422 | 2 | 5.8 × 10−4 |
| Kyokai no. 9 | 19.3 | 1,696 | 1 | 5.9 × 10−4 |
| Kyokai no. 10 | 13.4 | 3,046 | 2 | 6.6 × 10−4 |
| Kyokai no. 701 | 9.9 | 2,147 | 3 | 1.4 × 10−3 |
| Kyokai no. 901 | 8.1 | 2,998 | 6 | 2.0 × 10−3 |
| RAK1536 | 17.8 | 1,885 | 1 | 5.3 × 10−4 |
UV irradiation for 20 s at a distance of 35 cm.
Auxotrophic mutants obtained included His−, Leu−, Arg−, Ura−, Met−, and Trp− mutants (Table 5). The His− mutant RAK1536 derived from Kyokai no. 7 was used for mutagenesis and screening for constructing multiple auxotrophic mutants. The survival rate for RAK1536 exposed to UV for 20 s was 17.8%, similar to the first-round mutation frequency of 22.0% The Lys− mutant RAK1546 was isolated from RAK1536 at a frequency of 5 × 10−4, resulting in a double mutant (His− Lys−) derived from Kyokai no. 7. These results indicate that mutants carrying recessive mutations can be isolated by conventional replica plating even from diploid industrial yeast strains.
TABLE 5.
Nutrients required by mutants and complementing genes
| Strain(s)
|
Nutrient required | Genea | |
|---|---|---|---|
| Parental | Mutant | ||
| Kyokai no. 7 | RAK1536 | Histidine | HIS3 |
| Kyokai no. 7 | RAK1537 | Methionine | ND |
| RAK1536 | RAK1546 | Lysine | LYS4 |
| Kyokai no. 701 | RAK1763 | Leucine | LEU4 |
| Kyokai no. 701 | RAK1764 | Arginine | ND |
| Kyokai no. 701 | RAK1785 | Uracil | ND |
| Kyokai no. 10 | RAK1786 | Methionine | ND |
| Kyokai no. 10 | RAK1787 | Tryptophan | TRP3 |
| Kyokai no. 9 | RAK1788 | Tryptophan | ND |
| Kyokai no. 901 | RAK1922, RAK1923, RAK1924, RAK1925, RAK1927 | Tryptophan | TRP3 |
| Kyokai no. 901 | RAK1926 | Tryptophan | ND |
ND, not determined.
The alcohol fermentation ability of the mutants was examined with a small-scale rice-Koji mixture (Table 6). Except for the Met− mutant RAK1786, auxotrophic mutants produced alcohol concentrations equivalent to those produced by the parental strains, suggesting that UV mutagenesis did not affect fermentation ability significantly.
TABLE 6.
Alcohol fermentation of auxotrophic mutants
| Strain
|
Mutation(s) | % (vol/vol) Alcohol | |
|---|---|---|---|
| Tested | Parent | ||
| Kyokai no. 7 | Parent | 10.4 | |
| RAK1536 | Kyokai no. 7 | His3− | 12.3 |
| RAK1537 | Kyokai no. 7 | Met− | 12.3 |
| RAK1546 | Kyokai no. 7 | His3− Lys4− | 10.9 |
| Kyokai no. 701 | Parent | 9.5 | |
| RAK1763 | Kyokai no. 701 | Leu4− | 11.0 |
| RAK1764 | Kyokai no. 701 | Arg− | 11.9 |
| RAK1785 | Kyokai no. 701 | Ura− | 12.8 |
| Kyokai no. 9 | Parent | 9.7 | |
| RAK1788 | Kyokai no. 9 | Trp− | 11.2 |
| Kyokai no. 901 | Parent | 10.2 | |
| RAK1922 | Kyokai no. 901 | Trp3− | 10.3 |
| RAK1923 | Kyokai no. 901 | Trp3− | 8.6 |
| RAK1924 | Kyokai no. 901 | Trp3− | 9.6 |
| RAK1925 | Kyokai no. 901 | Trp3− | 9.9 |
| RAK1926 | Kyokai no. 901 | Trp− | 8.8 |
| RAK1927 | Kyokai no. 901 | Trp3− | 9.4 |
| Kyokai no. 10 | Parent | 12.2 | |
| RAK1786 | Kyokai no. 10 | Met− | 7.0 |
| RAK1787 | Kyokai no. 10 | Trp3− | 9.3 |
Cloning of genes complementing auxotrophic mutations.
Genes complementing auxotrophic mutations were cloned by transforming the auxotrophs with wild-type yeast genomic DNA libraries and plating transformants directly on MM or MM-His plates. Colonies were picked, and plasmids were extracted. Mutants were retransformed with cloned plasmids for confirmation, and plasmid inserts were partially sequenced. The sequence data indicated the presence of the HIS3, TRP3, and LYS4 genes in plasmids p1685 (RAK1536: pSMA112 library), p1688 (RAK1922: pRS318 library), and p1687 (RAK1546: YEp51B library), respectively. All Trp− mutants were transformed with the plasmid containing TRP3. RAK1787, RAK1923, RAK1924, RAK1925, and RAK1927 were complemented, but RAK1788 and RAK1926 were not, indicating that the former strains were trp3 (Table 5). Attempts to clone genes complementing other auxotrophic mutations were not successful.
The leucine biosynthetic pathway contains three enzymes, α-isopropylmalate synthase (Leu4), α-isopropylmalate isomerase (Leu1), and β-isopropylmalate dehydrogenase (Leu2). In order to identify the mutant gene in the Leu− mutant RAK1763, the strain was transformed first with a plasmid containing LEU2 and separately with LEU1 generated by PCR with primers LEU1-1 and LEU1-2 (Table 2). No transformants were obtained with either method. Strain RAK1763 then was transformed with a plasmid carrying PCR-generated LEU4 (Table 3). Transformants were obtained, indicating that strain RAK1763 was leu4.
Construction of plasmids with auxotrophic markers.
Because we obtained auxotrophic mutants of the Kyokai strains and identified the complementing genes, we constructed vectors for gene manipulations of the strains. TRP3 and LYS4 were cloned by PCR from Kyokai no. 7 chromosomal DNA and used to construct the counterselective vectors pGG121 and pGG122 (Table 3). These plasmids can be used for two-step gene replacement in the trp3 and lys4 mutants of the Kyokai strains (10, 26). In addition to the counterselective plasmids, we constructed plasmids for gene overexpression applicable to self-cloning. The plasmids contained TDH3p for constitutive overexpression, adjacent to the auxotrophic markers. Low-copy number (CEN/ARS) and high-copy-number (2μm) plasmids were also constructed (Table 3).
Construction of ATF1 overexpression strains by one-step cloning.
The overexpression of ATF1 was reported to produce a large amount of isoamyl acetate, a banana-like flavor, in Japanese sake and other alcoholic drinks (19, 34, 36). In our previous study (26), an ATF1-overexpressing sake strain was constructed by inserting TDH3p at the 5′ end of ATF1 by a two-step replacement protocol with a drug resistance marker. The constructed strain contained exclusively yeast DNA. However, the construction process was complicated due to the insertion and subsequent excision of the drug resistance marker.
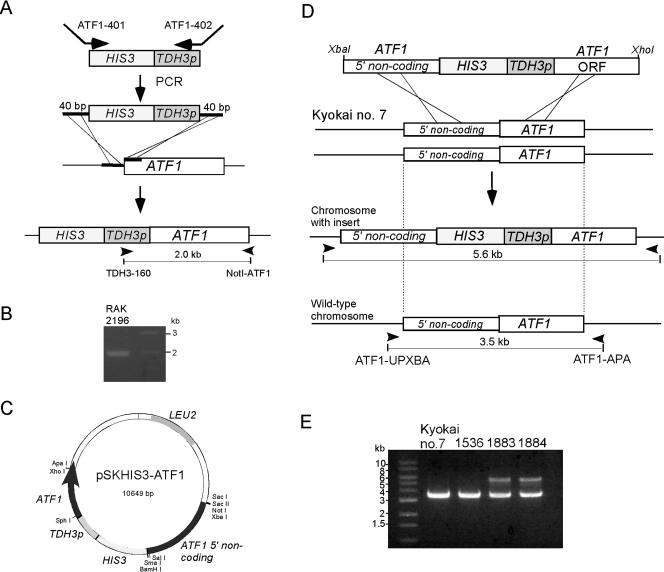
In the present study, similar TDH3p-ATF1 strains were constructed by the one-step protocol. First, a PCR fragment was amplified with primers ATF1-401 and ATF1-402 and with pScHIS3TDH3p as a template to generate 40-bp sequences homologous to ATF1 at both ends. One was from the 5′ upstream noncoding region, and the other was from the N-terminal coding region (Fig. 2A). RAK1536 was transformed with the fragment several times. Among 11 His+ transformants, only one (RAK2196) contained the correct insertion, as judged by colony PCR with primers TDH3p-160 and NotI-ATF1 (Fig. 2B). The low integration efficiency may have been due to the use of short homologous sequences. Therefore, we constructed pSKHIS3-ATF1, in which the yeast HIS3 marker was inserted between the 5′ noncoding ATF1 sequence and TDH3p-driven ATF1 of plasmid p305ATTDH3pATF1 (26) (Fig. 2C). An XbaI-XhoI fragment of pSKHIS3-ATF1 containing a long 5′ noncoding region and an ATF1 coding region at either end of HIS3-TDH3p was introduced into RAK1536 by transformation (Fig. 2D). Three transformants were obtained; two (RAK1883 and RAK1884) out of the three showed the correct insertion because two bands (3.5 and 5.6 kb) were seen by colony PCR with primers ATF1-UPXBA and ATF1-APA, whereas Kyokai no. 7 and RAK1536 showed only a 3.5-kb band (Fig. 2E). The 3.5-kb band corresponded to the wild-type ATF1 fragment, and the 5.6-kb band corresponded to the HIS3-TDH3p insert, indicating that correct insertion occurred in one chromosome of the diploid Kyokai no. 7 strain.
FIG. 2.
One-step cloning of TDH3p-ATF1. One-step cloning was performed by two protocols (A and D). (A) PCR amplification with primers ATF1-401 and ATF1-402 and with pScHIS3TDH3p as a template produced a fragment that contained 40 bp of ATF1 sequence at both ends. Transformation with the fragment resulted in an insertion of HIS3-TDH3p at the 5′ upstream region of ATF1. (B) A 2.0-kb band produced by colony PCR from transformant RAK2196 indicated the correct insertion. (C) pSKHIS3-ATF1 contained TDH3p upstream of the ATF1 open reading frame, the HIS3 marker, and 5′ noncoding ATF1 sequences. (D) Transformation with the XbaI-XhoI fragment resulted in an insertion of HIS3-TDH3p at the 5′ end of ATF1. (E) Transformants RAK1883 and RAK1884 produced two bands, one of which was 5.6 kb and the other of which was 3.5 kb, corresponding to the altered and wild-type chromosomes, respectively. Arrowheads in panels A and D indicate primers used for colony PCR.
Kyokai no. 7, RAK1536, and RAK1884 were incubated at 15°C for 7 days in Koji extract, and the flavor components were measured. The TDH3p-ATF1 integrant produced a larger amount of isoamyl acetate (15.4 ppm) than the parental strains (5.7 ppm in Kyokai no. 7 and 5.9 ppm in the his3/his3 mutant), suggesting that the overexpression of ATF1 occurred. The levels of alcohol production by these strains were 17.5% in the TDH3p-ATF1 integrant, 18.2% in Kyokai no. 7, and 18.1% in the his3/his3 mutant, indicating that fermentation ability was not affected by mutagenesis.
DISCUSSION
Due to the presumed diploidy or polyploidy of industrial yeast strains (12, 17, 21, 22), screening for recessive mutations by replica plating has not been routinely used as it has been for haploids. Instead, mutants of industrial yeast strains have been isolated by positive selection for drug or stress resistance phenotypes (8, 9, 11, 20, 27, 32, 37, 40, 41, 54) or by enrichment procedures (13, 33, 42, 43). Here we show that mutants carrying auxotrophic recessive mutations can be routinely isolated in industrial sake strains by replica plating at frequencies ranging from 5.3 × 10−4 to 2.0 × 10−3.
In order to explain the higher-than-expected frequencies for the diploid sake strains, we propose that following the initial induction of a mutation in one allele, a loss of heterozygosity (LOH) is likely to have occurred (1, 23, 25, 28). Alternatively, it is possible that the sake strains were heterozygous at some of the relevant loci prior to mutagenesis and that the mutagenesis simply induced LOH, perhaps through mitotic gene conversion or mitotic crossover (38).
To observe LOH under the conditions used in this study, a ura3− mutant was selected from URA3+ haploid strains, from a homozygous URA3+/URA3+ diploid strain, and from a heterozygous ura3−/URA3+ diploid strain by plating on 5-FOA following UV mutagenesis. The number of 5-FOA-resistant colonies obtained from the heterozygous diploid was more than 100-fold greater than that obtained from either the haploid strains or the homozygous diploid (data not shown), indicating that LOH was possible under our conditions. Mitotic crossover occurred more frequently in genes located far from the centromere (38). All identified auxotrophic mutations (his3, leu4, lys4, and trp3) in this study were located more than 200 kbp away from the centromere. Therefore, it is possible that the homozygous alleles were produced by mitotic crossover after UV mutagenesis. Although several studies have reported relatively high frequencies of recessive mutations in diploid strains of S. cerevisiae (1, 25, 28), the isolation of auxotrophic mutants of diploid strains has not been investigated systematically. To the best of our knowledge, this is the first report showing the isolation of mutants of diploid strains by conventional mutagenesis and screening procedures used for haploid strains.
If the Kyokai strains already contained heterozygous auxotrophic mutations, then induced mutations in the wild-type alleles and/or LOH would produce homozygous mutant alleles. Kyokai no. 901 seemed to be such a case. Among six independent auxotrophic mutants selected from Kyokai no. 901, five were trp3. However, further analysis is required to confirm this hypothesis.
We also determined mutation frequencies following sequential UV mutagenesis of Kyokai no. 7. If UV mutagenesis had caused significant damage to chromosomal DNA, then it likely would have impaired fermentation ability and caused significant lethality. RAK1536, a his3 mutant derived from Kyokai no. 7 by UV mutagenesis, was used as a parental strain for a second round of UV mutagenesis. The second mutagenesis resulted in similar survival rates and frequencies of auxotrophic mutants, indicating that the first round of UV mutagenesis may not have induced many mutations leading to heterozygosity.
Contamination of haploid cells in a population of diploid cells can explain elevated frequencies of induced auxotrophic mutants. However, this scenario was unlikely because no sporulation of any of the Kyokai strains was observed under the experimental conditions. Further, none of the selected mutants mated as either MATa or MATα (data not shown). In addition, transformants harboring the TDH3p-ATF1 fragment produced two PCR bands (Fig. 2E), one derived from the wild-type chromosome and the other derived from the chromosome containing the integrated fragment, indicating that the strains were diploid. Also, the fermentation ability of the mutants was similar to that of the parental strains (Table 6). Thus, the auxotrophic mutants were unlikely to have been haploid contaminants.
If gene manipulations of industrial yeast strains are performed with a drug resistance marker on a bacterial plasmid, then the removal of the drug resistance marker and bacterial DNA sequences from the yeast transformant is an essential step prior to commercial application; hence, we developed a method to counterselect for the loss of the unwanted sequences (3, 4, 10, 26). However, if yeast auxotrophic markers can be used in conjunction with auxotrophic industrial mutants, then the removal of the markers will be unnecessary. Based on this concept, we constructed a self-cloning yeast by inserting a HIS3-TDH3p fragment, containing only yeast DNA, into the 5′ upstream region of ATF1 in Kyokai no. 7. A PCR-amplified fragment containing 40 bp of homologous sequences at both ends of HIS3-TDH3p was used for the gene disruption (35, 57). However, the frequency of correct insertions of the PCR fragment was low; only 1 out of 11 insertions was correctly targeted. Integration may have occurred at one of the two his3 alleles in the his3/his3 host. On the other hand, two out of three transformants were correctly targeted when a longer plasmid fragment was used for transformation. The use of industrial strains harboring complete gene deletions for auxotrophic markers would greatly reduce undesirable recombination between the introduced markers and the mutant loci. The construction of such deletion strains, as already done for laboratory strains (15), would greatly facilitate the application of recombinant DNA technology in industrial yeasts.
In conclusion, this study challenges the view that it is difficult to isolate recessive mutants from industrial diploid strains of S. cerevisiae and shows that, surprisingly, traditional methods previously used with laboratory haploid strains are effective for mutational and molecular breeding of industrial strains.
Acknowledgments
We thank Shinji Amari for help in constructing the genomic library in pSMA112 and Yasunobu Matsuura and Koichi Kuroe for help in constructing plasmids.
REFERENCES
- 1.Acuña, G., F. E. Würgler, and C. Sengstag. 1994. Reciprocal mitotic recombination is the predominant mechanism for the loss of a heterozygous gene in Saccharomyces cerevisiae. Environ. Mol. Mutagen. 24:307-316. [DOI] [PubMed] [Google Scholar]
- 2.Akada, R. 2002. Genetically modified industrial yeast ready for application. J. Biosci. Bioeng. 94:536-544. [DOI] [PubMed] [Google Scholar]
- 3.Akada, R., K. Matsuo, K. Aritomi, and Y. Nishizawa. 1999. Construction of recombinant sake yeast containing a dominant FAS2 mutation without extraneous sequences by a two-step replacement protocol. J. Biosci. Bioeng. 87:43-48. [DOI] [PubMed] [Google Scholar]
- 4.Akada, R., I. Hirosawa, M. Kawahata, H. Hoshida, and Y. Nishizawa. 2002. Sets of integrating plasmids and gene disruption cassettes containing improved counter-selection markers designed for repeated use in budding yeast. Yeast 19:393-402. [DOI] [PubMed] [Google Scholar]
- 5.Akada, R., Y. Shimizu, Y. Matsushita, M. Kawahata, H. Hoshida, and Y. Nishizawa. 2002. Use of a YAP1 overexpression cassette conferring specific resistance to cerulenin and cycloheximide as an efficient selectable marker in the yeast Saccharomyces cerevisiae. Yeast 19:17-28. [DOI] [PubMed] [Google Scholar]
- 6.Akada, R., T. Murakane, and Y. Nishizawa. 2000. DNA extraction method for screening yeast clones by PCR. BioTechniques 28:668-674. [DOI] [PubMed] [Google Scholar]
- 7.Akada, R., J. Yamamoto, and I. Yamashita. 1997. Screening and identification of yeast sequences that cause growth inhibition when overexpressed. Mol. Gen. Genet. 254:267-274. [DOI] [PubMed] [Google Scholar]
- 8.Akita, O., T. Ida, T. Obata, and S. Hara. 1990. Mutants of Saccharomyces cerevisiae producing a large quantity of β-phenethyl alcohol and β-phenethyl acetate. J. Ferment. Technol. 69:125-128. [Google Scholar]
- 9.Arikawa, Y., M. Yamada, M. Shimosaka, M. Okazaki, and M. Fukuzawa. 2000. Isolation of sake yeast mutants producing a high level of ethyl caproate and/or isoamyl acetate. J. Biosci. Bioeng. 90:675-677. [DOI] [PubMed] [Google Scholar]
- 10.Aritomi, K., I. Hirosawa, H. Hoshida, M. Shiigi, Y. Nishizawa, S. Kashiwagi, and R. Akada. 2004. Self-cloning yeast strains containing novel FAS2 mutations produce a higher amount of ethyl caproate in Japanese sake. Biosci. Biotechnol. Biochem. 68:206-214. [DOI] [PubMed] [Google Scholar]
- 11.Ashida, S., E. Ichikawa, K. Suginami, and S. Imayasu. 1987. Isolation and application of mutants producing sufficient isoamyl acetate, a sake flavor component. Agric. Biol. Chem. 51:2061-2065. [Google Scholar]
- 12.Benitez, T., J. M. Gasent-Ramirez, F. Castrejon, and A. C. Codon. 1996. Development of new strains for the food industry. Biotechnol. Prog. 12:149-163. [Google Scholar]
- 13.Bilinski, C. A., A. M. Sills, and G. G. Stewart. 1984. Morphological and genetic effects of benomyl on polyploid brewing yeasts: isolation of auxotrophic mutants. Appl. Environ. Microbiol. 48:813-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 15.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 16.Chen, D. C., B. C. Yang, and T. T. Kuo. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 21:83-84. [DOI] [PubMed] [Google Scholar]
- 17.Dequin, S. 2001. The potential of genetic engineering for improving brewing, wine-making and baking yeasts. Appl. Microbiol. Biotechnol. 56:577-588. [DOI] [PubMed] [Google Scholar]
- 18.Falk, M. C., B. M. Chassy, S. K. Harlander, T. J. Hoban, M. N. McGloughlin, and A. R. Akhlaghi. 2002. Food biotechnology: benefits and concerns. J. Nutr. 132:1384-1390. [DOI] [PubMed] [Google Scholar]
- 19.Fujii, T., N. Nagasawa, A. Iwamatsu, T. Bogaki, Y. Tamai, and M. Hamachi. 1994. Molecular cloning, sequence analysis, and expression of the yeast alcohol acetyltransferase gene. Appl. Environ. Microbiol. 60:2786-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukuda, K., M. Watanabe, K. Asano, H. Ueda, and S. Ohta. 1990. Breeding of brewing yeast producing a large amount of β-phenylethyl alcohol and β-phenylethyl acetate. Agric. Biol. Chem. 54:269-271. [Google Scholar]
- 21.Guijo, S., J. C. Mauricio, J. M. Salmon, and J. M. Ortega. 1997. Determination of the relative ploidy in different Saccharomyces cerevisiae strains used for fermentation and ‘flor’ film ageing of dry sherry-type wines. Yeast 13:101-117. [DOI] [PubMed] [Google Scholar]
- 22.Hammond, J. R. 1995. Genetically-modified brewing yeasts for the 21st century. Progress to date. Yeast 11:1613-1627. [DOI] [PubMed] [Google Scholar]
- 23.Hannan, M. A., J. Calkins, and W. L. Lasswell. 1980. Recombinagenic and mutagenic effects of sunlamp (UV-B) irradiation in Saccharomyces cerevisiae. Mol. Gen. Genet. 177:577-580. [DOI] [PubMed] [Google Scholar]
- 24.Hill, J. E., A. M. Myers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2:163-167. [DOI] [PubMed] [Google Scholar]
- 25.Hiraoka, M., K. Watanabe, K. Umezu, and H. Maki. 2000. Spontaneous loss of heterozygosity in diploid Saccharomyces cerevisiae cells. Genetics 156:1531-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirosawa, I., K. Aritomi, H. Hoshida, S. Kashiwagi, Y. Nishizawa, and R. Akada. 2004. Construction of a self-cloning sake yeast that overexpresses alcohol acetyltransferase gene by a two-step gene replacement protocol. Appl. Microbiol. Biotechnol. 65:68-73. [DOI] [PubMed] [Google Scholar]
- 27.Ichikawa, E., N. Hosokawa, Y. Hata, Y. Abe, K. Suginami, and S. Imayasu. 1991. Breeding of a sake yeast with improved ethyl caproate productivity. Agric. Biol. Chem. 55:2153-2154. [Google Scholar]
- 28.James, A. P., and B. J. Kilbey. 1977. The timing of UV mutagenesis in yeast: a pedigree analysis of induced recessive mutation. Genetics 87:237-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jonas, D. A., I. Elmadfa, K. H. Engel, K. J. Heller, G. Kozianowski, A. König, D. Müller, J. F. Narbonne, W. Wackernagel, and J. Kleiner. 2001. Safety considerations of DNA in food. Ann. Nutr. Metab. 45:235-254. [DOI] [PubMed] [Google Scholar]
- 30.Kawahata, M., S. Amari, Y. Nishizawa, and R. Akada. 1999. A positive selection for plasmid loss in Saccharomyces cerevisiae using galactose-inducible growth inhibitory sequences. Yeast 15:1-10. [DOI] [PubMed] [Google Scholar]
- 31.Kitamoto, K., K. Oda, K. Gomi, and K. Takahashi. 1990. Construction of uracil and tryptophan auxotrophic mutants from sake yeasts by disruption of URA3 and TRP1 genes. Agric. Biol. Chem. 54:2979-2987. [Google Scholar]
- 32.Kitamoto, K., K. Oda-Miyazaki, K. Gomi, and C. Kumagai. 1993. Mutant isolation of non-urea producing sake yeast by positive selection. J. Ferment. Bioeng. 75:359-363. [Google Scholar]
- 32a.Kodama, Y., Y. Nakao, N. Nakamura, T. Fujimura, K. Shirahige, and T. Ashikari. 2003. Diversity of the chromosomal structure in lager brewing yeasts. Yeast 20:S276. [Google Scholar]
- 33.Kyogoku, Y., and K. Ouchi. 1995. Isolation of a cold-sensitive fermentation mutant of a baker's yeast strain and its use in a refrigerated dough process. Appl. Environ. Microbiol. 61:639-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lilly, M., M. G. Lambrechts, and I. S. Pretorius. 2000. Effect of increased yeast alcohol acetyltransferase activity on flavor profiles of wine and distillates. Appl. Environ. Microbiol. 66:744-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorenz, M. C., R. S. Muir, E. Lim, J. McElver, S. C. Weber, and J. Heitman. 1995. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene 158:113-117. [DOI] [PubMed] [Google Scholar]
- 36.Mason, A. B., and J. P. Dufour. 2000. Alcohol acetyltransferases and the significance of ester synthesis in yeast. Yeast 16:1287-1298. [DOI] [PubMed] [Google Scholar]
- 37.Mizoguchi, H., M. Watanabe, and A. Nishimura. 1999. Characterization of a PDR1 mutant allele from a clotrimazole-resistant sake yeast mutant with improved fermentative activity. J. Biosci. Bioeng. 88:20-25. [DOI] [PubMed] [Google Scholar]
- 38.Nakai, S., and R. K. Mortimer. 1969. Studies of the genetic mechanism of radiation-induced mitotic segregation in yeast. Mol. Gen. Genet. 103:329-338. [DOI] [PubMed] [Google Scholar]
- 39.Nakazawa, N., T. Ashikari, N. Goto, T. Amachi, R. Nakajima, S. Harashima, and Y. Oshima. 1992. Partial restoration of sporulation defect in sake yeasts, Kyokai no. 7 and no. 9, by increased dosage of the IME1 gene. J. Ferment. Bioeng. 73:265-270. [Google Scholar]
- 40.Oda, K., K. Kitamoto, K. Gomi, and K. Takahashi. 1990. Isolation of uracil auxotrophic mutants from sake yeast strains and sake brewing using the mutants. Hakkokogaku 68:399-403. [Google Scholar]
- 41.Oda, K., K. Kitamoto, K. Takahashi, and K. Yoshizawa. 1988. Isolation of lysine auxotrophic mutants from brewing yeast strains. J. Brew. Soc. Jpn. 83:614-617. [Google Scholar]
- 42.Ouchi, K., and H. Akiyama. 1971. Non-foaming mutants of sake yeasts. Agric. Biol. Chem. 35:1024-1032. [Google Scholar]
- 43.Ouchi, K., M. Shimoda, Y. Nakamura, Y. Kojima, and T. Nishiya. 1983. Isolation of auxotrophic mutans from industrial yeasts. Hakkokogaku 61:349-352. [Google Scholar]
- 44.Pretorius, I. S. 2000. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16:675-729. [DOI] [PubMed] [Google Scholar]
- 45.Rose, M. D., and J. R. Broach. 1991. Cloning genes by complementation in yeast. Methods Enzymol. 194:195-230. [DOI] [PubMed] [Google Scholar]
- 46.Rothstein, R. 1991. Targeting, disruption, replacement, and allele rescure: integrative DNA transformation in yeast. Methods Enzymol. 194:281-301. [DOI] [PubMed] [Google Scholar]
- 47.Schilter, B., and A. Constable. 2002. Regulatory control of genetically modified (GM) foods: likely developments. Toxicol. Lett. 127:341-349. [DOI] [PubMed] [Google Scholar]
- 48.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 49.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 50.Sherman, F., and P. Wakem. 1991. Mapping yeast genes. Methods Enzymol. 194:38-57. [DOI] [PubMed] [Google Scholar]
- 51.Sikorski, R. S., and J. D. Boeke. 1991. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 194:302-318. [DOI] [PubMed] [Google Scholar]
- 52.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sprague, G. F., Jr. 1991. Assay of yeast mating reaction. Methods Enzymol. 194:77-93. [DOI] [PubMed] [Google Scholar]
- 54.Teunissen, A., F. Dumortier, M. Gorwa, J. Bauer, A. Tanghe, A. Loïez, P. Smet, P. Van Dijck, and J. M Thevelein. 2002. Isolation and characterization of a freeze-tolerant diploid derivative of an industrial baker's yeast strain and its use in frozen doughs. Appl. Environ. Microbiol. 68:4780-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptonally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 56.Tsuboi, M., and T. Takahashi. 1988. Genetic analysis of the non-sporulating phenotype of brewer's yeasts. J. Ferment. Technol. 66:605-613. [Google Scholar]
- 57.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. El Dow, M. Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]