Abstract
In this study we investigated by using 16S rRNA-based methods the distribution and biomass of archaea in samples from (i) sediments above outcropping methane hydrate at Hydrate Ridge (Cascadia margin off Oregon) and (ii) massive microbial mats enclosing carbonate reefs (Crimea area, Black Sea). The archaeal diversity was low in both locations; there were only four (Hydrate Ridge) and five (Black Sea) different phylogenetic clusters of sequences, most of which belonged to the methanotrophic archaea (ANME). ANME group 2 (ANME-2) sequences were the most abundant and diverse sequences at Hydrate Ridge, whereas ANME-1 sequences dominated the Black Sea mats. Other seep-specific sequences belonged to the newly defined group ANME-3 (related to Methanococcoides spp.) and to the Crenarchaeota of marine benthic group B. Quantitative analysis of the samples by fluorescence in situ hybridization (FISH) showed that ANME-1 and ANME-2 co-occurred at the cold seep sites investigated. At Hydrate Ridge the surface sediments were dominated by aggregates consisting of ANME-2 and members of the Desulfosarcina-Desulfococcus branch (DSS) (ANME-2/DSS aggregates), which accounted for >90% of the total cell biomass. The numbers of ANME-1 cells increased strongly with depth; these cells accounted 1% of all single cells at the surface and more than 30% of all single cells (5% of the total cells) in 7- to 10-cm sediment horizons that were directly above layers of gas hydrate. In the Black Sea microbial mats ANME-1 accounted for about 50% of all cells. ANME-2/DSS aggregates occurred in microenvironments within the mat but accounted for only 1% of the total cells. FISH probes for the ANME-2a and ANME-2c subclusters were designed based on a comparative 16S rRNA analysis. In Hydrate Ridge sediments ANME-2a/DSS and ANME-2c/DSS aggregates differed significantly in morphology and abundance. The relative abundance values for these subgroups were remarkably different at Beggiatoa sites (80% ANME-2a, 20% ANME-2c) and Calyptogena sites (20% ANME-2a, 80% ANME-2c), indicating that there was preferential selection of the groups in the two habitats. These variations in the distribution, diversity, and morphology of methanotrophic consortia are discussed with respect to the presence of microbial ecotypes, niche formation, and biogeography.
The microbially mediated anaerobic oxidation of methane (AOM) is the major biological sink of the greenhouse gas methane in marine sediments (49) and serves as an important control for emission of methane into the hydrosphere. The AOM metabolic process is assumed to be a reversal of methanogenesis coupled to the reduction of sulfate to sulfide involving methanotrophic archaea (ANME) and sulfate-reducing bacteria (SRB) as syntrophic partners (7, 20, 21, 23, 69). Neither the ANME groups nor their sulfate-reducing partners have been isolated yet, and the enzymes and biochemical pathways involved in AOM remain unknown (18, 19). Very recently, however, Krüger et al. described a candidate enzyme (Ni-protein I) that may catalyze methane activation in a reverse terminal methyl-coenzyme M reductase reaction (27), supporting the hypothesis that reverse methanogenesis occurs in the ANME groups. Field and laboratory studies have provided ample evidence that AOM can be mediated by structured consortia consisting of archaea (ANME group 2 [ANME-2]) belonging to the order Methanosarcinales and SRB belonging to the Desulfosarcina-Desulfococcus branch (DSS) of the Deltaproteobacteria (7, 40); below these consortia are referred to as ANME-2/DSS aggregates. These consortia oxidize methane with sulfate, yielding equimolar amounts of carbonate and sulfide (37). A second archaeal group (ANME-1), which is distantly related to the Methanosarcinales and Methanomicrobiales, has also been shown to mediate AOM (34, 41). Using fluorescence in situ hybridization (FISH) combined with secondary ion mass spectrometry, Orphan and coworkers were able to measure carbon isotopic signatures of single aggregates of ANME-1 and ANME-2 cells (40, 41). Their δ13C values were extremely low and thus provided direct evidence for methanotrophy in both phylogenetic clusters. At hot spot sites of AOM in different marine environments phylogenetic analyses based on 16S rRNA gene sequencing of microbial communities showed that there was relatively low diversity of archaea compared to the bacterial diversity in these habitats (21, 26, 28, 39, 58) (GenBank accession numbers AY593257 to AY593349 and AF357889 to AF361694 [http://www.ncbi.nlm.nih.gov]) and indicated the co-occurrence of several ANME-1 and ANME-2 populations. However, such studies so far have not provided quantification of the biomasses of the different groups and their distribution in the environment.
The presence of several microbial populations with essentially the same function in a given environment is still a major puzzle in our concept of biodiversity and microbial ecology. In this study we dealt with methanotrophic archaea, which are limited to extreme environments with anoxic, methane-rich, and sulfate-containing sediments. Hence, the rest of the ocean represents a barrier to the dispersal of these organisms, making them an interesting case study for the central question of microbial biogeography, which is “Is everything everywhere?” (3, 5, 15, 71). To find clues to potential niche occupation by the different ANME groups, we investigated the phylogenetic diversity, distribution, and abundance of the methanotrophs at selected seep sites with high rates of AOM (34, 65), including sediments from three different types of chemosynthetic communities (Beggiatoa mats, Calyptogena fields, and Acharax fields) above outcropping methane hydrate at Hydrate Ridge (Cascadia margin off Oregon) and massive methanotrophic microbial mats at Black Sea methane seeps.
MATERIALS AND METHODS
Study sites, sampling, and geochemical description.
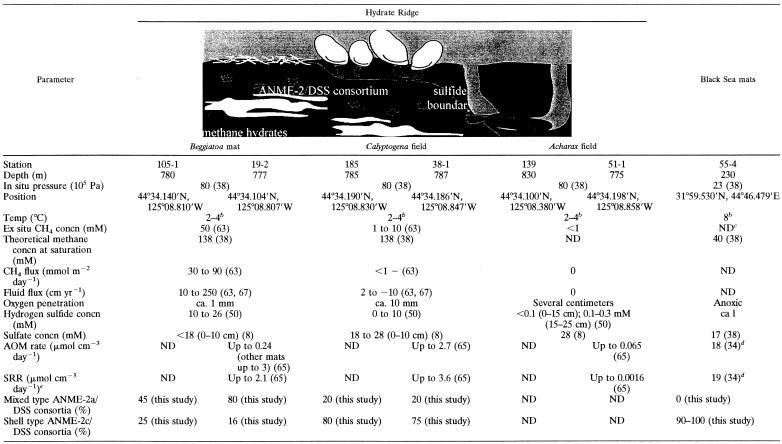
Sediment samples from Hydrate Ridge were obtained during R/V SONNE cruises SO143-2 in August 1999 (9) and SO148-1 in August 2000 (30) at the crest of southern Hydrate Ridge at the Cascadia convergent margin off the coast of Oregon. At Hydrate Ridge, discrete methane hydrate layers occur on the seafloor at a water depth of 600 to 800 m, which corresponds to the hydrate stability limit (50, 57). The hydrates are located a few centimeters beneath the sediment surface and form mounds that are several meters in diameter (56). The mounds are covered by sediment and are populated by sulfide-oxidizing communities, which benefit from large quantities of hydrogen sulfide generated as a by-product of AOM (4, 37, 50, 67). The sediments above the hydrates were covered by mats of giant filamentous sulfur-oxidizing bacteria belonging to the genus Beggiatoa extending several millimeters into the overlying bottom water (stations 105-1 and 19-2; referred to as Beggiatoa mats below) or by clam fields consisting of Calyptogena spp. (stations 185-1 and 38-1; referred to as Calyptogena fields below). Layers of gas hydrate were found in cores with Beggiatoa mats at sediment depths of >13 cm and in cores with Calyptogena fields at sediment depths of >16 cm. Strong degassing from decomposing hydrates was observed. Sediments not covered by either community often contained subsurface-dwelling chemosynthetic Acharax bivalves. The concentrations of methane and sulfide were low in the upper 15 cm in these Acharax fields. Sediment cores that were 20 to 40 cm long were obtained by using a video-guided multiple corer from gas hydrate-bearing sediments and from Acharax fields not enriched in methane in the surface sediments. Geochemical parameters are described in detail in Table 1. Upon recovery all sediment cores were immediately transferred to a cold room (4°C). Samples were processed within 4 h after sampling by using the methods described below.
TABLE 1.
Geochemical characterization of Hydrate Ridge and Black Sea stations and abundance of ANME-2a/DSS and ANME-2c/DSS aggregatesa
The diagram of the chemosynthetic communities at Hydrate Ridge is from reference 65. The numbers in parentheses are references.
Shipboard measurement.
ND, not determined.
The units are micromoles per gram (dry weight) per day.
SRR, sulfate reduction rate.
The second study area is located in the Black Sea and represents a field in which there is active seepage of free gas on the slope of the northwestern Crimea area. Here, a field of conspicuous microbial reefs forming chimney-like structures was discovered at a water depth of 230 m in anoxic waters (29, 34, 46). The reef structure consists of porous carbonate structures covered by microbial mats that are up to 10 cm thick. The chimneys are up to 4 m high and 1 m wide. They are composed mainly of magnesium calcite and aragonite with δ13C values of ca. −30‰, indicating that the carbonate is predominantly derived from AOM (59). Gas seeping from the seafloor at the Ukrainian shelf edge contains 95 to 99% methane and minor amounts of N2, CO2, and H2. In cross sections of the mat, the outside is dark gray to black. The interior part of the mat is pink to brownish (34). Major portions of the mat (ca. 30%; Knittel et al., unpublished data) consist of irregularly distributed cavities and channels containing seawater and gases. Nodules had the same morphology but contained less carbonate. The microbial mats were sampled by using the manned submersible JAGO during the R/V Prof. LOGACHEV cruise in July 2001 (Table 1, station 55-4). Furthermore, material from a nodule (station 14; 31°58.877′N, 44°46.616′E; depth, 182 m) and material from a gel within the mat cavities and channels (station 10; 31°59.559′N, 44°46.515′E; depth, 235 m) were used in this study.
DNA extraction, PCR amplification, and clone library construction.
Sediment cores from Hydrate Ridge were sectioned into 1-cm layers and deep frozen (−20°C) for DNA extraction at the home laboratory. Total community DNA was directly extracted from 2 g of sediment from station 19-2 (depths, 2 to 4 and 6 to 7 cm) and station 38 (depth, 2 to 6 cm) as described by Zhou et al. (72). The following two primer sets were used for PCR amplification of 16S rRNA genes from extracted chromosomal DNA: (i) primers ARCH20F (33) and Uni1392 (42), which resulted in an almost 1,400-bp product; and (ii) primers ARCH20F and ARCH958R (10), which resulted in a ca. 950-bp product. Both primer sets were used with an annealing temperature of 58°C. PCR products were purified with a QiaQuick PCR purification kit (QIAGEN, Hilden, Germany). Clone libraries in pGEM-T-Easy (Promega, Madison, Wis.) or the TOPO pCR4 vector (Invitrogen, Karlsruhe, Germany) were constructed as described previously (48).
For construction of Black Sea clone libraries, a fresh mat (station 55-4) was cut into the following sections: (i) surface biomass, which was dark gray to black (thickness, ca. 1 to 2 mm; clones having designations beginning with BS-S); (ii) material from the black-to-pink transition zone (ca. 2 to 4 mm below the mat surface; clones having designations beginning with BS-M); and (iii) the interior portion of the mat, which was pink (ca. 4 to 8 mm below the mat surface; clones having designations beginning with BS-R). DNA was extracted with a FASTDNA spin kit for soil (Bio 101 Inc., Carlsbad, Calif.). In addition, two clone libraries were constructed from formaldehyde-fixed samples from station 14 (clones having designations beginning with BS-K) and station 55-4 (clones having designations beginning with BS-SR). Aliquots (1 μl) of fixed cells were directly used for PCR and cloning as described above.
Amplified rRNA gene restriction analysis.
Clones known to contain the inserts that were the correct size (ca. 1.4 or 0.95 kb) were screened by amplified rRNA gene restriction analysis in order to identify clones with different inserts. Two restriction enzymes (HaeIII and RsaI) were used for screening as described previously (48).
Sequencing and phylogenetic analysis.
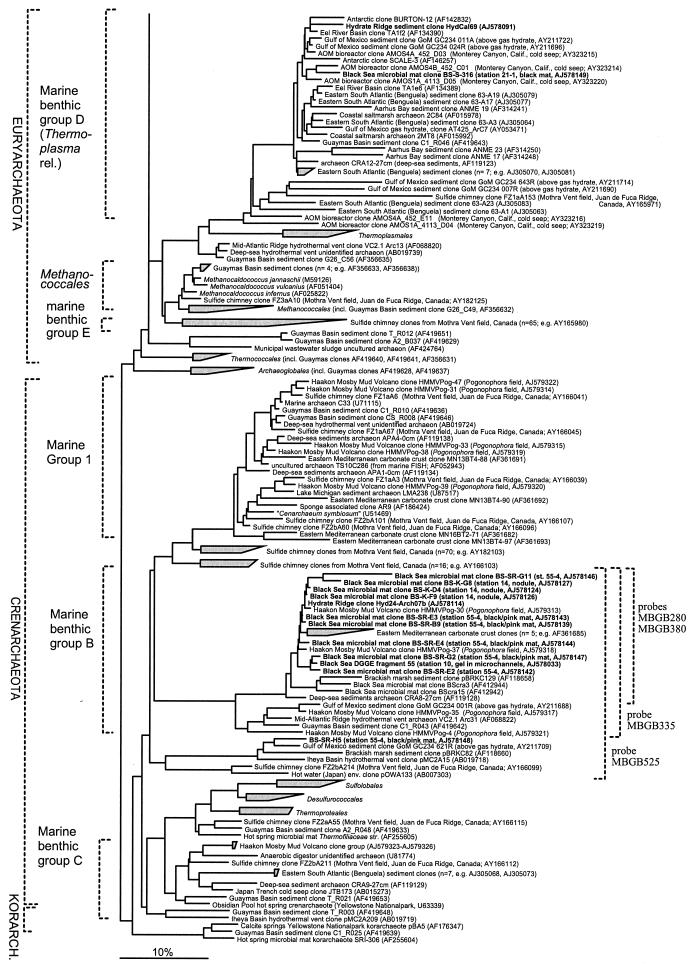
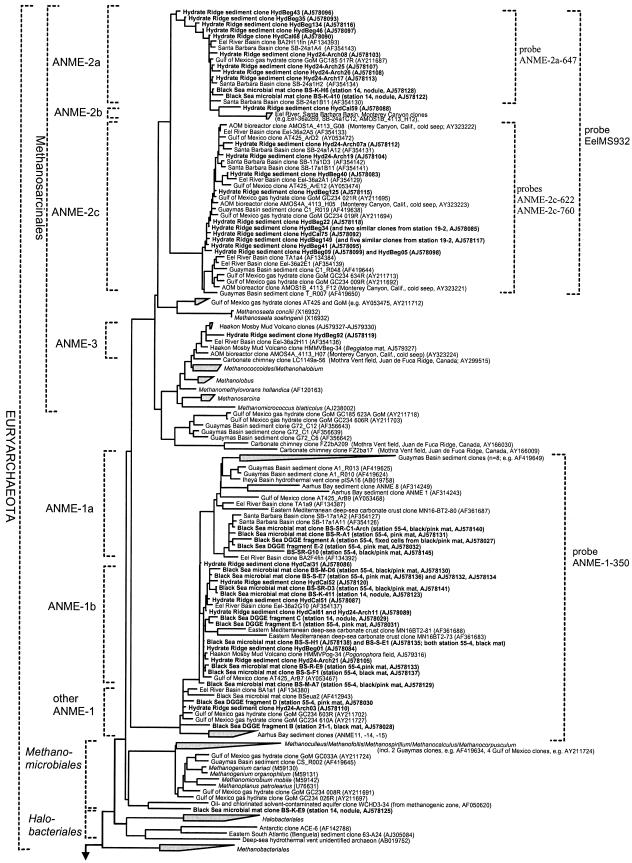
Taq cycle sequencing of plasmid DNAs from selected clones with vector primers and universal rRNA gene-specific primers was performed by GATC Biotech (Konstanz, Germany) and AGOWA (Berlin, Germany). Sequence data were analyzed with the ARB software package (31). Phylogenetic trees were calculated with sequences from Hydrate Ridge sediments and Black Sea microbial mats together with sequences which are available in the EMBL, GenBank, and DDJB databases by performing parsimony, neighbor-joining, and maximum-likelihood analyses with a subset of 200 nearly full-length sequences (>1,300 bp). Filters which excluded highly variable positions were used. In all cases, general tree topology and clusters were stable (a maximum-likelihood tree is shown in Fig. 1). Partial sequences (<1,300 bp) were inserted into the reconstructed tree by parsimony criteria with global-local optimization with no changes in the overall tree topology.
FIG. 1.
Phylogenetic tree showing the affiliations of 16S rRNA gene sequences retrieved from Hydrate Ridge sediments and Black Sea microbial mats (boldface type) with selected reference sequences of the domain Archaea. Besides cultivated organisms all previously published clone sequences from methane-rich sites are included as references (at least one representative per phylogenetic group). The tree was constructed by using maximum-likelihood analysis in combination with filters excluding highly variable positions in a subset of 200 nearly full-length sequences (>1,300 bp). Partial sequences were inserted into the reconstructed tree by using parsimony criteria with global-local optimization, without allowing changes in the overall tree topology. Probe specificity is indicated by brackets. Bar = 10% estimated sequence divergence.
Quantification and measurement of ANME-2/DSS aggregates.
4′,6′-Diamidino-2-phenylindole staining (DAPI) was used to measure sizes of ANME-2a/DSS and ANME-2c/DSS aggregates by epifluorescence microscopy of FISH-treated samples (Hydrate Ridge stations 19-2 and 38 [depth, 1 to 2 cm] and Black Sea mats from station 55-4). The numbers of cells in the aggregates were calculated based on the assumptions that the aggregates were completely surrounded by SRB and that both the aggregates and the cells within the aggregates were spheres.
FISH.
Subsamples of Hydrate Ridge sediment cores were sliced into 1-cm sections, fixed for 2 to 3 h in 3% formaldehyde, washed twice with 1× phosphate-buffered saline (130 mM NaCl, 10 mM sodium phosphate; pH 7.2), and finally stored in 1× phosphate-buffered saline-ethanol (1:1) at −20°C. A Black Sea microbial mat was fixed with paraformaldehyde, embedded in Tissue Tek OCT compound (Ted Pella Inc., Redding, Calif.), and sectioned with a cryostat (HM505 E; Microm, Walldorf, Germany) as previously described (52). Hybridization and microscopic counting of hybridized and DAPI-stained cells were performed as described previously (55). Means were calculated by using 10 to 40 randomly chosen fields for each filter section, which corresponded to 800 to 1,000 DAPI-stained cells. Cy3-, Cy5-, and fluorescein-monolabeled oligonucleotides were purchased from ThermoHybaid (Ulm, Germany). Probes and formamide concentrations used in this study are listed in Table 2. The background signals of samples, observed with the nonsense probe NON338, were negligible (<0.1%).
TABLE 2.
Oligonucleotide probes used in this study
| Probe | Specificity
|
Sequence (5′ to 3′) | Positiona | Formamide concn (%, vol/vol)b | Reference | |
|---|---|---|---|---|---|---|
| Group | Taxon | |||||
| DSS658 | Desulfosarcina-Desulfococcus | SRB of Delta-proteobacteria | TCC ACT TCC CTC TCC CAT | 658-685 | 60 | 32 |
| ARCH915 | Most archaea | Archaea | GTG CTC CCC CGC CAA TTC CT | 915-934 | 35 | 2 |
| ANME-1-350 | ANME-1 | Euryarchaeota | AGT TTT CGC GCC TGA TGC | 350-367 | 40 | 7 |
| EeIMS932 | ANME-2 | Euryarchaeota | AGC TCC ACC CGT TGT AGT | 932-949 | 65 | 7 |
| ANME-2a-647 | ANME-2a | Euryarchaeota | TCT TCC GGT CCC AAG CCT | 647-664 | 50 | This study |
| ANME-2c-622 | ANME-2c | Euryarchaeota | CCC TTG GCA GTC TGA TTG | 622-639 | 50 | This study |
| ANME-2c-760 | ANME-2c | Euryarchaeota | CGC CCC CAG CTT TCG TCC | 760-777 | 60 | This study |
| MBGB-280 | Marine benthic group B | Crenarchaeota | TCA CGG CCC CTA TCG ATT | 280-297 | 50 | This study |
| MBGB-335 | Marine benthic group B | Crenarchaeota | TGC GCC TCG TAA GGC CTG | 335-352 | 50-60 | This study |
| MBGB-380 | Marine benthic group B | Crenarchaeota | GTA ACC CCG TCA CAC TTT | 380-397 | 50 | This study |
| MBGB-525c | Marine benthic group B | Crenarchaeota | AGA GCT GGT TTT ACC GCG | 525-542 | 40-50 | This study |
Position in the 16S rRNA of Escherichia coli.
Formamide concentration in the hybridization buffer.
Probe MBGB-525 also targets several sequences from korarchaeota and other archaea due to a single terminal mismatch.
Scanning electron microscopy (SEM).
Black Sea microbial mat sections (treated as described above for FISH) were directly placed on aluminum specimen mounts. The OCT compound was washed out with sterile water three times for 3 to 5 min each time, and mat sections were dehydrated by covering the samples with 50, 80, and 96% ethanol for 3 min each. Air-dried samples were gold sputtered with a BAL-TEC SCD005 sputter coater for 100 s at 30 mA and were viewed with a Hitachi S-3200N scanning electron microscope operating at 20 kV.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper have been deposited in the EMBL, GenBank, and DDBJ nucleotide sequence database under accession numbers AJ578027 to AJ578033 and AJ578083 to AJ578150.
RESULTS
Archaeal diversity in Hydrate Ridge sediments and Black Sea microbial mats.
Compared to the previously discovered high bacterial diversity at methane seeps (6, 26, 28, 39, 58), the archaeal diversity was relatively low in the Hydrate Ridge sediments and Black Sea microbial mats. Seven different phylogenetic groups were detected; five of these groups belong to the Euryarchaeota, and two belong to the Crenarchaeota (Table 3 and Fig. 1). Archaea belonging to ANME-2 dominated the libraries from Hydrate Ridge sediments and accounted for 76% (n = 82) of all archaeal clones. These archaea could be assigned to subgroups ANME-2a (13%) and ANME-2c (62%). In contrast, only low numbers of ANME-2 representatives were retrieved from Black Sea microbial mats (station 14; n = 2). Both ANME-2 sequences belonged to ANME-2a. The levels of sequence similarity to cultivated species were in all cases less than 91%. However, the levels of similarity to sequences from uncultivated organisms were high and often exceeded 99%. Together with environmental sequences from the Eel River and the Santa Barbara Basin (21, 39), from Monterey Canyon (18), from the Guaymas Basin (58), and from the Gulf of Mexico (28, 35), ANME-2c and ANME-2a form a clade specific for AOM sites. Today, ANME-2b consists only of sequences from Eel River and Santa Barbara Basin sediment (39), as well as Monterey Canyon (18).
TABLE 3.
Phylogenetic affiliations of 16S rRNA gene clones retrieved from Hydrate Ridge sediments and Black Sea microbial mats
| Phylogenetic group | No. of clones
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Hydrate Ridge
|
Black Sea microbial mat
|
|||||||
| Beggiatoa mat (station 19-2) | Beggiatoa mat (station 19-2) | Calyptogena field (station 38-1) | PCR from extracted DNA
|
PCR from fixed cells
|
||||
| Station 55-4
|
Station 55-4 (black/pink) | Station 14 (nodule) | ||||||
| 2-4 cm | 6-7 cm | 2-6 cm | Black | Black/pink | Pink | |||
| Euryarchaeota | ||||||||
| ANME-1a | 1 | 13 | 1 | |||||
| ANME-1b | 4 | 3 | 14 | 29 | 25 | 32 | 8 | 2 |
| ANME-1 unaffiliated | 1 | |||||||
| ANME-2a | 7 | 5 | 2 | 2 | ||||
| ANME-2c | 20 | 46 | 1 | |||||
| ANME-2 unaffiliated | 1 | |||||||
| ANME-3 | 3 | |||||||
| Methanomicrobiales | 1 | |||||||
| Marine benthic group D | 1 | |||||||
| Crenarchaeota | ||||||||
| Marine benthic group B | 30 | 9 | ||||||
| Unaffiliated | 3 | |||||||
| Total no. of clones per library | 31 | 57 | 20 | 29 | 25 | 33 | 21 | 6 |
The second archaeal group mediating AOM is ANME-1 (34, 41), which is distantly related to the Methanosarcinales and Methanomicrobiales. ANME-1 can be divided into two subgroups, ANME-1a and ANME-1b, and other sequences which cannot be affiliated with these subgroups (Fig. 1). Like the subgroups of ANME-2, both of the ANME-1 subgroups do not contain any cultivated species but comprise only uncultivated species from methane seeps with levels of similarity up to 99% (21, 28, 39, 58, 62) (GenBank accession numbers AY593257 to AY593349, AF357889 to AF361694, AJ704631 to AJ704652, and AJ579313 to AJ579330 [http://www.ncbi.nlm.nih.gov]). Sequences belonging to ANME-1 were most abundant in Black Sea clone libraries (71% of all archaeal clones; n = 111) (Fig. 1 and Table 3). The major portion was affiliated with ANME-1b (61%), and only a minor fraction (10%) belonged to ANME-1a. At Hydrate Ridge ANME-1 sequences were the second most abundant phylotype recovered; they accounted for 20% of the sequences, all of which were affiliated with ANME-1b.
Few sequences from Hydrate Ridge (n = 3) could be affiliated with a third seep-specific clade, ANME-3. This group is phylogenetically closely related to methanogenic bacteria belonging to the Methanococcoides (94 to 96%). Besides Hydrate Ridge sequences this group comprises mainly sequences from Haakon Mosby Mud Volcano (GenBank accession numbers AJ704631 to AJ704652 and AJ579313 to AJ579330 [http://www.ncbi.nlm.nih.gov]) and some sequences from the Eel River Basin (39), from Monterey Canyon (18), and from a sulfide chimney from the Mothra Vent Field in Canada (53). A single sequence from the Black Sea was related to the Methanomicrobiales, and another sequence from Hydrate Ridge was affiliated with the Euryarchaeota of marine benthic group D (70), which is related to the order Thermoplasmales.
Marine benthic group B (MBGB) belonging to the Crenarchaeota was abundant in the clone libraries constructed by using fixed cells from Black Sea mat stations 55-4 (56%; n = 30) and 14 (60%; n = 9). None of the members of this group have been cultivated yet. The diversity within the group is high; the levels of sequence similarity are between 82 and 99%.
ANME-2 abundance.
Hydrate Ridge sediments were dominated by high numbers of ANME-2/DSS aggregates, which accounted for more than 90% of the total cells (i.e., aggregated cells plus single archaea and bacteria [7, 65]). Single ANME-2 cells accounted for only around 1% of the total cells, and this value did not vary significantly with depth (data not shown). We rarely detected aggregated ANME-2 cell clusters without any bacterial partner. These aggregates were smaller than average aggregates (7) and consisted of 4 to 36 cells. At the Acharax stations no single ANME-2 cells and only a few aggregated ANME-2 cells were found.
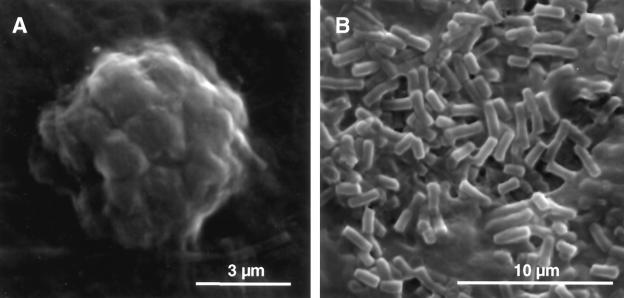
ANME-2 cells were also detected in Black Sea microbial mats (Fig. 2A and B); however, they were limited to microenvironments in regions that were 3 to 6 mm below the mat surface. Like the cells at Hydrate Ridge, the ANME-2 cells formed spherical aggregates associated with SRB belonging to the Desulfococcus-Desulfosarcina branch (Fig. 3B). SEM analysis showed high levels of these aggregates (Fig. 3A) restricted to specific areas of the mat section. In these areas almost no cylindrical ANME-1 cells were detected. An average ANME-2/DSS aggregate in the Black Sea mat consisted of an inner core of about 100 coccoid archaeal cells, similar to the Hydrate Ridge consortia (7). However, the archaeal cells were each 1.2 to 1.4 μm in diameter and thus substantially larger than those at Hydrate Ridge. The Black Sea aggregates were only partially surrounded by Desulfosarcina-Desulfococcus cells (diameter, 1 μm). The diameters of 60 ANME-2/DSS aggregates from the Black Sea ranged from 4 to 20 μm, and the average was 8.3 ± 4.0 μm; hence, these aggregates were significantly larger than the average consortium at Hydrate Ridge (diameter, ca. 3 μm [7, 26]). We also detected a few aggregates which seemed to have a bacterial partner other than DSS. A minimum of three ANME-2 cells together were observed, and a relatively large percentage of the ANME-2 aggregates did not have any tightly associated partner (ca. 10%). In total, we estimated that ANME-2 cells accounted for 5% of the total cells in the 3- to 6-mm subsurface mat horizons in which they were detected (ca. 1 × 1010 cells cm of mat−3). For the total mat they were insignificant in terms of biomass (<1%).
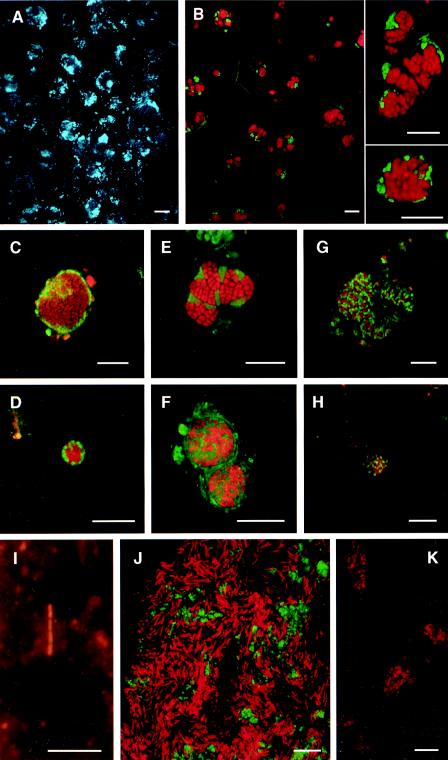
FIG. 2.
Single cells and cell aggregates of ANME-1 and ANME-2 archaea in sediments from Hydrate Ridge sediments and thin sections of Black Sea microbial mats, visualized by FISH. Scale bars = 10 μm. Panels A to H, J, and K are confocal laser scanning micrographs. Panel I is a regular epifluorescence micrograph. (A) DAPI staining showing the embedded aggregates in the bulk biomass in Black Sea microbial mats. (B) Corresponding FISH image of ANME-2/DSS aggregates (probe ANME-2c-760 labeled with Cy5 [red] and probe DSS658 labeled with Cy3 [green]). DSS cells surrounded the archaea partially or completely. In many aggregates the ANME cells had only a few or no DSS partners. (C to F) Different types of ANME-2c/DSS aggregates (probe ANME-2c-622 labeled with Cy3 [red] and probe DSS658 labeled with Cy5 [green]), including aggregates with an inner core of archaea surrounded by a shell of SRB (shell type). (G and H) ANME-2a/DSS aggregates (probe ANME-2a-647 labeled with Cy3 [red] and probe DSS658 labeled with Cy5) from Beggiatoa site station 38 (depth, 1 to 2 cm), showing cell-to-cell association of the partners (mixed type). (I) ANME-1 cells stained with Cy3-labeled probe ANME-1-350 in Hydrate Ridge sediments (Beggiatoa site station 19-2 [depth, 5 to 6 cm]) (J) Color overlay of ANME-1 cells targeted with probe ANME-1-350 labeled with Cy3 (red) and SRB of the Desulfosarcina-Desulfococcus branch labeled with fluorescein (probe DSS658; green) in a Black Sea mat section. (K) Marine benthic group B Crenarchaeota targeted with probe MBGB-380 labeled with Cy3 in a Black Sea mat section.
FIG. 3.
ANME-1 archaea and ANME-2/DSS aggregates in the Black Sea microbial mat: SEM images of a gold-sputtered formaldehyde-fixed mat section. (A) ANME-2/DSS aggregate, showing the typical spherical structure. (B) Cylindrical ANME-1 cells.
ANME-2 subgroup detection.
The ANME-2 cluster can be divided into three subgroups, ANME-2a, ANME-2b, and ANME-2c (39). To study the distribution and abundance of these subgroups, we developed several new oligonucleotide probes, one probe specific for ANME-2a (ANME-2a-647) and two probes specific for ANME-2c (ANME-2c-622 and ANME-2c-760). No probe was developed for ANME-2b since we did not obtain ANME-2b sequences from our samples. Sequences belonging to ANME-2c had at least four mismatches with the probe specific for ANME-2a; sequences belonging to ANME-2a had at least five mismatches with probe ANME-2c-622 and two mismatches with probe ANME-2c-760 (one central mismatch and one terminal mismatch). In sections from Black Sea microbial mats (station 55-4) only cells belonging to ANME-2c could be detected. ANME-2a cells were not detected in the sections; however, due to the great complexity of the mat they may have occurred in other regions.
In Hydrate Ridge sediments two different types of ANME-2/DSS aggregates were detected. At the Beggiatoa sites ANME-2a/DSS aggregates were the most abundant aggregates, and they accounted for ca. 80% of all aggregates at station 19-2 (depth, 1 to 2 cm) and 45% of all aggregates at station 105 (depth, 1 to 2 cm). At both Calyptogena sites (stations 38 and 185) ANME-2a/DSS aggregates accounted for only ca. 20% of all aggregates (depth, 1 to 2 cm). At these sites, ANME-2c/DSS aggregates were the most abundant aggregates, and they accounted for 75% of the aggregates at station 38 and 80% of the aggregates at station 185. At the Beggiatoa sites only 16% (station 19-2) and 25% (station 105-1) of the total ANME-2/DSS aggregates belonged to ANME-2c. In addition to the difference in phylogenetic origin, the two populations could be distinguished from each other by the morphology of the aggregates which they formed. The ANME-2c/DSS aggregates represented the common spherical type with an inner archaeal core which was partially or fully surrounded by an outer shelf of SRB (Fig. 2C and F) (referred to as the shell type below). In the shell type, sometimes the SRB cells grew into the inner ANME-2c core colony, but in general they were not completely mixed with the archaeal cells. In contrast, in the ANME-2a/DSS aggregates the SRB and archaea were completely mixed, and the resulting aggregates were not always spherical (Fig. 2G and H); below we refer to these consortia as the mixed type. Only a very few ANME-2a/DSS aggregates which were the shell type were detected.
ANME-1 abundance.
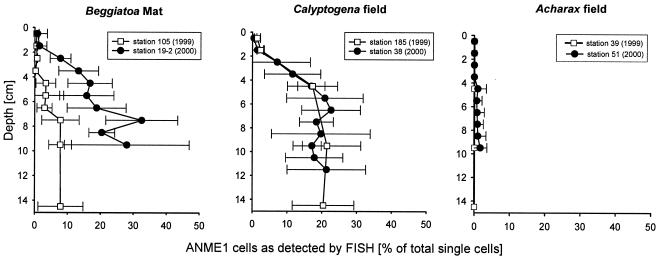
The ANME-1 cells were cylindrical and were, like ANME-2 cells, autofluorescent under UV light, a feature typical of methanogenic archaea containing coenzyme F420. The length varied between 1.5 and 3 μm, and the diameter was about 0.6 μm. At Hydrate Ridge these cells mostly occurred as single cells without any directly associated bacterial or archaeal partner. However, they often formed long chains of 2 to 10 cells (Fig. 2I). We rarely detected spherically aggregated ANME-1 cells. At the Beggiatoa and Calyptogena sites at Hydrate Ridge the FISH detection rates for ANME-1 cells were comparable and increased strongly with depth (Fig. 4). In the surface layers (depth, 0 to 2 cm) the ANME-1 cells accounted for between 1 and 2% of the total single cells (≪1% of the total cells). At the Beggiatoa sites the percentage of ANME-1 cells strongly increased to 8% of the total DAPI-detected single cells at a depth of 7 to 15 cm at station 105 and to 20 to 33% of the total DAPI-detected single cells at a depth of 7 to 10 cm at station 19-2. These percentages correspond to absolute ANME-1 cell numbers of 1 × 108 and 5 × 108 cells cm of sediment−3, respectively. In samples from Calyptogena sites (stations 185 and 38) the ANME-1 abundance was comparable; the highest percentages were 21 to 23% at a depth of 6 to 12 cm (up to 1.0 × 109 cells cm−3). The ANME-1 abundance at the Acharax sites ranged between 0 and 2% of the single cells in the upper 15 cm of the sediment. On the basis of the total cell numbers, including the cells in the aggregates (7, 65), ANME-1 comprised up to 5% of all archaea throughout the sediment column.
FIG. 4.
Detection and quantification of ANME-1 cells in Hydrate Ridge sediments by FISH.
In contrast to the Hydrate Ridge sediments, the Black Sea microbial mats (stations 14 and 55-4) were clearly dominated by ANME-1 cells (34). These cells were highly active, as indicated by a bright FISH signal, and had the same morphology as the cells at Hydrate Ridge (Fig. 2J). SEM analysis of mat sections revealed very abundant cylindrical microorganisms embedded in a thick extracellular polysaccharide matrix (Fig. 3B). On average, throughout the mat ANME-1 cells accounted for 20% of the total cells (ca. 4 × 1010 cells cm of mat−3); the maximum was ca. 50% (ca. 9 × 1010 cells cm of mat−3) in the samples analyzed here. Values up to 70% were reported for similar samples analyzed in a previous study (34).
Detection of MBGB.
Many clone sequences retrieved from the Black Sea microbial mats and other methane seeps were affiliated with Crenarchaeota belonging to MBGB (Fig. 1). To quantify the in situ abundance of these clones, we designed four FISH probes specific for the Black Sea sequences and several other sequences in this cluster. With these probes very small cocci (diameter, 0.2 to 0.4 μm) could be detected in the Black Sea mats (Fig. 2K). To our knowledge, this is the first in situ visualization of this group. The organisms occurred mostly in large clusters and were abundant in regions with lower cell densities (e.g., the channels through the mats). Clear signals were available but did not allow quantification because of the very small size of the cells. DAPI signals were also hardly detectable. However, dual hybridization of mat sections with the MBGB-specific probes in combination with the general archaeal probe ARCH915 showed a clear overlay of the two signals. In Hydrate Ridge sediments at Calyptogena site 38 (depths, 1 to 2 and 7 to 8 cm) very few cocci (<1%) showed weak signals with the new Cy3-monolabeled MBGB probes. Tourova et al. proposed that these organisms might be a new form of sulfate reducers (64). The metabolism of most Crenarchaeota other than the cultivated thermophilic extremophiles is not known yet (11, 25, 51). Their high abundance and association with seeps might indicate that the MBGB are also involved in AOM or may use specific carbon intermediates generated from AOM. However, so far, based on isotopic signatures of biomarker lipids there is no clear evidence for involvement of Crenarchaeota in AOM (51).
DISCUSSION
Diversity of methanotrophic guilds at cold seeps.
The overall archaeal diversity in the Black Sea and Hydrate Ridge clone libraries was low; these libraries contained only five and four phylogenetic groups, respectively. Similar low diversities (only two to five major groups) have been reported for all but one of the methane seeps investigated so far (1, 6, 21, 28, 35, 39, 62) (GenBank accession numbers AY593257 to AY593349 and AF357889 to AF361694 [http://www.ncbi.nlm.nih.gov]). The archaeal community of hydrothermally active sediments of Guaymas Basin (58) consists of 13 phylogenetic lineages. At this site, a variety of methanogens and other thermophilic archaea not related to ANME groups have been isolated. However, AOM appeared not be the dominant carbon cycling process in the Guaymas sediments (24), in contrast to the other methane seeps.
Compared to the overall low archaeal diversity at cold seeps, the 16S rRNA diversity within and between the individual ANME groups appears to be relatively high. ANME-1 and ANME-2 show <85% similarity to each other and hence are two different lineages. The phylogenetic distance is so large that members of ANME-1 and ANME-2 certainly belong to different orders or families, which, however, have apparently similar physiological properties. Based on 16S rRNA gene similarity values of <96%, ANME-1a and ANME-1b contain different species or even genera. The same applies to the ANME-2 subgroups ANME-2a and ANME-2c.
Co-occurrence of methanotrophic groups.
ANME-1 and ANME-2 co-occur in Hydrate Ridge sediments and in Black Sea microbial mats, as well as in many other methane-rich habitats. In situ quantification of these groups, however, showed that they were present at very different levels, indicating that there were different mechanisms of selection of the different groups by the habitat. ANME-1 dominated the Black Sea microbial mats (40 to 50% of the total cells), and ANME-2/DSS aggregates were restricted to small microenvironments and accounted for only 1% of the total cells. In contrast, ANME-2/DSS aggregates dominated the Hydrate Ridge sediments (>90% of the total cells), and ANME-1 cells were detected only in deeper sediment layers and the number of cells was at least 1 order of magnitude less. The environmental conditions at the Hydrate Ridge and Black Sea habitats were compared (Table 1) to identify possible factors that determined niches for ANME-1 and ANME-2. At both sites excess methane was available as the main carbon and energy source (the in situ concentrations were 10 to 60 and 20 mM). Sulfate at concentrations of >2 mM was found to be the only known electron acceptor in the horizons with abundant ANME biomass. Hence, the availability of an electron donor and an electron acceptor obviously did not directly exert selective pressure. The parameter that differed the most between the two locations is oxygen. The Black Sea microbial mats thrive in a permanently anoxic habitat because oxygen is depleted in the subsurface waters beneath a depth of ca. 120 m. In contrast, the bottom waters at Hydrate Ridge are oxic, and the surface sediments are sporadically flushed with oxygen by bottom water currents, bioturbation, and gas ebullition (67). Thus, ANME-1 may be more sensitive to oxygen than ANME-2. This hypothesis is supported by the very low ANME-1 abundance (≪1% of the total cells) in the surface sediments at Hydrate Ridge and the increase in the abundance with depth to 5% of the total cells despite the concurrent decrease in sulfate availability and the increase in sulfide concentrations. Furthermore, ANME-1 was completely absent in the oxygenated surface sediments at the Acharax site, although ANME-2/DSS aggregates were present in these layers (26).
Environmental niches of the ANME-2a/DSS and ANME-2c/DSS aggregates.
ANME-2a and ANME-2c co-occur in Hydrate Ridge sediments; however, the abundance varies greatly on a meter to decimeter scale according to the type of chemosynthetic community (the sulfide-oxidizing bacterium Beggiatoa or the symbiotic clam Calyptogena) populating the surface sediments. Both sites have steep gradients of pore water sulfate (7, 12), high hydrogen sulfide concentrations (50), and high rates of AOM and sulfate reduction (7, 65) (Table 1). However, the fluid flow and methane fluxes from the seafloor are substantially greater but also more variable at Beggiatoa sites than at Calyptogena sites (50, 63, 68). At Beggiatoa sites oxygen had a maximum penetration of a few millimeters. In contrast, at Calyptogena sites oxygen penetrated deeper into the sediments due to the bioturbating activity of the clams, which live with their feet in the sulfide-rich zones of the sediment. The dominance of ANME-2a/DSS and ANME-2c/DSS aggregates is clearly linked with a specific type of chemosynthetic communities populating the surface sediment for reasons that are not known yet.
Biogeography of the anaerobic methanotrophs (ANME).
Methanotrophic archaea were first detected in sediments of cold seeps of the northeast Pacific continental margin, such as the sediments from Eel River (21, 39, 41) and Hydrate Ridge (7). The data for lipid biomarker and DNA signatures indicate the global presence of these archaea in anoxic methane-rich sediments. So far, ANME signatures have been detected at all methane seeps investigated, including seeps in the north Pacific Ocean, north and south Atlantic Ocean, Mediterranean Sea, Black Sea, and North Sea (1, 7, 13, 14, 18, 21, 22, 26, 28, 34, 35, 40, 41, 43, 44, 53, 58, 60-62). Furthermore, ANME groups have also been found in the methane-sulfate transition zones of different coastal environments (23a, 66). An analysis of the current data set for the 16S rRNA genes of the ANME groups indicates that their occurrence is limited to the upper few meters of subsurface sediments, as no genes have been retrieved from any deep biosphere core yet. Since so far the presence of ANME-1, ANME-1, and ANME-3 is restricted to anoxic, methane-rich, and sulfate-containing sediments and the oxic bottom waters of the ocean present a barrier to their distribution, these groups are an interesting case for the study of microbial biogeography, as are all extremophiles. The occurrence of biogeographical trends in the distribution of microorganisms is debated (16, 17, 45, 71) because such trends contradict a basic paradigm of microbial ecology, that everything is everywhere—the environment selects (3, 5). Based on 16S rRNA genes as the evolutionary marker, the present data do not indicate the biogeography of ANME-1, ANME-1, and ANME-3 or subclusters of these groups, in contrast to various thermophilic microorganisms (45, 71). Other globally abundant groups of microbes, such as Alphaproteobacteria belonging to the SAR11 clade (36, 47) or the Roseobacter clade-affiliated cluster (54), do not have a biogeography based on oceanic regions or latitudes. Instead, as observed in this study, clusters appear to be formed by subgroups, which are specifically adapted to certain niches (e.g., water depth, light availability, and temperature). However, these results may be related to insufficient phylogenetic resolution of 16S rRNA sequences. Recently, Whitaker et al. (71) and Papke et al. (45) used high-resolution multilocus sequence analysis to show that the genetic distances between thermophilic prokaryotes having almost identical 16S rRNAs increased proportionally with geographic distance. These findings revealed that populations are isolated from one another by geographic barriers and have diverged over the course of their recent evolutionary history. Besides multilocus sequence analysis, other markers, such as functional genes specific for methanotrophs, or simply a clear increase in the size of the ANME 16S rRNA data set to several thousand sequences could be used to address the biogeography of ANME.
Conclusions.
A comparison of 16S rRNA gene sequences showed the ubiquitous presence of methanotrophic archaea in almost all methane environments investigated so far independent of the in situ temperature, depth, pressure, and methane and sulfate concentrations. ANME-1 and ANME-2, as well as their phylogenetic subgroups, co-occur at all seep sites that have been investigated quantitatively; however, microscopic analysis of the distribution of the subgroups has revealed the dominance of certain types within microniches in the environments. To single out the factors responsible for the selection of certain ecotypes of methanotrophs, environmental conditions and geochemical gradients need to be analyzed in situ with a high resolution relevant to the scale of the microenvironments detected in this study. This would require parallel investigations with microsensors in the field or experimental studies in flowthrough microcosms.
Acknowledgments
We thank the officers, crews, and shipboard scientific parties of R/V SONNE during TECFLUX cruises SO143 and SO148 (grants 03G0143A and 03G0148A) to Hydrate Ridge and during the GHOSTDABS cruise (grant 03G0559A) of R/V Prof. LOGACHEV in summer 2001 for their excellent support. We acknowledge GEOTECHNOLOGIEN projects OMEGA (grant 03G0566A), LOTUS (grant 03G0565), and GHOSTDABS for providing access to samples and infrastructure. We are indebted to Doug Bartlett, Sander Heijs, Brian Lanoil, and Andreas Teske for sharing their sequence data prior to publication for probe construction. Armin Gieseke is acknowledged for providing an introduction to laser scanning microscopy, and Julia Polansky and Andreas Lemke are acknowledged for technical assistance.
This study was part of the program MUMM (Mikrobielle Umsatzraten von Methan in gashydrathaltigen Sedimenten; grant 03G0554A) supported by the Bundesministerium für Bildung und Forschung (Germany). Further support was provided by the Max Planck Society, Germany.
Footnotes
Publication GEOTECH-85 of the GEOTECHNOLOGIEN program and no. 10 of the research program GHOSTDABS of the Bundesministerium für Bildung und Forschung and the DFG.
REFERENCES
- 1.Aloisi, G., I. Bouloubassi, S. K. Heijs, R. D. Pancost, C. Pierre, J. S. S. Damsté, J. C. Gottschal, L. J. Forney, and J.-M. Rouchy. 2002. CH4-consuming microorganisms and the formation of carbonate crusts at cold seeps. Earth Planet. Sci. Lett. 203:195-203. [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baas-Becking, L. G. M. 1934. Geobiologie of Inleiding Tot de Milieukunde. In W. P. van Stockum and N. V. Zoon (ed.), Diligentia Wetensch, serie 18/19. van Stockum’s Gravenhange, The Hague, The Netherlands.
- 4.Barry, J. P., R. E. Kochevar, and C. H. Baxter. 1997. The influence of pore-water chemistry and physiology in the distribution of vesicomyid clam at cold seeps in Monterey Bay: implications for patterns of chemosynthetic community organization. Limnol. Oceanogr. 42:318-328. [Google Scholar]
- 5.Beijerinck, M. W. 1913. De infusies en de ontdekking der backteriën. In Jaarboek van de Koninklijke Akademie v. Wetenschappen. Müller, Amsterdam, The Netherlands.
- 6.Bidle, K. A., M. Kastner, and D. H. Bartlett. 1999. A phylogenetic analysis of microbial communities associated with methane hydrate containing marine fluids and sediments in the Cascadia margin (ODP8 site 892B). FEMS Microbiol. Lett. 177:101-108. [DOI] [PubMed] [Google Scholar]
- 7.Boetius, A., K. Ravenschlag, C. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jørgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 8.Boetius, A., and E. Suess. 2004. Hydrate Ridge: a natural laboratory for the study of microbial life fueled by methane from near-surface gas hydrates. Chem. Geol. 205:291-310. [Google Scholar]
- 9.Bohrmann, G., P. Linke, P. Suess, and O. Pfannkuche. 2000. R.V. SONNE cruise report SO143: TECFLUX-I (June 29-September 6, 1999; Honolulu-Astoria-San Diego). GEOMAR Rep. 93:217. [Google Scholar]
- 10.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLong, E. F. 1998. Everything in moderation: Archaea as ‘non-extremophiles.’ Curr. Opin. Genet. Dev. 8:649-654. [DOI] [PubMed] [Google Scholar]
- 12.Elvert, M., J. Greinert, E. Suess, and M. J. Whiticar. 2001. Carbon isotopes of biomarkers derived from methane-oxidizing microbes at Hydrate Ridge, Cascadia convergent margin, p. 115-129. In C. K. Paull and W. P. Dillon (ed.), Natural gas hydrates: occurrence, distribution, and dynamics, vol. 124. American Geophysical Union, Washington, D.C. [Google Scholar]
- 13.Elvert, M., E. Suess, J. Greinert, and M. J. Whiticar. 2000. Archaea mediating anaerobic methane oxidation in deep-sea sediments at cold seeps of the eastern Aleutian subduction zone. Org. Geochem. 31:1175-1187. [Google Scholar]
- 14.Elvert, M., E. Suess, and M. J. Whiticar. 1999. Anaerobic methane oxidation associated with marine gas hydrates: superlight C-isotopes from saturated and unsaturated C20 and C25 irregular isoprenoids. Naturwissenschaften 86:295-300. [Google Scholar]
- 15.Fenchel, T. 2003. Biogeography for bacteria. Science 301:925-926. [DOI] [PubMed] [Google Scholar]
- 16.Fenchel, T. 2002. Microbial behavior in a heterogeneous world. Science 296:1068-1071. [DOI] [PubMed] [Google Scholar]
- 17.Finlay, B. J. 2002. Global dispersal of free-living microbial eukaryote species. Science 296:1061-1063. [DOI] [PubMed] [Google Scholar]
- 18.Girguis, P. R., V. J. Orphan, S. J. Hallam, and E. F. DeLong. 2003. Growth and methane oxidation rates of anaerobic methanotrophic archaea in a continuous-flow bioreactor. Appl. Environ. Microbiol. 69:5472-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hallam, S. J., P. R. Girguis, C. M. Preston, P. M. Richardson, and E. F. DeLong. 2003. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl. Environ. Microbiol. 69:5483-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen, L. B., K. Finster, H. Fossing, and N. Iversen. 1998. Anaerobic methane oxidation in sulfate depleted sediments: effects of sulfate and molybdate additions. Aquat. Microb. Ecol. 14:195-204. [Google Scholar]
- 21.Hinrichs, K. U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. DeLong. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 22.Hinrichs, K.-U., and A. Boetius. 2002. The anaerobic oxidation of methane: new insights in microbial ecology and biogeochemistry, p. 457-477. In G. Wefer, D. Billett, D. Hebbeln, B. B. Jørgensen, M. Schlüter, and T. Van Weering (ed.), Ocean margin systems. Springer-Verlag, Berlin, Germany.
- 23.Hoehler, T. M., M. J. Alperin, D. B. Albert, and C. S. Martens. 1994. Field and laboratory studies of methane oxidation in an anoxic marine sediment: evidence for a methanogen-sulfate reducer consortium. Glob. Biogeochem. Cycles 8:451-463. [Google Scholar]
- 23a.Ishii, K., M. Mußmann, B. J. MacGregor, and R. Amann. 2004. An improved fluorescence in situ hybridization protocol for the identification of bacteria and archaea in marine sediments. FEMS Microbiol. Ecol. 50:203-212. [DOI] [PubMed] [Google Scholar]
- 24.Kallmeyer, J., and A. Boetius. 2004. Effects of temperature and pressure on sulfate reduction and anaerobic oxidation of methane in hydrothermal sediments of Guaymas Basin. Appl. Environ. Microbiol. 70:1231-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karner, M. B., E. F. DeLong, and D. M. Karl. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507-510. [DOI] [PubMed] [Google Scholar]
- 26.Knittel, K., A. Boetius, A. Lemke, H. Eilers, K. Lochte, O. Pfannkuche, P. Linke, and R. Amann. 2003. Activity, distribution, and diversity of sulfate reducers and other bacteria in sediments above gas hydrate (Cascadia Margin, OR). Geomicrobiol. J. 20:269-294. [Google Scholar]
- 27.Krüger, M., A. Meyerdierks, F. O. Glöckner, R. Amann, F. Widdel, M. Kube, R. Reinhardt, J. Kahnt, R. Böcher, R. K. Thauer, and S. Shima. 2003. A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature 426:878-881. [DOI] [PubMed] [Google Scholar]
- 28.Lanoil, B. D., R. Sassen, M. T. La Duc, S. T. Sweet, and K. H. Nealson. 2001. Bacteria and Archaea physically associated with Gulf of Mexico gas hydrates. Appl. Environ. Microbiol. 67:5143-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lein, A. Y., M. V. Ivanov, N. V. Pimenov, and M. B. Gulin. 2002. Geochemical peculiarities of the carbonate constructions formed during microbial oxidation of methane under anaerobic conditions. Mikrobiologiya 71:78-90. (In Russian.) [PubMed]
- 30.Linke, P., and E. Suess. 2001. R.V. SONNE cruise report SO148 TECFLUX-II-2000 (Victoria-Victoria; July 20-August 12, 2000). GEOMAR Rep. 98:122. [Google Scholar]
- 31.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manz, W., M. Eisenbrecher, T. R. Neu, and U. Szewzyk. 1998. Abundance and spatial organization of gram negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol. Ecol. 25:43-61. [Google Scholar]
- 33.Massana, R., A. E. Murray, C. M. Preston, and E. F. DeLong. 1997. Vertical distribution and phylogenetic characterization of marine planktonic archaea in the Santa Barbara Channel. Appl. Environ. Microbiol. 63:50-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michaelis, W., R. Seifert, K. Nauhaus, T. Treude, V. Thiel, M. Blumenberg, K. Knittel, A. Gieseke, K. Peterknecht, T. Pape, A. Boetius, R. Amann, B. B. Jørgensen, F. Widdel, J. Peckmann, N. V. Pimenov, and M. B. Gulin. 2002. Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science 297:1013-1015. [DOI] [PubMed] [Google Scholar]
- 35.Mills, H. J., C. Hodges, K. Wilson, I. R. MacDonald, and P. A. Sobecky. 2003. Microbial diversity in sediments associated with surface-breaching gas hydrate mounds in the Gulf of Mexico. FEMS Microbiol. Ecol. 46:39-52. [DOI] [PubMed] [Google Scholar]
- 36.Morris, R. M., M. S. Rappé, S. A. Connon, K. L. Vergin, W. A. Siebold, C. A. Carlson, and S. J. Giovannoni. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806-810. [DOI] [PubMed] [Google Scholar]
- 37.Nauhaus, K., A. Boetius, M. Krüger, and F. Widdel. 2002. In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ. Microbiol. 4:296-305. [DOI] [PubMed] [Google Scholar]
- 38.Nauhaus, K., T. Treude, A. Boetius, and M. Krüger. 11 November 2004. Environmental regulation of the anaerobic oxidation of methane: a comparison of ANME-I- and ANME-II-communities. Environ. Microbiol. 10.1111/j.1462-2920.2004.00669.x. [DOI] [PubMed]
- 39.Orphan, V. J., K.-U. Hinrichs, W. Ussler III, C. K. Paull, L. T. Taylor, S. P. Sylva, J. M. Hayes, and E. F. DeLong. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67:1922-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orphan, V. J., C. H. House, K.-U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484-487. [DOI] [PubMed] [Google Scholar]
- 41.Orphan, V. J., C. H. House, K.-U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. USA 99:7663-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pace, N. R. 1996. New perspective on the natural microbial world: molecular microbial ecology. ASM News 62:463-470. [Google Scholar]
- 43.Pancost, R. D., E. C. Hopmans, and J. S. S. Damsté. 2001. Archaeal lipids in Mediterranean cold seeps: molecular proxies for anaerobic oxidation. Geochim. Cosmochim. Acta 65:1611-1627. [Google Scholar]
- 44.Pancost, R. D., S. J. Sinninghe Damsté, S. de Lint, M. J. E. C. van der Maarel, J. C. Gottschal, and T. M. S. S. Party. 2000. Biomarker evidence for widespread anaerobic methane oxidation in Mediterranean sediments by a consortium of methanogenic archaea and bacteria. Appl. Environ. Microbiol. 66:1126-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papke, R. T., N. B. Ramsing, M. M. Bateson, and D. M. Ward. 2003. Geographical isolation in hot spring cyanobacteria. Environ. Microbiol. 5:650-659. [DOI] [PubMed] [Google Scholar]
- 46.Pimenov, N. V., I. I. Rusanov, M. N. Poglazova, L. L. Mityushina, D. Y. Sorokin, V. N. Khmelenina, and Y. A. Trotsenko. 1997. Bacterial mats on coral-like structures at methane seeps in the Black Sea. Mikrobiologiya 66:354-360. (In Russian.) [Google Scholar]
- 47.Rappe, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 48.Ravenschlag, K., K. Sahm, J. Pernthaler, and R. Amann. 1999. High bacterial diversity in permanently cold marine sediments. Appl. Environ. Microbiol. 65:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reeburgh, W. S. 1996. “Soft spots” in the global methane budget, p. 334-342. In M. E. Lidstrom and F. R. Tabita (ed.), Microbial growth on C1 compounds. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 50.Sahling, H., D. Rickert, R. W. Lee, P. Linke, and E. Suess. 2002. Macrofaunal community structure and sulfide flux at gas hydrate deposits from the Cascadia convergent margin, NE Pacific. Mar. Ecol. Prog. Ser. 231:121-138. [Google Scholar]
- 51.Schouten, S., E. C. Hopmans, E. Schefuss, and J. S. S. Damsté. 2002. Distributional variations in marine crenarchaeotal membrane lipids: a new tool for reconstructing ancient sea water temperatures. Earth Planet. Sci. Lett. 204:265-274. [Google Scholar]
- 52.Schramm, A., L. H. Larsen, N. P. Revsbech, N. B. Ramsing, R. Amann, and K.-H. Schleifer. 1996. Structure and function of a nitrifying biofilm as determined by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 62:4641-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schrenk, M. O., D. S. Kelley, J. R. Delaney, and J. A. Baross. 2003. Incidence and diversity of microorganisms within the walls of an active deep-sea sulfide chimney. Appl. Environ. Microbiol. 69:3580-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Selje, N., M. Simon, and T. Brinkhoff. 2004. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 427:445-448. [DOI] [PubMed] [Google Scholar]
- 55.Snaidr, J., R. Amann, I. Huber, W. Ludwig, and K. H. Schleifer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63:2884-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suess, E., G. Bohrmann, D. Rickert, W. Kuhs, M. E. Torres, A. Trehu, and P. Linke. 2002. Properties and fabric of near-surface hydrates at Hydrate Ridge, Cascadia Margin, p. 740-744. In Proceedings of the Fourth International Conference on Gas Hydrates, Yokohama, Japan.
- 57.Suess, E., M. E. Torres, G. Bohrmann, R. W. Collier, J. Greinert, P. Linke, G. Rehder, A. Trehu, K. Wallmann, G. Winckler, and E. Zuleger. 1999. Gas hydrate destabilization: enhanced dewatering, benthic material turnover and large methane plumes at the Cascadia convergent margin. Earth Planet. Sci. Lett. 170:1-15. [Google Scholar]
- 58.Teske, A., K.-U. Hinrichs, V. Edgcomb, A. de Vera Gomez, D. Kysela, S. P. Sylva, M. L. Sogin, and H. W. Jannasch. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thiel, V., M. Blumenberg, T. Pape, R. Seifert, and W. Michaelis. 2003. Unexpected occurrence of hopanoids at gas seeps in the Black Sea. Org. Geochem. 34:81-87. [Google Scholar]
- 60.Thiel, V., J. Peckmann, H. H. Richnow, U. Luth, J. Reitner, and W. Michaelis. 2001. Molecular signals for anaerobic methane oxidation in Black Sea seep carbonates and a microbial mat. Mar. Chem. 73:97-112. [Google Scholar]
- 61.Thiel, V., J. Peckmann, R. Seifert, P. Wehrung, J. Reitner, and W. Michaelis. 1999. Highly isotopically depleted isoprenoids: molecular markers for ancient methane venting. Geochim. Cosmochim. Acta 63:3959-3966. [Google Scholar]
- 62.Thomsen, T. R., K. Finster, and N. B. Ramsing. 2001. Biogeochemical and molecular signatures of anaerobic methane oxidation in a marine sediment. Appl. Environ. Microbiol. 67:1646-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Torres, M. E., J. McManus, D. Hammond, M. A. de Angelis, K. Heeschen, S. Colbert, M. D. Tryon, K. M. Brown, and E. Suess. 2002. Fluid and chemical fluxes in and out of sediments hosting methane hydrate deposits on Hydrate Ridge, OR. I. Hydrological provinces. Earth Planet. Sci. Lett. 201:525-540. [Google Scholar]
- 64.Tourova, T. P., T. V. Kolganova, B. B. Kuznetsov, and N. V. Pimenov. 2002. Phylogenetic diversity of the archaeal component in microbial mats on coral-like structures associated with methane seeps in the Black Sea. Mikrobiologiya 71:230-236. (In Russian.) [PubMed] [Google Scholar]
- 65.Treude, T., A. Boetius, K. Knittel, K. Wallmann, and B. B. Jørgensen. 2003. Anaerobic oxidation of methane above gas hydrates at Hydrate Ridge, NE Pacific Ocean. Mar. Ecol. Prog. Ser. 264:1-14. [Google Scholar]
- 66.Treude, T., M. Krüger, A. Boetius, and B. B. Jørgensen. Unpublished data.
- 67.Tryon, M. D., K. M. Brown, and M. E. Torres. 2002. Fluid and chemical fluxes in and out of sediments hosting methane hydrate deposits on Hydrate Ridge, OR. II. Hydrological processes. Earth Planet. Sci. Lett. 201:541-557. [Google Scholar]
- 68.Tryon, M. D., K. M. Brown, M. E. Torres, A. M. Tréhu, J. McManus, and R. W. Collier. 1999. Measurements of transience and downward fluid flow near episodic methane gas vents, Hydrate Ridge, Cascadia. Geology 27:1075-1078. [Google Scholar]
- 69.Valentine, D. L., and W. S. Reeburgh. 2000. New perspectives on anaerobic methane oxidation. Environ. Microbiol. 2:477-484. [DOI] [PubMed] [Google Scholar]
- 70.Vetriani, C., H. W. Jannasch, B. J. MacGregor, D. A. Stahl, and A. L. Reysenbach. 1999. Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Whitaker, R. J., D. W. Grogan, and J. W. Taylor. 2003. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301:976-978. [DOI] [PubMed] [Google Scholar]
- 72.Zhou, J., M. A. Brunns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]