Abstract
This study used a genetic fingerprinting technique (automated ribosomal intergenic spacer analysis [ARISA]) to characterize microbial communities from a culture-independent perspective and to identify those environmental factors that influence the diversity of bacterial assemblages in Wisconsin lakes. The relationships between bacterial community composition and 11 environmental variables for a suite of 30 lakes from northern and southern Wisconsin were explored by canonical correspondence analysis (CCA). In addition, the study assessed the influences of ARISA fragment detection threshold (sensitivity) and the quantitative, semiquantitative, and binary (presence-absence) use of ARISA data. It was determined that the sensitivity of ARISA was influential only when presence-absence-transformed data were used. The outcomes of analyses depended somewhat on the data transformation applied to ARISA data, but there were some features common to all of the CCA models. These commonalities indicated that differences in bacterial communities were best explained by regional (i.e., northern versus southern Wisconsin lakes) and landscape level (i.e., seepage lakes versus drainage lakes) factors. ARISA profiles from May samples were consistently different from those collected in other months. In addition, communities varied along gradients of pH and water clarity (Secchi depth) both within and among regions. The results demonstrate that environmental, temporal, regional, and landscape level features interact to determine the makeup of bacterial assemblages in northern temperate lakes.
The application of molecular biological techniques to microbial ecology has led to a greatly increased appreciation of microbial diversity (58, 84) and has sparked interest in determining the factors that constrain microbial community composition and its variation in terrestrial and aquatic systems (37, 44, 45, 53, 82, 83). Numerous studies have described the tremendous variability in the composition of communities of bacterioplankton among lakes (13, 29, 39, 44, 45, 74, 82, 83), and a growing body of literature has begun to suggest possible causes of this variation (44, 45, 53, 82, 83). Previous work on 13 Wisconsin lakes suggested that, in addition to the influence of temperature and climate, two dominant forces might structure freshwater bacterial community composition (BCC): system productivity (as expressed by chlorophyll a concentration) and dissolved organic carbon (DOC) (83). While other studies have provided corroborating support for these influences (32, 44, 47, 82), the conclusions of the Wisconsin study may have been confounded by the geographic details of the study design. In particular, the differences in BCC attributed to lake productivity may have also been related, to a greater or lesser extent, to regional differences between northern (oligotrophic) and southern (eutrophic) Wisconsin lakes (83).
Geography and spatial autocorrelation can impart structure to ecological data, and this structure may coincide with other sources of environmental variability, leading to spurious correlations among biological and environmental variables (6, 42). Untangling the effects of environmental variation from those due to autocorrelation (i.e., purely spatial effects) represents a major challenge for microbial ecologists investigating patterns in BCC. Lindström and Leskinen (47) have suggested that regional differences among lakes can influence the community composition of the abundant bacteria. Recently Whitaker et al. (80) attributed all “Sulfolobus islandicus” genetic variance in hot springs to geographic variation rather than to environmental sources. These studies suggest that a regional perspective may be an important consideration for the assessment of BCC and its patterns and environmental correlates.
A geographical perspective may also be valuable within individual lake districts. Borrowing ideas from landscape ecology, limnologists have begun to consider how a lake's position in the overland and groundwater flow system (i.e., landscape position [LP]) can impact various features of lake ecology (41, 61, 64, 67). These studies have shown that a wide range of biological and environmental variables appear to correlate with a lake's LP and that some of these patterns may be general across different lake districts (61, 64, 67). In particular, it was shown that lakes higher in the landscape tend to be more isolated in terms of direct overland connections to other lakes, and these lakes express a greater temporal range of variation in a number of environmental characteristics than lakes lower in the landscape (40, 49). These patterns may be reflected in the composition and dynamics of lake bacterial communities.
Geological and ecological forces operate at a variety of hierarchical scales (1, 56) and can generate complicated associations and interactions that are reflected in community composition. The present study uses data from 30 Wisconsin lakes to evaluate the spatial, temporal, and environmental influences on BCC. The principal goals were to determine the extent to which similar bacterial communities could be found in lakes with similar environments and to identify the features that best explain variation in lake BCC. The study design was intended to allow for investigation at broad (i.e., regional) and local spatial scales and to capture some of the temporal variation that lake bacterial communities have been shown to exhibit over the course of the year (12, 16, 30, 66, 82). In addition, because conclusions of microbial ecological studies can be influenced by biases inherent in molecular methods (78) and by numerical artifacts arising from data transformations (28, 55), this study investigated the impact of various treatments and transformations applied to the bacterial data set. The questions addressed here are as follows. Do the detection limits of molecular analytical techniques or the data transformation applied to community data (or both) influence the conclusions and, if so, how much? Do similar environmental conditions produce similar bacterial communities? What are the most important environmental controls of BCC? Do regional, landscape level, or seasonal factors produce similar bacterial communities, and to what extent do these factors correlate to environmental controls?
MATERIALS AND METHODS
Field sampling.
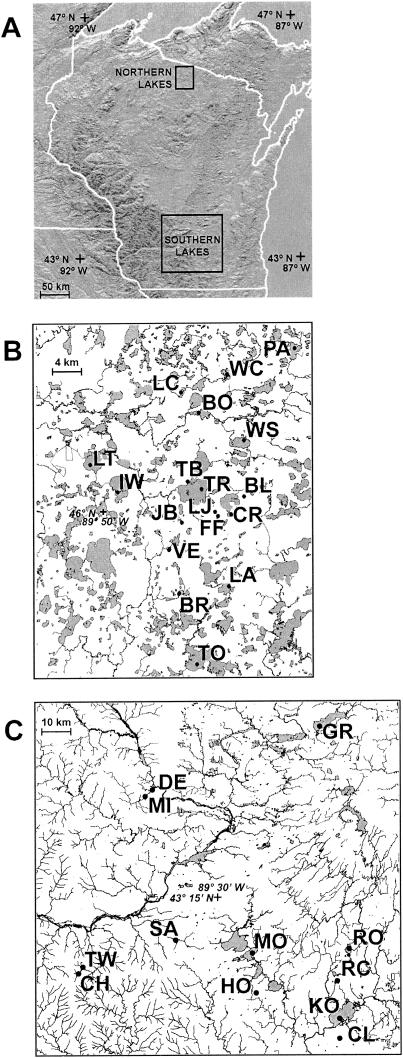
In 2002, 30 lakes were sampled for BCC and water chemistry parameters. These lakes were distributed throughout the Northern Highlands Lake District (Vilas and Oneida Counties) in Wisconsin and also throughout several counties in southern Wisconsin (Fig. 1). The lakes differed in a number of chemical and physical variables, including inorganic and organic nutrient content, pH, productivity, size, and relative position within the groundwater and overland flow systems (Table 1). Each lake was sampled on three separate occasions: late May to early June, late July, and early October. All water samples were collected over the deepest spot in the lake in cases where bathymetry was known or from the center of the lake otherwise.
FIG.1.
Map of study lakes. (A) State map of Wisconsin showing the locations of the northern and southern regions. (B) Detail of northern Wisconsin lakes. (C) Detail of southern Wisconsin lakes. Lake abbreviations are as listed in Table 1.
TABLE 1.
Lake detailsa
| Lake name (abbreviation) | Lat (N) | Long (W) | SA | z (m) | LPb | pH | Chl a (μg/liter) | DOC (mg/liter) | DP (μg/liter) | DN (μg/liter) |
|---|---|---|---|---|---|---|---|---|---|---|
| Blueberry (BL) | 46.02 | 89.59 | 1.8 | 5.8 | S | 6.6 (1.3) | 7.2 (7.2) | 5.3 (1.6) | 6.0 (2.1) | 375 (8) |
| Boulder (BO) | 46.12 | 89.67 | 216.3 | 7.6 | D | 6.3 (0.6) | 5.9 (1.7) | 6.6 (1.0) | 8.6 (5.2) | 440 (158) |
| Brandy (BR) | 45.91 | 89.70 | 44.5 | 12.9 | D | 7.2 (2.6) | 6.6 (7.0) | 4.3 (1.5) | 6.1 (3.3) | 340 (187) |
| Clear (CL) | 42.80 | 88.98 | 33.2 | 4.0 | S | 8.3 (1.2) | 5.6 (5.5) | 5.6 (1.9) | 10.0 (3.1) | 635 (126) |
| Cox Hollow (CH) | 43.01 | 90.11 | 38.5 | 6 | D | 8.5 (0.8) | 13.9 (13.9) | 4.0 (1.3) | 12.2 (9.8) | 724 (344) |
| Crystal (CR) | 46.00 | 89.61 | 36.7 | 21.2 | S | 7.3 (1.3) | 3.9 (3.5) | 5.4 (6.7) | 3.0 (3.2) | 235 (78) |
| Delton (DE) | 43.60 | 89.78 | 108.0 | 3 | D | 8.6 (0.6) | 31.2 (27.0) | 4.6 (0.8) | 17.6 (16.8) | 645 (139) |
| Firefly (FF) | 46.00 | 89.63 | 10.9 | 13.7 | S | 6.8 (1.7) | 11.7 (23.1) | 3.0 (0.9) | 3.6 (1.3) | 345 (240) |
| Green (GR) | 43.79 | 89.04 | 2,972.8 | 71.9 | D | 8.6 (0.4) | 7.4 (11.8) | 5.4 (1.2) | 8.7 (6.6) | 539 (86) |
| Hook (HO) | 42.94 | 89.34 | 50.6 | 1 | S | 5.4 (1.1) | 11.3 (5.5) | 12.2 (1.8) | 19.9 (6.3) | 754 (31) |
| Ike Walton (IW) | 46.03 | 89.81 | 576.3 | 64.4 | S | 6.2 (0.8) | 4.7 (4.0) | 3.4 (1.3) | 4.2 (1.3) | 402 (136) |
| Jimi Hendrix Bog (JB) | 45.99 | 89.70 | 1 | 3.5 | S | 7.0 (1.8) | 11.1 (8.6) | 6.8 (1.8) | 7.7 (5.4) | 532 (30) |
| Koshkonong (KO) | 42.86 | 88.97 | 4,233.0 | 2 | D | 8.5 (0.3) | 63.1 (95.3) | 10.4 (3.3) | 110.0 (155.1) | 1,407 (1029) |
| Little Arbor Vitae (LA) | 45.91 | 89.62 | 216.1 | 8.4 | D | 7.2 (2.0) | 10.5 (23.3) | 3.9 (2.3) | 9.3 (5.8) | 429 (221) |
| Little Crooked (LC) | 46.15 | 89.70 | 60.6 | 6.6 | D | 6.3 (0.7) | 9.7 (12.0) | 4.9 (0.8) | 6.7 (2.4) | 398 (294) |
| Little John, Jr. (LJ) | 46.01 | 89.64 | 10.1 | 6.8 | S | 7.1 (1.6) | 13.1 (3.6) | 6.4 (2.2) | 6.5 (7.3) | 496 (186) |
| Little Trout (LT) | 46.06 | 89.86 | 407.3 | 19.7 | D | 6.6 (0.3) | 6.6 (0.3) | 6.4 (0.9) | 5.2 (3.3) | 378 (144) |
| Mirror (MI) | 43.57 | 89.81 | 55.4 | 3.6 | D | 8.2 (0.6) | 12.3 (17.1) | 3.1 (0.7) | 30.4 (14.7) | 1,130 (810) |
| Monona (MO) | 43.07 | 89.35 | 1,324.0 | 15.8 | D | 8.1 (0.5) | 10.4 (19.6) | 5.2 (0.6) | 50.8 (56.1) | 90 (661) |
| Palmer (PA) | 46.20 | 89.50 | 256.9 | 3.9 | D | 7.1 (1.5) | 13.5 (14.7) | 9.3 (3.6) | 8.3 (8.6) | 506 (202) |
| Red Cedar (RC) | 42.98 | 88.98 | 145.3 | 1.5 | S | 7.2 (0.1) | 5.2 (5.9) | 11.8 (1.9) | 11.6 (12.5) | 1,196 (876) |
| Rock (RO) | 43.08 | 88.93 | 554.8 | 18.2 | D | 8.2 (0.5) | 4.5 (4.7) | 8.8 (1.8) | 8.9 (4.9) | 667 (82) |
| Salmo Pond (SA) | 43.12 | 89.69 | 1.1 | 4.5 | S | 7.6 (0.4) | 7.3 (9.6) | 2.0 (1.1) | 4.5 (1.5) | 645 (133) |
| Tomahawk (TO) | 45.82 | 89.67 | 1,494.6 | 25.4 | D | 6.6 (1.0) | 2.8 (2.6) | 3.3 (1.0) | 4.5 (1.0) | 291 (122) |
| Trout (TR) | 46.03 | 89.66 | 1,607.9 | 7.9 | D | 7.3 (1.8) | 4.2 (4.6) | 3.7 (2.0) | 4.5 (3.3) | 271 (83) |
| Trout Bog (TB) | 46.04 | 89.69 | 1.1 | 7.2 | S | 5.7 (1.0) | 29.4 (10.0) | 19.1 (18.1) | 12.4 (4.9) | 672 (16) |
| Twin Valley (TW) | 43.03 | 90.09 | 61.5 | 3.4 | D | 8.5 (0.7) | 11.9 (17.1) | 3.5 (0.3) | 9.2 (1.8) | 486 (312) |
| Verna (VE) | 45.96 | 89.72 | 31.2 | 18.3 | D | 7.2 (1.3) | 3.6 (2.2) | 7.0 (1.6) | 5.5 (1.1) | 540 (117) |
| White Sand (WS) | 46.09 | 89.59 | 497.4 | 21.5 | D | 6.3 (0.8) | 3.7 (2.1) | 3.2 (1.5) | 3.2 (1.3) | 279 (125) |
| Wildcat (WC) | 46.17 | 89.61 | 123.4 | 6.9 | D | 6.6 (0.5) | 7.6 (7.9) | 5.5 (0.2) | 6.1 (2.4) | 382 (138) |
Lat, latitude; Long, longitude; SA, surface area (hectares); z, maximum depth, Chl a, chlorophyll a; Numbers in parentheses are the ranges (maximum observed value − minimum observed value).
D, drainage; S, seepage.
DNA samples for BCC characterization were collected by submerging a length of plastic tubing (2.5-cm interior diameter) to the desired depth, plugging the end, and retrieving the sample. Field sampling was performed as described by Yannarell and Triplett (83) with the following modifications. Triplicate integrated water samples were collected from the surface of the lake down to the Secchi depth or to a depth of 5 m, depending on which was shallower. A 250- to 500-ml subsample of water from each replicate was vacuum filtered through a 0.2-μm-pore-size polyethersulfone membrane filter (Pall Supor-200; Gelman). Filters were place into cryovials, frozen immediately in liquid nitrogen, and transported back to the laboratory, where they were stored at −80°C. Water samples for DNA were not prefiltered to exclude organisms, and thus samples included DNA from heterotrophic bacterioplankton as well as cyanobacteria and small numbers of planktonic eukaryotes.
The Secchi depth, depth to the bottom of the lake, surface water temperature, and pH were recorded at each site. Up to 1 liter of water from each of the triplicate integrated samples was filtered through glass fiber filters (Whatman) to collect phytoplankton for determination of chlorophyll a concentration. Subsamples for analysis of DOC, color, specific absorbance of UV radiation A (SUVA) (79), dissolved nitrogen (DN), dissolved phosphorus (DP), and concentrations of nitrate plus nitrite (nitrite was oxidized to nitrate prior to detection [see below]) and ammonia were filtered from integrated water samples through 0.4-μm-pore-size polycarbonate membrane filters (Osmonics), placed immediately on ice, and transported back to the laboratory. Glass fiber filters and water samples for nitrate plus nitrite were kept frozen and in the dark until analyzed. Water samples for total nitrogen and phosphorus were acidified with 1 ml of Optima HCl and refrigerated. All other samples were refrigerated until analysis, which was performed no more than 60 days after collection.
Laboratory analyses. (i) ARISA.
BCC was assessed by automated ribosomal intergenic spacer analysis (ARISA) (7, 23). ARISA is a molecular technique that utilizes the length heterogeneity of the intergenic transcribed spacer (ITS) region of bacterial rRNA operons to construct bacterial community “fingerprint” profiles (Fig. 2) (23). Treating the elements of ARISA profiles as operational taxonomic units allows for “whole-community” ecological comparisons. It should be pointed out, however, that fingerprint-based assessments of BCC may overlook certain community members and may also misclassify community members by assigning ecologically identical organisms (e.g., members of the same species) to different operational taxonomic units or by assigning ecologically distinct organisms to the same operational taxonomic unit (7, 23). For the present study, ARISA profiles were assumed to be indicative of BCC, and differences in ARISA profiles were assumed to reflect variation in the composition of the respective bacterial communities.
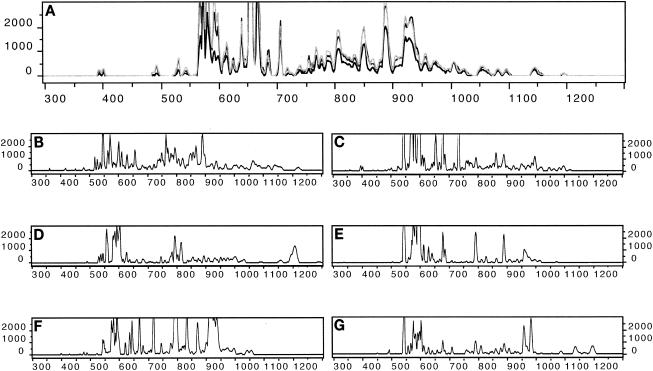
FIG. 2.
Examples of ARISA profiles. (A) Reproducibility of ARISA fingerprints, as exemplified by profiles from replicate filters. The profiles were generated from May samples of Koshkonong Lake (southern, drainage, hypereutrophic). Replicate 1, black; replicate 2, dark gray; replicate 3, light gray. (B) July profile from Koshkonong Lake. (C) October profile from Koshkonong Lake. (D) October profile from Firefly Lake (northern, seepage, oligotrophic). (E) October profile from Blueberry Lake (northern, seepage, mesotrophic). (F) October profile from Hook Lake (southern, seepage, humic). (G) October profile from White Sand Lake (northern, drainage, oligotrophic).
Bacterial DNA was extracted from filters with a FastPrep DNA purification kit (Bio 101, Inc.) by following the SPIN protocol. ARISA was performed as previously described (82) with the modification of Yannarell and Triplett (83). Visual alignment of ARISA profiles was performed with the help of Genotyper, version 2.1 (PE Applied Biosystems). The signal strength (i.e., peak heights and areas) of each ARISA peak was first normalized to account for run-to-run variations in signal detection. Normalized peak heights and areas were calculated by dividing each peak's height and area by a scale factor that related the total signal strength of the peak's profile (i.e., the sum of all peak areas in the profile) to the total signal strength of one of the replicate profiles that was designated (arbitrarily) as a standard. Only peaks sized between 390 and 1,250 bp were considered, and all peaks within 1.2 bp of higher peaks (i.e., shoulder peaks) were eliminated from profiles (83).
To investigate the relative impacts of smaller and larger peaks in the data analyses, four different ARISA data sets were created, each differing from the other in regard to the level of sensitivity applied to ARISA fragment analysis. These sensitivity levels were defined as follows: ARISA peaks were considered for analyses only if the normalized peak height exceeded 50, 100, 150, or 200 fluorescence units (FU). Following the application of these sensitivity levels, profiles from three replicate filters for each lake-date combination were merged into a single consensus profile by averaging the peak area (in FU) of each ARISA peak across the replicates. These consensus profiles were used as the basis for all data analyses.
(ii) Environmental data.
Analyses were conducted as outlined by the North Temperate Lakes Long-Term Ecological Research site (50), and more details about these protocols are available via the “online datasets” link at http://lter.limnology.wisc.edu/index.html. DOC was analyzed by high-temperature combustion on a Shimadzu TOC-5000 analyzer using potassium hydrogen phthalate standards. Color was determined with a Kontron 930 spectrophotometer, as was SUVA at 254 nm. DN and DP were analyzed on a segmented flow calorimeter following potassium persulfate-sodium hydroxide digestion. Ammonium and nitrate plus nitrite were analyzed on a segmented flow colorimeter with a copper-cadmium column to oxidize nitrite to nitrate for colorimetric detection. Chlorophyll a was extracted from glass fiber filters with Optima methanol following maceration and homogenization with a miniblender. Chlorophyll a concentration was determined with a Kontron 930 spectrophotometer based on the absorbance (of acidified extract) at 665 and 750 nm.
Numerical methods. (i) Data sets and data transformations: explanatory (environmental) variables.
The environmental data collected resulted in a data set with 11 quantitative environmental variables: water temperature, Secchi depth, pH, chlorophyll a, SUVA, color, DOC, dissolved phosphorus, dissolved nitrogen, ammonia, and nitrate plus nitrite. All of these variables except for water temperature, pH, and SUVA were log10 transformed to better conform to normality. In addition, all 11 variables were standardized by subtracting the mean and dividing by the standard deviation (SD; the so-called z-score standardization [43]). This had the effect of setting the mean of each variable equal to 0 and setting the SD (and hence variance) to 1 and making all quantitative variables dimensionless (43). All analyses utilized these z-scores.
In addition to the 11 quantitative variables, 3 qualitative variables were used in the analyses. These qualitative variables were coded as seven binary dummy variables (43). The study design yielded two sets of dummy variables: region (with two values, northern Wisconsin and southern Wisconsin) and month (with three values, May, July, and October). To assess the effect of lake LP (41), a third dummy variable was determined for each lake, and the lake order concept of Riera et al. (64) was applied to divide the lakes into two categories. Lakes with lake order ≤0 were classified as seepage lakes. This category included headwater lakes and lakes not connected to other lakes by permanent overland streams; lakes in this category are presumed to receive most of their water inputs through precipitation and groundwater flow. Lakes with positive lake order were classified as drainage lakes, and this category included all lakes possessing both inflowing and outflowing streams. Note that the drainage lakes in this study are not necessarily connected to each other. For example, Trout and Tomahawk are two northern Wisconsin drainage lakes, but they are located in different watersheds, the former draining into the Chippewa-Flambeau river system and the latter draining into the Wisconsin River (Fig. 1).
The effects of these three qualitative variables on ARISA profile richness were tested by three-way analysis of variance using the R-1.8.0 language and environment (R Foundation for Statistical Computing, 2003). Separate tests were performed for each ARISA sensitivity level, with Bonferroni correction for multiple testing applied.
(ii) Data sets and data transformations: response variables (ARISA).
PCR amplification is known to introduce bias in the ratio of amplified products in mixed-template reactions (60, 68, 69) and is not generally trusted to yield quantitative estimates of bacterial abundance in methods such as ARISA (23, 78). However, several studies have revealed that fingerprinting methods produce remarkably replicable results, even when profiles are produced from different replicate extractions or when they are based on different nucleic acids (Fig. 2) (23, 54, 62). Additionally, it is well known that different transformations of ecological data can lead to different conclusions, e.g., presence-absence of species versus species abundance (14, 15).
To assess the impacts of these different views of molecular data, three different transformations were applied to ARISA profiles to generate quantitative, semiquantitative, and presence-absence data sets. Quantitative ARISAs used the normalized peak areas (FU) of ARISA peaks as direct estimates of abundance. For semiquantitative ARISAs, the normalized peak areas (FU) of ARISA fragments were “relativized” through division by total (normalized) profile signal strength (FU), mimicking transformations for percent cover or percent dominance. Finally, presence-absence analyses were conducted by scoring ARISA peaks as 1 if they were present in a given profile and 0 if they were absent but observed elsewhere in the data set. These three transformations were applied to data from each of the four sensitivity levels previously mentioned. Thus, a total of 12 different ARISA data sets were used as the bases for analyses.
For each ARISA data set, the Bray-Curtis similarity (coefficient S17 = 1 − D14) of Lengendre and Legendre (43) was used to assess the degrees of similarity among ARISA profiles obtained from different lakes and times (hereafter referred to as sites) and to produce a site-by-site similarity matrix, Si (where i [1 ≤ i ≤ 12] refers to the sensitivity level or transformation of the particular ARISA data set). Note that for presence-or-absence-transformed data, the Bray-Curtis similarity is identical to the Sørensen's similarity (coefficient S8 of Lengendre and Legendre [43]). To determine the influence of ARISA sensitivity and data transformation on the structure of BCC data, a nonparametric form of the Mantel test (43) was implemented. The various matrices Si, which summarized the relationships in BCC observed between sites, were compared with the Spearman r rank correlation coefficient, and the results of these comparisons were summarized by a complete-linkage clustering. The production of Si, calculation of Spearman's r, and clustering were all performed with the software package PRIMER 5 for Windows, version 5.2.7 (PRIMER-E Ltd., 2001; routines SIMILARITY, TWO-STAGE, and CLUSTER).
(iii) CCA.
Canonical ordination can be regarded as an extension of linear regression analysis to the multivariate case (34, 43). To determine which environmental variables best explained patterns of similarity in ARISA profiles among sites, canonical correspondence analysis (CCA) (71, 73) was applied. CCA is an ordination technique that seeks the most prominent linear gradients in multivariate data sets, under the constraint that the gradients are linear combinations of a set of explanatory variables. Like multiple linear regression, CCA can use forward (or backward) selection to generate the most efficient model from a set of potential explanatory variables.
CCA was performed using the software package Canoco for Windows, version 4.51 (Biometris-Plant Research International; 1997 to 2003) according to the recommendations of the authors (72). The 11 quantitative and 7 dummy environment variables constituted the initial pool of explanatory variables, and the 12 different sensitivity levels or transformations of ARISA were used as response variables in separate runs of CCA. To determine the combination of explanatory variables that described the most influential gradients of ARISA profiles, the forward selection method outlined by ter Braak and Verdonschot (73) was used. Explanatory variables were added until the addition of further variables failed to contribute significant improvement to the model's explanatory power, as assessed by permutation test (499 permutations under the full model) with sequential Bonferroni adjustment applied to significance tests. Each CCA was conducted under the following additional conditions: biplot scaling with a focus on relationships among sites and down-weighting of rare ARISA fragments, which reduced the abundance of rare ARISA fragments (those occurring with a frequency less than 20% of the frequency of the most common ARISA fragment) in proportion to their frequency (72).
To further explore the patterns in BCC revealed by ARISA, partial CCA was employed. This type of analysis can discern patterns related to one set of variables while controlling for a different set of variables, called the covariables (43). For partial CCA, the analysis was first carried out for the covariables, and then the residual variation was subjected to a second CCA where the axes were constrained by the remaining variables (73). Partial CCA was done using Canoco for Windows, version 4.51, with the dummy variables for region and LP used as covariables.
RESULTS
Depending on the sensitivity level applied to ARISAs, the average richness of profiles from these lakes ranged from (means ± SD) 37.6 ± 15.4 to 55.5 ± 16.0 peaks per profile. Profile richness was affected by the ARISA sensitivity level increase (Fig. 3). Profile richness (Fig. 3) and pattern (Fig. 2) varied in a regular fashion between the two regions and among lakes occupying different LPs. Profiles from southern lakes were significantly richer than profiles from northern lakes, and drainage lakes were also significantly richer than seepage lakes (Table 2). There was a marginally significant tendency for the north-to-south richness drop to be more pronounced in drainage lakes than in seepage lakes (Table 2). The time of sample collection had no detectable influence on profile richness (Table 2), although the patterns of ARISA profiles for the lakes changed over time (Fig. 2).
FIG. 3.
Profile richness for northern versus southern and drainage versus seepage lakes. The summarized data include samples from all study months. Error bars show 1 SD. Bars are colored based on the sensitivity level used to determine the minimum peak height, in FU, of ARISA peaks included in the analysis as follows: 50 FU, white; 100 FU, light gray; 150 FU, dark gray; 200 FU, black. Asterisks indicate the significance level of comparisons (Table 2): **, significant at α = 0.01; ***, significant at α = 0.001.
TABLE 2.
Analysis of variance for ARISA profile richnessa
| Sensitivity cutoff and variable | dfb | MSc | F statistic | P |
|---|---|---|---|---|
| 50 FU | ||||
| Mo | 2 | 115.0 | 0.78 | 0.460 |
| Region | 1 | 8,020.4 | 54.69 | <<0.001*** |
| LP | 1 | 1,380.3 | 9.41 | 0.003* |
| Mo-region | 2 | 325.0 | 1.11 | 0.335 |
| Mo-LP | 2 | 448.1 | 1.53 | 0.223 |
| Region-LP | 1 | 556.3 | 3.86 | 0.053 |
| Mo-region-LP | 1 | 52.2 | 0.36 | 0.553 |
| Residual | 79 | 146.6 | ||
| 100 FU | ||||
| Mo | 2 | 121.4 | 0.86 | 0.458 |
| Region | 1 | 9,945.2 | 70.28 | <<0.001*** |
| LP | 1 | 1,253.6 | 8.86 | 0.004* |
| Mo-region | 2 | 140.1 | 0.99 | 0.376 |
| Mo-LP | 2 | 235.1 | 1.66 | 0.196 |
| Region-LP | 1 | 680.3 | 4.81 | 0.031 |
| Mo-region-LP | 1 | 5.7 | 0.04 | 0.842 |
| Residual | 79 | 141.5 | ||
| 150 FU | ||||
| Mo | 2 | 139.6 | 1.10 | 0.339 |
| Region | 1 | 9,608.6 | 75.55 | <<><0.001*** |
| LP | 1 | 1,391.5 | 10.94 | 0.001** |
| Mo-region | 2 | 280.5 | 1.10 | 0.337 |
| Mo-LP | 2 | 484.7 | 1.91 | 0.155 |
| Region-LP | 1 | 524.3 | 4.12 | 0.046* |
| Mo-region-LP | 1 | 9.2 | 0.07 | 0.789 |
| Residual | 79 | 10,046.8 | ||
| 200 FU | ||||
| Mo | 2 | 112.2 | 0.95 | 0.391 |
| Region | 1 | 9,252.6 | 78.36 | <<0.001*** |
| LP | 1 | 1,244.0 | 10.53 | 0.002** |
| Mo-region | 2 | 145.9 | 1.24 | 0.296 |
| Mo-LP | 2 | 233.4 | 1.98 | 0.145 |
| Region-LP | 1 | 358.4 | 3.04 | 0.085 |
| Mo-region-LP | 1 | 9.8 | 0.08 | 0.774 |
| Residual | 79 | 118.1 |
Tests were carried out for each level of ARISA sensitivity, using only peaks exceeding the reported fluorescence cutoff. All significance levels have been Bonferroni corrected (α′ = α/4). Mo refers to the month of sample collection (May, July, or October); region refers to north versus south; LP refers to seepage versus drainage. Significance levels: *, α = 0.05; **, α = 0.01; ***, α = 0.001.
df, degrees of freedom.
MS, mean square.
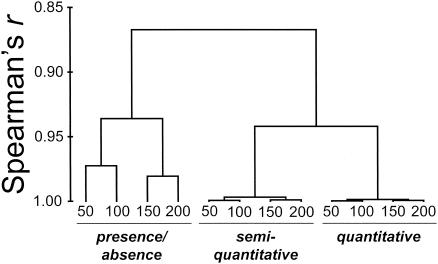
The combination of ARISA sensitivity level and data transformation had a pronounced effect on the relationships among sites, as revealed by ARISA (Fig. 4). The greatest impact was that of data transformation, and there was a tendency for data structures to be more similar to other data structures resulting from the same transformation than to those resulting from different transformations (Fig. 4). The effect of sensitivity level was noticeable only for presence-absence-transformed data sets, with both semiquantitative and quantitative analyses being insensitive to ARISA fragment sensitivity level (Fig. 4).
FIG. 4.
Complete linkage clustering scheme for ARISA data sets. The links in the tree are based on the results of nonparametric Mantel tests (Spearman's r). The branches represent similarity matrices generated from different transformations of ARISA profiles under different levels of sensitivity. Transformations are listed at the bottom, and the numbers at the branch ends indicate the minimum peak height, in FU, of ARISA peaks considered in generation of similarity matrices.
The influence of ARISA sensitivity level and data transformation was reflected in the outcomes of CCA (Table 3). Irrespective of the ARISA fragment sensitivity level, the semiquantitative and quantitative analyses each yielded four models that were functionally equivalent, while the presence-absence analyses produced different models for different ARISA sensitivity levels (Table 3). Despite the differences among data transformations, the following five variables were common to every model: region, LP, month (May), Secchi depth, and pH (Table 3). Overall, these models explained between 14.3 and 24.6% of the total variation in ARISA fragment distribution among sites.
TABLE 3.
Statistics for models from CCAa
| Transformation | Sensitivity (FU) | Model | Σλi | Trace | ARISA-env (%) |
|---|---|---|---|---|---|
| Presence or absence | 50 | Region, LP, May, temp, Secchi, pH | 0.347 | 2.419 | 54.4 |
| 100 | Region, LP, May, temp, Secchi, pH, DOC | 0.437 | 2.674 | 47.8 | |
| 150 | Region, LP, May, July, Oct, Secchi, pH, DOC | 0.490 | 2.979 | 47.6 | |
| 200 | Region, LP, May, July, Oct, Secchi, pH, DOC, TP | 0.586 | 3.181 | 45.85 | |
| Semiquantitative | 50 | Region, LP, May, temp, Secchi, pH | 0.799 | 4.419 | 58.6 |
| 100 | Region, LP, May, temp, Secchi, pH | 0.816 | 4.527 | 58.6 | |
| 150 | Region, LP, May, temp, Secchi, pH | 0.837 | 4.675 | 58.6 | |
| 200 | Region, LP, May, temp, Secchi, pH | 0.860 | 4.799 | 58.6 | |
| Partial | May, temp, Secchi, pH | 0.393 | 4.799 | 63.1 | |
| Quantitative | 50 | Region, LP, May, temp, Secchi, pH, DOC, DN, DP | 0.994 | 4.031 | 42.8 |
| 100 | Region, LP, May, temp, Secchi, pH, DOC, DN, DP | 1.003 | 4.077 | 42.8 | |
| 150 | Region, LP, May, temp, Secchi, pH, DOC, DN, DP | 1.020 | 4.166 | 42.8 | |
| 200 | Region, LP, May, temp, Secchi, pH, DOC, DN, DP | 1.039 | 4.238 | 42.7 | |
| Partial | May, temp, Secchi, pH, DOC, DN, DP | 0.629 | 4.238 | 41.1 |
Transformation refers to whether peak data from ARISA profiles were used quantitatively, semiquantitatively, or as presence or absence only. Only peaks exceeding the reported sensitivity fluorescence cutoffs were analyzed. Model, environmental variables included in the analysis following forward selection with sequential Bonferroni adjustments applied for significance. Underlined terms were determined to be significant in all 12 models. Partial CCA models used region and LP as covariables. Σλi is the sum of all canonical eigenvalues; trace is the sum of all eigenvalues (Σλi/trace = proportion of variance of ARISA peaks explained by the model). ARISA-env refers to the fraction of the ARISA fragment-environment relationship explained by the first two CCA axes (see Fig. 5 and 6).
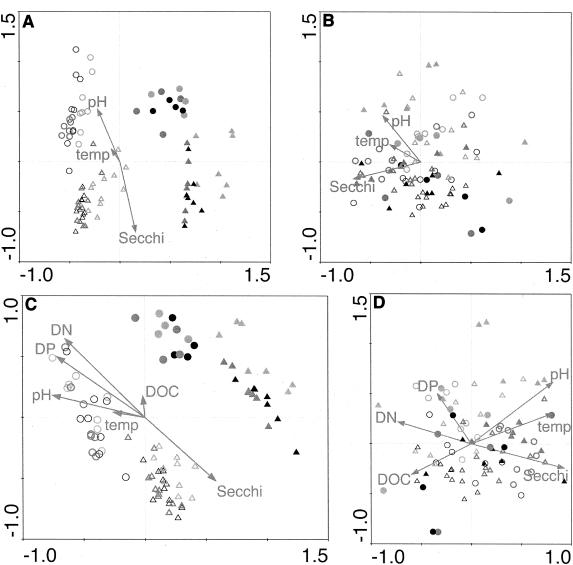
In general, the ARISA fragment-environment relationships were well characterized by the first two axes of CCA plots (Table 3), and these plots tended to be remarkably similar regardless of ARISA sensitivity level and data transformation (Fig. 5 and 6). Profiles from drainage lakes always plotted separately from seepage lake profiles, and northern and southern profiles were also easily distinguished (Fig. 5 and 6). Within each regional and LP grouping of points, the samples collected in May tended to stand out in relation to the other points. The effect of collection month was especially pronounced for the presence-absence analyses (Fig. 5), while only the northern lake May samples tended to stand out in the semiquantitative and quantitative plots (Fig. 6). The removal of the regional and LP effects through partial CCA did not change the set of explanatory variables comprising the remainder of the model (Table 3), but the relationships among these variables were more complicated and tended to describe the variation among ARISA profiles less well (Fig. 6B and D).
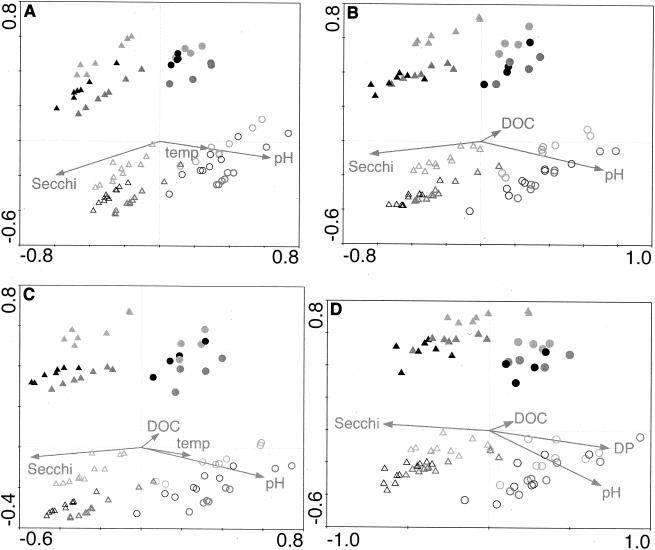
FIG. 5.
CCA biplots for presence-absence-transformed lake ARISA data. Points represent ARISA profiles, and the symbols indicate the values of qualitative variables from the models (Table 3): region (northern lakes, triangles; southern lakes, circles); LP (drainage lakes, open symbols; seepage lakes, filled symbols); month (May, light gray; July, dark gray; October, black). Arrows for quantitative variables show the direction of increase of each variable, and the length of the arrow indicates the degree of correlation with the ordination axes. Secchi, Secchi depth; temp, water temperature. CCA defined ARISA profiles with sensitivity levels of 50 (A), 100 (B), 150 (C), and 200 FU (D) to define the minimum size of analyzed ARISA peaks.
FIG. 6.
CCA biplots for semiquantitative and quantitative lake ARISA data. Points represent ARISA profiles, and symbols indicate the values of qualitative variables from the models (Table 3): region (northern lakes, triangles; southern lakes, circles); LP (drainage lakes, open symbols; seepage lakes, filled symbols); and month (May, light gray; July, dark gray; October, black). Arrows for quantitative variables show the direction of increase of each variable, and the length of the arrow indicates the degree of correlation with the ordination axes. Secchi, Secchi depth; temp, water temperature. A 150-FU sensitivity level was used to define the minimum size of ARISA fragments included in the analyses. (A) CCA for semiquantitative ARISA profile analysis. (B) Partial CCA for semiquantitative analysis. Here the effects of region and LP have been removed by including them as covariables in the model. (C) CCA for quantitative ARISA profile analysis. (D) Partial CCA for quantitative analysis (covariables are region and LP).
DISCUSSION
Use of molecular data sets in microbial ecology.
A researcher's ability to make inferences and evaluate patterns is limited by the quality of the available data. The application of PCR-based techniques to microbial ecology has opened up a promising avenue of inquiry (33, 58), but there is some uncertainty concerning what, exactly, the data from these studies reveal about the invisible microbial world. This uncertainty belongs to two broad categories: (i) the ecological relevance of operational taxonomic units (OTUs) and (ii) the quantitative-versus-binary (i.e., presence or absence) use of PCR-generated data sets.
ARISA provides no specific phylogenetic information about bacterial communities, so it is not possible to definitively address the ecological relevance of changes in OTUs described by ARISA profiles. The answer to this question will ultimately depend on the ability of ARISA-based OTUs to distinguish different species and also on whether bacterial species (as opposed to strains or polyphyletic functional groups) are the units responding to ecological change. Efforts are ongoing by the authors and their colleagues to link specific ARISA peaks from profiles to sequences of 16S rRNA genes from environmental clones, and these data should be invaluable in determining which bacterial species respond to different sources of environmental variation. In the meantime, it is possible to roughly gauge the relationship of ARISA-based OTUs to separate bacterial species. In a survey of intergenic transcribed spacer sequences from GenBank, Fisher and Triplett (23) found that more than 90% of species within the same genus, and 80% of species from different genera, would contribute unique ARISA peaks to profiles and that fewer than 30% of surveyed taxa would generate multiple ARISA peaks from the same individual. Thus, differences in ARISA profiles are likely to provide reasonable approximations of ecologically relevant variation in bacterial communities.
Another concern in the use of genetic fingerprints relates to the quantitative use of post-PCR data to describe the natural abundances of the organisms generating these patterns. It has been well documented that the application of PCR to mixed-template reactions, such as those from whole-community DNA extracts, can bias the ratio of products with respect to their starting ratio (60, 68, 69) and to the ratio of the organisms in the original community (19). There are numerous other potential sources of bias in PCR that could prevent its use in quantitative studies (78). It appears that a conservative application of PCR-based data for molecular ecology would involve simply looking at the presence or absence of the elements comprising profiles. However, presence-absence data analyses also have problems. First of all, these analyses are completely insensitive to community patterns arising from differences in species evenness and dominance, and this may distort ecological relationships in the data set. By simulating different sources of experimental error, including error due to PCR bias and error due to presence-absence data transformations, Muylaert et al. (55) found that the errors introduced by presence-absence data transformations were the most likely to obscure relationships between species and environment. Additionally, similarity indices such as Bray-Curtis (S17) can be disproportionately influenced by rare species when presence-absence transformations are applied, as these transformations weight all species identically (14, 15). This can result in analyses that are dramatically influenced by organisms generating signals right around the detection limits of molecular techniques (28). This effect is demonstrated in the present study by the influence of ARISA sensitivity levels on the presence-absence analyses and the lack of the same for the semiquantitative and quantitative analyses (Fig. 4 and 5; Table 3). This phenomenon illustrates the point that presence-absence studies are not immune to the effects of PCR bias, because this bias almost certainly plays a role in determining whether or not certain elements exceed the detection threshold of the analysis in the first place.
Ultimately, microbial ecologists should strive to use methods that are legitimately quantitative (70) or should strive to determine precisely how much PCR bias affects the results of molecular ecology studies. If PCR bias is such that it can make rare species appear to be very abundant or vice versa, then it has the potential to greatly influence the results of ecological studies involving molecular data. If, however, it is only the ratio of templates (or species) to products that is affected, then a semiquantitative approach (e.g., one that scores high-signal [FU] elements as abundant and weights them accordingly in analyses) may be appropriate.
The three data transformations applied to the datasets of the present study influenced the resulting CCA models (Fig. 4; Table 3). This was not entirely unexpected. The different transformations emphasized different aspects of the profiles, and these differences may be instructive in regard to the nature of the ecological response. For example, while DP and DN were significant variables for quantitative analyses, they were generally not significant in the presence-absence and semiquantitative analyses (Fig. 4 and 5; Table 3). This could indicate that these two variables were more important influences on the absolute dominance of different organisms between profiles, because this information would have been lost in both the presence-absence and semiquantitative transformations. However, the outstanding feature of this approach is that the various models had a set of explanatory variables in common, and this may indicate that the strategy adopted here is a useful one for dealing with issues arising from PCR-based barriers to quantitative analysis as well as the difficulties associated with presence-absence data sets. By analyzing samples with a variety of data transformations microbial ecologists may be able to identify the common features of the data structure that are insensitive to biases introduced by the data analyses and they may also be able to use the differences to identify how different parts of the community respond to different sources of variation.
Sources of variation in BCC.
In spite of the differences in data and model structure produced by different sensitivity levels and transformations, there were some remarkably consistent features of the CCA solutions. Five variables (region, LP, month [May], Secchi depth, and pH) were common to all models (Table 3). In addition, CCA plots (Fig. 5 and 6A and C) revealed the same overall patterns. Sites from drainage lakes and those from seepage lakes were arrayed along two parallel lines, with sites from northern lakes at one end and seepage lakes at the other. The quantitative environmental variables combined to describe gradients, also running from north to south or vice versa, that paralleled the major trends within the groupings of drainage and seepage lakes. The robustness of this pattern with respect to the original sensitivity level or data transformation applied to ARISA strongly suggests that these patterns describe predominant structures within the pelagic bacterial communities of these study lakes.
(i) Geographic sources of variation.
The main patterns revealed in all CCA plots were related to the statewide regional distribution of lakes and to the relative position of the lakes in the local overland and groundwater flow system (Fig. 5 and 6A and C; Table 3). Thus, bacterial communities responded to ecological variation on at least two different geographical scales. At the larger, regional scale, bacterial communities from northern Wisconsin lakes tended to be different from those in southern Wisconsin. This result echoes several recent studies demonstrating the important influence of regionalization on BCC (47, 80, 83). In addition, ARISA profiles from southern Wisconsin had, on average, more peaks than those from northern Wisconsin (Fig. 3; Table 2), which suggests that southern Wisconsin bacterial communities were more species rich. This was also noted in a previous study of Wisconsin lakes (83). There are several factors that could account for these regionalized effects. Northern Wisconsin and southern Wisconsin differ in many important aspects, including geology (51, 67), climate and vegetation (17), land use and land cover (63, 65), and anthropogenic impacts on lakes (4, 5, 26), and it is reasonable that these factors could influence lake BCC through a variety of mechanisms. It is also possible that regional differences in BCC reflect the biogeography of bacteria or of organisms that interact with bacteria. There is some current controversy concerning whether microbes even show biogeographical patterns of distribution (3, 27, 31, 80). Because ARISA provides no phylogenetic information (7, 23), it is not possible for the present study to address this question directly. However, depending on the ARISA sensitivity level, from 83 to 94% of all ARISA fragments in the present study were detected in both northern and southern lakes (data not shown). Under the assumption that each unique ARISA fragment represented a different species, most of the bacteria in the present study were distributed statewide, and thus it is not likely that bacterial biogeography provides a good explanation for the observed regional effect.
At the landscape scale, the drainage-versus-seepage classification of the lake of origin represented another prominent source of geographic variation in BCC of sites (Fig. 5 and 6A and B; Table 3). Lake LP is related to a variety of variables not measured in the present study, including concentrations of silica and major ions, susceptibility to acidification through acid precipitation, lake size and shape, richness and abundance of several vertebrate and invertebrate taxa, and intensity of human exploitation (61, 64), and any of these factors could potentially impact bacterial communities. This study used the drainage-versus-seepage classification of lakes as a rough surrogate for lake LP. Limnologists have long recognized the distinction between drainage lakes and seepage lakes (35, 67), and, even though the lake order concept of Riera et al. (64) represents a substantial refinement of the landscape context for lakes, the largest differences in variables noted by Riera et al. tended to be between seepage lakes and drainage lakes (64). These differences appear to be reflected in the composition of bacterial communities in Wisconsin lakes.
Another potential explanation for the landscape-level effect is related to the hydrology of these systems. In a study of two Swedish lakes, Lindström and Bergström (46) found that bacterial communities in the lake with short hydraulic retention time were similar to communities in the inlet and outlet streams, while those in the lake with long hydraulic retention time were distinct from the stream communities. Thus, bacterial communities in drainage lakes can be influenced by organisms washing in from streams, particularly when the water retention time of the lake is low. Unfortunately, the water retention time is not known for the lakes in the present study, nor were stream communities characterized for comparison. However, it is reasonable to generalize that water residence time in these regions of Wisconsin is higher for seepage lakes than for drainage lakes (9). Thus BCC in drainage lakes may be more susceptible to the influences of bacteria that wash in from outside, while seepage lakes may have a longer time in which to develop indigenous communities. ARISA richness was higher in drainage lakes than in seepage lakes (Fig. 3; Table 2), which could indicate the presence of greater numbers of “foreign” bacteria in these lakes. The influence of transient “wash-in” communities and water residence time on BCC bears further investigation.
(ii) Environmental sources of variation.
Two quantitative environmental variables, pH and Secchi depth, were consistently found to be significant in the CCA models (Table 3). These two variables tended to be strongly associated with the axis of regional variation (Fig. 5 and 6A and B), reflecting the fact that southern Wisconsin lakes tended to have a higher pH and northern Wisconsin lakes tended to have greater water clarity (Table 1). However, partial CCA, from which the effects of region and LP had been removed, showed that, within each region of Wisconsin, Secchi depth and pH were significantly related to variation in BCC (Fig. 6C and D; Table 3).
Secchi depth is a rough measure of water clarity, which itself represents a combination of several different causes. Thus, the identification of Secchi depth as important in these models does not suggest an immediate mechanistic explanation for variation in BCC. Secchi depth may indicate that light levels or spectral composition had a direct influence on BCC in these lakes, as shown in several studies (76, 77). However, water clarity can also be influenced by particulate (e.g., organisms) and dissolved (e.g., DOC) matter in the water. Water color, chlorophyll a, DOC, and SUVA were also measured in this study, and none of these variables was significantly correlated with Secchi depth (data not shown). However, when CCA was performed with Secchi depth removed from the pool of available variables, it was consistently replaced in the models by DOC and/or SUVA (data not shown). Thus, it is likely that the effect of Secchi depth properly reflected a composite effect of all of these sources of variation, but it was particularly associated with variables relating to the quantity and quality of carbon in the water. Regardless of the specific mechanism(s) responsible, it is clear that bacterial communities in these lakes were responding to environmental variation expressed along a water clarity gradient.
Another important source of variation in this study was pH, which has been implicated by other workers in studies of aquatic bacterial communities (44, 47, 53) and communities of organisms likely to influence BCC (21, 24, 25, 38). pH may reflect the influence of geology on water chemistry and is itself an important control of the biogeochemical transformations which can take place in a given environment. pH can also mediate the availability of ions and trace metals, which can have both inhibitory and growth-enhancing effects. Thus, pH may affect BCC through direct biological mechanisms and may also reflect the indirect influences of other unmeasured factors.
Previous work in Wisconsin lakes suggested that BCC might be structured by two principal ecological forces, one related to lake primary productivity and one related to organic carbon (83). It is well known that phosphorus or nitrogen or both are often limiting to bacterial and phytoplankton growth (10, 11, 18, 20, 57), and it has been demonstrated that BCC can also be influenced by nutrient enrichment (22). Both primary production and DOC in lakes represent sources of energy available to bacteria, and chlorophyll a and DOC may also to some extent represent potential interactions with phytoplankton, which have been identified by other workers as important influences on bacterial growth, production, and BCC (2, 16, 30, 36, 45, 52, 75, 81). Therefore it is somewhat surprising that these variables were not consistently identified as important in the present study (Table 3). Quantitative analyses identified DN, DP, and DOC as important variables (Fig. 6B; Table 3), and this may indicate that these variables determine which bacteria dominate particular communities. Additionally, the effects of these variables may have been subsumed by other explanatory variables in these models. In particular, nitrogen and phosphorus concentrations, and to some extent primary producer biomass (as assessed by measuring chlorophyll a content), were higher in southern lakes (Table 1), and, as has been discussed, DOC appeared to explain some of the variance associated with Secchi depth. That partial CCA did not enhance the significance of these variables (Table 3) demonstrates that they did not represent significant sources of variation within the two study regions, but they may have been influential in determining variation in BCC between northern and southern lakes.
(iii) Temporal variation in BCC.
Many studies have demonstrated that lake bacterial communities show considerable variation in time and may exhibit seasonal patterns (8, 30, 59, 82). Temporal variation was also apparent in the ARISA profiles of the present study (Fig. 2A to C). However, the role of sample month, especially as revealed by CCA, was not straightforward. There was no consistent temporal pattern of ARISA fragment richness (Table 2). While May was always determined to be an important descriptor in models, the other 2 months were only included in models based on presence-or-absence-transformed data (Table 3). Thus, samples collected in May were consistently different from samples collected in other months regarding both the set of ARISA fragments detected (i.e., presence or absence) and the dominance of communities by particular ARISA fragments, and this distinction is readily apparent from CCA plots (Fig. 5 and 6). Samples collected in July and October may have been unique in a subset of the ARISA fragments detected in these samples, as evidenced by the presence-or-absence analyses that weight all species equally (Fig. 5; Table 3). Therefore, while different ARISA fragments were consistently present and absent in these lakes at different times, the dominance of profiles by ARISA fragments varied from lake to lake in a fashion that was not consistent in July and October. If the semiquantitative and quantitative analyses are to be trusted, this indicates that the dominant community members in these lakes were determined by lake-specific factors that were not seasonally coherent (48).
This conclusion, however, is not consistent with another interpretation of the data. Table 3 indicates that water temperature was a significant explanatory factor in all of the models that did not include the full complement of months. July sample dates were consistently warmer than other sample dates (data not shown), and thus water temperature may mask some of the variation explained by the July dummy variable. Thus, if May was significant and water temperature was a proxy for July, then these models suggest that October samples were also consistently different given the colinearity of these three variables. This interpretation suggests that ARISA profiles showed coherent temporal variation in both the composition (presence or absence) and dominance of bacterial communities. The present study was conducted on a very coarse temporal scale, given the evidence that lake BCC can radically change over the course of a few weeks (30, 36, 82). This week-to-week variation in lakes may have made it difficult to detect any temporally coherent patterns in BCC in these lakes. However, the inclusion of either water temperature or the full complement of months in CCA models suggests that such temporally coherent behavior may exist, and studies with better temporal resolution over a large set of lakes could help sort this out.
Coherent behavior of BCC seems unquestionably apparent in the case of May samples (Fig. 5 and 6; Table 3). The dummy variable May was not related to any other variable measured in the present study (data not shown), so a mechanism is not readily apparent here. Previous work in Wisconsin lakes has demonstrated that, within the same lake, spring BCC is remarkably stable (82), and this suggests that the springtime ecology of bacterioplankton is controlled by a tight set of constraints. These constraints may include lower water temperatures; the presence or absence of large populations of grazers (i.e., the clear-water phase); the activity of primary producers; and the concentrations of limiting nutrients as determined by wintertime recycling, springtime biological uptake, and/or subsidy by elevated spring runoff. Additionally, temperate lakes stratify in the spring, and mixing dynamics may also help to structure bacterial communities. Thus, the significant influence of May samples here may be due to the proximity of the clear-water phase, the onset of stratification, or a variety of other factors that conspire to place bacterial communities in different lakes under a similar set of ecological pressures (30, 82).
Concluding remarks.
As microbial ecologists increasingly utilize numerical approaches to explain patterns in community composition, it will become more and more important to understand how the biases inherent in a molecular view of the world can influence these analyses. The present work showed that data transformations can affect the outcomes of multivariate analyses, but, despite the differences in models, it may still be possible to discern patterns that are insensitive to transformation-induced error. These robust patterns likely reflect real ecological variation in the underlying microbial communities.
This study revealed that lake BCC displayed coherent geographical variation on both regional and landscape scales, and bacterial communities in different lakes may also display coherent temporal variation. This highlights the importance of spatial and temporal structure in the community ecology of pelagic lake bacteria. Significant variation along water clarity and pH gradients both between northern and southern lakes and within each region of Wisconsin was also detected. It is to be expected that the considerable amount of unexplained variance in ARISA profiles (Table 3) represents individualistic community responses to grazing pressure, antagonistic and consortial interactions, and other factors that were not measured here but that have been shown to be important in other studies. Nevertheless, the present study demonstrates that geographic and temporal coherence, coupled with environmental influences, can generate patterns in BCC detectable across a variety of lake types.
Acknowledgments
This work was funded by NSF grants MCB 9977903, MCB 0401987, DEB 9632853, and DEB 0217533, awarded to the Center for Limnology at University of Wisconsin-Madison. The work was also supported by the Florida Agricultural Experiment Station.
We are grateful to A. Kent, K. Novakofski, T. Kratz, J. Rusak, G. Lauster, K. McMahon, R. Newton, and J. Thoyre for their valuable assistance with data collection and with the preparation of the manuscript. Special thanks are also offered to J. Chipman for assistance with the spatial data set and for graciously producing the graphics for Fig. 1 and other high-quality maps.
Footnotes
This is Journal Series no. R-10604 of the Florida Agricultural Experiment Station.
REFERENCES
- 1.Allen, T. F. H., and T. B. Starr. 1982. Hierarchy: perspectives for ecological complexity. University of Chicago Press, Chicago, Ill.
- 2.Arrieta, J., and G. Herndl. 2002. Changes in bacterial β-glucosidase diversity during a coastal phytoplankton bloom. Limnol. Oceanogr. 47:594-599. [Google Scholar]
- 3.Bass-Becking, L. G. M. 1934. Geobiologie of Inleiding Tot de Milieukunde. W. P. Van Stokum & Zoon N. V., The Hague, The Netherlands.
- 4.Bennett, E. M. 2003. Soil phosphorus concentrations in Dane County, Wisconsin, USA: an evaluation of the urban-rural gradient paradigm. Environ. Manag. 32:476-487. [DOI] [PubMed] [Google Scholar]
- 5.Bennett, E. M., T. Reed-Andersen, J. N. Houser, J. R. Gabriel, and S. R. Carpenter. 1999. A phosphorus budget for the Lake Mendota watershed. Ecosystems 2:69-75. [Google Scholar]
- 6.Borcard, D., P. Legendre, and P. Drapeau. 1992. Partialling out the spatial component of ecological variation. Ecology 73:1045-1055. [Google Scholar]
- 7.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosshard, P. P., R. Stettler, and R. Bachofen. 2000. Seasonal and spatial community dynamics in the meromictic Lake Cadagno. Arch. Microbiol. 174:168-174. [DOI] [PubMed] [Google Scholar]
- 9.Cardille, J. A., M. T. Coe, and J. A. Vano. Impacts of climate variation and catchment area on water balance and lake hydrologic type in groundwater-dominated systems: a generic lake model. Earth Interact., in press.
- 10.Carlsson, P., and D. A. Caron. 2001. Seasonal variation of phosphorus limitation of bacterial growth in a small lake. Limnol. Oceanogr. 46:108-120. [Google Scholar]
- 11.Caron, D. A. 1994. Inorganic nutrients, bacteria, and the microbial loop. Microb. Ecol. 28:295-298. [DOI] [PubMed] [Google Scholar]
- 12.Casamayor, E. O., G. Muyzer, and C. Pedrós-Alió. 2001. Composition and temporal dynamics of planktonic archaeal assemblages from anaerobic sulfurous environments studied by 16S rDNA denaturing gradient gel electrophoresis and sequencing. Aquat. Microb. Ecol. 25:237-246. [Google Scholar]
- 13.Casamayor, E. O., H. Schafer, L. Baneras, C. Pedrós-Alió, and G. Muyzer. 2000. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 66:499-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke, K. R. 1993. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18:117-143. [Google Scholar]
- 15.Clarke, K. R., and R. H. Green. 1988. Statistical design and analysis for a ‘biological effects’ study. Mar. Ecol. Prog. Ser. 46:213-226. [Google Scholar]
- 16.Crump, B. C., G. W. Kling, M. Bahr, and J. E. Hobbie. 2003. Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Appl. Environ. Microbiol. 69:2253-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curtis, J. T. 1971. The vegetation of Wisconsin: an ordination of plant communities. University of Wisconsin Press, Madison.
- 18.Downing, J. A., C. W. Osenberg, and O. Sarnelle. 1999. Meta-analysis of marine nutrient-enrichment experiments: variation in the magnitude of nutrient limitation. Ecology 80:1157-1167. [Google Scholar]
- 19.Farrelly, V., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felip, M., M. L. Pace, and J. J. Cole. 1996. Regulation of planktonic bacterial growth rates: the effects of temperature and resources. Microb. Ecol. 31:15-28. [DOI] [PubMed] [Google Scholar]
- 21.Fischer, J. M., J. L. Klug, A. R. Ives, and T. F. Frost. 2001. Ecological history affects zooplankton community responses to acidification. Ecology 82:2984-3000. [Google Scholar]
- 22.Fisher, M. M., J. L. Klug, G. Lauster, M. Newton, and E. W. Triplett. 2000. Effects of resources and trophic interactions on freshwater bacterioplankton diversity. Microb. Ecol. 40:125-138. [DOI] [PubMed] [Google Scholar]
- 23.Fisher, M. M., and E. W. Triplett. 1999. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 65:4630-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frost, T. M., P. K. Montz, and T. K. Kratz. 1998. Zooplankton community responses during recovery from acidification in Little Rock Lake, Wisconsin. Restor. Ecol. 6:336-342. [Google Scholar]
- 25.Frost, T. M., P. K. Montz, T. K. Kratz, T. Badillo, P. L. Brezonik, M. J. Gonzalez, R. G. Rada, C. J. Watras, K. E. Webster, J. G. Wiener, C. E. Williamson, and D. P. Morris. 1999. Multiple stresses from a single agent: Diverse responses to the experimental acidification of Little Rock Lake, Wisconsin. Limnol. Oceanogr. 44:784-794. [Google Scholar]
- 26.Gergel, S. E., M. G. Turner, and T. K. Kratz. 1999. Dissolved organic carbon as an indicator of the scale of watershed influence on lakes and rivers. Ecol. Appl. 9:1377-1390. [Google Scholar]
- 27.Hedlund, B. P., and J. T. Staley. 2003. Microbial endemism and biogeography, p. 225-231. In A. T. Bull (ed.), Microbial diversity and bioprospecting. ASM Press, Washington, D.C.
- 28.Hewson, I., and J. A. Fuhrman. 2004. Richness and diversity of bacterioplankton species along an estuarine gradient in Moreton Bay, Australia. Appl. Environ. Microbiol. 70:3425-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiorns, W. D., B. A. Methé, S. A. Nierzwicki-Bauer, and J. P. Zehr. 1997. Bacterial diversity in Adirondack mountain lakes as revealed by 16S rRNA gene sequences. Appl. Environ. Microbiol. 63:2957-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Höfle, M. G., H. Haas, and K. Dominik. 1999. Seasonal dynamics of bacterioplankton community structure in a eutrophic lake as determined by 5S rRNA analysis. Appl. Environ. Microbiol. 65:3164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horner-Devine, M. C., K. M. Carney, and B. J. M. Bohannan. 2004. An ecological perspective on bacterial biodiversity. Proc. R. Soc. Lond. Ser. B 271:113-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horner-Devine, M. C., M. A. Leibold, V. H. Smith, and B. J. M. Bohannan. 2003. Bacterial diversity patterns along a gradient of primary productivity. Ecol. Lett. 6:613-622. [Google Scholar]
- 33.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phyolgenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jongman, R. H. G., C. J. F. ter Braak, and O. F. R. Van Tongeren (ed.). 1995. Data analysis in community and landscape ecology. Cambridge University Press, Cambridge, United Kingdom.
- 35.Juday, C., and V. W. Meloche. 1943. Physical and chemical evidence relating to the lake basin seal in certain areas of the Trout Lake region of Wisconsin. Trans. Wis. Acad. Sci. Arts Lett. 35:157-174. [Google Scholar]
- 36.Kent, A. D., S. E. Jones, A. C. Yannarell, G. H. Lauster, J. H. Graham, T. K. Kratz, and E. W. Triplett. Annual patterns in bacterioplankton community variability in humic lake. Microb. Ecol., in press. [DOI] [PubMed]
- 37.Kent, A. D., and E. W. Triplett. 2002. Microbial communities and their interactions in soil and rhizosphere ecosystems. Annu. Rev. Microbiol. 56:211-236. [DOI] [PubMed] [Google Scholar]
- 38.Klug, J. L., J. M. Fischer, A. R. Ives, and B. Dennis. 2000. Compensatory dynamics in planktonic community responses to pH perturbations. Ecology 81:387-398. [Google Scholar]
- 39.Konopka, A., T. Bercot, and C. Nakatsu. 1999. Bacterioplankton community diversity in a series of thermally stratified lakes. Microb. Ecol. 38:126-135. [DOI] [PubMed] [Google Scholar]
- 40.Kratz, T. K., J. J. Magnuson, P. Bayley, B. J. Benson, C. W. Berish, C. S. Bledsoe, E. R. Blood, C. J. Bowser, S. R. Carpenter, G. L. Cunningham, R. A. Dahlgren, T. M. Frost, J. C. Halfpenny, J. D. Hansen, D. Heisey, R. S. Inouye, D. W. Kaufman, A. McKee, and J. Yarie. 1995. Temporal and spatial variability as neglected ecosystem properties: lessons learned from 12 North American ecosystems, p. 359-383. In D. Rapport and P. Calow (ed.), Evaluating and monitoring the health of large-scale ecosystems, vol. 128. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 41.Kratz, T. K., K. E. Webster, C. J. Bowser, J. J. Magnuson, and B. J. Benson. 1997. The influence of landscape position on lakes in northern Wisconsin. Freshw. Biol. 37:209-217. [Google Scholar]
- 42.Legendre, P. 1993. Spatial autocorrelation: trouble or new paradigm? Ecology 74:1659-1673. [Google Scholar]
- 43.Legendre, P., and L. Legendre. 1998. Numerical ecology, 2nd English ed. Elsevier Science BV, Amsterdam, The Netherlands.
- 44.Lindström, E. S. 2000. Bacterioplankton community composition in five lakes differing in trophic status and humic content. Microb. Ecol. 40:104-113. [DOI] [PubMed] [Google Scholar]
- 45.Lindström, E. S. 2001. Investigating influential factors on bacterioplankton community composition: results from a field study of five mesotrophic lakes. Microb. Ecol. 42:598-605. [DOI] [PubMed] [Google Scholar]
- 46.Lindström, E. S., and A. K. Bergström. 2004. Influence of inlet bacteria on bacterioplankton assemblage composition in lakes of different hydraulic retention time. Limnol. Oceanogr. 49:125-136. [Google Scholar]
- 47.Lindström, E. S., and E. Leskinen. 2002. Do neighboring lakes share common taxa of bacterioplankton? Comparison of 16S rDNA fingerprints and sequences from three geographic regions. Microb. Ecol. 44:1-9. [DOI] [PubMed] [Google Scholar]
- 48.Magnuson, J. J., B. J. Benson, and T. K. Kratz. 1990. Temporal coherence in the limnology of a suite of lakes in Wisconsin, USA. Freshw. Biol. 23:145-159. [Google Scholar]
- 49.Magnuson, J. J., and T. K. Kratz. 2000. Lakes in the landscape: approaches to regional limnology. Verh. Int. Verein. Limnol. 27:74-87. [Google Scholar]
- 50.Magnuson, J. J., T. K. Kratz, T. F. Allen, D. E. Armstrong, B. J. Benson, C. J. Bowser, D. W. Bolgrien, S. R. Carpenter, T. F. Frost, S. T. Gower, T. M. Lillesand, J. A. Pike, and M. G. Turner. 1997. Regionalization of long-term ecological research (LTER) on north temperate lakes. Verh. Int. Verein Limnol. 26:522-528. [Google Scholar]
- 51.Martin, L. 1965. The physical geography of Wisconsin, 3rd ed. University of Wisconsin Press, Madison.
- 52.Mayer, J., M. T. Dokulil, M. Salbrechter, M. Berger, T. Posch, G. Pfister, A. K. T. Kirschner, B. Velimirov, A. Steitz, and T. Ulbricht. 1997. Seasonal successions and trophic relations between phytoplankton, zooplankton, ciliate and bacteria in a hypertrophic shallow lake in Vienna, Austria. Hydrobiologia 342:165-174. [Google Scholar]
- 53.Methé, B. A., and J. P. Zehr. 1999. Diversity of bacterial communities in Adirondack lakes: do species assemblages reflect lake water chemistry? Hydrobiologia 401:77-96. [Google Scholar]
- 54.Moeseneder, M. M., C. Winter, and G. J. Herndl. 2001. Horizontal and vertical complexity of attached and free-living bacteria of the eastern Mediterranean Sea, determined by 16S rDNA and 16s rRNA fingerprints. Limnol. Oceanogr. 46:95-107. [Google Scholar]
- 55.Muylaert, K., K. Van der Gucht, N. Vloemans, L. De Meester, M. Gillis, and W. Vyverman. 2002. Relationship between bacterial community composition and bottom-up versus top-down variables in four eutrophic shallow lakes. Appl. Environ. Microbiol. 68:4740-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O'Neill, R. V., D. L. DeAngelis, J. B. Waide, and T. F. H. Allen. 1986. A hierarchical concept of ecosystems. Princeton University Press, Princeton, N.J.
- 57.Pace, M. L., and J. J. Cole. 1996. Regulation of bacteria by resources and predation tested in whole-lake experiments. Limnol. Oceanogr. 41:1448-1460. [Google Scholar]
- 58.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 59.Pernthaler, J., F. O. Glöckner, S. Unterholzner, A. Alfreider, R. Psenner, and R. Amann. 1998. Seasonal community and population dynamics of pelagic bacteria and archaea in a high mountain lake. Appl. Environ. Microbiol. 64:4299-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quinlan, R., A. M. Paterson, R. I. Hall, P. J. Dillon, A. N. Wilkinson, B. F. Cumming, M. S. V. Douglas, and J. P. Smol. 2003. A landscape approach to examining spatial patterns of limnological variables and long-term environmental change in a southern Canadian lake district. Freshw. Biol. 48:1676-1697. [Google Scholar]
- 62.Riemann, L., and M. Middleboe. 2002. Stability and viral community compositions in Danish coastal waters as depicted by DNA fingerprinting techniques. Aquat. Microb. Ecol. 27:219-232. [Google Scholar]
- 63.Riera, J., P. R. Voss, S. R. Carpenter, T. K. Kratz, T. M. Lillesand, J. A. Schnaiberg, M. G. Turner, and M. W. Wegener. 2001. Nature, society and history in two contrasting landscapes in Wisconsin, USA: interactions between lakes and humans during the twentieth century. Land Use Policy 18:41-51. [Google Scholar]
- 64.Riera, J. L., J. J. Magnuson, T. K. Kratz, and K. E. Webster. 2000. A geomorphic template for the analysis of lake districts applied to the Northern Highland Lake District, Wisconsin, USA. Freshw. Biol. 43:301-318. [Google Scholar]
- 65.Schnaiberg, J., J. Riera, M. G. Turner, and P. R. Voss. 2002. Explaining human settlement patterns in a recreational lake district: Vilas County, Wisconsin, USA. Environ. Manag. 30:24-34. [DOI] [PubMed] [Google Scholar]
- 66.Sommaruga, R., and D. Conde. 1997. Seasonal variability of metabolically active bacterioplankton in the euphotic zone of a hypertrophic lake. Aquat. Microb. Ecol. 13:241-248. [Google Scholar]
- 67.Soranno, P. A., K. E. Webster, J. L. Riera, T. K. Kratz, J. S. Baron, P. A. Bukaveckas, G. W. Kling, D. S. White, N. Caine, R. C. Lathrop, and P. R. Leavitt. 1999. Spatial variation among lakes within landscapes: ecological organization along lake chains. Ecosystems 2:395-410. [Google Scholar]
- 68.Suzuki, M., M. S. Rappé, and S. J. Giovannoni. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5 ′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.ter Braak, C. J. F. 1986. Canonical correspondence analysis: a new eigenvector technique for multivariate direct gradient analysis. Ecology 67:1167-1179. [Google Scholar]
- 72.ter Braak, C. J. F., and P. Šmilauer. 2002. CANOCO reference manual and CanoDraw for Windows user's guide: software for canonical community ordination, version 4.5. Microcomputer Power, Ithaca, N.Y.
- 73.ter Braak, C. J. F., and P. F. M. Verdonschot. 1995. Canonical correspondence analysis and related multivariate methods in ecology. Aquat. Sci. 57:1015-1621. [Google Scholar]
- 74.Van der Gucht, K., K. Sabbe, L. De Meester, N. Vloemans, G. Zwart, M. Gillis, and W. Vyverman. 2001. Contrasting bacterioplankton community composition and seasonal dynamics in two neighbouring hypertrophic freshwater lakes. Environ. Microbiol. 3:680-690. [DOI] [PubMed] [Google Scholar]
- 75.van Hannen, E. J., W. Mooij, M. P. van Agterveld, H. J. Gons, and H. J. Laanbroek. 1999. Detritus-dependent development of the microbial community in an experimental system: qualitative analysis by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2478-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vila, X., C. A. Abella, J. B. Figueras, and J. P. Hurley. 1998. Vertical models of phototrophic bacterial distribution in the metalimnetic microbial communities of several freshwater North-American kettle lakes. FEMS Microbiol. Ecol. 25:287-299. [Google Scholar]
- 77.Vila, X., X. P. Cristina, C. A. Abella, and J. P. Hurley. 1999. Effects of gilvin on the composition and dynamics of metalimnetic communities of phototrophic bacteria in freshwater North-American lakes. J. Appl. Microbiol. 85(Suppl.):138S-150S. [DOI] [PubMed] [Google Scholar]
- 78.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 79.Weishaar, J. L., G. R. Aiken, B. A. Bergamaschi, M. S. Fram, R. Fujii, and K. Mopper. 2003. Evaluation of specific ultraviolet absorbance as an indicator of the chemical composition and reactivity of dissolved organic carbon. Environ. Sci. Technol. 37:4702-4708. [DOI] [PubMed] [Google Scholar]
- 80.Whitaker, R. J., D. W. Grogan, and J. W. Taylor. 2003. Geographic barriers isolate endemic populations of hyperthermophilic Archaea. Science 301:976-978. [DOI] [PubMed] [Google Scholar]
- 81.White, P. A., J. Kalff, J. B. Rasmussen, and J. M. Gasol. 1991. The effect of temperature and algal biomass on bacterial production and specific growth rate in freshwater and marine habitats. Microb. Ecol. 21:99-115. [DOI] [PubMed] [Google Scholar]
- 82.Yannarell, A. C., A. D. Kent, G. L. Lauster, T. K. Kratz, and E. W. Triplett. 2004. Temporal patterns in bacterial communities in three temperate lakes of different trophic status. Microb. Ecol. 46:391-405. [DOI] [PubMed] [Google Scholar]
- 83.Yannarell, A. C., and E. W. Triplett. 2004. Within- and between-lake variability in the composition of bacterioplankton communities: investigations using multiple spatial scales. Appl. Environ. Microbiol. 70:214-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zehr, J. P., and M. A. Voytek. 1999. Molecular ecology of aquatic communities: reflections and future directions. Hydrobiologia 401:1-8. [Google Scholar]