Abstract
A PCR-based approach was developed to detect ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) form I large-subunit genes (cbbL) as a functional marker of autotrophic bacteria that fix carbon dioxide via the Calvin-Benson-Bassham cycle. We constructed two different primer sets, targeting the green-like and red-like phylogenetic groups of cbbL genes. The diversity of these cbbL genes was analyzed by the use of three differently managed agricultural soils from a long-term field experiment. cbbL gene fragments were amplified from extracted soil DNAs, and PCR products were cloned and screened by restriction fragment length polymorphism analysis. Selected unique cbbL clones were sequenced and analyzed phylogenetically. The green-like cbbL sequences revealed a very low level of diversity, being closely related to the cbbL genes of Nitrobacter winogradskyi and Nitrobacter vulgaris. In contrast, the red-like cbbL gene libraries revealed a high level of diversity in the two fertilized soils and less diversity in unfertilized soil. The majority of environmental red-like cbbL genes were only distantly related to already known cbbL sequences and even formed separate clusters. In order to extend the database of available red-like cbbL sequences, we amplified cbbL sequences from bacterial type culture strains and from bacterial isolates obtained from the investigated soils. Bacterial isolates harboring the cbbL gene were analyzed phylogenetically on the basis of their 16S rRNA gene sequences. These analyses revealed that bacterial genera such as Bacillus, Streptomyces, and Arthrobacter harbor red-like cbbL genes which fall into the cbbL gene clusters retrieved from the investigated soils.
The Calvin-Benson-Bassham cycle is the major and most abundant pathway for CO2 fixation (30). In addition to its essential capacity as the mechanism of primary production in nearly all ecosystems, the Calvin cycle plays a major role in effecting the concentration of atmospheric CO2. The Calvin cycle exists in diverse organisms, from bacteria to algae to green plants. The most abundant protein on earth, ribulose-1,5-biphosphate carboxylase/oxygenase (RubisCO), catalyzes the first, rate-limiting step in the Calvin cycle (5). RubisCO is a bifunctional enzyme that controls the reduction of CO2 and the oxygenolysis of ribulose-1,5-bisphophate. Since RubisCO is responsible for the overwhelming amount of carbon fixation by plants, nearly all primary production is linked to the function of this enzyme. RubisCO is a well-studied enzyme because of its extensive agricultural and environmental significance (2, 14, 22).
RubisCO exists in multiple natural forms which differ in structure, catalytic property, and O2 sensitivity (33). Form I RubisCO is a hexadecamer composed of eight large and eight small subunits (L8S8) and occurs in photo- and chemoautotrophic organisms. The form II protein consists only of large subunits (Ln), with 25 to 30% amino acid sequence identity to form I (33), and is found in photo- and chemoautotrophs. It is assumed that the common ancestor of RubisCO was similar to the form II enzyme because it operates successfully under low-O2 and high-CO2 concentrations, which resemble the conditions that existed in the early earth's atmosphere (12, 33). The form I RubisCO protein evolved as the atmospheric CO2 concentration decreased and the O2 concentration increased (18, 27). Recently, it was discovered that some members of Archaea possess a form III RubisCO (3, 16). Forms I, II, and III of RubisCO contain catalytic active amino acid residues that are necessary for carboxylation as well as oxygenation (8). Form IV RubisCO, which lacks several of the required amino acid residues for the catalytic activity of RubisCO (8), has been discovered in Bacillus subtilis (17), Chlorobium tepidum (8), and Archaeoglobus fulgidus (16). Form IV is designated a RubisCO-like protein, as its sequence is most closely related to RubisCO but it is not involved in the Calvin cycle.
The large subunit of form I RubisCO is encoded by the cbbL gene (18). The cbbL gene is 1,400 bp long and thus is large enough for use in meaningful phylogenetic analyses, for which there are sufficient sequences of cbbL genes deposited in public databases. Phylogenetic studies based on these cbbL sequences revealed that form I RubisCO proteins can be subdivided into two major groups, the green-like and red-like groups (36). The green-like group contains cbbL sequences from plants, algae, and α-, β-, and γ-Proteobacteria as well as from Cyanobacteria. The red-like type occurs in nongreen algae and α- and β-Proteobacteria. Form I RubisCO can be considered an enzyme that is predominantly found in photosynthetic and aerobic chemolithoautotrophic organisms. Clearly, the cbbL phylogeny disagrees with the phylogeny based on rRNA gene sequences (4). Thus, for instance, Rhodobacter capsulatus contains a green-like cbbL gene, whereas Rhodobacter sphaeroides harbors a red-like enzyme. Additionally, both organisms have a closely related form II RubisCO (4). Studies with Rhodobacter azotoformans demonstrated for the first time that a single bacterial cell can contain green-like as well as red-like cbbL genes (34).
Microorganisms play a key role in our understanding of regional and global carbon ecology, as they are involved in almost all processes of the carbon cycle due to their abundance and high metabolic diversity. Soils are significant compartments of the terrestrial carbon cycle and act as a source or sink for different carbon compounds, such as CO2 or methane. However, the diversity of CO2 fixation in soil bacteria has not yet been studied in detail. We focused on the apparently most important functional gene (cbbL) involved in CO2 fixation and developed distinct cbbL-specific primer sets to detect bacterial green-like and red-like types of the large-subunit gene of form I RubisCO. The aim of this study was to determine the diversity and composition of these cbbL types in soil bacteria isolated from differently managed agricultural soils from a long term, so-called eternal rye experiment in Halle/Saale, Germany. For a period of more than 125 years, these soils have been planted with rye and have not received fertilizer, mineral fertilizer (N, P, or K), or farmyard manure.
MATERIALS AND METHODS
Bacterial strains.
All bacterial strains used for this study are listed in Table 1. They were cultured as recommended by the Deutsche Sammlung von Mikroorganismen und Zellkulturen type culture collection (Braunschweig, Germany). These bacterial cultures were used to evaluate the specificities of the newly designed cbbL primers and to check for the presence of cbbL genes.
TABLE 1.
PCR amplification of genomic DNAs from reference organisms by the use of selected cbbL primer pairs
| cbbL type | Species | Straina | PCR resultb | Presence of database sequencec |
|---|---|---|---|---|
| Red-like | Azospirillum brasilense Sp 7 | ATCC 29145 | − | − |
| Azospirillum lipoferum | GSF 19 | + | − | |
| Azospirillum doebereinerae | GSF 21 | − | − | |
| Herbaspirillum seropedicae | DSM 6445 | − | − | |
| Ochrobactrum anthropi | DSM 6882 | + | − | |
| Ochrobactrum tritici | DSM 13340 | − | − | |
| Ralstonia eutropha | DSM 531 | + | + | |
| Ralstonia picketti | DSM 6297 | − | − | |
| Rhizobium leguminosarum bv. trifolii | ATCC 53912 | + | − | |
| Sinorhizobium fredii | ATCC 35423 | + | − | |
| Sinorhizobium meliloti | DSM 30135 | + | + | |
| Sinorhizobium terangae | DSM 11282 | + | − | |
| Sinorhizobium xinjiangense | DSM 5852 | − | − | |
| Xanthobacter agilis | DSM 3770 | + | − | |
| Xanthobacter autotrophicus | DSM 432 | + | − | |
| Xanthomonas campestris | DSM 1350 | − | − | |
| Green-like | Nitrobacter vulgaris | DSM 10236 | + | + |
| Nitrobacter winogradskyi | DSM 10237 | + | + |
ATCC, American Type Culture Collection; DSM, Deutsche Sammlung von Mikroorganismen; GSF, National Research Center for Environment and Health.
+, PCR product of expected size; −, no amplification.
Data are from the literature.
Soil samples.
Topsoil samples (from a depth of 0 to 10 cm) were taken in autumn 2000 from an agricultural long-term field experiment in Halle (Saale), Germany (eternal rye cultivation). The soil is a typical haplic phaeozem, and the matrix consists of 8% clay, 23% silt, and 69% sand (23). The experiment started in 1878 and exploits the sustainable applicability of different fertilization treatments on the cultivation of rye. For this study, samples from the following three treatment groups were analyzed: (i) HKO, a plot that remained unfertilized; (ii) HSM, a plot that received farmyard manure (12 tons ha−1 year−1); and (iii) HNPK, a plot that received mineral fertilizer (60 kg of N, 24 kg of P, and 75 kg of K ha−1 year−1). After sampling, soil samples were passed through a 2-mm-pore-size sieve to remove plant material. DNA extraction was performed immediately, or samples were frozen at −20°C after sample collection.
Isolation of bacterial strains.
In order to recover soil bacteria with hitherto unknown red-like cbbL genes, we isolated bacteria from the HNPK soil sample. One gram of soil was mixed with 9 ml of an extraction solution (0.1 g of NaCl liter−1, 0.02 g of CaCl2 · 2H2O liter−1, 0.2 g of MgSO4 · 7H2O liter−1, 5 g of Tween 80 liter−1) and homogenized for 5 min in an oscillating mixer (Retsch, Haan, Germany). The soil suspension was serially diluted to a factor of 10−6. Aliquots (100 μl) were spread on Rhizobium medium (21) and incubated for 2 days at 22°C. Sixty-four colonies were picked randomly, streaked on Rhizobium medium, and incubated for 2 days at 30°C. For DNA extraction, colonies from bacterial isolates were cultured in 3 ml of liquid Rhizobium medium overnight at 30°C. The cells were centrifuged and used for DNA extraction as described below.
Primer design.
All cbbL nucleotide sequences which were available from the National Center for Biotechnology Information sequence database were used to establish a cbbL database by use of the ARB software package (20; http://www.arb-home.de). The sequences were translated into amino acids, and the deduced amino acid sequences were aligned with the GDE sequence editor implemented in the ARB software package. Amino acid alignments were performed manually, and nucleotide sequences were aligned accordingly. Based on these data, we designed two primer sets specific for selected cbbL sequences of the red-like and green-like groups. The primers cbbLR1F and cbbLR1R, which were used for amplification of the red-like RubisCO form I cbbL gene, were designed from sequence alignment data given for the cbbL genes of Ralstonia eutropha H16, the Ralstonia eutropha megaplasmid pHG1, and Sinorhizobium meliloti WSM419. The primers cbbLG1F and cbbLG1R, which were used for amplification of the green-like cbbL genes, were designed from multiple sequence alignment data for the cbbL genes of Nitrobacter vulgaris T3, Nitrobacter winogradskyi IFO14297, N. winogradskyi ATCC 14123, Hydrogenophaga pseudoflava DSM1083, Thiobacillus denitrificans ATCC 25259, and Nitrospira sp. strain TCH716. The primers that were designed and used for this study are listed in Table 2.
TABLE 2.
Selected primers used for amplification of cbbL genes
| Primera | Positionsb (nt) | Primer sequence (5′-3′)c |
|---|---|---|
| cbbLR1F | 634-651 | AAG GAY GAC GAG AAC ATC |
| cbbLR1R | 1435-1454 | TCG GTC GGS GTG TAG TTG AA |
| cbbLG1F | 397-416 | GGC AAC GTG TTC GGS TTC AA |
| cbbLG1R | 1413-1433 | TTG ATC TCT TTC CAC GTT TCC |
Primers were named so that cbbLR1 primers targeted genes with red-like cbbL sequences and cbbLG1 primers targeted genes with green-like cbbL sequences; forward and reverse primers are indicated with an “F” or “R” as the last letter.
Positions correspond to the red-like cbbL gene of Ralstonia eutropha H16 (U20584) and the green-like cbbL gene of N. vulgaris (L22885).
Y = C or T; S = G or C.
Extraction of chromosomal DNAs from soil.
Genomic DNAs from pure bacterial cultures and soil samples were extracted and purified by use of a FastDNA spin kit for soil (Qbiogene Inc., Carlsbad, Calif.) according to the manufacturer's protocol.
Amplification of cbbL genes.
Amplification of the RubisCO genes from extracted DNAs via PCR was performed with the newly designed primer pairs described above. Amplification from 100 ng of extracted DNA was performed in 50-μl reaction mixtures containing 50 pmol each of a forward and reverse primer, a 200 μM concentration of each deoxynucleoside triphosphate (Fermentas GmbH, St. Leon-Rot, Germany), 1.5 mM MgCl2, and 1 U of Taq polymerase (Fermentas GmbH) in the 1× reaction buffer provided with the enzyme. The cycle conditions for cbbL-specific PCRs were as follows: 4 min of initial denaturation at 95°C, followed by 32 cycles of 1 min of denaturation at 95°C, 1 min of annealing at 57°C for the red-like and 62°C for the green-like cbbL primers, and 1 min of elongation at 72°C. The reaction was completed by a final extension for 10 min at 72°C. Aliquots of the PCR products were analyzed in 1.5% (wt/vol) agarose gels (PeqLab Biotechnology GmbH, Erlangen, Germany) by horizontal gel electrophoresis. DNAs were visualized by UV excitation after staining with ethidium bromide (0.5 mg liter−1).
Cloning and screening of environmental clones.
PCR products of the expected sizes (1,100 bp for green-like and 800 bp for red-like genes) from soil samples as well as from bacterial cultures were eluted from agarose gels by use of a NucleoSpin extraction kit (Macherey & Nagel, Düren, Germany). Eluted PCR products were ligated into the vector pCR2.1-TOPO (Invitrogen, San Diego, Calif.) and transformed into competent Escherichia coli cells provided with a TA cloning kit (Invitrogen) according to the manufacturer's protocol. Plasmids from the cbbL libraries were isolated by use of a NucleoSpin plasmid kit (Macherey & Nagel). Clones containing putative cbbL genes were screened by EcoRI restriction endonuclease digestion. Each 10-μl digestion reaction mixture consisted of 2 μl of purified plasmid, 1 μl of buffer O+ (Fermentas GmbH), and 2 U of EcoRI (Fermentas GmbH) and was incubated at 37°C for 2 h. Clones which harbored a correctly sized red-like cbbL insert were screened by restriction fragment length polymorphism (RFLP). Ten microliters of each PCR product was hydrolyzed with 2 U of the restriction endonuclease BbvI (MBI Fermentas). Restriction fragments were analyzed in 3.5% (wt/vol) agarose gels (PeqLab) and visualized as described previously.
Amplification and cloning of 16S rRNA genes.
To obtain the corresponding 16S rRNA gene sequences of cbbL-positive bacterial isolates, we performed PCRs to amplify the 16S rRNA gene from each isolate by using the primer pair 616-Forward (5′-AGA-GTT-TGA-TYM-TGG-CTC-AG-3′) and 630-Reverse (5′-CAK-AAA-GGA-GGT-GAT-CC-3′) (13), resulting in a full-length PCR product of about 1,500 bp. The PCR products were eluted from agarose gels and cloned as described above. Plasmids were extracted, and the presence of inserts with the correct size was checked as described above.
Sequencing reactions.
Plasmids containing cbbL inserts from soil DNA or from pure bacterial cultures as well as 16S rRNA gene inserts from pure cultures were used directly for sequencing. Both strands were sequenced by use of the vector-specific primers M13 reverse and T7 promoter. The plasmids were sequenced in an ABI Prism 377 automated sequencer (Applied Biosystems, Weiterstadt, Germany) by use of a Big Dye Terminator sequencing kit (Applied Biosystems).
Phylogenetic analysis.
The newly obtained cbbL nucleotide sequences were added to the established cbbL database implemented in the ARB software package (20; http://www.arb-home.de). The sequences were translated into amino acids, and the deduced amino acid sequences were aligned with GDE 2.2 editor software. Nucleic acid sequences were aligned according to the amino acid alignments. Phylogenetic analyses based on amino acid and nucleotide sequences were performed by applying maximum likelihood, maximum parsimony, and neighbor-joining methods by use of the respective tools in the ARB software package. The 16S rRNA gene sequences obtained from the isolates were added to an existing database of about 20,000 small-subunit rRNA gene sequences by use of the fast alignment tool of the ARB software package. Alignments were checked visually. Phylogenetic analyses based on 16S rRNA gene sequences were performed by the methods described above.
Statistical analysis of red-like cbbL libraries.
To evaluate richness and evenness, we calculated diversity indices for the red-like cbbL libraries by using different patterns from the RFLP analysis as representations of different operational taxonomic units (OTUs) in a sample. The diversity indices included (i) species richness (S), or the total number of OTUs; (ii) library coverage (C), or the portion of a clone library of infinite size that was sampled (6); (iii) the Shannon-Weaver diversity index, calculated by use of the equation H = −∑(pi)(log2 pi), where p is the proportion of an individual OTU relative to the total number of all RFLP patterns (29); (iv) the Simpson's index, calculated by use of the equation D = 1 − ∑(pi)2 (32); and (v) evenness, calculated from the Shannon-Weaver diversity function by use of the equation E = H/Hmax, where Hmax = log2 S. The diversity of the clones was analyzed by rarefaction analysis (31). Rarefaction curves were produced by use of the analytical approximation algorithm described by Hurlbert (11), and 95% confidence intervals were estimated as described by Heck et al. (9). Calculations were performed with the Analytic Rarefaction freeware program (http://www.uga.edu/∼strata/software/Software.html).
Nucleotide sequence accession numbers.
The sequences determined in this study are available at GenBank under accession no. AY572110 to AY572155 (red-like cbbL sequences from environmental clones), AY572169 to AY572192 (green-like cbbL sequences from environmental clones), AY572156 to AY572168 (cbbL sequences from isolates), AY572464 to AY572473 (cbbL sequences from reference strains), and AY572474 to AY572486 (16S rRNA gene sequences from isolates).
RESULTS
Primer design.
The high degree of variability of cbbL sequences made it impossible for us to design a universal PCR primer set to target all cbbL genes that were available in public databases. Sequence similarities calculated from distance matrices of all pairwise comparisons of cbbL nucleotide sequences ranged from 22 to 100%. The sequence similarities within the two cbbL clusters, the red-like group (57.8 to 100%) and the green-like group (60.7 to 99.2%), were significantly higher. Thus, we were able to construct primer sets that were specific for selected cbbL sequences of the two distantly related red-like and green-like cbbL clusters (Table 2). The red-like primers were derived from cbbL sequences of Ralstonia eutropha H16, Ralstonia eutropha megaplasmid pHG1, and S. meliloti WSM419, and the green-like primers were derived from cbbL sequences of N. vulgaris T3, N. winogradskyi strain IFO14297, N. winogradskyi strain ATCC 14123, H. pseudoflava DSM1083, T. denitrificans ATCC 25259, and Nitrospira sp. strain TCH716. Comparisons of the chosen primer sequences to all sequences available in public sequence databases indicated significant sequence similarities to cbbL genes only.
Amplification of cbbL genes from pure cultures and environmental samples.
To evaluate the efficiency of the newly designed cbbL-specific primers, we performed gene amplification with DNAs extracted from Ralstonia eutropha and S. meliloti as representatives of the red-like group. Amplification with the primer combination cbbLR1F and cbbLR1R yielded the expected 800-bp size and generated specific products which were visible as a single band on stained agarose gels. We also successfully amplified the 800-bp cbbL gene fragment from DNAs extracted from all three different soil samples.
The efficiency of the green-like gene-specific cbbL primers cbbLG1F and cbbLG1R was determined by the use of reference DNAs extracted from N. vulgaris and N. winogradskyi. A distinct gene fragment of 1,100 bp was obtained. In contrast to amplification with the red-like gene-specific primers, positive amplification of RubisCO genes was only possible with DNAs from the HKO and HNPK soil samples. No PCR products were observed with DNAs from the HSM soil sample, which had received fertilization with farmyard manure every year. In order to exclude the effect of inhibiting contaminants in the soil, we performed the green-like gene-specific PCR with DNAs from HSM soil spiked with DNA from N. vulgaris. Since the obtained PCR product showed the expected cbbL fragment size of about 1,100 bp, inhibition effects could be excluded. Various efforts to optimize the PCR conditions were not successful at yielding the expected fragment size of green-like cbbL sequences from HSM soil.
To extend the data set of available red-like cbbL genes, we amplified cbbL genes from different bacterial strains from culture collections (Table 1). cbbL sequences from Xanthobacter agilis DSM3770, Xanthobacter autotrophicus DSM432, Ochrobactrum anthropi DSM6882 (19), Sinorhizobium fredii ATCC35423, Sinorhizobium terangae DSM11282, Rhizobium leguminosarum bv. Trifolii ATCC 53912, and Azospirillum lipoferum GSF19 were determined. Moreover, cbbL gene fragments of bacterial isolates from the HNPK soil sample were amplified by use of the red-like cbbL primer set. A total of 13 of 64 investigated bacterial isolates revealed the correct PCR product size of 800 bp.
Green-like cbbL clone libraries.
PCR products with the correct size that were amplified from the HKO and HNPK soil samples by the use of green-like gene-specific cbbL primers were used to establish clone libraries. A total of 59 clones obtained from HKO soil were analyzed, and 12 clones were identified as positive for cbbL after restriction of the plasmids with EcoRI. For the library that was established from the HNPK soil sample, 155 clones were analyzed, but again, only 12 clones showed the correct fragment size after EcoRI restriction. Sequence analyses of selected cbbL clones with inserts that were shorter or longer than expected revealed that these clones were not related to cbbL. Due to the fact that we obtained an insufficient number of positive green-like cbbL clones, we did not perform RFLP analysis but sequenced all 24 clones.
Red-like cbbL clone libraries.
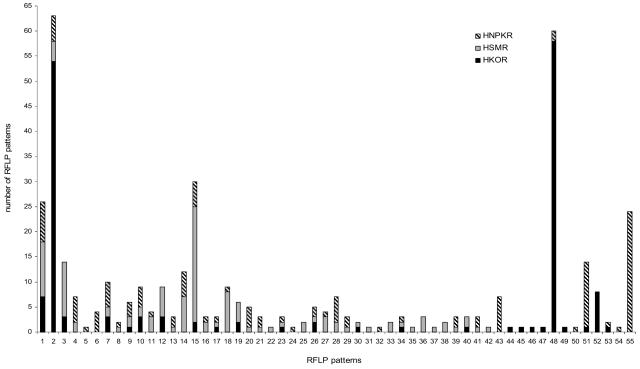
The PCR products that were amplified by use of the red-like gene-specific cbbL primers from the three differently managed soil samples, HKO, HSM, and HNPK, were used to construct clone libraries of red-like cbbL genes. A total of 405 of 624 clones from the different gene libraries showed the correct fragment size. The inserts were restricted with EcoRI and screened by RFLP analysis. The number and abundance of restriction patterns were used as measures of cbbL diversity in the different soil samples. Fifty-five different patterns (designated 1 to 55) were found in the libraries, and the pattern types were not distributed evenly among the different soil clone libraries. We identified 24 different restriction patterns for 158 screened clones from the HKO gene library. A larger number of different patterns was detected in the gene libraries derived from HSM and HNPK soils, although a smaller number of clones was screened by RFLP analysis. We obtained 38 different patterns for 121 HSM clones and 35 different patterns for 126 HNPK clones. Thus, HKO soil contained relatively limited cbbL diversity compared to the HSM and HNPK soils, which showed similar high levels of diversity. The distribution of the different RFLP patterns from the three cbbL libraries is shown in Fig. 1. RFLP types 2 (34%) and 48 (37%) dominated among the HKO clones, while RFLP type 15 accounted for the majority of the HSM clones (19%) and RFLP type 55 accounted for the majority of the HNPK clones (43%). Additionally, all three investigated clone libraries contained RFLP types which were less abundant and were represented by only a single clone. Several RFLP patterns were only present in one of the three established libraries. Thirty-eight clones with RFLP types that occurred more than once and eight representatives with unique RFLP patterns were selected from the three cbbL gene libraries and then sequenced.
FIG. 1.
Distribution of RFLP patterns of cbbL gene fragments from the red-like cbbL gene libraries of the HKO, HSM, and HNPK soil samples. cbbL PCR products were digested with the restriction endonuclease BbvI.
Diversity indices of red-like cbbL clone libraries.
In order to extrapolate similarities or differences among the red-like cbbL clone libraries, we performed statistical analyses. Biodiversity analyses were possible because the clone libraries were created under almost identical conditions. Table 3 shows the diversity indices that were used to compare the gene libraries. The diversity indices of the HSM and HNPK libraries were high and similar to each other, in contrast to the indices obtained for the HKO clone library. The Shannon-Weaver, Simpson's, and evenness values indicated that the diversity of cbbL sequences from HSM and HNPK soils differed drastically from that of sequences obtained from HKO soil. In addition, rarefaction analysis was performed on different RFLP patterns from each red-like cbbL gene library as representations of OTUs, with full coverage of a library expected to give a plateau-shaped curve (Fig. 2). Rarefaction analysis confirmed the results obtained with the diversity indices. Rarefaction curves of HSM and HNPK soils represented nonasymptotic curves. The high levels of diversity of the HSM and HNPK clone libraries were also reflected in the larger numbers of different RFLP types. An underestimation of species diversity from HSM and HNPK soils is expected, as the coverage of the libraries was estimated to be 86 or 89%, respectively (Table 3). The coverage of the HKO library was higher (92%) than that of the other two soil cbbL libraries.
TABLE 3.
Diversity indices obtained for red-like cbbL libraries from HKO, HSM, and HNPK soil samples
FIG. 2.
Rarefaction curves for the expected number of OTUs represented by different RFLP patterns from the red-like cbbL gene libraries.
Phylogenetic analysis based on cbbL sequence data. (i) Green-like cbbL sequences from soil.
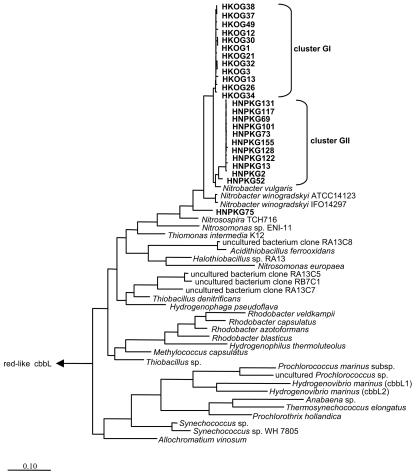
Sequences were designated beginning with the soil sample name, with an added “G” for sequences from the green-like library or “R” for sequences from the red-like library, followed by the clone number in the library. Comparisons with the National Center for Biotechnology Information database by BLAST searches revealed that all sequences were clearly related to known cbbL sequences. A total of 24 partial green-like cbbL gene sequences (ca. 1,100 bp) were identified, including 12 from HKO soil and 12 from HNPK soil. The sequence similarities from pairwise comparisons of all available bacterial green-like cbbL sequences, including the soil clones, ranged from 60.7 to 100%. The similarity values for all 24 cbbL clones from the Halle soils were 90.7 to 100% and formed two subclusters (Fig. 3). The GI cluster includes sequences from the HKO soil, and the GII cluster contains sequences from the HNPK soil only. The sequence HNPKG75 is the most distantly related green-like cbbL clone and exhibits a sequence similarity value of 91.7% for the closest related soil clone, HKOG13. A phylogenetic tree for green-like cbbL nucleotide sequences (Fig. 3) shows the highest degree of relatedness for the soil clones and the sequences from N. vulgaris and N. winogradskyi (97 to 99%).
FIG. 3.
Phylogenetic analysis of green-like cbbL genes. A consensus tree was constructed by distance (neighbor-joining), maximum parsimony, and maximum likelihood methods. The red-like cbbL sequence of Ralstonia eutropha was used as an outgroup for tree calculations. The bar indicates 10% estimated sequence divergence. Clones obtained from Halle soil samples were designated HKO or HNPK, followed by their number in the clone library. These sequences are shown in bold.
(ii) Red-like cbbL sequences from soil.
A total of 46 red-like cbbL soil clones from the three differently managed soil samples, HKO, HSM, and HNPK, were analyzed. In contrast to the green-like cbbL sequences from Halle soil, the red-like cbbL sequences were much more diverse, with similarities ranging from 57.8 to 100%. The soil clone sequences were distributed all over the red-like cbbL cluster (Fig. 4). An influence of soil management on the distribution of red-like cbbL sequences was not found. Soil clones that showed the same RFLP patterns also had the same cbbL sequences, which is represented by a nucleotide similarity value of 100%. Most red-like cbbL clone sequences obtained from soil were only distantly related to already published red-like cbbL sequences. Moreover, we observed the formation of three new monophyletic clusters, namely, RI, RII, and RIII. The RI and RIII clusters contained only environmental cbbL sequences from the investigated agricultural soils.
FIG. 4.
Phylogenetic analysis of red-like cbbL genes. A consensus tree was constructed by distance (neighbor-joining), maximum parsimony, and maximum likelihood methods. The green-like cbbL sequence of N. vulgaris was used as an outgroup for tree calculations. The bar represents 0.1 changes per nucleotide or amino acid. Clones obtained from Halle soil samples were designated HKO, HSM, or HNPK, followed by their number in the clone library. The cbbL sequences amplified from bacterial strains and the environmental clone sequences are shown in bold. The cbbL sequences of bacterial isolates are designated with an “R” followed by a number and a “c.” These sequences are shaded in gray.
(iii) Red-like cbbL sequences of bacterial strains.
In order to extend the data set of red-like cbbL gene sequences, we examined collection cultures for the presence of cbbL genes by using the same primer set as that used for soil samples. The cbbL sequences amplified from X. agilis and X. autotrophicus grouped with already known sequences from Xanthobacter sp. strain COX and Xanthobacter flavus and showed high similarity values for these sequences (>90%). The cbbL sequences of S. fredii and S. terangae clustered with the cbbL sequence of S. meliloti. The O. anthropi cbbL sequence completed this Sinorhizobium cluster, with a very high similarity value of 99.3% for S. fredii. Surprisingly, the cbbL sequences of Azospirillum lipoferum and R. leguminosarum bv. Trifolii showed low similarity values for other known red-like cbbL sequences. The most closely related sequence of Azospirillum lipoferum was that from Bradyrhizobium sp. strain CPP, with a similarity value of 86.3%. R. leguminosarum bv. Trifolii had the highest nucleotide similarity to the cbbL sequence of the environmental clone F38 (81.9%). Interestingly, the red-like cbbL clone HNPKR7 from the RII cluster showed 100% sequence similarity to the cbbL sequence of R. leguminosarum bv. Trifolii.
To further fill the gaps of red-like cbbL sequences, especially in the three new clusters, RI, RII, and RIII, we isolated bacteria from the HNPK soil sample by using Rhizobium medium. From a total of 64 isolates, 13 contained detectable cbbL sequences. The cbbL sequences of the isolates were designated with an “R,” followed by the isolate number and a “c” to designate the cbbL gene. The nucleotide similarities of the cbbL sequences retrieved from the isolated bacteria ranged from 73 to 99.6%. The sequences R36c and R47c grouped into the RI cluster, and the next related cbbL sequence was that of HKOR22, with a similarity value of 93%. R39c and R37c joined the RII cluster. The sequence R39c exhibited the highest similarity value (90.3%) for the environmental clone HSMR29. The most related cbbL sequence to R37c was the soil clone sequence HNPKR1 (89.3%). The sequences R43c, R40c, and R46c showed high nucleotide similarities (>99.3%) and grouped into the RIII cluster (Fig. 4). The cbbL sequence of the R45 isolate completed cluster RIII, with an average similarity value of 81.7% for the other sequences within this cluster. The other nucleotide similarity values within the RIII cluster ranged from 99.6 to 100%. The remaining cbbL sequences of the bacterial isolates were distributed singly all over the red-like cbbL tree. Two isolates were obtained whose cbbL sequences were 100% identical to those from soil clones: R46c was identical to HKOR3D and R33c was identical to HNPKR16.
(iv) Phylogenetic analysis of cbbL-positive bacterial isolates based on 16S rRNA gene sequences.
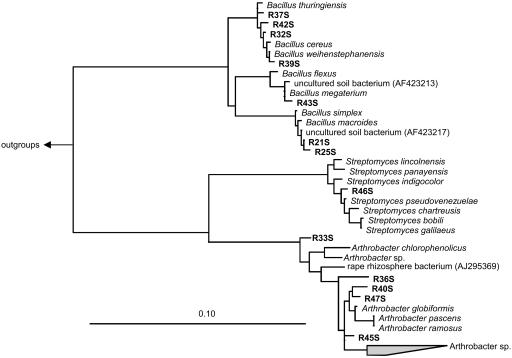
The phylogenetic positions of bacterial isolates harboring red-like cbbL genes were examined by use of the 16S rRNA gene as a phylogenetic marker. The 16S rRNA gene sequences were designated with an “R,” followed by the isolate number and “S.” The 16S rRNA gene sequence similarity values of all isolates ranged from 80.2 to 99.6%. The similarity values of the 16S rRNA gene sequences of the isolates for already known sequences were remarkably high, ranging from 98 to 99.7%. The R33S sequence had a 98% similarity to the sequences of Arthrobacter pascens and Arthrobacter ramosus. Phylogenetic analysis revealed (Fig. 5) that 7 of 13 sequences grouped with Bacillus species sequences, with very high similarity values (>99.5%). Furthermore, five 16S rRNA gene sequences (R33S, R36S, R40S, R45S, and R47S) were affiliated with a monophyletic cluster of Arthrobacter species. The nucleotide similarity values of the soil isolates and of cultured representatives of this group ranged from 98 to 99.5%, with the highest similarity between the sequences from isolate R47S and Arthrobacter globiformis. Interestingly, the 16S rRNA gene sequence of the R46 isolate was affiliated with the Streptomyces pseudovenezuelae sequence, with a nucleotide similarity of 99.5%.
FIG. 5.
Phylogenetic 16S rRNA gene sequence consensus tree reflecting the relationships of red-like cbbL-containing bacterial isolates. The sequences of the isolates are designated with an “R” followed by a number and an “S,” and they are shown in bold. An encompassing collection of organisms representing all major lineages of the Archaea and Bacteria were used as outgroups for tree calculations. The bar indicates 10% estimated sequence divergence.
DISCUSSION
This is the first reported investigation of the molecular diversity of RubisCO form I large-subunit cbbL genes in agricultural soils. Light-dependent CO2 fixation by plants, algae, and cyanobacteria is restricted in terrestrial environments to the top few millimeters of the soil profile. In deeper soil layers, light-independent bacterial CO2 fixation may play a hitherto underestimated important role. Only a few soil bacteria, including some heterotrophic, mostly root-associated bacteria and the chemolithoautotrophic nitrifying bacteria, have been known to harbor cbbL genes. We showed here that all green-like cbbL sequences obtained from agricultural soils by the use of our primer system were closely related to N. vulgaris and N. winogradskyi. In contrast, the red-like cbbL genes retrieved from three different soil samples were unexpectedly highly diverse. In fact, we found only one terrestrial red-like cbbL clone (HNPKR7) that was identical to a cbbL gene of a cultivated bacterium (R. leguminosarum bv. Trifolii). However, this does not necessarily imply that the environmental sequence HNPKR7 was retrieved from R. leguminosarum bv. Trifolii, since the cbbL phylogeny is incongruent with the phylogeny based on 16S rRNA genes (4). The incongruity of 16S rRNA and cbbL gene sequences became obvious when we compared phylogenetic affiliations based on 16S rRNA genes of the red-like cbbL RA cluster with red-like cbbL similarity values. For example, the RA cluster contains cbbL sequences from bacteria which belong to both α- and β-Proteobacteria but whose cbbL sequences are identical. Delwiche and Palmer (4) postulated that multiple processes are involved in the incongruity of the cbbL phylogeny, including horizontal gene transfer and gene duplication associated with differential gene loss. Thus, they suggested that at least four independent horizontal gene transfers are responsible for the green-like-red-like split (4). Another example is now given by the RIII cluster, which contains closely related cbbL sequences from Bacillus, Streptomyces, and Arthrobacter isolates. However, there are some surprising conservative exceptions, since the red-like cbbL genes of X. agilis and X. autotrophicus as well as those of S. fredii and S. terangae cluster together with already known cbbL sequences of phylogenetically closely related Xanthobacter and Sinorhizobium species, respectively. In these cases, there is congruency with the 16S rRNA gene phylogeny.
Unexpectedly, the isolation experiments revealed cbbL sequences in hitherto unknown autotrophic bacteria. The phylogenetic positions of red-like cbbL-containing isolates were determined by 16S rRNA gene sequence analyses. We identified bacteria belonging to the gram-positive genera Bacillus, Streptomyces, and Arthrobacter. A RubisCO-like protein, classified as form IV RuBisCO, was previously detected in B. subtilis (17). Recently, Ashida et al. (1) clearly demonstrated that this RubisCO-like protein is involved in the methionine pathway, which had been predicted already by studies of other groups (7, 26). However, based on gene sequence data, the form IV RubisCO-like gene ykrW of B. subtilis (17) is clearly different from the form I red-like cbbL sequences of the gram-positive isolates shown in this study.
Examinations of the evolutionary relationships of specific functional bacterial groups by use of the 16S rRNA gene and a corresponding functional marker gene have been performed before in several instances (10, 15, 28, 35). Nitrogen-fixing bacteria can be detected by use of the nifH gene encoding the nitrogenase reductase (35), and methanotrophic bacteria can be detected by use of the pmoA gene, which encodes the α subunit of the particulate methane monooxygenase (10). The phylogenies of nifH and pmoA are (with only a few exceptions) congruent with the phylogenies based on 16S rRNA genes (10, 35). Another example of a functional gene which is used as a functional marker is the amoA gene (28). The evolutionary relationships of ammonia-oxidizing bacteria with the 16S rRNA gene phylogeny are similar (28). In contrast, the detection of dsrAB, which is used as a functional gene of sulfate-reducing bacteria, shows a partial inconsistency with the corresponding 16S rRNA gene phylogeny (15).
The discovery of the RI, RII, and RIII clusters clearly demonstrates that soils harbor an unprecedented high level of diversity among red-like cbbL sequences. In addition, these sequences were only distantly related to known cbbL sequences from public databases (Fig. 4). However, it is not possible to extrapolate from this high genetic diversity of red-like RubisCO proteins in terrestrial environments to their physiological and ecological roles because the present data are only based on cbbL gene diversity. However, it was previously shown by the use of 14CO2 labeling experiments of soils that 14CO2 is fixed to a level of 3 to 5% of the respiration rate (25). This activity was stimulated by the addition of an organic substrate and was completely abolished after soil fumigation (25). In addition, preliminary results have demonstrated that cbbL mRNAs can be detected in the soils studied, especially after the addition of a substrate. This points to the possible importance of mixotrophic CO2 fixation in soils that are activated with carbon substrates. Interestingly, the diversity of red-like cbbL genes was increased in soils which were regularly fertilized (HNPK and HSM), although no obvious correlation could be found between the type of fertilization and the cbbL sequence types. We assume that fertilization favors the diversity of cbbL-bearing bacteria by directly or indirectly providing more available energy substrates for soil microbes.
Whether it is a general phenomenon that heterotrophic bacteria such as Arthrobacter, Bacillus, and Streptomyces use cbbL genes and RubisCO under certain ecophysiological conditions still needs to be studied in detail. Facultative chemoautotrophic or mixotrophic bacteria are capable of utilizing a wide range of substrates. Therefore, they may have an improved niche quality which would be advantageous in diverse soil habitats with rapidly changing conditions.
In contrast to the red-like sequences, the diversity of the green-like cbbL sequences seems rather limited in agricultural soils. Using our primer system, we detected cbbL sequences which were closely affiliated exclusively with the nitrite-oxidizing bacteria N. vulgaris and N. winogradskyi. The phylogenetic tree based on green-like cbbL sequences contains predominantly sequences of bacteria living in aquatic habitats, such as different Cyanobacteria and Rhodobacter species. These microorganisms are highly diverse and are distributed all over the green-like group (Fig. 3). Since we investigated soil samples, the low diversity of the identified green-like cbbL sequences, limited to the nitrifying bacterium Nitrobacter, can be explained. In the soil that was treated with 12 tons of manure ha−1 year−1, green-like cbbL genes were not found. This heavy manuring resulted in a 30% increase in organic carbon in the soil (24). Nitrifiers are possibly underrepresented in this soil, with an increased heterotrophic microbial community. The content of organic carbon in the soil was not influenced much by mineral fertilization, and in the unfertilized HKO soil, organic carbon declined to the range of 10% (24). The lower content of organic carbon in HKO soil may provide limiting conditions for the diversity of red-like cbbL gene-carrying bacteria and may favor chemolithoautotrophic bacteria.
In conclusion, this communication revealed a huge unprecedented red-like cbbL diversity in agricultural soils under three different long-term soil management practices. In a related study, a very similar diversity pattern of cbbL genes was found in an agricultural soil of a different type (loamy cambisol from Loess at Scheyern, upper Bavaria, Germany) (I. M. Pattis, unpublished data). This new knowledge about RubisCO form I genes in soil bacteria is certainly dependent on the primer systems used and should be further supplemented by studying more terrestrial habitats or other types of RubisCO genes, such as those corresponding to form II RubisCO. Future functional studies of the newly discovered red-like cbbL genes are necessary to understand the ecophysiological role of cbbL genes in soil bacteria such as Bacillus, Arthrobacter, and Streptomyces. Experiments to study the activities of RubisCO proteins in these bacteria are currently being performed. Moreover, more knowledge about the in situ activities of bacteria carrying cbbL genes is necessary. cbbL mRNAs can be retrieved from soils, and the diversity of these mRNAs is expected to reveal functionally active, cbbL-carrying bacteria. This will eventually contribute to a better understanding of the ecological role of the autotrophy of soil bacteria for CO2 dynamics in soils.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft in the framework of the project network “Soils as source and sink for CO2—mechanisms and regulation of organic matter stabilisation in soils” (SPP 1090).
We thank Angelika Schulz for excellent technical support.
REFERENCES
- 1.Ashida, H., Y. Saito, C. Kojima, K. Kobayashi, N. Ogasaware, and A. Yokota. 2003. A functional link between RuBisCO-like protein of Bacillus and photosynthetic RuBisCO. Science 302:286-290. [DOI] [PubMed] [Google Scholar]
- 2.Bainbridge, G., P. Madgwick, S. Parmar, R. Mitchell, M. Paul, J. Pitts, A. J. Keys, and M. A. J. Parry. 1995. Engineering rubisco to change its catalytic properties. J. Exp. Bot. 46:1269-1276. [Google Scholar]
- 3.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. Fitzgerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. Geoghagen, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 4.Delwiche, C. F., and J. D. Palmer. 1996. Rampant horizontal transfer and duplication of Rubisco genes in eubacteria and plastids. Mol. Biol. Evol. 13:873-882. [DOI] [PubMed] [Google Scholar]
- 5.Ellis, R. J. 1979. The most abundant protein in the world. Trends Biochem. Sci. 4:241-244. [Google Scholar]
- 6.Good, I. J. 1958. The population frequency of species and the estimation of the population parameters. Biometrics 40:237-246. [Google Scholar]
- 7.Grundy, F. J., and T. H. Henkin. 2002. Biosynthesis of serine, glycine, cysteine, and methionine, p. 245-254. In A. L. Sonenshein, R. M. Losick, and J. A. Hoch (ed.), Bacillus subtilis and its relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 8.Hanson, T. E., and F. R. Tabita. 2001. A ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO)-like protein from Chlorobium tepidum that is involved with sulfur metabolism and the response to oxidative stress. Proc. Natl. Acad. Sci. USA 98:4397-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heck, K. L., G. van Belle, and D. Simberloff. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56:1459-1461. [Google Scholar]
- 10.Horz, H. P., M. Tchawa Yimga, and W. Liesack. 2001. Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling. Appl. Environ. Microbiol. 67:4177-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurlbert, S. H. 1971. The nonconcept of species diversity: a critique and alternative parameters. Ecology 52:577-586. [DOI] [PubMed] [Google Scholar]
- 12.Jordan, D. B., and W. L. Ogren. 1983. Species variation in kinetic properties of ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch. Biochem. Biophys. 227:425-433. [DOI] [PubMed] [Google Scholar]
- 13.Juretschko, S., G. Timmermann, M. Schmid, K. H. Schleifer, A. Pommerening-Röser, H. P. Koops, and M. Wagner. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellogg, E. A., and N. D. Juliano. 1997. The structure and function of RuBisCO and their implications for systematic studies. Am. J. Bot. 84:413-428. [PubMed] [Google Scholar]
- 15.Klein, M., M. Friedrich, A. J. Roger, P. Hugenholtz, S. Fishbain, H. Abicht, L. L. Blackall, D. A. Stahl, and M. Wagner. 2001. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 183:6028-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, J. C. Venter, et al. 1997. The complete genome of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 17.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 18.Kusian, B., and B. Bowien. 1997. Organization and regulation of cbb CO2 assimilation genes in autotrophic bacteria. FEMS Microbiol. Rev. 21:135-155. [DOI] [PubMed] [Google Scholar]
- 19.Lebuhn, M., W. Achouak, M. Schloter, O. Berge, H. Meier, M. Barakat, A. Hartmann, and T. Heulin. 2000. Taxonomic characterization of Ochrobactrum sp. isolates from soil samples and wheat roots, and description of Ochrobactrum tritici sp. nov. and Ochrobactrum grignonense sp. nov. Int. J. Syst. Evol. Microbiol. 6:2207-2223. [DOI] [PubMed] [Google Scholar]
- 20.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüssmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malik, K. A., and H. G. Schlegel. 1981. Chemolithoautotrophic growth of bacteria able to grow under N2-fixing conditions. FEMS Microbiol. Lett. 11:63-67. [Google Scholar]
- 22.McFadden, B. A., J. Torres-Ruiz, H. Daniell, and G. Sarojini. 1986. Interaction, functional relations and evolution of large and small subunits in RuBisCO from prokaryota and eukaryota. Philos. Trans. R. Soc. Lond. Biol. Sci. 313:347-358. [DOI] [PubMed] [Google Scholar]
- 23.Merbach, W., L. Schmidt, and L. Wittenmeyer. 1999. Die Dauerdüngungsversuche in Halle (Saale). Teubner, Stuttgart, Germany.
- 24.Merker, J. 1956. Untersuchungen an den Ernten und Böden des Versuches “Ewiger Roggenbau” in Halle. Kühn-Arch. 70:154-215. [Google Scholar]
- 25.Miltner A., F. D. Kopinke, R. Kindler, D. Selesi, A. Hartmann, and M. Kästner. Non-phototrophic CO2 fixation by soil microorganisms. Plant Soil, in press.
- 26.Murphy, B. A., F. J. Grundy, and T. M. Henkin. 2002. Prediction of gene function in methylthioadenosine recycling from regulatory signals. J. Bacteriol. 184:2314-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paoli, G. C., N. S. Morgan, F. R. Tabita, and J. M. Shively. 1995. Expression of the cbbL, cbbS and cbbM genes and distinct organization of the cbb Calvin cycle structural genes of Rhodobacter capsulatus. Arch. Microbiol. 164:396-405. [PubMed] [Google Scholar]
- 28.Purkhold, U., A. Pommerening-Röser, S. Juretschko, M. C. Schmid, H. P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon, C. E., and W. Weaver. 1963. The mathematical theory of communication, p. 117. University of Illinois Press, Urbana, Ill.
- 30.Shively, J. M., W. Devore, L. Stratford, L. Porter, L. Medlin, and S. E. Stevens. 1986. Molecular evolution of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO). FEMS Microbiol. Lett. 37:251-257. [Google Scholar]
- 31.Simberloff, D. 1978. Use of rarefaction and related methods, p. 150-165. In K. L. Dickson et al. (ed.), Biological data in water pollution assessment: quantitative and statistical analyses. American Society for Testing and Materials, Philadelphia, Pa.
- 32.Simpson, E. H. 1949. Measurement of diversity. Nature 163:688. [Google Scholar]
- 33.Tabita, F. R. 1988. Molecular and cellular regulation of autotrophic carbon fixation in microorganisms. Microbiol. Rev. 52:155-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uchino, Y., and A. Yokota. 2003. “Green-like” and “red-like” RubisCO cbbL genes in Rhodobacter azotoformans. Mol. Biol. Evol. 20:821-830. [DOI] [PubMed] [Google Scholar]
- 35.Ueda, T., Y. Suga, N. Yahiro, and T. Matsuguchi. 1995. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J. Bacteriol. 177:1414-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watson, G. M., and F. R. Tabita. 1997. Microbial ribulose-1,5-bisphosphate carboxylase/oxygenase: a molecule for phylogenetic and enzymological investigations. FEMS Microbiol. Lett. 146:13-22. [DOI] [PubMed] [Google Scholar]